Abstract
Glycine betaine is an effective osmoprotectant for Bacillus subtilis. Its import into osmotically stressed cells led to the buildup of large pools, whose size was sensitively determined by the degree of the osmotic stress imposed. The amassing of glycine betaine caused repression of the formation of an osmostress-adaptive pool of proline, the only osmoprotectant that B. subtilis can synthesize de novo. The ABC transporter OpuA is the main glycine betaine uptake system of B. subtilis. Expression of opuA was upregulated in response to both sudden and sustained increases in the external osmolarity. Nonionic osmolytes exerted a stronger inducing effect on transcription than ionic osmolytes, and this was reflected in the development of corresponding OpuA-mediated glycine betaine pools. Primer extension analysis and site-directed mutagenesis pinpointed the osmotically controlled opuA promoter. Deviations from the consensus sequence of SigA-type promoters serve to keep the transcriptional activity of the opuA promoter low in the absence of osmotic stress. opuA expression was downregulated in a finely tuned manner in response to increases in the intracellular glycine betaine pool, regardless of whether this osmoprotectant was imported or was newly synthesized from choline. Such an effect was also exerted by carnitine, an effective osmoprotectant for B. subtilis that is not a substrate for the OpuA transporter. opuA expression was upregulated in a B. subtilis mutant that was unable to synthesize proline in response to osmotic stress. Collectively, our data suggest that the intracellular solute pool is a key determinant for the osmotic control of opuA expression.
INTRODUCTION
In the microbial world, the amassing of compatible solutes is a widely used stress response to fend off the detrimental effects of high osmolarity on cellular water content, turgor, and physiology (1). The intracellular accumulation of compatible solutes strongly stimulates cell proliferation under otherwise growth-inhibiting high-osmolarity conditions. Compatible solutes can be scavenged by bacteria from environmental sources or synthesized. Genes encoding uptake systems for compatible solutes or enzymes for their synthesis are typically subjected to osmotic control at the transcriptional level (1–4). In addition, the activity of transporters for these compounds is often regulated in response to cytoplasmic or membrane-derived signals emanating from osmotic stress (5–8). Both regulatory mechanisms allow the microbial cells to sensitively adjust their compatible solute content in response to the osmotic conditions prevalent in their surroundings at a given time (5, 6, 8–11).
The soil bacterium Bacillus subtilis lives in a taxing habitat where desiccation processes frequently create high-salinity and high-osmolarity microniches, thereby exposing B. subtilis cells to sustained osmotic stress (12). B. subtilis makes extensive use of compatible solutes to cope with this challenge. Its initial cellular response to acute osmotic stress relies on the uptake of large amounts of potassium ions (13, 14). In the subsequent phase of acclimatization to sustained high-osmolarity surroundings, compatible solutes such as proline and glycine betaine are amassed via synthesis and uptake (9, 15–17). This leads, in turn, to a reduction in the cellular potassium content through the activities of K+ efflux systems (13, 16, 18), which allows the cell to reduce the ionic strength of the cytoplasm without compromising its overall osmotic potential.
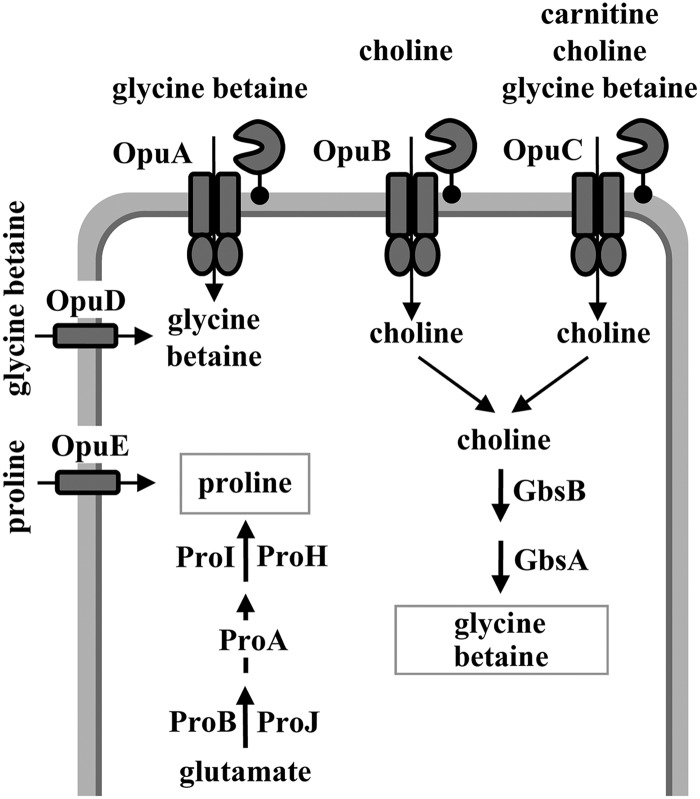
Proline is the only compatible solute that B. subtilis can synthesize de novo in response to osmotic stress, and B. subtilis possesses a dedicated biosynthetic pathway for the production of proline as an osmoprotectant (Fig. 1) (9, 16, 19). Predictably, the genetic disruption of this osmostress-adaptive proline biosynthetic route causes osmotic sensitivity (9). B. subtilis is also equipped with a set of five osmotically inducible osmoprotectant uptake systems, the Opu family of transporters (Fig. 1), which allow B. subtilis to scavenge a broad spectrum of compatible solutes as a cellular defense against osmotic stress (12) and also against high- and low-temperature challenges (20, 21). With the exception of proline (22), none of the osmoprotectants used by B. subtilis can be exploited as nutrients; they are therefore bona fide stress protectants.
Fig 1.
Compatible-solute uptake and synthesis in B. subtilis. Uptake systems and synthesis pathways for compatible solutes are summarized schematically. Detailed descriptions of the Opu transport systems and their substrate specificities have been provided elsewhere (12, 20). The synthesis of glycine betaine from the precursor choline (taken up via the OpuB and OpuC transporters) is catalyzed through a two-step oxidation process by the GbsB and GbsA enzymes, with glycine betaine aldehyde as the intermediate (24, 25). The anabolic and osmoadaptive proline biosynthetic routes and their genetic control have been described elsewhere (9, 19). In addition to the Δ1-pyrroline-5-carboxylase reductases ProI and ProH, shown here, a third protein (ProG) with Δ1-pyrroline-5-carboxylase reductase activity operates in B. subtilis (94); its physiological role is unclear.
Among the compatible solutes employed, glycine betaine assumes an especially important role for B. subtilis, since it can be both synthesized and imported via three high-affinity and osmotically inducible transport systems. Synthesis of glycine betaine relies on the uptake of the precursor molecule choline (via OpuB and OpuC) (23), which then undergoes a two-step oxidation reaction catalyzed by the GbsB and GbsA enzymes to produce glycine betaine (Fig. 1) (24, 25). The ABC transporters OpuA and OpuC and the BCCT (betaine-choline-carnitine-transporter)-type carrier OpuD mediate the import of glycine betaine (Fig. 1) (23, 26, 27). An assessment of the kinetic properties of the OpuA, OpuC, and OpuD transporters revealed that the OpuA system is the dominant glycine betaine uptake system (27). OpuA consists of the ATPase OpuAA, the integral membrane protein OpuAB, and the solute receptor OpuAC, an extracellular high-affinity glycine betaine binding protein that is tethered to the membrane via a lipid anchor (Fig. 1) (26, 28–31).
The expression of the B. subtilis opuA gene cluster (opuAA-opuAB-opuAC) is inducible in response to an increase in external salinity (26). In addition, OpuA transport activity might also be osmotically controlled (32) in a fashion analogous to that discovered by B. Poolman and coworkers for the corresponding glycine betaine uptake system OpuA of Lactococcus lactis. In the latter system (33), two cystathionine-beta-synthase (CBS) domains present near the carboxy-terminal end of the OpuAA ATPase subunit serve as sensors for changes in the ion strength of the cytoplasm and set the overall transport activity of the OpuA transport system in response to external osmotic changes (34). A repressor protein (BusR) encoded by a gene positioned upstream of the L. lactis opuA locus participates in the osmotic induction of opuA transcription (35). Interestingly, the in vitro DNA-binding activity of BusR is responsive to ionic strength (36). No BusR-related regulatory protein is present in B. subtilis, and the mechanism(s) underlying the osmotic control of opuA gene expression (26) in this microorganism is unresolved.
Here we have addressed the osmotic control of opuA expression in response to both ionic and nonionic solutes under acute and sustained osmotic stress conditions and in response to intracellularly accumulated compatible solutes. We assessed the buildup of glycine betaine pools in osmotically challenged B. subtilis cells and studied the influence of glycine betaine accumulation on the size of the osmostress-adaptive proline pool attained via de novo synthesis. Our data show that there is a linear relationship between the degree of the osmotic stress imposed on the B. subtilis cell and the size of the glycine betaine pool generated via import. As the glycine betaine content of the cell increases, a concomitant decrease in the size of the proline pool ensues. With respect to opuA expression, our data collectively indicate that the B. subtilis cell sensitively monitors its intracellular solute pool to precisely set the level of opuA transcription under high-osmolarity stress conditions.
MATERIALS AND METHODS
Chemicals.
Antibiotics, compatible solutes, the ninhydrin agent for the quantification of proline, and the chromogenic substrate for TreA enzyme assays (para-nitrophenyl-α-d-glucopyranoside [PNPG]) were obtained from Sigma-Aldrich (Steinheim, Germany). Radiolabeled [1-14C]glycine betaine (55 mCi mmol−1) was purchased from American Radiolabeled Chemicals Inc. (St. Louis, MO).
Bacterial strains.
The Escherichia coli strain DH5α (Invitrogen, Carlsbad, CA) was used for all routine cloning experiments. The B. subtilis strain JH642 (trpC2 pheA1) and its various mutant derivatives (Table 1) were used throughout this study. Strain JH642 is a member of the domesticated lineage of B. subtilis laboratory strains (37) and was kindly provided to us by James A. Hoch (Scripps Research Institute, La Jolla, CA).
Table 1.
Bacterial strains used in this study
| Straina | Relevant genotype | Construction or reference |
|---|---|---|
| JH642 | trpC2 pheA1 | J. Hoch |
| MBB9 | amyE::[Φ(opuAA′-treA)1 cat] Δ(treA::neo) | This study |
| JSB8 | Δ(proHJ::tet)1 | 19 |
| MBB12 | Δ(opuA::tet)2 Δ(opuD::neo)2 opuC-20::Tn10(spc) | This study |
| MBB13 | Δ(opuA::tet)2 Δ(opuD::neo)2 opuC-20::Tn10(spc) ΔtreA::ery | This study |
| MBB14 | amyE::[Φ(opuAA′-treA)1 cat] Δ(opuA::tet)2 opuC-20::Tn10 (spc) Δ(opuD::neo)2 ΔtreA::ery | This study |
| MBB16 | amyE::[Φ(opuAA′-treA)1 cat] Δ(degS-degU::aphA3)1 ΔtreA::ery | This study |
| MBB18 | Δ(opuA::tet)2 Δ(opuD::neo)2 opuB-20::Tn10(spc) | This study |
| MBB19 | amyE::[Φ(opuAA′-treA)1 cat] Δ(sigB2::cat) ΔtreA::ery | This study |
| MBB20 | ΔtreA::ery Δ(opuA::tet)2 Δ(opuD::neo)2 opuB-20::Tn10(spc) | This study |
| MBB21 | amyE::[Φ(opuAA′-treA)1 cat] Δ(opuA::tet)2 Δ(opuD::neo)2 opuB-20::Tn10(spc) Δ(treA::neo) | This study |
| GNB37 | Δ(treA::ery) | G. Nau-Wagner |
| FSB1 | Δ(treA::neo) | 54 |
| BLOB22 | sigBΔ2::cat | 17 |
| RMKB34 | opuBD::tet opuC-20::Tn10(spc) Δ(opuD::neo)2 | 23 |
| RMKB30 | opuC-20::Tn10(spc) Δ(opuD::neo)2 | R. Kappes |
| TMB117 | amyE::[Φ(opuAA′-treA)1 cat] Δ(treA::neo) Δ(proHJ::tet)1 | This study |
| TMB150 | amyE::[Φ(opuAA′-treA)1 cat] Δ(treA::neo) Δ(yceK::spc) | This study |
| QB4238 | trpC2 Δ(degS-degU::aphA3)1 | 95 |
All strains, except for QB4283, are derivatives of the B. subtilis wild-type laboratory strain JH642 (37) and therefore carry the trpC2 and pheA1 mutations in addition to the genetic markers indicated. Strain QB4283 is a derivative of B. subtilis strain 168.
