Abstract
Most previously reported methods for purifying DNA-origami nanostructures rely on agarose-gel electrophoresis (AGE) for separation. Although AGE is routinely used to yield 0.1–1 µg purified DNA nanostructures, obtaining >100 µg of purified DNA-origami structure through AGE is typically laborious because of the post-electrophoresis extraction, desalting and concentration steps. Here, we present a readily scalable purification approach utilizing rate-zonal centrifugation, which provides comparable separation resolution as AGE. The DNA nanostructures remain in aqueous solution throughout the purification process. Therefore, the desired products are easily recovered with consistently high yield (40–80%) and without contaminants such as residual agarose gel or DNA intercalating dyes. Seven distinct three-dimensional DNA-origami constructs were purified at the scale of 0.1–100 µg (final yield) per centrifuge tube, showing the versatility of this method. Given the commercially available equipment for gradient mixing and fraction collection, this method should be amenable to automation and further scale up for preparation of larger amounts (e.g. milligram quantities) of DNA nanostructures.
INTRODUCTION
Self-assembly of a long, single-stranded circular DNA (scaffold strand) together with many synthetic oligonucleotides (staple strands)—the method known as DNA origami (1–7)—has proven an efficient way of fabricating nanostructures with customizable and well-defined geometry. In addition, the DNA-origami nanostructures feature addressable surfaces, thus offering a powerful technique of organizing material with up to sub-nanometer precision (8–10). Despite continued efforts in optimizing the structural design and folding conditions (11), the assembly yield of DNA origami is in most cases far <100%. However, well-folded and contamination-free DNA nanostructures are required for many applications. Therefore, a purification step after folding has become a standard procedure for many DNA-origami studies. Most widely used purification methods are based on agarose-gel electrophoresis (AGE), through which the well-folded objects are resolved as a distinct band and separated from slower migrating by-products (e.g. misfolded structures and aggregates) as well as the faster migrating non-integrated staple strands. After electrophoresis, the well-folded structures are extracted from the gel through homogenization (3,4,6,11) or electro-elution (12), reconstituted into the desired buffer and adjusted to desired concentration. As effective as the AGE-based separation is, the following two problems remain: the extraction step requires significant user input and quickly becomes laborious as the purification scales up and the DNA nanostructures recovered in this way usually co-purify with contaminants such as agarose-gel residues and ethidium bromide (or other staining reagents). There is therefore a pressing need for a scalable purification method that reduces labour cost and contaminants while maintaining the high separation resolution of AGE.
Here, we report a scalable, cost-effective and contamination-free method of purifying DNA-origami nanostructures through rate-zonal centrifugation, which separates molecular species by subjecting them to high centrifugal force in a density gradient media (13). Similar approaches have been applied to purify macromolecules such as proteins (14), protein–RNA complexes (e.g. ribosomes (15)) and metal colloids (16). In our case, well-folded DNA-origami structures are separated from other unwanted species (e.g. non-integrated staple strands, misfolded structures and aggregates) due to their distinct and well-defined mass and shape. A scheme that depicts the purification workflow is illustrated in Figure 1. In brief, a folded DNA-origami sample is loaded to the top of a linear quasi-continuous gradient of 15–45% glycerol and spun at ∼300 000g for 1–3 h. Fractions are then collected from the glycerol gradient and analysed using AGE. Those containing desired products are then combined, buffer-exchanged and concentrated.
