Abstract
Cyclic di-GMP (c-di-GMP) is a secondary messenger that controls a variety of cellular processes, including the switch between a biofilm and a planktonic bacterial lifestyle. This nucleotide binds to cellular effectors in order to exert its regulatory functions. In Salmonella, two proteins, BcsA and YcgR, both of them containing a c-di-GMP binding PilZ domain, are the only known c-di-GMP receptors. BcsA, upon c-di-GMP binding, synthesizes cellulose, the main exopolysaccharide of the biofilm matrix. YcgR is dedicated to c-di-GMP-dependent inhibition of motility through its interaction with flagellar motor proteins. However, previous evidences indicate that in the absence of YcgR, there is still an additional element that mediates motility impairment under high c-di-GMP levels. Here we have uncovered that cellulose per se is the factor that further promotes inhibition of bacterial motility once high c-di-GMP contents drive the activation of a sessile lifestyle. Inactivation of different genes of the bcsABZC operon, mutation of the conserved residues in the RxxxR motif of the BcsA PilZ domain, or degradation of the cellulose produced by BcsA rescued the motility defect of ΔycgR strains in which high c-di-GMP levels were reached through the overexpression of diguanylate cyclases. High c-di-GMP levels provoked cellulose accumulation around cells that impeded flagellar rotation, probably by means of steric hindrance, without affecting flagellum gene expression, exportation, or assembly. Our results highlight the relevance of cellulose in Salmonella lifestyle switching as an architectural element that is both essential for biofilm development and required, in collaboration with YcgR, for complete motility inhibition.
INTRODUCTION
Bis-(3′–5′)-cyclic dimeric GMP (c-di-GMP), discovered by Benziman and colleagues in 1987 as an allosteric activator of the cellulose synthase in Gluconacetobacter xylinus (1), is now widely recognized as a ubiquitous bacterial second messenger and a key regulator in bacterial transition from a motile and planktonic to a sessile and biofilm lifestyle (reviewed in references 2 to 7). High intracellular c-di-GMP levels promote extracellular matrix production and subsequent biofilm formation and repress motility, whereas low intracellular c-di-GMP levels suppress matrix production and promote single-cell motility (8, 9). In recent years, significant progress in elucidating the enzymology of c-di-GMP turnover has been achieved. The levels of this signaling molecule are regulated through the action of diguanylate cyclases (DGCs) and c-di-GMP-specific phosphodiesterasas (PDEs) (10). The cyclase activity, which converts two molecules of GTP to c-di-GMP, is encoded in the GGDEF protein domain (11, 12), while phosphodiesterase activity, which hydrolyzes c-di-GMP to linear 5′-pGpG or two GMP molecules, is encoded in the EAL (13–15) and HD-GYP (16) domains. Many GGDEF domains also contain a conserved RxxD motif, called the I site, located 5 amino acids upstream of the GGDEF motif, that acts as a secondary binding site for c-di-GMP, allowing noncompetitive product inhibition by dimeric c-di-GMP (17–19).
Apart from modulating lifestyle switching, recent studies have revealed that c-di-GMP controls numerous cellular functions, including cell cycle progression, virulence of animal and plant pathogens, and cell-cell signaling (5, 20, 21). To exert this global activity, c-di-GMP has to bind to and allosterically alter the structure and output function of specific effectors. In agreement with the variety of c-di-GMP-related outputs, the nature of the c-di-GMP binding molecules is diverse. These include the PilZ domain-containing proteins (22), enzymatically inactive GGDEF proteins that contain a degenerate GGDEF domain but a conserved intact I site (23, 24), enzymatically inactive EAL proteins which retain the ability to bind c-di-GMP but no longer hydrolyze it (25–27), PelD from P. aeruginosa, which binds c-di-GMP through an RxxD motif resembling an I site found in active DGCs (28), transcription factors that do not share a predictable c-di-GMP binding site (29–36), the Escherichia coli polynucleotide phosphorylase (37), and riboswitches (38–41).
Among them, the best-studied type of effectors is the PilZ protein family. In 2006, Amikam and Galperin first suggested, based on bioinformatics analyses, that PilZ domains may function as c-di-GMP effectors (42). A study carried out the same year demonstrated that YcgR, an E. coli PilZ domain protein, and the PilZ domain of the Gluconacetobacter xylinus cellulose synthase, BcsA, bind c-di-GMP tightly and specifically. Also, the function of YcgR as a c-di-GMP-dependent inhibitor of motility was established (22). Later, other members of the PilZ protein family, present in several species of bacteria, were experimentally shown to interact directly with c-di-GMP (43–51). These studies confirmed the role of PilZ domain proteins in the synthesis of exopolysaccharides, biofilm formation (44, 46, 52), and motility control (43, 44, 51, 53) and identified novel contributions of these proteins in virulence regulation of animal and plant pathogens (44, 47, 51, 53). Binding of c-di-GMP to the PilZ domain causes structural changes in the protein that initiate the downstream signal transduction cascade (22, 48, 49).
The phyletic distribution of the PilZ domain is generally similar to those of the GGDEF and EAL domains (42). With respect to the number of PilZ domain proteins encoded in each genome, some bacterial species, such as Pseudomonas aeruginosa, Vibrio cholerae, and Borrelia burgdorferi, present a correlation between the number of enzymes for controlling the levels of c-di-GMP and that of PilZ domain proteins (44, 46, 51). Remarkably, for most enterobacteria, YcgR and BcsA are the only PilZ domain proteins encoded in the genome, although these usually contain multiple genes encoding GGDEF and EAL domain proteins. In the case of Salmonella, YcgR and BcsA are not simply the sole PilZ proteins encoded in its genome but also are the only c-di-GMP receptors described until now. BcsA of Salmonella is encoded in one of the bacterial cellulose synthesis (bcs) operons (54, 55), and its cytoplasmic PilZ domain is thought to regulate the enzymatic activity of a periplasmic cellulose synthesis domain, based on its binding to c-di-GMP. Thus, high c-di-GMP cellular levels posttranslationally promote the synthesis of cellulose, a β-1-4-d-glucose polymer, which is a main component of the extracellular matrix of the Salmonella biofilm (8, 56). Several studies have shown the crucial importance of this exopolysaccharide during biofilm development under laboratory conditions (54, 55), on epithelial cell surfaces (57), and on glass coverslips (58).
On the other hand, and as stated above, YcgR is dedicated to regulation of motility. Three recent studies have shown that upon c-di-GMP binding, YcgR reduces motility by interacting directly with flagellar motor proteins, slowing down flagellar rotor speed and altering the frequency of the rotational switch (59–61). To assess the role of YcgR, these studies and others have used a strain carrying a mutation in yhjH, a gene encoding a standalone EAL domain protein with a predicted phosphodiesterase activity. Its mutation is expected to result in elevated c-di-GMP levels, and thus, a yhjH mutant shows a defect in swimming motility both in E. coli and in Salmonella enterica serovar Typhimurium. The motility defect of the yhjH mutant can be rescued by deleting the effector-encoding gene, ycgR (22, 59–62). Importantly, however, the motility recuperation of the yhjH ycgR double mutant shown in most of these reports is not total (22, 60–62), suggesting that in the absence of YcgR there is still an additional element that mediates inhibition of motility in the presence of high c-di-GMP intracellular levels. Here we present evidence that the exopolysaccharide cellulose works in cooperation with YcgR to stop bacteria and mediate the c-di-GMP-dependent transition between motility and sessility in Salmonella. On one hand, YcgR works as a backstop brake, inducing flagellar rotational bias and reducing motor function (61), and on the other, cellulose synthesized by BcsA completely stops bacteria, probably by means of interfering with flagellum functionality through steric hindrance.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this work are described in Table S1 in the supplemental material. Escherichia coli XL1-Blue, S. enterica subsp. enterica serovar Enteritidis (S. Enteritidis), and S. enterica subsp. enterica serovar Typhimurium (S. Typhimurium) cells were grown in LB broth and on LB agar with appropriate antibiotics at the following concentrations: kanamycin (Km), 50 μg ml−1; ampicillin (Am), 100 μg ml−1; chloramphenicol (Cm), 20 μg ml−1; and tetracycline (Tc), 20 μg ml−1.
DNA manipulations.
Routine DNA manipulations were performed using standard procedures unless otherwise indicated. Plasmid DNA from E. coli was purified using a Quantum Prep plasmid kit (Bio-Rad). Plasmids were transformed into E. coli and S. Enteritidis by electroporation. Transformants carrying the Red helper plasmid were made electrocompetent as described in reference 54. Restriction enzymes were purchased from New England BioLabs and used according to the manufacturer's instructions. Oligonucleotides were obtained from Thermo Scientific and are listed in Table S2 of the supplemental material. Phage P22 HT105/1 int-201 (63) was used to carry out transductions between strains according to recommended protocols (64).
One-step inactivation of chromosomal genes.
For disruption of the yhjH, ycgR, and bcsA genes in S. Enteritidis 3934, PCR-generated linear DNA fragments in combination with the helper plasmid pKOBEGA were used (54, 65). A selectable antibiotic resistance gene was generated by PCR using primer pairs of 80-nucleotide (nt)-long primers that included 60-nt homology extensions for the targeted locus and 20-nt priming sequences for the kanamycin, tetracycline, and chloramphenicol resistance genes as templates from a freshly isolated colony of E. coli MC4100 ybeW::Km, S. Typhimurium TT3699 ara651::Tn10, and S. Typhimurium SV4406 rcsB::MudQ, respectively. Primer pairs yhjH Km Fw and yhjH Km Rv, ycgR Tet Fw and ycgR Tet Rv, and bcsA Clo Fw and bcsA Clo Rv were used for disruption of the yhjH, ycgR, and bcsA genes, respectively. Electroporation (25 μF, 200 Ω, 2.5 kV) was carried out according to the manufacturer's instructions (Bio-Rad) using 50 μl of cells and 1 to 5 μg of purified and dialyzed (0.025-μm nitrocellulose filters; Millipore) PCR product. Shocked cells were added to 1 ml of LB broth, incubated overnight at 30°C, and then spread on LB agar with Km, Tc, or Cm to select Kmr, Tcr, or Cmr transformants after incubation at 30°C for 24 h. Mutants were then grown on LB broth with Km, Tc, or Cm at 43°C for 24 h and incubated overnight on LB agar with Am at 30°C to test for the loss of the helper plasmid.
