Abstract
Bacterial degradation of steroids is widespread, but the metabolic pathways have rarely been explored. Previous studies with Pseudomonas sp. strain Chol1 and the C24 steroid cholate have shown that cholate degradation proceeds via oxidation of the A ring, followed by cleavage of the C5 acyl side chain attached to C-17, with 7α,12β-dihydroxy-androsta-1,4-diene-3,17-dione (12β-DHADD) as the product. In this study, the pathway for degradation of the acyl side chain of cholate was investigated in vitro with cell extracts of strain Chol1. For this, intermediates of cholate degradation were produced with mutants of strain Chol1 and submitted to enzymatic assays containing coenzyme A (CoA), ATP, and NAD+ as cosubstrates. When the C24 steroid (22E)-7α,12α-dihydroxy-3-oxochola-1,4,22-triene-24-oate (DHOCTO) was used as the substrate, it was completely transformed to 12α-DHADD and 7α-hydroxy-androsta-1,4-diene-3,12,17-trione (HADT) as end products, indicating complete removal of the acyl side chain. The same products were formed with the C22 steroid 7α,12α-dihydroxy-3-oxopregna-1,4-diene-20-carboxylate (DHOPDC) as the substrate. The 12-keto compound HADT was transformed into 12β-DHADD in an NADPH-dependent reaction. When NAD+ was omitted from assays with DHOCTO, a new product, identified as 7α,12α-dihydroxy-3-oxopregna-1,4-diene-20S-carbaldehyde (DHOPDCA), was formed. This aldehyde was transformed to DHOPDC and DHOPDC-CoA in the presence of NAD+, CoA, and ATP. These results revealed that degradation of the C5 acyl side chain of cholate does not proceed via classical β-oxidation but via a free aldehyde that is oxidized to the corresponding acid. The reaction leading to the aldehyde is presumably catalyzed by an aldolase encoded by the gene skt, which was previously predicted to be a β-ketothiolase.
INTRODUCTION
Steroids constitute a large and diverse class of natural compounds. While steroids serve various functions in eukaryotic organisms, such as membrane constituent and hormonal functions, they occur only rarely in prokaryotic organisms. Nevertheless, many bacteria are capable of transforming steroids, and this property is used for the biotechnological production of steroid drugs (1–3). In addition, bacteria from diverse phylogenetic groups can degrade naturally occurring steroids completely and use them as sources of carbon and energy. Bacteria are also able to degrade synthetic steroids, which is relevant for public health, because the occurrence of steroid drugs in the environment is believed to influence the fertility of animals and humans (4). Compared to the degradation of other natural compounds, such as aromatic compounds, relatively little is known about bacterial degradation of steroids. To date, only one bacterial metabolic pathway for aerobic degradation of steroids, the 9,10-seco pathway, has been described in detail (5–9).
The 9,10-seco pathway starts with oxidative reactions at the A ring of the steroid that lead to the formation of Δ1,4-3-keto structures. Subsequently, the side chain attached to the D ring at C-17 is removed in a stepwise fashion, leading to the production of androsta-Δ1,4-diene-3,17-diones (ADDs), which are the central intermediates of the 9,10-seco pathway (8). ADDs are further degraded by a monooxygenase-catalyzed hydroxylation at C-9, which causes the opening of the B ring concomitantly with the aromatization of the A ring (10). The resulting 9,10-seco steroids are further degraded by the breakdown of the aromatic A ring (11, 12). The remaining C and D rings form acidic perhydroindane derivatives that are further degraded by as-yet-unknown reactions (13). Recently, it was shown that the degradation of rings A and B can also precede side chain degradation in Rhodococcus jostii (14).
While the reactions leading to Δ1,4-3-keto structures and the breakdown of ADDs have been well studied, the removal of the steroid side chain has not yet been explored in great detail. We investigated this process by studying the degradation of the C5 acyl side chain of the bile salt cholate (compound I in Fig. 1) by Pseudomonas sp. strain Chol1 (15). Bile salts are surface-active steroid compounds that aid the digestion of lipophilic nutrients in the digestive tracts of vertebrates and enter the environment in significant amounts by excretion (16, 17).
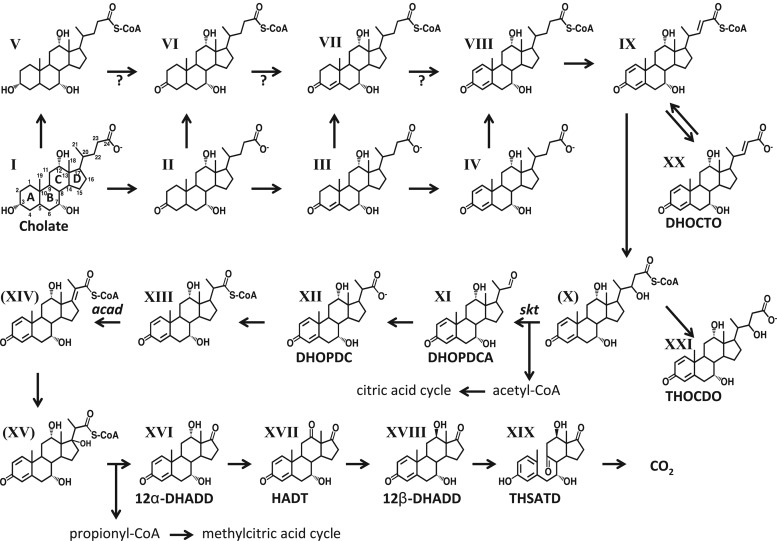
Fig 1.
Section of the proposed pathway of cholate (compound I) degradation in Pseudomonas sp. strain Chol1. The following compounds have been identified: II, 3-ketocholate; III, Δ1/4-3-ketocholate (the position of the double bond has not been identified yet; for simplicity, only the more probable Δ4 isomer was chosen); IV, Δ1,4-3-ketocholate; V, cholyl-CoA; VI, 3-ketocholyl-CoA; VII, Δ1/4-3-ketocholyl-CoA; VIII, Δ1,4-3-ketocholyl-CoA; IX, CoA ester of (22E)7α,12α-dihydroxy-3-oxochola-1,4-triene-24-oate (DHOCTO); XI, 7α,12α-dihydroxy-3-oxopregna-1,4-diene-20S-carbaldehyde (DHOPDCA); XII, 7α,12α-dihydroxy-3-oxopregna-1,4-diene-20-carboxylate (DHOPDC); XIII, CoA ester of DHOPDC; XVI, 7α,12α-dihydroxy-androsta-1,4-diene-3,17-dione (12α-DHADD); XVII, 7α-hydroxy-androsta-1,4-diene-3,12,17-trione (HADT); XVIII, 12β-DHADD; XIX, 3,7,12-trihydroxy-9,10-secoandrosta-1,3,5(10)triene-9,17-dione (THSATD); XX, DHOCTO; and XXI, 7α,12α22-trihydroxy-3-oxochola-1,4-diene-24-oate (THOCDO). The compounds shown in brackets are plausible intermediates that have not been detected yet.
The degradation of the C5 acyl side chain is believed to proceed via the stepwise removal of an acetyl and a propionyl residue from the steroid skeleton. This pathway is supported by the analysis of two transposon mutants of strain Chol1 that have defects in the degradation of the acyl side chain of cholate.
The first mutant, strain G12, is defective in the gene skt, which encodes a putative thiolase (18). When this mutant is incubated with cholate, two steroid compounds with a complete C5 acyl side chain accumulate in culture supernatants, namely, (22E)-7α,12α-dihydroxy-3-oxochola-1,4,22-triene-24-oate (DHOCTO, XX) and 7α,12α,22-trihydroxy-3-oxochola-1,4-diene-24-oate (THOCDO, XXI). The second mutant, strain R1, is defective in the gene acad, which encodes a putative acyl coenzyme A (CoA) dehydrogenase (19). When this mutant is incubated with cholate, a steroid compound with a C3 acyl side chain, namely, 7α,12α-dihydroxy-3-oxopregna-1,4-diene-20-carboxylate (DHOPDC, XII), accumulates in culture supernatants as a dead-end product.
The phenotypes and genotypes of these mutants strongly suggest that the first part of side chain degradation proceeds via a classical β-oxidation, in which the putative thiolase Skt would catalyze the thiolytic cleavage of acetyl-CoA from the side chain with the CoA ester of DHOPDC as a product (8). For this, a β-keto-acyl-CoA substrate for Skt originating from oxidation of the C-22 hydroxyl group of THOCDO-CoA (X) has to be postulated. DHOPDC-CoA would then be the substrate for Acad that would be responsible for introducing a double bond into the acyl side chain by an oxidative reaction. After hydration of the double bond, the C3 acyl side chain is believed to be cleaved from the steroid skeleton by an aldolase reaction (8).
While this pathway for degradation of the C5 acyl side chain appears biochemically plausible, it has never been confirmed in vitro and, thus, is still hypothetical. In particular, the putative β-keto-acyl-CoA substrate for Skt has never been detected, and the last two reactions of the removal of the C3 acyl side chain have also not yet been verified.
Thus, the goal of our study was to elucidate the pathway for degradation of the C5 acyl side chain of cholate in vitro by using cell extracts of strain Chol1 and intermediates of cholate degradation (such as DHOCTO and DHOPDC) as substrates. In addition, we wanted to elucidate the fate of the acetyl-CoA and propionyl-CoA residues, which are released in the course of the side chain degradation of cholate.
MATERIALS AND METHODS
Cultivation of bacteria.