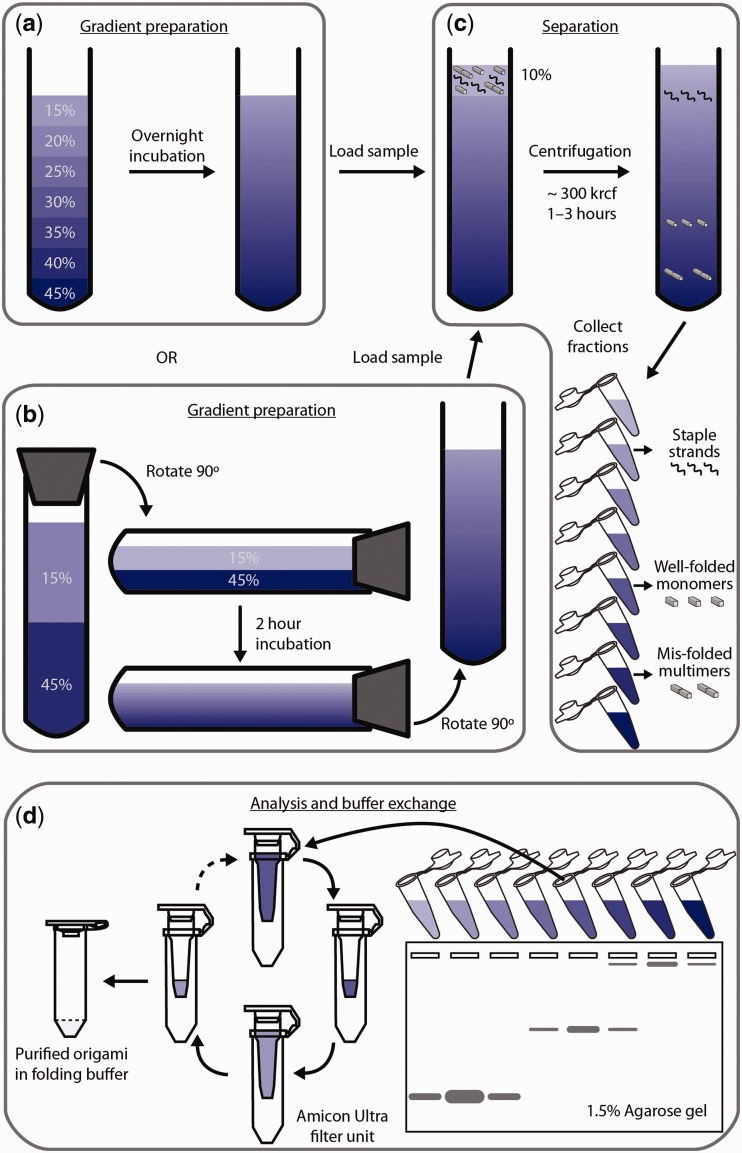
Figure 1.
Scheme of the rate-zonal centrifugation purification. (a) Preparation of glycerol gradient through overnight incubation of seven layers of glycerol solution from 15% to 45% with 5% increment per layer. (b) An alternative way of preparing the same gradient as shown in (a): a capped tube containing two layers of glycerol solution (15% and 45%) is laid down, incubated for 2 h and returned to vertical position. (c) Separation of different species (free staple strands, well-folded monomers and misfolded multimers) in the DNA-origami folding mixture through centrifugation. (d) Post-centrifugation process to first identify the fractions containing correctly folded nanostructures and then reconstitute such fractions into DNA-origami folding buffer.
MATERIALS AND METHODS
DNA oligonucleotides were purchased from Bioneer Inc. (Alameda, CA, USA). Scaffold strands p3024 and p7308 were cloned and amplified in-house following standard molecular biology protocols (17). Ultracentrifuge tubes and adaptors were purchased from Beckman-Coulter Inc. (Miami, FL, USA). Gel-loading tips were purchased from USA Scientific (Orlando, FL, USA). Rubber stoppers were purchased from VWR (Radnor, PA, USA). SYBR Safe DNA gel stain solution was purchased from Life Technologies (Carlsbad, CA, USA). Amicon filters were purchased from Millipore (Billerica, MA, USA). All other reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA).
The assembly of the DNA-origami nanostructures was accomplished in a one-pot reaction by mixing 50 nM scaffold strands with a pool of oligodeoxyribonucleotide staple strands (250 or 500 nM of each; reverse-phase cartridge purified) in a folding buffer containing 5 mM Tris (pH 8), 1 mM ethylenediaminetetraacetic acid (EDTA) and 10–20 mM MgCl2 (exact concentration depends on the target nanostructure) and subjecting the mixture to a thermal-annealing ramp that cooled from 80°C to 24°C over the course of 15–72 h (exact annealing time depends on the target nanostructure).
