Abstract
Brucella spp. and Sinorhizobium meliloti are alphaproteobacteria that share not only an intracellular lifestyle in their respective hosts, but also a crucial requirement for cell envelope components and their timely regulation for a successful infectious cycle. Here, we report the characterization of Brucella melitensis mucR, which encodes a zinc finger transcriptional regulator that has previously been shown to be involved in cellular and mouse infections at early time points. MucR modulates the surface properties of the bacteria and their resistance to environmental stresses (i.e., oxidative stress, cationic peptide, and detergents). We show that B. melitensis mucR is a functional orthologue of S. meliloti mucR, because it was able to restore the production of succinoglycan in an S. meliloti mucR mutant, as detected by calcofluor staining. Similar to S. meliloti MucR, B. melitensis MucR also represses its own transcription and flagellar gene expression via the flagellar master regulator ftcR. More surprisingly, we demonstrate that MucR regulates a lipid A core modification in B. melitensis. These changes could account for the attenuated virulence of a mucR mutant. These data reinforce the idea that there is a common conserved circuitry between plant symbionts and animal pathogens that regulates the relationship they have with their hosts.
INTRODUCTION
The bacterial envelope is a bacterium's first point of contact with its challenging environment, and in the case of symbionts or pathogens, it is the main target for antibacterial host defenses (1). These multilayered structures (cytoplasmic membrane and cell wall and possibly the outer membrane, polysaccharide capsule, and proteinaceous S layer) have both protective and adaptive functions that require tightly regulated gene expression to control their biosynthesis and to adjust their properties in response to changing environments (2–4).
In the alphaproteobacteria, Sinorhizobium meliloti is the paradigmatic model for studying the crucial role surface components play and how their production is finely tuned to respond appropriately to the environmental signals (e.g., carbon and nitrogen sources, phosphate starvation, and plant signals) and various stresses (osmolality, ionic strength, and oxidation) related to either their free-living state or their partnership with leguminous hosts. For example, the establishment of rhizobium-legume symbiosis requires the timely and spatially regulated bacterial synthesis (or modification) of four classes of envelope-associated polysaccharides: outer membrane lipopolysaccharides (LPS), periplasmic cyclic β-(1,2)-glucans and external capsular polysaccharides (K), and the exopolysaccharides (EPS) (succinoglucan [EPS I] and galactoglucan [EPS II]) (5–7). The regulated production of EPS is particularly well described and involves an intricate regulatory network (for a review, see reference 2). Briefly, the inner-membrane sensor histidine kinase ExoS and the cytoplasmic transcriptional regulator ChvI constitute a two-component system (TCS) that controls succinoglycan production (8). A third partner, the periplasmic protein ExoR (believed to sense calcium and ammonium), is involved in this regulatory cascade (9, 10). Both ExoR and ExoS are involved in regulating EPS I production, and S. meliloti mutants with mutations of the genes involved in LPS sulfatation and flagellum biosynthesis (11–13), exoR and exoS, overproduce EPS I and are symbiotically deficient (14–16).
Finally, the zinc finger protein MucR appears to couple the two EPS biosynthetic pathways by positively regulating succinoglycan biosynthesis genes and repressing the synthesis of galactoglucan (17–20). It remains to be determined how mucR becomes active, but it has also been shown to repress flagellar-gene expression (21). In addition, most of these signaling pathways are influenced by the quorum-sensing (QS) hierarchy of S. meliloti (22). S. meliloti belongs to the order Rhizobiales in the α-2 subdivision of the class Proteobacteria and, although a plant symbiont, is very closely related to the animal pathogens belonging to the genus Brucella (23). Brucella spp. are considered to be facultative intracellular parasites that cause brucellosis, a chronic globally widespread zoonotic disease that affects a broad range of mammals, including livestock and humans (24).
Most Brucella virulence determinants have been associated with the bacterial surface as either permanent or transient structural components (e.g., the envelope and its appendices, the virB type IV secretion system [25], the flagellum [26], or their respective regulators [27–29]). Sinorhizobium and Brucella not only lead to similar intracellular chronic infections within a host-derived membrane-bound compartment of their respective hosts (30), but also share similar requirements for establishing a relationship with their dedicated hosts (31). In Brucella spp., cell envelope-associated polysaccharides and their regulated production also play a crucial role during the interaction with the host. First, the LPS O chain is required to resist complement-mediated lysis (32), avoid intracellular killing, mediate early steps in vacuolar trafficking (33), and inhibit host cell apoptosis (34). Second, cyclic glucans allow the bacteria to prevent phagosome-lysosome fusion and reach their final replicative compartment (35). Third, the BvrS/BvrR TCS, which is orthologous to ExoS/ChvI, is also critical to the infectious cycle and is clearly involved in the homeostasis of the outer membrane (OM) (36, 37). Fourth, among the targets of this TCS, which were identified by transcriptomic analysis (38), is the QS regulator VjbR, which was previously demonstrated to be a major regulator of outer-membrane organization (OM proteins, flagellum, and type IV secretion system) (39). Notably, in medium containing yeast extract, VjbR mutants have an aggregative phenotype that has been suggested to be linked to EPS production (29). Fifth, a transcriptional regulator with 61% identity to S. meliloti MucR was identified during a screen of Brucella melitensis transposon mutants (40). The mucR transposon mutant had decreased virulence in both cellular and mouse models of infection but was otherwise uncharacterized. More recently, the protective efficiency of this mutant was evaluated as a live attenuated vaccine (41).
