Abstract
In all species of the genus Shewanella, the redox-sensing Arc two-component system consists of the response regulator ArcA, the sensor kinase ArcS, and the separate phosphotransfer protein HptA. Compared to its counterpart ArcB in Escherichia coli, ArcS has a significantly different domain structure. Resequencing and reannotation revealed that in the N-terminal part, ArcS possesses a periplasmic CaChe-sensing domain bracketed by two transmembrane domains and, moreover, that ArcS has two cytoplasmic PAS-sensing domains and two receiver domains, compared to a single one of each in ArcB. Here, we used a combination of in vitro phosphotransfer studies on purified proteins and phenotypic in vivo mutant analysis to determine the roles of the different domains in ArcS function. The analysis revealed that phosphotransfer occurs from and toward the response regulator ArcA and involves mainly the C-terminal RecII domain. However, RecI also can receive a phosphate from HptA. In addition, the PAS-II domain, located upstream of the histidine kinase domain, is crucial for function. The results support a model in which phosphorylation of RecI stimulates histidine kinase activity of ArcS in order to maintain an appropriate level of phosphorylated ArcA according to environmental conditions. In addition, the study reveals some fundamental mechanistic differences between ArcS/HptA and ArcB with respect to signal perception and phosphotransfer despite functional conservation of the Arc system in Shewanella and E. coli.
INTRODUCTION
In many natural environments, bacteria are challenged by a variety of rapidly changing conditions that have to be sensed in order for the bacteria to respond appropriately. To this end, most bacteria employ two-component phosphotransfer systems comprising a sensor histidine kinase (HK) and a response regulator (RR) (1). In the signaling process, signal perception first leads to an autophosphorylation of the HK at a conserved histidine residue. In a second step, the phosphoryl group is transferred to an aspartate residue within a receiver domain of the cognate RR (2). The RR then elicits an appropriate response, often by DNA binding and up- and/or downregulation of corresponding genes (3, 4). Reversibility of these phosphotransfer reactions (5–7) as well as the modular organization of the components further increases the adaptivity of two-component signal transduction (TCS) and allows the assembly of complex multistep phosphorelay systems involving extra phosphoaspartate receiver (Rec) and histidine phosphotransfer (Hpt) domains (8). These can be localized on separate distinct proteins (9) but often occur as Rec and Hpt domains within the HK, which is then referred to as hybrid HK (7).
In Escherichia coli, one of the best-studied TCS systems employing such a hybrid HK, the anoxic redox control (Arc) system, mediates the cellular response to changes in environmental oxygen levels (4, 10). The E. coli Arc system consists of the sensor kinase ArcB, localized in the cytoplasmic membrane, and the DNA-binding RR ArcA (Fig. 1, upper panel) (11–14). Notably, the environmental oxygen level is not directly sensed by ArcB. Instead, the sensor kinase responds to the redox state of the ubiquinone and menaquinone pools within the inner membrane (15, 16). Under anaerobic conditions, the quinone pools are in a reduced state, and two conserved cysteine residues within the sensory PAS domain of ArcB remain in the thiol form (17). In this state, ArcB is highly active as a kinase and allows phosphotransfer to the RR ArcA. More specifically, upon autophosphorylation of ArcB at a conserved histidine residue in the histidine kinase cassette (HKH292), the phosphoryl group is transferred first to an aspartate in the receiver domain (RecD576) and, finally, via a histidine residue within the transfer domain of the HK (HptH717) to an aspartate residue located in the receiver domain (RecD54) of the RR ArcA (11, 18). Phosphorylated ArcA then binds to the promoter regions of its target genes or operons to activate or repress transcription (4, 19, 20). Under aerobic conditions, the quinone pool in the membrane shifts to its oxidized state and allows the formation of intermolecular disulfide bonds between the sensory cysteine residues of two ArcB monomers, shutting down the protein kinase activity (17). Instead, a reverse phosphorelay from E. coli ArcA (ArcAEc) RecD54 to ArcB RecD576 via HptH717 leads to an effective switch-off of the system (21).
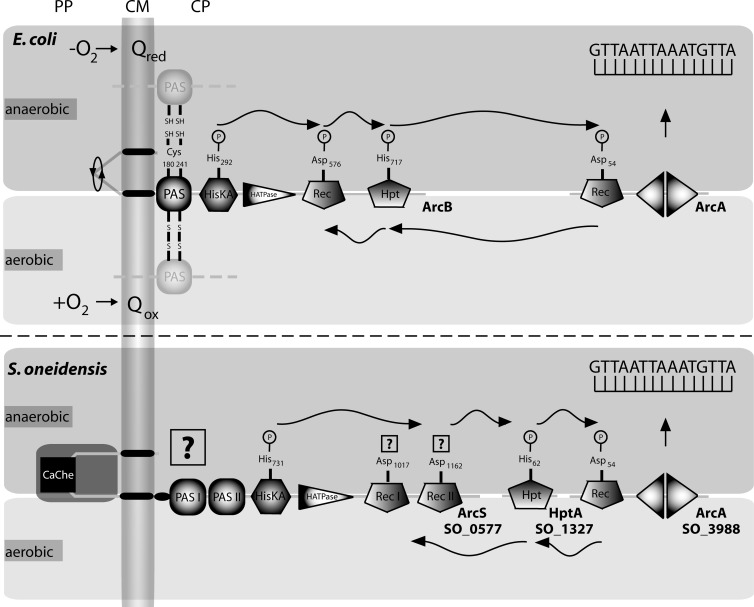
Fig 1.
Simplified comparative model of the domain architectures of and phosphoflow in the Arc systems of E. coli and S. oneidensis. The E. coli system (upper panel) consists of the membrane-integrated histidine sensor kinase (HK) ArcB and the response regulator (RR) ArcA. A reduced state of the quinone pool preserves the sulfhydryl groups of two highly conserved cysteine residues located within the PAS sensor domain. Under these conditions, the histidine kinase is highly active and a phosphorelay to the receiver domain of the RR ArcA occurs. Phosphorylated ArcA then binds to its target DNA sequences as indicated. Under aerobic conditions, the oxidized state of the quinone pool allows formation of interprotein disulfide bonds between two ArcB monomers, resulting in a rotational switch that decreases ArcB kinase activity. Under these conditions, reverse phosphotransfer from the RR to the HK occurs, resulting in signal decay. In Shewanella (lower panel), the Arc system is formed by the HK ArcS, the phosphotransfer protein HptA, and the RR ArcA. ArcB and ArcS have fundamentally different domain architectures. In previous studies, we and others have established that autophosphorylation of ArcS occurs and that a phosphoryl group can be transferred from the HK to the RR and back. However, the mechanisms of signal perception and the exact route of the phosphotransfer between the components remain elusive so far. Transmembrane regions are represented by black bars. CaChe, CaChe-sensing core domain; PAS, PAS sensing domain; HisKA, histidine kinase dimerization protein; HATPase, histidine kinase ATPase domain; Hpt, histidine transfer domain; Rec, receiver domain; Qred, reduced quinone pool; Qox, oxidized quinone pool. In ArcS, a putative signaling helix is located between the transmembrane and the PAS-I domains.
The versatile Gram-negative gammaproteobacterium Shewanella oneidensis MR-1 possesses a highly conserved ortholog of the RR ArcA of E. coli, and like ArcAEc, S. oneidensis ArcA (ArcASo; SO_3988) is involved in the response to changing oxygen levels (Fig. 1, lower panel). Like ArcAEc, ArcASo has been implicated in directly or indirectly regulating numerous genes or operons (22). However, the overlap in the ArcA regulons of the two species is surprisingly small. While ArcAEc has a major role under anaerobic conditions, e.g., by repressing the tricarboxylic acid (TCA) cycle (12, 13), loss of ArcASo has a significant impact on aerobic growth of S. oneidensis MR-1 in complex media (23). Recent studies on proteomic changes upon deletion of ArcASo indicate that this decrease in growth rate is due to impaired peptide utilization under these conditions (24). The role of the Arc system in regulation of metabolism also is rather limited in S. oneidensis MR-1. A major exception to this is regulation of dimethyl sulfoxide (DMSO) reduction, which was demonstrated to be strongly dependent on the Arc system (22).
