Abstract
Spinal muscular atrophy is a severe motor neuron disease caused by reduced levels of the ubiquitous Survival of MotoNeurons (SMN) protein. SMN is part of a complex that is essential for spliceosomal UsnRNP biogenesis. Signal recognition particle (SRP) is a ribonucleoprotein particle crucial for co-translational targeting of secretory and membrane proteins to the endoplasmic reticulum. SRP biogenesis is a nucleo-cytoplasmic multistep process in which the protein components, except SRP54, assemble with 7S RNA in the nucleolus. Then, SRP54 is incorporated after export of the pre-particle into the cytoplasm. The assembly factors necessary for SRP biogenesis remain to be identified. Here, we show that 7S RNA binds to purified SMN complexes in vitro and that SMN complexes associate with SRP in cellular extracts. We identified the RNA determinants required. Moreover, we report a specific reduction of 7S RNA levels in the spinal cord of SMN-deficient mice, and in a Schizosaccharomyces pombe strain carrying a temperature-degron allele of SMN. Additionally, microinjected antibodies directed against SMN or Gemin2 interfere with the association of SRP54 with 7S RNA in Xenopus laevis oocytes. Our data show that reduced levels of the SMN protein lead to defect in SRP steady-state level and describe the SMN complex as the first identified cellular factor required for SRP biogenesis.
INTRODUCTION
The SMN protein was discovered because decreased levels of this protein correlate with the phenotypic severity of spinal muscular atrophy (SMA) (1,2), a neuromuscular disease characterized by the degeneration of the lower motor neurons, leading to muscular weakness and atrophy [reviewed in (3)]. The disease is due to recessive mutations or deletions affecting the survival of motor neuron (SMN1) gene (4). Two genes, SMN1 and SMN2, code for the SMN protein in humans, the copy number of SMN2 being a determinant of disease severity (1,5). Indeed, while SMN1 produces full-length transcripts, SMN2 mainly produces an alternatively spliced messenger RNA (mRNA) lacking exon 7 (SMNΔEx7) (6,7). As the SMNΔExon7 protein is unstable and rapidly degraded (8,9), SMN2 cannot fully compensate for the loss of SMN1 in SMA. The SMN protein is ubiquitously expressed and essential in all eukaryotes that have been tested so far, including Schizosaccharomyces pombe. In Saccharomyces cerevisiae, an orthologous protein-coding gene has not yet been found. In vertebrates, the SMN protein is predominantly associated with Gemin2–8 and Unrip proteins to form a large stable complex called the SMN complex (10–13). SMN and Gemin2 orthologs but no other components of the SMN complex have been found in the fungus S. pombe (14–17). The metazoan SMN complex has been proposed to function in universal eukaryotic processes related to RNA metabolism, including transcription, splicing, ribonucleoprotein (RNP) biogenesis and in neuron-specific functions, like neurite and axon outgrowth, growth cone excitability, mRNA transport and the function of the neuromuscular junction [reviewed in (18–21)]. The most well characterized mechanism of action of the SMN complex is in the assembly of the spliceosomal U-rich small nuclear RNP (UsnRNP) (18,19,22–27). Accordingly, SMN deficiency would alter the stoichiometry of snRNAs that might cause widespread and tissue-specific pre-mRNA splicing defects in SMA mice models (28,29), as well as in the S. pombe model organism carrying a temperature-degron allele of the SMN protein (30). More recently, the splicing of some, but not all, minor U12-type introns was reported to be inhibited in cells derived from SMA patients, and in mammalian cells and Drosophila larvae expressing low levels of SMN, demonstrating a link between SMN deficiency and alterations of splicing events mediated by the minor spliceosome (31,32). However, the identity of the impaired or altered SMN function(s) responsible for SMA is still a matter of debate. This is reinforced by the fact that although the SMN complex has been called the master ribonucleoprotein assembler (33), there is no direct proof of its involvement in assembly mechanisms other than UsnRNPs. For instance, it has been previously suggested that the SMN complex may play a role in box C/D and H/ACA RNP assembly based on its interaction with Fibrillarin, a core component of the C/D box RNPs and with Gar1, a common component of the H/ACA box RNPs (34,35). In addition, a decrease of the levels of U3 small nucleolar RNA (a C/D box snoRNA) was found upon reduction of SMN levels in HeLa cells by RNAi (36). However, whether box C/D and H/ACA RNP assembly depends on the SMN complex, has only been poorly studied. Several data also suggest that the SMN protein alone or the SMN complex may assist the assembly of specific mRNAs into mRNP particles in neurons, as well as their stability, their targeting to the neuronal transport system along neurites and their localized translation in synapses and axonal growth cones (37–40) [reviewed in (20)].
Interestingly, while the signal recognition particle (SRP) is one of the most abundant RNPs in eukaryotic cells, a possible involvement of the SMN complex in its assembly had not been proposed yet. Here, we bring very strong arguments in favor of a role of the SMN complex in its stability and biogenesis. SRP is an ubiquitous RNP that co-translationally delivers most membrane and secretory proteins to the plasma membrane in prokaryotes and to the endoplasmic reticulum in eukaryotes [for reviews, (41–43)]. Mammalian SRP consists of six proteins, SRP9, 14, 19, 54, 68, 72 and a single RNA molecule, i.e. the 7S RNA (Figure 1A). The RNA secondary structure possesses extensive base paired regions, which form a prominent central helix flanked by a small (or Alu) and a large (or S) domain (Figure 1A) (44). The S-domain, which corresponds to the central region of the RNA, associates with the SRP19 and SRP54 proteins, and a SRP68/72 heterodimer. The Alu-domain comprises both the 3′ and 5′ terminal RNA regions and binds a SRP9/14 heterodimer. Fungal SRP resembles its mammalian counterpart in that it also consists of six proteins (SRP72p, SRP68p, SRP54p, SRP14p and Sec65p, which are homologous to the mammalian proteins, and Srp21) and a single RNA molecule called srp7 in S. pombe (45–48). Studies in yeasts (49,50), Xenopus laevis oocytes (51,52) and mammalian cells (53,54) support a model in which all SRP proteins, except SRP54, are imported to the nucleolus for assembly with 7S RNA. The resulting pre-particle is then exported to the cytoplasm where it incorporates SRP54, leading to the formation of a mature and functional RNP. None of the factors required for the in vivo biogenesis of SRP has yet been described.
Figure 1.
Immunoselected native SMN complexes associate in vitro with 7S RNA. (A) Human 7S RNA sequence. The secondary structure was established by previous studies (75,76). Stems are numbered 2 to 8 according to the criteria of previous study (76). (B) Native SMN complexes (SMN complex) were purified by immunoprecipitation with anti-Flag antibodies using total extracts prepared from HeLa TET-off cells that stably expressed Flag-tagged Gemin2. The protein composition of the complexes was analyzed by SDS–PAGE followed by silver staining. The non-specific proteins immunoselected from HeLa TET-off cells that do not express Flag-Gemin2 are shown as a negative control (Control). Proteins were identified according to their molecular weight as referred to molecular weight markers (MW) and to (39) and (68). The identity of the proteins was confirmed by western blotting and mass spectrometry (data not shown). (C) The purified native SMN complex (SMN complex) shown in (B) or the non-specific proteins purified from parental cells (Control) were incubated with in vitro transcribed [32P]UTP-labeled 7S RNA, U1 snRNA and U1ΔSL1 in the constant presence of U6 snRNA as an internal negative control. Subsequently, bound RNAs were isolated after washing and analyzed by electrophoresis on denaturing polyacrylamide gels and autoradiography. Lanes labeled ‘Total’ represent 10% of input.
By screening for possible interactions of in vitro transcribed RNAs with purified human SMN complex, we identified the 7S RNA as a possible partner of this complex. We observed that the binding of 7S RNA to purified SMN complexes was strongly competed by U1 and U2 snRNAs. The use of 7S RNA domains taken individually and RNase protection experiments identified the RNA determinants required for interaction with the SMN complex. Moreover, we showed that Gemin5, which is the snRNA binding protein of the SMN complex, interacts in vitro with the 7S RNA. These observations motivated us to investigate a possible role of the SMN complex in SRP biogenesis, and accordingly, we detected an association of SRP with the SMN complex in different systems, i.e. X. laevis oocytes, HeLa cells and yeast S. pombe. Importantly, our analysis of 7S RNA levels by real-time polymerase chain reaction (PCR) in tissues of SMN-deficient mice revealed a significant reduction of 7S RNA in the spinal cord, whereas it remained the same in the brain and the heart. We also observed a decrease in the 7S RNA levels and in the amount of SRP in a S. pombe mutant strain carrying a temperature-degron allele of SMN. Altogether, we bring strong data in favor of an in vivo association of the SMN complex with SRP and show that the SMN protein may be required for SRP stability. As we show that antibodies directed against either SMN or Gemin2 interfere with the cytoplasmic association of SRP54 with 7S RNA in X. laevis oocytes, we propose that the SMN complex functions in SRP biogenesis. The possibility that SRP biogenesis may be altered upon SMN protein deficiency in SMA is discussed.
MATERIALS AND METHODS
DNA constructs and antibodies
Plasmids encoding vertebrate wild-type UsnRNAs, as well as U1ΔSL1 and U4ΔSm, have been described previously (55–57) and so have the construct for production of Flag-tagged Gemin5 (58) and production of GST-SMN (59). For production of GST-tagged Gemin5 in bacteria, a DNA corresponding to the ORF of Gemin5 was generated by PCR amplification and cloned into the pDEST-15 vector with Gateway technology (Invitrogen). As human and X. laevis 7S RNA share 87% sequence homology (60), we used human 7S RNA in our experiments. DNA fragment corresponding to the sequence encoding the human 7S RNA was generated by RT–PCR amplification using specific primers and total RNAs from HeLa cells, and cloned downstream of the T7 promoter into the pUC18 vector. The 7S RNA transcript obtained by in vitro transcription using this plasmid contains an additional G at the 5′-end. Construction of deletion mutants of 7S RNA into the pUC18-7S was carried out by PCR amplification using specific primers. The RNA 7S Alu+long stem comprises nucleotides 1–128 and nucleotides 222–300 linked by a GUAA sequence; the RNA 7S Alu+short stem contains nucleotides 1–98 and nucleotides 252–300 linked by an UA sequence; the RNA 7S Alu contains nucleotides 1–65 and nucleotides 282–300 linked by an UA sequence. The S-domain+long stem and the S-domain RNAs comprise nucleotides 48–300 and 101–250, respectively. All the mutated RNAs contain an additional G at the 5′-end. All the constructs were analyzed by automated sequencing.