Construction of amino acid substitutions in the BcsA and CheY proteins.
To perform a BcsA RxxxR substitution for BcsA RxxxD in order to disrupt the c-di-GMP binding motif, a protocol described previously was carried out with some modifications (66). In a first step, primers PilZ Clo Fw and PilZ Clo Rv, with 60-bp homology extensions, were used to amplify a chloramphenicol resistance cassette and an I-SceI recognition site from plasmid pWRG100. This construct was integrated within bcsA, next to the mutation site, via λ Red-mediated recombination using plasmid pWRG99, a temperature-sensitive plasmid for independent inducible expression of the λ Red recombinase and I-SceI endonuclease. After confirming proper insertion of the resistance cassette by colony PCR, phosphorylated 80-mer double-stranded DNAs (dsDNAs), derived from oligonucleotides PilZ Dimer Fw and PilZ Dimer Rv, were electroporated into the mutant strain still containing the pWRG99 plasmid. The 80-mer DNA fragment was a synthetic 80-mer mutant dsDNA fragment that included the desired mutation and part of the homologous regions used for integration of the resistance cassette. After 1 h of incubation at 28°C, 100 μl of a 10−2 dilution was plated on LB agar plates containing 500 ng/ml anhidrotetracycline, which induced expression of I-SceI endonuclease. After overnight incubation at 28°C, single colonies were purified, and successful recombination was checked by monitoring absence of antibiotic resistance, colony PCR with oligonucleotides bcsA-A and bcsA-B, and sequencing of the resulting fragment. Finally, pWRG99 was cured by incubating selected colonies at 37°C.
To construct Salmonella strains carrying the D57A substitution and the D13K and Y106W substitutions in CheY (CheY_D57A and CheY_D13K/Y106W, respectively), two separate PCR products with overlapping sequences, including the targeted sequence, were combined. The reverse oligonucleotide of the PCR necessary to generate the first fragment and the forward oligonucleotide needed to generate the second fragment were complementary to allow PCR products to anneal. Specifically, primer pairs cheY Fw and cheY Mut1 Rv and cheY Mut1 Fw and cheY Rv were used to generate the two DNA fragments required for CheY_D13K substitution. Genomic DNA from the CheY_D13K mutant strain was used as the template to generate the CheY_D13K/Y106W double mutant strain. For that, primer pairs cheY Fw and cheY Mut2 Rv and cheY Mut2 Fw and cheY Rv were used. Primer pairs cheY Fw and cheY Mut3 Rv and cheY Mut3 Fw and cheY Rv were used to generate the two DNA fragments required for CheY_D57A substitution. In each case, 1-μl volumes of the two purified PCR products were mixed, and a second PCR using cheY Fw and cheY Rv primers was performed to obtain a single fragment. The fusion product was purified and cloned using the Zero Blunt Topo PCR kit (Promega). Once the construction was confirmed by sequencing, the fragment was cloned into the NotI and SacI sites of plasmid pKO3blue (67), which was electroporated into S. Enteritidis 3934. The following steps of integration and excision of the plasmid were performed as described previously (67). CheY_D13K/Y106W and CheY_D57A modifications were tested by PCR using primer pairs cheY Mut1 Comp and cheY-E, cheY Mut2 Comp and cheY-F, and cheY Mut3 Comp and cheY-E, respectively. Also, the CheY_D13K/Y106W and CheY_D57A alleles were amplified by PCR using the cheY-E and cheY-F oligonucleotides and sequenced.
Isolation of motile suppressor mutants and genetic mapping.
To isolate spontaneous motile suppressor mutants that recuperate the motility defect of a strain containing high levels of c-di-GMP independently of YcgR, a previous protocol was performed (59), with some modifications. Approximately 108 cells of strain ΔXII ΔycgR Psen4316::hmsT pBR328::stm1987, which expresses two c-di-GMP sources and contains a ycgR mutation, were placed in the center of a soft agar motility plate and incubated for 20 h at 37°C. After that time, six suppressor mutants became visible as flares emanating from the center of the immotile colony. To identify the spontaneous mutations causing suppression of the motility defect, each suppressor was subjected to random transposon mutagenesis with MudJ using phage P22 (68). Fifty thousand transposon mutants of each suppressor were independently pooled and used to grow a P22 phage lysate. A small amount of phages were expected to contain a stretch of chromosomal DNA harboring the suppressor mutation plus a nearby transposon insertion. Pool lysates were transduced to the original strain, and a pool of transductants of each transduction was incubated in the middle of a rectangular plate (Nunc) containing swimming medium. After 16 h of incubation at room temperature, motile mutants were able to escape the central part of the plate and swim. These mutants were isolated, and new individual P22 lysates were grown. Clonal lysates were used to transduce the ΔycgR pBR328::stm1987 and ΔXII ΔycgR Psen4316::hmsT strains, and transductants were checked for motility. Mutants showing a cotransduction frequency of the selected kanamycin resistance and the suppressor mutation higher than 90% were selected. To identify genes containing MudJ fusions, chromosomal DNA from each mutant was purified, and the DNA sequence of the region adjacent to the transposon insertion site was determined at Secugen (Spain), using primer MuL (see Table S2 in the supplemental material), specific to the left arm of the MudJ transposon, as described previously (68).
Motility assay.
Swimming plates were made using LB 0.3% agar. Bacterial strains were grown in LB broth at 37°C. Then, 1 μl of the stationary cultures was inoculated into swimming plates that were incubated at 23°C for 16 h. Medium was supplemented with 70 μM IPTG (isopropyl-β-d-thiogalactopyranoside) (Sigma) when strains overexpressing sen3440 (bcsZ) were tested. Ampicillin (100 μg/ml) was added if necessary. All strains assayed for a particular experiment were grown on the same swimming plate. Images were taken in a GBox Chemi HR16 system (Syngene). Different parts of a photograph were cut and put together to assemble horizontal figures showing swimming behavior.
Quantitative measurement of motility was performed by calculating the area of the motility halo of each strain with the use of the image-processing software program ImageJ. The percent motility relative to that of the parental strain was then calculated. Experiments were performed in triplicate on three separate days.
Tethering assay.
For tether analysis, a previously described method was carried out (69). First, bacteria were grown in LB broth at 28°C for 6 h, and 400 μl of the cell suspension was passed through a 28 gauge needle 40 to 60 times to shear the flagella. The cells were harvested and washed with an equal volume of motility medium [7.6 mM (NH4)2SO4, 2 mM MgSO4, 20 μM FeSO4, 0.1 mM EDTA, and 60 mM potassium phosphate (pH 6.8)]. A 20-μl drop of the suspension was placed under a coverslip mounted on a glass slide and sealed with paraffin. The coverslip was pretreated with anti-FliC antibodies for 30 min and then washed with motility medium. The cells were allowed to attach to the coverslip for 30 min. The tethered cells were observed with an Eclipse TE300 microscope (Nikon).
Western blotting.
Samples for Western analysis were harvested as follows. Bacteria were grown overnight in LB (supplemented with Am if necessary) at 37°C, and then a 1:100 dilution of the overnight culture was prepared. This inoculum was further incubated at 28°C for 6 h, with the exception of part of the samples used to analyze hmsT expression, which were incubated at 23°C for 16 h. For green fluorescent protein (GFP) and 3×Flag detection, cells were harvested, washed, and finally resuspended in 50 μl of phosphate-buffered saline (PBS). An equal volume of Laemmli sample buffer was added to each sample, and they were boiled at 100°C for 5 min. To detect exported flagellin, 11 ml of culture was harvested. The optical density at 600 nm (OD600) of each sample was measured in order to level all samples to the same OD600 by adding sterile LB. Samples were vortexed for a complete minute to release the flagella to the medium. Cells were pelleted, and 8 ml of the supernatant was transferred to a 10,000 nominal molecular weight limit (NMWL) centrifugal filter device (Millipore). Volume was concentrated to 500 μl. The resulting sample was centrifuged at 14,000 rpm for 5 min in order to avoid contamination from cells that remained in the sample. Twenty microliters of the final supernatant was mixed with an equal volume of Laemmli sample buffer and boiled at 100°C for 5 min.
Proteins were separated on SDS-polyacrylamide gels (12% to 5%) and stained with Coomassie brilliant blue R250 (0.25%; Sigma). For Western blotting, proteins were transferred to Hybond-ECL nitrocellulose membranes (Amersham Biosciences) by electroblotting. Probing was carried out with anti-GFP (Living Colors A.v. monoclonal antibody [JL-8]; Clontech), anti-FliC (generously provided by Carlos Gamazo, Universidad de Navarra, Pamplona, Spain), monoclonal anti-Flag M2 (Sigma), or anti-GroEL (Sigma) antibodies diluted 1:10,000 for 60 min at room temperature. Nitrocellulose membranes were washed with 0.1% PBS-Tween and then incubated with alkaline phosphatase-conjugated secondary antibodies (goat anti-mouse for GFP, FliC, and 3×Flag and goat anti-rabbit for GroEL; Sigma) diluted 1:2,500 for 60 min at room temperature. Bound ligands were detected using the ECL Plus Western blotting substrate (Pierce).
Flagellin immunofluorescence.
Bacteria were grown overnight in LB (supplemented with ampicillin if necessary) at 37°C. Then, a 1:100 dilution of the overnight culture was prepared, and incubation continued at 28°C for 6 h. Two hundred fifty microliters of liquid culture samples were pelleted at 10,000 × g, washed once with PBS, and fixed with an equal volume of 2.4% paraformaldehyde–0.01% glutaraldehyde. Then, 10 μl of the cell suspension was transferred onto a polylysine (Sigma)-treated coverslip and incubated for 30 min. Coverslips were immersed in ice-cold methanol for 5 min and washed with acetone. Fixed cells were treated with 20 μl of 1% bovine serum albumin (BSA) in PBS for 30 min to block unspecific binding, followed by incubation with 20 μl of anti-FliC antibodies diluted 1:200 for 30 min and then with 20 μl of goat anti-rabbit Alexa Fluor 488 (Invitrogen) diluted 1:200 and 2 mg Hoechst 33342 stain (Invitrogen) for 30 min. The slide was washed with PBS between each treatment. All reagents were diluted in 1% BSA in PBS, and all incubations were carried out at room temperature in a wet and dark chamber. After a final wash with water, the slide was glued to a microscope slide with Aqua Poly/Mount medium (Polysciences) overnight. Samples were viewed and images were captured using an Eclipse TE300 microscope (Nikon).