Media and growth conditions.
B. subtilis was cultivated in Spizizen's minimal medium (SMM) (38), with 0.5% (wt/vol) glucose as the carbon source and l-tryptophan (20 mg liter−1) and l-phenylalanine (18 mg liter−1) to satisfy the auxotrophic growth requirements of strain JH642 (trpC2 pheA1) (Table 1). A solution of trace elements was added to SMM (38). The osmolarity of the growth medium was increased by the addition of NaCl (5 M stock solution), KCl (1.6 M stock solution), glycerol (2.72 M stock solution), lactose (1.22 M stock solution), or sucrose (1.24 M stock solution). The osmolarities of the growth media were determined with a Wescor 5500 vapor pressure osmometer (Wescor Inc., Logan, UT). Glycine betaine, choline, and carnitine solutions were sterilized by filtration and added to the growth media from 100 mM stock solutions. All B. subtilis cultures were inoculated from precultures growing exponentially in prewarmed minimal medium to an optical density at 578 nm (OD578) of 0.1, and the cultures were propagated at 37°C in a shaking water bath set to 220 rpm. Typically, for the determination of TreA activity in opuA-treA fusion strains, RNA preparations, or growth curves, the B. subtilis cells were grown in 20-ml or 100-ml culture volumes in 100-ml or 500-ml Erlenmeyer flasks, respectively. The B. subtilis strains were routinely maintained on Luria-Bertani (LB) agar plates. The antibiotics chloramphenicol (5 μg ml−1), kanamycin (5 μg ml−1), erythromycin-lincomycin (0.4 μg ml−1 and 15 μg ml−1), spectinomycin (100 μg ml−1), and tetracycline (10 μg ml−1) were used for the selection of gene disruption mutations in B. subtilis after the transformation of appropriate recipient strains with chromosomal DNA.
Construction of B. subtilis mutant strains.
B. subtilis mutants with defects in the various opu loci were constructed by transformation with chromosomal DNA of previously described strains carrying antibiotic resistance cassettes in various opu genes (23, 26, 27). Transformants were selected on LB agar plates containing the appropriate antibiotics. The preparation of chromosomal DNA and the transformation of B. subtilis cells with chromosomal or plasmid DNA followed routine procedures (39); all the mutant strains constructed are listed in Table 1. A deletion of the presumptive yceK regulatory gene (40, 41) was constructed by amplifying the 5′ and 3′ regions bordering the yceK locus via PCR and connecting them by long-flanking region PCR (42) with a gene encoding a spectinomycin (spc) resistance cassette. The spc gene was derived from plasmid pDG1726 (43). The fused DNA fragment resulting from the long-flanking region PCR was used to transform competent cells of the opuA-treA reporter fusion strain MBB9 (Table 1), with subsequent selection for spectinomycin-resistant colonies. The proper insertion of the Δ(yceK::spc) gene disruption into the chromosome of the resulting strain TMB150 (Table 1) was verified by PCR using DNA primers flanking the authentic yceK locus. Production of the extracellular α-amylase AmyE by B. subtilis strains was tested by flooding the colonies grown on LB plates containing 1% starch with Gram's iodide stain (0.5% [wt/vol] iodide, 1% potassium iodide) for 1 min and scoring for zones of clearing around the colonies after the stain was decanted (39).
Construction of plasmids and site-directed mutagenesis.
To clone the wild-type opuA promoter, we amplified a 976-bp opuA DNA fragment from the chromosome of strain JH642 by PCR using primers containing artificial EcoRV and BamHI restriction sites at their 5′ ends (forward primer, EcoRV-opuA [5′-GCGCGATATCGAATTCAATTCCAATAAAAG-3′]; reverse primer, BamHI-opuA rev [5′-CGCGCGGATCCTTTGAACGATGTTGCCG-3′]). This DNA fragment was then inserted into a pBluescript SK(−) vector (Stratagene, Heidelberg, Germany) that had been cut with EcoRV and BamHI. This yielded plasmid pTMB1, and the nucleotide sequence of the inserted opuA fragment was verified by DNA sequence analysis. In parallel, we cloned the same PCR fragment carrying the opuA promoter and part of the opuAA coding region into the low-copy-number treA operon fusion vector pJMB1 (M. Jebbar and E. Bremer, unpublished results), thereby producing plasmid pMBB10 (amyE::opuA-treA-cat::amyE). This fusion construct was inserted into the chromosome of B. subtilis after linearization of the plasmid DNA by restriction digestion, transformation of an appropriate B. subtilis strain {e.g., strain FSB1 [Δ(treA::neo)]} (Table 1), and selection of chloramphenicol-resistant colonies on LB agar plates. This integrated the amyE::opuA-treA-cat::amyE construct, via a double homologous recombination event, stably into the nonessential chromosomal amyE locus without integrating the plasmid backbone of the pJMB1 reporter gene fusion vector as well. The integration of the opuA-treA reporter gene fusions into the B. subtilis chromosome at the amyE site was verified by scoring the AmyE-negative phenotype of such transformants. Mutations in the B. subtilis opuA promoter were isolated using the QuikChange site-directed mutagenesis kit (Stratagene, Heidelberg, Germany) and a series of custom-synthesized mutagenic DNA primers. Plasmid pTMB1, carrying the wild-type opuA promoter, served as the template for the mutagenesis experiments. The mutant opuA promoter fragments were recovered from these pTMB1-derived plasmids by digestion with EcoRV and BamHI and were inserted into the treA operon fusion vector pJMB1. After linearization of the resulting plasmids by restriction digestion, the B. subtilis strain FSB1 [Δ(treA::neo)] (Table 1) was transformed with the mutant opuA-treA fusion constructs, and their integration into the chromosomal amyE locus was selected for by chloramphenicol resistance and subsequent scoring of the AmyE phenotype. To construct a plasmid for primer extension analysis, we cloned the 976-bp opuA DNA fragment (derived from plasmid pMBB10) as an EcoRI-BamHI restriction fragment into the E. coli-B. subtilis shuttle vector pRB373 (44), thereby producing plasmid pMBB39.
Northern blot analysis.
Total RNA was isolated from exponentially growing B. subtilis JH642 cultures (OD578, approximately 1.0) by the acidic-phenol method (45). Two micrograms of total RNAs were denatured by heating (70°C, 5 min) in the presence of 50% formamide. The denatured RNA sample was then electrophoretically separated in a 1.4% agarose gel buffered with 20 mM morpholinepropanesulfonic acid (MOPS) (pH 7.0). Northern blot analysis was performed as described previously (21). Antisense RNA probes for opuAA and trnB-Ala (a constitutively expressed gene for a B. subtilis tRNA) were prepared using an in vitro transcription system (Strip-EZ kit; Ambion, Austin, TX). DNA templates for in vitro transcription reactions were generated by PCR using the following primer pairs, with the reverse primer carrying an artificial T7 promoter sequence in addition to either the opuAA- or the trnB-Ala-specific sequence: opuAA-for195 (5′-GGTCTATCAGGGAGCGG-3′) and opuA-rev969-T7 (5′-TAATACGACTCACTATAGGGAGGCTGTCGCCGATCCGCAACGC-3′) for the opuAA probe and primers trnB-Ala-for (5′-GGGGCCTTAGCTCAGCT-3′) and trnB-Ala-rev-T7 (5′-TAATACGACTCACTATAGGGAGGAAGTGGAGCCTAGCGGG-3′). By using the resulting PCR products as templates for in vitro transcription reactions, hybridization probes internal to the opuAA gene (519 nucleotides) or covering the entire trnB-Ala gene (76 nucleotides) were generated.
Primer extension analysis.
Strain JH642 (pMBB39-opuAA′) was grown either in SMM alone or in SMM with 0.4 M NaCl in the presence of 5 μg ml−1 kanamycin to select for the presence of the plasmid. RNA was prepared from these cultures after they had reached the mid-exponential-growth phase (OD578, 0.8 to 1.0). Total RNA was isolated, and an opuA-specific antisense DNA primer (opuA-PE [5′-CCGCTCCCTGATAGACCC-3′]) labeled at its 5′ end with the fluorescent dye IRD800 (purchased from MWG, Ebersberg, Germany) was hybridized to the opuA mRNA and was extended with avian myeloblastosis virus (AMV) reverse transcriptase (Promega, Madison, WI). The same primer was used for a DNA sequence reaction using plasmid pMBB39 as a template to allow the identification of the 5′ end of the opuA mRNA. DNA sequencing was performed using the dideoxy chain termination method with the “Thermo Sequenase fluorescent-labeled primer cycle sequencing kit with 7-deaza-GTP” (Amersham Pharmacia Biotech, Freiburg, Germany). The products of the primer extension and DNA sequencing reactions were analyzed using a DNA sequencer (model 4000; Li-COR Biosiences, Bad Homburg, Germany).
TreA reporter enzyme assays.
The expression of chromosomal opuA-treA reporter gene fusions was monitored by assaying TreA [phospho-α-(1,1)-glucosidase] enzyme activity (46) by using the chromogenic substrate para-nitrophenyl-α-d-glucopyranoside as detailed previously (19). One unit of TreA activity is defined as 1 μmol of substrate converted per min per mg of protein. The protein concentrations of the samples were estimated from the optical density of the B. subtilis cell culture. In all of the opuA-treA gene fusion strains used, the natural treA gene in the chromosome of B. subtilis (47) was disrupted (Table 1), so that the measured TreA enzyme activity reflected only the activity of the TreA reporter enzyme produced by the cell in response to the transcriptional activity of the opuA promoter.
Determination of intracellular glycine betaine pools.
To determine the sizes of intracellular glycine betaine pools, B. subtilis cultures were grown to the mid-exponential-growth phase (OD578, about 1.5) in SMM with varying salinities in the presence of 1 mM glycine betaine; the glycine betaine sample used for this experiment was spiked with 0.64 μM radiolabeled [1-14C]glycine betaine. Aliquots (300 μl) of the cultures were withdrawn. The cells were collected by filtration onto filter discs (pore size, 0.45 μM; Schleicher & Schuell, Dassel, Germany) and were then carefully washed with isoosmotic growth medium to remove extracellular [1-14C]glycine betaine. The radioactivity in the B. subtilis cells collected on the filters was then measured by scintillation counting, and the intracellular pool size of glycine betaine was calculated as described previously (21) using a B. subtilis cell volume of 0.67 μl per 1 OD578 unit of cell culture (S. Moses, E. P. Bakker, and E. Bremer, unpublished data). The B. subtilis cell volumes were calculated from the determination of the internal and total water spaces. To this end, aliquots of the cultures were incubated with 3H2O and inulin-[14C]carboxylic acid (Amersham Life Science, Freiburg, Germany), and the distribution of these radiolabeled compounds was measured by scintillation counting after centrifugation through silicon oil (48).