Pseudomonas sp. strain Chol1 and the transposon mutant strains G12 (defective in skt) and R1 (defective in acad) were grown in the phosphate-buffered mineral medium MMChol as described previously (15, 18, 19). Strain Chol1 was grown with cholate (2 mM), and strains G12 and R1 were grown with mixtures of cholate (2 mM) and succinate (12 mM) in the presence of kanamycin (10 μg ml−1). To induce strain G12 for the transformation of cholate, precultures were grown with 12β-DHADD (XVIII) (ca. 2 mM) as described previously (18). For the production of 12β-DHADD, strain Chol1 was grown with cholate under anoxic conditions with nitrate as the electron acceptor, as described previously (15).
Preparation of cell suspensions and cell extracts.
Cells of strains Chol1, R1, and G12 were harvested in the late-exponential growth phase by centrifugation at 9,000 × g for 10 min at 4°C. For cell suspension experiments, cells were washed with MMChol medium without a carbon source and finally resuspended in MMChol with cholate (2 mM) to an optical density at 600 nm (OD600) of 1. For the preparation of cell extracts, cells were washed in 50 mM K-Na-phosphate buffer (pH 7.0), resuspended in a small volume of the buffer used for the respective enzyme assay, and broken by three passages through a cooled French press (Aminco) at 138 MPa. Homogenates were centrifuged at 17,900 × g for 30 min at 4°C to separate the cell extracts from cell debris. To remove molecules with molecular masses of <5,000 Da, cell extracts were subjected to a desalting step using a Sephadex G-25 matrix (PD-10; GE Healthcare). Protein concentration was determined by the BCA assay (Pierce, USA) with bovine serum albumin as the standard. All cell extracts were immediately used for enzyme assays or stored at −20°C.
Enzyme assays.
All enzyme assays were performed at 30°C. Enzyme assays for acyl-CoA ligase activities contained 50 mM morpholinepropanesulfonic acid (MOPS) buffer (pH 7.8), 1 mM CoA, 1 mM ATP, 2.5 mM MgCl2, and cell extracts (0.6 to 0.8 mg protein ml−1) and were started by adding 1 mM cholate or 100 to 200 μM Δ1/4- and Δ1,4-3-ketocholate, DHOCTO, or DHOPDC. Electron acceptors were added in final concentrations of 1 mM [NAD+, K3Fe(CN)6] or 25 μM phenazine methosulfate (PMS). Samples were withdrawn immediately after the reaction was started (t0 = 0 min) and at defined time intervals thereafter and subsequently analyzed by high-performance liquid chromatography (HPLC) or liquid chromatography-mass spectrometry (LC-MS). To hydrolyze CoA-ester bonds, samples were incubated with 500 mM NaOH at 30°C for at least 30 min.
The activity of citrate synthase (EC 2.3.3.1) was measured by monitoring the acetyl-CoA-, oxaloacetate-, and 5,5′-dithiobis-2-nitrobenzoic acid (DTNB)-dependent formation of thionitrobenzoate in a spectrophotometer at 412 nm (ε = 13.6 mM−1 cm−1). Assays contained HEPES buffer (pH 8.0), 1 mM DTNB, 0.1 mM acetyl-CoA, and cell extracts (0.5 to 0.6 mg protein ml−1) and were started by adding 2 mM oxaloacetate. The activity of 2-methylcitrate synthase (EC 2.3.3.5) was measured in the same way as for citrate synthase, except propionyl-CoA was used instead of acetyl-CoA.
The activity of isocitrate dehydrogenase (EC 1.1.1.42) was measured by monitoring the d-isocitrate-dependent formation of NADPH in a spectrophotometer at 365 nm (ε = 3.4 mM−1 cm−1). Assays contained 100 mM Tris-HCl buffer (pH 8.0), 10 mM MgCl2, 2.5 mM NADP+, and cell extracts (0.5 to 0.6 mg protein ml−1) and were started by adding 1 mM d-isocitrate.
The activity of isocitrate lyase (EC 4.1.3.1) was measured in a coupled assay by determining the isocitrate-dependent oxidation of NADH in a spectrophotometer at 340 nm (ε = 6.2 mM−1 cm−1). The assay is based on the NADH-dependent reduction of glyoxylate to glycolate by lactate dehydrogenase. The assays contained 50 mM HEPES buffer (pH 7.0), 2.5 mM MgCl2, 0.3 mM NADH, 1 U lactate dehydrogenase, and cell extracts (0.4 to 0.5 mg protein ml−1) and were started by adding 5 mM d-isocitrate.
The activity of pyruvate dehydrogenase (EC 1.2.4.1) was measured by the pyruvate- and CoA-dependent formation of NADH in a spectrophotometer at 365 nm (ε = 3.4 mM−1 cm−1). The assays contained 50 mM Tris-HCl buffer (pH 7.8), 2 mM MgCl2, 2.5 mM dithiothreitol, 0.4 mM thiamine pyrophosphate, 0.1 mM CoA, 5 mM NAD+, and cell extracts (0.4 to 0.5 mg protein ml−1) and were started by adding 5 mM pyruvate.
HPLC.
All steroid compounds were analyzed with a reversed-phase HPLC system equipped with a UV-visible (UV-Vis) light diode array detector as described previously (15, 20). K-Na-phosphate buffer (10 mM, pH 7.1) (eluent A) and acetonitrile (eluent B) were used as eluents with a total flow rate of 0.8 ml min−1. For purification of steroid compounds, a semipreparative reversed-phase column was used (250 by 8 mm, Eurosphere II, 100-5 C18 H [Knauer]), with a flow rate of 2 ml min−1. For the detection and purification of P4, P6, 12β-DHADD, and DHOPDC, a gradient method was used, starting with 20% eluent B for 2 min, increasing to 70% eluent B within 9 min, and returning to 20% eluent B within 1 min, followed by an equilibration of 6 min. For the detection and purification of Δ1/4- and Δ1,4-3-ketocholate and DHOCTO, a gradient method was used, starting with 20% eluent B for 2 min, increasing to 34% eluent B within 9 min, and returning to 20% eluent B within 1 min, followed by an equilibration of 6 min. For analysis of the CoA activation assays and purification of P5 and P7, a gradient method was used, starting with 10% eluent B for 2 min, increasing to 56% eluent B within 23 min, and returning to 10% eluent B within 1 min, followed by an equilibration of 6 min.
Purification of steroid compounds.
Steroid compounds from culture supernatants and from the CoA activation assays were purified by organic extraction with ethylacetate followed by semipreparative HPLC as described previously (19) using the HPLC gradient methods described above and by solid-phase extraction as described below. The purity of the steroid compounds was assessed by HPLC analysis. For some steroid compounds, the purity was also assessed by LC-MS analysis and nuclear magnetic resonance (NMR) spectroscopy.
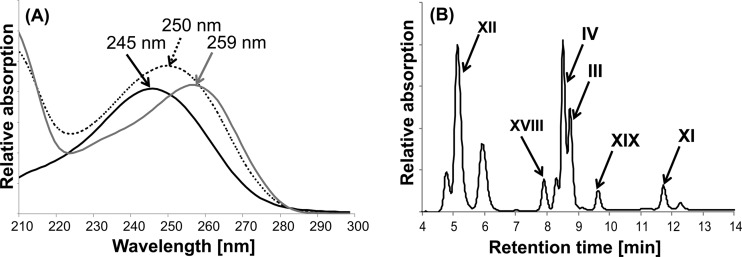
For the purification of DHOCTO (XX), DHOPDC (XII), and Δ1/4- and Δ1,4-3-ketocholate, the mutant strains G12 and R1 were cultivated as described above. In the supernatants of these cultures, either DHOCTO (strain G12) or DHOPDC (strain R1) accumulated along with Δ1/4- and Δ1,4-3-ketocholate (III and IV, respectively) in both cases (18, 19). Those supernatants were used for the purification of Δ1/4- and Δ1,4-3-ketocholate by semipreparative HPLC. For the further purification of DHOCTO and DHOPDC, cultures of the respective mutant strains in the late-exponential growth phase were diluted 1:10 (vol/vol) with fresh MMChol containing succinate (12 mM) and kanamycin (10 μg ml−1) for a second growth passage. Under these conditions, the residual Δ1/4- and Δ1,4-3-ketocholate were completely transformed into DHOCTO (strain G12) or DHOPDC (strain R1), which remained in the culture supernatants as dead-end products. After organic extraction, DHOCTO- and DHOPDC-containing extracts were submitted to semipreparative HPLC or to solid-phase extraction using Chromabond C18 ec columns (Macherey-Nagel). The columns were washed and equilibrated with 1 column volume of methanol followed by 1 volume of MOPS buffer (50 mM, pH 7.8) before DHOCTO- or DHOPDC-containing extracts were loaded on the columns. After washing the columns with 3 volumes of MOPS buffer, DHOCTO or DHOPDC was eluted with absolute methanol, which was evaporated under low pressure (Eppendorf concentrator 5301). Both compounds were resuspended in MOPS buffer, and the purity was controlled by HPLC analysis. Product patterns of CoA activation assays showed that there was no difference between the DHOCTO and DHOPDC purifications that were generated by semipreparative HPLC or by solid-phase extraction. The concentrations of DHOCTO and DHOPDC in the resulting solutions were estimated by measuring their absorbance at 245 nm, which is a characteristic maximum for steroid compounds with a Δ1/4- and Δ1,4-3-keto structure (21) (Fig. 2A). According to calculations based on an averaged molar extinction coefficient of ε245 nm of 14.7 cm−1 mM−1, the final concentrations were in a range of 3 to 5 mM.
Fig 2.
UV spectroscopic and chromatographic properties of the intermediates of cholate degradation. (A) UV spectra of Δ1,4-3-ketocholate (IV in Fig. 1) (black line), Δ1,4-3-ketocholyl-CoA (VIII in Fig. 1) (dotted black line), and cholyl-CoA (V in Fig. 1) (gray line) as characteristic examples for steroid compounds with a Δ1/4- or Δ1,4-3-keto structure of the A ring (absorption maximum at 245 nm) and for their acyl-CoA esters (absorption maximum at 250 nm), respectively. (B) HPLC chromatogram of a culture supernatant of Pseudomonas sp. strain Chol1 growing with cholate after 8 h of incubation. The analysis wavelength was 245 nm.