The entire coding region of fission yeast smn1+ was amplified by PCR and cloned into the pREP41-NTAP vector (obtained from K. Gould) using standard methods. The plasmid encoding the TAP-SpSMN fusion was transformed into a diploid heterozygous strain carrying a deletion of the smn1+ gene (BG_1375; Bioneer Corporation) together with plasmid pON177, which allows sporulation. Transformed cells were selected on Edinburgh minimal medium (EMM) plates with appropriate amino acids supplements. After sporulation and dissection, clones bearing a smn1+ deletion and the pREP41-TAP-SpSMN plasmid were identified by diagnostic PCR and western blot analysis.
The antibodies used in these experiments were as follows: anti-SMN [2B1, (61); 4B3, (62)], anti-Gemin2 (2E17) (23), anti-Gemin3 (12H12) (63), anti-Gemin4 (17D10) (64), anti-Gemin5 (10G11) (58), anti-Gemin6 (20H8) (65), anti-SRP9 (PTG lab), anti-SRP14 (PTG lab and kind gift from Dr Dobberstein), anti-SRP54 (BD transduction laboratories and PTG lab), anti-SRP68 (PTG lab), anti-tubuline (Sigma), anti-Y12 (66), anti-Sox2 (PTG lab), anti-Lin28 (PTG lab) and non-immune (SP2/O) (67).
In vitro transcription of RNAs
In vitro transcription and [32P]UTP labeling of RNAs were carried out at 37°C for 3 h on 1 µg of linearized plasmid in 15 µl of transcription buffer (TB) (80 mM HEPES–KOH, pH 7.5, 24 mM MgCl2, 2 mM spermidine, 40 mM DTT) containing 40 U RNasin, 30 U T7 RNA polymerase, 1.3 mM of ATP, CTP, GTP, 66 µM UTP and 10 µCi αUTP [32P] (GE Healthcare, 3000 Ci/mmol). In vitro transcription of unlabeled RNAs was carried out at 37°C for 3 h on 1 µg of linearized plasmid in 30 µl of TB containing 60 U T7 RNA polymerase and 7.5 mM of ATP, CTP, GTP and UTP. RNAs were then purified by electrophoresis on 7 M urea–8% acrylamide gels and precipitated with ethanol.
In vitro binding of RNAs to immunoselected SMN complexes
Native SMN complexes were affinity purified using a HeLa Tet-Off cell line stably expressing Flag-tagged Gemin2 as described previously (39,68). In vitro RNA-binding experiments and competition experiments were performed as described earlier (56), except that 20 000 cpm of radioactive RNAs were used instead of 10 000. For competition experiments, purified native SMN complexes were pre-incubated with 250 nM of unlabeled RNAs for 30 min at 4°C. Subsequently, 20 000 cpm of [32P]UTP-labeled 7S RNA was added to the pre-incubated mixtures and further incubated for 1 h at 4°C. Bound RNAs were isolated and analyzed by electrophoresis on 7 M urea–10% polyacrylamide gels.
RNase protection assay
In vitro transcribed 7S RNA bound or not to purified SMN complexes was incubated in 50 µl of Resuspension buffer (RSB) 100 buffer (10 mM Tris–HCl, pH 7.5, 100 mM NaCl, 2.5 mM MgCl2) in the presence of 5 µg of tRNA. Digestion was for 6 min at 20°C, in the presence of 2 U of T1 RNase (Ambion), 5 U of T2 RNase (Mobitec) or 0.002 U of V1 RNase (Roche). RNase reactions were stopped by addition of 50 µl of water, 4 µl of EDTA 0.1 M, pH 8 and 20 µg of tRNA, followed by phenol extraction. For production of a ladder, an alkaline hydrolysis of the naked RNA was performed for 3 min at 95°C, using 20 000 cpm of RNA dissolved in 1 µl of 100 mM sodium carbonate, pH 9.2. To identify the nucleotides in the ladder, an RNase T1 digestion was also performed in denaturing conditions. To do so, 20 000 cpm of RNA was incubated with 2 µg of tRNA in Citrate Buffer (20 mM Citrate NaOH, 1 mM EDTA, 7 M urea, 0.05% Xylene cyanol blue, 0.05% bromophenol blue) for 10 min at 65°C; 2 U of RNase T1 was then added and the incubation was followed for 10 min at 65°C.
In vitro binding of RNAs to Gemin5
HeLa cells were transfected with plasmid expressing Flag-tagged Gemin5 using the Calphos Mammalian transfection kit (Clontech) according to the manufacturer’s recommendation. Immunoprecipitation of the SMN complex or Flag-Gemin5 alone using anti-Flag antibodies were carried out as described previously (39,68), in the presence or in the absence of 1% Empigen-BB detergent, respectively. Recombinant GST, GST-SMN and GST-Gemin5 proteins were expressed in Escherichia coli and purified by affinity chromatography on glutathione-Sepharose beads, as described (39). In vitro RNA binding experiments on Flag-Gemin5 or GST-Gemin5 were performed as described previously (56).
Xenopus laevis oocyte microinjections
Pieces of X. laevis ovary were dissected, treated with 5000 U collagenase and individual stage VI oocytes were prepared for microinjection as described (69). Injections and culture of oocytes were carried out in OR2 medium (5 mM HEPES–KOH, pH 7.8, 82.5 mM NaCl, 2.5 mM KCl, 1 mM MgCl2, 1 mM Na2HPO4,) containing 1 mM CaCl2 as described previously (70). For nuclear injection, oocytes were previously centrifuged for 15 min at 1000g to allow the visualization of the germinal vesicle (71). In a typical injection experiment, 18.4 nl of [32P] UTP-labeled RNA (1 × 106 cpm/µl for each RNA) were injected into the nucleus of the oocytes. After a 6-h incubation, oocytes were manually dissected to prepare nuclear and cytoplasmic fractions in RSB 150 G buffer [10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 2.5 mM MgCl2, glycerol 10% (v/v)] for further analysis. For the antibody inhibition experiments, oocytes were first injected with 18.4 nl of antibody (1 µg/µl) into the cytoplasm 1 h before they received a second injection of 18.4 nl of [32P] UTP-labeled RNA into the nucleus.
Immunoprecipitation experiments and analysis
For a typical immunoprecipitation experiment using X. laevis extracts, nuclear, cytoplasmic or total fractions from 20 oocytes were homogenized in 300 µl of RSB 150 buffer (10 mM Tris–HCl, pH 7.5, 150 mM NaCl, 2.5 mM MgCl2) and the insoluble material was pelleted by centrifugation. The supernatant was incubated with antibodies bound to protein A or G-Sepharose (Amersham). Immunoprecipitations and analysis of bound RNAs were performed as previously described (55). For immunoprecipitation using HeLa cells, cytoplasmic extracts were prepared as previously described (72) and incubated with specific antibodies bound to protein A or G-Sepharose (Amersham) for 2 h at 4°C in RSB 100 buffer (10 mM Tris–HCl, pH 7.5, 100 mM NaCl, 2.5 mM MgCl2) containing 0.01% NP-40. The beads were washed with the same buffer and the immunoprecipitated proteins were analyzed by SDS–PAGE and western blotting as previously described (23). The immunoprecipitated RNAs were analyzed by RT–PCR using the oligonucleotides described in (73) for analysis of the mRNA coding GAPDH, and the following oligonucleotides for the analysis of 7S RNA: forward 5′-GGAGTTCTGGGCTGTAGTGC-3′, reverse 5′-GATCAGCACGGGAGTTTT-3′.
Fission yeast in vivo cross-linking, extract preparation and immunoprecipitation
Fission yeast cells bearing a smn1+ deletion and the pEP41-TAP-SpSMN plasmid were grown in EMM2-Leu medium and cross-linked by addition of formaldehyde (1% final concentration) for 30 min at 28°C with continuous shaking. Adding glycine to 125 mM final concentration terminated the reaction. After incubation for 5 min, cells were washed twice with PBS and frozen in liquid nitrogen. Cells were resuspended in RNA IP Buffer (50 mM Tris–HCl, pH 8, 1 mM EDTA, 1% NP40, 0.5% Na-deoxycholate, 150 mM NaCl) and broken using a FreezerMill SPEX6770 grinder. After thawing on ice and homogenization with a tight dounce, the extract was centrifuged at 15 000g for 20 min. TAP-tag purification was performed as described previously (74) with an overnight Tobbaco Etch Virus (TEV) protéase (Promega) incubation at 4°C. After digestion with RQ1 RNase free DNAse (Promega) according to manufacturer’s procedure and incubation with proteinase K for 30 min at 37°C, the reverse cross-linking reaction was carried out at 70°C for 45 min. The samples were phenol–chloroform extracted and the RNA ethanol-precipitated. Yeast extract preparation and native gel electrophoresis, as well as RT–PCR analysis were performed as previously described [(30) and (31), respectively]. Primer sequences and PCR regimes are available upon request.
RNA isolation from mice and real-time RT–PCR
Ten-day-old SMNΔ7 mice (n = 5) and age-matched wild-type controls (n = 5) were anesthetized (10 mg/kg xylazine, 100 mg/kg ketamine) and perfused intracardially with 0.1 mol/l PBS. The brain, the liver and the spinal cord tissues were removed and total RNA was extracted using the Nucleospin RNA II kit (Macherey Nagel). Total RNA was treated for DNA contamination by DNase RQ1 (Promega) (1 U/µg of RNA) for 30 min at 37°C. RNAs were then phenol/chloroform extracted and ethanol precipitated. First strand cDNA synthesis was carried out using 1 µg of DNA-free RNA. This was added to 0.5 µg random hexamer in nuclease-free water. This mixture was heated at 65°C for 10 min and then cooled on ice for 10 min to allow primers to anneal. A master mix was added that contained 0.4 mM dNTPs, 40 U RNAsin (Fermentas), 1 x M-MLV reaction buffer and 200 U of M-MLV (Promega), in a total reaction of 25 µl. The samples were then incubated at 37°C for 1 h. About 1 µl of diluted cDNA (2- to 100-fold dilution) was used for each real-time PCR reaction. The reactions were carried out on a LightCycler (Roche) using SYBR Green I Master mix (Roche). Specific primers were used to generate cDNA: 7S forward 5′-GGAGTTCTGGGCTGTAGTGC-3′, 7S reverse 5′-GATCAGCACGGGAGTTTT-3′, RP29 forward 5′-GCCTATGTCCTTCGCGTACT-3′ and RP29 reverse 5′-CTGAAGGCAAGATGGGTCAC-3′. The efficacy of each primer pairs was determined and was comprised between 1.8 and 2. As a control of the amplification specificity, melting curve analysis was performed for each PCR experiments. Each run included negative controls. Quantification was performed using LightCycler software (Roche). Data were calculated according to the ΔΔCT method. Results were expressed as mean ± standard error of the mean of three measurements. Differences between 2ΔCT values for the RNA target versus 2ΔCT values for the RNA reference were analyzed using unpaired Student’s t-test; P < 0.05 was considered significant.
