Abstract
Electrostatic interactions between the stator protein MotA and the rotor protein FliG are important for bacterial flagellar motor rotation. Arg90 and Glu98 of MotA are required not only for torque generation but also for stator assembly around the rotor, but their actual roles remain unknown. Here we analyzed the roles of functionally important charged residues at the MotA-FliG interface in motor performance. About 75% of the motA(R90E) cells and 45% of the motA(E98K) cells showed no fluorescent spots of green fluorescent protein (GFP)-MotB, indicating reduced efficiency of stator assembly around the rotor. The FliG(D289K) and FliG(R281V) mutations, which restore the motility of the motA(R90E) and motA(E98K) mutants, respectively, showed reduced numbers and intensity of GFP-MotB spots as well. The FliG(D289K) mutation significantly recovered the localization of GFP-MotB to the motor in the motA(R90E) mutant, whereas the FliG(R281V) mutation did not recover the GFP-MotB localization in the motA(E98K) mutant. These results suggest that the MotA-Arg90-FliG-Asp289 interaction is critical for the proper positioning of the stators around the rotor, whereas the MotA-Glu98-FliG-Arg281 interaction is more important for torque generation.
INTRODUCTION
Salmonella enterica can swim in liquid environments by rotating several flagella, which arise randomly over the cell surface. The flagellum consists of at least three parts: the basal body (rotary motor), the hook (universal joint), and the filament (helical propeller). The Salmonella flagellar motor, which is embedded within the cell membranes, is powered by proton motive force (PMF) across the cytoplasmic membrane and can rotate in both counterclockwise and clockwise directions (1, 2).
The flagellar motor converts the energy of proton flow by the PMF across the cytoplasmic membrane into the mechanical work required for toque generation. Five flagellar proteins—MotA, MotB, FliG, FliM, and FliN—are involved in motor performance (1, 2). MotA and MotB form the stator complex with four copies of MotA and two copies of MotB in the cytoplasmic membrane (3–5). The stator complex forms a proton channel to couple the proton flow with torque generation (6–11). At least 11 stators surround the rotor in a fully functional motor (12). FliG, FliM, and FliN form the C ring on the cytoplasmic face of the basal body MS ring (13). The MS-C ring complex acts as the rotor. FliG, FliM, and FliN are responsible not only for torque generation but also for switching the direction of motor rotation and flagellar assembly (14). The N-terminal region of FliG directly binds to the MS ring (15). Because a specific interaction of the C-terminal domain of FliG (FliGC) with MotA are required not only for flagellar motor rotation (16, 17) but also for stator assembly around the rotor (18–20), FliGC is believed to be located at the upper part of the C ring (Fig. 1). The FliG ring is believed to have the 26-fold rotational symmetry (21), in agreement with direct observations that the motor containing a single stator unit rotates with 26 steps per revolution (22, 23). FliM and FliN form a stable complex with one copy of FliM and four copies of FliN in solution (24). The FliM-FliN ring complex has the rotational symmetry that varies from 32- to 36-fold (25). The crystal structures of FliG (26–29), FliM (30), and FliN (24) and a crystal structure of the FliG-FliM complex (31) have been solved and possible models for their organization have been proposed (28–32). However, because the resolution of the MS-C ring structure obtained by electron cryomicroscopy and single particle image analysis is still limited (25), these models still remain ambiguous.
Fig 1.
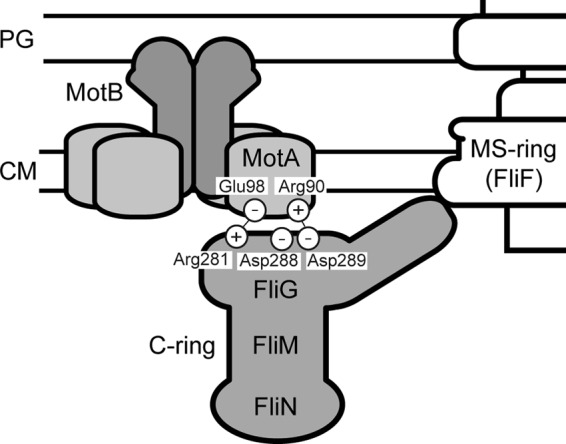
Schematic diagram of possible electrostatic interactions at the rotor-stator interface. The stator complex consists of four copies of MotA and two copies of MotB. FliG is a rotor protein and is postulated to occupy the upper part of the C ring. The interactions between MotA-Arg90 and FliG-Asp289 and between MotA-Glu98 and FliG-Arg281 are important for motor function (17).
Arg90 and Glu98, which are located in a cytoplasmic loop between transmembrane helices 2 and 3 of MotA, are highly conserved residues among the MotA orthologues and are of primary importance for motor function (16). The MotA(R90E) mutation is partially suppressed by the FliG(D289A) and FliG(D289K) mutations but not by replacement with the other amino acids. The MotA(E98K) mutation is partially suppressed by the FliG(R281V) and FliG(R281W) mutations but not by replacement with the others (17). Thus, these suppressor mutations display allele specificity, suggesting that the interactions between MotA-Arg90 and FliG-Asp289 and those between MotA-Glu98 and FliG-Arg281 are important for motor function (Fig. 1) (17). It has been shown that the R90E and E98K mutations considerably affect the assembly of green fluorescent protein (GFP)-labeled MotA/B complexes around the rotor, suggesting that these electrostatic interactions are critical for stator assembly to activate the motor (18). This opens the question of whether these interactions are directly involved in the torque generation process.