P5 and P6 were purified from the respective enzyme assay mixtures by solid-phase extraction and subsequent semipreparative HPLC as described above before using them as the substrates for the enzymatic assays.
For NMR analysis, steroid compounds were purified from the respective in vitro assays (P4, P5, P7) or from culture supernatants (12β-DHADD) using organic extraction, solid-phase extraction, and semipreparative HPLC purification as described previously (19) but without acidification of the aqueous phase for the second organic extraction step after HPLC purification. Residues were resuspended in deuterated solvents as indicated in Table 1.
Table 1.
1H and 13C chemical shifts of 12α-DHADD, 12β-DHADD, 12α-20S-DHOPDCA, and 12α-20R-DHOPDCAa
| Atom no. | 12α-DHADD (XVI) in MeOD |
12β-DHADD (XVIII) in D2O |
12α-20S-DHOPDCA (XI) in MeOD |
12α-20R-DHOPDCA in MeOD |
||||
|---|---|---|---|---|---|---|---|---|
| δC (ppm) | δH (ppm), 2,3,4JHHb | δC (ppm) | δH (ppm), 2,3,4JHH | δC (ppm) | δH (ppm), 2,3,4JHH | δC (ppm) | δH (ppm), 2,3,4JHH | |
| 1 | 157.6 | 7.29, d, 10.4 Hz | 159.5 | 7.38, d, 10.4 Hz | 157.6 | 7.29, m | 157.6 | 7.29, d, 10.6 Hz |
| 2 | 126.0 | 2.26, dd, 10.4/1.7 Hz | 125.7 | 6.28, dd, 10.4/2.0 Hz | 125.9 | 6.24, d, 9.8 Hz | 125.9 | 6.24, d, 10.6 Hz |
| 3 | 186.9 | —c | 188.5 | — | — | — | — | — |
| 4 | 125.8 | 6.15, td, 3.0/1.7 Hz | 125.3 | 6.16, t, 1.8 Hz | 125.3 | 6.12, s | 125.3 | 6.12, s |
| 5 | 168.3 | — | 170.1 | — | 168.5 | — | 168.5 | — |
| 6 | 40.2 | 2.58, dd, 14.2/3.1 Hz | 40.0 | 2.54, dd, 14.3/3.2 Hz | 40.4 | 2.52, dd, 13.7/2.0 Hz | 40.4 | 2.52, dd, 13.5/2.8 Hz |
| 2.87, m | 2.83, ddd, 14.3/3.2/1.8 Hz | 2.80, m, 13.7 Hz | 2.80, m, 13.5 Hz | |||||
| 7 | 67.7 | 4.20, q, 2.7 Hz | 67.8 | 4.22, q, 3.2 Hz | 68.8 | 4.05, m | 68.7 | 4.05, m |
| 8 | 38.8 | 2.08, m | 37.3 | 1.99, m | 39.7 | 1.78, m | 39.5 | 1.80, m |
| 9 | 39.0 | 2.05, m | 42.6 | 1.54, m | 38.5 | 1.97, m | 38.1 | 2.00, m |
| 10 | 43.3 | — | 43.8 | — | 43.2 | — | 43.4 | — |
| 11 | 29.2 | 1.94, m | 29.9 | 1.69axial, td, 12.9/11.2 Hz | — | — | — | — |
| 2.01, m | 1.95equatorial, m | — | — | — | — | |||
| 12 | 68.2 | 4.09, m | 70.5 | 3.71, dd, 11.2/4.7 Hz | — | — | — | — |
| 13 | 52.3 | — | 51.5 | — | 53.6 | — | 50.2 | — |
| 14 | 37.8 | 2.44, m | 43.2 | 1.53, m | 38.6 | 1.95, m | 41.3 | 1.98, m |
| 15 | 20.3 | 1.68, m | 20.3 | 1.77, tt, 12.8/9.2 Hz | 24.6 | 1.36, m | 24.8 | 1.36, m |
| 2.17, m | 2.00, m | 1.62, m | 1.62, m | |||||
| 16 | 36.2 | 2.13, m | 35.4 | 2.13, dt, 20.2/9.2 Hz | 28.5 | 1.36, m | 28.7 | 1.35, m |
| 2.44, m | 2.49, dd, 20.2/9.0 Hz | 1.63, m | 1.63, m | |||||
| 17 | 220.1 | — | 224.5 | — | 42.7 | 2.31, m | 41.9 | 2.27, t, 7.4 Hz |
| 18 | 13.1 | 0.95, s | 7.2 | 0.97, s | 7.2 | 0.90, m | 9.1 | 1.05, m |
| 19 | 17.3 | 1.31, s | 17.4 | 1.25, s | 17.2 | 1.27, s | 17.2 | 1.28, s |
| 20 | 48.4 | 2.31, m | 48.9 | 2.35, m | ||||
| 21 | 12.3 | 1.07, d, 5.3 Hz | 11.2 | 1.19, d, 7.0 Hz | ||||
| 22 | 206.8 | 9.56, d, 4.8 Hz | 205.3 | 9.56, d, 2.9 Hz | ||||
12α-DHADD, XVI in Fig. 1 and P4 in Fig. 4 and 5; 12β-DHADD, XVIII in Fig. 1; 12α-20S-DHOPDCA, XI in Fig. 1 and P5 in Fig. 4; 12α-20R-DHOPDCA, P7.
s, singlet; d, doublet; t, triplet; q, quartet; m, multiplet; dd, doublet of a doublet; td, triplet of a doublet; tt, triplet of a triplet; ddd, doublet of a doublet of a doublet.
—, no proton at given position, or proton resonance could not be assigned.
UV-Vis spectroscopy.
UV-Vis spectra of steroid compounds were recorded with a diode array detector during HPLC analyses and were used for a first classification of the configuration of the A ring as well as for determination of CoA ester formation. Degradation products of cholate with a Δ1/4- or Δ1,4-3-keto structure of the A ring exhibit the aforementioned distinct absorption maxima around 245 nm, while the CoA moieties of CoA esters exhibit maxima around 260 nm (Fig. 2A). If an intermediate has both structural elements, the absorption of the Δ1/4- or Δ1,4-3-keto structure of the A ring is added to the absorption of the CoA moiety, resulting in an absorption maximum that is shifted to 250 nm (Fig. 2A).
Mass spectrometry.
Mass spectra were obtained on an LTQ Orbitrap XL LC-MS/MS instrument (Thermo Scientific) using electrospray in the positive-ion mode. Chromatographic separation was performed using an HTS PAL autosampler together with a Rheos 2200 HPLC and a Macherey-Nagel AG 125/2 Nucleosil 120.3 C18 column. Acetic acid (0.5‰ [vol/vol] in H2O) was added postcolumn at 20 μl min−1 using a Bischoff 2200 HPLC pump. The following gradient system was used: 10 mM ammonium acetate (pH 7) (eluent A) and acetonitrile (eluent B) at a flow rate of 250 μl min−1, starting with 5% eluent B for 2 min and followed by a linear increase to 90% eluent B in 30 min. The gradient composition was kept at 90% eluent B for 4 min and then returned to initial conditions in 1 min, followed by 3 min of reequilibration, giving a total run time of 40 min. The injection volume was 50 μl. The scan range was 300 to 1,200 Da at a mass resolution of 100,000 at m/z 400. The needle voltage was set to 5,000 V, the capillary voltage to 42 V, the tube lens to 135 V, and the capillary temperature to 275°C. The normalized collision energy was set to 35 V for the MS/MS experiments, with an isolation width of 1 Da. Mass accuracies determined with the calibration mixture from Thermo were better than 1.7 ppm (0.2 ppm at 195 Da, 0.5 ppm at 524 Da, 1.2 ppm at 1,222 Da, 1.4 ppm at 1,422 Da, and 1.6 ppm at 1,622 Da).
For measurements in the negative-ion mode, mass spectra were obtained on a LXQ LC-MS/MS instrument (Thermo Scientific) using electrospray in the negative mode. Chromatographic separation was performed using a Dionex Ultimate 3000 HPLC system equipped with a Dionex Acclaim 120 reversed-phase column (C18, 5 μm, 300/4.6). The following gradient system was used: 50 mM ammonium acetate (pH 5) (eluent A) and methanol (eluent B) at a flow rate of 600 μl min−1, starting with 10% eluent B for 2 min and followed by a linear increase to 90% eluent B in 20 min. The gradient composition was kept at 90% eluent B for 30 min and then returned to initial conditions in 1 min. The injection volume was 50 μl. The scan range was from 50 to 1,300 Da. The needle voltage was set to 4,000 V, the capillary voltage to −7 V, the tube lens to −5.1 V, and the capillary temperature to 300°C. The normalized collision energy was set to 35 V for MS/MS experiments, with an isolation width of 1 Da.
NMR spectroscopy.
NMR spectra were acquired at 300K on a Bruker Avance III 600 MHz spectrometer equipped with a 5-mm TCI-H/C/N triple-resonance cryoprobe with an actively shielded Z gradient. HPLC-purified samples were dissolved in D2O or methanol-d4. The proton one-dimensional (1D) spectra were acquired with 32,768 data points and a spectral width of 16 ppm. Solvent signals were suppressed using the excitation-sculpting sequence (22). The 2D correlated spectroscopy (COSY) experiments were recorded with 256 increments and 2,000 detected complex points. The heteronuclear single quantum correlation (HSQC) and heteronuclear multiple-bond correlation (HMBC) spectra were recorded with 256 increments and 2,000 and 4,000 detected complex points and with spectral widths of 200 ppm and 250 ppm in the indirect dimension, respectively. The spectra were processed by applying zero filling to double the number of real points in both dimensions and using a squared sine bell function with a sine bell shift of π/4 and π/3 in F2 and F1, respectively. Chemical shifts of the 13C nuclei were assigned indirectly from the HSQC and HMBC spectra. Acquisition, processing, and analyses of all spectra were performed with the Bruker Topspin (v3.0) software. In general, diastereotopic protons were not assigned stereospecifically, since high spectral overlap prevented a sufficient resolution of some of the decisive coupling patterns.