A linear glycerol gradient (15–45%, v/v) was prepared in one of the following two ways. In the first way, seven layers of glycerol solution in 1 × TE-Mg buffer (5 mM Tris–HCl, 1 mM EDTA and 10 mM MgCl2, pH 8), 80 or 400 μl per layer, were laid carefully into a 0.8-ml (Beckman #344090) or 3.5-ml (Beckman #349622) ultracentrifuge tube using longneck gel-loading tips with 45% glycerol solution at the bottom and 5% concentration decrement per layer (Figure 1a). The tube was then incubated overnight at 4°C to form a quasi-continuous gradient. Alternatively, two layers of glycerol solution in 1 × TE-Mg buffer, 1.4 ml per layer, were laid carefully into a 3.5-ml (Beckman #349622) ultracentrifuge tube with 45% glycerol solution at the bottom and 15% glycerol solution on top. The tube was capped with a rubber stopper (00m, VWR #59580-069), laid flat slowly and incubated at room temperature for 2 h. The tube was then returned to its vertical position (Figure 1b). The gradients prepared in the above two ways have been proven to yield almost the same separation efficiency for at least one structure. New or thoroughly cleaned tubes (interior brushed with mild detergent) should be used when preparing gradients in the latter way.
Desired amount of folded DNA-origami nanostructures containing 10% glycerol were loaded on top of the glycerol gradient (Figure 1c). In a typical experiment, 50 μl (or 400 μl) of 50 nM nanostructure was purified per 0.8 ml (or 3.5 ml) tube. The centrifuge tubes were then placed inside the centrifuge buckets (Beckman #356860 adapters were used to fit the 0.8 ml tubes), suspended on a swinging-bucket rotor (Beckman SW 55 Ti) and spun at 50 000 rpm (∼300 000g max) for 1 (Figure 2b–g) or 3 h (Figure 2a) at 4°C. The optimal centrifugation time depends on the exact shape and mass of the nanostructure and should be experimentally determined. (Excessive centrifugation time may lead to sample pelleting at the tube bottom and thus negatively impact the purification efficiency.) At this point, the well-folded nanostructures will have separated from free staple strands and unwanted multimers due to their different sedimentation rates. Twelve to sixteen equal-volume fractions were collected from top to bottom of the centrifuge tube using longneck gel-loading tips.
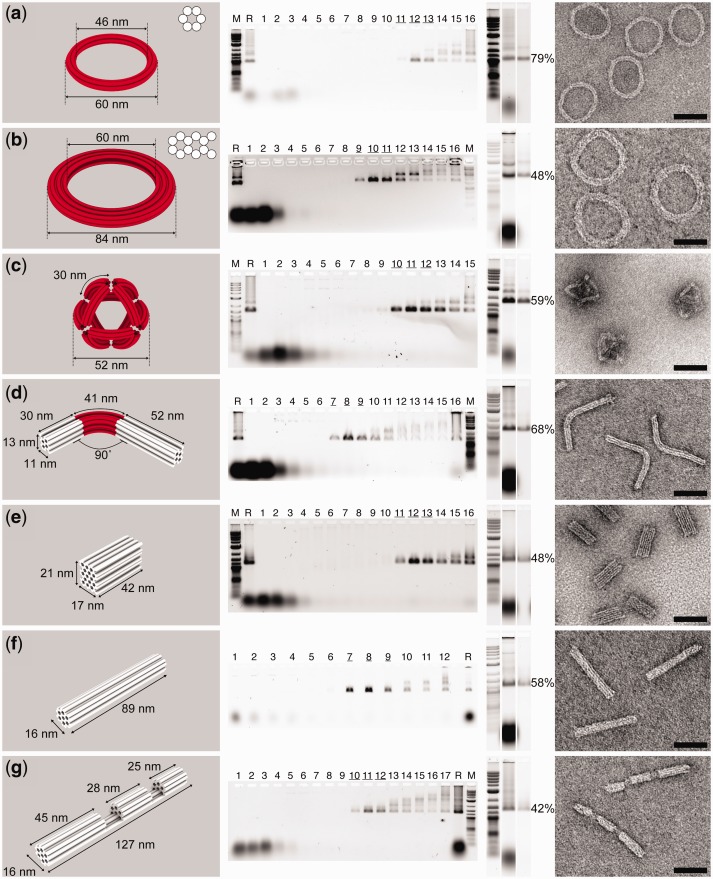
Figure 2.