The motA(R90E) and motA(E98K) mutants are nonmotile (17, 18). However, an increase in the expression level of the MotA(R90E)/B complex by >10-fold makes 70% of the cells motile, and their swimming speed reaches ca. 60% of the wild-type level. The motA(E98K) allele is still nonfunctional even under high-expression conditions (18). These results raise the possibility that MotA-Arg90 and MotA-Glu98 would play distinct roles in motor function. To investigate the roles of these residues in motor rotation in more detail, we characterized the motA(R90E) and motA(E98K) mutants in the presence of the extragenic FliG(D289K) and FliG(R281V) suppressor mutations, respectively. We show that the FliG(D289K) mutation significantly improves the subcellular localization of GFP-MotB in the motA(R90E) mutant, suggesting that the restoration of the swimming motility is due to the improvement in the stator assembly process. However, it is not the case for the motility restoration of the motA(E98K) mutant by the FliG(R281V) mutation. We will discuss the distinct roles of these residues in flagellar motor rotation.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids used in the present study are listed in Table 1. To introduce the MotA(R90E), MotA(E98K), FliG(R281D), FliG(R281V), or FliG(D289K) mutation into the Salmonella gfp-motB strain, the fliG or motA gene on the chromosome was replaced by the motA(R90E), motA(E98K), fliG(R281D), fliG(R281V), or fliG(D289K) allele, respectively, using the λ Red homologous recombination system (37). P22-mediated transduction was carried out as described by Yamaguchi et al. (33). Procedures for DNA manipulations were carried out as described previously (38). QuikChange site-directed mutagenesis was performed as described in the manufacturer's instructions (Stratagene). DNA sequencing was carried out as described previously (39). L broth (LB) and motility medium were prepared as described previously (40, 41).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Salmonella strains | ||
| SJW1103 | Wild type for motility and chemotaxis | 33 |
| SJW2241 | ΔmotA-motB | 34 |
| YSC2123 | ΔmotA-motB ΔcheY ΔfimA fliC(Δ204-292) | This study |
| YSC2302 | ΔmotA-motB ΔfliC::pHluorin | This study |
| YVM003 | gfp-motB | 18 |
| YVM012 | gfp-motB fliC(Δ204-292) | This study |
| YVM031 | motA(R90E) gfp-motB | 18 |
| YVM032 | motA(E98K) gfp-motB | This study |
| YVM033 | gfp-motB fliG(D289K) | This study |
| YVM034 | gfp-motB fliG(R281V) | This study |
| YVM035 | motA(R90E) gfp-motB fliG(D289K) | This study |
| YVM036 | motA(E98K) gfp-motB fliG(R281V) | This study |
| YVM037 | gfp-motB fliG(R281D) | This study |
| YVM039 | motA(E98K) gfp-motB fliG(R281D) | This study |
| YVM046 | fliG(R281V) | This study |
| YVM047 | motA(E98K) | This study |
| YVM048 | motA(E98K) fliG(R281V) | This study |
| Plasmids | ||
| pBAD24 | Expression vector | 35 |
| pYC20 | pBAD24/MotA+MotB | 36 |
| pYC20(R90E) | pBAD24/MotA(R90E)+MotB | 18 |
| pYC20(E98K) | pBAD24/MotA(E98K)+MotB | 18 |
| pYC109 | pBAD24/MotA+MotB(Δ52-71) | 36 |
| pYC109(R90E) | pBAD24/MotA(R90E)+MotB (Δ52-71) | This study |
| pYC109(E98K) | pBAD24/MotA(E98K)+MotB (Δ52-71) | This study |
Bead assays.
Bead assays using polystyrene beads with diameters of 1.5, 1.0, 0.8, and 0.5 μm (Invitrogen) were performed as described previously (42). Torque calculations were done as described previously (42, 43). To produce speed histograms, the rotation rate of each motor labeled with 1.0-μm bead was sampled at 1 kHz for 10 s, and the average speed was determined from a power spectrum using 1-s data windows at 0.1-s intervals as described before (12, 18, 23).
Intracellular pH measurement.
The intracellular pH of Salmonella YSC2302 strain transformed with pYC20, pYC20(R90E), or pYC20(E98K) was measured using the pHluorin probe, which is a ratiometric pH indicator with excitation wavelengths at 410 and 470 nm and an emission wavelength at 508 nm (44), as described previously (36).
Preparation of whole-cell proteins and immunoblotting.
Salmonella cells were grown overnight at 30°C in LB with shaking. Cell pellets were suspended in sodium dodecyl sulfate (SDS) loading buffer and normalized by cell density to give a constant amount of cells. After SDS-PAGE, immunoblotting with polyclonal anti-FliG or anti-MotB antibody was carried out as described previously (40).
Measurements of free-swimming speeds of motile Salmonella cells.
For measurements of swimming speeds, Salmonella cells were observed under a phase-contrast microscope (CH40; Olympus) at room temperature. The swimming speed of individual cells was analyzed as described previously (41).
Fluorescence microscopy.
Salmonella cell bodies and the epifluorescence of GFP were observed as described previously (18). GFP fluorescence images were captured by an electron-multiplying charge-coupled device camera (C9100-02; Hamamatsu Photonics) with a 2-s exposure. Fluorescence image processing was carried out with the ImageJ version 1.43 software (National Institutes of Health). To quantify the fluorescence intensity of each GFP-MotB spot, integral fluorescence of a certain region containing a single fluorescent spot was measured, and then the intensity of a nearby spot-less cellular region of the same cell was subtracted as the background intensity. The number of GFP-MotB spots was counted above the arbitrary threshold value in each cell.
RESULTS
Effect of the MotA(R90E) mutation on torque generation.
The Salmonella motA(R90E) mutant is nonmotile. However, the overexpression of the MotA(R90E)/B complex makes most of the mutant cells swim in liquid media at ca. 60% of the wild-type swimming speed (18). In contrast, the motA(E98K) mutant is nonmotile even under high-induction conditions (18). This raises the possibility that the E98K mutation abolishes an actual torque generation step at the MotA-FliG interface, whereas the R90E mutation simply reduces the number of stators around the rotor. To test this hypothesis, we analyzed the torque-speed relationship of fully induced wild-type MotA/B and MotA(R90E)/B motor (Fig. 2A). The R90E mutation reduced both torque and speed only slightly for a wide range of load, suggesting that MotA-Arg90 is not so critical for torque generation.
Fig 2.