Here, we report a detailed characterization of the transcriptional regulator MucR and show that it modulates bacterial surface properties and resistance to environmental stresses (i.e., oxidative stress, cationic peptide, and detergents). Using heterospecific complementation, we show that B. melitensis mucR is a functional orthologue of S. meliloti mucR based on its ability to restore succinoglycan production in the S. meliloti Rm101 mucR mutant. In addition, similar to S. meliloti, B. melitensis MucR inhibits flagellar-gene expression via the flagellar master regulator and negatively regulates its own transcription. More surprisingly, we demonstrate that MucR regulates a lipid A core modification in B. melitensis. In addition to the BvrS/BvrR TCS (37), this is the second transcriptional regulator of Brucella spp. shown to modulate the lipid A core component of LPS. Considering the strong conservation of the mucR gene in alphaproteobacteria and the link generally established between the gene and altered host-bacterial interaction, it would be worthwhile to examine LPS alterations associated with mucR mutations in other alphaproteobacteria.
MATERIALS AND METHODS
Strains, plasmids, and culture conditions.
All strains and plasmids used in this study are listed in Table 1.
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Characteristicsa | Source or reference |
|---|---|---|
| B. melitensis | ||
| 16M | WT; Nalr; parental strain | A. Macmillan, Central Veterinary Laboratory, Weybridge, UK |
| 16M pBBR-gfp(ASV) | Nalr Cmr; WT carrying pBBR-gfp(ASV) | This study |
| 16M pBBRPmucRgfp(ASV) | Nalr Cmr; WT carrying pBBRPmucRgfp(ASV) | This study |
| ΔmucR | Nalr; deletion strain for mucR gene | This study |
| ΔmucR KI | Nalr; chromosomal insertion of the mucR gene in the ΔmucR strain | This study |
| ΔmucR pBBRMCS1 | Nalr Cmr; ΔmucR strain carrying the plasmid pBBRMCS1 | This study |
| ΔmucR pBBRmucR | Nalr Cmr; ΔmucR strain carrying the complementation plasmid pBBRmucR | This study |
| ΔmucR pBBRPmucRgfp(ASV) | Nalr Cmr; ΔmucR strain carrying pBBRPmucRgfp(ASV) | This study |
| ΔmucRKI pBBRPmucRgfp(ASV) | Nalr Cmr; ΔmucR KI strain carrying pBBRPmucRgfp(ASV) | This study |
| ΔvjbR | Nalr ΔvjbR::Kanr (referred to as CD100 in reference 29) | 27 |
| S. meliloti | ||
| Rm1021 | Nalr Smr; WT | 42 |
| Rm1021::exoY | Rm1021 exoY::Tn5 (formerly Rm7210); Nalr Smr Neor | 43 |
| Rm2011 | Nalr Smr; wild type | 44 |
| Rm101 | Rm2011 Spcr cassette inserted into the PmacI site of mucR | 45 |
| E. coli | ||
| DH10B | F− mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara leu)7697 galU galK rpsL nupG λ− (Smr) | Gibco BRL |
| S17-1 | recA thi pro hsdR− M+ RP4:2-Tc:Mu:Km Tn7 λpir; allows plasmid mobilization | 46 |
| Plasmids | ||
| pBBR1MCS1 | Broad-host-range cloning vector; high copy number; Cmr | 47 |
| pGEM-Teasy | Ampr | Promega |
| pJQ200-uc1 | sacB Gtmr | 48 |
| pBBR-gfp(ASV) | Cmr | M. Terwagne |
| pJQΔmucR | sacB Gtmr strain containing the ΔmucR fragment; used to construct the deletion strain | This study |
| pJQmucR | sacB Gtmr strain containing the mucR gene flanked by its upstream and downstream regions | This study |
| pBBRmucR | Cmr; complementation plasmid | This study |
| pBBRPmucRgfp(ASV) | Cmr; reporter plasmid bearing the transcriptional fusion PmucRgfp(ASV) | This study |
Nalr, nalidixic acid resistance; Smr, streptomycin resistance; Neor, neomycin resistance; Cmr, chloramphenicol resistance; Spcr, spectinomycin resistance; Ampr, ampicillin resistance; Gtmr, gentamicin resistance.