RESULTS
Transformation of Δ1/4- and Δ1,4-3-ketocholate using nondesalted cell extracts.
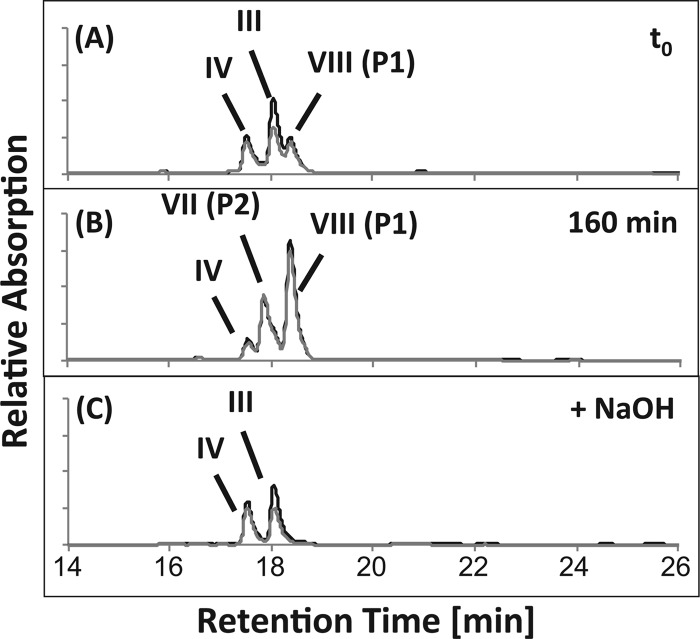
To test whether Δ1/4- and Δ1,4-3-ketocholate (Fig. 1, III and IV, respectively) can be activated with CoA and subsequently be transformed into other intermediates, these compounds were purified from culture supernatants of the mutant strains G12 and R1 and submitted to in vitro assays for CoA activation using nondesalted cell extracts of strain Chol1. HPLC analyses of these assays revealed cell extract-, ATP-, and CoA-dependent consumption of Δ1/4- and Δ1,4-3-ketocholate and the concomitant formation of two products, P1 and P2 (Fig. 3A and B), which showed typical UV absorption spectra of CoA esters with a λmax around 250 nm (Fig. 2A). After alkaline treatment of the respective assay mixtures, these products disappeared, while Δ1/4- and Δ1,4-3-ketocholate reappeared (Fig. 3C). These results indicate that the products P1 and P2 were the CoA esters of Δ1/4- and Δ1,4-3-ketocholate (VII and VIII), respectively. No further steroid compounds with UV spectra characteristic for a Δ1/4- or Δ1,4-3-keto structure of the A ring (λmax around 245 nm) (Fig. 2A) were detected in these assays. Further steroid compounds were also not detected when the assays were supplied with K3Fe(CN)6 as an electron acceptor, indicating that the respective CoA esters could not be further degraded by cell extracts of strain Chol1 under the applied conditions.
Fig 3.
HPLC chromatograms of a CoA activation assay with cell extracts of Pseudomonas sp. strain Chol1 containing a mixture of Δ1/4- and Δ1,4-3-ketocholate (III and IV in Fig. 1) as the substrates incubated for 0 min (A) and 160 min (B). P1 and P2 represent the CoA esters of Δ1/4- and Δ1,4-3-ketocholate (VII and VIII in Fig. 1), respectively, which are hydrolyzed completely after treatment with NaOH (C). The analysis wavelengths were 245 nm (black) and 260 nm (gray).
Transformation of DHOCTO and DHOPDC using nondesalted cell extracts.
In the next step, we investigated whether DHOCTO (XX) and DHOPDC (XII) could be activated with CoA and subsequently transformed into other intermediates. For this, DHOCTO and DHOPDC were purified from culture supernatants of the skt mutant strain G12 and the acad mutant strain R1, respectively, and submitted to in vitro assays for CoA activation.
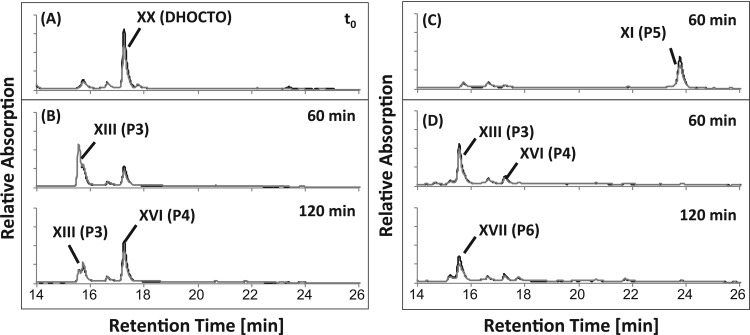
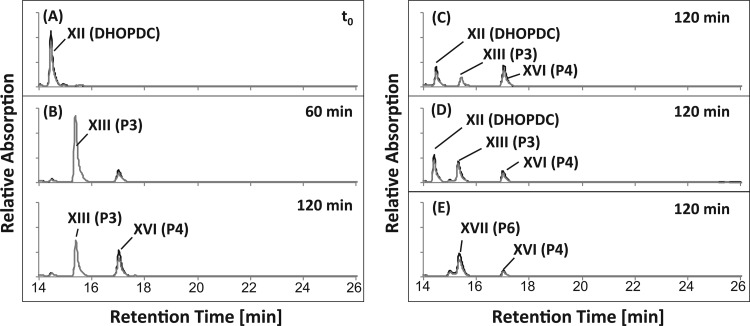
HPLC analyses of assays containing nondesalted cell extracts and DHOCTO as the substrate revealed cell extract-, ATP-, and CoA-dependent consumption of DHOCTO, the transient formation of a new product, P3, with a UV spectrum typical for CoA esters, and the formation of an accumulating new product, P4 (Fig. 4B) with a λmax around 245 nm. LC-MS analyses of these products revealed ions with m/z [M+H]+ of 1,124.2 and 317.1 for P3 and P4, respectively. The mass of the P3 ion indicated a molecular mass of 1,123 Da (C43H64N7O20P3S), which exactly matches the mass of the CoA ester of DHOPDC (19) (Fig. 1, XIII). In agreement with that, the same product was also formed in identical assays with DHOPDC as the substrate (Fig. 5B). These results indicate that P3 was DHOPDC-CoA. Interestingly, DHOPDC-CoA was not hydrolyzed after treatment with NaOH. P4 also accumulated in CoA activation assays with DHOPDC as the substrate (Fig. 5B). The mass of the P4 ion indicated a molecular mass of 316 Da (C19H24O4), which exactly matches the mass of DHADD (XVIII), the end product of anaerobic nitrate-dependent cholate degradation by strain Chol1 (15). However, chromatographic comparisons of DHADD showed that P4 and DHADD had different retention times. When cell extracts of succinate-grown cells were used, which are not induced for cholate degradation (15), DHOCTO and DHOPDC were not converted at all (data not shown).
Fig 4.
HPLC chromatograms of CoA activation assays with cell extracts of Pseudomonas sp. strain Chol1 containing DHOCTO (XX in Fig. 1) as the substrate. (A) Representative t0 for all assays. (B) Assay with nondesalted cell extracts incubated for 60 and 120 min. (C) Assay with desalted cell extracts incubated for 60 min. (D) Assay with desalted cell extracts in the presence of NAD+ incubated for 60 and 120 min. P3 was identified as the CoA ester of DHOPDC (XIII), P4 was identified as 12α-DHADD (XVI), P5 was identified as DHOPDCA (XI), and P6 was identified as HADT (XVII). The analysis wavelengths were 245 nm (black) and 260 nm (gray).
Fig 5.
HPLC chromatograms of CoA activation assays with cell extracts of Pseudomonas sp. strain Chol1 containing DHOPDC (XII in Fig. 1) as the substrate. (A) Representative t0 for all assays. (B) Assay with nondesalted cell extracts incubated for 60 and 120 min. (C) Assay with desalted cell extracts incubated for 120 min. (D) Assay with desalted cell extracts in the presence of NAD+ incubated for 120 min. (E) Assay with desalted cell extracts in the presence of NAD+ and PMS incubated for 120 min. P3 was identified as the CoA ester of DHOPDC (XIII), P4 was identified as 12α-DHADD (XVI), and P6 was identified as HADT (XVII). The analysis wavelengths were 245 nm (black) and 260 nm (gray).
Identification of P4 as 12α-DHADD.
To elucidate the structure of P4 (Fig. 1, XVI) and to compare it to DHADD (XVIII), both compounds were purified from assay mixtures and culture supernatants of anaerobically grown cells, respectively, and submitted to NMR analysis. Both compounds were fully characterized by 1D and 2D NMR spectroscopy, and all proton and carbon resonances could be assigned (Table 1).
While the chemical shifts of rings A, B, and D were very similar for both compounds, ring C and its direct vicinity showed significantly different chemical shifts. Analysis employing HSQC, HMBC, and COSY spectra revealed the exact same chemical connectivities for both compounds, corresponding to DHADD. The differences in the chemical shifts turned out to arise from the configuration of the stereogenic center at C-12. In DHADD, the carbon-bound H12 exhibits a double doublet with 3JHH couplings of 11.2 Hz and 4.7 Hz. The two coupling constants correspond to an axial-axial and an axial-equatorial coupling, respectively. This coupling pattern is in agreement with an equatorial (β) position of the C-12 OH group. In addition, the upfield-shifted resonances of the axial ring protons H9 and H14 at 1.54 ppm and 1.53 ppm, respectively, are characteristic of a distant β-OH group at C-12.