Purification result of a series of 3D DNA-origami structures: (a) 6-helix-bundle (6-hb) ring, (b) 12-hb ring, (c) octahedron with curved 6-hb edges, (d) 18-hb rod bent by 90°, (e) 48-hb brick, (f) 24-hb rod and (g) 24-hb rod with two cavities. Computer-rendered 3D models of the structures are shown in the leftmost column, where each straight or curved DNA helix is shown as a white or red cylinder, respectively. Cross-sections of 6-hb and 12-hb rings are shown on the upper right corner of the 3D models in (a) and (b). AGE analyses of glycerol-gradient fractions collected after centrifugation are shown in the second column from the left. M: 0.1–10.0 kb DNA ladder (New England Biolabs); R: Raw DNA-origami assemblies before purification; consecutive Arabic numbers denote the fractions collected from top to bottom of the gradient, with fraction 1 the lightest. Underscored fractions are those enriched for well-folded DNA-origami structures. AGE characterizations of the purified products are shown in the third column from the left. Lanes (from left to right) are loaded with 0.1–10.0 kb DNA ladder, unpurified DNA-origami folding mixture and DNA-origami structures purified through centrifugation, respectively. Recovery yield of each structure (calculated from band intensities measured using ImageJ) is presented to the right of the corresponding gel image. Representative TEM images of the purified structures are shown in the rightmost column. Scale bars: 50 nm.
Aliquots of each fraction (5–10 μl per fraction) were loaded into separate wells of a non-denaturing, 1.5% agarose gel containing 1× SYBR Safe gel stain and separated by electrophoresis at 2.5 V/cm in 0.5× TBE buffer (45 mM Tris-borate, 1 mM EDTA (disodium salt), pH 8.3) containing 10 mM MgCl2 for 2.5 h at room temperature (Figure 1d). The gels were then scanned on a Typhoon FLV 9000 laser scanner. The fractions containing the desired products were combined and reconstituted into native folding buffer of the nanostructure or 1 × TE-Mg buffer using Amicon Ultra-0.5 ml centrifugal filters (MWCO 30 or 100 kDa) following the manufacturer’s manual. Centrifugal force <4500g was used in this step to minimize sample damage and loss. Typically 50–100 μl of solution containing purified DNA nanostructures were obtained. Alternatively, gel-filtration columns could be used for buffer exchange if the final DNA concentration is not the primary concern, as this process may dilute the sample slightly. The purified structures were subjected to AGE and transmission electron microscopy (TEM) to determine their purity and recovery yield.