Classically, Brucella strains were grown with shaking at 37°C in 2YT medium (10 g liter yeast extract−1, 16 g liter peptone−1, 5 g liter NaCl−1) containing the appropriate antibiotics from a stationary-phase overnight culture (2YT; 10 ml) back diluted to an optical density at 600 nm (OD600) of 0.05.
For RNA extraction, 10 ml of bacteria was harvested from a 200-ml 2YT culture grown to mid-log phase (OD600 = 0.5). The 10-ml cultures were used to follow GFP(ASV) (green fluorescent protein) production from B. melitensis pBBRPmucRgfp(ASV) and from B. melitensis harboring the vector pBBR-gfp(ASV). GFP(ASV) is an unstable variant of GFPmut3 and is a useful reporter gene for monitoring transient gene expression because of the reduced half-life of the reporter gene (49). Escherichia coli strains were routinely grown in Luria-Bertani (LB) medium with appropriate antibiotics at 37°C. S. meliloti strains were cultivated in LB broth with 2.5 mM MgSO4 and 2.5 mM CaCl2 at 30°C. Matings were performed by mixing E. coli S17-1 donor cells with Brucella or S. meliloti recipient strains on 2YT or LB medium, respectively, for 3 to 4 h. The mixed population was plated on medium containing the appropriate antibiotics to select for B. melitensis and S. meliloti conjugants. Chloramphenicol, gentamicin, and nalidixic acid were used at 20, 50, and 25 (8 for S. meliloti) μg ml−1, respectively. Growth curves were monitored using a Bioscreen system (Thermo Fisher, Erembodegem-Aalst, Belgium), which continuously monitors OD600 readings in a multiwell format.
Molecular techniques.
DNA manipulations were performed using standard molecular techniques (50). Restriction enzymes were purchased from Roche, and primers were purchased from Eurogentec. Primer sequences are listed in Table S1 in the supplemental material.
Mutant construction.
The B. melitensis 16M ΔmucR deletion mutant was constructed by allelic replacement using a two-step strategy. Briefly, 500-bp upstream and downstream regions flanking the mucR gene were amplified by PCR from B. melitensis genomic DNA using the primers PmucRF and PmucRR and TmucRF and TmucRR, respectively. For each construction, a second PCR was used to join the two PCR products using the primer pairs PmucRF and TmucRR. Finally, the ΔmucR fragment was cloned into pGEM-T Easy (Promega) to generate the intermediate vector pGEMTΔmucR. The ΔmucR fragment was excised by NotI restriction, subcloned into the final vector pJQ200-uc1, and used to construct a Brucella mutant following a previously described strategy (51). Gene replacement was confirmed by PCR using the following primers: mucR upstream and mucR downstream. To construct the strain ΔmucRKI, we used the same recombination strategy using a plasmid carrying the mucR gene with its upstream and downstream regions.
Construction of the complementation plasmid pBBRmucR.
The mucR gene was amplified by PCR from B. melitensis genomic DNA using the primers (Eurogentec) mucR XhoI F and mucR ClaI R, which contain XhoI and ClaI restriction sites, respectively. The PCR product was cloned into pGEM-T Easy (Promega) to generate the intermediate vector pGEMTmucR. After sequencing, the fragment mucR was excised by a ClaI and XhoI double restriction digest and subcloned into a previously XhoI-ClaI-restricted plasmid, pBBRMCS1, to obtain pBBRmucR. The vector was then transferred to the ΔmucR strain to obtain the complemented ΔmucR pBBRmucR strain.
Construction of the reporter plasmid pBBRPmucRgfp(ASV).
The region containing the putative mucR promoter was amplified by PCR from B. melitensis genomic DNA using the primers XhoIpmucRR and BamHIpmucRF, which contain XhoI and BamHI restriction sites, respectively. The PCR product was first cloned into the vector pGEM-T Easy. The fragment was then inserted in frame upstream of the promoterless gfp(ASV) reporter gene in pBBR1MCS to generate the plasmid pBBRPmucRgfp(ASV).
Cellular infection.
Evaluation of the intracellular survival of B. melitensis wild-type (WT) and ΔmucR strains in RAW 264.7 murine macrophages was performed as previously described (52). Briefly, bacterial strains were grown overnight in 2YT medium and then inoculated at a multiplicity of infection (MOI) of 300 into cell monolayers in 24-well plates. After a 10-min centrifugation at 1,000 rpm at room temperature, the plates were placed in a 5% CO2 atmosphere at 37°C for 1 h. Afterward, the cells were washed with phosphate-buffered saline (PBS) and incubated in medium containing 50 μg ml gentamicin−1 at 37°C under 5% CO2 until the end of the infection (48 h). The cells were then washed and lysed in sterile MilliQ water for 10 min, and serial dilutions of lysates were plated on 2YT solid medium to enumerate CFU. The data are expressed as CFU per well on a logarithmic scale.
Mouse infections.