While the ArcA ortholog was readily identified in Shewanella species, a corresponding sensor kinase orthologous to ArcB is lacking. Instead, we and others have recently demonstrated that, in addition to ArcASo, the Shewanella Arc system consists of the hybrid sensor histidine kinase ArcS (SO_0577) and the phosphotransfer protein HptA (SO_1327) (23, 25, 26). HptA is highly similar to the C-terminal Hpt domain of ArcB of E. coli, and, interestingly, the corresponding hptA is located in the same genetic context as arcB. Accordingly, we proposed HptA to be a relic of the original sensor kinase (23). In contrast, the sensor kinase ArcS differs fundamentally from its E. coli counterpart ArcB with respect to both sequence homology and domain organization, and thus we concluded that it had a phylogenetically distinct origin. Compared to ArcB with its short periplasmic region and a single cytoplasmic PAS domain, the sensory part of ArcS consists of a large periplasmic region with an embedded CaChe (as found in calcium channels and chemotaxis receptors [27]) domain and two cytoplasmic PAS sensor domains. Moreover, the catalytic part of ArcS lacks the Hpt domain typical of ArcB but possesses two receiver domains instead of a single one as in ArcB. While the second receiver domain of ArcS (ArcS-RecII) shares significant sequence similarities to ArcB, the first receiver (ArcS-RecI) does not. Strikingly, ArcS in concert with HptA can complement an E. coli ArcB mutant with respect to the cellular response to anaerobic conditions. Vice versa, growth and gene expression phenotypes caused by the absence of arcS and hptA in S. oneidensis MR-1 can be restored by ectopic expression of E. coli arcB (23, 25, 26). Thus, despite the significant differences in domain architecture, ArcB and ArcS/HptA appear to be functionally conserved.
The extended sensing and catalytic regions of ArcS compared to those of ArcB raise the question of the domains' functional roles, the nature of signals that can be perceived, and the phosphoflow from or toward the sensor kinase. In this study, we used phenotypic and gene expression analysis approaches with defined mutant strains and in vitro analyses with purified proteins to identify domains crucial for function of ArcS and to further determine the molecular mechanism of the ArcS/HptA/ArcA signaling pathway. The results support a model in which the ArcS histidine kinase activity is stimulated by phosphorylation of an adjacent receiver domain.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Strains and plasmids used in this study are listed in Table 1. S. oneidensis MR-1 and E. coli were routinely grown in lysogeny broth (LB) (28, 29) at 30°C (for S. oneidensis) or 37°C (for E. coli), unless indicated otherwise. All aerobic growth analyses were performed in 24-well plates with orbital shaking at 30°C in a Tecan Infinite M200 plate reader (Tecan, Männedorf, Switzerland). This procedure resulted in increased doubling times for ArcS mutants compared to those published earlier (23). This was probably due to differences in oxygen availability, as we previously measured aerobic growth in shaking Erlenmeyer flasks. If required, media were solidified by using 1.5% (wt/vol) agar. Anaerobic growth was assayed in 4 M minimal medium (30) adjusted to pH 7.5 (instead of LB [28]) and supplemented with 20 mM lactate as a carbon source and 40 mM (instead of 220 mM [28]) dimethyl sulfoxide (DMSO) as a terminal electron acceptor. To remove oxygen from culture tubes, they were stoppered, sealed, and flushed with nitrogen gas for several minutes with periodic shaking prior to autoclaving (25). The optical density at 600 nm (OD600) of the cultures was monitored. If necessary, media were supplemented with 10 μg · ml−1 chloramphenicol, 50 μg · ml−1 kanamycin sulfate, 100 μg · ml−1 ampicillin sodium salt, or 10% (wt/vol) sucrose. To allow growth of the conjugation strain E. coli WM3064, meso-diaminopimelic acid (DAP) was added to a final concentration of 300 μM. For promoter induction of pBAD plasmids, l-arabinose was added to a final concentration of 0.2% (wt/vol) in liquid medium.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotypea | Source or reference |
|---|---|---|
| Escherichia coli strains | ||
| DH5αλpir | recA1 gyrA Δ(lacIZYA-argF) (ϕ80d lac [lacZ] M15) pir RK6 | 50 |
| WM3064 | thrB1004 pro thi rpsL hsdS lacZ ΔM15 RP4-1360 Δ(araBAD) 567ΔdapA 1341::[erm pir(wt)] | W. Metcalf, University of Illinois, Urbana-Champaign |
| Shewanella oneidensis MR-1 strains | ||
| S79 | Wild type | 51 |
| ΔarcS mutant | ΔSO_0577 (arcS) | 23 |
| KI-His6-arcS | Chromosomal replacement of native arcS by a mutated version of arcS encoding an N-terminal His6 tag | This study |
| KI-His6-arcS PcsgAB-lux | (KI-His6-arcS); csgAB::PcsgAB-lux | This study |
| KI-arcS-HKH731A | Chromosomal replacement of native arcS by a mutated version encoding arcS (H731A) | This study |
| KI-arcS-HKH731A PcsgAB-lux | arcS (H731A); csgAB::PcsgAB-lux | This study |
| KI-arcS-RecID1017N | Chromosomal replacement of native arcS by a mutated version encoding arcS (D1017N) | This study |
| KI-arcS-RecID1017N PcsgAB-lux | arcS (D1017N); csgAB::PcsgAB-lux | This study |
| KI-arcS-RecIID1162N | Chromosomal replacement of native arcS by a mutated version encoding arcS (D1162N) | This study |
| KI-arcS-RecIID1162N PcsgAB-lux | arcS (D1162N); csgAB::PcsgAB-lux | This study |
| KI-arcS-RecI+IID1017/1162N | Chromosomal replacement of native arcS by a mutated version encoding arcS (D1017N; D1162N) | This study |
| KI-arcS-RecI+IID1017/1162N PcsgAB-lux | arcS (D1017N; D1162N); csgAB::PcsgAB-lux | This study |
| KI-arcS-PASI464-573 | Chromosomal replacement of native arcS by a mutated version encoding arcS (ΔPAS464-573) | This study |
| KI-arcS-PASI464-573 PcsgAB-lux | arcS (ΔPAS464-573); csgAB::PcsgAB-lux | This study |
| KI-arcS-PASII594-698 PcsgAB-lux | Chromosomal replacement of native arcS by a mutated version encoding arcS (ΔPAS594-698) | This study |
| KI-arcS-PASII594-698 | arcS (ΔPAS594-698); csgAB::PcsgAB-lux | This study |
| ΔarcA mutant | ΔSO_3988 (arcA) | 23 |
| ΔcsgAB mutant | ΔSO_0865-SO_0866 (csgAB) | This study |
| Plasmids | ||
| pGP704Sac28Km | mobRP4 ori-R6KsacB, suicide plasmid for generating in-frame deletions or integrations; Kmr | Chengyen Wu, unpublished |
| pGP704Sac28Km-KI-arcS-HKH731A | arcSH731A in pGP704Sac28Km | This study |
| pGP704Sac28Km-KI-arcS-RecID1017N | arcSD1017N in pGP704Sac28Km | This study |
| pGP704Sac28Km-KI-arcS-RecIID1162N | arcSD1126N in pGP704Sac28Km | This study |
| pGP704Sac28Km-KI-arcS-RecI+IID1017/1162N | arcSD1017N/D1126N in pGP704Sac28Km | This study |
| pBBR1-MCS5-TT-RBS-lux | oriT mobRK2 oriR lacZα, luxCDABE followed by terminators lambda T0 rrnB1 T1, reporter system for plasmid-based transcriptional fusions; Gmr | 38 |
| pBBR1-MCS5-TT-RBS-lux-PcsgAB | PcsgAB promoter fragment in pBBR1-MCS5-TT-RBS-lux | |
| pNPTS138-R6KT | oriT ori-R6K sacB, suicide plasmid for generating in-frame deletions or integrations; Kmr | 23 |
| pNPTS138R6KT-Lux-PcsgAB | PcsgAB-lux fragment from pBBR1-MCS5-TT-RBS-lux-PcsgAB in pNPTS138-R6KT | This study |
| pNPTS138-R6KT-KI-His6-arcS | ATG-His6-arcS fragment in pNPTS138-R6KT | This study |
| pNPTS138-R6KT-KI-arcS-PASI464-573 | arcSΔPAS-I (Δ aa 464–573) in pNPTS138-R6KT | This study |
| pNPTS138-R6KT-KI-arcS-PASII594-698 | arcSΔPAS-II (Δ aa 594–698) in pNPTS138-R6KT | This study |
| pNPTS138-R6KT-ΔcsgAB | ΔcsgAB fragment in pNPTS138-R6KT | This study |
| pBAD-HisA-arcS-694 | arcS (aa 694–1236) in pBAD-HisA | 23 |
| pBAD-HisA-arcS-694H731A | arcSH731A (aa 694–1236) in pBAD-HisA | This study |
| pBAD-HisA-arcS-694H731A/D1017N | arcSH731A/D1017N (aa 694–1236) in pBAD-HisA | This study |
| pBAD-HisA-arcS-694 H731A/D1162N | arcS H731A/D1162N (aa 694–1236) in pBAD-HisA | This study |
| pBAD-HisA-arcS-694 | arcSH731A/D1017/1162N (aa 694–1236) in pBAD-HisA | This study |
| pBAD-HisA-arcA | arcA in pBAD-HisA | 23 |
| pGEX4T-1 | ori pBR322 lacI gst, vector for overproduction of GST-fused proteins; Apr | GE Healthcare |
| pGEX4T-1-hptA | hptA in pGEX4T-1 | 23 |
| pBAD33-HisA | ori-p15A araC PBAD, vector for arabinose-inducible production of His-tagged proteins; Cmr | N. Hess, unpublished |
| pBAD33-HisA-arcS-HKH731A | arcSH731A in pBAD33-HisA | This study |
| pBAD33-HisA-arcS-RecID1017N | arcSD1017N in pBAD33-HisA | This study |
| pBAD33-HisA-arcS-RecIID1162N | arcSD1162N in pBAD33-HisA | This study |
| pBAD33-HisA-arcS-RecI+IID1017/1162N | arcSD1017/1162N in pBAD33-HisA | This study |
Apr, ampicillin resistance; Cmr, chloramphenicol resistance; Gmr, gentamicin resistance; Kmr, kanamycin resistance.