In contrast, P4 exhibits resonances around 2 ppm for protons H9 and H14, characteristic of a proximate axial (α) 12-OH group, leading to deshielding of the axial protons H9 and H14. In the case of P4, the coupling constants of protons H11 and H12 could not be resolved due to the overlapping signals of impurities.
Thus, the only difference between DHADD and P4 is the conformation of the OH group at C-12, which is in agreement with their identical molecular masses. P4 was therefore identified as 12α-DHADD (XVI), while the end product of anaerobic cholate degradation by strain Chol1 was identified as 12β-DHADD (XVIII). This difference in stereochemistry must be the reason for the different retention times of 12α-DHADD and 12β-DHADD during HPLC analysis.
Transformation of DHOCTO and DHOPDC using desalted cell extracts.
To further dissect the reaction sequence of the side chain degradation leading from DHOCTO to 12α-DHADD, we used desalted cell extracts in the DHOCTO and DHOPDC activation assays. HPLC analyses of assay mixtures containing desalted cell extracts and DHOCTO as the substrate revealed cell extract-, CoA-, and ATP-dependent consumption of DHOCTO and the formation and accumulation of a new product, P5, with a higher retention time (Fig. 4C). None of the products found with nondesalted cell extracts were formed in those assays. P5 had a UV spectrum with a λmax around 245 nm, and LC-MS analysis revealed an ion with m/z [M+H]+ of 359.2. Based on the fact that steroid compounds containing a Δ1,4-3-keto structure exhibit similar molar extinction coefficients at 245 nm (21), the concentration of accumulated P5 estimated from its peak area is in the same range as the initial substrate concentration, indicating a complete conversion of DHOCTO into P5.
When the assay mixtures were supplied with NAD+ as an electron acceptor, P5 was not produced, while transient formation of DHOPDC-CoA and 12α-DHADD as observed with nondesalted cell extracts was restored. In addition, a new product, P6, accumulated and had a λmax around 245 nm (Fig. 4D). According to the peak area at 245 nm, DHOCTO was completely converted into P6.
This result suggested that further degradation of P5 was NAD+ dependent. To test this possibility, assay mixtures for DHOCTO activation were supplied with NAD+ after P5 had been formed. In these assays, P5 was consumed completely, while DHOPDC-CoA and 12α-DHADD were formed transiently, and P6 accumulated as the end product (data not shown). When NADP+ (1 mM) was added as an electron acceptor, P5 was also transformed, but the reaction time was significantly lower, and only DHOPDC-CoA and 12α-DHADD were formed.
When desalted cell extracts were used in the DHOPDC activation assays, the conversion of DHOPDC into DHOPDC-CoA and 12α-DHADD was significantly slower and incomplete (Fig. 5C). Supplementation of those assays with NAD+ (Fig. 5D) or with PMS alone (data not shown) did not restore the original reaction kinetics. The addition of NAD+ and PMS simultaneously led to the formation and accumulation of P6 (Fig. 5E). According to the peak area at 245 nm, DHOPDC was completely converted into P6. Notably, formation of P5 was not observed in any of the assays with DHOPDC as the substrate.
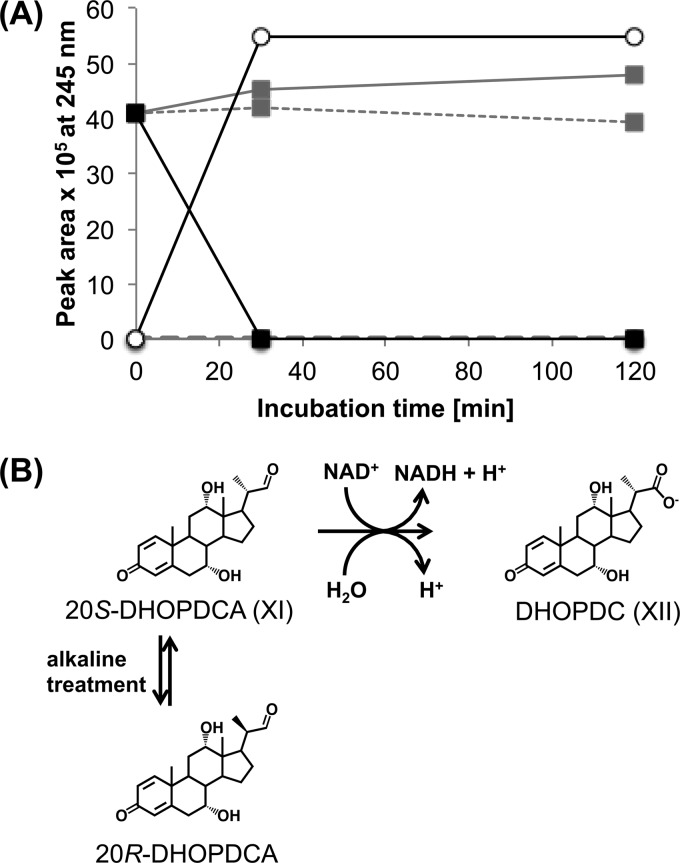
Identification of P5 as 7α,12α-dihydroxy-3-oxopregna-1,4-diene-20S-carbaldehyde (20S-DHOPDCA).
To elucidate the structure of P5, it was purified and submitted to NMR analysis. The side chain of P5 was identified by the distinct resonance of the aldehyde group and its vicinal branch H20, which exhibits correlations to the ring proton H17, the methyl group H21, and the aldehyde proton H22 in the 2D COSY spectrum. CH/CH2 groups 11 and 12 could not be assigned due to high spectral overlap and the presence of impurities. Nevertheless, the configuration of the 12-OH group could be assigned to be in the α position according to the characteristic resonance patterns of CH groups 9 and 14. The proton resonances around 2 ppm and carbon resonances around 40 ppm of these groups would be shifted to ca. 1.5 ppm and 44 ppm, respectively, in the case of a β-OH group at C-12 (Table 1). The downfield-shifted proton resonances arise from the axial 12-OH group, which has an influence on the axial protons H9 and H14.
When the pH values of solutions containing P5 were raised to 10 to 12 with NaOH, a second product, P7, was formed chemically from P5 with a very similar UV spectrum but a higher retention time. P7 exhibits the same chemical connectivities, with slightly shifted NMR resonances in the proximity of the stereogenic center at C-20, like those of P5 (Table 1). Therefore, P5 was identified as 7α,12α-dihydroxy-3-oxopregna-1,4-diene-20S-carbaldehyde (20S-DHOPDCA) (Fig. 1, XI), and P7 was identified as the respective 20R diastereomer. P7 could be formed from P5 by abstraction of the acidic proton H20 under alkaline conditions, leading to the respective enolate and subsequent nonstereospecific reprotonation at C-20, yielding a 1:1 mixture of the two diastereomers.
In supernatants of strain Chol1 growing with cholate, a product which coeluted with and had the same UV spectrum as 20S-DHOPDCA accumulated transiently (Fig. 2B). The same product was also detected in culture supernatants of strain R1 during growth with succinate in the presence of cholate. These results indicate that the aldehyde intermediate 20S-DHOPDCA is also formed in vivo during the degradation of cholate.
Oxidation of 20S-DHOPDCA to DHOPDC in desalted cell extracts.
To further characterize 20S-DHOPDCA, it was purified from the respective DHOCTO activation assays and was used as the substrate for enzymatic tests containing desalted cell extracts of strain Chol1 and NAD+. Under these conditions, 20S-DHOPDCA was immediately transformed into DHOPDC (Fig. 6) as the only product. This reaction was cell extract and NAD+ dependent. The identity of DHOPDC was confirmed by coelution experiments with purified DHOPDC (data not shown).
Fig 6.
(A) Transformation of DHOPDCA (squares) into DHOPDC (circles) with a desalted cell extract of Pseudomonas sp. strain Chol1 in the presence of NAD+ (black lines). Controls without cell extract (dashed gray lines) or without NAD+ (solid gray line) showed no transformation of DHOPDCA and no formation of DHOPDC. (B) Chemical and biochemical reactions of DHOPDCA: NAD+-dependent oxidation of 20S-DHOPDCA to DHOPDC and chemical transformation of 20S-DHOPDCA to 20R-DHOPDCA under alkaline conditions; the 20R isomer was not oxidized by cell extracts of strain Chol1.
When a mixture of 20S- and 20R-DHOPDCA was used as the substrate, only the 20S stereoisomer reacted, while the 20R isomer remained unchanged (data not shown), indicating that further enzymatic transformation of DHOPDCA is stereospecific regarding the configuration at C-20.
Transformation of DHOCTO and DHOPDC in cell extracts of the mutant strains G12 and R1.
The same CoA activation assays as those carried out with the wild-type strain Chol1 were used to investigate the fate of DHOCTO and DHOPDC in CoA activation assays with cell extracts of the mutant strains G12 and R1, both of which have defects in the degradation of the acyl side chain of cholate.