Cellulose staining and fluorescence microscopy.
Direct cellulose staining was performed by following a modified protocol from reference 70. Bacteria were grown and fixed as explained above. The coverslips containing fixed cells were incubated with PBS containing 0.001% calcofluor (Fluorescent Brightener 28; Sigma). A fresh stock of 1% calcofluor in 20% glycerol containing 25 mM NaOH was prepared prior to the experiment. Bacterial cell membranes were stained with FM4-64 stain (Molecular Probes/Invitrogen) diluted in PBS to a final concentration of 2 μg/ml.
Fluorescence images were acquired using a Zeiss Axiokop 2 Plus microscope equipped with an HBO 50/AC camera (Zeiss).
Overexpression of bcsZ and endoglucanase assay.
The sen3440 (bcsZ) coding region was amplified from S. Enteritidis 3934 chromosomal DNA using bcsZ NdeI Fw and bcsZ NotI Rv oligonucleotides containing NdeI and NotI restriction sites. The PCR product was cloned into the pGEM-T Easy (Promega) vector and confirmed by sequencing. An NdeI-NotI bcsZ fragment was obtained by digestion and cloned into pUA1108 (71), yielding plasmid pUA1108::bcsZ, so that bcsZ expression was under the control of the tac promoter. Plasmid pUA1108::bcsZ was transformed in the S. Enteritidis 3934 strain, and IPTG-mediated overexpression of bcsZ was confirmed in crude extracts by SDS-PAGE (data not shown).
To assay the endoglucanase activity of bcsZ, bacterial colonies were grown on LB agar plates containing 0.1% of carboxymethylcellulose (CMC) (Sigma) and 70 μM IPTG. After 16 h of incubation at 37°C, the agar medium was flooded with an aqueous solution of 1 mg/ml Congo red (Sigma) for 15 min. The Congo red solution was then poured off, and plates were further treated by flooding them with 1 M NaCl for 15 min. White halos around the colonies became visible if CMC was degraded by bacteria (72).
Construction and characterization of ΔXII strains and derivatives.
The inability to generate an rpoS mutant by double crossover over the ΔXII strain described in reference 67 revealed a deletion of the rpoS gene in this strain. Precise identification of the complete deleted region in this strain was carried out through primer walking using chromosomal DNA as the template. The PCR product obtained with primers pcm Fw and sen2747 Rv was sequenced to determine the missing region (see Fig. S1 in the supplemental material).
Construction of the new ΔXII strain was carried out, taking as a basis the ΔIX strain (67) and then changing the order of gene deletions (see Fig. S2). ΔXII + sen4316-3×Flag, ΔXII Psen4316::hmsT, ΔXII Psen4316::hmsT-GS, and ΔXII Psen4316::hmsT-3×Flag derivatives were obtained from the new ΔXII strain, following the same procedure described in reference 67. Phenotypic assays were performed as described previously (67).
RESULTS
High c-di-GMP levels can inhibit motility independently of YcgR.
Inactivation of the YhjH phosphodiesterase has been routinely used to investigate how elevated levels of c-di-GMP interfere with bacterial motility through the PilZ protein YcgR (22, 59–62). Most of these studies reached the same conclusion, that mutation of ycgR was able to substantially but not totally restore the bacterial motility defect of an yhjH mutant, suggesting that an additional element might cooperate in the motility inhibition caused by the presence of high levels of c-di-GMP in the cell. To investigate the YcgR-independent mechanisms of motility inhibition in S. Enteritidis, we first constructed single ΔyhjH and ΔycgR mutants and a double ΔyhjH ΔycgR mutant using the clinical isolate S. Enteritidis 3934 and analyzed their motility behavior. In agreement with findings of previous studies, deletion of yhjH impaired swimming motility, whereas the additional mutation of ycgR only partially restored bacterial motility (Fig. 1). To analyze if inhibition of bacterial motility in the absence of YcgR could be attributed to any source of high levels of c-di-GMP and not only to the specific absence of YhjH, we complemented S. Enteritidis 3934 and its corresponding ΔycgR derivative with a plasmid expressing stm1987, a gene that encodes an active diguanylate cyclase able to promote cellulose synthesis under different environmental conditions (56). Results showed that complementation of not only the wild type but also the ΔycgR strain with pBR328::stm1987 completely blocked bacterial motility (Fig. 1). This inhibition was dependent on c-di-GMP synthesis, since complementation of the wild-type strain with the same plasmid producing a modified STM1987 protein with a degenerate GGGSF motif did not alter swimming behavior (Fig. 1). These results indicated that high levels of c-di-GMP can completely inhibit Salmonella motility independently of YcgR.
Fig 1.
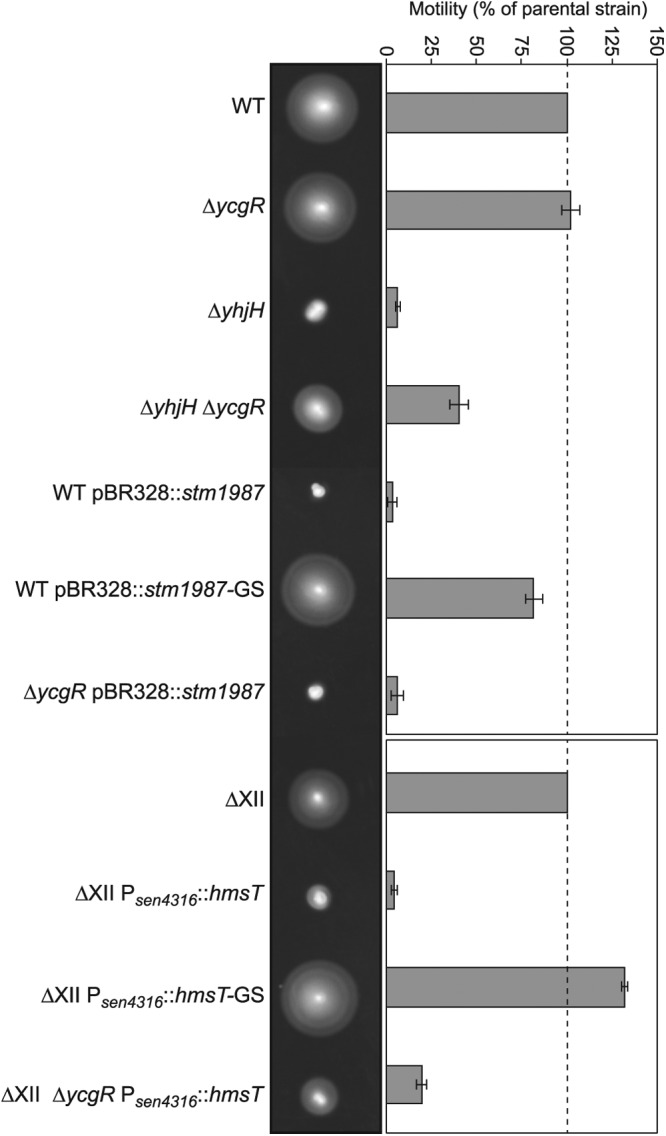
High c-di-GMP levels inhibit Salmonella motility in the absence of YcgR. Representative swimming motility plates after incubation at 23°C for 16 h are shown. Quantitative measurement of motility is also shown. The total area of growth was measured, and the percent motility relative to that of the parental strain was calculated. Means and standard deviations of results from three repeats on three separate days are shown. Overexpression of the DGC-encoding gene stm1987 in a wild-type S. Enteritidis strain (WT pBR328::stm1987) completely inhibited motility even in the absence of YcgR (ΔycgR pBR328::stm1987). Overexpression of a heterologous DGC-encoding gene, hmsT, in a multiple S. Enteritidis mutant in all genes encoding GGDEF domain proteins (ΔXII Psen4316::hmsT) also resulted in motility blockage that was only slightly restored when the ycgR gene was mutated (ΔXII ΔycgR Psen4316::hmsT). Inhibition of motility depended on the capacity of STM1987 and HmsT to synthesize c-di-GMP (see WT pBR328::stm1987-GS and ΔXII Psen4316::hmsT-GS strains, respectively).
Ectopic activation of c-di-GMP synthesis inhibits Salmonella motility in the absence of the GGDEF family of proteins.
Many GGDEF domain proteins harbor a c-di-GMP binding I site that serves as a control point of their own activity through a product inhibition mechanism. Also, enzymatically inactive GGDEF domain proteins may function as c-di-GMP receptors through their I sites, leading to a spatial sequestration of the protein (23, 24). Thus, we hypothesized that one or more GGDEF domain proteins of Salmonella might bind c-di-GMP under excess conditions of this nucleotide and then act as direct negative effectors, leading to a blockage of cell motility in the absence of YcgR. To test this, we made use of an S. Enteritidis 3934 derivative, called ΔXII, which is a multiple mutant carrying mutations in all genes encoding GGDEF domain proteins and thus incapable of synthesizing c-di-GMP (67). Its use permits restoration of individual DGC-encoding genes and thus manipulation of c-di-GMP intracellular levels without the interference of other GGDEF domain proteins. Strain ΔXII was generated again for this study because our previously described ΔXII strain (67) was found to contain a chromosomal deletion (see Fig. S1 in the supplemental material) that makes the strain unable to swim. The new ΔXII strain presents a swimming behavior similar to that of the wild-type strain, indicating that the presence of c-di-GMP is not required for bacterial motility. Phenotypic characterization of the new ΔXII strain is presented in Fig. S2 and will be further extended elsewhere. Taking the new ΔXII strain as a basis, we then constructed a derivative, ΔXII Psen4316::hmsT, in which the hmsT gene from Yersinia pestis was inserted in the site corresponding to the deletion of the DGC-encoding gene sen4316. As a consequence, the heterologous hmsT gene was expressed under the control of sen4316 promoter. hmsT encodes a very active DGC that reaches maximum levels at ambient temperatures below 30°C (73, 74). To confirm HmsT production from this genetic location, a 3×Flag epitope coding sequence was added to hmsT, leading to strain ΔXII Psen4316::hmsT-3×Flag and Western blot analyses were carried out. Results showed very high production of the HmsT protein in comparison to the expression of the original DGC, SEN4316, under its own promoter (Fig. 2). Thus, we were able to analyze motility function in a strain (ΔXII Psen4316::hmsT) with a single, heterologous and very active source of c-di-GMP. Expression of HmsT in ΔXII totally impaired swimming motility. HmsT diguanylate cyclase activity was essential for motility blockage, since ΔXII expressing HmsT with a degenerate GGGSF motif showed a swimming size similar to that of ΔXII. Mutation of ycgR in ΔXII Psen4316::hmsT only slightly restored motility (Fig. 1). These results confirmed the fact that elevated c-di-GMP levels can inhibit Salmonella motility even in the absence of YcgR and also ruled out Salmonella GGDEF domain proteins as direct repressors of motility.