Measurements of cellular proline pools.
The intracellular proline contents of B. subtilis strains were determined by a colorimetric assay detecting proline as a colored proline-ninhydrin complex that can be quantified by measuring the absorption of the solution at 480 nm (49). Cells of the B. subtilis strain studied were grown in SMM in the absence or presence of glycine betaine, using NaCl and glycine betaine concentrations specified in the figures, until they reached an OD578 of about 1.5. The cells (8 ml) were then harvested by centrifugation and were frozen at −20°C until further use. The soluble cellular content was extracted, and the amount of proline was assessed according to the procedure detailed by Bates et al. (49). Proline concentrations were determined by establishing a standard curve with l-proline for the colorimetric proline assay, and intracellular proline concentrations of B. subtilis were then calculated by using a B. subtilis cell volume of 0.67 μl per 1 OD578 unit of cell culture.
RESULTS
Glycine betaine and proline pools in osmotically stressed B. subtilis cells.
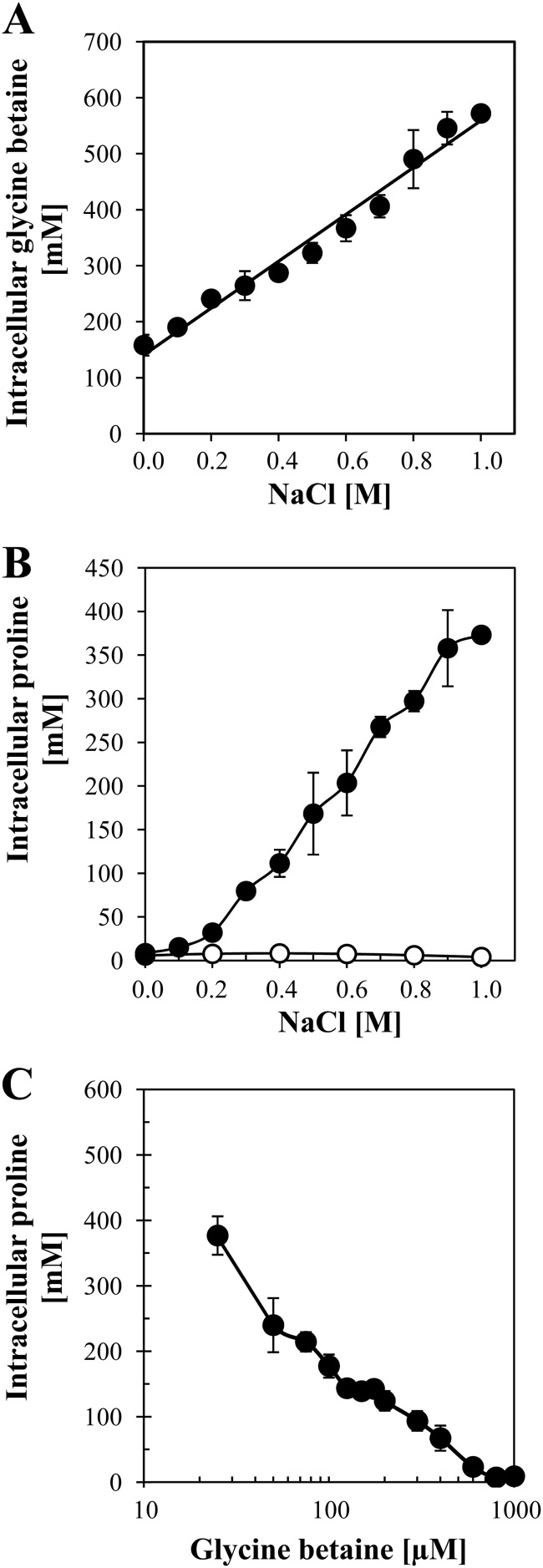
To monitor the buildup of an intracellular glycine betaine pool in osmotically stressed cells from an exogenous supply of glycine betaine, we grew the B. subtilis wild-type strain JH642 (OpuA+ OpuC+ OpuD+) (Fig. 1) in SMM containing various concentrations of NaCl in the presence of 1 mM glycine betaine. The glycine betaine solution used was spiked with 0.64 μM radiolabeled [1-14C]glycine betaine, which allowed us to determine the glycine betaine content of the cells by scintillation counting after recovering the cells by filtration. We found that the concentration of the glycine betaine pool increased from about 160 mM in cells grown in SMM (about 350 mosmol kg−1) to about 570 mM in cells grown in SMM with 1 M NaCl (about 2,190 mosmol kg−1) (Fig. 2A). There was a perfect linear relationship between the glycine betaine content of the cells and the strength of the osmotic stress imposed (Fig. 2A). Notably, even in cells grown in SMM, a growth medium widely used for physiological studies of B. subtilis (38) and considered not to impose a significant degree of osmotic stress (15), a considerable glycine betaine pool could be detected (Fig. 2A) when this medium was supplemented with 1 mM glycine betaine. The buildup of the glycine betaine pool in cells grown in SMM in the absence of additional NaCl is probably largely dependent on the transport activity of the OpuA system, since its transport capacity (Vmax, about 110 nmol min−1 mg protein−1) well exceeds those of the OpuC and OpuD glycine betaine importers, which exhibit Vmax values of about 41 nmol min−1 mg protein−1 and 16 nmol min−1 mg protein−1, respectively, under these growth conditions (27). All three glycine betaine transporters possess similar Km values in the low micromolar range (27).
Fig 2.
Adjustment of the intracellular compatible-solute pool of B. subtilis to external salinity and glycine betaine availability. (A) Cultures of the wild-type strain JH642 were grown in SMM to the mid-exponential-growth phase (OD578, about 1.5). The medium contained the NaCl concentrations indicated and 1 mM glycine betaine spiked with 0.64 μM radiolabeled [1-14C]glycine betaine. Intracellular glycine betaine (●) was quantified by scintillation counting. (B) For the analysis of cytoplasmic proline pools, cells of the B. subtilis wild-type strain JH642 were grown to the mid-exponential-growth phase (OD578, about 1.5) in SMM with the NaCl concentrations indicated in the absence (●) or presence (○) of 1 mM glycine betaine. After extraction of the soluble solute pools from these cells, their proline content was determined by a colorimetric assay (49). (C) Cells of strain JH642 were grown to the mid-exponential-growth phase (OD578, about 1.5) in SMM containing 1 M NaCl and the indicated concentrations of glycine betaine. Their proline content (●) was determined by a colorimetric assay (49). All data presented in this figure are the means for two independent cultures that were assayed for their internal solute concentrations in duplicate.
Brill et al. (9) have recently reported that the proline pool of osmotically stressed B. subtilis cells increases linearly in response to the degree of the osmotic stress imposed once the salinity of the growth medium, SMM, exceeds 0.2 M NaCl. We reconfirmed this pattern of proline accumulation and now provide quantitative measurements of the sizes of the cellular proline pools attained in osmotically stressed B. subtilis cells (Fig. 2B). The content of free proline rose from 8.5 mM in cells cultivated in SMM to about 373 mM in cells grown in SMM containing 1 M NaCl (Fig. 2B). Strikingly, when 1 mM glycine betaine was present in the growth medium, the buildup of an osmoadaptive proline pool was completely prevented. These cells contained a proline pool of just 8 mM, despite the fact that they were subjected to sustained osmotic stress by growth in SMM with 1 M NaCl (Fig. 2B). Consequently, it is clear that B. subtilis cells prefer to take up externally provided glycine betaine rather than engage in the de novo synthesis of the compatible solute proline to attain relief from osmotic stress.
In natural ecosystems, B. subtilis is unlikely to be exposed frequently to the glycine betaine concentration (1 mM) that we have used in our growth assays. Not only does the provision of this compatible solute through root exudates, decaying plant material, and osmotically downshocked microbial cells yield differing supplies of glycine betaine in natural settings, but also the concentrations typically should not exceed micromolar values (50–53). As a consequence of such low concentrations of glycine betaine in the environment, an osmoprotective pool made up of this compatible solute alone cannot be built up in fast-growing cells. We derive this conclusion from a calculation of the potential glycine betaine content of B. subtilis cells grown to an OD578 of 1.5 (corresponding to a cytoplasmic volume of 0.975 μl per 1 ml culture) that suggests that in the presence of 25 μM, 75 μM, or 600 μM glycine betaine, an intracellular pool size of 26 mM, 77 mM, or 615 mM, respectively, can be generated. It is assumed for this calculation that the externally provided glycine betaine is completely taken up by the cells. We realize that in natural settings (e.g., B. subtilis cells growing in the soil next to root hairs exuding glycine betaine or proline), the situation might be much more complex, since the cells do not grow fast and their population density is much lower than under laboratory conditions. Nevertheless, the estimate above presented suggests that the B. subtilis cell facing limiting concentrations of osmoprotectants in its environment builds up an intracellular compatible-solute pool consisting of a mixture of imported osmoprotectants and newly synthesized proline.
We therefore assessed the effects of various concentrations of glycine betaine on the size of the intracellular proline pool formed via de novo synthesis in osmotically stressed B. subtilis cells (grown in SMM with 1 M NaCl). For this experiment, we titrated the glycine betaine concentration present in the growth medium from 0 μM to 1,000 μM. Our results revealed a finely tuned influence of exogenously provided glycine betaine on the intracellular proline pool (Fig. 2C). Even very low (25 μM) glycine betaine concentrations notably reduced the proline content of the cells, and the presence of this compatible solute at 75 μM was sufficient to reduce the proline pool by about 50%. A supply of 600 μM glycine betaine reduced the proline content to a basal level of 21 mM (Fig. 2C). Within the framework of the calculations presented above, an external supply of 600 μM glycine betaine would theoretically be sufficient to attain a pool size of 615 mM. In agreement with these calculations, we actually measured a glycine betaine pool of about 570 mM in cells challenged with 1 M NaCl and grown in the presence of 1 mM glycine betaine (Fig. 2A).
Induction of opuA expression in response to an osmotic upshock and sustained high salinity.
The OpuA uptake system is the dominating transporter for glycine betaine in B. subtilis (27). We therefore focus below on the regulation of the opuA operon in response to osmotic stress and the availability of compatible solutes. To provide a genetic tool for these studies, we constructed an opuA-treA operon fusion and ectopically inserted this reporter construct stably as a single copy into the B. subtilis chromosome at the nonessential amyE locus, yielding the B. subtilis strain MBB9. The treA gene encodes a highly salt tolerant phospho-α-(1,1)-glucosidase (46), and the TreA enzyme is therefore a reporter well suited for transcriptional studies involving high-salinity-stressed B. subtilis cells (9, 54, 55).