Virulence assays using BALB/c mice were performed as described previously (26). Briefly, 8-week-old mice were inoculated intraperitoneally with 500 μl of a suspension containing 4 × 104 CFU of the appropriate bacterial strain. At 1 and 4 weeks postinoculation, mice from each group were sacrificed, and spleens were collected. The spleens were homogenized in 2 ml of PBS–0.1% Triton X-100, and serial dilutions of the homogenates were plated on 2YT solid medium to determine the bacterial load. The lysis of spleen homogenates with 0.1% Triton X-100 does not affect the survival of mucR mutants, as similar results were obtained with a lysis protocol using distilled water (data not shown). The data are expressed as the log10 CFU per spleen. Data were statistically analyzed via a Mann-Whitney statistical test using the program Prism. The animal-handling and study procedures were in accordance with the current European legislation (directive 86/609/EEC) and in agreement with the corresponding Belgian law, Arrêté royal relatif à la protection des animaux d'expérience du 6 avril 2010 publié le 14 mai 2010. The complete protocol was reviewed and approved by the Animal Welfare Committee of the Facultés Universitaires Notre-Dame de la Paix (FUNDP), Belgium (permit number 05-558).
Oxidative-resistance assay.
Oxidation resistance assays were performed according to previously described protocols with some modifications (53). Cells were grown overnight in 10 ml of 2YT medium with shaking and adjusted to a concentration of 5 × 105 CFU ml−1 in PBS.
In 96-well plates, 50 μl of the bacterial suspension was supplemented with 50 μl of H2O2 (freshly diluted in PBS) at final concentrations of 1 mM, 2.5 mM, and 5 mM. A negative-control experiment was performed by adding 50 μl of PBS (without H2O2) to the same bacterial suspension. After exposure for 1 h in a 37°C shaking incubator, the cells were rapidly diluted with PBS and plated onto 2YT medium. After 5 days at 37°C, CFU were enumerated, and the survival of each bacterial strain was determined as a percentage of the negative control.
Detergent sensitivity, polymyxin B sensitivity, and Congo red staining.
Bacteria from an overnight culture in 2YT medium were spotted on tryptic soy broth (TSB) agar medium (Difco) containing 2% SDS, 0.1% Triton X-100, or 0.01% Congo red in triplicate (20 μl per spot) and incubated at 37°C for 4 days. Images were captured using a Canon A430 camera, and the contrast and brightness of the complete image were optimized with the correction tool of PowerPoint software. Polymyxin B sensitivity was determined using an Etest containing a preformed gradient covering a continuous MIC range from 0.064 to 1,024 μg ml−1 (bioMérieux). Brucella was adjusted to an OD750 of 0.109 (1 McFarland standard) in 2YT medium. The suspension was spread onto Mueller-Hinton II (cation-adjusted) broth (BD Difco) plates using a cotton swab, and the Etest strips were then applied. The plates were incubated for 72 h at 37°C. For each strain, the MIC was determined as the concentration at which the ellipse intersects the concentration scale printed on the Etest strip. Three independent tests were performed.
Quantitative real-time reverse transcription-PCR (qRT-PCR).
Total RNA samples were prepared as previously described (39) for B. melitensis 16M, the ΔmucR mutant, and the complemented ΔmucR pBBRmucR mutant. DNA was removed from the samples using DNase (Kit Fermentas), and samples were reverse transcribed with SuperScript II Reverse Transcriptase (Invitrogen) using random oligonucleotide hexamers, as recommended in the manufacturer's protocol. RNA and cDNA quantities were measured using a NanoDrop spectrophotometer (ND-1000; Thermo Fisher Scientific). The resulting cDNA samples were used as the template in real-time PCRs. Primers were designed using PrimerExpress 2.0 software (Applied Biosystems) to generate PCR products ranging from 80 to 100 bp and are listed in Table S2 in the supplemental material. Reactions performed without reverse transcriptase were used as negative controls to test for DNA contamination. Real-time PCRs were performed with SYBR green mix (Applied Biosystems) in 96-well optical reaction plates (Applied Biosystems). Ratios were calculated using the ΔΔCT method for each primer in an Applied Biosystems Step One Plus real-time PCR instrument. The results for each target mRNA were normalized to 16S rRNA transcript levels and averaged.
Total extraction and SDS-proteinase K extraction of LPS.
Total extracts were prepared from B. melitensis strains cultivated in 2YT medium. Bacteria were concentrated to obtain an OD600 equivalent of 10 and inactivated at 80°C for 1 h. Total extracts were used for SDS-proteinase K extraction of LPS as previously described (54). Samples were loaded onto a 16% or 15% polyacrylamide gel for SDS-PAGE analysis. The gels were then silver stained or transferred onto a nitrocellulose membrane (Amersham) for Western blotting (54). In the silver-stained gel, some contaminating (or LPS-linked) proteins were also observed, and the most common contaminants had masses of 25,000 to 27,000 Da (35).
Western blotting.