RESULTS
Association of in vitro produced 7S RNA with immunoselected SMN complexes
First we tested whether the 7S RNA component of human SRP may interact with the SMN complex. To this end, we carried out in vitro binding assays using various [32P]UTP-labeled RNAs and immunoselected SMN complexes, in conditions that were previously established to study the SMN complex–UsnRNA interactions (55). To isolate SMN complexes, we used a cell line that stably expressed Flag-tagged Gemin2. A total cell extract was incubated with anti-Flag antibodies linked to agarose beads as previously described (39,68). We verified that the material retained on the beads contained all the known components of the SMN complex, namely Gemin2–8 and unrip, and that no Sm proteins and other proteins known to interact with the SMN complex were present in detectable amounts (39). The purified SMN complexes bound to agarose beads (SMN complex) and, as a negative control, agarose beads incubated with a cellular extract prepared from the parental HeLa Tet-Off cells (Control) (Figure 1B) were incubated with in vitro transcribed RNAs. In addition to the tested [32P]UTP-labeled 7S RNA, U1 snRNA and U1ΔSL1, which does not contain the stem–loop 1 necessary for specific interaction of U1 snRNA with the SMN complex (55), were used as positive and negative controls, respectively. In vitro transcribed U6 snRNA was added in each of the assays as an internal negative control (Figure 1C). As expected, the immunoselected SMN complexes associated with U1 snRNA (Figure 1C, lane 4), but not with U1ΔSL1 (Figure 1C, lane 5). Interestingly, 7S RNA was retained on the SMN complexes (Figure 1C, lane 6). This interaction was likely to be specific, because our negative control U6 snRNA was not immunoselected in any of the assays (Figure 1C, lanes 4–6), and also because 7S RNA did not bind to the beads pre-incubated with extracts from Hela Tet-Off cells (Figure 1C, lane 9). Therefore, these first results indicated that 7S RNA may interact with the SMN complex.
U1 and U2 snRNAs compete with 7S RNA for binding to immunoselected SMN complexes
Although all UsnRNAs bind to the SMN complex in the course of UsnRNP assembly, competition experiments have shown that all of them do not bind to the same site on this complex (56). Therefore, having demonstrated that 7S RNA can interact in vitro with the SMN complex, it was important to test whether one or several UsnRNAs can compete with 7S RNA for binding to the SMN complex. Competition binding experiments were performed in conditions previously established for UsnRNAs (56). Purified SMN complexes bound to agarose beads (SMN complex) were incubated with trace amounts of [32P]UTP-labeled 7S RNA and an excess of non-radioactive U1, U1ΔSL1, U2, U4, U5 or 7S RNAs. After incubation, bound RNAs were analyzed by denaturing polyacrylamide gel electrophoresis. As shown in Figure 2, non-radioactive 7S RNA competed with itself and non-radioactive U1 and U2 snRNAs significantly competed with labeled 7S RNA. This competition was specific because U1ΔSL1 was unable to fully compete with 7S RNA. The binding level of labeled 7S RNA in the presence of non-radioactive U4 or U5 snRNA was almost comparable with the one in the presence of non-radioactive U1ΔSL1, indicating that U4 and U5 snRNAs did not significantly compete with labeled 7S RNA. These data suggested that U1, U2 and 7S RNAs share the same binding site or have partially overlapping binding sites on the SMN complex, whereas U4 and U5 snRNAs have different binding sites.
Figure 2.

U1 and U2 snRNAs compete with 7S RNA for binding to the SMN complex. Purified native SMN complexes (SMN complex) were pre-incubated with an excess of unlabeled U1, U1ΔSL1, U2, U4, U5 and 7S. A pre-incubation was also done without the presence of any RNA (/) as a control. Non-specific proteins purified from parental cells (Control) were also pre-incubated. Then, in vitro transcribed [32P]UTP-labeled 7S RNA was added to the pre-incubated mixtures and further incubated. Bound RNAs were isolated after washing and analyzed by electrophoresis on denaturing polyacrylamide gels and autoradiography. Lanes labeled ‘Total’ represent 10% of input. The intensity of signal in each condition was quantified using the Typhoon 9410 (Amersham).
The overall 7S RNA is important for efficient interaction with the SMN complex
In order to delineate the region(s) of 7S RNA necessary for its interaction with the SMN complex, we individually produced 7S RNA domains (Figure 3A) and tested their ability to interact with purified SMN complex. As shown in Figure 3B, the RNAs corresponding to the Alu domain alone or the Alu domain extended with helix 5 did not bind to purified SMN complex. RNAs corresponding to the S-domain and the S-domain extended with helix 5 had the capability to interact albeit at very low level compared with wild-type RNA. Therefore, the entire 7S RNA molecule seems to be required for efficient interaction. To go into more details, we analyzed the regions of 7S RNA that are protected against RNase activities when it is bound to the SMN complex (Figure 4). In vitro transcribed 7S RNA bound to the purified SMN complex or free 7S RNA were digested by T1, T2 or V1 RNases. RNase protections were observed in three regions of the RNA: in the upper loop of the Alu domain, in the helix 5d region (nucleotides 86–103) and in one strand of helix 8 (nucleotides 174–198). It is interesting to note that when the 7S RNA was bound to the SMN complex, the sensitivity of residues G76, G209 and G210 to T1 RNase digestion was increased. These data suggest that upon binding to the SMN complex, the 7S RNA undergoes a conformational change leading to destabilization of the two base pairs G209-C190 and G210-C189.
Figure 3.
The overall 7S RNA molecule is required for efficient interaction with the purified SMN complex in vitro. (A) Schematic representation of the 7S RNA domains used in the study. (B) The purified native SMN complex (SMN complex) or the non-specific proteins purified from parental cells (Control) were incubated with in vitro transcribed [32P]UTP-labeled 7S RNA WT or the individual 7S RNA domains, in the constant presence of U6 snRNA as an internal negative control. Subsequently, bound RNAs were isolated after washing and analyzed by electrophoresis on denaturing polyacrylamide gels and autoradiography. Lanes labeled ‘Total’ represent 10% of input.
Figure 4.
The SMN complex protects three regions of the 7S RNA. (A) In vitro transcribed 7S RNA was incubated in the absence (Transcript) or in the presence (SMN complex) of the purified SMN complex. The RNA was then digested by RNases T1, T2 and V1 as described in the ‘Materials and Methods’ section. The cleavage products were analyzed by electrophoresis on denaturing polyacrylamide gels. An alkaline hydrolysis is shown (H). Nucleotides are numbered on the left and were designed by the use of RNase T1 in denaturing conditions and by a longer fractionation on polyacrylamide gel to identify residues in the 5′-end (data not shown). (B) The protected regions within the 7S RNA are indicated in red, and the G residues that were more sensitive to T1 RNase cleavages are indicated by blue arrows.
Gemin5 directly binds 7S RNA in vitro
Even though multiple binding sites for UsnRNAs seem to exist within the SMN complex (56), Gemin5 had been shown to be the protein of the SMN complex that directly interacts with all the UsnRNAs (57). Therefore, we tested whether Gemin5 was also able to interact with 7S RNA. To this end, we prepared total extracts from 293 T cells that were transiently expressing Flag-tag Gemin5 (Flag-Gemin5). These extracts were incubated with anti-FLAG antibodies bound to agarose beads in the presence of Empigen (Figure 5A). SMN complexes are disrupted under these conditions, Gemin5 being therefore efficiently dissociated from them (57) (Figure 5A). As previously described, Empigen-treated Flag-Gemin5 was able to bind to U1 and U4 snRNAs, but not to U4ΔSm and U6 snRNA (57), and the data revealed the binding of 7S RNA to Flag-Gemin5 (Figure 5B). To exclude the possibility that some minor components in cell extracts that had been co-immunoselected with Flag-Gemin5 confered this binding specificity to 7S RNA, GST-tagged Gemin5 was expressed and purified from E. coli by immobilization on Glutathione sepharose beads. In contrast to a GST–SMN fusion protein and GST alone, the bound GST-tagged Gemin5 was able to interact with 7S RNA (Figure 5C), indicating that Gemin5 binds directly to 7S RNA without requiring any other components of the SMN complex or other cellular factors.
Figure 5.
Gemin5 binds to 7S RNA. (A) Immunoprecipitation experiments using anti-Flag antibody were carried out on total extracts prepared from HeLa cells transiently expressing Flag-tagged Gemin5 in the absence (−) or in the presence (+) of 1% Empigen-BB detergent. The immunoprecipitated proteins were analyzed by SDS–PAGE and western blotting using antibodies directed against the indicated proteins. In total, 5% of the extracts used in the experiments is shown. (B) After immunoprecipitations as in (A) in the presence of 1% Empigen-BB detergent, purified Flag-Gemin5 was incubated with [32P]UTP-labeled 7S RNA, U4 snRNA or U1 snRNA, in the constant presence of U6 snRNA used as an internal negative control. (C) Recombinant GST-Gemin5, GST-SMN or GST were expressed in E. coli and bound to glutathione-Sepharose beads. The proteins were incubated with [32P]UTP-labeled 7S RNA or U1 snRNA, in the constant presence of U6 snRNA, used as an internal negative control. The recombinant proteins used in the study have been analyzed by SDS–PAGE and coomassie staining. In panels B and C, bound RNAs were isolated after washing and analyzed by electrophoresis on denaturing polyacrylamide gels. Lanes labeled ‘Total’ represent 10% of the input.
The SMN complex associates with SRP in Hela cell extracts
To determine whether the 7S interaction with the SMN complex has a biological significance, we next tested whether the SMN complex was able to associate with SRP in cellulo. Our strategy was to immunoprecipitate HeLa cell extracts with anti-SRP9 (Figure 6A) or anti-SRP54 (Figure 6B) antibodies and to test for the presence of proteins of the SMN complex in the immunoprecipitated material. We also used an anti-SMN antibody 4B3 and tested whether SRP co-immunoprecipitated with SMN (Figure 6C). The immunoprecipitated proteins (IP) and proteins from an aliquot of the respective extracts (Total) were fractionated by SDS–PAGE and analyzed by western blotting. The anti-SRP9 and anti-SRP54 antibodies efficiently immunoprecipitated endogenous SRP9 and SRP54 proteins, respectively, as well as other SRP proteins (SRP68 and SRP14), indicating that the entire SRP particle was immunoprecipitated. Importantly, consistent with an association of SRP with the SMN complex in vivo, all the components of the SMN complex that were tested (SMN and Gemin2, 3, 4 and 5) were found to co-immunoprecipitate with SRP9 (Figure 6A) and SRP54 (Figure 6B). In the reverse experiment, the anti-SMN antibody immunoprecipitated the SRP proteins that were tested, i.e. SRP9, SRP19 and SRP68. The 7S RNA was also associated with SMN, whereas the mRNA coding for Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) did not and was used as a negative control (Figure 6C). Our results support the conclusion that the SMN complex associates with the SRP in HeLa cells.