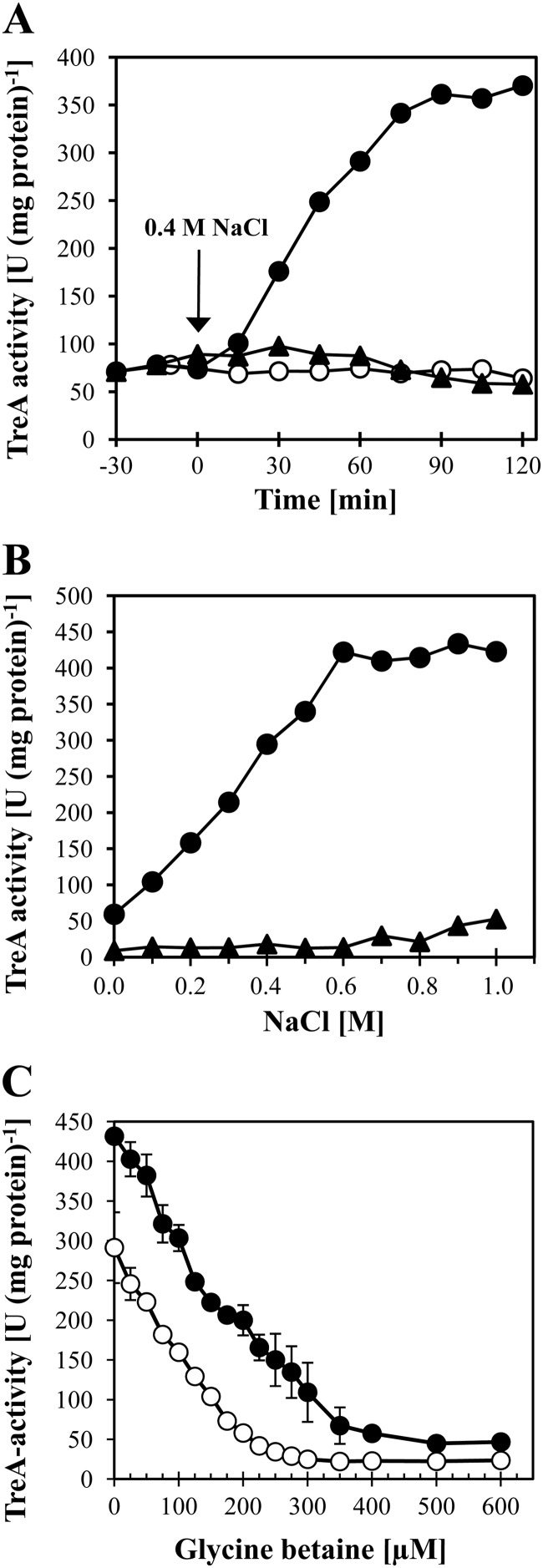
A previous study by Kempf and Bremer (26) established that enhanced expression of a plasmid-encoded opuA-lacZ translational fusion could be triggered in a high-salinity complex growth medium (LB medium with 0.5 M NaCl). To study the transcriptional response of opuA in response to increases in external salinity in greater detail and under better-controlled conditions, we grew the opuA-treA reporter strain MBB9 in a chemically defined medium (SMM) to early-log phase and then subjected it to a sudden osmotic upshift with 0.4 M NaCl. This increase in external salinity caused a rapid and strong transcriptional response of the opuA-treA fusion and increased the activity of the TreA reporter enzyme from a basal level of 75 U (mg protein)−1 to 361 U (mg protein)−1 within 90 min subsequent to the osmotic upshift (Fig. 3A). In addition to its ability to perceive sudden increases in external salinity, the B. subtilis cell was also able to react to incremental increases in external salinity when the opuA-treA reporter strain MBB9 was continuously grown in media containing different concentrations of NaCl. The strength of opuA-treA expression increased linearly as the external salinity increased up to 0.6 M NaCl and then leveled off at higher salinities (Fig. 3B). Osmotic induction of opuA expression was also evident when we assessed opuA transcription by Northern blot analysis (Fig. 4A). This experiment also proved that the opuA locus is transcribed as an operon, since the size of the opuA mRNA observed (about 3,000 nucleotides) (Fig. 4A) closely matches that calculated for the coding and intragenic regions (2,985 bp) of the opuAA-opuAB-opuAC gene cluster (26).
Fig 3.
Osmotic control of opuA expression and its response to glycine betaine availability. (A) A culture of the opuA-treA fusion strain MBB9 was grown in SMM to the early-exponential-growth phase (OD578, approximately 0.5) and was divided into three portions. Two portions were subjected to an osmotic upshift with 0.4 M NaCl either in the absence (●) or in the presence (▲) of 1 mM glycine betaine; the third portion was not subjected to an osmotic upshift and did not receive glycine betaine (○). Aliquots of the cultures were withdrawn at the indicated times and were assayed for TreA reporter enzyme activity. This experiment was conducted in three biological replicates, and the data shown represent a typical data set. (B) The opuA-treA fusion strain MBB9 was grown in SMM with various salinities in the absence (●) or presence (▲) of 1 mM glycine betaine. Cells that had reached the same optical density (OD578, 0.6 to 0.9) were assayed for TreA reporter enzyme activity. The data shown represent two independently grown cultures with two replicates of the TreA assays each. (C) The opuA-treA fusion strain MBB9 was grown in SMM with either 0.4 M NaCl (○) or 0.8 M NaCl (●) and the indicated concentrations of glycine betaine. Cells that had reached the same optical density (OD578, about 1.5) were assayed for TreA reporter enzyme activity. The data shown represent two independently grown cultures with two replicates of the TreA assays each.
Fig 4.
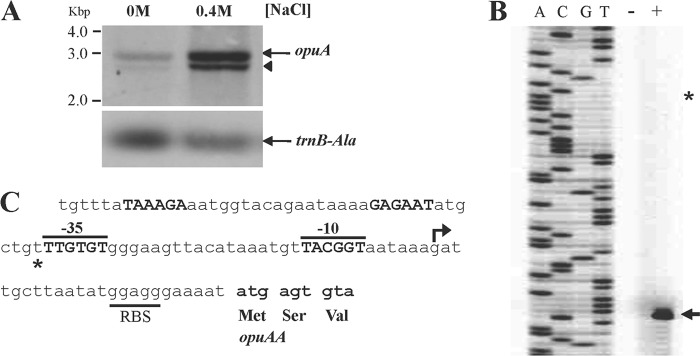
Osmoregulated transcription of the opuA operon occurs from a SigA-type promoter. (A) Northern blot analysis of the opuAA transcript (opuAA-opuAB-opuAC) (26) in cells grown either in SMM alone or in SMM containing 0.4 M NaCl. opuA mRNA was detected by hybridization using a digoxigenin-labeled opuAA antisense RNA probe. The full-length opuA mRNA is indicated by an arrow; the shorter opuA-specific hybridization signal (indicated by an arrowhead) is believed to represent either a degradation or a premature termination product of the opuAA-opuAB-opuAC transcript. As a control for the total RNA quantities blotted onto the membrane in this experiment, an antisense RNA probe against the transcript of a constitutively expressed tRNA gene (trnB-Ala) was used. (B) Primer extension analysis of the opuA transcript in cells of strain JH642 (pMBB39 opuAA′) grown either in SMM alone (−) or in SMM with 0.4 M NaCl (+) to the mid-exponential-growth phase (OD578, approximately 1). The arrow indicates the position of the 5′ end of the opuA mRNA; the position where the second opuA mRNA species had been identified previously (26) is marked by a star. (C) DNA sequence of the opuA promoter region. The mapped transcriptional start site for the osmoregulated opuA mRNA is marked by an arrow, and the corresponding −35 and −10 regions of the osmotically controlled opuA promoter are indicated (marked in boldface letters and overlined). The position of the promoter that was previously thought to direct a constitutively produced second opuA mRNA species (26) is shown in boldface letters, and the corresponding previously suggested transcriptional start site is indicated by a star. The ribosome binding site (RBS) and the beginning of the opuAA coding region are indicated.
Salt-induced opuA expression is not controlled by SigB, the DegS-DegU two-component regulatory system, or the ArsR-type regulator YceK.
Salt stress (56, 57) is one of the strongest inducers of the SigB-controlled general stress regulon of B. subtilis (58, 59); however, upregulation of the transcription of its members in response to salt stress is only transient (54, 57, 60). As expected from the data shown in Fig. 3B, which record high levels of opuA-treA expression under sustained salt stress, salt stress-mediated induction of opuA-treA expression was not prevented in a sigB mutant background (Table 2).
Table 2.
Salt induction of opuA transcription is independent of the alternative transcription factor SigB, the two-component DegSU system, and the ArsR-type YceK regulator
| Straina | TreA activity (U [mg protein]−1) |
|
|---|---|---|
| Without NaCl | With NaCl | |
| MBB9 (wild type) | 75 ± 4 | 431 ± 37 |
| MBB16 {Δ([degS-degU::aphA3]1)} | 81 ± 1 | 373 ± 3 |
| MBB19 [Δ(sigB2::cat)] | 66 ± 2 | 338 ± 2 |
| ΤΜΒ150 [Δ(yceK::spc)1] | 69 ± 5 | 460 ± 39 |
Cells of the various opuA-treA fusion strains were grown to the mid-exponential-growth phase (OD578, approximately 1) in SMM in the absence or presence of 0.4 M NaCl and were assayed for TreA reporter enzyme activity.
Several studies have implicated the two-component DegS-DegU regulatory system of B. subtilis in sensing salt stress (61, 62) and in developing osmostress resistance (63). There is also considerable overlap between high-salinity-induced B. subtilis genes and members of the DegS-DegU regulon (64). We therefore wondered whether the salt-stress-mediated expression of opuA was under the control of this two-component regulatory system and tested the influence of a degSU gene disruption on opuA-treA expression. None was observed (Table 2).
The yceK locus is transcribed divergently from the opuA operon and encodes a presumptive regulatory protein belonging to the ArsR family; it contains a helix-turn-helix DNA-binding motif (40, 41). Prompted by the vicinity of the yceK and opuA loci, we tested for a possible role of the predicted YceK transcriptional regulator in the osmotic control of opuA expression. None was observed (Table 2).
The level of opuA induction differs in response to ionic and nonionic solutes.
To determine whether the observed increase in opuA expression (Fig. 3A and B) depends exclusively on high concentrations of NaCl or other salts, or whether it reflects a true osmotic response, we grew the opuA-treA reporter fusion strain MBB9 in a minimal medium (SMM) in the absence or presence of isoosmotic concentrations of both ionic (NaCl, KCl) and nonionic (lactose, sucrose) osmolytes. As documented in Table 3, an increased level of opuA transcription results from an increase in external osmolarity and therefore does not reflect a salt-specific response. The signal(s) that is perceived by the B. subtilis cell and induces opuA expression requires the establishment of an osmotically effective gradient across the cytoplasmic membrane. This conclusion can be inferred from an experiment where glycerol was used to increase the osmolarity of the growth medium. Glycerol, a compound that can permeate membranes at high concentrations and thus fails to impose osmotic stress on microbial cells, did not trigger a significantly enhanced level of opuA transcription (Table 3).
Table 3.
Osmotic induction of opuA transcription
| Medium | Osmolarity (mosmol kg−1) | TreA activity (U [mg protein]−1)a |
Intracellular glycine betaine pool (mM)b | |
|---|---|---|---|---|
| Without glycine betaine | With glycine betaine (1 mM) | |||
| SMM | 356 | 55 ± 6 | 12 ± 1 | 126 ± 1 |
| SMM with 0.68 M glycerol | 1,100 | 62 ± 10 | 20 ± 2 | 162 ± 33 |
| SMM with 0.4 M NaCl | 1,188 | 251 ± 17 | 25 ± 2 | 304 ± 78 |
| SMM with 0.4 M KCl | 1,178 | 246 ± 33 | 25 ± 3 | 299 ± 5 |
| SMM with 0.61 M lactose | 1,143 | 987 ± 73 | 68 ± 11 | 776 ± 67 |
| SMM with 0.62 M sucrose | 1,118 | 947 ± 87 | 63 ± 13 | 785 ± 89 |
Cells of the opuA-treA fusion strain MBB9 were grown to the mid-exponential-growth phase (OD578, approximately 1) in SMM alone or SMM with increased osmolarities that were generated by the addition of the indicated ionic or nonionic osmolyte.
Cultures of strain RMKB30 (OpuA+ OpuC− OpuD−) were grown to the mid-exponential-growth phase (OD578, approximately 1) in SMM alone or SMM with increased osmolalities in the presence of 1 mM glycine betaine spiked with 0.64 μM radiolabeled [1-14C]glycine betaine.