Nitrocellulose membranes were blocked overnight in PBS containing 5% nonfat dry milk. After being washed three times in PBS-0.05% Tween 20 for 10 min, the membranes were incubated for 1 h with the primary antibodies diluted in PBS (0.05% Tween 20, 1% dry milk), washed three times in PBS-0.05% Tween 20 for 10 min, incubated for 1 h with the secondary antibody diluted in PBS (0.05% Tween 20, 1% dry milk), and finally washed three times for 10 min in PBS-0.05% Tween 20. The blots were developed using an enhanced chemiluminescence (ECL) system (100 mM Tris-HCl, pH 8.5, 0.009% H2O2, 0.2 mM coumaric acid, and 1.25 mM luminol).
Immunodetection.
Immunodetection was performed with primary mouse monoclonal antibodies (MAbs) (undiluted hybridoma culture supernatant) against the O antigen of Brucella (A76/12G12/F12) (55) and against the LPS core (A68/24G12/A8 and A68/24D8/G9) (56) for LPS detection. Flagellar protein was detected using anti-FliC (diluted 1:3,000) or anti-FlgE (1:5,000) rabbit polyclonal antibodies (26). PrlR, which was used as a loading control, was detected using anti-PrlR polyclonal rabbit antibodies (1:1,000) (57). Secondary antibodies consisted of horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Amersham catalog no. NA931; 1:10,000 dilution) and HRP-conjugated donkey anti-rabbit IgG, HRP-linked whole antibody (Amersham catalog no. NA934; 1:5,000 dilution).
Fluorescence microscopy.
Bacteria were spotted onto a microscope slide layered with a 1% agarose pad containing PBS (58). These slides were placed on a microscope stage at room temperature. The samples were observed on a Nikon i80 fluorescence microscope through a 100× differential interference contrast (DIC) (Nomarski) or phase-contrast objective with a Hamamatsu Orca-ER LCD camera. Image acquisition and processing were performed with NIS element software (Nikon).
Fluorescence-activated cell sorter (FACS) analysis.
Bacterial suspensions were washed in PBS and fixed in 2% paraformaldehyde (PFA), pH 7.4, at room temperature for 20 min. After an additional wash in PBS, the bacteria were used for flow cytometric analysis of GFP(ASV) production with a FACScalibur using CellQuest software (Becton Dickinson) as previously described (59). Bacteria were gated according to size and scatter to eliminate debris from analysis. Then, 50,000 individual events were excited with a 488-nm argon ion laser, and emission light was detected through a 530-nm bandpass filter.
Scanning electron microscopy.
Bacterial aggregates of B. melitensis overexpressing mucR (pBBRmucR) and WT B. melitensis were observed by scanning electron microscopy. The two strains were grown for 72 h in 2YT medium. Glass coverslips were treated with poly-l-lysine (0.05 mg ml−1; Sigma) for 1 h at 4°C. For each strain, 1-ml cell suspensions were centrifuged onto the pretreated coverslips at 1,000 rpm for 10 min at room temperature. The medium was removed, and adherent cells were fixed for 20 min with 2% PFA in PBS. After the incubation, samples were washed with PBS and dehydrated twice 10 min in 25, 50, 75, 85, and 100% ethanol at room temperature. Dehydrated samples were then prepared by critical-point drying (Balzer; CPD 030) and covered with a thin layer of gold (25 nm). Examinations were performed using a scanning electron microscope (Jeol 7500F) at the University of Namur, Namur, Belgium.
Statistical analysis.
For the mouse experiments, we used a Mann-Whitney test included in the program GraphPad Prism to statistically analyze our results. P values of <0.05 were considered to represent a significant difference.
For the cellular infections, oxidative-resistance assays, and qRT-PCR, after testing for homogeneity of variance (Bartlett test), one-way analysis of variance (ANOVA) was performed on the log10 CFU per well, on the survival percentage values, or on the ΔCT values, respectively. When needed, a Scheffe's comparison test was performed, and statistical significance at a P value of <0.05 was accepted.
RESULTS
MucR is required for a successful B. melitensis infection in both RAW264.7 macrophages and BALB/c mice.