Fig 2.
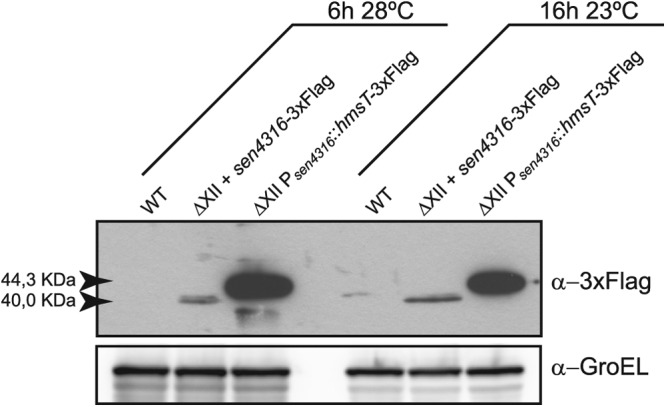
Expression of hmsT gene under the sen4316 promoter results in a high accumulation of the HmsT protein. Western blot analysis of HmsT and SEN4316 expression in ΔXII derivative strains in which the hmsT or sen4316 genes with a 3×Flag epitope coding sequence were chromosomally restored under the sen4316 promoter is shown. Strains were grown under tethering assay conditions, that is, at 28°C for 6 h, or under swimming conditions, that is, at 23°C for 16 h. HmsT was highly produced under both conditions compared to the expression of the original Salmonella DGC-encoding gene, sen4316. A Western blot using anti-GroEL antibodies was used as a loading control.
High c-di-GMP concentrations render Salmonella cells completely immobile without affecting flagellum synthesis.
c-di-GMP can impact motility at the level of transcription, posttranscription, and function (reviewed in reference (75). To establish the level at which high c-di-GMP contents repress Salmonella motility, we first used a plasmid-borne transcriptional fusion of the promoter of the flagellin gene (fliC) to an unstable variant of GFP, GFP(LVA) (76). Plasmid pPfliCGFP was introduced into the wild type, ΔXII, and ΔXII Psen4316::hmsT strains, and expression levels were assessed by Western blotting using anti-GFP antibodies (Fig. 3A). Results showed no differences in GFP expression between samples, indicating that elevated c-di-GMP levels in Salmonella do not affect transcriptional regulation of flagella. Next, we investigated regulation at the posttranscriptional level by analyzing the amount of exported flagellin from the wild-type (WT), WT pBR328::stm1987, ΔXII, and ΔXII Psen4316::hmsT strains. Results revealed that the amounts of flagellin were equal between strains (Fig. 3B). We then tested by an immunodetection assay whether c-di-GMP-mediated inhibition of motility was a result of alterations in flagellum assembly. As shown in Fig. 3C, flagella were correctly assembled, regardless of the c-di-GMP levels inside the cell. Altogether, these results pointed out to an interference of high c-di-GMP levels with flagellar function.
Fig 3.
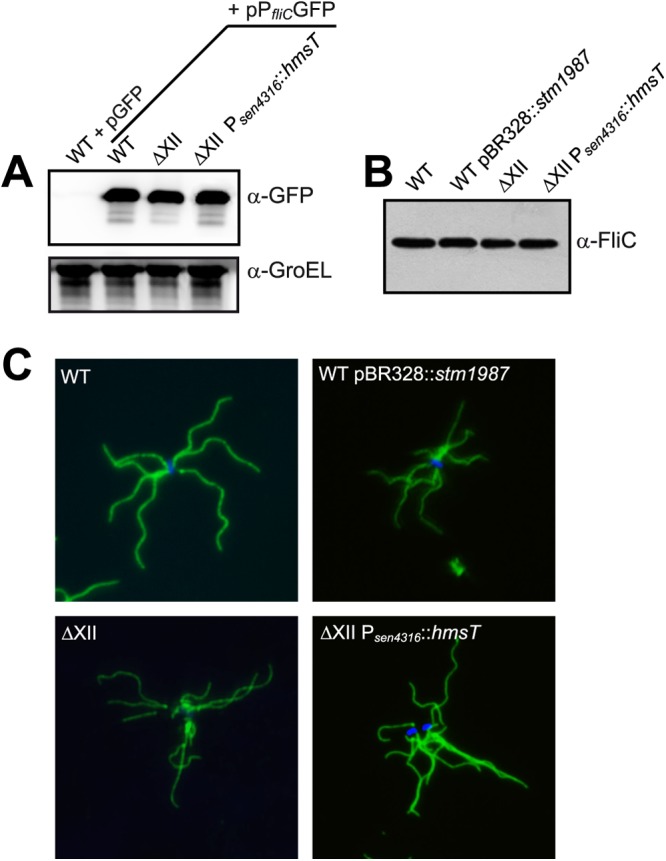
High c-di-GMP levels do not regulate flagellum gene expression, flagellar exportation, and assembly. (A) Analysis of fliC promoter activity. Samples of the wild-type strain, ΔXII, and ΔXII Psen4316::hmsT harboring gfp[LVA] transcriptional fusions to the fliC promoter were analyzed by Western blotting using anti-GFP antibodies. A Western blot using anti GroEL antibodies was used as a loading control. (B) Western blot analysis of exported flagellin from the wild type, ΔXII, and derivative strains overexpressing stm1987 and hmsT, respectively. (C) Flagellin immunofluorescence of the wild type, ΔXII, and derivative strains overexpressing a DGC. Blue fluorescence corresponds to the Hoechst 33342 dsDNA stain. In all cases, bacteria were grown under tethering assay conditions.
It has been shown that YcgR, when bound to c-di-GMP, interacts with flagellar motor proteins and results in the induction of a counterclockwise (CCW) motor bias and thus inhibition of motility (60, 61). To assess whether blockage of motility under elevated c-di-GMP levels and in the absence of YcgR was also due to an alteration in rotation behavior, we performed a tethering assay that permits measurement of flagellar rotation through the observation under the microscope of Salmonella cells bound to a glass surface by a single flagellum. It is known that the switch from counterclockwise to clockwise (CW) rotation requires interaction of the phosphorylated form of the signaling protein CheY with FliM, a protein of the flagellar rotor (77). Thus, two CheY mutants of S. Enteritidis 3934 were constructed to be used as controls of the assay. The first one contained a point mutation in the aspartate residue (D57A) in order to prevent phosphorylation of CheY, leading to a counterclockwise-bias rotation (78). The second one contained two point mutations (D13K and Y106W) which mimic the constitutive phosphorylation state of CheY, resulting in clockwise-bias rotation (79). Results of the tethering assay are summarized in Table 1. Wild-type and ΔXII cells rotated primarily in the CCW direction and displayed intervals of CW rotation. Their corresponding ycgR mutants showed the same rotational behavior. In agreement with previously published results, the ΔyhjH mutant showed a CCW rotation bias, while the ΔyhjH ΔycgR strain fully recuperated wild-type motility (60, 61). Notably, cells that highly expressed a very active DGC, namely, WT pBR328::stm1987 and ΔXII Psen4316::hmsT cells, did not rotate in any direction, remaining completely immobile. Deletion of ycgR in these cells was unable to restore rotation. Overall, these results indicated that under elevated c-di-GMP intracellular levels, flagellum gene expression, translation, and assembly processes are not affected, while an unknown YcgR-independent mechanism negatively regulates flagellum function, impeding its rotation.
Table 1.
Swimming and predominant flagellum rotation behavior of Salmonella wild type, ΔXII, strains with high c-di-GMP levels, and their corresponding ycgR mutants
| Strain name or description | Swimming behavior | Rotation behavior |
|---|---|---|
| WT | Motile | CCW with reversals |
| ΔycgR | Motile | CCW with reversals |
| ΔyhjH | Nonmotile | CCW bias |
| ΔyhjH ΔycgR | Partially motile | CCW with reversals |
| WT pBR328::stm1987 | Nonmotile | No rotation |
| ΔycgR pBR328::stm1987 | Nonmotile | No rotation |
| ΔXII | Motile | CCW with reversals |
| ΔXII ΔycgR | Motile | CCW with reversals |
| ΔXII Psen4316::hmsT | Nonmotile | No rotation |
| ΔXII ΔycgR Psen4316::hmsT | Slightly motile | No rotation |
| CheY_D57A | Nonmotile | CCW bias |
| CheY_D13K/Y106W | Nonmotile | CW bias |
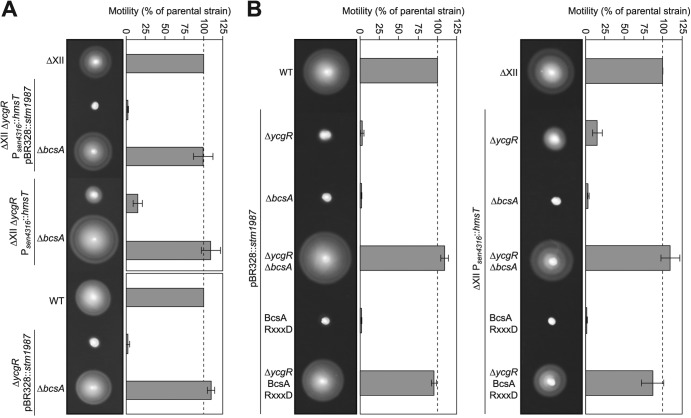
Inhibition of motility under high-c-di-GMP conditions and in the absence of ycgR is reversed by mutations in the bcsABZC operon.