Strain constructions.
Molecular methods were performed using standard protocols (31) or according to the manufacturers' instructions. Kits for the isolation of plasmids and purification of PCR products were purchased from HISS Diagnostics GmbH (Freiburg, Germany). Enzymes were purchased from New England BioLabs (Frankfurt, Germany) and Fermentas (St. Leon-Rot, Germany). Replicative plasmids were transferred into E. coli strains by transformation using chemically competent cells (32) and into Shewanella spp. by electroporation (33). Generation of in-frame deletions or genomic introduction of arcS mutant versions was performed using a kanamycin resistance/sucrose sensitivity counterselection procedure as described earlier (34, 35). arcS variants bearing either point mutations (arcS-H731A, arcS-D1017N, or arcS-D1162N) or deletions were constructed by overlap extension PCR using mismatched primer pairs or oligonucleotides bracketing the designated gene region, respectively; the corresponding primers are listed in Table S1 in the supplemental material (36). The final PCR products were isolated from agarose gels, digested with the appropriate restriction enzymes, and ligated into the suicide vector pGP704Sac28Km or pNPTS138-R6KT. Sequential crossover was performed using S. oneidensis ΔarcS as a parental strain. Integration mutants were identified by colony PCR using primers framing arcS and were further confirmed by sequencing. Arabinose-inducible arcS mutant derivatives were generated by cloning the corresponding arcS′ sequence into pBAD33-HisA using the appropriate suicide construct as a template. For further phenotypic analyses, the resulting constructs were transferred by electroporation into the respective ΔarcS mutant strain.
RNA extraction and qRT-PCR.
Cells growing exponentially under aerobic conditions in LB (OD600 of 0.5 to 0.7) were harvested by centrifugation at 4,600 × g for 15 min at 4°C, and the cell sediments were immediately frozen in liquid nitrogen and stored at −80°C. Total RNA was extracted from S. oneidensis MR-1 cells by using the hot-phenol method (37). Residual chromosomal DNA was removed by using Turbo DNA-Free (Applied Biosystems, Darmstadt, Germany) according to the manufacturer's instructions. Extracted DNA-free total RNA was applied as the template for random-primed first-strand cDNA synthesis by using the RevertAid first-strand cDNA synthesis kit (Thermo Scientific/Fermentas, St. Leon-Rot, Germany) according to the manufacturer′s instructions. cDNA was then used as the template for quantitative RT-PCR (qRT-PCR) (real-time 7300 PCR machine; Applied Biosystems) by using the Sybr green detection system (Applied Biosystems). Samples were assayed at least in duplicate. The signal was standardized to recA, where the cycle threshold (CT) was determined automatically by use of real-time 7300 PCR software (Applied Biosystems) after 40 cycles. The efficiency of each primer pair was determined by using five different concentrations of S. oneidensis MR-1 chromosomal DNA (100 μg · ml−1, 10 μg · ml−1, 1 μg · ml−1, 100 ng · ml−1, and 10 ng · ml−1) as templates for qRT-PCR.
Measuring transcriptional activities with lux fusions.
To generate Arc-dependent luciferase activity, we first constructed vector pBBR1-MCS5-TT-RBS-lux-PcsgAB by introducing the csgAB promoter into pBBR1-MCS5-TT-RBS-lux (38). In a second step, we transferred the PcsgAB-luxCDABE fusion into pNPTS138-R6KT (23) using BamHI and SphI. Finally, the resultant construct was introduced into S. oneidensis MR-1 genome by single homologous recombination. Recombination-positive clones were selected by kanamycin resistance and were further confirmed by PCR. For phenotypic analysis, 100 μl of cells growing exponentially under aerobic conditions in LB (OD600 of 0.5 to 0.7) was transferred to white polystyrene plates (Greiner Bio One, Frickenhausen, Germany) and measured in a Tecan Infinite M200 plate reader. Luminescence was further normalized to the culture density and is presented in relative luminescence units (RLU).
Overproduction and purification of recombinant proteins.
His6-ArcS and His6-ArcA as well as the corresponding derivatives were overproduced in E. coli DH5αλpir cells harboring the corresponding expression plasmids in superoptimal broth (SOB) medium. Expression was induced by the addition of l-arabinose at a final concentration of 0.2% (wt/vol) to exponentially growing cultures followed by incubation for 4 h at 37°C. To induce the production of glutathione S-transferase (GST)-HptA proteins, 1 mM isopropyl-1-thio-β-d-galactopyranoside (IPTG) was added to the corresponding cultures. For the purification of recombinant proteins (39), cells were resuspended in lysis buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 0.2 mM phenylmethylsulfonyl fluoride [PMSF], 0.5 mg/ml lysozyme) and lysed by three passages through a French press (SLM-Aminco; Spectronic) at 18,000 lb/in2.The lysate was clarified by centrifugation at 100,000 × g for 1 h at 4°C, and the supernatant was filtered (0.45 μm). The purification was performed by affinity chromatography at 4°C, according to the manufacturers' instructions, in a batch procedure using either 1 ml Ni-nitrilotriacetic acid (NTA) Superflow (Qiagen, Hilden, Germany) for His6 proteins or 1 ml GST-Bind resin (Novagen) for GST fusion proteins. Elution fractions containing purified protein were pooled and dialyzed against TGMNKD buffer (named after its components, i.e., 50 mM Tris-HCl [pH 8.0], 10% [vol/vol] glycerol, 5 mM MgCl2, 150 mM NaCl, 50 mM KCl, and 1 mM dithiothreitol) overnight at 4°C prior to use for further assays (40).
In vitro autophosphorylation and phosphotransfer assays.
To autophosphorylate ArcA, 10 μM protein was incubated in TGMNKD buffer with an equivalent volume of acetyl [32P]phosphate for 30 min at room temperature (39). The reaction was quenched with 5× Laemmli sample buffer (0.125 M Tris-HCl [pH 6.8], 20% glycerol, 4% SDS, 10% β-mercaptoethanol, 0.02% bromophenol blue). Radioactive acetyl phosphate was generated by incubating the following reaction mixture at room temperature for 2 h: 1.5 units of acetate kinase (Sigma, Schnelldorf, Germany), TKM buffer (2.5 mM Tris-HCl [pH 7.6], 6 mM potassium acetate, 1 mM MgCl2), and 10 μl of [γ-32P]ATP (14.8 GBq/mmol; Amersham Biosciences/GE, Freiburg, Germany) in a total volume of 100 μl. To remove the acetate kinase, the reaction was subjected to centrifugation in a Microcon YM-10 centrifugal filter unit (Millipore, Schwalbach, Germany) for 1 h. The flowthrough was collected and stored at 4°C. Phosphotransfer reactions with purified ArcA, HptA, ArcS, and corresponding point mutations were performed by first autophosphorylating 10 μM ArcA for 30 min as described above. An aliquot was removed for an autophosphorylation control. Subsequently, an equivalent volume containing the response regulator at an equal concentration was added to the reaction mixture and incubated for 1 min. Both reactions were then quenched with 5× Laemmli sample buffer (kinase final concentration, 1 μM). For analysis of the autophosphorylation or the phosphotransfer reaction, 10-μl samples were loaded without prior heating on a 15% polyacrylamide gel and separated by denaturing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (41). Subsequently, gels were exposed to a PhosphorImager screen overnight, and images were detected on a Typhoon Trio PhosphorImager (Amersham Biosciences). Gels were subsequently stained with Coomassie dye (Carl Roth, Karlsruhe, Germany) to visualize proteins.