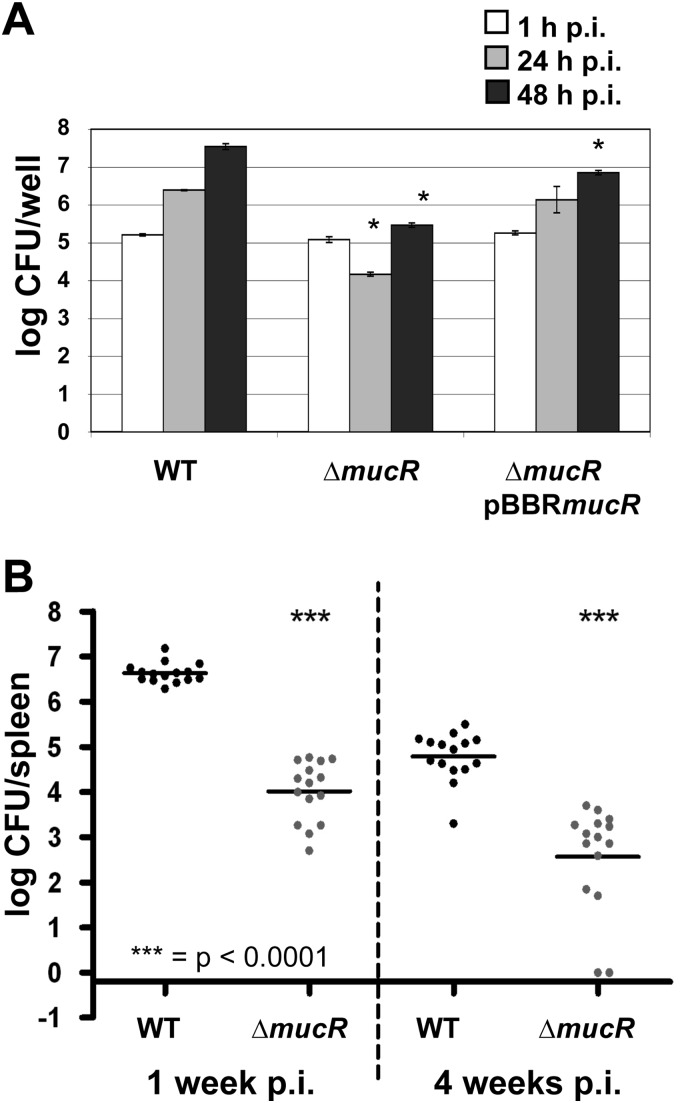
To characterize the role of mucR in virulence, we constructed a deletion mutant of BMEI1364 (referred as ΔmucR) in B. melitensis 16M by allelic replacement and characterized it phenotypically. We first analyzed the ability of the newly constructed mutant to multiply within cultured macrophages. As shown in Fig. 1A, the intracellular bacterial load of the ΔmucR strain was significantly reduced at 24 h and 48 h postinfection (p.i.) in murine RAW 264.7 macrophages compared to the WT control. The intracellular replication of the ΔmucR strain was almost restored to WT levels in the complemented ΔmucR strain. These results are consistent with the reduced intracellular growth of a transposon mutant described previously using the mouse macrophage-like cell line J774.A1 (40). The difference between the ΔmucR and WT strains cannot be due to the mutant's reduced capacity to invade the cells, because the number of intracellular bacteria at 1 h p.i. was the same for each strain. On the other hand, all strains displayed the same growth rate between 24 h p.i. and 48 h p.i. (Fig. 1A). The ΔmucR strain also showed reduced virulence in HeLa cells (data not shown). Taken together, these results confirm the importance of MucR in B. melitensis 16M intracellular infection, especially during the first hours of infection.
Fig 1.
MucR is crucial for the virulence of B. melitensis in both cellular and mouse models of infection. (A) Intracellular replication of WT, ΔmucR, and ΔmucR pBBRmucR B. melitensis strains in Raw 264.7 murine macrophages. The data are the average log10 CFU per well. The error bars represent the standard deviations of triplicates in one of three representative experiments. Significant differences between the strains are noted by an asterisk (P < 0.05). (B) Virulence of WT and ΔmucR B. melitensis strains in the spleens of BALB/c mice. The graph represents pooled data from two independent experiments and indicates the log10 CFU per spleen in each mouse. The lines indicate the mean log10 CFU for each group (n = 15). Statistical analysis was performed using the Mann-Whitney test (***, P < 0.0001).
To verify that, as previously described for the transposon mutant (40), the mucR gene is involved in the successful infection process of B. melitensis in vivo, we compared the behaviors of the ΔmucR and WT strains in a mouse model of infection at 1 and 4 weeks p.i. (Fig. 1B). We observed a large reduction (more than a 2-log-unit difference) in the splenic bacterial load at just 1 week p.i. in BALB/c mice infected with the ΔmucR strain. Decreased persistence of the ΔmucR strain was also consistently observed at 4 weeks p.i. (Fig. 1B). Moreover, this virulence attenuation was associated with reduced splenomegaly (data not shown).
The mucR mutant enters prematurely into stationary phase in bacterial cultures.