However, inspection of the data summarized in Table 3 revealed that the degree of opuA induction was much more pronounced when sugars were used to increase the osmolarity of the growth medium. The addition of lactose or sucrose increased the level of opuA expression about 17-fold, whereas NaCl and KCl enhanced opuA expression only about 5-fold. Dissimilar effects of isoosmotic ionic and nonionic osmolytes on potassium pools and osmotically regulated gene expression in E. coli (65, 66) and Lactobacillus plantarum have been reported before (67). In addition, studies on the osmotic induction of the proHJ promoter of B. subtilis revealed a considerably stronger effect of nonionic osmolytes than of ionic osmolytes on the transcription of this operon as well (9). Nevertheless, the differences in the degree of opuA-treA induction in response to isoosmotic concentrations of salts and sugars are unusual; we therefore sought independent confirmation for our observation. To this end, we measured the glycine betaine pools in strain RMKB20, a mutant with defective OpuC and OpuD glycine betaine transporters (Fig. 1) but with an intact OpuA system (27), in response to different types of osmolytes.
Since OpuA is the only functional glycine betaine transporter operating in strain RMKB20, the glycine betaine pools attained reflect only the steady-state transport activity of the OpuA system. In agreement with the transcriptional data on opuA-treA expression reported above (Table 3), the sugars lactose and sucrose triggered the buildup of glycine betaine pools substantially larger than those elicited by the addition of the salts NaCl and KCl to the growth medium (Table 3). Consequently, there can be no doubt that isosmotic concentrations of ionic and nonionic compounds exert different effects on OpuA-mediated glycine betaine import by eliciting transcriptional responses of different magnitudes from the opuA operon (Table 3). The distinct effects of ionic and nonionic osmolytes on the OpuA-mediated buildup of glycine betaine pools are smaller than those found for the corresponding transcriptional activity of the opuA-treA reporter fusion (Table 3). Our data do not address the possibility that sugars and salts might also have disparate effects on the transport activity of the OpuA system in cells challenged by high osmolarity on a long-term basis.
The compatible solute glycine betaine exerts a major influence on opuA expression.
Previous studies with different microorganisms have shown that in addition to functioning as an osmoprotectant, glycine betaine can also influence gene expression. Genes that are involved in the synthesis or uptake of various types of compatible solutes are particularly strongly affected, and their transcriptional activity is downregulated under osmotic stress conditions (for examples, see references 9, 35, 54, and 68 to 72). To test whether glycine betaine would also influence opuA expression, we included 1 mM glycine betaine in the growth medium of the opuA-treA reporter strain MBB9, in which all three glycine betaine transport systems (OpuA, OpuC, and OpuD) operating in B. subtilis are intact (Fig. 1). The presence of this concentration of glycine betaine strongly downregulated the osmotic induction of the reporter opuA-treA fusion in response to either ionic or nonionic osmolytes (Table 3). Likewise, the presence of 1 mM glycine betaine fully prevented the induction of opuA-treA expression subsequent to a sudden osmotic upshift with 0.4 M NaCl (Fig. 3A) and under sustained high-salinity growth conditions (Fig. 3B). Hence, the presence of glycine betaine dramatically influences the pattern of opuA expression under osmotic stress conditions.
To further evaluate the influence of glycine betaine on opuA expression, we titrated its concentration in media containing either 0.4 M or 0.8 M NaCl. Even the addition of very small concentrations of glycine betaine (e.g., 25 μM) to the growth medium had measurable effects on the transcriptional activity of the opuA-treA reporter fusion (Fig. 3C). Incremental increases in the glycine betaine content of the growth medium successively decreased opuA-treA expression down to a basal level (Fig. 3C). The amount of glycine betaine required to fully shut down opuA transcription was dependent on the salinity of the growth medium. Approximately 300 μM glycine betaine was required to prevent osmotic induction of opuA-treA expression in cells grown in the presence of 0.4 M NaCl, whereas about 500 μM glycine betaine was required to accomplish this in cells grown in the presence of 0.8 M NaCl (Fig. 3C).
Role of the uptake and intracellular accumulation of glycine betaine in controlling opuA expression.
Since glycine betaine apparently exerts a strong influence on opuA expression, we investigated this regulatory phenomenon in greater detail. We asked the following four questions. (i) Does the regulatory influence of glycine betaine on opuA expression require its uptake? (ii) Is uptake of glycine betaine via the OpuA system a prerequisite for the reduction of opuA expression, or can this effect also be exerted when glycine betaine is imported by a different Opu transport system? (iii) Can glycine betaine synthesis from the precursor choline exert regulatory effects on opuA gene expression as well? (iv) Is the repressing effect on opuA gene expression specifically caused by glycine betaine, or can it also be conferred by other compatible solutes that function as effective osmoprotectants for B. subtilis?
To test whether the uptake of glycine betaine by osmotically stressed cells is required for the reduction in opuA expression, we used strain MBB14, which is defective in the OpuA, OpuC, and OpuD glycine betaine transporters (Fig. 1). In this strain, glycine betaine is no longer able to repress opuA-treA expression (Table 4), demonstrating that glycine betaine needs to be taken up by the cell in order to exert its repressive effect on opuA transcription.
Table 4.
Repression of opuA transcription by compatible solutes
| Compatible solute in growth mediuma | TreA activity (U [mg protein]−1)b |
|||||
|---|---|---|---|---|---|---|
| Without NaCl |
With 0.4 M NaCl |
|||||
| MBB9 (wild type) | MBB14 (opuB+) | MBB21 (opuC+) | MBB9 (wild type) | MBB14 (opuB+) | MBB21 (opuC+) | |
| None | 49 ± 9 | 48 ± 10 | 49 ± 6 | 218 ± 25 | 205 ± 9 | 204 ± 24 |
| Glycine betaine | 12 ± 5 | 50 ± 12 | 16 ± 2 | 19 ± 3 | 210 ± 6 | 59 ± 4 |
| Choline | 23 ± 12 | 18 ± 3 | 27 ± 1 | 17 ± 5 | 38 ± 7 | 57 ± 8 |
| Carnitine | 15 ± 8 | 39 ± 4 | 11 ± 3 | 32 ± 4 | 203 ± 17 | 28 ± 2 |
Cells of the various opuA-treA fusion strains were grown to the mid-exponential-growth phase (OD578, approximately 1) in SMM alone or in SMM containing 0.4 M NaCl. The presence in the growth medium of the compatible solutes glycine betaine and carnitine and the precursor for glycine betaine synthesis (choline) is indicated. The final concentration of the various osmoprotectants was 1 mM.
The presence of osmoprotectant uptake systems operating in the wild-type strain MBB9 (OpuA+ OpuB+ OpuC+ OpuD+) and its mutant derivatives strain MBB14 (OpuA− OpuB+ OpuC− OpuD−) and strain MBB21 (OpuA− OpuB− OpuC+ OpuD−) is indicated.
Strain MBB21 is defective in the OpuA and OpuD glycine betaine transport systems but allows glycine betaine uptake via the OpuC transporter (Fig. 1). The addition of glycine betaine to high-salinity-grown cells of MBB21 reduced opuA-treA expression to a degree similar to that observed for strain MBB9, which can acquire glycine betaine through the OpuA, OpuC, and OpuD transporters (Table 4). Consequently, the repressing effect of glycine betaine on opuA expression is independent of its uptake via the OpuA transporter.
Choline, the biosynthetic precursor for glycine betaine synthesis by B. subtilis (Fig. 1) (24, 25), does not function as a compatible solute by itself; its osmoprotective effects rely entirely on its enzymatic conversion to glycine betaine (24). The addition of choline to the growth medium repressed opuA-treA expression to an extent similar to that observed for the addition of preformed glycine betaine (Table 4). Hence, glycine betaine does not need to be imported in order to influence opuA transcription; it can also exert its effect on opuA transcription when it is newly produced.
Repression of opuA-treA expression was also caused by the addition to the growth medium of carnitine (Table 4), a highly effective osmoprotectant for B. subtilis that is taken up exclusively via the OpuC transport system (Fig. 1) (73). It should be noted in this context that carnitine is not metabolized by B. subtilis to glycine betaine (73). As observed for glycine betaine, carnitine needs to enter the B. subtilis cell in order to repress opuA-treA expression, as evidenced by the fact that in the opuC mutant strain MBB14, carnitine was no longer able to reduce opuA transcription.
Taken together, these data show that the repressing effect of glycine betaine on opuA-treA expression was independent of the way (transport or synthesis) it was amassed by the osmotically stressed B. subtilis cell, that this regulatory effect was independent of the functioning of the OpuA uptake system, and that a reduction in opuA expression can also be accomplished by other compatible solutes used by B. subtilis as osmoprotectants.
The strength of opuA expression is modulated by the proline pool of osmotically stressed cells.
Since the intracellular accumulation of the compatible solutes glycine betaine and carnitine downregulated opuA expression at high salinity (Table 4), we wondered if the production of proline, the only compatible solute that B. subtilis can synthesize de novo under osmotic stress conditions (9, 16), would affect the level of opuA transcription as well. To test this notion, we introduced the opuA-treA fusion into a B. subtilis strain that is defective in the ProJ-ProA-ProH-mediated osmoadaptive proline biosynthesis route (Fig. 1) due to the presence of the Δ(proHJ::tet)1 mutation (9). We then compared the expression level of the reporter gene fusion present in the Δ(proHJ::tet)1 genetic background with that present in an isogenic strain that is proficient in osmoadapative proline production. The expression of the opuA-treA fusion was osmotically inducible in both strains, but the level of opuA-treA expression was considerably higher in the strain carrying the Δ(proHJ::tet)1 mutation (Fig. 5). Thus, the activity of the opuA promoter is sensitive to the compatible-solute pool present in osmotically stressed B. subtilis cells. Osmotic induction of opuA-treA expression was fully repressible by the addition of 1 mM glycine betaine to the growth medium, regardless of whether the reporter strain was able to synthesize osmoadaptive levels of proline or not (Fig. 5).
Fig 5.
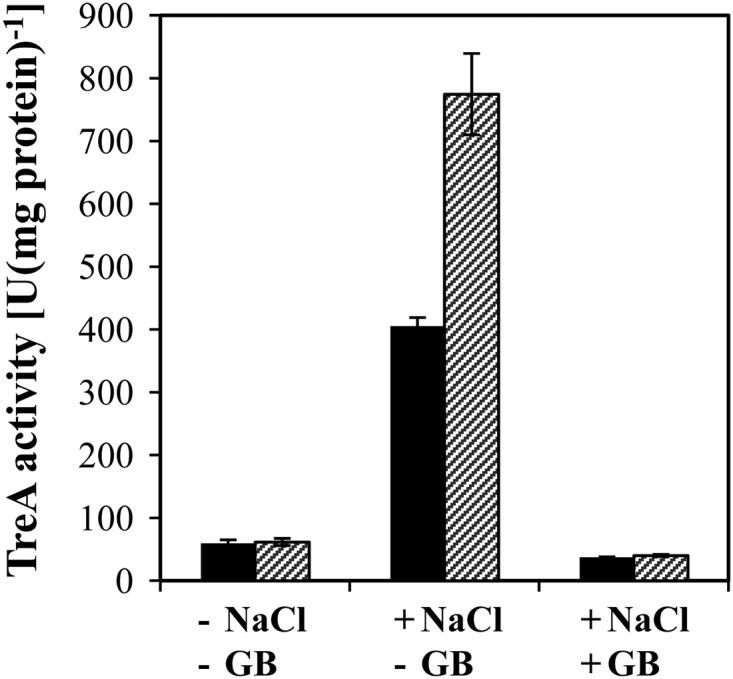
Osmoregulated expression of opuA-treA expression is responsive to the cellular proline pool. Cells of the opuA-treA reporter strains MBB9, carrying a genetically intact proHJ operon (filled bars), and TMB117, carrying a disruption [Δ(proHJ::tet)1] of the proHJ locus (hatched bars), were grown in SMM alone or in SMM with 0.6 mM NaCl in the absence (−) or presence (+) (1 mM) of glycine betaine (GB). The TreA activities given are means from two independently grown cultures whose TreA activities were determined in duplicate.
opuA transcription depends on a single, osmotically inducible promoter.