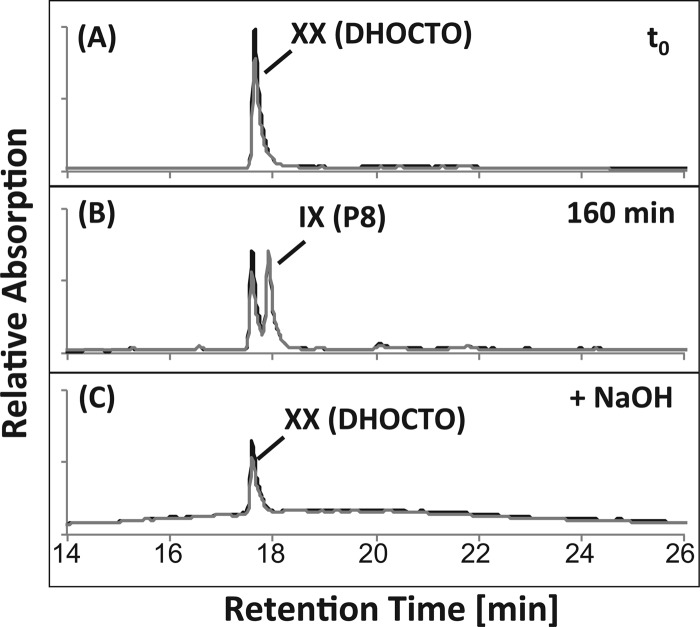
When DHOCTO was used as the substrate for CoA activation with desalted or nondesalted cell extracts of the skt mutant strain G12, HPLC analysis showed the formation of only one product (P8) with a UV spectrum characteristic for a CoA ester (λmax around 250 nm), which disappeared after alkaline treatment, while DHOCTO remained (Fig. 7). LC-MS analyses revealed an ion with m/z [M+H]+ of 1,150.2, indicating a molecular mass of 1,149 Da (C43H64N7O20P3S). These results show that P8 was the CoA ester of DHOCTO (Fig. 1, IX). Furthermore, the LC-MS analyses revealed an ion with m/z [M−H]− of 417.15. This ion indicates a molecular mass of 418 Da (C24H34O6), which exactly matches the mass of THOCDO (XXI). Formation of DHOPDCA was not observed in those assays. These results indicate that further degradation of THOCDO is prevented in the absence of the enzyme encoded by the skt gene.
Fig 7.
HPLC chromatograms of a CoA activation assay with nondesalted cell extracts of the skt mutant Pseudomonas sp. strain G12 containing DHOCTO (XX in Fig. 1) as the substrate incubated for 0 min (A) and 160 min (B). The analysis wavelengths were 245 nm (gray) and 260 nm (black). P8 was identified as the CoA ester of DHOCTO (IX) and was hydrolyzed completely after treatment with NaOH (C).
When DHOPDC was used as the substrate for CoA activation with desalted or nondesalted cell extracts of strain G12, HPLC analysis showed the formation of DHOPDC-CoA (XIII), while other products, such as 12α-DHADD, were not formed, even after the addition of NAD+ alone or with PMS (data not shown). This result indicated that strain G12 was obviously not induced for further degradation of DHOPDC-CoA.
When DHOCTO was used as the substrate for CoA activation with desalted cell extracts of the acad mutant strain R1, HPLC analysis showed the consumption of DHOCTO and the formation of DHOPDCA (XI) as an end product. When NAD+ was supplied, DHOPDC (XII) and DHOPDC-CoA (XIII) were formed. When DHOPDC was used as the substrate for CoA activation with a desalted or nondesalted cell extract of strain R1, DHOPDC-CoA (XIII) was the only product. Neither 12α-DHADD nor P6 was formed in assays with cell extracts of strain R1 (data not shown).
Transformation of 12α-DHADD to 12β-DHADD in desalted cell extracts and identification of P6 as 7α-hydroxy-androsta-1,4-diene-3,12,17-trione (HADT).
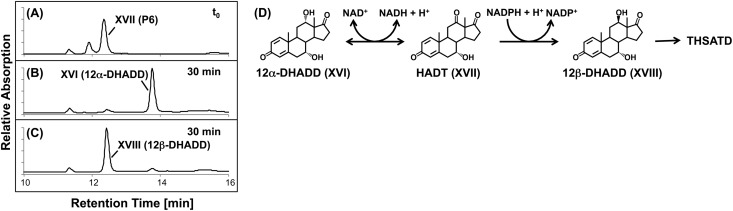
To analyze the reactions involved in the stereoinversion of the 12-hydroxyl group, 12α-DHADD was purified from the respective CoA activation assays and used as the substrate for enzymatic tests with desalted cell extracts of strain Chol1. When assay mixtures were supplied with NAD+, 12α-DHADD was transformed into a product that coeluted with, and had the same UV spectrum as, P6. Transformation of 12α-DHADD into P6 was NAD+ dependent and could be accelerated by the addition of PMS (data not shown).
To further characterize P6, it was purified from DHOPDC activation assays (see above) and used as the substrate in assays containing desalted cell extracts of strain Chol1. When assay mixtures were supplied with 1 mM NADH, P6 was transformed into a compound that coeluted with, and had the same UV spectrum as, 12α-DHADD (Fig. 8B). When the assay mixtures were supplied with 1 mM NADPH, P6 was transformed into a compound that coeluted with, and had the same UV spectrum as, 12β-DHADD (Fig. 8C). Without the addition of an electron donor, P6 did not react at all.
Fig 8.
Chromatograms of enzyme assays with desalted cell extracts of Pseudomonas sp. strain Chol1 containing P6 as the substrate. (A) Representative t0 for all assays. (B) Assay with NADH incubated for 30 min. (C) Assay with NADPH incubated for 30 min. P6 was identified as HADT (XVII in Fig. 1). The analysis wavelength was 245 nm. (D) Isomerization of 12α-DHADD to 12β-DHADD via HADT. The oxidation to HADT is NAD+ dependent, and the back reaction could be measured in vitro; the reduction of HADT to 12β-DHADD is NADPH dependent; 12β-DHADD is further degraded to THSATD.
MS analysis of purified P6 revealed an ion with m/z [M+H]+ of 315.1, indicating a molecular mass of 314 Da and a molecular formula of C19H22O4. This molecular mass and the redox reactions leading to the different isomers of DHADD indicate that P6 is 7α-hydroxy-androsta-1,4-diene-3,12,17-trione (HADT) (Fig. 1, XVII). Obviously, the stereoinversion of the 12-hydroxyl group of DHADD proceeds via the 12-keto intermediate HADT.
Degradation of acetyl-CoA and propionyl-CoA.
During degradation of the acyl side chain of cholate, one acetyl-CoA and one propionyl-CoA are released. To investigate the fate of these two CoA esters, we checked for respective enzyme activities in cell extracts of strain Chol1.
Acetyl-CoA can be processed through the citric acid cycle or through the glyoxylate cycle. To investigate whether the glyoxylate cycle was induced in cholate-grown cells of strain Chol1, we determined the activity of the isocitrate lyase. In extracts of cholate-grown cells, specific isocitrate lyase activities were about 44-fold lower than in extracts of acetate-grown cells, which served as a positive control (Table 2). In addition, pyruvate dehydrogenase activity was not detectable in extracts of acetate-grown cells, while it was present in extracts of cholate-grown cells. In contrast, there were no significant differences between the extracts of cholate- and acetate-grown cells regarding the specific activities of citrate synthase and isocitrate dehydrogenase (Table 1). These results indicate that acetyl-CoA is processed via the citric acid cycle and not via the glyoxylate cycle during growth of strain Chol1 with cholate.
Table 2.
Specific activities of enzymes in cell extracts of cholate- and acetate-grown cells of Pseudomonas sp. strain Chol1
| Enzyme | Sp act (mU [mg protein]−1)a |
|
|---|---|---|
| Cholate-grown cells | Acetate-grown cells | |
| Isocitrate lyase | 12 ± 1 | 538 ± 40 |
| Pyruvate dehydrogenase | 12 ± 4 | Not detectable |
| Citrate synthase | 687 ± 103 | 339 ± 14 |
| Isocitrate dehydrogenase | 387 ± 22 | 655 ± 100 |
Values after ± indicate standard deviations (n = 3).
Propionyl-CoA can be degraded via several pathways in bacteria, with the methylmalonyl-CoA pathway and the methylcitric acid cycle being the most prominent metabolic routes (23). A functional methylcitric acid cycle was recently shown to be essential for the growth of Mycobacterium tuberculosis with cholesterol (24). To investigate whether the 2-methylcitric acid cycle was induced in cholate-grown cells of strain Chol1, we determined the activity of the 2-methylcitrate synthase. In extracts of cholate-grown cells, specific 2-methylcitrate activity levels (551 ± 78 mU [mg protein]−1) were in the same range as those in extracts of propionate-grown cells (723 ± 117 mU [mg protein]−1), which served as a positive control. In extracts of succinate-grown cells, which are not induced for cholate degradation (15), specific 2-methylcitrate synthase activity levels were about 6-fold lower (87 ± 38 mU [mg protein]−1). These results indicate that the degradation of propionyl-CoA proceeded via the 2-methylcitric acid cycle during growth of strain Chol1 with cholate.
DISCUSSION
The goal of this study was to elucidate the degradation of the acyl side chain of cholate in Pseudomonas sp. strain Chol1 by an in vitro approach. The key to this approach was the availability of DHOCTO (XX), which has an unsaturated acyl side chain, as a substrate. In cell extracts of cholate-grown strain Chol1 cells, DHOCTO was completely converted into 12β-DHADD (XVIII) in a CoA- and ATP-dependent manner. Thus, all reactions required for the cleavage of the acyl side chain of DHOCTO are operative in vitro. The original hypothesis was that the degradation of the acyl side chain proceeds via a classical β-oxidation pathway. According to this hypothesis, the CoA ester of THOCDO (X) should be oxidized to a 22-keto-acyl-CoA compound, which would be the substrate for a β-ketothiolase reaction catalyzed by Skt, yielding the CoA ester of DHOPDC (XIII) as a product. However, our in vitro analysis revealed the formation of a free aldehyde instead of a β-keto-acyl-CoA compound as an intermediate in the transformation of DHOCTO to DHOPDC. In desalted extracts of cholate-grown strain Chol1 cells, DHOCTO was completely converted into the aldehyde DHOPDCA (XI) in a CoA- and ATP-dependent manner. There is clear evidence that DHOPDCA is also a true intermediate of cholate degradation in vivo.