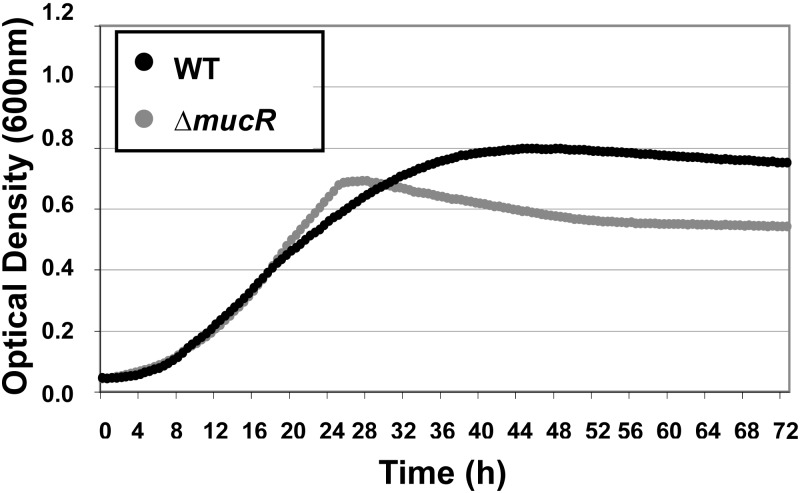
The attenuation reported above could be caused by an in vitro growth defect that is exacerbated in intracellular environments; we therefore examined the growth rates of the ΔmucR and WT strains in rich 2YT broth.
The ΔmucR mutant had a growth rate similar to that of the WT until 22 h (log phase) (Fig. 2). At this time point, the ΔmucR mutant transitioned into stationary phase earlier, more abruptly, and at a lower bacterial density than the WT (Fig. 2). One possible hypothesis is that the ΔmucR mutant is poorly adapted to cope with the stresses linked to stationary phase, which would be relevant to its reduced virulence, as previously suggested for other Brucella mutants whose adaptations upon entering stationary phase are affected (60).
Fig 2.
Growth curves of WT and ΔmucR B. melitensis strains in 2YT rich medium. Cultures were inoculated from a preculture to an initial OD600 of 0.05 in a Bioscreen plate. Bacteria were grown for 72 h with continuous shaking in 2YT medium. The optical density was measured every 30 min. The graph represents the average OD of technical triplicates for each condition from one of two representative experiments.
The mucR mutant is more sensitive to H2O2, detergents, and polymyxin B.
As a consequence of their intracellular lifestyle, Brucella spp. have to withstand various harsh environmental conditions in their phagosomal compartments within host macrophages, including exposure to reactive oxygen species (ROS) and nutrient deprivation. The successful adaptation of Brucella to these stresses has been correlated with stationary-phase physiology (60). Moreover, it has been shown in other bacteria that the limited nutrient availability associated with entry into stationary phase triggers a general stress response that can be cross-protective against heat and oxidative challenges (61, 62).
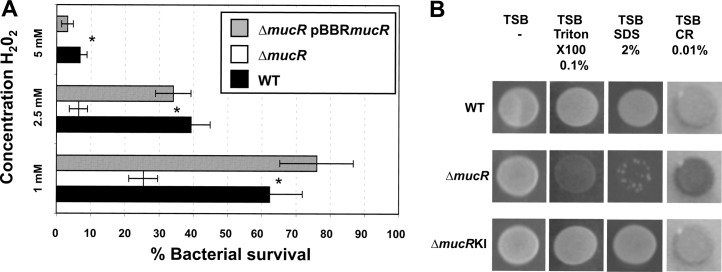
We therefore evaluated the ability of the ΔmucR mutant to resist exposure to exogenous hydrogen peroxide (H2O2). After a 1-hour exposure to various concentrations of H2O2, the survival rates of the ΔmucR strain were significantly reduced compared to the WT strain under the same conditions (Fig. 3A). The sensitivity of the ΔmucR mutant to this exogenous oxidative stress was complemented in trans with the plasmid pBBRmucR (Fig. 3A).
Fig 3.
Sensitivity to oxidative stress and detergents. (A) Survival of WT, ΔmucR mutant, and ΔmucR pBBRmucR B. melitensis at various concentrations of H2O2. The data are the averages of log10 CFU per well. The error bars represent the standard deviations of triplicates from one of three representative experiments. Significant differences between the strains are indicated by asterisks (P < 0.05). (B) Susceptibility of B. melitensis to surfactants and Congo red. Strains from an overnight preculture were spotted onto TSB plates containing the detergent Triton X-100 (0.1%), SDS (2%), or Congo red (CR) (0.01%) in triplicate. The plates were incubated for 4 days at 37°C. One representative spot is shown for each strain. The ΔmucR pBBRmucR strain gave the same results as the ΔmucRKI strain (data not shown).
We further hypothesized that a mucR mutation would generate sensitivity to other stresses. To test this idea, we evaluated bacterial growth on media containing detergents, including 0.1% Triton X-100 and 2% sodium dodecyl sulfate (SDS). While these concentrations did not inhibit the growth of the WT strain, the ΔmucR strain had impaired growth/survival in the presence of both detergents (Fig. 3B).
Furthermore, on a medium containing 0.01% Congo red, we observed that the ΔmucR strain retains much more of the dye, resulting in a more intense dark-red appearance (Fig. 3B). All these growth or staining phenotypes were restored by supplying mucR either on a plasmid or via chromosomal insertion (Fig. 3). Together, these observations suggest that there has been a major cell envelope alteration that affects the susceptibility to these compounds in a B. melitensis strain lacking mucR.