RESULTS
Sample quality after purification
Seven DNA-origami nanostructures (see Supplementary Figure S1 for strand diagrams)—a 6-helix-bundle (6-hb) ring (Figure 2a), a 12-hb ring (Figure 2b), an octahedron with curved 6-hb edges (Figure 2c), an 18-hb rod bent by 90° (Figure 2d), a 48-hb brick (Figure 2e), a 24-hb rod (Figure 2f) and a 24-hb rod with two cavities (Figure 2g)—were purified using the method described above. The effectiveness of rate-zonal centrifugation to separate desired product from non-integrated staple strands and misfolded structures was evaluated by resolving post-centrifugation gradient fractions through AGE (schematics in Figure 1d; gel images in Figure 2). Staple strands have the slowest sedimentation rate (i.e. resides in the top fractions of the gradient) and segregate far away from most folded materials. Well-folded DNA nanostructures have distinct mobilities that allow them to be separated from other misfolded structures (i.e. oligomeric nanostructures and aggregates), which usually travel faster in the gradient. Further quantitative analysis of gel images revealed that ∼85% of the well-folded nanostructures are enriched in 10–20% volume of the gradient. The effective separation resulted in final products with excellent purity, which is supported by both AGE and TEM study of purified DNA-origami samples (Figure 2 and Supplementary Figure S2). The AGE analysis revealed enriched well-folded structures with greatly reduced misfolded structures and almost completely eliminated free staple strands. The minor impurities could be attributed to disturbance and diffusion of the gradient during sample handling (e.g. pipetting fractions or transferring tubes in and out of the centrifuge) and slight structural deformation of purified DNA-origami molecules. The already low level of remaining free staple strands can be further reduced to gel-undetectable levels by post-processing through Amicon filtration (Figure 2, all gel slices to the left of the noted percentage yields). For comparison, we purified the 6-hb rings (Figure 2a) from the same folding batch through either electrophoresis (band electro-eluted) or this centrifugation method. Similar DNA nanostructure purity was observed by resolving the final products side-by-side on an agarose gel (Supplementary Figure S3). In addition, the DNA-origami structures purified through the centrifugation method were never in contact with agarose gel or DNA intercalating reagent (e.g. ethidium bromide or SYBR Safe), which benefits downstream applications involving TEM or fluorescence study by providing cleaner background or baseline signal.
Scalability and recovery yield
Purification scale
The purification was performed on 0.5 pmol or 20 pmol folded scaffold strand for each structure (Figure 2) using one 0.8 or 3.5 ml centrifuge tube. It is important to note that these numbers are far below the maximum purification capacity of each tube. The 0.8 and 3.5 ml tubes have been successfully used to purify 8 and 120 pmol of raw DNA-origami structures (i.e. starting material), respectively, without compromising separation resolution or recovery yield. A purification trial using one centrifuge tube per sample generated 0.05–50 pmol (0.1–100 µg) enriched correctly folded product, depending on the purification scale, DNA-origami folding efficiency and recovery yield. One centrifuge rotor (Beckman SW55) accommodates six centrifuge tubes and therefore can be used to purify six different DNA-origami species or larger amount of a single species in one purification trial.
Recovery yield
Another valuable feature of the centrifugation-based purification method is the reproducible, high recovery yield. In contrast to the AGE-based purification, the DNA always stays in the aqueous phase and there is no need to extract them from the gel matrix. Therefore, losses of material are greatly reduced. In addition, there is less batch-to-batch variation in the recovery yield, which is a common problem associated with the AGE-based purification method due to the experimental inconsistency (e.g. electro-elution time, homogenization temperature, etc.). As shown in Figure 2, ∼40–80% properly folded DNA-origami objects were recovered after centrifugation separation and post-processing (measured by ImageJ). Subsequent TEM analyses also confirmed the strong enrichment of nanostructures with designated geometry (Figure 2).
DISCUSSION
DNA nanotechnology is leading to various applications in structural biology (17), biophysics (18–20), biosensing (21–24) and therapeutic delivery (25–28), many of which entail the preparation of micrograms to milligrams of enriched high-quality DNA nanostructures. Traditional electrophoresis-based methods excel at separation resolution but often fall short in scalability. The rate-zonal centrifugation method presented here is an effective complement to the current state-of-the-art in DNA-origami purification—properly folded DNA structures are well separated in the glycerol gradient with similar resolution provided by AGE and can be recovered in large quantity, with high efficiency and ease. Furthermore, we demonstrated that purified 6-hb rings carrying staple extensions (handles) were able to host 5-nm gold nanoparticles through handle/anti-handle hybridization [Supplementary Figure S4, with design and protocol adapted from (29,30)], confirming the well-preserved activity of the single-stranded handles after purification.