First, DHOPDCA is also found in supernatants of strain Chol1 (Fig. 2B) and strain R1 during growth with and in the presence of cholate, respectively, while the aforementioned β-keto-acyl compound, which is the predicted intermediate for the β-thiolytic conversion of the CoA ester of THOCDO (X) to DHOPDC-CoA (XIII), has been detected neither in vitro nor in vivo. Thus, there is no evidence of the originally proposed intermediate, while an alternative intermediate was formed in vitro and in vivo. Second, the 20S isomer of DHOPDCA is specifically converted to DHOPDC in an NAD+-dependent reaction, and DHOPDC is further degraded to DHADD both in vivo and in vitro. Thus, DHOPDCA is not a dead-end intermediate of the side chain degradation but a substrate of a stereospecific enzymatic conversion. Third, the transient accumulation of DHOPDC in culture supernatants during growth of strain Chol1 with cholate (Fig. 2B) is in agreement with the formation of DHOPDCA and its oxidation to DHOPDC. If cholate degradation had proceeded via a 22-keto-acyl-CoA compound, DHOPDC-CoA would have been the direct product, and it would have been unlikely that free DHOPDC would be released into the supernatant.
In the skt mutant DHOPDCA was formed neither in vitro in cell extracts nor in vivo in growing cultures. Instead, DHOCTO and traces of THOCDO accumulated in vivo as well as in vitro. These results strongly suggest that the CoA ester of THOCDO (X) is the substrate of Skt and that DHOPDCA and acetyl-CoA are the products. Thus, Skt may catalyze an aldolase reaction rather than a β-ketothiolase reaction as originally proposed (18). A very similar aldolase reaction is proposed to be the last step of the side chain degradation of cholesterol and cholate (reaction leading from XV to XVI). A candidate gene for this reaction is located in the igr gene cluster in M. tuberculosis strain H37Rv, which is responsible for the removal of the propionyl-CoA side chain from the steroid skeleton (25). In this cluster, the enzyme encoded by ltp2 is proposed to catalyze the final aldolytic cleavage step of this reaction sequence by removing the propionyl-CoA side chain from the steroid skeleton. Interestingly, BLAST analysis predicts that Ltp2, just like Skt, belongs to the SCP-x-type thiolase family. However, from a mechanistic viewpoint, the release of the propionyl side chain from the steroid skeleton cannot proceed via a β-ketothiolase reaction. In summary, the mechanistic consideration for Ltp2 and the phenotype of the skt mutant call attention to the existence of aldolases that act on β-hydroxyl-acyl-CoA esters and have structural features that resemble SCP-x-type thiolases. A recent study showed that two further Ltp homologues from Rhodococcus rhodochrous, which are essential for the side chain degradation of C-24-branched phytosterols, are also aldolases rather than ketothiolases (26). Both enzymes lack three amino acid residues (Cys125, His375, and Cys403) characteristic for ketothiolases, which are also missing in Skt (18) and Ltp2 (25). Thus, these recent results from a cholesterol-degrading bacterium fully support our hypothesis about Skt. Unfortunately, THOCDO was not available as a substrate for further studies on the function of Skt, because it was produced in only very small amounts by the skt mutant strain G12.
Our in vitro studies indicate that the aldehyde DHOPDCA is transformed to DHOPDC-CoA by an NAD+-dependent aldehyde dehydrogenase and a DHOPDC acyl-CoA ligase. Thus, this pathway leads to the same product as the previously proposed β-oxidation. A clear disadvantage of this pathway in comparison to β-oxidation is the additional expenditure of ATP for CoA activation of DHOPDC. A possible advantage, however, could be that the formation of the β-keto acid is avoided, which tends to decarboxylate spontaneously, thereby leading to a potential dead-end metabolite. Such an alternative strategy for the degradation of an acyl side chain via a free aldehyde intermediate has been reported so far only for the degradation of ferulic acid by Delftia acidovorans (27) and two Pseudomonas strains (28, 29).
With wild-type cell extracts, DHOPDC-CoA (XIII) was further transformed into 12α-DHADD (XVI). The respective reaction sequence should comprise a dehydrogenation of the side chain followed by a hydration of the double bond and an aldolytic cleavage of propionyl-CoA from the steroid skeleton. As no reaction intermediates between DHOPDC-CoA and 12α-DHADD accumulated in our in vitro assays, the respective reaction steps obviously occur at a very high rate. The fact that DHOPDC-CoA was not further degraded in assays with cell extracts of strain R1 supports the idea that acad encodes the acyl-CoA dehydrogenase required for initiating the further conversion of DHOPDC-CoA and, thus, confirmed the phenotype of the mutant strain R1 in vitro. Within the aforementioned igr gene cluster of M. tuberculosis, the proteins encoded by fadE28 and fadE29 were shown to catalyze the dehydrogenation of the C3 side chain (25). The enzyme encoded by acad therefore has a function analogous to that of the heterodimeric complex of enzymes FadE28 to FadE29 and shows a similarity of 49% to FadE29 but only 12% to FadE28. In proximity to acad is a gene (30) (ORF_11377) that encodes a protein with high similarity to the aforementioned Ltp2 (63% identity on an amino acid level), suggesting that ORF_11377 is responsible for the reaction leading from compound XV to 12α-DHADD and propionyl-CoA.
While the dehydrogenation of DHOPDC-CoA was possible with cell extracts of strain Chol1, the dehydrogenation of Δ1,4-3-ketocholyl-CoA (VIII) was not. Possible reasons for this difference could be that the respective acyl-CoA dehydrogenases require different electron acceptors or that the electron acceptor for the dehydrogenation of DHOPDC-CoA was not limiting in cell extracts of strain Chol1. The electron acceptors for both reactions are not known, but according to analyses of other acyl-CoA dehydrogenases, it appears that a flavine cofactor is likely to be involved (31). The dehydrogenation of Δ1,4-3-ketocholyl-CoA may also be a bottleneck of the whole reaction sequence in vivo, because Δ1,4-3-ketocholate is always transiently accumulating in large amounts in the wild-type culture supernatants. (Fig. 2B).
The isomerization of 12α-DHADD (XVI) to 12β-DHADD (XVIII) via HADT (XVII) has also been observed during cholate degradation by Comamonas testosteroni strain TA441 and is catalyzed by the consecutive action of a dehydrogenase (SteA) and a reductase (SteB) (32). Genes (ORF_11442 and ORF_11447) encoding predicted proteins with high similarities to SteA (58%) and SteB (70%), respectively, are also found in the genome of strain Chol1 (30). Our in vitro studies revealed an interesting specificity for electron acceptors and donors involved in this inversion. In the presence of NAD+, 12α-DHADD is oxidized to HADT (XVII). With NADH, HADT reacted back to 12α-DHADD, while in the presence of NADPH the 12-keto group is reduced to the stereoinverted 12β-hydroxyl group, yielding 12β-DHADD. The function of this specificity for the reduction of HADT is not known. In C. testosteroni, the formation of 12β-DHADD is necessary for the subsequent hydroxylation at C-9 and the formation of THSATD (32). The same might be true for strain Chol1, because 12β-DHADD, the end product of anaerobic cholate degradation, is immediately transformed into THSATD by cell suspensions upon aeration (15). With respect to this, it can be assumed that high intracellular levels of NADPH pull the reaction sequence toward the degradation of the steroid skeleton.
Our study revealed that all acidic intermediates of the side chain degradation of cholate available to us, namely, Δ1,4-3-ketocholate (IV) and the respective monoene (III), DHOCTO (XX) and DHOPDC (XII), could be activated with CoA in vitro. Nevertheless, it is still unclear whether substrate-specific CoA ligases act on the free acids or whether only one or two ligases with broad substrate specificities are involved. Recently, two CoA ligases, CasG and CasI, specific for the CoA activation of cholate and of DHOPDC-like steroids, respectively, were identified in R. jostii (33). As CoA activation is an energy-consuming process, it is also reasonable that CoA transferases could be involved, reducing the demand for ATP during the side chain degradation of steroid compounds. It is also still unclear whether the activated cholyl-CoA molecule or only the free acidic molecule can be the substrate for A ring-oxidizing enzymes. The resistance of the CoA ester of DHOPDC toward hydrolysis at alkaline pH (11 to 12) might be caused by steric hindrance of the branched side chain and the D ring, as observed for other β-branched thioesters in native chemical ligation reactions (34).
Our in vitro study has also revealed the fate of the acetyl-CoA and the propionyl-CoA residues that are released from the steroid skeleton during degradation of the acyl side chain. The acetyl residue was channeled into the energy metabolism via the tricarboxylic acid (TCA) cycle and not into the assimilatory metabolism via the glyoxylate cycle. This could be the reason for the fact that strain R1 is not able to grow with cholate as the sole carbon source, despite its ability to release one acetyl-CoA residue from the substrate (19). The propionyl residue was further degraded via the methylcitric acid cycle, which is supported by the fact that all genes for this pathway are present in the genome of strain Chol1, while those for the methylmalonyl-CoA pathway are not (32).
To our knowledge, our study is the first detailed in vitro reconstitution of the complete side chain degradation of a steroid compound in bacteria. In earlier studies, degradation of the steroid side chain was shown with cell extracts of different Mycobacterium strains leading from 3-oxo-24-ethylcholest-4-en-26-oic acid and lithocholic acid to the respective 17-keto products (35, 36). However, there were no CoA esters or other intermediates detected in those earlier studies. Based on this in vitro study and on the genome information, molecular and biochemical studies for the full elucidation of the cholate degradation pathway in strain Chol1 are on the way in our laboratory. Given the high similarity of Acad and Skt to predicted proteins in Comamonas testosteroni, it is likely that the same pathway is present in this well-characterized steroid-degrading proteobacterium as well.