Localization and quantification of ArcS.
For the quantification and localization of ArcS in the membrane, S. oneidensis strains encoding amino-terminally His6-tagged ArcS were cultivated in 50 ml LB to exponential growth phase (OD600 of 0.4 to 0.6). Subsequently, cells were fractionated as previously described (42, 43). Membrane vesicle fractions of 2 × 109 cells and cytosolic fractions of 2 × 108 cells were separated by SDS-PAGE using 12.5% gels (41). ArcS was detected by Western immunoblotting using anti-His6 tag antibody (horseradish peroxidase [HRP] conjugated) (Abcam, Cambridge, United Kingdom) and CDP-Star (Roche Applied Science, Mannheim, Germany) following the manufacturers' instructions. Quantification analysis was performed with the ImageJ software (http://rsbweb.nih.gov/ij/).
RESULTS
Reannotation of S. oneidensis arcS.
A bioinformatic analysis readily identifies orthologs to ArcS in all of 21 fully sequenced Shewanella species, and all of these orthologous proteins share the typical ArcS domain architecture of S. oneidensis MR-1, with an important exception: the proteins encoded by the annotated arcS open reading frames of S. oneidensis MR-1, S. sediminis, S. halifaxensis, and S. piezotolerans lack a second N-terminal transmembrane domain present in ArcS of all other Shewanella species (see Fig. S1 in the supplemental material). Protein sequence comparisons strongly indicate that this is caused by a misannotation of ArcS in S. sediminis, S. halifaxensis, and S. piezotolerans, likely due to the occurrence of a potential ATG start codon further downstream. In contrast to the case for these three species, a similar N-terminal expansion of the S. oneidensis arcS open reading frame was not possible, suggesting either that ArcS of this species in fact has only a single transmembrane domain or that the corresponding DNA sequence is incorrect. To test the second hypothesis, we resequenced arcS, including a 500-bp region upstream of the annotated open reading frame. By this we identified an additional thymidine encoded at a position 80 bp upstream of the annotated start codon, which prevented the extension of the arcS reading frame by a frameshift mutation. The corrected sequence revealed that, in accordance with all other Shewanella ArcS orthologs, the arcS reading frame of S. oneidensis MR-1 likely starts 144 bp upstream of the originally annotated start codon, extending the annotated protein by 48 residues. Thus, arcS is 3,601 bp in length and encodes a protein of 1,236 amino acids (aa) with a predicted molecular mass of 140.5 kDa. A putative Shine-Dalgarno sequence (AAGGG) was identified 6 bp upstream of the newly identified start codon. Similar to the case for arcS in other Shewanella species, the corrected gene is predicted to encode a protein with a second N-terminal transmembrane domain (see Fig. S1 in the supplemental material). To test whether this prediction is correct, a sequence encoding 6 histidine residues was inserted behind the putative start codon, and production of the protein was determined by immunoblotting using antibodies targeted against the His6 tag. Membrane preparation was performed to enrich His6-ArcS to levels detectable after Western immunoblotting. As expected, a signal corresponding to a protein of the predicted molecular mass was obtained in the membrane fraction (Fig. 2). In addition, His6-ArcS was fully active and supported the function of the Shewanella Arc system with respect to growth and csgAB expression (Table 2; see Fig. S2 in the supplemental material). Based on these results we concluded that ArcS sensor kinases of Shewanella possess two transmembrane domains that bracket a periplasmic CaChe-sensing domain.
Fig 2.

ArcS Western blot analysis of localization and production. Membrane (M) and cytosolic (C) fractions of S. oneidensis MR 1 cells bearing a copy of plasmid-encoded His6-ArcS (ΔarcS + pBAD33-HisA-ArcS, left lanes), lacking ArcS (ΔarcS, middle lanes), or with a chromosomally encoded His6-ArcS version (KI-His6-arcS; right lanes) were analyzed for the presence of ArcS by SDS-PAGE using either Coomassie stain (upper panel) or His6-specific antibodies (lower panel). The exposure time for chemiluminescence development is given in minutes.
Table 2.
Doubling times of Arc mutants of S. oneidensis MR-1
| Conditions | Mean doubling time (min) ±SD |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Wild-type strain MR-1 | Strain MR-1 carrying mutation: |
||||||||
| ΔarcS | KI-His6-arcS | arcS-HKH731A | arcS-RecID1017N | arcS-RecIID1162N | arcS-RecI+IID1017N/D1162N | arcS-ΔPASI | arcS-ΔPASII | ||
| Aerobic | 42 ± 4 | 95 ± 8 | 42 ± 6 | 82 ± 10 | 53 ± 3 | 77 ± 11 | 89 ± 9 | 49 ± 4 | 88 ± 9 |
| Anaerobic | 110 ± 12 | NGa | 128 ± 9 | NG | NG | NG | NG | 92 ± 13 | NG |
NG, no growth after 40 h.
Molecular in vitro analysis of phosphoreception by ArcS.
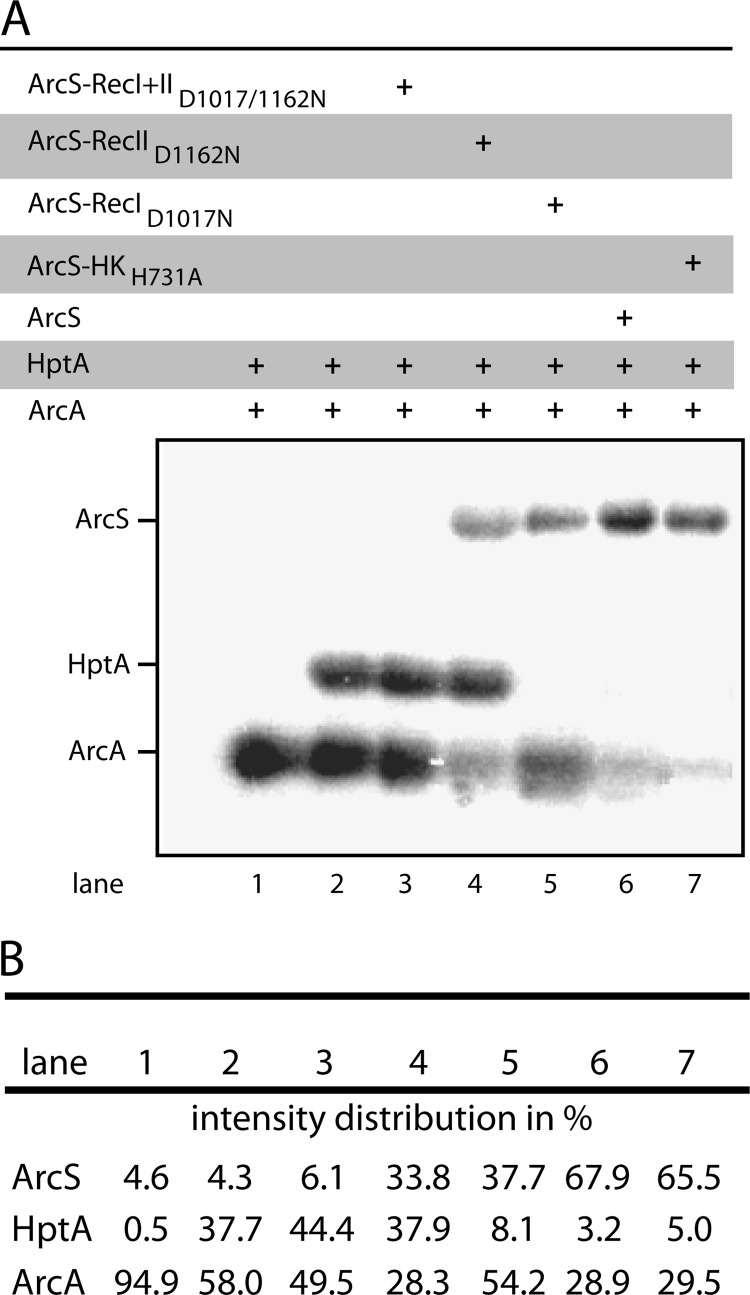
Our previous studies demonstrated that phosphotransfer occurs from ArcA to ArcS exclusively via HptA, and the corresponding phosphorylation sites in ArcA (Asp54) and HptA (His62) were identified (Fig. 1) (23). However, the route of the phosphoryl group from HptA to ArcS remained unknown. ArcS has two predicted receiver domains with conserved aspartic acid residues at positions 1017 (RecI) and 1162 (RecII), both of which might serve as potential phosphoacceptors (Fig. 1). To investigate the roles of ArcS-RecID1017 and ArcS-RecIID1162 in the phosphotransfer from HptA to ArcS, we conducted an in vitro study on purified proteins. To this end, N-terminal His6 fusions of ArcS and corresponding substitution mutants ArcS-HKH731A, ArcS-RecID1017N, ArcS-RecIID1162N, and ArcS-RecID1017N-RecIID1162N (ArcS-RecI+IID1017/1162N) were produced as soluble variants (aa 694 to 1236) that