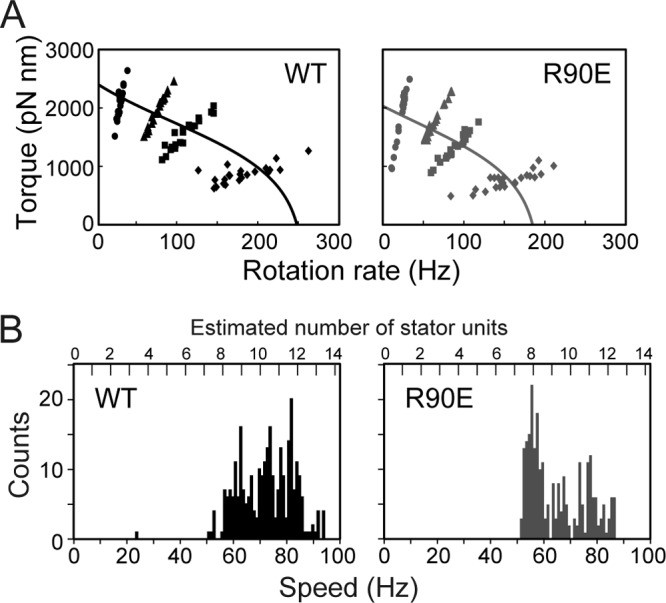
Effect of the MotA(R90E) mutation on torque generation by the flagellar motor. (A) Torque-speed curve of fully induced wild-type and MotA(R90E) mutant motor. The Salmonella YSC2122 strain [ΔmotA-motB, fliC(Δ204-292)] was transformed with pYC20 (wild-type MotA/B [WT]) or pYC20(R90E) [MotA(R90E)/B, abbreviated as R90E], and the resulting transformants were grown in LB containing 100 μg of ampicillin/ml and 100 μM arabinose for 5 h at 30°C. Rotation measurements of individual flagellar motors were carried out by tracking the position of 1.5-μm (circle), 1.0-μm (triangle), 0.8-μm (square), or 0.5-μm (diamond) beads attached to the sticky flagellar filament. All of the measurements were made at around 23°C. (B) Speed histogram of fully induced wild-type (left) or MotA(R90E) mutant motor (right). Rotation rate of each flagellar motor labeled with 1.0-μm bead was determined using 1-s data windows at an interval of 0.1 s. The number labels above the data represent the units corresponding to multiples of 7 Hz.
It has been shown that a single stator unit rotates 1.0-μm bead attached to a motor at ∼7 Hz (12, 23). To estimate the stator number in the MotA(R90E) mutant motor, the averaged rotation rates were determined (Fig. 2B). The wild-type motor showed a distribution in the speed histogram with the speed ranging from 63 to 84 Hz. The mutant motor also rotated at the speed ranging from 56 to 84 Hz, but most of them showed the highest peak at 56 Hz, indicating that the stator number is slightly less in the fully induced MotA(R90E) motor than in the wild-type motor. These results suggest that the R90E mutation significantly reduces the efficiency of stator assembly into the motor. It is also possible that the MotA(R90E) mutant stator turns the motor at ca. 80% of the wild-type speed and that the stator numbers are similar in fully induced wild-type and MotA(R90E) mutant motors.
Effect of the MotA(R90E) and MotA(E98K) mutations on proton conductivity.
It has been reported that several mutations of charged residues in a cytoplasmic loop of PomA, which is a MotA homolog, reduce the sodium ion channel activity of the unplugged PomA/B stator complex of the sodium-driven flagellar motor in Vibrio alginolyticus (20). To test whether the MotA(R90E) and MotA(E98K) mutations affect the proton conductivity of the MotA/B stator complex, we measured the intracellular pH of the Salmonella ΔmotA-motB ΔfliC::pHluorin strain expressing either MotA/B, MotA/B(Δ52-71), MotA(R90E)/B(Δ52-71), or MotA(E98K)/B(Δ52-71) using the pHluorin probe (44).
Residues 52 to 71 of MotB are postulated to act as a plug that prevent the MotA/B complex from leaking protons when it is not assembled into a motor, and the MotA/B(Δ52-71) complex made by deleting these residues is unplugged to show full proton channel activity, causing proton leakage and decreasing intracellular pH considerably, whereas the MotB(D33N) mutation suppresses proton leakage through the unplugged, activated MotA/B(Δ52-71) proton channel (36, 45). Since the proton conductivity of the MotA/B(Δ52-71) complex is estimated to be 2 orders of magnitude higher than that of the wild-type MotA/B complex (36), it is easy to examine the effect of MotA mutations on proton conductivity by using MotB(Δ52-71) as the partner.
In agreement with a previous report (36, 45), the expression of the MotA/B(Δ52-71) complex decreased the intracellular pH by ca. 0.7 pH units after arabinose induction compared to the wild-type MotA/B complex (Fig. 3). The R90E and E98K mutations still allowed the MotA/B(Δ52-71) complex to decrease the intracellular pH by ∼0.7 pH units (Fig. 3). This indicates that these MotA mutant proteins still form a proton channel complex along with MotB(Δ52-71) to conduct protons through the unplugged, activated proton channel.
Fig 3.
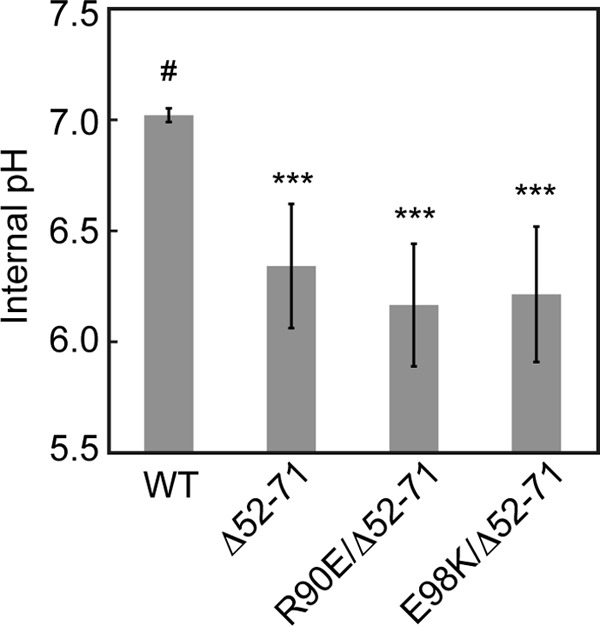
Effect of MotA(R90E) and MotA(E98K) mutations on the proton conductivity of the unplugged, activated MotA/B(Δ52-71) proton channel. Intracellular pH of YSC2123 cells (ΔmotA-motB ΔfliC::pHluorin) was measured using pHluorin expressed in the cytoplasm after the induction of MotA/B (WT), MotA/B(Δ52-71) (Δ52-71), MotA(R90E)/B(Δ52-71) (R90E/Δ52-71), or MotA(E98K)/B(Δ52-71) (E98K/Δ52-71) at an external pH of 5.5. The data that exhibited a statistically significant differences (***, P < 0.001) compared to the wild-type MotA/B (#) are highlighted with asterisks.