We note that the reported method may be improved further in the future to achieve better consistency, greater end-product purity and higher throughput, particularly through the automation of the gradient-mixing and fraction-collection processes using commercially available liquid-handling equipment. It is worth pointing out that in this work a universal gradient (15–45% linear glycerol gradient) was used for all structures. Therefore, when applied to a variety of DNA-origami structures, such a gradient may be more effective in purifying certain ones versus others. Users should be able to fine-tune the gradient to find the optimal conditions for each DNA structure of interest. Finally, higher capacity centrifuge tubes and rotor sets could be used to increase the purification scale by another order of magnitude (e.g. Beckman SW32 rotor accommodates up to 38.5-ml tubes). The rate-zonal centrifugation method, as an effective and easy-to-adapt purification approach, could help addressing the technical challenge of large-scale DNA nanostructure preparation and in turn, enable the development of many applications in the field of structural DNA nanotechnology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4.
FUNDING
National Institutes of Health (NIH) [1DP2OD004641, 1U54GM094608]; Office of Naval Research [N000141010241, N000014091118]; Wyss Institute for Biologically Inspired Engineering Faculty Award (to W.M.S.); Canadian Institutes for Health Research (to S.D.P.); Netherlands Organization for Scientific Research (NWO-Rubicon) (to M.K.). Funding for open access charge: NIH [1DP2OD004641].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the Imaging Core and General Equipment Facility of Wyss Institute for Biologically Inspired Engineering for the use of TEM and ultracentrifuge. They thank Dr Weiming Xu at Yale School of Medicine for inspiring discussion at the early stage of the work.
REFERENCES
- 1.Rothemund P. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 2.Andersen E, Dong M, Nielsen M, Jahn K, Subramani R, Mamdouh W, Golas M, Sander B, Stark H, Oliveira C, et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 3.Douglas S, Dietz H, Liedl T, Högberg B, Graf F, Shih WM. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature. 2009;459:414–418. doi: 10.1038/nature08016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dietz H, Douglas S, Shih WM. Folding DNA into twisted and curved nanoscale shapes. Science. 2009;325:725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han D, Pal S, Liu Y, Yan H. Folding and cutting DNA into reconfigurable topological nanostructures. Nat. Nanotech. 2010;5:712–717. doi: 10.1038/nnano.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liedl T, Högberg B, Tytell J, Ingber D, Shih WM. Self-assembly of three-dimensional prestressed tensegrity structures from DNA. Nat. Nanotech. 2010;5:520–524. doi: 10.1038/nnano.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han D, Pal S, Nangreave J, Deng Z, Liu Y, Yan H. DNA origami with complex curvatures in three-dimensional space. Science. 2011;332:342–346. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 8.Nangreave J, Han D, Liu Y, Yan H. DNA origami: a history and current perspective. Curr. Opin. Chem. Biol. 2010;14:608–615. doi: 10.1016/j.cbpa.2010.06.182. [DOI] [PubMed] [Google Scholar]
- 9.Shih WM, Lin C. Knitting complex weaves with DNA origami. Curr. Opin. Struct. Biol. 2010;20:276–282. doi: 10.1016/j.sbi.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tørring T, Voigt N, Nangreave J, Yan H, Gothelf K. DNA origami: a quantum leap for self-assembly of complex structures. Chem. Soc. Rev. 2011;40:5636–5646. doi: 10.1039/c1cs15057j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ke Y, Bellot G, Voigt NV, Fradkov E, Shih WM. Two design strategies for enhancement of multilayer–DNA-origami folding: underwinding for specific intercalator rescue and staple-break positioning. Chem. Sci. 2012;3:2587–2597. doi: 10.1039/C2SC20446K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bellot G, McClintock MA, Lin C, Shih WM. Recovery of intact DNA nanostructures after agarose gel-based separation. Nat. Methods. 2011;8:192–194. doi: 10.1038/nmeth0311-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Churchill L, Banker G, Cotman C. Gradient design to optimize rate zonal separations. Anal. Biochem. 1973;56:370–382. doi: 10.1016/0003-2697(73)90203-0. [DOI] [PubMed] [Google Scholar]