were lacking the periplasmic, transmembrane, and PAS sensor domains. ArcA and HptA were produced as an N-terminal His6 fusion and a GST fusion, respectively. Our previous study revealed that no phosphotransfer occurred when phosphorylated sensor kinase ArcS was incubated with purified GST-HptA and/or ArcA. In contrast, when phosphorylated ArcA was incubated with GST-HptA and ArcS, the phosphoryl group was rapidly transferred via GST-HptA to ArcS (23). Therefore, our in vitro experiments on ArcS substitution mutants were conducted under similar conditions. To this end, purified ArcA was incubated with acetyl [γ32P]phosphate, resulting in a signal on ArcA (Fig. 3, lane 1). As previously demonstrated, a reverse phosphorelay from ArcA via HptA (Fig. 3A, lane 2) to ArcS (Fig. 3A, lane 7) could be readily established. No difference in phosphotransfer occurred when ArcS was replaced with ArcS-HKH731A (Fig. 3A, lane 6), clearly demonstrating that the phosphate accumulates not within the histidine kinase domain in ArcS but on ArcS-RecID1017 and/or ArcS-RecIID1162. Therefore, we tested the phosphorelay from ArcA via HptA to ArcS employing mutant proteins ArcS-RecID1017N, ArcS-RecIID1162N, and ArcS-RecI+IID1017/1162N. While no signal was detected for the double substitution mutant protein (Fig. 3A, lane 3), both ArcS-RecID1017N and ArcS-RecIID1162N accumulated radioactivity (Fig. 3A, lanes 4 and 5). From these observations we conclude that both ArcS receiver domains are involved in reverse phosphotransfer from HptA. Interestingly, phosphotransfer from HptA to the individual ArcS receiver substitution mutants occurred in a different fashion. When ArcS-RecID1017N was used in the phosphotransfer reactions, no residual signal remained on HptA, and a significant signal on ArcA was readily detected (Fig. 3A, lane 5). In contrast, a strong residual signal remained on HptA when ArcS-RecIID1162N was used in the phosphotransfer reaction, while the signal intensity on ArcA was about 30% lower than in the reaction with ArcS-RecID1017N. In addition, the combined signal intensity of ArcS in ArcS-RecID1017N and ArcS-RecIID1162N corresponded to the signal intensity detected for ArcS or ArcS-HKH731A (Fig. 3B). Taken together, these observations suggest that HptA interacts with both receivers, albeit with different specificities. Based on our results, we speculate that the phosphoryl group may flow freely between ArcA-Rec, HptA, and ArcS-RecII in a way that leaves no remaining signal on HptA. Our previous studies have already provided evidence that no functional interaction occurs between ArcS and ArcA (28). In contrast, phosphotransfer from ArcA to ArcS-RecI may occur only unidirectionally, strongly suggesting a lower specificity of HptA for ArcS-RecI. The different behavior of both ArcS receiver domains during phosphotransfer raises fundamental questions with respect to their functional differentiation.
Fig 3.
In vitro studies of phosphotransfer from ArcA to ArcS. (A) Autoradiographic analysis of phosphotransfer between the indicated variants of ArcSaa694-1236, GST-HptA, and ArcA. Phosphorylated ArcA was incubated for 1 min with the indicated components, and the samples were then separated by SDS-PAGE and analyzed for their radioactive signal intensities. (B) Corresponding signal intensities were calculated using ImageJ and are given as normalized numbers, setting each lane as 100%.
In vivo analysis of ArcS substitution mutants.
Our in vitro analysis suggested different roles of the two ArcS receiver domains in phosphotransfer reactions. Therefore, we further determined whether the observed differences in phosphoflow have an impact on the function of the Shewanella Arc system in vivo. To this end, either the wild-type copy of arcS was replaced on the chromosome by the corresponding substitution mutant version or the mutated genes were overexpressed in a ΔarcS background strain. Previous publications reported that deletions in components of the S. oneidensis Arc system result in severe growth defects under aerobic conditions as well as under anaerobic conditions with DMSO as a terminal electron acceptor (22, 23, 25, 26). To determine whether the introduced substitutions result in similar growth phenotypes, the corresponding mutant strains were cultivated under similar conditions (Table 2). In addition, as a phenotypic marker at the transcriptional level, we determined differences in expression of csgAB (SO_0866/SO_0865), which is predicted to encode structural subunits of type I fimbriae (curli) in S. oneidensis. The corresponding csgAB promoter was previously demonstrated to be directly bound by ArcA (22) and is under strict negative control of the Arc regulation pathway. Hence, in the absence of arcS or arcA, csgA and csgB are highly upregulated (22, 23). Previous experiments revealed that, rather unexpectedly, upregulation of csgAB does not result in any phenotypic changes related to growth or cell-cell interactions such as clumping or biofilm formation (data not shown). To simplify further analysis of the effect of substitutions or deletions in the S. oneidensis Arc system on csgB regulation by the S. oneidensis MR-1 Arc system, we developed transcriptional reporter fusions. To this end, the luxCDABE operon was integrated directly into the native locus on the chromosome. Expression from the csgAB promoter was then measured as a function of relative light units (RLU). In all strains tested, the RLU values correlated well with the differences in expression levels as determined by qRT-PCR (Fig. 4; see Fig. S3 in the supplemental material). Thus, the PcsgAB::lux reporter fusion was suitable for analyzing the regulatory effects of mutations introduced into arcS.
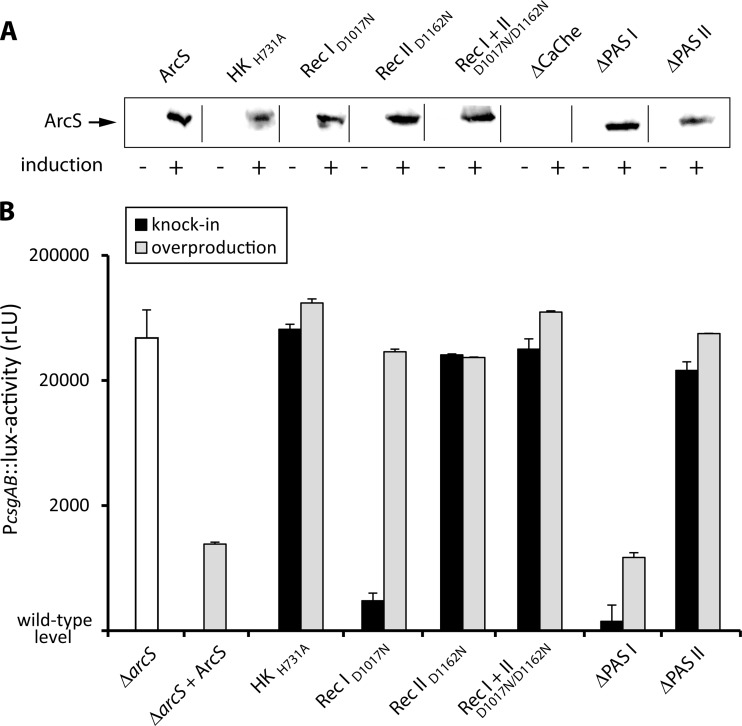
Fig 4.