Effect of the FliG(D289K) and FliG(R281V) mutations on motility and stator assembly.
The FliG(D289K) and FliG(R281V) mutations partially suppress the loss-of-function phenotype of the motA(R90E) and motA(E98K) mutants, respectively, suggesting that MotA-Arg90 and MotA-Glu98 interact with FliG-Asp289 and FliG-Arg281, respectively (17). MotA-Arg90 and MotA-Glu98 are required for stator assembly (18), raising the possibility that FliG-Asp289 and FliG-Arg281 are involved in stator assembly around the rotor. To test this, we introduced the FliG(D289K) and FliG(R281V) mutations to a Salmonella gfp-motB strain using the λ Red homologous recombination system (37). These mutations did not affect either protein stability of FliG (see Fig. S1A, upper panel, lanes 1 to 3, in the supplemental material) or flagellar formation (see Fig. S1B in the supplemental material).
E. coli fliG(D289K) and fliG(R281V) mutants are nonmotile (17). However, because there are some differences between E. coli and Salmonella flagellar motors (46), we measured free-swimming speeds of the fliG(D289K) and fliG(R281V) mutants in liquid media (Fig. 4). The FliG(D289K) and FliG(R281V) mutations did not affect the expression level of GFP-MotB, as judged by immunoblotting with polyclonal anti-MotB antibody (see Fig. S1A, lower panel, lanes 1 to 3, in the supplemental material). The Salmonella fliG(D289K) gfp-motB strain was nonmotile as in E. coli. In contrast to the E. coli fliG(R281V) mutant, ca. 90% of the Salmonella fliG(R281V) gfp-motB cells were motile, and their swimming speed was about two-thirds that of the wild-type cells (Fig. 4). To test whether the GFP tag affects the function of FliG(R281V), we carried out P22-mdeiated transduction to replace the gfp-motB allele by the wild-type motB gene. The fliG(R281V) mutant also swam in soft agar, although not at the wild-type level (see Fig. S2A in the supplemental material). Because the E. coli fliG(R281V) mutant is nonmotile (17), the Salmonella motor is more robust against the FliG(R281V) mutation than the E. coli one.
Fig 4.
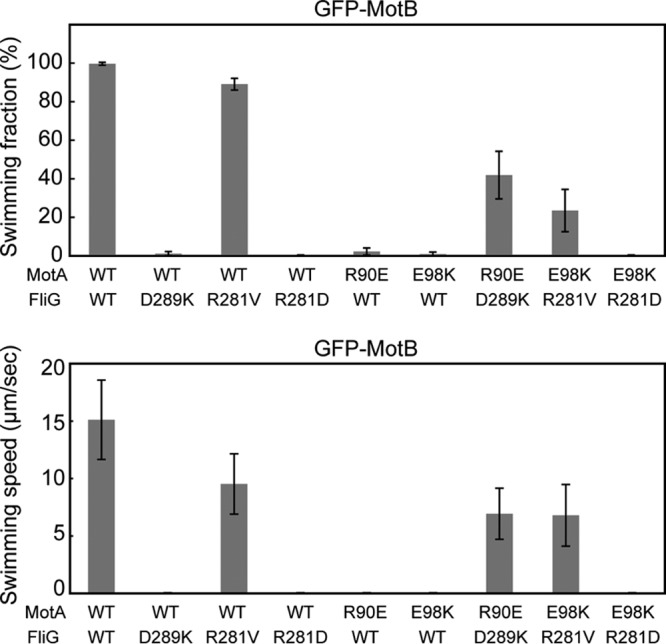
Effect of FliG and MotA mutations on free-swimming motility. The fraction and speed of free-swimming cells are indicated for the following strains: YVM003 (WT/WT), YVM033 (WT/D289K), YVM034 (WT/R281V), YVM037 (WT/R281D), YVM031 (R90E/WT), YVM032 (E98K/WT), YVM035 (R90E/D289K), YVM036 (E98K/R281V), and YVM039 (E98K/R281D). The swimming fraction is the fraction of swimming cells. The swimming speed is the average speed of >40 cells, and vertical lines indicate the standard deviations. If the fraction of motile cells was <5% of the total cells, the swimming speed is presented as zero. Measurements were made at around 23°C.