ACKNOWLEDGMENTS
Initial parts of this work were performed at the University of Konstanz (Germany) in the group of Bernhard Schink, who is acknowledged for his support. We thank Karin Niermann for technical assistance. Mass spectrometric measurements by Marc Schürmann (Münster) are highly acknowledged.
H.M.M. was responsible for the NMR analysis.
This work was funded by a grant of the Deutsche Forschungsgemeinschaft (DFG, PH71/3-1) (to B.P.) and by a grant of the Deutscher Akademischer Austauschdienst (DAAD) (to V.S.).
Footnotes
Published ahead of print 30 November 2012
REFERENCES
- 1. Bortolini O, Medici A, Poli S. 1997. Biotransformations on steroid nucleus of bile acids. Steroids 62: 564–577 [DOI] [PubMed] [Google Scholar]
- 2. Donova MV, Egorova OV. 2012. Microbial steroid transformations: current state and prospects. Appl. Microbiol. Biotechnol. 94: 1423–1447 [DOI] [PubMed] [Google Scholar]
- 3. Mahato SB, Garai S. 1997. Advances in microbial steroid biotransformation. Steroids 62: 332–345 [DOI] [PubMed] [Google Scholar]
- 4. Silva CP, Otero M, Esteves V. 2012. Processes for the elimination of estrogenic steroid hormones from water: a review. Environ. Pollut. 165: 38–58 [DOI] [PubMed] [Google Scholar]
- 5. Hayakawa S. 1982. Microbial transformation of bile acids. A unified scheme for bile acid degradation and hydroxylation of bile acids. Z Allg. Mikrobiol. 22: 309–326 [DOI] [PubMed] [Google Scholar]
- 6. Horinouchi M, Hayashi T, Kudo T. 2012. Steroid degradation in Comamonas testosteroni. J. Steroid Biochem. Mol. Biol. 129: 4–14 [DOI] [PubMed] [Google Scholar]
- 7. Kieslich K. 1985. Microbial side-chain degradation of sterols. J. Basic Microbiol. 25: 461–474 [DOI] [PubMed] [Google Scholar]
- 8. Philipp B. 2011. Bacterial degradation of bile salts. Appl. Microbiol. Biotechnol. 89: 903–915 [DOI] [PubMed] [Google Scholar]
- 9. Yam KC, Okamoto S, Roberts JN, Eltis LD. 2011. Adventures in Rhodococcus—from steroids to explosives. Can. J. Microbiol. 57: 155–168 [DOI] [PubMed] [Google Scholar]
- 10. Petrusma M, Hessels G, Dijkhuizen L, Van Der Geize R. 2011. Multiplicity of 3-ketosteroid-9α-hydroxylase enzymes in Rhodococcus rhodochrous DSM43269 for specific degradation of different classes of steroids. J. Bacteriol. 193: 3931–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dresen C, Lin L, D'Angelo I, Tocheva E, Strynadka N, Eltis L. 2010. A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism. J. Biol. Chem. 285: 22264–22275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yam K, D'Angelo I, Kalscheuer R, Zhu H, Wang J, Snieckus V, Ly L, Converse P, Jacobs W, Jr, Strynadka N. 2009. Studies of a ring-cleaving dioxygenase illuminate the role of cholesterol metabolism in the pathogenesis of Mycobacterium tuberculosis. PLoS Pathog. 5: e1000344 doi:10.1371/journal.ppat.1000344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lack NA, Yam KC, Lowe ED, Horsman GP, Owen RL, Sim E, Eltis LD. 2010. Characterization of a carbon-carbon hydrolase from Mycobacterium tuberculosis involved in cholesterol metabolism. J. Biol. Chem. 285: 434–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Swain K, Casabon I, Eltis LD, Mohn WW. 28 September 2012. Two transporters essential for the reassimilation of novel cholate metabolites by Rhodococcus jostii RHA1. J. Bacteriol. doi:10.1128/JB.01167-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Philipp B, Erdbrink H, Suter MJ, Schink B. 2006. Degradation of and sensitivity to cholate in Pseudomonas sp. strain Chol1. Arch. Microbiol. 185: 192–201 [DOI] [PubMed] [Google Scholar]
- 16. Hofmann AF, Mysels KJ. 1987. Bile salts as biological surfactants. Colloids Surf. 30: 145–173 [Google Scholar]
- 17. Ridlon J, Kang D, Hylemon P. 2006. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47: 241–259 [DOI] [PubMed] [Google Scholar]
- 18. Birkenmaier A, Möller H, Philipp B. 2011. Identification of a thiolase gene essential for β-oxidation of the acyl side chain of the steroid compound cholate in Pseudomonas sp. strain Chol1. FEMS Microbiol. Lett. 318: 123–130 [DOI] [PubMed] [Google Scholar]
- 19. Birkenmaier A, Holert J, Erdbrink H, Möller HM, Friemel A, Schoenenberger R, Suter MJ, Klebensberger J, Philipp B. 2007. Biochemical and genetic investigation of initial reactions in aerobic degradation of the bile acid cholate in Pseudomonas sp. strain Chol1. J. Bacteriol. 189: 7165–7173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jagmann N, Brachvogel H, Philipp B. 2010. Parasitic growth of Pseudomonas aeruginosa in co-culture with the chitinolytic bacterium Aeromonas hydrophila. Environ. Microbiol. 12: 1787–1802 [DOI] [PubMed] [Google Scholar]
- 21. Tenneson M, Baty J, Bilton R, Mason A. 1979. The degradation of cholic acid by Pseudomonas sp. NCIB 10590. Biochem. J. 184: 613–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hwang T, Shaka A. 1995. Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J. Magn. Reson. 112: 275–279 [Google Scholar]
- 23. Textor S, Wendisch V, Graaf A, Müller U, Linder M, Linder D, Buckel W. 1997. Propionate oxidation in Escherichia coli: evidence for operation of a methylcitrate cycle in bacteria. Arch. Microbiol. 168: 428–436 [DOI] [PubMed] [Google Scholar]
- 24. Griffin JE, Pandey AK, Gilmore SA, Mizrahi V, McKinney JD, Bertozzi CR, Sassetti CM. 2012. Cholesterol catabolism by Mycobacterium tuberculosis requires transcriptional and metabolic adaptations. Chem. Biol. 19: 218–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thomas ST, VanderVen BC, Sherman DR, Russell DG, Sampson NS. 2011. Pathway profiling in Mycobacterium tuberculosis: elucidation of cholesterol-derived catabolite and enzymes that catalyze its metabolism. J. Biol. Chem. 286: 43668–43678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wilbrink MH, Van Der Geize R, Dijkhuizen L. 2012. Molecular characterization of ltp3 and ltp4, essential for C24-branched chain sterol side chain degradation in Rhodococcus rhodochrous DSM43269. Microbiology doi:10.1099/mic.0.059501-0 [DOI] [PubMed] [Google Scholar]
- 27. Plaggenborg R, Steinbüchel A, Priefert H. 2001. The coenzyme A-dependent, non-beta-oxidation pathway and not direct deacetylation is the major route for ferulic acid degradation in Delftia acidovorans. FEMS Microbiol. Lett. 205: 9–16 [DOI] [PubMed] [Google Scholar]
- 28. Narbad A, Gasson MJ. 1998. Metabolism of ferulic acid via vanillin using a novel CoA-dependent pathway in a newly-isolated strain of Pseudomonas fluorescens. Microbiology 144: 1397–1405 [DOI] [PubMed] [Google Scholar]
- 29. Overhage J, Priefert H, Steinbüchel A. 1999. Biochemical and genetic analyses of ferulic acid catabolism in Pseudomonas sp. strain HR199. Appl. Environ. Microbiol. 65: 4837–4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Holert J, Alam I, Larsen M, Antunes A, Bajic V, Stingl U, Philipp B. 2013. Genome sequence of Pseudomonas sp. strain Chol1, a model organism for the degradation of bile salts and other steroid compounds. Genome Announc. doi:10.1128/genomeA.00014-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ghisla S, Thorpe C. 2004. Acyl-CoA dehydrogenases. Eur. J. Biochem. 271: 494–508 [DOI] [PubMed] [Google Scholar]
- 32. Horinouchi M, Hayashi T, Koshino H, Malon M, Yamamoto T, Kudo T. 2008. Identification of genes involved in inversion of stereochemistry of a C-12 hydroxyl group in the catabolism of cholic acid by Comamonas testosteroni TA441. J. Bacteriol. 190: 5545–5554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mohn WW, Wilbrink MH, Casabon I, Stewart GR, Liu J, van der Geize R, Eltis LD. 28 September 2012. A gene cluster encoding cholate catabolism in Rhodococcus spp. J. Bacteriol. doi:10.1128/JB.01169-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pollock SB, Kent SBH. 2011. An investigation into the origin of the dramatically reduced reactivity of peptide-prolyl-thioesters in native chemical ligation. Chem. Commun. (Camb.) 47: 2342–2344 [DOI] [PubMed] [Google Scholar]
- 35. Fujimoto Y, Chen C, Szeleczky Z, Ditullio D, Sih C. 1982. Microbial degradation of the phytosterol side chain. I. Enzymic conversion of 3-oxo-24-ethylcholest-4-en-26-oic acid into 3-oxochol-4-en-24-oic acid and androst-4-ene-3,17-dione. J. Am. Chem. Soc. 104: 4718–4720 [Google Scholar]
- 36. Lee K, Park H. 1991. The conversion of lithocholic acid into 5β-androstan-3,17-dione in the cell-free system of Mycobacterium sp. NRRL B-3805. Arch. Pharm. Res. 14: 261–265 [Google Scholar]