- 14.Patsch J, Sailer S, Kostner G, Sandhofer F, Holasek A, Braunsteiner H. Separation of the main lipoprotein density classes from human plasma by rate-zonal ultracentrifugation. J. Lipid Res. 1974;15:356–366. [PubMed] [Google Scholar]
- 15.Eikenberry E, Bickle T, Traut R, Price C. Separation of large quantities of ribosomal subunits by zonal ultracentrifugation. Eur. J. Biochem. 1970;12:113–116. doi: 10.1111/j.1432-1033.1970.tb00827.x. [DOI] [PubMed] [Google Scholar]
- 16.Steinigeweg D, Schütz M, Salehi M, Schlücker S. Fast and cost-effective purification of gold nanoparticles in the 20-250 nm size range by continuous density gradient centrifugation. Small. 2011;7:2443–2448. doi: 10.1002/smll.201100663. [DOI] [PubMed] [Google Scholar]
- 17.Douglas S, Chou J, Shih WM. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc. Natl Acad. Sci. USA. 2007;104:6644–6648. doi: 10.1073/pnas.0700930104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu J, Liu M, Liu Y, Woodbury N, Yan H. Interenzyme substrate diffusion for an enzyme cascade organized on spatially addressable DNA nanostructures. J. Am. Chem. Soc. 2012;134:5516–5519. doi: 10.1021/ja300897h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilner O, Weizmann Y, Gill R, Lioubashevski O, Freeman R, Willner I. Enzyme cascades activated on topologically programmed DNA scaffolds. Nat. Nanotech. 2009;4:249–254. doi: 10.1038/nnano.2009.50. [DOI] [PubMed] [Google Scholar]
- 20.Liu C, Kim E, Demple B, Seeman N. A DNA-based nanomechanical device used to characterize the distortion of DNA by Apo-SoxR protein. Biochemistry. 2012;51:937–943. doi: 10.1021/bi201196s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ke Y, Lindsay S, Chang Y, Liu Y, Yan H. Self-assembled water-soluble nucleic acid probe tiles for label-free RNA hybridization assays. Science. 2008;319:180–183. doi: 10.1126/science.1150082. [DOI] [PubMed] [Google Scholar]
- 22.Lin C, Liu Y, Yan H. Self-assembled combinatorial encoding nanoarrays for multiplexed biosensing. Nano Lett. 2007;7:507–512. doi: 10.1021/nl062998n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bell N, Engst C, Ablay M, Divitini G, Ducati C, Liedl T, Keyser U. DNA origami nanopores. Nano Lett. 2012;12:512–517. doi: 10.1021/nl204098n. [DOI] [PubMed] [Google Scholar]
- 24.Wei R, Martin T, Rant U, Dietz H. DNA origami gatekeepers for solid-state nanopores. Angew. Chem. Int. Ed. 2012;51:4864–4867. doi: 10.1002/anie.201200688. [DOI] [PubMed] [Google Scholar]
- 25.Douglas S, Bachelet I, Church G. A logic-gated nanorobot for targeted transport of molecular payloads. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 26.Jiang Q, Song C, Nangreave J, Liu X, Lin L, Qiu D, Wang Z-G, Zou G, Liang X, Yan H, et al. DNA origami as a carrier for circumvention of drug resistance. J. Am. Chem. Soc. 2012;134:13396–13403. doi: 10.1021/ja304263n. [DOI] [PubMed] [Google Scholar]
- 27.Walsh A, Yin H, Erben C, Wood M, Turberfield A. DNA cage delivery to mammalian cells. ACS Nano. 2011;5:5427–5432. doi: 10.1021/nn2005574. [DOI] [PubMed] [Google Scholar]
- 28.Liu X, Xu Y, Yu T, Clifford C, Liu Y, Yan H, Chang Y. A DNA nanostructure platform for directed assembly of synthetic vaccines. Nano Lett. 2012;12:4254–4259. doi: 10.1021/nl301877k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding B, Deng Z, Yan H, Cabrini S, Zuckermann R, Bokor J. Gold nanoparticle self-similar chain structure organized by DNA origami. J. Am. Chem. Soc. 2010;132:3248–3249. doi: 10.1021/ja9101198. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Z, Jacovetty E, Liu Y, Yan H. Encapsulation of gold nanoparticles in a DNA origami cage. Angew. Chem. Int. Ed. 2011;50:2041–2044. doi: 10.1002/anie.201006818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.