Figure 6.
The SMN complex associates with SRP in HeLa cell extracts. IP experiments were carried out on HeLa cytoplasmic extracts using anti-SRP9 (A), anti-SRP54 (B), or anti-SMN 4B3 (C) antibodies bound to A-sepharose [in (A) and (B)] or G-dynabeads [in (C)]. The unrelated anti-Sox2 and anti-Lin28 antibodies were used as negative controls in (A) (Controls 1 and 2, respectively). A-sepharose alone was used as control in (B) and negative control mouse IgG (DAKO) in (C). The immunoprecipitated proteins were analyzed by SDS–PAGE and western-blotting with antibodies to the indicated proteins; 3% of the inputs are shown (Total). The immunoprecipitated RNAs were analyzed by RT–PCR in (C).
SMN and Gemin2 associate with the 7S RNA in the cytoplasm of X. laevis oocytes
Our observations that 7S RNA can interact with purified SMN complexes and that SRP associates with the SMN complex in human cells suggested a possible involvement of the SMN complex in SRP biogenesis. As microinjection of vertebrate UsnRNAs in X. laevis oocytes had been one key approach to demonstrate the role of the SMN complex in UsnRNP assembly (22,55), we decided to use this approach to test our hypothesis. This was as much justified as human and X. laevis 7S RNA microinjections in X. laevis oocytes had also been previously used for in cellulo study of SRP assembly (51,52). Previous microinjection of 7S RNA in the nucleus of X. laevis oocytes, showed that it is first localized in the nucleolus before being exported to and stably accumulated in the cytoplasm (51,52). Importantly, in X. laevis oocytes, the SMN and Gemin2 proteins are detected in the cytoplasm but not in nuclei (22). As a first step in the demonstration of our hypothesis, we had to test whether the X. laevis SMN complex can associate with 7S RNA in oocytes. To this end, we produced an in vitro transcribed [32P]UTP-labeled human 7S RNA that we injected into the nucleus of oocytes and after 6 h of incubation, the oocytes were manually dissected. More than 50% of the injected 7S RNA was found to be transported to the cytoplasm (C), whereas the remaining part was still in the nucleus (N) (Figure 7A, lanes 1 and 2). We performed IPs on both the nuclear and cytoplasmic fractions, using antibodies that were able to recognize specifically the endogenous SMN, Gemin2 and SRP54 proteins and non-immune antibodies (IP control). We used nuclear fractions as negative controls in these assays. In addition to 7S RNA, we microinjected two other radiolabeled RNAs: (i) X. laevis U1 snRNA was used as a positive control because it has been shown to associate with the SMN complex in the cytoplasm of X. laevis oocytes (22) and (ii) a truncated version of human 7S RNA (Alu+short stem) was used as a negative control, because this RNA failed to bind efficiently to the purified SMN complexes in our in vitro experiments (Figure 3). In agreement with previous data (51), a portion of the injected 7S Alu+short stem was transported to the cytoplasm (Figure 7A, lanes 1 and 2). Microinjected human U6 snRNA, which is known to be retained in the nucleus (77), was used to control the quality of the dissections. As expected, (i) anti-SMN and anti-Gemin2 antibodies immunoprecipitated U1 snRNA from the cytoplasmic fraction (Figure 7A, lanes 4 and 6, respectively), but not the nuclear fraction (Figure 7A, lanes 3 and 5, respectively) and (ii) anti-SRP54 and non-immune antibodies did not immunoprecipitate U1 snRNA (Figure 7A, lanes 7–10). In contrast, we found that the radiolabeled 7S RNA, which had been transported to the cytoplasm, was immunoprecipitated by anti-SRP54 antibodies (Figure 7A, lane 8), indicating the interaction of SRP54 with the injected RNA. Radiolabeled 7S RNA was also efficiently immunoprecipitated from the cytoplasmic fraction when using anti-SMN and anti-Gemin2 antibodies, whereas the 7S Alu+short stem was not (Figure 7A, lanes 4 and 6). These results demonstrated a cytoplasmic association of 7S RNA with SMN and Gemin2 proteins after its injection in the nuclei of X. laevis oocytes, which suggested a possible role of the SMN complex in the cytoplasmic step of SRP assembly.
Figure 7.
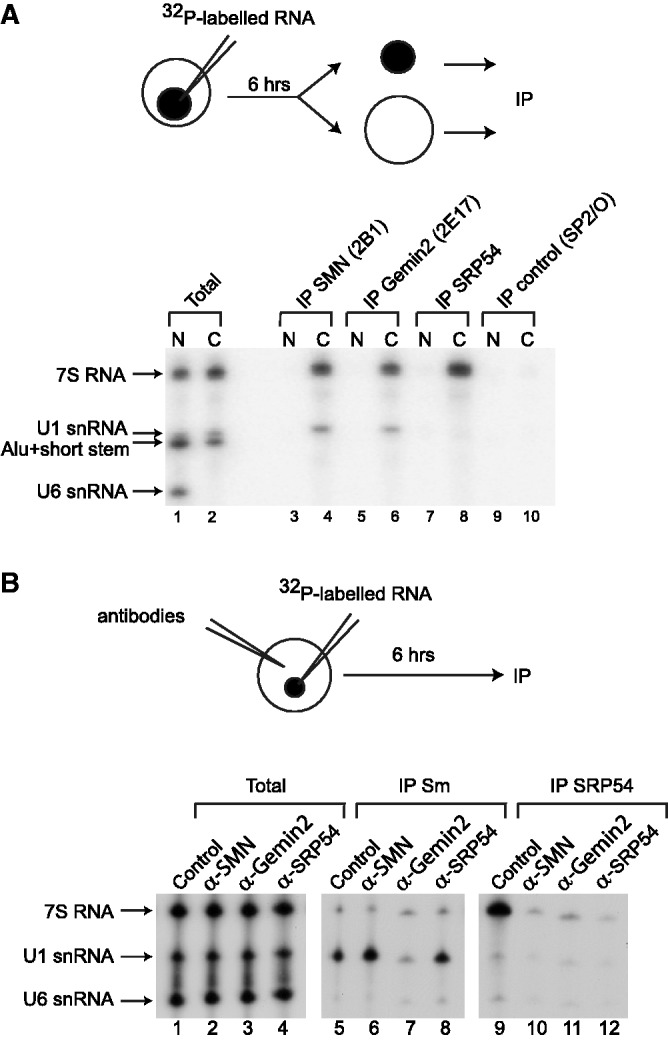
Anti-SMN and anti-Gemin2 antibodies interfere with the binding of SRP54 on 7S RNA in the cytoplasm of X. laevis oocytes. (A) A mixture of [32P]UTP-labeled U1, U6, 7S and 7S 5′-3′-regions RNAs was injected into the nucleus of X. laevis oocytes. After incubation for 6 h, the oocytes were manually dissected into nuclear (N) and cytoplasmic (C) fractions. IPs were then carried out from both fractions with the anti-SMN (2B1), anti-Gemin2 (2E17), anti-SRP54 or control non-immune (SP2/O) antibodies. Bound RNAs were analyzed by electrophoresis on a denaturing polyacrylamide gel and autoradiography. About 10% of each fraction was loaded on the gel (Total). (B) Either non-immune (SP2/O), anti-SRP54, anti-Gemin2 (2E17) or anti-SMN (2B1) antibodies were injected into the cytoplasm of X. laevis oocytes. The same oocytes were nuclear injected 1 h later with a mixture of [32P]UTP-labeled U6 snRNA and 7S RNA. After 6 h, total cellular extracts were prepared from these oocytes. Immunoprecipitations were then carried out with the anti-Sm core (Y12) (IP Y12) or anti-SRP54 (IP SRP54) antibodies. RNAs were analyzed by electrophoresis on a denaturing polyacrylamide gel and autoradiography. About 10% of each injected RNA was loaded on the gel (Total).
Anti-SMN and anti-Gemin2 antibodies strongly interfere with SRP54 association to 7S RNA in X. laevis oocytes
The cytoplasmic step of SRP assembly consists in SRP54 association to 7S RNA. To test whether the SMN complex might be involved in this step in the cytoplasm of X. laevis oocytes, we microinjected anti-SMN (2B1), anti-Gemin2 (2E17) or anti-SRP54 antibodies in the cytoplasm of oocytes and tested their effects on the association of SRP54 with 7S RNA which was microinjected 1 h later in the nucleus. Microinjection of non-immune antibodies (SP2/O) was used as a negative control, and U6 snRNA and U1 snRNA were co-injected with 7S RNA. After 6 h of incubation, total extracts were prepared and immunoprecipitations were carried out using either the anti-Sm core protein antibody (Y12) (66) to monitor the assembly of Sm proteins on U1 snRNA, or anti-SRP54 antibodies to test for SRP54 binding to the 7S RNA. As expected, Y12 antibodies did not immunoprecipitate U6 snRNA and 7S RNA, which both do not associate with Sm proteins (Figure 7B, lanes 5–8) and anti-SRP54 antibodies did not immunoprecipitate U6 and U1 snRNAs that do not interact with SRP54 (Figure 7B, lanes 9–12). Pre-injection of oocytes with non-immune (SP2/O, Control) or anti-SRP54 antibodies, did not alter Sm protein association with nuclear-injected U1 snRNA as evidenced by its efficient immunoprecipitation with Y12 antibodies (Figure 7B, lanes 5 and 8). As expected (22), the anti-Gemin2 (2E17) antibody inhibited the Sm core assembly on U1 snRNA (Figure 7B, lane 7), while as previously described (22), the anti-SMN antibody (2B1) unexpectedly slightly stimulated the assembly (Figure 7B, lane 6). When the oocytes were pre-injected with non-immune antibody (Control) (Figure 7B, lane 9), the injected 7S RNA was efficiently immunoprecipitated with anti-SRP54 antibody. In contrast, after pre-injection of anti-SRP54 antibody in the cytoplasm of oocytes (Figure 7B, lane 12), the injected 7S RNA was not immunoprecipitated with anti-SRP54 antibody. Thus, upon binding to its SRP54 epitope, the anti-SRP54 antibody abolished the association of SRP54 with 7S RNA. We observed that the association of 7S RNA with SRP54 was inhibited in oocytes that were pre-injected with anti-SMN (2B1) or anti-Gemin2 (2E17) antibodies (Figure 7B, lanes 10 and 11). Thus, we could conclude that occlusion of SMN or Gemin2 epitopes by antibodies interfered with the association of SRP54 to microinjected 7S RNA. Taken together, these data were strong evidence in favor of a role of the SMN complex in the last step of SRP biogenesis, i.e. association of SRP54 to 7S RNA in the cytoplasm.