To find out the genetic element involved in the negative regulation of flagellum function under elevated c-di-GMP cytoplasmic levels independent of YcgR, we followed a strategy, described elsewhere (59), which consists in the isolation of spontaneous motile suppressors, defective in the potential target of c-di-GMP and thus able to move as does the wild-type strain. For that, we again made use of strain ΔXII and constructed a derivative strain that harbors two unique sources of c-di-GMP and a mutation in ycgR. The resulting strain (ΔXII ΔycgR Psen4316::hmsT pBR328::stm1987) expressed the hmsT gene from the chromosome and stm1987 from a plasmid. This strain was completely nonmotile on swimming plates (Fig. 4A) and did not rotate in any direction. The double source of c-di-GMP prevented isolation of false suppressor mutations in hmsT or stm1987. This genetic approach, fully described in Materials and Methods, allowed us to isolate three different mutants in which blockage of motility was suppressed. The insertion site of the MudJ transposon was identified by sequencing to be the bcsC locus in two of the suppressor mutants and the bcsB locus in the other mutant. Since these two genes belong to one of the bcs operons (bcsABZC), involved in cellulose synthesis, and the BcsA protein is a direct target of c-di-GMP, we suspected that restoration of motility might actually be due to transposon insertion into these loci instead of cotransduction of spontaneous mutations. To test this, we mutated bcsA in strain ΔXII ΔycgR Psen4316::hmsT pBR328::stm1987. The mutation completely restored motility to the levels for ΔXII (Fig. 4A). Transduction of the bcsA mutation into the initial working strains, the ΔycgR pBR328::stm1987 strain and ΔXII ΔycgR Psen4316::hmsT, also led to a total motility recuperation (Fig. 4A) and to normal rotational behavior (CCW with reversals), confirming that mutation of genes responsible for cellulose synthesis suppresses motility blockage of ycgR mutants that present high levels of c-di-GMP.
Fig 4.
C-di-GMP binding to BcsA is responsible for motility inhibition in the absence of YcgR. Representative swimming motility plates and quantitative measurement of motility after incubation at 23°C for 16 h are shown. (A) Swimming motility of a strain that expresses two unique sources of c-di-GMP and presents a mutation in ycgR was completely rescued by means of an additional mutation of the cellulose synthase-encoding gene, bcsA. The same result was obtained when the bcsA mutation was transduced to a ycgR single mutant overexpressing stm1987 and to ΔXII strain containing a ycgR mutation and overexpressing hmsT. (B) Deletion of both ycgR and bcsA in strains that present high levels of c-di-GMP is needed to recuperate swimming behavior. Restoration of motility is also achieved by mutating the c-di-GMP binding motif of the PilZ domain of BcsA.
Next, we asked whether a bcs mutation was sufficient to recuperate Salmonella motility in cells with an elevated c-di-GMP content and harboring a wild-type allele of ycgR. For this, we transduced the bcsA mutation into WT pBR328::stm1987 and ΔXII Psen4316::hmsT cells and tested swimming and microscopic rotational behavior. Resulting cells were incapable of swimming (Fig. 4B) and showed a CCW bias, indicating that both bcs and ycgR mutations are needed to recuperate wild-type motility and suggesting that targeting of very high c-di-GMP levels to one of its effectors, YcgR or BcsA, is enough to inhibit Salmonella motility. To provide support for the role of BcsA in motility inhibition and to discard polar effects of the bcsA mutation on other genes of the operon, a point mutation of the c-di-GMP binding motif (RxxxR) present in the PilZ domain of BcsA was generated. The ΔycgR BcsA RxxxD pBR328::stm1987 and ΔXII ΔycgR BcsA RxxxD Psen4316::hmsT strains recuperated swimming motility (Fig. 4B), demonstrating that BcsA activation by c-di-GMP binding is needed for flagellar motility inactivation.
Early cellulose synthesis as a consequence of high c-di-GMP cellular levels is responsible for motility blockage.
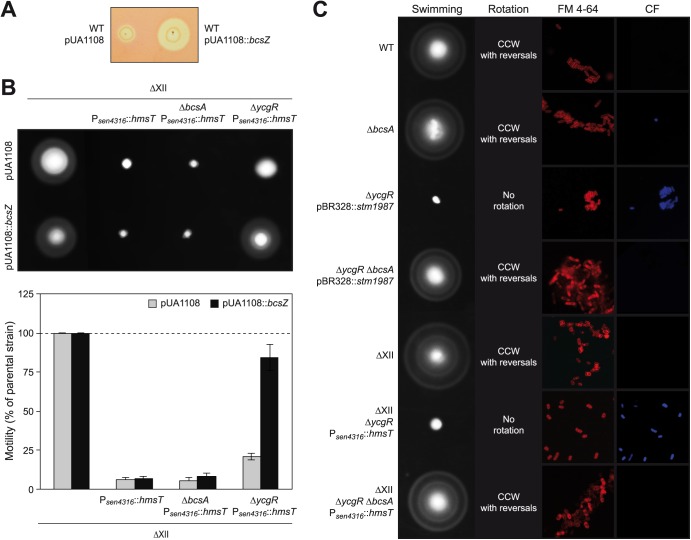
The role of the cellulose synthase BcsA in inhibition of motility might be either direct, through the inactivation of the flagellar rotor by an unknown mechanism, or indirect, by means of the synthesis of cellulose, which might be responsible per se for the motility blockage. An example of a glycosyltransferase that has a direct effect on motility is EpsE from Bacillus subtilis, which acts as a flagellar clutch through its interaction with the flagellar rotor (80). To assess the level at which BcsA inhibits motility, we evaluated whether an ectopic degradation of cellulose was able to restore swimming motility, which would indicate that cellulose is the element that blocks Salmonella motility. To achieve cellulose degradation, the putative endoglucanase gene bcsZ, present in one of the bcs operons, was cloned in a plasmid under a strong inducible promoter. We first validated the use of this plasmid by analyzing endoglucanase activity of BcsZ on carboxymethylcellulose (CMC)- and IPTG-containing plates (Fig. 5A). A CMC degradation halo was observed surrounding the wild-type strain containing plasmid pUA1108::bcsZ. This plasmid was then introduced into the strains ΔXII, ΔXII Psen4316::hmsT, ΔXII ΔbcsA Psen4316::hmsT, and ΔXII ΔycgR Psen4316::hmsT, and swimming motility was examined. Degradation of cellulose synthesized by strain ΔXII Psen4316::hmsT was not sufficient to recuperate motility function, suggesting again that binding of very high levels of c-di-GMP to one of its effectors, in this case YcgR, is enough to block Salmonella motility. On the other hand, hydrolysis of cellulose synthesized by a strain containing high levels of c-di-GMP but lacking YcgR, ΔXII ΔycgR Psen4316::hmsT, relieved motility impairment (Fig. 5B). Thus, this result implicated cellulose in immobilization of flagella.
Fig 5.
A premature cellulose production that has taken place as a consequence of high intracellular c-di-GMP levels in the cell is responsible for motility inhibition. (A) Endoglucanase activity of BcsZ was confirmed by assessing carboxymethylcellulose (CMC)-degrading activity of the wild-type S. Enteritidis strain harboring the bcsZ-overexpressing plasmid pUA1108::bcsZ. As a control, the endoglucanase phenotype of the wild-type strain harboring an empty pUA1108 overexpression vector is also presented. (B) Degradation of the cellulose produced by a strain that overexpresses a unique and very active source of c-di-GMP and lacks YcgR is enough to recuperate swimming motility. Strains assayed were transformed with an empty pUA1108 plasmid or with the overexpressing plasmid pUA1108::bcsZ. Representative swimming motility plates and quantitative measurement of motility after incubation at 23°C for 16 h are shown. (C) Correlation between swimming motility, rotation behavior in the tethering assay, and cellulose production. Early cellulose synthesis was detected in strains that overexpressed a DGC and were grown under tethering assay conditions, that is, at 28°C for 6 h. Detection of cellulose production by calcofluor staining (CF) is shown in the right panel. Membrane staining with FM4-64 is shown in the intermediate panel.
If cellulose matrix per se is somehow impeding flagellar rotation, then this exopolysaccharide should be surrounding the cells that presented a null rotation behavior during the tethering assay. To confirm this assumption, bacteria grown under the same conditions used for tethering experiments were stained with calcofluor, a dye that stains cellulose, and also cell membranes were stained with the probe FM4-64. We could observe cellulose surrounding the totality of ΔycgR bacteria that express a very active source of c-di-GMP, the ΔycgR pBR328::stm1987 and ΔXII ΔycgR Psen4316::hmsT strains, while no fluorescence was detected around the wild-type strain and negative controls of cellulose production, that is, the ΔXII and ΔbcsA strains (Fig. 5C). Overall, these results indicate that an early cellulose production achieved as a consequence of high c-di-GMP cellular levels can inhibit Salmonella motility by impeding flagellar rotation.
Inactivation of cellulose production totally recuperates the motility defect of a double ΔyhjH ΔycgR mutant.
As stated previously, recent reports have revealed that under the presence of increased levels of c-di-GMP achieved by the absence of the phosphodiesterase YhjH, E. coli and S. Typhimurium cells show a defect in swimming motility that can be rescued, although not completely, by deleting ycgR (22, 59–62). To determine whether in these cases cellulose is also responsible for the partial motility deficiency of ΔyhjH ΔycgR double mutants, we first mutated bcsA in our S. Enteritidis 3934 ΔyhjH ΔycgR strain. The resultant cells wholly recuperated wild-type motility. Then, a single ΔyhjH mutant, double ΔyhjH ΔycgR and ΔyhjH ΔbcsA mutants, and a triple ΔyhjH ΔycgR ΔbcsA mutant were constructed in S. Typhimurium 14028 and S. Typhimurium UMR1, two of the strains that have been used in the reports mentioned above. Swimming behavior resembled all the results obtained with the S. Enteritidis strain, confirming that under high c-di-GMP conditions both YcgR and cellulose synthesized by BcsA contribute to the inhibition of Salmonella motility (Fig. 6).
Fig 6.