In vivo analysis of the effect of ArcS substitution mutants on csgB expression. (A) Overproduction of His6-ArcS and its mutated variants (indicated at the top). Extracts of cells in which plasmid-borne expression was induced or not induced (as indicated at the bottom) were separated by SDS-PAGE. The proteins were specifically detected using antibodies raised against the N-terminal His tag. (B) csgB promoter activity depending on various levels of ArcS and its mutated variants under aerobic conditions. Promoter activity was determined by a chromosomally integrated PcsgB-lux fusion as light emission (RLU) relative to the OD600. Wild-type expression levels were almost below background levels (not shown). The white bar displays the activity in the absence of arcS. The black bars display the activity when arcS and corresponding mutants were expressed at chromosomal levels, and the gray bars display the activity when the corresponding proteins were overproduced. The error bars display the standard deviations from three independent experiments.
A differential role in vivo for the arcS receiver domains.
In a first set of experiments, arcS was chromosomally replaced by its corresponding mutant versions encoding ArcS-HKH731A, ArcS-RecID1017N,ArcS-RecIID1162N, and ArcS-RecI+IID1017/1162N and the phenotypes were compared to those of the wild type and a ΔarcS mutant. To determine whether the mutated proteins are stable, the ArcS variants were produced by ectopic expression of the corresponding genes from a plasmid in a ΔarcS background. The proteins were then detected using antibodies raised against a His6 tag that was added to the C termini. All mutated proteins could be readily and stably overproduced in S. oneidensis MR-1 at similar levels (Fig. 4A), and we therefore concluded that the observed phenotypes were caused by the introduced mutations and not by rapid degradation or instability of the mutated proteins. In line with previous results, aerobic cultures of ΔarcS mutants had an increased doubling time (95 min) compared to that of the wild type (42 min). No growth was observed under anaerobic conditions with DMSO as a terminal electron acceptor (Table 2). Accordingly, expression levels of csgB were highly increased in ΔarcS mutants compared to those of the wild type (Fig. 4B). Overexpression of arcS in a ΔarcS mutant restored the wild-type phenotype, indicating that the ArcS protein level has only minor effects on the function (Fig. 4B).
Our phosphotransfer analysis with purified proteins demonstrated that a substitution of ArcS-HKH731A had no effect on the reverse phosphorelay in vitro. However, under in vivo conditions, a severe phenotype occurred with ArcS-HKH731A with respect to growth and csgB gene expression (Table 2; Fig. 4B). Under aerobic conditions, the doubling time increased from 42 ± 4 min to 82 ± 10 min, and no growth occurred with DMSO as an alternative electron acceptor after 40 h under anaerobic conditions (Table 2). In addition, csgB expression increased to levels similar to those in an arcS mutant. Thus, the histidine kinase function of ArcS is crucial under both aerobic and anaerobic conditions. This effect was independent of the protein levels, since overproduction of ArcS-HKH731A did not result in a different phenotype.
Our in vitro data have led to the conclusion that ArcS-RecIID1162 is involved primarily in the phosphoflow from or toward ArcA. Accordingly, both substitution mutations affecting RecII, arcS-RecIID1162N and arcS-RecI+IID1017/1162N, result in similar growth phenotypes that are comparable to those of a ΔarcS mutant. Under aerobic conditions, the doubling time increased in both mutants (to 77 ± 10 min and 89 ± 9 min, compared to 95 ± 8 min for a ΔarcS mutant), and no growth occurred under anaerobic conditions on DMSO (Table 2). csgB expression also increased to a level similar to that in ΔarcS. As previously observed with ArcS, overproduction of ArcS-RecIID1162 and arcS-RecI+IID1017/1162N did not affect the corresponding phenotypes. However, in sharp contrast to substitutions that affect the RecII domain, the phenotypes of an arcS-RecID1017N substitution were strikingly different depending on the conditions. Under aerobic conditions, the doubling time of this strain (53 ± 3 min) was not as highly increased as was observed for the RecII substitution mutants but did not support growth under anaerobic conditions (Table 2).
According to the observed aerobic growth behavior, when expressed at the chromosomal level, arcSD1017N displayed wild-type-like csgB expression (Fig. 4; see Fig. S2 in the supplemental material). In contrast, overproduction of this sensor kinase variant resulted in a significant increase of the PcsgB activity to a level similar to that found for ΔarcS. Taken together, our findings on the mutated receiver domains support the results of the in vitro phosphotransfer studies and strongly indicate a functional differentiation of both ArcS receiver domains. While the RecII domain appears to play a major role in mediating phosphotransfer from and toward the response regulator ArcA under all conditions, the RecI domain rather exerts its function under anaerobic conditions when primarily ArcS kinase activity is required.
In vivo analysis of the ArcS sensory domains.
Compared to ArcB in E. coli, Shewanella ArcS possesses two instead of a single PAS domain and an additional periplasmic CaChe domain, all of which might be involved in signal perception and in proper function. To determine a potential role of these sensing domains, we performed a similar analysis as on the phosphotransmission domains in ArcS. To this end, deletions of the corresponding gene regions were introduced into arcS. In addition, the mutated versions of the genes were overexpressed in a ΔarcS background to determine the protein stability. While ArcS deleted in the cytoplasmic PAS-I (PAS464-573) or PAS-II (PAS594-698) domains was stably produced, a deletion in the periplasmic CaChe domain resulted in unstable protein. Accordingly, the ΔCaChe mutant exhibited a ΔarcS phenotype (data not shown). The deletion of PAS-I had only a minor effect on the aerobic growth rate, and the expression of csgB was not significantly affected. In contrast to the deletion of PAS-I, deletion of PAS-II resulted in a phenotype that strongly resembled the ΔarcS phenotype with respect to growth under both aerobic and anaerobic conditions and with respect to csgB expression.
DISCUSSION
The Arc two-component system is present in numerous species of the gammaproteobacteria and is involved in mediating the response to changing environmental oxygen levels. In most cases, the corresponding components of this system, the response regulator (RR) ArcA and its cognate sensor histidine kinase (HK), can be readily identified by simple homology comparison within the orders Vibrionales, Pasteurellales, Aeromonadales, Enterobacteriales, and Alteromonadales (23). However, the Shewanella Arc system represents an exception within the Alteromonadales. While ArcASo is characterized by high identity levels (81%) compared to its counterpart in E. coli, an ArcB ortholog could not be identified in Shewanella. Previous studies by our group and other groups strongly indicate that the original arcB was lost from the chromosome, except for the region encoding the C-terminal phosphotransfer domain, hptA (23, 25, 26). Instead, ArcS constitutes the cognate sensor kinase for the Shewanella Arc system (23, 26). Both the sensor and the phosphotransfer regions differ significantly between ArcS and ArcB. In this study, we provide first insights into the roles of the different domains in ArcS function.
In E. coli as well as in S. oneidensis MR-1, ArcA was demonstrated to be active under anaerobic as well as aerobic conditions (4, 10, 19, 22). It is thought that ArcA DNA-binding efficiency depends on dimerization and multimerization occurring in response to the phosphorylation levels (44). Thus, ArcB adjusts the level of phosphorylated ArcA according to the redox level of the quinone pools. In the Arc model system of E. coli, the histidine kinase domain is activated under anaerobic conditions (Fig. 1). This results in phosphoflow toward ArcA, which acts as a repressor or activator on the corresponding genes or operons (11, 18, 45). Under aerobic conditions, the activity of the histidine kinase activity is decreased, enabling a reverse phosphotransfer from ArcA to ArcB. ArcB also acts as a phosphatase, leading to signal decay (21). Previous studies demonstrated that, despite the significant differences in domain structure, ArcS/HptA can functionally complement the loss of arcB in E. coli upon ectopic production and that, vice versa, arcB expression restores the wild-type phenotype of an arcS/hptA deletion in S. oneidensis MR-1 (23, 25, 26). Although our in vitro phosphotransfer studies so far demonstrated only a reverse phosphotransfer from ArcA to ArcS via HptA, these cross complementation studies demonstrate that ArcS/HptA is similarly able to mediate forward and reverse phosphotransfer from and toward the response regulator. This also indicates that the activity of the ArcS histidine kinase domain can be tuned similarly to that of ArcB.
Role of the ArcS HK domain.