The transcriptional landscape of the opuA regulatory region has been analyzed previously by Kempf and Bremer (26) via primer extension analysis, and two opuA mRNA species with 5′ ends spaced 38 bp apart were found (Fig. 4C). The longer transcript was constitutively produced at high levels in cells cultivated in LB medium alone and in LB medium containing 0.5 M NaCl, whereas the amount of the shorter transcript was strongly inducible when increased amounts of NaCl were present in the growth medium. We reevaluated the transcriptional start site(s) of the opuA operon by primer extension analysis with total RNA isolated from cells of strain JH642(pMBB39) grown in a chemically defined medium (either in SMM alone or in SMM with 0.4 M NaCl) to early-log phase. In contrast to the previously reported primer extension analysis data (26), our new primer extension experiment shows that there is only a single opuA promoter and that the activity of this promoter is responsive to increases in the salinity of the growth medium (Fig. 4B). This conclusion is fully consistent with data from the recently reported genomewide transcriptional profiling study of Nicolas et al. (74) on the condition-dependent transcriptome of B. subtilis, where only a single opuA mRNA start site was detected under a large number of growth conditions (74).
We assessed the functionality and validity of the proposed osmostress-responsive opuA promoter (26) by site-directed mutagenesis. The first T · A base pair in the −10 regions of SigA-type promoters (Fig. 6) is highly conserved and is typically critical for promoter activity (75). We changed this A · T base pair in the opuA promoter into a C · G base pair via site-directed mutagenesis and found that this point mutation abolished osmotic induction of opuA expression and allowed only a very basal level of promoter activity (Fig. 6). The properties of this mutant opuA promoter are therefore fully consistent with our finding in the new primer extension experiment that there is only a single promoter driving opuA transcription (Fig. 4B and C). We do not have any satisfying explanation for the differences between the previous primer extension data (26) and our current primer extension data (Fig. 4B).
Fig 6.
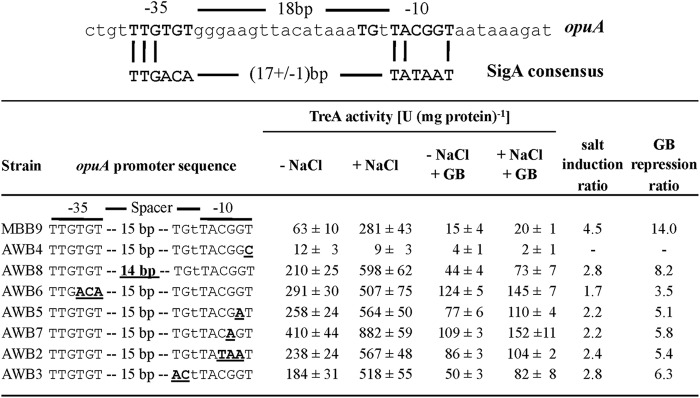
Mutational analysis of the osmotically controlled opuA promoter. (Top) Comparison of the DNA sequence of the opuA promoter to the the consensus sequence of the −10 and −35 regions of SigA-type promoters (75, 83). (Bottom) Transcriptional data of opuA-treA fusion strains with the indicated promoter mutations. Cells were grown either in SMM alone (− NaCl) or in SMM with 0.4 M NaCl (+ NaCl) and in the presence (1 mM) or absence of glycine betaine (GB). The salt induction ratio compares the values of TreA activities in cells grown in the presence versus the absence of NaCl. The GB repression ratio compares TreA activities in cells grown at high salinity in the presence versus the absence of GB. The TreA activities given are means from two independently grown cultures whose TreA activities were determined in duplicate.
Determinants of the osmotically controlled opuA promoter.
The opuA promoter deviates in both its −35 and −10 sequences from the consensus sequence of SigA-type promoters, and it also possesses a spacer length of 18 bp (Fig. 4C and 6), which is suboptimal spacing for this group of promoters (75). Since only a few osmotically controlled SigA-type promoters from B. subtilis have been analyzed in some detail (9, 54, 55), and since osmoregulation of gene expression might proceed without a classical regulatory protein and might instead depend on solute-mediated interactions between the RNA polymerase and a given promoter (76–79), we studied the contributions of key elements of the opuA promoter to osmotic control and functionality via site-directed mutagenesis.
First, we shortened the length of the spacer segment separating the −10 and −35 regions from 18 to 17 nucleotides, a configuration most frequently found in SigA-type promoters (75). In this promoter mutant, both the basal and osmotically induced levels of opuA expression increased considerably, but with a concomitant reduction in the degree of osmotic induction of transcription (Fig. 6). The −10 regions of SigA-type promoters from B. subtilis are typically very AT rich (75), but the corresponding region of the opuA promoter possesses an unusual enrichment of G · C base pairs (Fig. 6). The B. subtilis opuA promoter shares this feature with other osmotically controlled promoters from B. subtilis, E. coli, and Salmonella enterica serovar Typhimurium (54, 55, 78, 80–82). Mutations that improved the match of the opuA −10 region, or of the −35 region, with the consensus sequence of SigA-type promoters substantially increased both the basal and osmotically induced levels of opuA expression. Each of these opuA promoter variants still permitted osmotic control of gene expression to some extent (Fig. 6).
A TG motif at position −16 is frequently found in B. subtilis SigA-type promoters (75), and mutations affecting this motif typically strongly decrease promoter activity (83). In contrast to expectations, the change of the TG motif present in the wild-type opuA promoter (Fig. 6) to an AC variant did not inactivate the promoter. Instead, this promoter mutant displayed increased promoter activity both at low salinity and at high salinity, with some remaining degree of osmotic induction (Fig. 6).
Since the accumulation of glycine betaine strongly affects the strength of opuA expression (Fig. 2C and 3C), we also tested the susceptibility of all newly constructed opuA promoter variants to this repressing effect. Each opuA promoter variant retained control by glycine betaine to some extent (Fig. 6).
DISCUSSION
The nearly linear relationship between the salinity of the environment and the pools of proline (formed by de novo synthesis) and glycine betaine (formed by import) implies that the B. subtilis cell can perceive incremental increases in external salinity. This allows it to modulate the intracellular levels of these two compatible solutes over a broad range of external salinities and to keep them within rather confined boundaries at a given salinity. B. subtilis achieves this through coordinated adjustments in the transcriptional activity of the proHJ proline biosynthetic genes (9) and of the loci (opuA, opuC, and opuD) encoding glycine betaine uptake systems (23, 26, 27), and probably also through posttranscriptional control mechanisms (32, 34) regulating the activity of these transporters. In natural ecosystems, B. subtilis is exposed to different types and mixtures of compatible solutes (50). The B. subtilis cell can take advantage of a wide spectrum of osmoprotectants via their import through the Opu transport systems (12, 20). The data reported here for glycine betaine show that the B. subtilis cell is able to physiologically integrate compatible-solute import with its osmostress-adaptive proline biosynthetic activities (9, 16).
The molecular mechanism(s) that allows B. subtilis to detect osmotic changes in its environment and then set the level of gene expression on a genomewide scale (56, 64) is rather poorly understood. Also unclear are the genetic processes that control opuA transcription in response to increases in external osmolarity elicited by either ionic or nonionic compounds. Our data reveal a major impact on opuA expression of the intracellular solute pools formed through the synthesis of proline and glycine betaine (formed by the oxidation of choline) (Fig. 1) and through the uptake of preformed osmoprotectants. By linking opuA transcription to the intracellular pool sizes of glycine betaine and proline, B. subtilis prevents uncontrolled and wasteful overaccumulation of glycine betaine, and probably also of several other compatible solutes (31), via the OpuA transport system.
A process of release and recapture of compatible solutes by continuously osmotically stressed B. subtilis cells, which we have recently discovered for newly produced proline (84), also exists for glycine betaine synthesized from the precursor choline (S. Moses and E. Bremer, unpublished data). This activity will aid the cell in finely tuning the glycine betaine pools at a given salinity. Nothing is currently known about the nature of the observed release of glycine betaine from osmotically stressed B. subtilis cells and its influence on the steady-state pool of this compatible solute, but the existence of specific betaine efflux systems has been suggested from experiments conducted with S. enterica serovar Thyphimurium (85) and Lactobacillus plantarum (86). Release of glycine betaine by a MscS-type channel has been observed in Corynebacterium glutamicum (87), and such channels also exist in B. subtilis (88, 89). However, any role of MscL- or MscS-type channels in the release of proline from continuously osmotically challenged B. subtilis cells has been ruled out (84).
The size of the intracellular proline pool of B. subtilis is dependent not only on external salinity but also on the presence of the osmoprotectant glycine betaine in the growth medium. Optimal use of energetic resources available to salt-stressed cells might underlie this phenomenon (90), but the preference of glycine betaine accumulation via import over de novo synthesis of proline might also be rooted in the cells' aim to adjust and optimize cytoplasmic solvent properties by accumulating different kinds of compatible solutes (91). Hints that such processes might operate in microbial cells come from studies where salinity-dependent switching of the type of compatible solute synthesized is observed (11, 92, 93).
The amassing of compatible solutes impinges on the steady-state intracellular levels of potassium in osmotically stressed B. subtilis cells (13, 16). Given that potassium, and its counterion glutamate, is involved in the osmotic stress response of many bacterial species (1–4, 8, 76, 91), we consider it likely that these solute species also participate in setting the level of opuA transcription. Whether this is accomplished through yet unrecognized regulatory proteins (e.g., proteins such as the ion-responsive BusR repressor of L. lactis [35, 36]) or through solute-modulated interactions of the RNA polymerase (76, 77, 79) at the B. subtilis opuA promoter region remains to be determined. Mutational analysis of the opuA promoter revealed that its deviations from the consensus sequence of SigA-type B. subtilis promoters (75, 83) tune and optimize it both with respect to the osmotic inducibility of its transcriptional activity and with respect to its repressibility in response to intracellular compatible-solute pools.
The reduction in the level of osmotically induced transcription of genes involved in the synthesis or uptake of compatible solutes in response to the presence of these compounds in the growth medium is a well-known phenomenon (for examples, see references 9, 35, 54, and 68 to 72). In contrast to most of the previously reported studies, we not only evaluated the effects of substantial concentrations of glycine betaine (typically 1 mM) on gene expression but also used glycine betaine at concentrations (micromolar) that in all likelihood are more representative of the amounts of compatible solutes found in natural ecosystems (50–53). This experiment revealed a very finely tuned response of the level of opuA transcription to intracellular glycine betaine pools, which, to the best of our knowledge, has not been recorded before in any study addressing the influence of compatible solutes on gene expression.
We find it remarkable that the expression of opuA can be completely shut down by intracellular glycine betaine pools despite the fact that the cells remained exposed to high-salinity surroundings. This finding implies that the B. subtilis cell does not sense environmental osmolarity per se when it sets the level of opuA expression but reacts to the degree of osmotic stress it perceives. The accumulation of glycine betaine alleviates osmotic stress (15), and this either blinds the B. subtilis cell to the environmentally imposed osmotic signal or renders the cell unable to process this information into an output response leading to enhanced opuA expression. Mutants that will allow opuA expression in the presence of glycine betaine might thus pry open the door to a clearer picture of the inner workings of the osmotic sensory and response system(s) of B. subtilis.