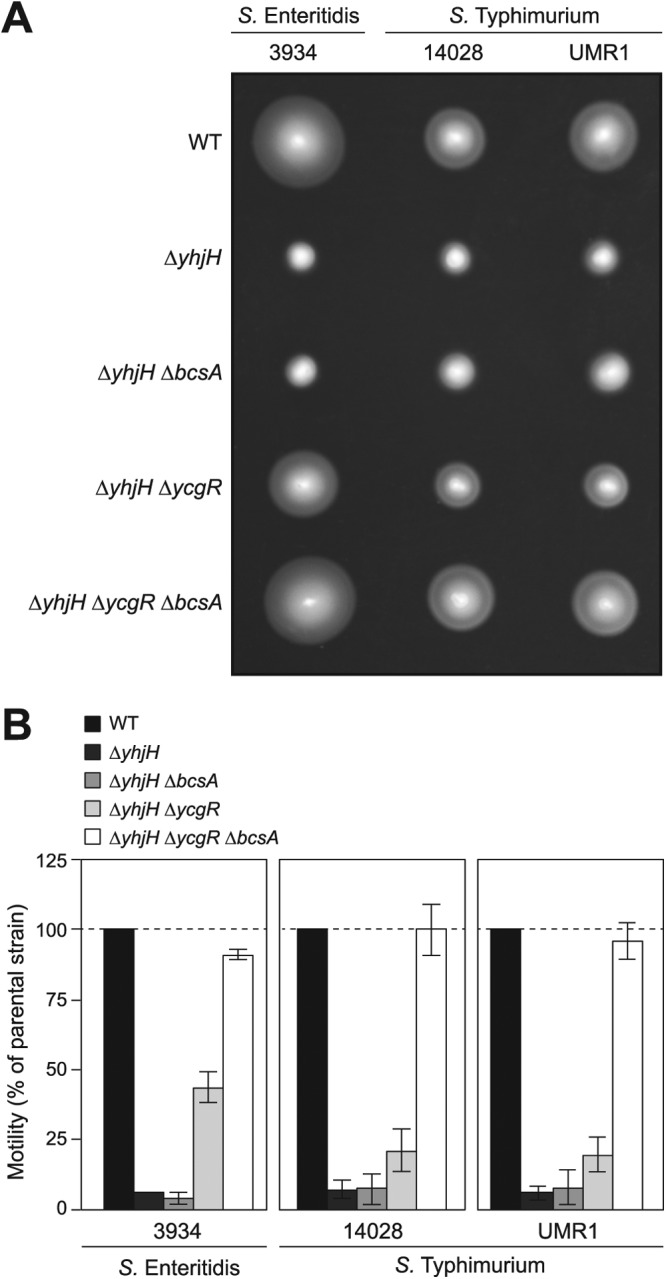
The motility defect of double yhjH and ycgR mutants is rescued by inactivation of cellulose production. (A) Representative swimming motility plates after incubation at 23°C for 16 h. (B) Quantitative measurement of motility. The total area of growth was measured, and the percent motility relative to that of the parental strain was calculated. Means and standard deviations of results from three repeats on three separate days are shown. Inactivation of ycgR is able to substantially but not totally restore the bacterial motility defect of S. Enteritidis or S. Typhimurium ΔyhjH mutants. An additional mutation of the cellulose synthase-encoding gene, bcsA, leads to a total recuperation of wild-type motility in both Salmonella species.
DISCUSSION
Salmonella follows a cyclic lifestyle in which host colonization is alternated with periods of survival outside the host. A major factor that contributes to this way of life is the capacity of Salmonella to form biofilms. Salmonella biofilms, encountered on many biotic and abiotic surfaces, are more resistant to several environmental stress factors and likely contribute to survival in nonhost environments, transmission to new hosts, and establishment and persistence of infections in appropriate hosts (reviewed in reference 81). A major role of the bacterial second messenger c-di-GMP is precisely the regulation of the transition from planktonic growth to biofilm formation, whereby high c-di-GMP levels promote the production of adhesins and exopolysaccharides of the biofilm extracellular matrix and inhibit motility (8, 82). In this study, we have shown that cellulose, a β-1-4-d-glucose polymer encoded by the bcsABZC and bcsEFG operons, is not only a main component of the biofilm extracellular matrix produced by Salmonella under different environmental conditions but also plays an additional role in Salmonella lifestyle switching as a blocking agent of motility through the impairment of flagellar rotation.
The aim of the present work, that is, the identification of the c-di-GMP effector that inhibits Salmonella motility in the absence of YcgR, was initially considered taking into account previous studies that observed that inactivation of the ycgR gene only partially reversed the immotile phenotype of strains containing elevated intracellular c-di-GMP levels (22, 60–62). These high c-di-GMP levels were accounted by deleting the yhjH gene, encoding a PDE. We first confirmed these observations with S. Enteritidis and then analyzed the effects of overexpressing other sources of c-di-GMP in the absence of YcgR. For this, on one hand, we complemented a ΔycgR mutant with stm1987, which encodes a very active DGC of S. Typhimurium, and on the other, we overexpressed the hmsT gene of Y. pestis from the chromosome of a ΔycgR mutant carrying mutations in all genes encoding GGDEF proteins. This way, high production of this protein was achieved, and effects on motility could be analyzed in a strain with a sole and heterologous source of c-di-GMP. High HmsT expression levels can be explained because the hmsT sequence inserted in ΔXII comprised a fragment from the translation start site to the stop codon and thus was not subjected to its known transcriptional and posttranscriptional regulation (83, 84). Overexpression of stm1987 or hmsT resulted in bacteria that could not rotate at all and thus were unable to swim. C-di-GMP can inhibit motility by affecting several mechanisms that include the transcription, translation, and assembly of flagella (reviewed in reference 75). As regards c-di-GMP effectors and in addition to YcgR posttranslational regulation of flagellar motility, two transcription factors, VpsT of V. cholerae and FleQ of P. aeruginosa, have been described to control flagellum gene expression upon c-di-GMP binding (29, 30). However, our work revealed that in Salmonella, high c-di-GMP intracellular levels do not influence any of the aforementioned processes, except the YcgR-related mechanism. These results stand in opposition to the ones described by Lamprokostopoulou et al., which showed that high c-di-GMP concentrations in S. Typhimurium promote the presence of cell-associated flagellin but inhibit secretion of monomeric flagellin into the culture supernatant (85). One possible explanation for this is that these authors analyzed flagellin levels in strains grown in an invasion-inducing environment, and thus, other regulatory elements might come into action that might not be present under our experimental conditions.
In order to find out the element responsible for inhibition of motility in the absence of YcgR, we carried out a spontaneous mutagenesis that resulted in the identification of cellulose synthesis-encoding genes. In support of the implication of cellulose per se in flagellar rotation impairment, we firstly showed that double mutants in ycgR and different genes of the bcsABZC operon recuperated wild-type swimming behavior. Second, mutation of the conserved PilZ domain residue RxxxR of BcsA, implicated in c-di-GMP binding (22) and critical for cellulose synthase activity, restored the motility of ΔycgR mutants. Third, degradation of cellulose by the BcsZ endoglucanase caused the same effect. Last, cellulose could be visualized surrounding the cells whose flagella could not rotate at all during the tethering assay. It is important to note that the cellulase activity of the putative endoglucanase-encoding gene bcsZ had not been reported until now. Thus, our findings demonstrate that the overexpression of active DGCs results in a very premature cellulose production that impedes flagellar rotation and thus swimming motility even in the absence of YcgR.
In a very recent study, Chen et al. demonstrated that mutation of yuxH, a gene encoding an EAL protein of B. subtilis, leads to a defect in motility that can be reversed by an additional mutation of ypfA, a gene encoding a putative c-di-GMP receptor containing a PilZ domain. Interestingly, motility restoration was substantial but not total (86). This observation resembles motility control in Salmonella, and according to our results, this analogy provides evidence for a putative role of B. subtilis biofilm matrix components in motility inhibition.
It is noteworthy that deletion of ycgR in strains overexpressing stm1987 did not relieve motility impairment at all. In the case of strains overexpressing hmsT, the ycgR deletion only slightly rescued swimming motility. This exacerbated effect on flagellar rotation and swimming can be explained because cells overexpressing any of these two DGCs supposedly contain very high levels of c-di-GMP, greater than the ones present in a yhjH mutant. As a consequence, production of cellulose is triggered, which obstructs flagellar rotation without the need of YcgR interaction with flagellar motor proteins. In a contrary situation, when these two active DGCs were expressed in a ycgR+ strain but unable to synthesize cellulose, motility was also completely inactivated. These results indicate that when the bacterial cytoplasm is overloaded with c-di-GMP, binding of the nucleotide to any of its PilZ domain-containing effectors, YcgR or BcsA, is enough to inhibit Salmonella motility. We hypothesize that in a wild-type background and during biofilm development, c-di-GMP orchestrates the Salmonella motile-to-sessile transition by means of the activity of these two receptors. YcgR, which is expressed at the post-exponential growth phase (3), would be used first in this transition, and then cellulose accumulation would ensure that in a Salmonella biofilm, flagella stay paralyzed but ready to rotate again upon degradation of the exopolysaccharide. In this regard, the bcs operon has been shown to be transcriptionally regulated in the phytopathogenic bacterium Dickeya dadantii (87) and Burkholderia cenocepacia (33) but appears to be transcribed constitutively in Salmonella (55).
Polysaccharides are a major fraction of the EPS (extracellular polymeric substances) matrix that is secreted by biofilm cells and play several roles in the architecture of the biofilm, such as allowing adhesion to abiotic and biotic surfaces, enabling aggregation of bacterial cells, mediating the cohesion of biofilms, retaining water and maintaining a highly hydrated microenvironment, conferring a protective barrier, promoting the sorption of organic and inorganic compounds, providing a source of nutrients for utilization by the biofilm community, storing excess carbon, and allowing the accumulation, retention, and stabilization of enzymes (reviewed in reference 88). Here we have provided evidence for an additional role of the polysaccharide cellulose as a blocking agent of bacterial motility that would maintain bacterial cells immotile inside a biofilm, but still a question remains opened as to the exact mechanism by which cellulose impedes flagellar rotation. We hypothesize that cellulose accumulation outside the cell might sterically hinder rotation of flagella, but we cannot rule out the possibility that an unknown molecule might sense the cellulose produced and then somehow stop flagellar rotation. This issue will have to be elucidated in future studies.