7S RNA associates with SMN in S. pombe and its level is decreased in a mutant carrying a temperature-degron allele of SMN
Another approach to test for a possible role of the SMN complex in SRP assembly was to use yeast genetic methods. As mentioned in the ‘Introduction’ section, although the SMN complex is not present in S. cerevisiae, an SMN protein is present in S. pombe and a tdSMN strain carrying a thermosensitive-degron SMN allele had been produced (30). On the other hand, SRP is highly conserved in evolution and the sequence of the S. pombe 7S RNA gene (srp7) had been reported (45,46). The S. pombe 7S RNA is 254-nt long and shows ∼50% of sequence identity to its human counterpart, its secondary structure being strikingly similar to the proposed structure of human 7S RNA (46). Therefore, S. pombe turned to be an excellent model to investigate the possible involvement of the SMN complex in SRP biogenesis. First, we determined whether SMN also associates with 7S RNA in S. pombe. To this end, after in vivo RNA–protein cross-linking with formaldehyde of a S. pombe strain expressing a TAP-tagged version of the fission yeast SMN protein, we performed immunoprecipitation experiments under stringent conditions using IgG Sepharose beads (78). RT–PCR analysis of RNAs covalently cross-linked to and co-purified with TAP-SMN revealed that 7S RNA and, as expected, U2 snRNA were present in the pellet (Figure 8A), whereas other RNAs, such as U3 snoRNA and tRNASer showed no significant co-purification. These observations revealed an association of the SMN protein with the RNA component of SRP in S. pombe cells.
Figure 8.
The SMN protein associates with the 7S RNA in vivo in fission yeast cells and is required for the stability of the SRP complex. (A) Quantitative RT–PCR experiments were performed for the indicated RNA species using RNA affinity-purified from extracts prepared from fission yeast cells carrying a TAP-SMN construct or an empty TAP vector as control. Experiments were done in duplicate. (B) Northern blot analysis of RNA isolated from wild-type and temperature-sensitive tdSMN cells before and after shift for 8 h at 37°C. The RNA was separated on 6% denaturing polyacrylamide gels, subjected to northern blot and hybridized with oligonucleotide probes for the indicated RNA. (C) Analysis of RNP complexes in wild-type and temperature-sensitive tdSMN cells by native gel electrophoresis. Extracts were prepared from cells grown before and after a 8-h shift at 37°C and 20 µg of extract were separated on 4% native gels. The RNA was subjected to northern blot analysis and hybridized with probes for the indicated RNAs. The arrow points to the position of the U2/U5/U6 tri-snRNP.
Next, as SRP assembly was expected to be required for 7S RNA stability, we tested whether SMN was required for the accumulation of the S. pombe 7S RNA. To do so, northern blot analyses were performed on RNA extracted from a S. pombe wild-type strain and the tdSMN strain carrying a temperature-degron allele of SMN (30). After a temperature shift from 25°C to 37°C for 8 h, the 7S RNA level decreased by 40% in the mutant strain, whereas no temperature-related change was found in the wild-type strain (Figure 8B). As expected, a decrease of the spliceosomal U2 snRNA was also observed upon temperature shift, whereas the level of U3 snoRNA appeared unchanged (Figure 8B) (30). RNP levels were also analyzed using native gel electrophoresis that has previously been used to study RNP complexes in fission yeast (30,79). As shown in Figure 8C (panels at right), the U2 snRNP, as well as the U2/U5/U6 tri-snRNP (indicated by an arrow) can be observed in extracts prepared from wild-type and tdSMN cells at 25°C, but were strongly destabilized after a 8-h shift to 37°C in the tdSMN strain (lane 11), as expected from the requirement of SMN for spliceosomal snRNPs assembly (30). A decrease in the level of the SRP particle was also observed in the tdSMN cells upon shift to 37° (Figure 8C, compare lanes 1 and 3), whereas no change in the amount of the 7S RNP occurs in wild-type cells (compare lanes 2 and 4). Finally, similar ratios of U3 snoRNP were found in extracts prepared from the tdSMN and wild-type cells before and after shift to the non-permissive temperature (Figure 8C, middle panels), suggesting that the assembly of this snoRNP does not depend on the SMN complex in S. pombe. It should be noted that the level of the U3 particle appeared to increase slightly after shift to 37°C in the tdSMN background (Figure 8C, lane 7). Given that splicing is strongly inhibited at 37°C in the tdSMN cells (30), this might represent a compensatory response to defective splicing of pre-mRNAs coding for U3-associated proteins. Altogether, our data demonstrated that the SMN protein associates with 7S RNA and has an essential role in the steady-state level of SRP in S. pombe cells, which reinforces the idea of its possible implication in SRP biogenesis.
The steady-state level of 7S RNA is significantly decreased in spinal cord of SMA mice
Based on the data we obtained in both X. laevis oocytes and in S. pombe, we hypothesized that reduced SMN levels in mammalian cells might also affect the accumulation of 7S RNA. Therefore, we tested whether 7S RNA level is affected in an animal model of SMA. To this end, we analyzed total RNA from three different tissues of 10-day-old SMN-deficient mice (SMN2+/+, SMNdelta7+/+, Smn−/−). This severe SMA mouse model (corresponding to a SMA type II phenotype, Jackson’s laboratory, #5025) has a mean life expectancy of 13 days (80). Study on the spinal cord was particularly of high interest because: (i) it contains the motor neurons, which are the cells mainly affected in SMA, and (ii) due to a specific regulation of exon 7 inclusion in the SMN2 mRNA of motor neurons, expression of the full-length functional SMN protein is strongly reduced in SMA motor neurons as compared with other cells (81). Therefore, we compared the levels of 7S RNA in total RNA from spinal cord, brain and heart of five model mice and five controls corresponding to transgenic mice (SMN2+/+, SMNdelta7+/+, Smn+/+), by using reverse transcription with specific oligonucleotides followed by real-time PCR. Interestingly, the 7S level was markedly decreased in spinal cord (∼60%), but not in brain and heart (Figure 9). Our results support the conclusion that a marked decrease of the 7S RNA levels is linked to SMN deficiency in spinal cord of SMA mouse model.
Figure 9.

Analysis of endogenous 7S RNA level in tissues of SMA mice. Total RNA from the spinal cord, brain and heart of post-natal Day 10 control and SMA mice were analyzed by real-time RT–PCR. The level of 7S RNA was monitored by five independent biological replicates (n = 5). The relative amount of 7S RNA in SMA mice tissues is plotted as percent of the controls. RPS29 mRNA was used to normalize the RNA input. The level of 7S RNA in spinal cord was significantly reduced in SMA mice, whereas its levels in brain and heart remain the same (P < 0.05).
DISCUSSION
7S RNA and SRP associate specifically with the SMN complex
In this study, we found that the 7S RNA can bind to purified SMN complexes. The specificity of this 7S interaction is strongly supported by our finding of an association of 7S RNA with the SMN protein in both S. pombe and human cells and the specific association of microinjected 7S RNA with the SMN and Gemin2 proteins in X. laevis oocytes. As we also detected an association of SRP proteins with the SMN complex in HeLa cells, not only 7S but the entire SRP particle is likely able to interact with the SMN complex.
Studies performed on UsnRNAs have revealed a great complexity of the SMN complex RNA recognition mechanisms. Indeed, in spite of the presence of two highly conserved motifs in all UsnRNAs, which bind the Sm proteins (82), namely the Sm site (AUUUUUG) and the 3′ terminal stem–loop structure, the SMN complex carries distinct determinants for the selective recognition of the various UsnRNAs (55,56,83,84). More precisely, the two common UsnRNA motifs are required for recognition of U4 and U5 snRNAs. The 3′ terminal stem–loop structure but not the Sm site is required for binding of U2 snRNA, and the high-affinity binding site for the SMN complex on U1 snRNA lies outside of the two conserved motifs, i.e. in stem–loop 1. A 3′ stem–loop structure is also crucial for minor UsnRNA binding (84). Competition experiments using the entire SMN complex revealed at least two UsnRNA binding sites on the SMN complex, one for U4 snRNA and one for U1 snRNA. In addition, both U1 and U4 snRNAs can fully compete with U2 and U5 bindings, revealing an even higher complexity of the organization of the UsnRNA binding sites within the SMN complex (56). Our study extends this level of complexity, because we showed that the SMN complex can specifically recognize 7S RNA, which contains none of the UsnRNA canonical motifs. Moreover, the binding site for 7S RNA at least partially overlaps the U1 and U2 binding sites, as these two RNAs, but not U4 and U5, specifically compete with 7S binding on the SMN complex. RNAs corresponding to the Alu domain alone or the Alu domain extended with helix 5 did not bind to purified SMN complex, whereas RNAs corresponding to the S-domain and the S-domain extended with helix 5 had the capability to interact albeit at very low level compared with wild-type RNA. These results indicate that the recognition determinants depend on either the entire 7S RNA structure or on multiple domains spread over the RNA molecule. This is confirmed by the fact that three regions spanning the overall 7S RNA (one in the Alu domain, a second in helix 5 and a third in helix 8) were buried within the SMN complex when the RNA was bound.
Gemin5 interacts with all the RNA targets of the SMN complex containing the Sm or Lsm motif (57,84,85). The binding of 7S RNA to the SMN complex is probably also mediated by Gemin5, because we showed the specific interaction of Gemin5 with 7S RNA. Nevertheless, we cannot exclude the possibility that other proteins of the SMN complex contribute to 7S recognition.