ACKNOWLEDGMENTS
We are grateful to Jutta Gade for expert technical assistance and appreciate the kind help of Vickie A. Koogle in the language editing of our manuscript. We are grateful to the late Frank Kunst for providing us with a B. subtilis strain carrying a defect in the DegS-DegU system. We thank former members of our laboratory Gabriele Nau-Wagner and Reinhard Kappes for constructing various bacterial strains used in this study and Evert P. Bakker for advice and help in determining the volume of B. subtilis cells.
Financial support for this work was generously provided by grants from the BMBF via the Bacell-SysMo2 consortium (to U.V. and E. B.), the Fonds der Chemischen Industrie (to E.B.), and the Universities of Marburg and Greifswald.
Footnotes
Published ahead of print 23 November 2012
REFERENCES
- 1. Kempf B, Bremer E. 1998. Uptake and synthesis of compatible solutes as microbial stress responses to high osmolality environments. Arch. Microbiol. 170: 319–330 [DOI] [PubMed] [Google Scholar]
- 2. Csonka LN, Hanson AD. 1991. Prokaryotic osmoregulation: genetics and physiology. Annu. Rev. Microbiol. 45: 569–606 [DOI] [PubMed] [Google Scholar]
- 3. Bremer E, Krämer R. 2000. Coping with osmotic challenges: osmoregulation through accumulation and release of compatible solutes, p 79–97 In Storz G, Hengge-Aronis R. (ed), Bacterial stress responses. ASM Press, Washington, DC [Google Scholar]
- 4. Wood JM, Bremer E, Csonka LN, Krämer R, Poolman B, van der Heide T, Smith LT. 2001. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 130: 437–460 [DOI] [PubMed] [Google Scholar]
- 5. Ziegler C, Bremer E, Krämer R. 2010. The BCCT family of carriers: from physiology to crystal structure. Mol. Microbiol. 78: 13–34 [DOI] [PubMed] [Google Scholar]
- 6. Poolman B, Spitzer JJ, Wood JM. 2004. Bacterial osmosensing: roles of membrane structure and electrostatics in lipid-protein and protein-protein interactions. Biochim. Biophys. Acta 1666: 88–104 [DOI] [PubMed] [Google Scholar]
- 7. Poolman B, Blount P, Folgering JH, Friesen RH, Moe PC, van der Heide T. 2002. How do membrane proteins sense water stress? Mol. Microbiol. 44: 889–902 [DOI] [PubMed] [Google Scholar]
- 8. Krämer R. 2010. Bacterial stimulus perception and signal transduction: response to osmotic stress. Chem. Rec. 10: 217–229 [DOI] [PubMed] [Google Scholar]
- 9. Brill J, Hoffmann T, Bleisteiner M, Bremer E. 2011. Osmotically controlled synthesis of the compatible solute proline is critical for cellular defense of Bacillus subtilis against high osmolarity. J. Bacteriol. 193: 5335–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kuhlmann AU, Bremer E. 2002. Osmotically regulated synthesis of the compatible solute ectoine in Bacillus pasteurii and related Bacillus spp. Appl. Environ. Microbiol. 68: 772–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Saum SH, Müller V. 2008. Growth phase-dependent switch in osmolyte strategy in a moderate halophile: ectoine is a minor osmolyte but major stationary phase solute in Halobacillus halophilus. Environ. Microbiol. 10: 716–726 [DOI] [PubMed] [Google Scholar]
- 12. Bremer E. 2002. Adaptation to changing osmolarity, p 385–391 In Sonenshein AL, Hoch JA, Losick R. (ed), Bacillus subtilis and its closest relatives. ASM Press, Washington, DC [Google Scholar]
- 13. Whatmore AM, Reed RH. 1990. Determination of turgor pressure in Bacillus subtilis: a possible role for K+ in turgor regulation. J. Gen. Microbiol. 136: 2521–2526 [DOI] [PubMed] [Google Scholar]
- 14. Holtmann G, Bakker EP, Uozumi N, Bremer E. 2003. KtrAB and KtrCD: two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. J. Bacteriol. 185: 1289–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boch J, Kempf B, Bremer E. 1994. Osmoregulation in Bacillus subtilis: synthesis of the osmoprotectant glycine betaine from exogenously provided choline. J. Bacteriol. 176: 5364–5371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Whatmore AM, Chudek JA, Reed RH. 1990. The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. J. Gen. Microbiol. 136: 2527–2535 [DOI] [PubMed] [Google Scholar]
- 17. von Blohn C, Kempf B, Kappes RM, Bremer E. 1997. Osmostress response in Bacillus subtilis: characterization of a proline uptake system (OpuE) regulated by high osmolarity and the alternative transcription factor sigma B. Mol. Microbiol. 25: 175–187 [DOI] [PubMed] [Google Scholar]
- 18. Fujisawa M, Ito M, Krulwich TA. 2007. Three two-component transporters with channel-like properties have monovalent cation/proton antiport activity. Proc. Natl. Acad. Sci. U. S. A. 104: 13289–13294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brill J, Hoffmann T, Putzer H, Bremer E. 2011. T-box-mediated control of the anabolic proline biosynthetic genes of Bacillus subtilis. Microbiology 157: 977–987 [DOI] [PubMed] [Google Scholar]
- 20. Hoffmann T, Bremer E. 2011. Protection of Bacillus subtilis against cold stress via compatible-solute acquisition. J. Bacteriol. 193: 1552–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Holtmann G, Bremer E. 2004. Thermoprotection of Bacillus subtilis by exogenously provided glycine betaine and structurally related compatible solutes: involvement of Opu transporters. J. Bacteriol. 186: 1683–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moses S, Sinner T, Zaprasis A, Stöveken N, Hoffmann T, Belitsky BR, Sonenshein AL, Bremer E. 2012. Proline utilization by Bacillus subtilis: uptake and catabolism. J. Bacteriol. 194: 745–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kappes RM, Kempf B, Kneip S, Boch J, Gade J, Meier-Wagner J, Bremer E. 1999. Two evolutionarily closely related ABC transporters mediate the uptake of choline for synthesis of the osmoprotectant glycine betaine in Bacillus subtilis. Mol. Microbiol. 32: 203–216 [DOI] [PubMed] [Google Scholar]
- 24. Boch J, Kempf B, Schmid R, Bremer E. 1996. Synthesis of the osmoprotectant glycine betaine in Bacillus subtilis: characterization of the gbsAB genes. J. Bacteriol. 178: 5121–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nau-Wagner G, Opper D, Rolbetzki A, Boch J, Kempf B, Hoffmann T, Bremer E. 2012. Genetic control of osmoadaptive glycine betaine synthesis in Bacillus subtilis through the choline-sensing and glycine betaine-responsive GbsR repressor. J. Bacteriol. 194: 2703–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kempf B, Bremer E. 1995. OpuA, an osmotically regulated binding protein-dependent transport system for the osmoprotectant glycine betaine in Bacillus subtilis. J. Biol. Chem. 270: 16701–16713 [DOI] [PubMed] [Google Scholar]
- 27. Kappes RM, Kempf B, Bremer E. 1996. Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J. Bacteriol. 178: 5071–5079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Horn C, Sohn-Bösser L, Breed J, Welte W, Schmitt L, Bremer E. 2006. Molecular determinants for substrate specificity of the ligand-binding protein OpuAC from Bacillus subtilis for the compatible solutes glycine betaine and proline betaine. J. Mol. Biol. 357: 592–606 [DOI] [PubMed] [Google Scholar]
- 29. Kempf B, Gade J, Bremer E. 1997. Lipoprotein from the osmoregulated ABC transport system OpuA of Bacillus subtilis: purification of the glycine betaine binding protein and characterization of a functional lipidless mutant. J. Bacteriol. 179: 6213–6220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Horn C, Bremer E, Schmitt L. 2003. Nucleotide dependent monomer/dimer equilibrium of OpuAA, the nucleotide-binding protein of the osmotically regulated ABC transporter OpuA from Bacillus subtilis. J. Mol. Biol. 334: 403–419 [DOI] [PubMed] [Google Scholar]
- 31. Smits SH, Höing M, Lecher J, Jebbar M, Schmitt L, Bremer E. 2008. The compatible-solute-binding protein OpuAC from Bacillus subtilis: ligand binding, site-directed mutagenesis, and crystallographic studies. J. Bacteriol. 190: 5663–5671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen C, Beattie GA. 2007. Characterization of the osmoprotectant transporter OpuC from Pseudomonas syringae and demonstration that cystathionine-beta-synthase domains are required for its osmoregulatory function. J. Bacteriol. 189: 6901–6912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Biemans-Oldehinkel E, Poolman B. 2003. On the role of the two extracytoplasmic substrate-binding domains in the ABC transporter OpuA. EMBO J. 22: 5983–5993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Karasawa A, Erkens GB, Berntsson RP, Otten R, Schuurman-Wolters GK, Mulder FA, Poolman B. 2011. Cystathionine beta-synthase (CBS) domains 1 and 2 fulfill different roles in ionic strength sensing of the ATP-binding cassette (ABC) transporter OpuA. J. Biol. Chem. 286: 37280–37291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Romeo Y, Obis D, Bouvier J, Guillot A, Fourcans A, Bouvier I, Gutierrez C, Mistou MY. 2003. Osmoregulation in Lactococcus lactis: BusR, a transcriptional repressor of the glycine betaine uptake system BusA. Mol. Microbiol. 47: 1135–1147 [DOI] [PubMed] [Google Scholar]
- 36. Romeo Y, Bouvier J, Gutierrez C. 2007. Osmotic regulation of transcription in Lactococcus lactis: ionic strength-dependent binding of the BusR repressor to the busA promoter. FEBS Lett. 581: 3387–3390 [DOI] [PubMed] [Google Scholar]
- 37. Srivatsan A, Han Y, Peng J, Tehranchi AK, Gibbs R, Wang JD, Chen R. 2008. High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 4: e1000139 doi:10.1371/journal.pgen.1000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Harwood CR, Archibald AR. 1990. Growth, maintenance and general techniques, p 1–26 In Harwood CR, Cutting SM. (ed), Molecular biological methods for Bacillus. John Wiley & Sons Inc., Chichester, United Kingdom [Google Scholar]
- 39. Cutting SM, Vander Horn PB. 1990. Genetic analysis, p 27–74 In Harwood CR, Cutting SM. (ed), Molecular biological methods for Bacillus. John Wiley & Sons, Inc., Chichester, United Kingdom [Google Scholar]
- 40. Mäder U, Schmeisky AG, Florez LA, Stülke J. 2012. SubtiWiki—a comprehensive community resource for the model organism Bacillus subtilis. Nucleic Acids Res. 40: D1278–D1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barbe V, Cruveiller S, Kunst F, Lenoble P, Meurice G, Sekowska A, Vallenet D, Wang T, Moszer I, Medigue C, Danchin A. 2009. From a consortium sequence to a unified sequence: the Bacillus subtilis 168 reference genome a decade later. Microbiology 155: 1758–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kuwayama H, Obara S, Morio T, Katoh M, Urushihara H, Tanaka Y. 2002. PCR-mediated generation of a gene disruption construct without the use of DNA ligase and plasmid vectors. Nucleic Acids Res. 30: E2 doi:10.1093/nar/30.2.e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Guerout-Fleury AM, Shazand K, Frandsen N, Stragier P. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335–336 [DOI] [PubMed] [Google Scholar]
- 44. Brückner R. 1992. A series of shuttle vectors for Bacillus subtilis and Escherichia coli. Gene 122: 187–192 [DOI] [PubMed] [Google Scholar]
- 45. Majumdar D, Avissar YJ, Wyche JH. 1991. Simultaneous and rapid isolation of bacterial and eukaryotic DNA and RNA: a new approach for isolating DNA. Biotechniques 11: 94–101 [PubMed] [Google Scholar]