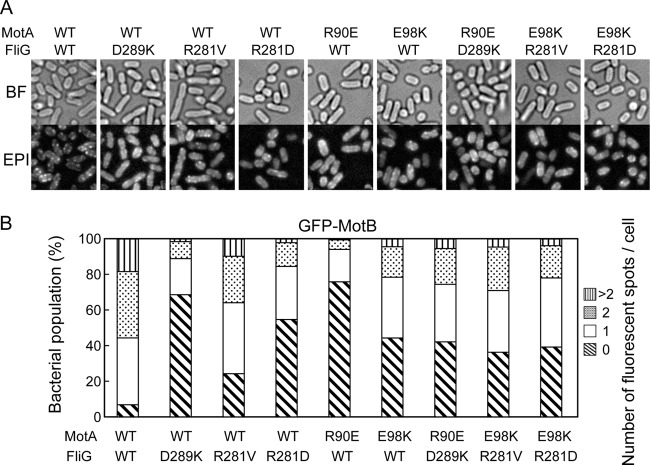
To test whether the FliG(D289K) and FliG(R281V) mutations affect stator assembly into a motor, we analyzed the subcellular localization of GFP-MotB by epi-illumination fluorescence microscopy (Fig. 5A) and measured the number and intensity of fluorescent spots of GFP-MotB (Fig. 5B). More than 90% of the wild-type fliG gfp-motB cells had more than one fluorescent spot, and the average intensity was 50 ± 20 arbitrary units (AU) (n = 50). With the FliG(D289K) mutation, ca. 70% of the cells showed no spots, and only 30% had the spots with an average intensity of 20 ± 11 AU (n = 50). With the FliG(R281V) mutation, ca. 75% of the cells had the spots but the average intensity was 28 ± 17 AU (n = 50). Such drastic reduction in both the number and intensity of GFP-MotB spots indicates that both FliG-Asp289 and FliG-Arg281 are required for stator assembly into the motor. Therefore, the impaired motility of these fliG mutants seems to be due to poor stator assembly around the rotor at a relatively low expression level of the MotA/B complex from the chromosome.
Fig 5.
Effect of FliG and MotA mutations on the subcellular localization of GFP-MotB. (A) Bright-field (BF) and epifluorescence (EPI) images of YVM003 (WT/WT), YVM033 (WT/D289K), YVM034 (WT/R281V), YVM037 (WT/R281D), YVM031 (R90E/WT), YVM032 (E98K/WT), YVM035 (R90E/D289K), YVM036 (E98K/R281V), and YVM039 (E98K/R281D). The cells were incubated at 30°C for 16 h in LB and then observed by fluorescence microscopy. Measurements were made at around 23°C. (B) Population fraction of the cells with different numbers of GFP-MotB spots in each strain described above. More than 400 cells were counted.
Multicopy effect of the MotA/B complex on motility of the fliG(D289K) and fliG(R281V) mutants.
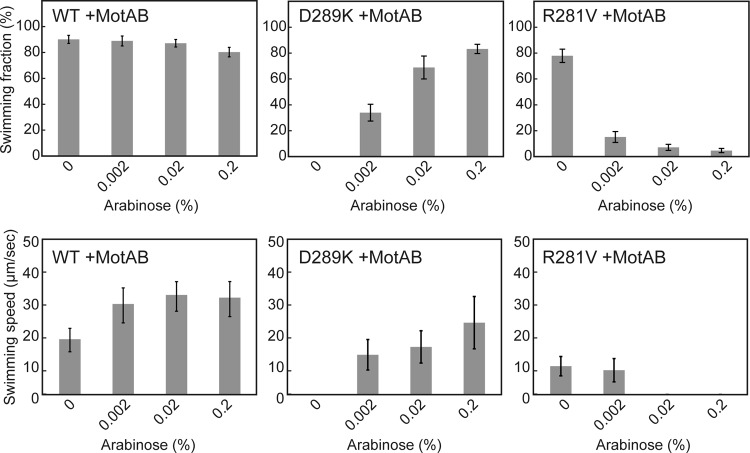
It has been shown that the overexpression of the MotA(R90E)/B complex significantly improves motor performance while that of the MotA(E98K)/B complex does not (18). Therefore, we investigated whether the overexpression of the MotA/B complex affects motility of the fliG(D289K) and fliG(R281V) mutants. We introduced a pBAD24-based plasmid encoding wild-type MotA and MotB into the fliG(D289K) gfp-motB and fliG(R281V) gfp-motB strains and measured free-swimming speeds of the resulting transformants in the presence of arabinose. We used the gfp-motB strain as a positive control. It has been shown that GFP-MotB is less functional than wild-type MotB (18). In agreement with this, bead assays revealed that the GFP tag reduces not only the zero-speed torque by half of the wild-type level but also the zero-torque speed by ca. 60% of the wild-type speed (see Fig. S3 in the supplemental material). An increment in the expression level of the MotA/B complex increased the swimming speed of the gfp-motB strain from about 20 to 30 μm/s (Fig. 6). Although most of the fliG(D289K) gfp-motB cells were nonmotile (Fig. 4), when the expression level of the MotA/B complex was increased by adding arabinose, >80% of the cells were motile, and their swimming speeds reached to ca. 70% of the wild-type levels (Fig. 6). This suggests that the impaired motility of the fliG(D289K) mutant is most likely due to a drastic decrease in the number of the stator units around the rotor. In contrast, for the fliG(R281V) gfp-motB strain, an increase in the expression level of the MotA/B complex markedly reduced the fraction of swimming cells as well as the swimming speed (Fig. 6). The overexpression of the MotA/B complex did not affect flagellar formation (data not shown). These results suggest that loss of function by the FliG(R281V) mutation is observed only when the motor has more than a critical number of stator units. Somehow, the MotA/B stator complexes around the rotor interfere with one another to impair the torque generation process when FliG has the R281V mutation.
Fig 6.
Multicopy effect of the wild-type MotA/B complex on free-swimming motility of fliG mutants. The fraction and speed of free-swimming cells of YVM003 (WT) transformed with pYC20 (MotAB), YVM033 (D289K) harboring pYC20, and YVM034 (R281V) carrying pYC20. The swimming fraction is the fraction of swimming cells. The cells were incubated at 30°C for 5 h in LB with 0.2, 0.02, or 0.002% arabinose. Measurements were made at around 23°C.
To investigate the effect of GFP-MotB on torque generation by the fliG(R281V) mutant, we analyzed free-swimming motility of the fliG(R281V) mutant in liquid media (see Fig. S2B in the supplemental material). Like the fliG(R281V) gfp-motB strain, ca. 75% of the fliG(R281V) mutant cells swam at ca. 40% of the wild-type speed (see Fig. S2B in the supplemental material). However, the overexpression of the MotA/B complex did not reduce the swimming speed of the fliG(R281V) mutant (see Fig. S2C in the supplemental material). These results suggest that the GFP directly affects the torque generation process when many wild-type MotA/B complexes are installed around the FliG(R281V) rotor.