Decreased SMN amounts can generate a reduction of the 7S RNA levels
Our observation of an association of both the 7S RNA and the SRP with the SMN complex could reflect an implication of the SMN complex in either SRP biogenesis or activity. It is now clearly proven that the SMN complex plays an essential role for assembly of Sm- and Lsm-containing RNPs, i.e. minor and major UsnRNPs, U7 snRNP and Herpesvirus saimiri HSURs (18,19,22–27,86–88). As a result, SMN deficiency was found to alter the repertoire of UsnRNAs in cells and tissues derived from SMA mice, in HeLa cells depleted of SMN by RNAi, as well as in the S. pombe mutant strain carrying a temperature-degron allele of SMN (28–30,32,89,90). Similarly, an implication of the SMN complex in SRP biogenesis was expected to result in SRP assembly defects in SMN-deficient cells. Furthermore, due to the degradation of misassembled 7S RNA molecule, a reduced level of 7S was expected in SMN-deficient organisms. Accordingly, we found that the expression of the degron-SMN allele in S. pombe decreased the 7S RNA cellular level. Using a mouse model for severe SMA, we also observed a marked decrease of 7S RNA in the spinal cord, a mainly affected tissue in SMA, whereas its level was unchanged in the brain and heart. It should be pointed out that in mice, a low level of SMN does not also cause a uniform reduction of UsnRNAs. Indeed, the reduction is rather cell and/or snRNA-specific (28,29,32,89). Thus, our studies report for the first time a defect in the level of another type of RNA than UsnRNAs in SMN-deficient model organisms. This observation favors the hypothesis of a role of the SMN complex in SRP assembly versus the hypothesis for a role of the SMN complex in SRP activity. Consistent with this hypothesis, we found that microinjection of anti-SMN (2B1) or anti-Gemin2 (2E17) antibodies in the cytoplasm of X. laevis oocytes strongly interfere with the cytoplasmic association of 7S RNA with SRP54. Therefore, we propose that the SMN complex plays a yet-to-be determined role in the cytoplasmic incorporation of SRP54 in SRP. Interestingly, our results suggest that upon 7S RNA binding to the SMN complex, helix 8 is in interaction with the complex and may undergo a conformational change. Indeed, G209 and G210 that form two side-by-side base pairs in helix 8 were more sensitive to cleavages by T1 RNase (Figure 4). This might indicate that the two base pairs are slightly destabilized, or become more accessible in the RNA tertiary structure, when the RNA is bound to the SMN complex. Interestingly, helix 8 is important for individual binding of SRP19 and SRP54 to 7S RNA in vitro (91–94). Moreover, these two base pairs are crucial for the SRP19-dependent binding of SRP54 (95). Therefore, the recognition of helix 8 by the SMN complex and its subsequent conformational change may be important for the SMN complex-mediated SRP assembly.
A defect in SRP biogenesis may contribute to SMA
Although reduced levels of SMN cause SMA, it is still unclear why a defect in a ubiquitously expressed protein causes a selective death of the alpha-motor neurons. A defect in the assembly of UsnRNPs is proposed to cause disease onset leading to abnormal splicing of specific mRNAs and disease progression (28–32,87,96,97). Due to the reduction in the levels of minor UsnRNAs observed in several tissues of SMA mice and to the defect in the assembly of the minor tri-snRNP observed in SMA-derived lymphocytes, one can reasonably think that minor splicing pathway could be specifically affected in SMA, and this may concern gene(s) of high importance for motor neurons activity (28,29,31,89). However, as microarray studies revealed splicing changes in various tissues from SMA mice, it is still not clear whether these alterations have a direct pathogenic role in SMA, or are indirect or secondary changes due to disease progression (29,98). Recently, it has been shown that SMN deficiency perturbs splicing of specific U12 intron-containing genes in mammalian cells and in Drosophila larvae (32).
Western blot analysis indicated that the amounts of SMN in different tissues of the SMNΔex7 mouse model used in this study are very low and barely detectable (29,80,99). Therefore, this low level makes difficult a precise comparison of SMN quantities between tissues. However, a recent study (81) proposed that one reason for the preferential vulnerability of motor neurons to SMA could be a lower expression of full-length SMN from the SMN2 gene in motor neurons compared with other cell types that would be due to a less efficient retention of exon7 in these cells. Thus, a lower level of SMN protein in this tissue might explain the reduction of 7S RNA levels in the spinal cord that we detected in the SMA mice. The SMN levels, albeit very low, might be sufficient to maintain normal 7S RNA levels in other tissues tested.
Our observations open the possibility that reduced SRP levels may contribute to SMA onset and/or severity. The effect of reduced levels of SRP in mammalian tissues, including the spinal cord, has not been studied yet in animal models. However, a crucial importance of SRP has been shown for several microorganisms. SRP is essential for growth in bacteria and in S. pombe (100,101). It is not essential in S. cerevisiae, but its absence considerably slows down cell growth, represses protein synthesis and induces a heat shock response (47,102,103). Tests on the effects of SRP deficiency were performed in mammalian cells using the possibility to reduce SRP protein expression. Reduced SRP14 expression in cultured mammalian cells has been shown to limit protein secretion and cell growth, but was not lethal (104). Furthermore, no growth defect was observed after silencing of SRP54 and SRP72 expression (105,106). However, due to the high stability and high level of expression of SRP, the level of SRP remaining in the cell in such RNA interference experiments and the existence of SRP-independent pathways (107–109) might have been sufficient to ensure protein secretion. Although synthesis, secretion and trafficking of neurotransmitters, neurotrophic factors, ligands and membrane receptors in neurons are crucial mechanisms for neuronal morphogenesis, synapse formation and for the regulation of the synaptic strength, little is currently known on the abundance and the functional importance of SRP in neurons. Most membrane and secreted proteins are synthesized in the cell body of neurons via protein translocation into the endoplasmic reticulum (ER) and are subsequently transported into the dendrites and axons. Neuronal proteins that are synthesized locally in dendrites and axons likely make use of the ER- and Golgi-like structures that have been detected within neurites, so that their synthesis likely also depends upon SRP [for recent reviews, (110,111)]. Hence, the reduced level of SRP that we detected in the spinal cord of the SMA mice model is expected to have a strong negative effect on motorneuronal development and function and consequently on the SMA pathology.
In conclusion, the human SMA disease forms a continuum of severity that may be explained by the reduced levels of several macromolecular machineries having diverse functions, including UsnRNP and SRP. Our data open a new area of research in the perspective of understanding the causes of the SMA pathology, as well as a new conceptual base in the view to decipher how the essential SRP is assembled in vivo.
FUNDING
The CNRS, the ‘Ministère de l’Enseignement Supérieur et de la Recherche’ ; the Association Française contre les Myopathies (AFM) ; pre-doctoral fellowship from the ‘Ministère de l’Enseignement Supérieur et de la Recherche’ and from AFM (to N.P.); pre-doctoral fellowship from the Ministère de l’Enseignement Supérieur et de la Recherche (to M.D.). Funding for open access charge: CNRS, University of Lorraine.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to K. Gould, D. Dobberstein, G. Dreyfuss and L. Pellizzoni for the gift of antibodies and plasmids. We thank Romain Carcenac, Elisa Dominguez and Stéphanie Astord for technical help (maintenance of SMA mice, tissue sample collecting and RNA extraction). S.L. is an investigator from INSERM.
REFERENCES
- 1.Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 2.Coovert DD, Le TT, McAndrew PE, Strasswimmer J, Crawford TO, Mendell JR, Coulson SE, Androphy EJ, Prior TW, Burghes AH. The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 3.Melki J. Spinal muscular atrophy. Curr. Opin. Neurol. 1997;10:381–385. doi: 10.1097/00019052-199710000-00005. [DOI] [PubMed] [Google Scholar]
- 4.Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy- determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 5.Elsheikh B, Prior T, Zhang X, Miller R, Kolb SJ, Moore D, Bradley W, Barohn R, Bryan W, Gelinas D, et al. An analysis of disease severity based on SMN2 copy number in adults with spinal muscular atrophy. Muscle Nerve. 2009;40:652–656. doi: 10.1002/mus.21350. [DOI] [PubMed] [Google Scholar]
- 6.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, Burghes AH, McPherson JD. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 8.Vitte J, Fassier C, Tiziano FD, Dalard C, Soave S, Roblot N, Brahe C, Saugier-Veber P, Bonnefont JP, Melki J. Refined characterization of the expression and stability of the SMN gene products. Am. J. Pathol. 2007;171:1269–1280. doi: 10.2353/ajpath.2007.070399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho S, Dreyfuss G. A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev. 2010;24:438–442. doi: 10.1101/gad.1884910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paushkin S, Gubitz AK, Massenet S, Dreyfuss G. The SMN complex, an assemblyosome of ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:305–312. doi: 10.1016/s0955-0674(02)00332-0. [DOI] [PubMed] [Google Scholar]
- 11.Meister G, Eggert C, Fischer U. SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol. 2002;12:472–478. doi: 10.1016/s0962-8924(02)02371-1. [DOI] [PubMed] [Google Scholar]
- 12.Gubitz AK, Feng W, Dreyfuss G. The SMN complex. Exp. Cell Res. 2004;296:51–56. doi: 10.1016/j.yexcr.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 13.Otter S, Grimmler M, Neuenkirchen N, Chari A, Sickmann A, Fischer U. A comprehensive interaction map of the human survival of motor neuron (SMN) complex. J. Biol. Chem. 2007;282:5825–5833. doi: 10.1074/jbc.M608528200. [DOI] [PubMed] [Google Scholar]
- 14.Hannus S, Buhler D, Romano M, Seraphin B, Fischer U. The Schizosaccharomyces pombe protein Yab8p and a novel factor, Yip1p, share structural and functional similarity with the spinal muscular atrophy-associated proteins SMN and SIP1. Hum. Mol. Genet. 2000;9:663–674. doi: 10.1093/hmg/9.5.663. [DOI] [PubMed] [Google Scholar]
- 15.Owen N, Doe CL, Mellor J, Davies KE. Characterization of the Schizosaccharomyces pombe orthologue of the human survival motor neuron (SMN) protein. Hum. Mol. Genet. 2000;9:675–684. doi: 10.1093/hmg/9.5.675. [DOI] [PubMed] [Google Scholar]