- 46. Gotsche S, Dahl MK. 1995. Purification and characterization of the phospho-α-(1,1)-glucosidase (TreA) of Bacillus subtilis 168. J. Bacteriol. 177: 2721–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schöck F, Dahl MK. 1996. Expression of the tre operon of Bacillus subtilis 168 is regulated by the repressor TreR. J. Bacteriol. 178: 4576–4581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bakker EP, Mangerich WE. 1981. Interconversion of components of the bacterial proton motive force by electrogenic potassium transport. J. Bacteriol. 147: 820–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bates SL, Waldren RP, Teare ID. 1973. Rapid determination of free proline for water-stress studies. Plant Soil 39: 205–207 [Google Scholar]
- 50. Welsh DT. 2000. Ecological significance of compatible solute accumulation by micro-organisms: from single cells to global climate. FEMS Microbiol. Rev. 24: 263–290 [DOI] [PubMed] [Google Scholar]
- 51. Vila-Costa M, Simo R, Harada H, Gasol JM, Slezak D, Kiene RP. 2006. Dimethylsulfoniopropionate uptake by marine phytoplankton. Science 314: 652–654 [DOI] [PubMed] [Google Scholar]
- 52. Bruce SJ, Guy PA, Rezzi S, Ross AB. 2010. Quantitative measurement of betaine and free choline in plasma, cereals and cereal products by isotope dilution LC-MS/MS. J. Agric. Food. Chem. 58: 2055–2061 [DOI] [PubMed] [Google Scholar]
- 53. Cosquer A, Pichereau V, Pocard JA, Minet J, Cormier M, Bernard T. 1999. Nanomolar levels of dimethylsulfoniopropionate, dimethylsulfonioacetate, and glycine betaine are sufficient to confer osmoprotection to Escherichia coli. Appl. Environ. Microbiol. 65: 3304–3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Spiegelhalter F, Bremer E. 1998. Osmoregulation of the opuE proline transport gene from Bacillus subtilis: contributions of the sigma A- and sigma B-dependent stress-responsive promoters. Mol. Microbiol. 29: 285–296 [DOI] [PubMed] [Google Scholar]
- 55. Fischer KE, Bremer E. 2012. Activity of the osmotically regulated yqiHIK promoter from Bacillus subtilis is controlled at a distance. J. Bacteriol. 194: 5197–5208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hahne H, Mäder U, Otto A, Bonn F, Steil L, Bremer E, Hecker M, Becher D. 2010. A comprehensive proteomics and transcriptomics analysis of Bacillus subtilis salt stress adaptation. J. Bacteriol. 192: 870–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nannapaneni P, Hertwig F, Depke M, Hecker M, Mäder U, Völker U, Steil L, van Hijum SA. 2012. Defining the structure of the general stress regulon of Bacillus subtilis using targeted microarray analysis and random forest classification. Microbiology 158: 696–707 [DOI] [PubMed] [Google Scholar]
- 58. Hecker M, Pane-Farre J, Völker U. 2007. SigB-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Annu. Rev. Microbiol. 61: 215–236 [DOI] [PubMed] [Google Scholar]
- 59. Price CW. 2011. General stress responses in Bacillus subtilis and related Gram-positive bacteria, p 301–318 In Storz G. (ed), Bacterial stress responses. 2nd ed. ASM Press, Washington, DC [Google Scholar]
- 60. Boylan SA, Redfield AR, Brody MS, Price CW. 1993. Stress-induced activation of the σb transcription factor of Bacillus subtilis. J. Bacteriol. 175: 7931–7937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kunst F, Rapoport G. 1995. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J. Bacteriol. 177: 2403–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dartois V, Debarbouille M, Kunst F, Rapoport G. 1998. Characterization of a novel member of the DegS-DegU regulon affected by salt stress in Bacillus subtilis. J. Bacteriol. 180: 1855–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ruzal SM, Sanchez-Rivas C. 1998. In Bacillus subtilis DegU-P is a positive regulator of the osmotic response. Curr. Microbiol. 37: 368–372 [DOI] [PubMed] [Google Scholar]
- 64. Steil L, Hoffmann T, Budde I, Völker U, Bremer E. 2003. Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. J. Bacteriol. 185: 6358–6370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shabala L, Bowman J, Brown J, Ross T, McMeekin T, Shabala S. 2009. Ion transport and osmotic adjustment in Escherichia coli in response to ionic and non-ionic osmotica. Environ. Microbiol. 11: 137–148 [DOI] [PubMed] [Google Scholar]
- 66. Shabala L. 2009. Organic vs inorganic: what makes the major contribution to osmotic adjustment in bacteria? Commun. Integr. Biol. 2: 74–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Glaasker E, Tjan FS, Ter Steeg PF, Konings WN, Poolman B. 1998. Physiological response of Lactobacillus plantarum to salt and nonelectrolyte stress. J. Bacteriol. 180: 4718–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Eshoo MW. 1988. lac fusion analysis of the bet genes of Escherichia coli: regulation by osmolarity, temperature, oxygen, choline, and glycine betaine. J. Bacteriol. 170: 5208–5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sutherland L, Cairney J, Elmore MJ, Booth IR, Higgins CF. 1986. Osmotic regulation of transcription: induction of the proU betaine transport gene is dependent on accumulation of intracellular potassium. J. Bacteriol. 168: 805–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Barron A, May G, Bremer E, Villarejo M. 1986. Regulation of envelope protein composition during adaptation to osmotic stress in Escherichia coli. J. Bacteriol. 167: 433–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Spanheimer R, Hoffmann M, Kögl S, Schmidt S, Pflüger K, Müller V. 2008. Differential regulation of Ota and Otb, two primary glycine betaine transporters in the methanogenic archaeon Methanosarcina mazei Gö1. J. Mol. Microbiol. Biotechnol. 15: 255–263 [DOI] [PubMed] [Google Scholar]
- 72. Gunasekera TS, Csonka LN, Paliy O. 2008. Genome-wide transcriptional responses of Escherichia coli K-12 to continuous osmotic and heat stresses. J. Bacteriol. 190: 3712–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kappes RM, Bremer E. 1998. Response of Bacillus subtilis to high osmolarity: uptake of carnitine, crotonobetaine and γ-butyrobetaine via the ABC transport system OpuC. Microbiology 144: 83–90 [DOI] [PubMed] [Google Scholar]
- 74. Nicolas P, Mäder U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, Becher D, Bisicchia P, Botella E, Delumeau O, Doherty G, Denham EL, Fogg MJ, Fromion V, Goelzer A, Hansen A, Hartig E, Harwood CR, Homuth G, Jarmer H, Jules M, Klipp E, Le Chat L, Lecointe F, Lewis P, Liebermeister W, March A, Mars RA, Nannapaneni P, Noone D, Pohl S, Rinn B, Rugheimer F, Sappa PK, Samson F, Schaffer M, Schwikowski B, Steil L, Stülke J, Wiegert T, Devine KM, Wilkinson AJ, van Dijl JM, Hecker M, Völker U, Bessieres P, Noirot P. 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335: 1103–1106 [DOI] [PubMed] [Google Scholar]
- 75. Helmann JD. 1995. Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 23: 2351–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cagliero C, Jin DJ. 23 October 2012. Dissociation and re-association of RNA polymerase with DNA during osmotic stress response in Escherichia coli. Nucleic Acids Res. [Epub ahead of print.] doi:10.1093/nar/gks988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gralla JD, Vargas DR. 2006. Potassium glutamate as a transcriptional inhibitor during bacterial osmoregulation. EMBO J. 25: 1515–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mellies J, Brems R, Villarejo M. 1994. The Escherichia coli proU promoter element and its contribution to osmotically signaled transcription activation. J. Bacteriol. 176: 3638–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Nagarajavel V, Madhusudan S, Dole S, Rahmouni AR, Schnetz K. 2007. Repression by binding of H-NS within the transcription unit. J. Biol. Chem. 282: 23622–23630 [DOI] [PubMed] [Google Scholar]
- 80. Lucht JM, Bremer E. 1991. Characterization of mutations affecting the osmoregulated proU promoter of Escherichia coli and identification of 5′ sequences required for high-level expression. J. Bacteriol. 173: 801–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Mellies J, Wise A, Villarejo M. 1995. Two different Escherichia coli proP promoters respond to osmotic and growth phase signals. J. Bacteriol. 177: 144–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Zhang X, Fletcher SA, Csonka LN. 1996. Site-directed mutational analysis of the osmotically regulated proU promoter of Salmonella typhimurium. J. Bacteriol. 178: 3377–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Voskuil MI, Chambliss GH. 1998. The −16 region of Bacillus subtilis and other gram-positive bacterial promoters. Nucleic Acids Res. 26: 3584–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hoffmann T, von Blohn C, Stanek A, Moses S, Barzantny S, Bremer E. 2012. Synthesis, release, and recapture of the compatible solute proline by osmotically stressed Bacillus subtilis cells. Appl. Environ. Microbiol. 78: 5753–5762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Koo SP, Higgins CF, Booth IR. 1991. Regulation of compatible solute accumulation in Salmonella typhimurium: evidence for a glycine betaine efflux system. J. Gen. Microbiol. 137: 2617–2625 [DOI] [PubMed] [Google Scholar]
- 86. Glaasker E, Konings WN, Poolman B. 1996. Glycine betaine fluxes in Lactobacillus plantarum during osmostasis and hyper- and hypo-osmotic shock. J. Biol. Chem. 271: 10060–10065 [DOI] [PubMed] [Google Scholar]
- 87. Börngen K, Battle AR, Moker N, Morbach S, Marin K, Martinac B, Krämer R. 2010. The properties and contribution of the Corynebacterium glutamicum MscS variant to fine-tuning of osmotic adaptation. Biochim. Biophys. Acta 1798: 2141–2149 [DOI] [PubMed] [Google Scholar]
- 88. Hoffmann T, Boiangiu C, Moses S, Bremer E. 2008. Responses of Bacillus subtilis to hypotonic challenges: physiological contributions of mechanosensitive channels to cellular survival. Appl. Environ. Microbiol. 74: 2454–2460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wahome PG, Cowan AE, Setlow B, Setlow P. 2009. Levels and localization of mechanosensitive channel proteins in Bacillus subtilis. Arch. Microbiol. 191: 403–414 [DOI] [PubMed] [Google Scholar]
- 90. Oren A. 2011. Thermodynamic limits to microbial life at high salt concentrations. Environ. Microbiol. 13: 1908–1923 [DOI] [PubMed] [Google Scholar]
- 91. Wood JM. 2011. Bacterial osmoregulation: a paradigm for the study of cellular homeostasis. Annu. Rev. Microbiol. 65: 215–238 [DOI] [PubMed] [Google Scholar]
- 92. Saum SH, Müller V. 2007. Salinity-dependent switching of osmolyte strategies in a moderately halophilic bacterium: glutamate induces proline biosynthesis in Halobacillus halophilus. J. Bacteriol. 189: 6968–6975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kuhlmann AU, Bursy J, Gimpel S, Hoffmann T, Bremer E. 2008. Synthesis of the compatible solute ectoine in Virgibacillus pantothenticus is triggered by high salinity and low growth temperature. Appl. Environ. Microbiol. 74: 4560–4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Belitsky BR, Brill J, Bremer E, Sonenshein AL. 2001. Multiple genes for the last step of proline biosynthesis in Bacillus subtilis. J. Bacteriol. 183: 4389–4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Msadek T, Kunst F, Klier A, Rapoport G. 1991. DegS-DegU and ComP-ComA modulator-effector pairs control expression of the Bacillus subtilis pleiotropic regulatory gene degQ. J. Bacteriol. 173: 2366–2377 [DOI] [PMC free article] [PubMed] [Google Scholar]