To test whether FliG-Arg281 is required for an actual torque generation step by the stator-rotor interactions, we introduced another mutation, FliG(R281D), into the gfp-motB strain and analyzed the subcellular localization of GFP-MotB. The R281D mutation did not affect either its protein stability (see Fig. S1A, lane 4, in the supplemental material) or flagellar formation (see Fig. S1B in the supplemental material). In contrast to the fliG(R281V) gfp-motB strain, the R281D mutation allowed no motility (Fig. 4), indicating that this mutation totally interferes with motor function. About 50% of the fliG(R281D) gfp-motB cells had more than one GFP-MotB spots (Fig. 5), and the average fluorescence intensity of the GFP-MotB spots was 29 ± 12 AU (n = 50). Thus, the R281D mutation affects stator assembly more severely than the R281V mutation, but this reduction in the efficiency of stator assembly do not explain the total loss of swimming motility. These observations suggest that FliG-Arg281 is more important for torque generation than FliG-Asp289.
Effect of the fliG mutations on the subcellular localization of GFP-labeled mutant stators.
To test how the FliG(D289K) and FliG(R281V) mutations suppress the MotA(R90E) and MotA(E98K) defects, respectively, we introduced the motA(R90E) and motA(E98K) alleles into the fliG(D289K) gfp-motB and fliG(R281V) gfp-motB strains, respectively. We also constructed the motA(R90E) gfp-motB and motA(E98K) gfp-motB strains as controls. In agreement with a previous report (18), the motA(R90E) gfp-motB and motA(E98K) gfp-motB strains were nonmotile in liquid media (Fig. 4), although the expression level of GFP-MotB was not changed by these motA mutations (see Fig. S1A, lower panel, lanes 5 and 6, in the supplemental material). Like the fliG(D289K) gfp-motB strain, ca. 75% of the motA(R90E) gfp-motB cells showed no fluorescent spots of GFP-MotB (Fig. 5), and the rest had one or two spots with an average intensity of 17 ± 10 AU (n = 50). When the FliG(D289K) mutation was present in the motA(R90E) gfp-motB strain, this FliG mutation allowed 40% of the cells to swim at ca. 50% of the wild-type speed (Fig. 4). Epifluorescent images revealed that more than 60% of the cells had more than one GFP-MotB spots with the average intensity increased to 21 ± 10 AU (n = 50) (Fig. 5). These results suggest that the FliG(D289K) mutation increases the probability of assembly of the MotA(R90E)/B mutant stator complex into the motor.
For the motA(E98K) gfp-motB strain, ca. 45% of the cells showed no fluorescent spots, and the rest had more than one spot with an average intensity of 19 ± 15 AU (n = 50) (Fig. 5). This indicates that the effect of the MotA(E98K) mutation on the subcellular localization of GFP-MotB was milder than that of the MotA(R90E) mutation. The FliG(R281V) suppressor mutation significantly improved the motility of the motA(E98K) gfp-motB strain (Fig. 4), but neither the number nor the intensity of GFP-MotB spots was increased (Fig. 5). The FliG(R281D) mutation recovered neither the motility (Fig. 4) nor the subcellular localization of GFP-MotB of the motA(E98K) gfp-motB strain (Fig. 5). Interestingly, the number and intensity of the GFP-MotB spots seen in the motA(E98K) fliG(R281D) gfp-motB cells were essentially the same as those observed in the motA(E98K) fliG(R281V) gfp-motB cells (Fig. 5). This indicates that stator assembly occurs at the same level in these two mutant cells. Therefore, we suggest that the FliG(R281V) mutation restores torque generation by the motA(E98K) mutant, whereas FliG(R281D) does not.
DISCUSSION
In MotA, Arg90 and Glu98 are the most important charged residues for motor function, as are Arg281, Asp288, and Asp289 in FliG (16, 17). Electrostatic interactions between MotA-Arg90 and FliG-Asp289 and between MotA-Glu98 and FliG-Arg281 are required for motor function (Fig. 1) (17). However, because the MotA(R90E) and MotA(E98K) mutations significantly reduce the efficiency of stator assembly into a motor (18), it remained unclear whether these interactions are directly involved in the torque generation process. Here, we analyzed the torque-speed curve of the motor with fully induced MotA(R90E)/B mutant stator complex and found that this MotA mutation does not affect motor performance very substantially (Fig. 2A). The MotA(R90E) mutation significantly reduced the subcellular localization of GFP-MotB to the flagellar motor (Fig. 5), a finding in agreement with a previous report (18). We also found that the FliG(D289K) mutation, which suppresses the MotA(R90E) defect, reduced the number and intensity of GFP-MotB spots in a way similar to the MotA(R90E) mutation (Fig. 5), indicating that the D289K mutation affects stator assembly into the motor as well. This is supported by our observation that the overexpression of the MotA/B complex significantly improves the free-swimming motility of the fliG(D289K) mutant, although not to the wild-type level (Fig. 6). These results suggest that the FliG(D289K) mutation significantly reduces the binding affinity of FliG for the MotA/B complex. Because the fliG(D289K) mutant was nonmotile (Fig. 4), its loss of motility seems to be the consequence of poor assembly of the MotA/B complex around the rotor when the MotA/B complex is expressed from the chromosome. When both MotA(R90E) and FliG(D289K) mutations were present, the number and intensity of fluorescent spots of GFP-MotB were significantly increased, and the motility was restored considerably (Fig. 4 and 5). Taken together, we propose that an electrostatic interaction between MotA-Arg90 and FliG-Asp289 is important for efficient process of stator assembly into the motor and is clearly not of primary importance for torque generation but could nevertheless contribute something.