In conclusion, the mechanisms of action of c-di-GMP as a lifestyle switch regulator that have been hitherto established are further broadened by our results, which indicate the existence of a double checkpoint involving YcgR and cellulose that ensures motility inhibition once a sessile lifestyle has been adopted, making the two processes mutually exclusive.
Supplementary Material
ACKNOWLEDGMENTS
We express our gratitude to Carlos Gamazo for providing the anti-FliC antibodies.
J.V. was supported by a Ramón y Cajal contract from the Ministerio de Economía y Competitividad, Spain. A JAE predoctoral research contract for V.Z. from the Consejo Superior de Investigaciones Científicas (CSIC, Spain) is gratefully acknowledged. This research was supported by a grant from the Departamento de Salud (Resolución 1312/2010), Gobierno de Navarra, Spain.
Footnotes
Published ahead of print 16 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01789-12.
REFERENCES
- 1. Ross P, Weinhouse H, Aloni Y, Michaeli D, Weinberger-Ohana P, Mayer R, Braun S, de Vroom E, van der Marel GA, van Boom JH, Benziman M. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279–281 [DOI] [PubMed] [Google Scholar]
- 2. Sondermann H, Shikuma NJ, Yildiz FH. 2012. You've come a long way: c-di-GMP signaling. Curr. Opin. Microbiol. 15:140–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Povolotsky TL, Hengge R. 2012. “Life-style” control networks in Escherichia coli: signaling by the second messenger c-di-GMP. J. Biotechnol. 160:10–16 [DOI] [PubMed] [Google Scholar]
- 4. Mills E, Pultz IS, Kulasekara HD, Miller SI. 2011. The bacterial second messenger c-di-GMP: mechanisms of signalling. Cell. Microbiol. 13:1122–1129 [DOI] [PubMed] [Google Scholar]
- 5. Römling U. 2012. Cyclic di-GMP, an established secondary messenger still speeding up. Environ. Microbiol. 14:1817–1829 [DOI] [PubMed] [Google Scholar]
- 6. Hengge R. 2009. Principles of c-di-GMP signalling in bacteria. Nat. Rev. Microbiol. 7:263–273 [DOI] [PubMed] [Google Scholar]
- 7. Jenal U, Malone J. 2006. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 40:385–407 [DOI] [PubMed] [Google Scholar]
- 8. Simm R, Morr M, Kader A, Nimtz M, Römling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123–1134 [DOI] [PubMed] [Google Scholar]
- 9. Römling U, Gomelsky M, Galperin MY. 2005. c-di-GMP: the dawning of a novel bacterial signalling system. Mol. Microbiol. 57:629–639 [DOI] [PubMed] [Google Scholar]
- 10. Tal R, Wong HC, Calhoon R, Gelfand D, Fear AL, Volman G, Mayer R, Ross P, Amikam D, Weinhouse H, Cohen A, Sapir S, Ohana P, Benziman M. 1998. Three cdg operons control cellular turnover of cyclic di-GMP in Acetobacter xylinum: genetic organization and occurrence of conserved domains in isoenzymes. J. Bacteriol. 180:4416–4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Paul R, Weiser S, Amiot NC, Chan C, Schirmer T, Giese B, Jenal U. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ryjenkov DA, Tarutina M, Moskvin OV, Gomelsky M. 2005. Cyclic diguanylate is a ubiquitous signaling molecule in bacteria: insights into biochemistry of the GGDEF protein domain. J. Bacteriol. 187:1792–1798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schmidt AJ, Ryjenkov DA, Gomelsky M. 2005. The ubiquitous protein domain EAL is a cyclic diguanylate-specific phosphodiesterase: enzymatically active and inactive EAL domains. J. Bacteriol. 187:4774–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Christen M, Christen B, Folcher M, Schauerte A, Jenal U. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 280:30829–30837 [DOI] [PubMed] [Google Scholar]
- 15. Tamayo R, Tischler AD, Camilli A. 2005. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 280:33324–33330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ryan RP, Fouhy Y, Lucey JF, Crossman LC, Spiro S, He Y-W, Zhang LH, Heeb S, Cámara M, Williams P, Dow JM. 2006. Cell-cell signaling in Xanthomonas campestris involves an HD-GYP domain protein that functions in cyclic di-GMP turnover. Proc. Natl. Acad. Sci. U. S. A. 103:6712–6717 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17. Chan C, Paul R, Samoray D, Amiot NC, Giese B, Jenal U, Schirmer T. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. U. S. A. 101:17084–17089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schirmer T, Jenal U. 2009. Structural and mechanistic determinants of c-di-GMP signalling. Nat. Rev. Microbiol. 7:724–735 [DOI] [PubMed] [Google Scholar]
- 19. Christen B, Christen M, Paul R, Schmid F, Folcher M, Jenoe P, Meuwly M, Jenal U. 2006. Allosteric control of cyclic di-GMP signaling. J. Biol. Chem. 281:32015–32024 [DOI] [PubMed] [Google Scholar]
- 20. Tamayo R, Pratt JT, Camilli A. 2007. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 61:131–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cotter PA, Stibitz S. 2007. c-di-GMP-mediated regulation of virulence and biofilm formation. Curr. Opin. Microbiol. 10:17–23 [DOI] [PubMed] [Google Scholar]
- 22. Ryjenkov DA, Simm R, Römling U, Gomelsky M. 2006. The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 281:30310–30314 [DOI] [PubMed] [Google Scholar]
- 23. Petters T, Zhang X, Nesper J, Treuner-Lange A, Gomez-Santos N, Hoppert M, Jenal U, Søgaard-Andersen L. 2012. The orphan histidine protein kinase SgmT is a c-di-GMP receptor and regulates composition of the extracellular matrix together with the orphan DNA binding response regulator DigR in Myxococcus xanthus. Mol. Microbiol. 84:147–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Duerig A, Abel S, Folcher M, Nicollier M, Schwede T, Amiot N, Giese B, Jenal U. 2009. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 23:93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Newell PD, Monds RD, O'Toole GA. 2009. LapD is a bis-(3, ″5″)-cyclic dimeric GMP-binding protein that regulates surface attachment by Pseudomonas fluorescens Pf0-1. Proc. Natl. Acad. Sci. U. S. A. 106:3461–3466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qi Y, Chuah MLC, Dong X, Xie K, Luo Z, Tang K, Liang Z-X. 2011. Binding of cyclic diguanylate in the non-catalytic EAL domain of FimX induces a long-range conformational change. J. Biol. Chem. 286:2910–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Navarro MVAS, De N, Bae N, Wang Q, Sondermann H. 2009. Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure 17:1104–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee VT, Matewish JM, Kessler JL, Hyodo M, Hayakawa Y, Lory S. 2007. A cyclic-di-GMP receptor required for bacterial exopolysaccharide production. Mol. Microbiol. 65:1474–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hickman JW, Harwood CS. 2008. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol. Microbiol. 69:376–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krasteva PV, Fong JCN, Shikuma NJ, Beyhan S, Navarro MVAS, Yildiz FH, Sondermann H. 2010. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327:866–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ferreira RBR, Chodur DM, Antunes LCM, Trimble MJ, McCarter LL. 2012. Output targets and transcriptional regulation by a cyclic dimeric GMP-responsive circuit in the Vibrio parahaemolyticus Scr network. J. Bacteriol. 194:914–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Srivastava D, Harris RC, Waters CM. 2011. Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J. Bacteriol. 193:6331–6341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fazli M, O'Connell A, Nilsson M, Niehaus K, Dow JM, Givskov M, Ryan RP, Tolker-Nielsen T. 2011. The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol. Microbiol. 82:327–341 [DOI] [PubMed] [Google Scholar]
- 34. Tao F, He YW, Wu DH, Swarup S, Zhang LH. 2010. The cyclic nucleotide monophosphate domain of Xanthomonas campestris global regulator Clp defines a new class of cyclic di-GMP effectors. J. Bacteriol. 192:1020–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Leduc JL, Roberts GP. 2009. Cyclic di-GMP allosterically inhibits the CRP-like protein (Clp) of Xanthomonas axonopodis pv. citri. J. Bacteriol. 191:7121–7122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chin K-H, Lee Y-C, Tu Z-L, Chen C-H, Tseng Y-H, Yang J-M, Ryan RP, McCarthy Y, Dow JM, Wang AHJ, Chou S-H. 2010. The cAMP receptor-like protein CLP is a novel c-di-GMP receptor linking cell-cell signaling to virulence gene expression in Xanthomonas campestris. J. Mol. Biol. 396:646–662 [DOI] [PubMed] [Google Scholar]
- 37. Tuckerman JR, Gonzalez G, Gilles-Gonzalez M-A. 2011. Cyclic di-GMP activation of polynucleotide phosphorylase signal-dependent RNA processing. J. Mol. Biol. 407:633–639 [DOI] [PubMed] [Google Scholar]
- 38. Smith KD, Shanahan CA, Moore EL, Simon AC, Strobel SA. 2011. Structural basis of differential ligand recognition by two classes of bis-(3′-5′)-cyclic dimeric guanosine monophosphate-binding riboswitches. Proc. Natl. Acad. Sci. U. S. A. 108:7757–7762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee ER, Baker JL, Weinberg Z, Sudarsan N, Breaker RR. 2010. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science 329:845–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. 2008. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321:411–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. 2009. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat. Struct. Mol. Biol. 16:1218–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Amikam D, Galperin MY. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6 [DOI] [PubMed] [Google Scholar]
- 43. Christen M, Christen B, Allan MG, Folcher M, Jenö P, Grzesiek S, Jenal U. 2007. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 104:4112–4117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pratt JT, Tamayo R, Tischler AD, Camilli A. 2007. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 282:12860–12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ramelot TA, Yee A, Cort JR, Semesi A, Arrowsmith CH, Kennedy MA. 2007. NMR structure and binding studies confirm that PA4608 from Pseudomonas aeruginosa is a PilZ domain and a c-di-GMP binding protein. Proteins 66:266–271 [DOI] [PubMed] [Google Scholar]