- 16.Paushkin S, Charroux B, Abel L, Perkinson RA, Pellizzoni L, Dreyfuss G. The survival motor neuron protein of Schizosacharomyces pombe. Conservation of survival motor neuron interaction domains in divergent organisms. J. Biol. Chem. 2000;275:23841–23846. doi: 10.1074/jbc.M001441200. [DOI] [PubMed] [Google Scholar]
- 17.Kroiss M, Schultz J, Wiesner J, Chari A, Sickmann A, Fischer U. Evolution of an RNP assembly system: a minimal SMN complex facilitates formation of UsnRNPs in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2008;105:10045–10050. doi: 10.1073/pnas.0802287105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolb SJ, Battle DJ, Dreyfuss G. Molecular functions of the SMN complex. J. Child Neurol. 2007;22:990–994. doi: 10.1177/0883073807305666. [DOI] [PubMed] [Google Scholar]
- 19.Chari A, Paknia E, Fischer U. The role of RNP biogenesis in spinal muscular atrophy. Curr. Opin. Cell Biol. 2009;21:387–393. doi: 10.1016/j.ceb.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Rossoll W, Bassell GJ. Spinal muscular atrophy and a model for survival of motor neuron protein function in axonal ribonucleoprotein complexes. Results Probl. Cell Differ. 2009;48:289–326. doi: 10.1007/400_2009_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burghes AH, Beattie CE. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat. Rev. Neurosci. 2009;10:597–609. doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 23.Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- 24.Meister G, Buhler D, Pillai R, lottspeich F, Fisher U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 2001;3:945––949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- 25.Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- 26.Chari A, Golas MM, Klingenhager M, Neuenkirchen N, Sander B, Englbrecht C, Sickmann A, Stark H, Fischer U. An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell. 2008;135:497–509. doi: 10.1016/j.cell.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 27.Pellizzoni L. Chaperoning ribonucleoprotein biogenesis in health and disease. EMBO Rep. 2007;8:340–345. doi: 10.1038/sj.embor.7400941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gabanella F, Butchbach ME, Saieva L, Carissimi C, Burghes AH, Pellizzoni L. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One. 2007;2:e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campion Y, Neel H, Gostan T, Soret J, Bordonne R. Specific splicing defects in S. pombe carrying a degron allele of the Survival of Motor Neuron gene. EMBO J. 2010;29:1817–1829. doi: 10.1038/emboj.2010.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boulisfane N, Choleza M, Rage F, Neel H, Soret J, Bordonne R. Impaired minor tri-snRNP assembly generates differential splicing defects of U12-type introns in lymphoblasts derived from a type I SMA patient. Hum. Mol. Genet. 2011;20:641–648. doi: 10.1093/hmg/ddq508. [DOI] [PubMed] [Google Scholar]
- 32.Lotti F, Imlach WL, Saieva L, Beck ES, Hao le T, Li DK, Jiao W, Mentis GZ, Beattie CE, McCabe BD, et al. An SMN-dependent U12 splicing event essential for motor circuit function. Cell. 2012;151:440–454. doi: 10.1016/j.cell.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terns MP, Terns RM. Macromolecular complexes: SMN–the master assembler. Curr. Biol. 2001;11:R862–R864. doi: 10.1016/s0960-9822(01)00517-6. [DOI] [PubMed] [Google Scholar]
- 34.Pellizzoni L, Baccon J, Charroux B, Dreyfuss G. The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr. Biol. 2001;11:1079–1088. doi: 10.1016/s0960-9822(01)00316-5. [DOI] [PubMed] [Google Scholar]
- 35.Jones KW, Gorzynski K, Hales CM, Fischer U, Badbanchi F, Terns RM, Terns MP. Direct interaction of the spinal muscular atrophy disease protein SMN with the small nucleolar RNA-associated protein fibrillarin. J. Biol. Chem. 2001;276:38645–38651. doi: 10.1074/jbc.M106161200. [DOI] [PubMed] [Google Scholar]
- 36.Watkins NJ, Lemm I, Ingelfinger D, Schneider C, Hossbach M, Urlaub H, Luhrmann R. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol. Cell. 2004;16:789–798. doi: 10.1016/j.molcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 37.Rossoll W, Jablonka S, Andreassi C, Kroning AK, Karle K, Monani UR, Sendtner M. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J. Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tadesse H, Deschenes-Furry J, Boisvenue S, Cote J. KH-type splicing regulatory protein interacts with survival motor neuron protein and is misregulated in spinal muscular atrophy. Hum. Mol. Genet. 2008;17:506–524. doi: 10.1093/hmg/ddm327. [DOI] [PubMed] [Google Scholar]
- 39.Piazzon N, Rage F, Schlotter F, Moine H, Branlant C, Massenet S. In vitro and in cellulo evidences for association of the survival of motor neuron complex with the fragile X mental retardation protein. J. Biol. Chem. 2008;283:5598–5610. doi: 10.1074/jbc.M707304200. [DOI] [PubMed] [Google Scholar]
- 40.Hubers L, Valderrama-Carvajal H, Laframboise J, Timbers J, Sanchez G, Cote J. HuD interacts with survival motor neuron protein and can rescue spinal muscular atrophy-like neuronal defects. Hum. Mol. Genet. 2011;20:553–579. doi: 10.1093/hmg/ddq500. [DOI] [PubMed] [Google Scholar]
- 41.Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu. Rev. Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 42.Cross BC, Sinning I, Luirink J, High S. Delivering proteins for export from the cytosol. Nat. Rev. Mol. Cell Biol. 2009;10:255–264. doi: 10.1038/nrm2657. [DOI] [PubMed] [Google Scholar]
- 43.Saraogi I, Shan SO. Molecular mechanism of co-translational protein targeting by the signal recognition particle. Traffic. 2011;12:535–542. doi: 10.1111/j.1600-0854.2011.01171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rosenblad MA, Larsen N, Samuelsson T, Zwieb C. Kinship in the SRP RNA family. RNA Biol. 2009;6:508–516. doi: 10.4161/rna.6.5.9753. [DOI] [PubMed] [Google Scholar]
- 45.Ribes V, Dehoux P, Tollervey D. 7SL RNA from Schizosaccharomyces pombe is encoded by a single copy essential gene. EMBO J. 1988;7:231–237. doi: 10.1002/j.1460-2075.1988.tb02804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brennwald P, Liao X, Holm K, Porter G, Wise JA. Identification of an essential Schizosaccharomyces pombe RNA homologous to the 7SL component of signal recognition particle. Mol. Cell Biol. 1988;8:1580–1590. doi: 10.1128/mcb.8.4.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brown JD, Hann BC, Medzihradszky KF, Niwa M, Burlingame AL, Walter P. Subunits of the Saccharomyces cerevisiae signal recognition particle required for its functional expression. EMBO J. 1994;13:4390–4400. doi: 10.1002/j.1460-2075.1994.tb06759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Selinger D, Brennwald P, Althoff S, Reich C, Hann B, Walter P, Wise JA. Genetic and biochemical analysis of the fission yeast ribonucleoprotein particle containing a homolog of Srp54p. Nucleic Acids Res. 1994;22:2557–2567. doi: 10.1093/nar/22.13.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ciufo LF, Brown JD. Nuclear export of yeast signal recognition particle lacking Srp54p by the Xpo1p/Crm1p NES-dependent pathway. Curr. Biol. 2000;10:1256–1264. doi: 10.1016/s0960-9822(00)00743-0. [DOI] [PubMed] [Google Scholar]
- 50.Grosshans H, Deinert K, Hurt E, Simos G. Biogenesis of the signal recognition particle (SRP) involves import of SRP proteins into the nucleolus, assembly with the SRP-RNA, and Xpo1p-mediated export. J. Cell Biol. 2001;153:745–762. doi: 10.1083/jcb.153.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He XP, Bataille N, Fried HM. Nuclear export of signal recognition particle RNA is a facilitated process that involves the Alu sequence domain. J. Cell Sci. 1994;107(Pt 4):903–912. doi: 10.1242/jcs.107.4.903. [DOI] [PubMed] [Google Scholar]
- 52.Sommerville J, Brumwell CL, Politz JC, Pederson T. Signal recognition particle assembly in relation to the function of amplified nucleoli of Xenopus oocytes. J. Cell Sci. 2005;118:1299–1307. doi: 10.1242/jcs.01726. [DOI] [PubMed] [Google Scholar]
- 53.Jacobson MR, Pederson T. Localization of signal recognition particle RNA in the nucleolus of mammalian cells. Proc. Natl Acad. Sci. USA. 1998;95:7981–7986. doi: 10.1073/pnas.95.14.7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Politz JC, Yarovoi S, Kilroy SM, Gowda K, Zwieb C, Pederson T. Signal recognition particle components in the nucleolus. Proc. Natl Acad. Sci. USA. 2000;97:55–60. doi: 10.1073/pnas.97.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yong J, Pellizzoni L, Dreyfuss G. Sequence-specific interaction of U1 snRNA with the SMN complex. EMBO J. 2002;21:1188–1196. doi: 10.1093/emboj/21.5.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yong J, Golembe TJ, Battle DJ, Pellizzoni L, Dreyfuss G. snRNAs contain specific SMN-binding domains that are essential for snRNP assembly. Mol. Cell Biol. 2004;24:2747–2756. doi: 10.1128/MCB.24.7.2747-2756.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Battle DJ, Lau CK, Wan L, Deng H, Lotti F, Dreyfuss G. The Gemin5 protein of the SMN complex identifies snRNAs. Mol. Cell. 2006;23:273–279. doi: 10.1016/j.molcel.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 58.Gubitz AK, Mourelatos Z, Abel L, Rappsilber J, Mann M, Dreyfuss G. Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J. Biol. Chem. 2002;277:5631–5636. doi: 10.1074/jbc.M109448200. [DOI] [PubMed] [Google Scholar]
- 59.Friesen WJ, Dreyfuss G. Specific sequences of the Sm and Sm-like (Lsm) proteins mediate their interaction with the spinal muscular atrophy disease gene product (SMN) J. Biol. Chem. 2000;275:26370–26375. doi: 10.1074/jbc.M003299200. [DOI] [PubMed] [Google Scholar]
- 60.Ullu E, Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984;312:171–172. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- 61.Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- 62.Burlet P, Huber C, Bertrandy S, Ludosky MA, Zwaenepoel I, Clermont O, Roume J, Delezoide AL, Cartaud J, Munnich A, et al. The distribution of SMN protein complex in human fetal tissues and its alteration in spinal muscular atrophy. Hum. Mol. Genet. 1998;7:1927–1933. doi: 10.1093/hmg/7.12.1927. [DOI] [PubMed] [Google Scholar]
- 63.Charroux B, Pellizzoni L, Perkinson RA, Shevchenko A, Mann M, Dreyfuss G. Gemin3: A novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J. Cell Biol. 1999;147:1181–1194. doi: 10.1083/jcb.147.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Charroux B, Pellizzoni L, Perkinson RA, Yong J, Shevchenko A, Mann M, Dreyfuss G. Gemin4. A novel component of the SMN complex that is found in both gems and nucleoli. J. Cell Biol. 2000;148:1177–1186. doi: 10.1083/jcb.148.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carissimi C, Saieva L, Gabanella F, Pellizzoni L. Gemin8 is required for the architecture and function of the survival motor neuron complex. J. Biol. Chem. 2006;281:37009–37016. doi: 10.1074/jbc.M607505200. [DOI] [PubMed] [Google Scholar]