The MotA(E98K) mutation also reduced the number and intensity of GFP-MotB spots (Fig. 5), in agreement with a previous report (18). The FliG(R281V) mutation, which suppresses the MotA(E98K) defect, reduced the number and intensity of GFP-MotB spots as well (Fig. 5). These results suggest that an interaction between MotA-Glu98 and FliG-Arg281 is required for efficient stator assembly around the rotor as well. Careful comparison of these mutants indicated that the effects of the MotA(E98K) and FliG(R281V) mutations on the subcellular localization of GFP-MotB are much smaller than those of the MotA(R90E) and FliG(D289K) mutations (Fig. 5B). This implies that the electrostatic interaction between MotA-Glu98 and FliG-Arg281 may play a secondary role in stator assembly. The FliG(R281V) mutation restored the motility of the motA(E98K) mutant (Fig. 4 and see Fig. S2B in the supplemental material) but not the subcellular localization of GFP-MotB at all (Fig. 5). This indicates that the FliG(R281V) mutation recovers the torque generation process rather than stator assembly around the rotor. Therefore, we suggest that the MotA-Arg90-FliG-Asp289 interaction is more important for torque generation.
The E. coli motA fliG double mutant expressing MotA and FliG(R281V) from plasmids was nonmotile (17). In contrast, the Salmonella fliG(R281V) mutant was motile (Fig. 4 and see Fig. S2 in the supplemental material), and the overexpression of the MotA/B complex did not abolish the motility of the fliG(R281V) mutant (see Fig. S2 in the supplemental material). However, when the MotA/B complex was overexpressed in the gfp-motB fliG(R281V) strain, the motility was decreased considerably (Fig. 6). The GFP tag reduced both the zero-speed torque at high loads and the zero-torque speed at low loads (see Fig. S3 in the supplemental material), suggesting that the GFP causes some misalignment of the stator complex relative to the rotor, affecting the function of the stator complex. Therefore, it is possible that the loss of motility of the gfp-motB fliG(R281V) strain caused by the overexpression of the MotA/B complex results from a failure of the MotA/GFP-MotB complex to cooperatively work along with the wild-type one when the FliG(R281V) mutation is present.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Kihara-Macnab and S. Kojima for critical reading of the manuscript and helpful comments, Y.-S. Che for constructing the Salmonella YSC2123 and YSC2302 strains, N. Kami-ike for technical assistance, and J. Rothman for the gift of the pHluorin probe.
Y.V.M. was a research fellow of the Japan Society for the Promotion of Science. This research has been supported in part by Grants-in-Aid for Scientific Research (10J02448 to Y.V.M. and 21227006 to K.N.) and a Grant-in-Aid for Scientific Research on Innovative Areas “Spying Minority in Biological Phenomena” (23115008 to T.M.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
Published ahead of print 16 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01971-12.
REFERENCES
- 1. Berg HC. 2003. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72:19–54 [DOI] [PubMed] [Google Scholar]
- 2. Sowa Y, Berry RM. 2008. Bacterial flagellar motor. Q. Rev. Biophys. 41:103–132 [DOI] [PubMed] [Google Scholar]
- 3. Braun T, Blair DF. 2001. Targeted disulfide cross-linking of the MotB protein of Escherichia coli: evidence for two H+ channels in the stator complex. Biochemistry 40:13051–13059 [DOI] [PubMed] [Google Scholar]
- 4. Kojima S, Blair DF. 2004. Solubilization and purification of the MotA/MotB complex of Escherichia coli. Biochemistry 43:26–34 [DOI] [PubMed] [Google Scholar]
- 5. Braun TF, Al-Mawasawi LQ, Kojima S, Blair DF. 2004. Arrangement of core membrane segments in the MotA/MotB protein-channel complex of Escherichia coli. Biochemistry 43:35–45 [DOI] [PubMed] [Google Scholar]
- 6. Block SM, Berg HC. 1984. Successive incorporation of force-generating units in the bacterial rotary motor. Nature 309:470–472 [DOI] [PubMed] [Google Scholar]
- 7. Blair DF, Berg HC. 1988. Restoration of torque in defective flagellar motors. Science 242:1678–1681 [DOI] [PubMed] [Google Scholar]
- 8. Blair DF, Berg HC. 1990. The MotA protein of Escherichia coli is a proton-conducting component of the flagellar motor. Cell 60:439–449 [DOI] [PubMed] [Google Scholar]
- 9. Wilson ML, Macnab RM. 1990. Co-overproduction and localization of the Escherichia coli motility proteins MotA and MotB. J. Bacteriol. 172:3932–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stolz B, Berg HC. 1991. Evidence for interactions between MotA and MotB, torque-generating elements of the flagellar motor of Escherichia coli. J. Bacteriol. 173:7033–7037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tang H, Braun TF, Blair DF. 1996. Motility protein complexes in the bacterial flagellar motor. J. Mol. Biol. 261:209–221 [DOI] [PubMed] [Google Scholar]
- 12. Reid SW, Leake MC, Chandler JH, Lo CJ, Armitage JP, Berry RM. 2006. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc. Natl. Acad. Sci. U. S. A. 103:8066–8071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Francis NR, Sosinsky GE, Thomas D, DeRosier DJ. 1994. Isolation, characterization, and structure of bacterial flagellar motors containing the switch complex. J. Mol. Biol. 235:1261–1270 [DOI] [PubMed] [Google Scholar]
- 14. Yamaguchi S, Aizawa Kihara S-IM, Isomura M, Jones CJ, Macnab RM. 1986. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J. Bacteriol. 168:1172–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kihara M, Miller GU, Macnab RM. 2000. Deletion analysis of the flagellar switch protein FliG of Salmonella. J. Bacteriol. 182:3022–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhou J, Blair DF. 1997. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J. Mol. Biol. 273:428–439 [DOI] [PubMed] [Google Scholar]
- 17. Zhou J, Lloyd SA, Blair DF. 1998. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. U. S. A. 95:6436–6441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Morimoto YV, Nakamura S, Kami-ike N, Namba K, Minamino T. 2010. Charged residues in the cytoplasmic loop of MotA are required for stator assembly into the bacterial flagellar motor. Mol. Microbiol. 78:1117–1129 [DOI] [PubMed] [Google Scholar]