- 46. Merighi M, Lee VT, Hyodo M, Hayakawa Y, Lory S. 2007. The second messenger bis-(3′-5′)-cyclic-GMP and its PilZ domain-containing receptor Alg44 are required for alginate biosynthesis in Pseudomonas aeruginosa. Mol. Microbiol. 65:876–895 [DOI] [PubMed] [Google Scholar]
- 47. Freedman JC, Rogers EA, Kostick JL, Zhang H, Iyer R, Schwartz I, Marconi RT. 2010. Identification and molecular characterization of a cyclic-di-GMP effector protein, PlzA (BB0733): additional evidence for the existence of a functional cyclic-di-GMP regulatory network in the Lyme disease spirochete, Borrelia burgdorferi. FEMS Immunol. Med. Microbiol. 58:285–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Benach J, Swaminathan SS, Tamayo R, Handelman SK, Folta-Stogniew E, Ramos JE, Forouhar F, Neely H, Seetharaman J, Camilli A, Hunt JF. 2007. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 26:5153–5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Habazettl J, Allan MG, Jenal U, Grzesiek S. 2011. Solution structure of the PilZ domain protein PA4608 complex with cyclic di-GMP identifies charge clustering as molecular readout. J. Biol. Chem. 286:14304–14314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ko J, Ryu KS, Kim H, Shin JS, Lee JO, Cheong C, Choi BS. 2010. Structure of PP4397 reveals the molecular basis for different c-di-GMP binding modes by Pilz domain proteins. J. Mol. Biol. 398:97–110 [DOI] [PubMed] [Google Scholar]
- 51. Pitzer JE, Sultan SZ, Hayakawa Y, Hobbs G, Miller MR, Motaleb MA. 2011. Analysis of the Borrelia burgdorferi cyclic-di-GMP-binding protein PlzA reveals a role in motility and virulence. Infect. Immun. 79:1815–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wilksch JJ, Yang J, Clements A, Gabbe JL, Short KR, Cao H, Cavaliere R, James CE, Whitchurch CB, Schembri MA, Chuah MLC, Liang Z-X, Wijburg OL, Jenney AW, Lithgow T, Strugnell RA. 2011. MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 7:e1002204 doi:10.1371/journal.ppat.1002204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McCarthy Y, Ryan RP, O'Donovan K, He Y-Q, Jiang B-L, Feng J-X, Tang J-L, Dow JM. 2008. The role of PilZ domain proteins in the virulence of Xanthomonas campestris pv. campestris. Mol. Plant Pathol. 9:819–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Solano C, García B, Valle J, Berasain C, Ghigo Gamazo J-MC, Lasa I. 2002. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol. Microbiol. 43:793–808 [DOI] [PubMed] [Google Scholar]
- 55. Zogaj X, Nimtz M, Rohde M, Bokranz W, Romling U. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452–1463 [DOI] [PubMed] [Google Scholar]
- 56. García B, Latasa C, Solano C, García-del Portillo F, Gamazo C, Lasa I. 2004. Role of the GGDEF protein family in Salmonella cellulose biosynthesis and biofilm formation. Mol. Microbiol. 54:264–277 [DOI] [PubMed] [Google Scholar]
- 57. Ledeboer NA, Jones BD. 2005. Exopolysaccharide sugars contribute to biofilm formation by Salmonella enterica serovar Typhimurium on HEp-2 cells and chicken intestinal epithelium. J. Bacteriol. 187:3214–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Prouty AM, Gunn JS. 2003. Comparative analysis of Salmonella enterica serovar Typhimurium biofilm formation on gallstones and on glass. Infect. Immun. 71:7154–7158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Boehm A, Kaiser M, Li H, Spangler C, Kasper CA, Ackermann M, Kaever V, Sourjik V, Roth V, Jenal U. 2010. Second messenger-mediated adjustment of bacterial swimming velocity. Cell 141:107–116 [DOI] [PubMed] [Google Scholar]
- 60. Fang X, Gomelsky M. 2010. A post-translational, c-di-GMP-dependent mechanism regulating flagellar motility. Mol. Microbiol. 76:1295–1305 [DOI] [PubMed] [Google Scholar]
- 61. Paul K, Nieto V, Carlquist WC, Blair DF, Harshey RM. 2010. The c-di-GMP binding protein YcgR controls flagellar motor direction and speed to affect chemotaxis by a “backstop brake” mechanism. Mol. Cell 38:128–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Girgis HS, Liu Y, Ryu WS, Tavazoie S. 2007. A comprehensive genetic characterization of bacterial motility. PLoS Genet. 3:1644–1660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schmieger H. 1972. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119:75–88 [DOI] [PubMed] [Google Scholar]
- 64. Maloy SR, Nunn WD. 1981. Selection for loss of tetracycline resistance by Escherichia coli. J. Bacteriol. 145:1110–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chaveroche MK, Ghigo JM, d'Enfert C. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97 doi:10.1093/nar/28.22.e97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Blank K, Hensel M, Gerlach RG. 2011. Rapid and highly efficient method for scarless mutagenesis within the Salmonella enterica chromosome. PLoS One 6:e15763 doi:10.1371/journal.pone.0015763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Solano C, GarcíA B, Latasa C, Toledo-Arana A, Zorraquino V, Valle J, Casals J, Pedroso E, Lasa I. 2009. Genetic reductionist approach for dissecting individual roles of GGDEF proteins within the c-di-GMP signaling network in Salmonella. Proc. Natl. Acad. Sci. U. S. A. 106:7997–8002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cano DA, Martinez-Moya M, Pucciarelli MG, Groisman EA, Casadesus J, Garcia-del Portillo F. 2001. Salmonella enterica serovar Typhimurium response involved in attenuation of pathogen intracellular proliferation. Infect. Immun. 69:6463–6474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Alon U, Camarena L, Surette MG, Aguera Y, Arcas B, Liu Y, Leibler S, Stock JB. 1998. Response regulator output in bacterial chemotaxis. EMBO J. 17:4238–4248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Grantcharova N, Peters V, Monteiro C, Zakikhany K, Römling U. 2010. Bistable expression of CsgD in biofilm development of Salmonella enterica serovar typhimurium. J. Bacteriol. 192:456–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Medina-Ruiz L, Campoy S, Latasa C, Cardenas P, Alonso JC, Barbé J. 2010. Overexpression of the recA gene decreases oral but not intraperitoneal fitness of Salmonella enterica. Infect. Immun. 78:3217–3225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Teather RM, Wood PJ. 1982. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl. Environ. Microbiol. 43:777–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Perry RD, Bobrov AG, Kirillina O, Jones HA, Pedersen L, Abney J, Fetherston JD. 2004. Temperature regulation of the hemin storage (Hms+) phenotype of Yersinia pestis is posttranscriptional. J. Bacteriol. 186:1638–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Simm R, Fetherston JD, Kader A, Römling U, Perry RD. 2005. Phenotypic convergence mediated by GGDEF-domain-containing proteins. J. Bacteriol. 187:6816–6823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wolfe AJ, Visick KL. 2007. Get the message out: cyclic-di-GMP regulates multiple levels of flagellum-based motility. J. Bacteriol. 190:463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ibarra JA, Knodler LA, Sturdevant DE, Virtaneva K, Carmody AB, Fischer ER, Porcella SF, Steele-Mortimer O. 2010. Induction of Salmonella pathogenicity island 1 under different growth conditions can affect Salmonella-host cell interactions in vitro. Microbiology 156:1120–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Welch M, Oosawa K, Aizawa S, Eisenbach M. 1993. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc. Natl. Acad. Sci. U. S. A. 90:8787–8791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Bourret RB, Hess JF, Simon MI. 1990. Conserved aspartate residues and phosphorylation in signal transduction by the chemotaxis protein CheY. Proc. Natl. Acad. Sci. U. S. A. 87:41–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Dyer CM, Quillin ML, Campos A, Lu J, McEvoy MM, Hausrath AC, Westbrook EM, Matsumura P, Matthews BW, Dahlquist FW. 2004. Structure of the constitutively active double mutant CheYD13K Y106W alone and in complex with a FliM peptide. J. Mol. Biol. 342:1325–1335 [DOI] [PubMed] [Google Scholar]
- 80. Guttenplan SB, Blair KM, Kearns DB. 2010. The EpsE flagellar clutch is bifunctional and synergizes with EPS biosynthesis to promote Bacillus subtilis biofilm formation. PLoS Genet. 6:e1001243 doi:10.1371/journal.pgen.1001243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Steenackers H, Hermans K, Vanderleyden J, de Keersmaecker SC. 2012. Salmonella biofilms: an overview on occurrence, structure, regulation and eradication. Food Res. Int. 45:502–531 [Google Scholar]
- 82. Römling U, Amikam D. 2006. Cyclic di-GMP as a second messenger. Curr. Opin. Microbiol. 9:218–228 [DOI] [PubMed] [Google Scholar]
- 83. Sun Y-C, Guo X-P, Hinnebusch BJ, Darby C. 2012. The Yersinia pestis Rcs phosphorelay inhibits biofilm formation by repressing transcription of the diguanylate cyclase gene hmsT. J. Bacteriol. 194:2020–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Bellows LE, Koestler BJ, Karaba SM, Waters CM, Lathem WW. 2012. Hfq-dependent, co-ordinate control of cyclic diguanylate synthesis and catabolism in the plague pathogen Yersinia pestis. Mol. Microbiol. 86:661–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lamprokostopoulou A, Monteiro C, Rhen M, Römling U. 2010. Cyclic di-GMP signalling controls virulence properties of Salmonella enterica serovar Typhimurium at the mucosal lining. Environ. Microbiol. 12:40–53 [DOI] [PubMed] [Google Scholar]
- 86. Chen Y, Chai Y, Guo Losick J-HR. 2012. Evidence for cyclic di-GMP-mediated signaling in Bacillus subtilis. J. Bacteriol. 194:5080–5090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Prigent-Combaret C, Zghidi-Abouzid O, Effantin G, Lejeune P, Reverchon S, Nasser W. 2012. The nucleoid-associated protein Fis directly modulates the synthesis of cellulose, an essential component of pellicle-biofilms in the phytopathogenic bacterium Dickeya dadantii. Mol. Microbiol. 86:172–186 [DOI] [PubMed] [Google Scholar]
- 88. Flemming H-C, Wingender J. 2010. The biofilm matrix. Nat. Rev. Microbiol. 8:623–633 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.