Our in vitro studies on purified proteins demonstrated that an inactivation of the HK domain in ArcS did not inhibit reverse phosphotransfer from the response regulator ArcA. However, the in vivo experiments revealed pronounced mutant phenotypes under both aerobic and anaerobic conditions in the corresponding substitution mutant. This was expected for anaerobic environments, as under these conditions, phosphoflow from ArcS to ArcA is likely to depend on the ArcS-HKH731 autophosphorylation site (18, 23, 45). In contrast, under aerobic conditions, a reverse ArcB-HKH292-independent phosphoflow is postulated for E. coli (21), and thus a wild-type like phenotype was expected for S. oneidensis ArcS-HKH731A in the presence of oxygen. The decreased growth rate observed for this mutant under aerobic conditions appears to be inconsistent with this model. However, recent studies on the E. coli Arc system revealed that ArcA phosphorylation not only occurs under fully anaerobic conditions but is already observed at 90% aerobiosis (10). Notably, in line with our finding on phenotypic effects of ArcS-HKH731A in S. oneidensis, Iuchi and Lin also reported a mutant phenotype under aerobic conditions upon ectopic production of ArcB-HKH292E in a ΔarcB E. coli strain (18). These and our findings suggest mechanistic convergence of the two sensor kinases despite evolutionary divergence.
Differential roles of the ArcS PAS domains.
The HK activity of ArcB in E. coli is thought to depend mainly on the formation of intermolecular disulfide bridges between the PAS domains of an ArcB homodimer in response to the redox state of the quinone pools in the membrane (15–17). The PAS domain of ArcB exhibits pronounced similarities to the PAS-I domain (57%) but not to the PAS-II domain of ArcS. PAS-I also harbors a conserved cysteine residue (C483) that might be involved in perception of the signal. However, deletion of the PAS-I domain (or replacement of C483 by alanine [data not shown]) resulted in only minor growth effects and did not effect csgB expression. In contrast, deletion of the PAS-II domain resulted in a phenotype which resembled that of an arcS deletion. Thus, it can be hypothesized that although both ArcS and ArcB respond to different environmental oxygen levels, the signal perception occurs in different fashions. Notably, the ArcB sensor kinases of Haemophilus influenzae and Mannheimia succiniciproducens, both of which lack the PAS domain, were demonstrated to be capable of partly complementing a ΔarcB phenotype in E. coli under different redox conditions (46, 47). A PAS sensory domain is also absent in ArcB of Actinobacillus and Pasteurella (23), strongly indicating that sensing of the environmental oxygen or cellular redox levels might occur independently of the PAS domains and cysteine residues. However, the exact mechanism remains obscure so far. It has been speculated that the presence of the PAS domain in E. coli ArcB might enable a faster response to rapidly changing conditions, which may not be required in the rather constant environment of H. influenzae (11). Thus, the expanded sensory region of ArcS in Shewanella species might contribute to appropriate responses to a wider range of environmental signals that is not restricted to cellular redox sensing. Further studies are required to elucidate the roles of the three potential sensing domains in ArcS and how this might affect the histidine kinase activity of this sensor kinase.
Functional differentiation of the ArcS Rec domains.
Compared to ArcB, ArcS comprises two receiver domains (RecI and RecII). The C-terminal RecII domain exhibits higher sequence similarity to the Rec domain in ArcB (64%) than RecI (44%) (23). Our in vitro phosphotransfer studies demonstrated that both RecI and RecII can receive a phosphoryl group from HptA; however, RecII appears to be the preferred phosphoacceptor. Accordingly, in vivo studies with ArcS in which the RecII domain was inactivated revealed that this mutation results in a phenotype that is strikingly similar to that of a ΔarcS mutant with respect to aerobic and anaerobic growth as well as csgB regulation. This also applies to mutants in which both RecI and RecII have been inactivated. Taken together, these findings strongly indicate that phosphotransfer in vivo from and toward ArcS is mediated almost exclusively via RecII. Inactivation of the ArcS RecI domain by substitution of the conserved phosphoaccepting aspartate resulted in an ArcS sensor kinase that was partly functional under aerobic conditions, when the sensor kinase is thought to act primarily as a phosphatase on ArcA (17, 21). In contrast, the arcS-RecID1017N mutant was severely impaired in function under anaerobic conditions, when kinase activity and phosphoflow toward the RR ArcA are required. This differential phenotypic behavior can be explained by the hypothesis that phosphorylated ArcS-RecID1017 stimulates ArcS kinase function and that, accordingly, nonphosphorylated ArcS-RecID1017 favors phosphatase activity. Interestingly, we also recognized a reverse phosphoflow from ArcA via HptA to ArcS-RecID1017 in vitro. However, we do not assume that ArcS-RecID1017 significantly contributes to the phosphorelay from and toward ArcA. Instead, we favor a model in which phosphorylation of ArcS-RecID1017 rather prevents complete dephosphorylation of ArcA and, hence, adjusts a physiological ratio of ArcA to ArcA∼P. This idea is further corroborated as, in contrast to native expression levels, overproduction of ArcS-RecID1017N results in significantly increased csgB expression. Here, the RecII domain might serve as a phosphate sink to result in a complete dephosphorylation of ArcA, which would lead to the observed phenotypes. A stimulating effect on kinase activity was previously reported for the single-receiver protein DivK of Caulobacter crescentus. Upon phosphorylation, DivK allosterically regulates DivJ and PleC activity and thus determines a sessile or motile lifestyle (48). Similarly, ArcS-RecI might tune ArcS kinase activity in a phosphorylation-dependent manner, thereby mediating the response to oxygen gradients. However, the respective phosphodonor for ArcS-RecI remains enigmatic. We speculate that this role is fulfilled by the ArcS kinase itself, resulting in a feedback activation loop. If this is correct, the missing forward phosphorelay in our in vitro experiments also becomes plausible. Another attractive hypothesis would be that ArcS-RecI is directly affected by levels of intracellular acetyl phosphate and thus acts as a kind of sensor. This is conceivable for Shewanella, as for this species energy conservation during anaerobic growth depends mainly on substrate-level phosphorylation with acetate as the major final product (49). Further studies are required to elucidate this potential role of acetyl phosphate for phosphorylation of RecI and stimulation of the HK activity in ArcS.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kirsten Jung for providing technical assistance that allowed finalization of necessary experiments for the study. We are also grateful to Kerstin Lassak for critical reading and helpful discussions.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG TH-831/3-1) and the Max-Planck-Gesellschaft.
Footnotes
Published ahead of print 16 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01715-12.