- 19. Kojima S, Nonoyma N, Takekawa N, Fukuoka H, Homma M. 2011. Mutations targeting the C-terminal domain of FliG can disrupt motor assembly in the Na+-driven flagella. J. Mol. Biol. 414:62–74 [DOI] [PubMed] [Google Scholar]
- 20. Takekawa N, Li N, Kojima S, Homma M. 2012. Characterization of PomA mutants defective in the functional assembly of the Na+-driven flagellar motor in Vibrio alginolyticus. J. Bacteriol. 194:1934–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Suzuki H, Yonekura K, Namba K. 2004. Structure of the rotor of the bacterial flagellar motor revealed by electron cryomicroscopy and single-particle image analysis. J. Mol. Biol. 337:105–113 [DOI] [PubMed] [Google Scholar]
- 22. Sowa Y, Rowe AD, Leake MC, Yakushi T, Homma M, Ishijima A, Berry RM. 2005. Direct observation of steps in rotation of the bacterial flagellar motor. Nature 437:916–919 [DOI] [PubMed] [Google Scholar]
- 23. Nakamura S, Kami-ike N, Yokota JP, Minamino T, Namba K. 2010. Evidence for symmetry in the elementary process of bidirectional torque generation by the bacterial flagellar motor. Proc. Natl. Acad. Sci. U. S. A. 107:17616–17620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brown PN, Mathews MA, Joss LA, Hill CP, Blair DF. 2005. Crystal structure of the flagellar rotor protein FliN from Thermotoga maritima. J. Bacteriol. 187:2890–2902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thomas DR, Francis NR, Chen X, DeRosier DJ. 2006. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:7039–7048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lloyd SA, Whitby FG, Blair DF, Hill CP. 1999. Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature 400:472–475 [DOI] [PubMed] [Google Scholar]
- 27. Brown PN, Hill CP, Blair DF. 2002. Crystal structure of the middle and C-terminal domains of the flagellar rotor protein FliG. EMBO J. 21:3225–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee LK, Ginsburg MA, Crovace C, Donohoe M, Stock D. 2010. Structure of the torque ring of the flagellar motor and the molecular basis for rotational switching. Nature 466:996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Minamino T, Imada K, Kinoshita M, Nakamura S, Morimoto YV, Namba K. 2011. Structural insight into the rotational switching mechanism of the bacterial flagellar motor. PLoS Biol. 9:e1000616 doi:10.1371/journal.pbio.1000616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Park SY, Lowder B, Bilwes AM, Blair DF, Crane BR. 2006. Structure of FliM provides insight into assembly of the switch complex in the bacterial flagella motor. Proc. Natl. Acad. Sci. U. S. A. 103:11886–11891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Paul K, Gonzalez-Bonet G, Bilwes AM, Crane BR, Blair D. 2011. Architecture of the flagellar rotor. EMBO J. 30:2962–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brown PN, Terrazas M, Paul K, Blair DF. 2007. Mutational analysis of the flagellar rotor protein FliG: sites of interaction with FliM and implications for organization of the switch complex. J. Bacteriol. 189:305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yamaguchi S, Fujita H, Sugata K, Taira T, Iino T. 1984. Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella. J. Gen. Microbiol. 130:255–265 [DOI] [PubMed] [Google Scholar]
- 34. Komoriya K, Shibano N, Higano T, Azuma N, Yamaguchi SAizawa S-I. 1999. Flagellar proteins and type III-exported virulence factors are the predominant proteins secreted into the culture media of Salmonella typhimurium. Mol. Microbiol. 34:767–779 [DOI] [PubMed] [Google Scholar]
- 35. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morimoto YV, Che Minamino Y-ST, Namba K. 2010. Proton-conductivity assay of plugged and unplugged MotA/B proton channel by cytoplasmic pHluorin expressed in Salmonella. FEBS Lett. 584:1268–1272 [DOI] [PubMed] [Google Scholar]
- 37. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saijo-Hamano Y, Minamino T, Macnab RM, Namba K. 2004. Structural and functional analysis of the C-terminal cytoplasmic domain of FlhA, an integral membrane component of the type III flagellar protein export apparatus in Salmonella. J. Mol. Biol. 343:457–466 [DOI] [PubMed] [Google Scholar]
- 39. Hara N, Namba K, Minamino T. 2011. Genetic characterization of conserved charged residues in the bacterial flagellar type III export protein FlhA. PLoS One 6:e22417 doi:10.1371/journal.pone.0022417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Minamino T, Macnab RM. 1999. Components of the Salmonella flagellar export apparatus and classification of export substrates. J. Bacteriol. 181:1388–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Minamino T, Imae Y, Oosawa F, Kobayashi Y, Oosawa K. 2003. Effect of intracellular pH on rotational speed of bacterial flagellar motors. J. Bacteriol. 185:1190–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Che Y-S, Nakamura S, Kojima S, Kami-ike N, Namba K, Minamino T. 2008. Suppressor analysis of the MotB(D33E) mutation to probe the bacterial flagellar motor dynamics coupled with proton translocation. J. Bacteriol. 190:6660–6667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakamura S, Kami-ike N, Yokota JP, Kudo S, Minamino T, Namba K. 2009. Effect of intracellular pH on the torque-speed relationship of bacterial proton-driven flagellar motor. J. Mol. Biol. 386:332–338 [DOI] [PubMed] [Google Scholar]
- 44. Miesenböck G, De Angelis DA, Rothman JE. 1998. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394:192–195 [DOI] [PubMed] [Google Scholar]
- 45. Hosking ER, Vogt C, Bakker EP, Manson MD. 2006. The Escherichia coli MotAB proton channel unplugged. J. Mol. Biol. 364:921–937 [DOI] [PubMed] [Google Scholar]
- 46. Nakamura S, Morimoto YV, Kami-ike N, Minamino T, Namba K. 2009. Role of a conserved prolyl residue (Pro-173) of MotA in the mechanochemical reaction cycle of the proton-driven flagellar motor of Salmonella. J. Mol. Biol. 393:300–307 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.