- 66.Lerner EA, Lerner MR, Janeway CA, Jr, Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc. Natl Acad. Sci. USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Choi YD, Dreyfuss G. Monoclonal antibody characterization of the C proteins of heterogeneous nuclear ribonucleoprotein complexes in vertebrate cells. J. Cell Biol. 1984;99:1997–1204. doi: 10.1083/jcb.99.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pellizzoni L, Baccon J, Rappsilber J, Mann M, Dreyfuss G. Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J. Biol. Chem. 2002;277:7540–7545. doi: 10.1074/jbc.M110141200. [DOI] [PubMed] [Google Scholar]
- 69.Chesnel F, Bonnec G, Tardivel A, Boujard D. Comparative effects of insulin on the activation of the Raf/Mos-dependent MAP kinase cascade in vitellogenic versus postvitellogenic Xenopus oocytes. Dev. Biol. 1997;188:122–133. doi: 10.1006/dbio.1997.8631. [DOI] [PubMed] [Google Scholar]
- 70.Hamon S, Le Sommer C, Mereau A, Allo MR, Hardy S. Polypyrimidine tract-binding protein is involved in vivo in repression of a composite internal/3′-terminal exon of the Xenopus alpha-tropomyosin Pre-mRNA. J. Biol. Chem. 2004;279:22166–22175. doi: 10.1074/jbc.M313809200. [DOI] [PubMed] [Google Scholar]
- 71.Le Sommer C, Lesimple M, Mereau A, Menoret S, Allo MR, Hardy S. PTB regulates the processing of a 3′-terminal exon by repressing both splicing and polyadenylation. Mol. Cell. Biol. 2005;25:9595–9607. doi: 10.1128/MCB.25.21.9595-9607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Siomi MC, Eder PS, Kataoka N, Wan L, Liu Q, Dreyfuss G. Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J. Cell Biol. 1997;138:1181–1192. doi: 10.1083/jcb.138.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Labourier E, Allemand E, Brand S, Fostier M, Tazi J, Bourbon HM. Recognition of exonic splicing enhancer sequences by the Drosophila splicing repressor RSF1. Nucleic Acids Res. 1999;27:2377–2386. doi: 10.1093/nar/27.11.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gould KL, Ren L, Feoktistova AS, Jennings JL, Link AJ. Tandem affinity purification and identification of protein complex components. Methods. 2004;33:239–244. doi: 10.1016/j.ymeth.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 75.Gundelfinger ED, Di Carlo M, Zopf D, Melli M. Structure and evolution of the 7SL RNA component of the signal recognition particle. EMBO J. 1984;3:2325–2332. doi: 10.1002/j.1460-2075.1984.tb02134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Larsen N, Zwieb C. SRP-RNA sequence alignment and secondary structure. Nucleic Acids Res. 1991;19:209–215. doi: 10.1093/nar/19.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vankan P, McGuigan C, Mattaj IW. Domains of U4 and U6 snRNAs required for snRNP assembly and splicing complementation in Xenopus oocytes. EMBO J. 1990;9:3397–3404. doi: 10.1002/j.1460-2075.1990.tb07541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26:182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 79.Huang T, Vilardell J, Query CC. Pre-spliceosome formation in S.pombe requires a stable complex of SF1-U2AF(59)-U2AF(23) EMBO J. 2002;21:5516–5526. doi: 10.1093/emboj/cdf555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Le TT, Pham LT, Butchbach ME, Zhang HL, Monani UR, Coovert DD, Gavrilina TO, Xing L, Bassell GJ, Burghes AH. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 81.Ruggiu M, McGovern VL, Lotti F, Saieva L, Li DK, Kariya S, Monani UR, Burghes AH, Pellizzoni L. A role for SMN exon 7 splicing in the selective vulnerability of motor neurons in spinal muscular atrophy. Mol. Cell Biol. 2011;32:126–138. doi: 10.1128/MCB.06077-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Branlant C, Krol A, Ebel JP, Lazar E, Haendler B, Jacob M. U2 RNA shares a structural domain with U1, U4, and U5 RNAs. EMBO J. 1982;1:1259–1265. doi: 10.1002/j.1460-2075.1982.tb00022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Golembe TJ, Yong J, Dreyfuss G. Specific sequence features, recognized by the SMN complex, identify snRNAs and determine their fate as snRNPs. Mol. Cell Biol. 2005;25:10989–11004. doi: 10.1128/MCB.25.24.10989-11004.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yong J, Kasim M, Bachorik JL, Wan L, Dreyfuss G. Gemin5 delivers snRNA precursors to the SMN complex for snRNP biogenesis. Mol. Cell. 2010;38:551–562. doi: 10.1016/j.molcel.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lau CK, Bachorik JL, Dreyfuss G. Gemin5-snRNA interaction reveals an RNA binding function for WD repeat domains. Nat. Struct. Mol. Biol. 2009;16:486–491. doi: 10.1038/nsmb.1584. [DOI] [PubMed] [Google Scholar]
- 86.Pillai RS, Grimmler M, Meister G, Will CL, Luhrmann R, Fischer U, Schumperli D. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 2003;17:2321–2333. doi: 10.1101/gad.274403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Golembe TJ, Yong J, Battle DJ, Feng W, Wan L, Dreyfuss G. Lymphotropic Herpesvirus saimiri uses the SMN complex to assemble Sm cores on its small RNAs. Mol. Cell Biol. 2005;25:602–611. doi: 10.1128/MCB.25.2.602-611.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Azzouz TN, Pillai RS, Dapp C, Chari A, Meister G, Kambach C, Fischer U, Schumperli D. Toward an assembly line for U7 snRNPs: interactions of U7-specific Lsm proteins with PRMT5 and SMN complexes. J. Biol. Chem. 2005;280:34435–34440. doi: 10.1074/jbc.M505077200. [DOI] [PubMed] [Google Scholar]
- 89.Workman E, Saieva L, Carrel TL, Crawford TO, Liu D, Lutz C, Beattie CE, Pellizzoni L, Burghes AH. A SMN missense mutation complements SMN2 restoring snRNPs and rescuing SMA mice. Hum. Mol. Genet. 2009;18:2215–2229. doi: 10.1093/hmg/ddp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jodelka FM, Ebert AD, Duelli DM, Hastings ML. A feedback loop regulates splicing of the spinal muscular atrophy-modifying gene, SMN2. Hum. Mol. Genet. 2010;19:4906–4917. doi: 10.1093/hmg/ddq425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wild K, Sinning I, Cusack S. Crystal structure of an early protein-RNA assembly complex of the signal recognition particle. Science. 2001;294:598–601. doi: 10.1126/science.1063839. [DOI] [PubMed] [Google Scholar]
- 92.Hainzl T, Huang S, Sauer-Eriksson AE. Structure of the SRP19 RNA complex and implications for signal recognition particle assembly. Nature. 2002;417:767–771. doi: 10.1038/nature00768. [DOI] [PubMed] [Google Scholar]
- 93.Kuglstatter A, Oubridge C, Nagai K. Induced structural changes of 7SL RNA during the assembly of human signal recognition particle. Nat. Struct. Biol. 2002;9:740–744. doi: 10.1038/nsb843. [DOI] [PubMed] [Google Scholar]
- 94.Oubridge C, Kuglstatter A, Jovine L, Nagai K. Crystal structure of SRP19 in complex with the S domain of SRP RNA and its implication for the assembly of the signal recognition particle. Mol. Cell. 2002;9:1251–1261. doi: 10.1016/s1097-2765(02)00530-0. [DOI] [PubMed] [Google Scholar]
- 95.Yin J, Yang CH, Zwieb C. Two strategically placed base pairs in helix 8 of mammalian signal recognition particle RNA are crucial for the SPR19-dependent binding of protein SRP54. RNA. 2004;10:574–580. doi: 10.1261/rna.5232404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wan L, Battle DJ, Yong J, Gubitz AK, Kolb SJ, Wang J, Dreyfuss G. The survival of motor neurons protein determines the capacity for snRNP assembly: biochemical deficiency in spinal muscular atrophy. Mol. Cell. Biol. 2005;25:5543–5551. doi: 10.1128/MCB.25.13.5543-5551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Winkler C, Eggert C, Gradl D, Meister G, Giegerich M, Wedlich D, Laggerbauer B, Fischer U. Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes Dev. 2005;19:2320–2330. doi: 10.1101/gad.342005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baumer D, Lee S, Nicholson G, Davies JL, Parkinson NJ, Murray LM, Gillingwater TH, Ansorge O, Davies KE, Talbot K. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet. 2009;5:e1000773. doi: 10.1371/journal.pgen.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Le TT, McGovern VL, Alwine IE, Wang X, Massoni-Laporte A, Rich MM, Burghes AH. Temporal requirement for high SMN expression in SMA mice. Hum. Mol. Genet. 2011;20:3578–3591. doi: 10.1093/hmg/ddr275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Phillips GJ, Silhavy TJ. The E. coli ffh gene is necessary for viability and efficient protein export. Nature. 1992;359:744–746. doi: 10.1038/359744a0. [DOI] [PubMed] [Google Scholar]
- 101.Althoff SM, Stevens SW, Wise JA. The Srp54 GTPase is essential for protein export in the fission yeast Schizosaccharomyces pombe. Mol. Cell. Biol. 1994;14:7839–7854. doi: 10.1128/mcb.14.12.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hann BC, Walter P. The signal recognition particle in S. cerevisiae. Cell. 1991;67:131–144. doi: 10.1016/0092-8674(91)90577-l. [DOI] [PubMed] [Google Scholar]
- 103.Mutka SC, Walter P. Multifaceted physiological response allows yeast to adapt to the loss of the signal recognition particle-dependent protein-targeting pathway. Mol. Biol. Cell. 2001;12:577–588. doi: 10.1091/mbc.12.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lakkaraju AK, Mary C, Scherrer A, Johnson AE, Strub K. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell. 2008;133:440–451. doi: 10.1016/j.cell.2008.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lakkaraju AK, Luyet PP, Parone P, Falguieres T, Strub K. Inefficient targeting to the endoplasmic reticulum by the signal recognition particle elicits selective defects in post-ER membrane trafficking. Exp. Cell Res. 2007;313:834–847. doi: 10.1016/j.yexcr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 106.Ren YG, Wagner KW, Knee DA, Aza-Blanc P, Nasoff M, Deveraux QL. Differential regulation of the TRAIL death receptors DR4 and DR5 by the signal recognition particle. Mol. Biol. Cell. 2004;15:5064–5074. doi: 10.1091/mbc.E04-03-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rapoport TA, Matlack KE, Plath K, Misselwitz B, Staeck O. Posttranslational protein translocation across the membrane of the endoplasmic reticulum. Biol. Chem. 1999;380:1143–1150. doi: 10.1515/BC.1999.145. [DOI] [PubMed] [Google Scholar]
- 108.Kraut-Cohen J, Gerst JE. Addressing mRNAs to the ER: cis sequences act up! Trends Biochem. Sci. 2011;35:459–469. doi: 10.1016/j.tibs.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 109.del Alamo M, Hogan DJ, Pechmann S, Albanese V, Brown PO, Frydman J. Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. PLoS Biol. 2011;9:e1001100. doi: 10.1371/journal.pbio.1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Valenzuela JI, Jaureguiberry-Bravo M, Couve A. Neuronal protein trafficking: emerging consequences of endoplasmic reticulum dynamics. Mol. Cell. Neurosci. 2011;48:269–277. doi: 10.1016/j.mcn.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 111.Ramirez OA, Couve A. The endoplasmic reticulum and protein trafficking in dendrites and axons. Trends Cell Biol. 2011;21:219–227. doi: 10.1016/j.tcb.2010.12.003. [DOI] [PubMed] [Google Scholar]