REFERENCES
- 1. Wuichet K, Cantwell BJ, Zhulin IB. 2010. Evolution and phyletic distribution of two-component signal transduction systems. Curr. Opin. Microbiol. 13:219–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183–215 [DOI] [PubMed] [Google Scholar]
- 3. Forst SA, Roberts DL. 1994. Signal transduction by the EnvZ-OmpR phosphotransfer system in bacteria. Res. Microbiol. 145:363–373 [DOI] [PubMed] [Google Scholar]
- 4. Salmon KA, Hung SP, Steffen NR, Krupp R, Baldi P, Hatfield GW, Gunsalus RP. 2005. Global gene expression profiling in Escherichia coli K12: effects of oxygen availability and ArcA. J. Biol. Chem. 280:15084–15096 [DOI] [PubMed] [Google Scholar]
- 5. Egger LA, Park H, Inouye M. 1997. Signal transduction via the histidyl-aspartyl phosphorelay. Genes Cells 2:167–184 [DOI] [PubMed] [Google Scholar]
- 6. Carmany DO, Hollingsworth K, McCleary WR. 2003. Genetic and biochemical studies of phosphatase activity of PhoR. J. Bacteriol. 185:1112–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tomenius H, Pernestig AK, Mendez-Catala CF, Georgellis D, Normark S, Melefors O. 2005. Genetic and functional characterization of the Escherichia coli BarA-UvrY two-component system: point mutations in the HAMP linker of the BarA sensor give a dominant-negative phenotype. J. Bacteriol. 187:7317–7324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang W, Shi L. 2005. Distribution and evolution of multiple-step phosphorelay in prokaryotes: lateral domain recruitment involved in the formation of hybrid-type histidine kinases. Microbiology 151:2159–2173 [DOI] [PubMed] [Google Scholar]
- 9. Burbulys D, Trach KA, Hoch JA. 1991. Initiation of sporulation in Bacillus subtilis is controlled by a multicomponent phosphorelay. Cell 64:545–552 [DOI] [PubMed] [Google Scholar]
- 10. Rolfe MD, Ter Beek A, Graham AI, Trotter EW, Asif HM, Sanguinetti G, de Mattos JT, Poole RK, Green J. 2011. Transcript profiling and inference of Escherichia coli K-12 ArcA activity across the range of physiologically relevant oxygen concentrations. J. Biol. Chem. 286:10147–10154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Malpica A, Pena-Sandoval GR, Rodriguez C, Franco B, Georgellis D. 2006. Signaling by the Arc two-component system provides a link between the redox state of the quinone pool and gene expression. Antioxid. Redox Signal. 8:781–795 [DOI] [PubMed] [Google Scholar]
- 12. Iuchi S, Lin EC. 1988. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc. Natl. Acad. Sci. U. S. A. 85:1888–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iuchi S, Cameron DC, Lin EC. 1989. A second global regulator gene (arcB) mediating repression of enzymes in aerobic pathways of Escherichia coli. J. Bacteriol. 171:868–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iuchi S, Matsuda Z, Fujiwara T, Lin EC. 1990. The arcB gene of Escherichia coli encodes a sensor-regulator protein for anaerobic repression of the arc modulon. Mol. Microbiol. 4:715–727 [DOI] [PubMed] [Google Scholar]
- 15. Bekker M, Alexeeva S, Laan W, Sawers G, Teixeira de Mattos J, Hellingwerf K. 2010. The ArcBA two-component system of Escherichia coli is regulated by the redox state of both the ubiquinone and the menaquinone pool. J. Bacteriol. 192:746–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Georgellis D, Kwon O, Lin EC. 2001. Quinones as the redox signal for the arc two-component system of bacteria. Science 292:2314–2316 [DOI] [PubMed] [Google Scholar]
- 17. Malpica R, Franco B, Rodriguez C, Kwon O, Georgellis D. 2004. Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc. Natl. Acad. Sci. U. S. A. 101:13318–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Iuchi S, Lin EC. 1992. Mutational analysis of signal transduction by ArcB, a membrane sensor protein responsible for anaerobic repression of operons involved in the central aerobic pathways in Escherichia coli. J. Bacteriol. 174:3972–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu X, De Wulf P. 2004. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J. Biol. Chem. 279:12588–12597 [DOI] [PubMed] [Google Scholar]
- 20. Lynch AS, Lin EC. 1996. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: characterization of DNA binding at target promoters. J. Bacteriol. 178:6238–6249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Georgellis D, Kwon O, De Wulf P, Lin EC. 1998. Signal decay through a reverse phosphorelay in the Arc two-component signal transduction system. J. Biol. Chem. 273:32864–32869 [DOI] [PubMed] [Google Scholar]
- 22. Gao H, Wang X, Yang ZK, Palzkill T, Zhou J. 2008. Probing regulon of ArcA in Shewanella oneidensis MR-1 by integrated genomic analyses. BMC Genomics 9:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lassak J, Henche AL, Binnenkade L, Thormann KM. 2010. ArcS, the cognate sensor kinase in an atypical Arc system of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 76:3263–3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yuan J, Wei B, Lipton MS, Gao H. 2012. Impact of ArcA loss in Shewanella oneidensis revealed by comparative proteomics under aerobic and anaerobic conditions. Proteomics 12:1957–1969 [DOI] [PubMed] [Google Scholar]
- 25. Gralnick JA, Brown CT, Newman DK. 2005. Anaerobic regulation by an atypical Arc system in Shewanella oneidensis. Mol. Microbiol. 56:1347–1357 [DOI] [PubMed] [Google Scholar]
- 26. Shroff NP, Charania MA, Saffarini DA. 2010. ArcB1, a homolog of Escherichia coli ArcB, regulates dimethyl sulfoxide reduction in Shewanella oneidensis MR-1. J. Bacteriol. 192:3227–3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Anantharaman V, Aravind L. 2000. Cache—a signaling domain common to animal Ca(2+)-channel subunits and a class of prokaryotic chemotaxis receptors. Trends Biochem. Sci. 25:535–537 [DOI] [PubMed] [Google Scholar]
- 28. Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miller JH. (ed). 1972. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 30. Gescher JS, Cordova CD, Spormann AM. 2008. Dissimilatory iron reduction in Escherichia coli: identification of CymA of Shewanella oneidensis and NapC of E. coli as ferric reductases. Mol. Microbiol. 68:706–719 [DOI] [PubMed] [Google Scholar]
- 31. Sambrook K, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 32. Inoue H, Nojima H, Okayama H. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23–28 [DOI] [PubMed] [Google Scholar]
- 33. Myers CR, Myers JM. 1997. Cloning and sequence of cymA, a gene encoding a tetraheme cytochrome c required for reduction of iron(III), fumarate, and nitrate by Shewanella putrefaciens MR-1. J. Bacteriol. 179:1143–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thormann KM, Duttler S, Saville RM, Hyodo M, Shukla S, Hayakawa Y, Spormann AM. 2006. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J. Bacteriol. 188:2681–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thormann KM, Saville RM, Shukla S, Pelletier DA, Spormann AM. 2004. Initial phases of biofilm formation in Shewanella oneidensis MR-1. J. Bacteriol. 186:8096–8104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51–59 [DOI] [PubMed] [Google Scholar]
- 37. Aiba H, Adhya S, de Crombrugghe B. 1981. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 256:11905–11910 [PubMed] [Google Scholar]
- 38. Gödeke J, Heun M, Bubendorfer S, Paul K, Thormann KM. 2011. Roles of two Shewanella oneidensis MR-1 extracellular endonucleases. Appl. Environ. Microbiol. 77:5342–5351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jagadeesan S, Mann P, Schink CW, Higgs PI. 2009. A novel “four-component” two-component signal transduction mechanism regulates developmental progression in Myxococcus xanthus. J. Biol. Chem. 284:21435–21445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rasmussen AA, Wegener-Feldbrügge S, Porter SL, Armitage JP, Søgaard-Andersen L. 2006. Four signalling domains in the hybrid histidine protein kinase RodK of Myxococcus xanthus are required for activity. Mol. Microbiol. 60:525–534 [DOI] [PubMed] [Google Scholar]
- 41. Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 42. Tetsch L, Koller C, Haneburger I, Jung K. 2008. The membrane-integrated transcriptional activator CadC of Escherichia coli senses lysine indirectly via the interaction with the lysine permease LysP. Mol. Microbiol. 67:570–583 [DOI] [PubMed] [Google Scholar]
- 43. Jung K, Tjaden B, Altendorf K. 1997. Purification, reconstitution, and characterization of KdpD, the turgor sensor of Escherichia coli. J. Biol. Chem. 272:10847–10852 [DOI] [PubMed] [Google Scholar]
- 44. Jeon Y, Lee YS, Han JS, Kim JB, Hwang DS. 2001. Multimerization of phosphorylated and non-phosphorylated ArcA is necessary for the response regulator function of the Arc two-component signal transduction system. J. Biol. Chem. 276:40873–40879 [DOI] [PubMed] [Google Scholar]
- 45. Georgellis D, Lynch AS, Lin EC. 1997. In vitro phosphorylation study of the arc two-component signal transduction system of Escherichia coli. J. Bacteriol. 179:5429–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Georgellis D, Kwon O, Lin EC, Wong SM, Akerley BJ. 2001. Redox signal transduction by the ArcB sensor kinase of Haemophilus influenzae lacking the PAS domain. J. Bacteriol. 183:7206–7212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jung WS, Jung YR, Oh DB, Kang HA, Lee SY, Chavez-Canales M, Georgellis D, Kwon O. 2008. Characterization of the Arc two-component signal transduction system of the capnophilic rumen bacterium Mannheimia succiniciproducens. FEMS Microbiol. Lett. 284:109–119 [DOI] [PubMed] [Google Scholar]
- 48. Paul R, Jaeger T, Abel S, Wiederkehr I, Folcher M, Biondi EG, Laub MT, Jenal U. 2008. Allosteric regulation of histidine kinases by their cognate response regulator determines cell fate. Cell 133:452–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hunt KA, Flynn JM, Naranjo B, Shikhare ID, Gralnick JA. 2010. Substrate-level phosphorylation is the primary source of energy conservation during anaerobic respiration of Shewanella oneidensis strain MR-1. J. Bacteriol. 192:3345–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Miller VL, Mekalanos JJ. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Venkateswaran K, Moser DP, Dollhopf ME, Lies DP, Saffarini DA, MacGregor BJ, Ringelberg DB, White DC, Nishijima M, Sano H, Burghardt J, Stackebrandt E, Nealson KH. 1999. Polyphasic taxonomy of the genus Shewanella and description of Shewanella oneidensis sp. nov. Internat. J. Syst. Bacteriol. 2:705–724 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.