Because envelope defects in Brucella have been associated with sensitivity to antimicrobial peptides, such as polymyxin B (28), we used a polymyxin B Etest (bioMérieux) strip to determine the MICs of polymyxin B for all the strains. The WT had a MIC of 64 units, whereas the MIC for ΔmucR was 32 units. This result confirms the previous indications that the ΔmucR strain has an altered cell envelope.
The mucR mutant displays an altered LPS profile.
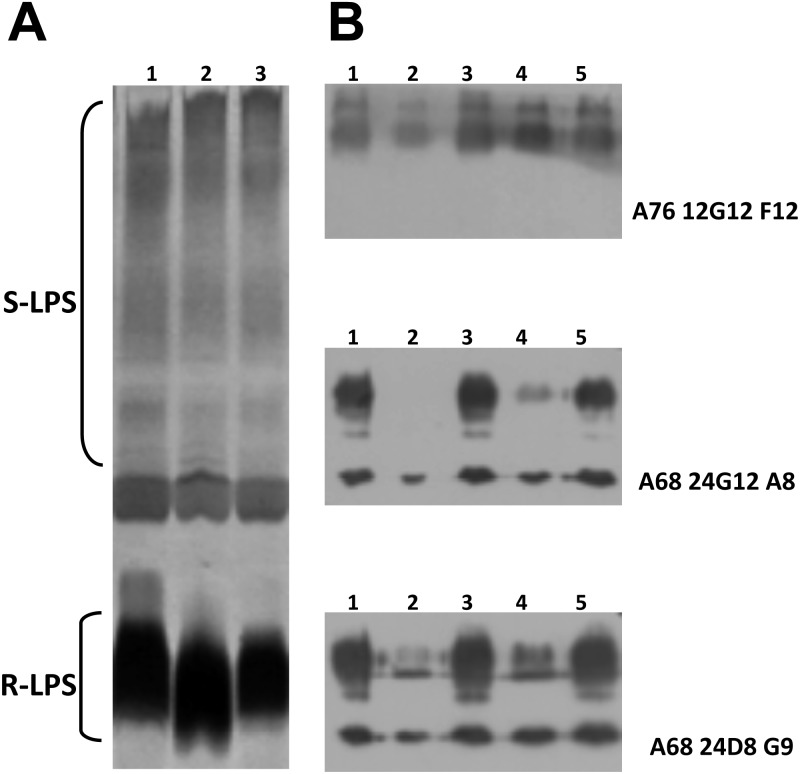
In the brucellae, LPS is a major virulence factor, and LPS alterations can generally impede the successful infection process (63) and, in particular, resistance to antimicrobial compounds (64). Like the WT strain, the ΔmucR strain was smooth, as determined by crystal violet staining (Fig. 4 and data not shown). Nevertheless, to detect some subtle LPS alterations, we examined the LPS migration pattern and the reactivity of the ΔmucR strain to anti-LPS MAbs compared to the WT strain. Figure 4A shows the SDS-PAGE profiles of SDS-proteinase K-treated extracts of WT B. melitensis (lane 1) and the ΔmucR mutant (lane 2) strains. As expected, the two strains showed similar migration patterns for their smooth LPS (S-LPS). Similarly, their reactivities with the anti-O-antigen MAb A76/12G12/F12 in a Western blot were comparable (Fig. 4B). However, the lipid A core fraction of the ΔmucR mutant migrated faster than the corresponding WT fraction. This observation suggests that there is a modification of the lipid A core. This hypothesis was corroborated by differences in the reactivities of the extracts with anti-core MAbs A68/24G12/A8 and A68/24D8/G9 (Fig. 4B) in a Western blot analysis. Normal SDS profiles and epitope detection were restored upon complementation in trans or via chromosomal insertion of the mucR gene (Fig. 4).
Fig 4.
Modification of LPS in the ΔmucR strain. (A) Silver staining of SDS-proteinase K-treated extracts following electrophoresis on a 16% acrylamide gel. Lane 1, WT; lane 2, ΔmucR; lane 3, ΔmucRKI. (B) Western blot with anti-LPS MAbs. Total extracts were separated on a 15% acrylamide gel by electrophoresis, transferred to nitrocellulose membranes, and probed using anti-O-antigen MAb A76/12G12/F12 and anti-core MAbs A68/24G12/A8 and A68/24D8/G9. Lane 1, WT; lane 2, ΔmucR; lane 3, ΔmucRKI; lane 4, ΔmucR pBBRMCS1; lane 5, ΔmucR pBBRmucR. S-LPS, smooth LPS; R-LPS, rough LPS.
The promoter activity of mucR is induced in 2YT medium containing 400 mM NaCl, correlating with morphological alterations of B. melitensis.
We have reported previously that Brucella spp. form bacterial aggregates over prolonged culture times when grown in 2YT medium containing 400 mM NaCl (57). Because MucR in S. meliloti regulates EPS I biosynthesis (18, 65) and is somehow involved in environmental-stress response (Fig. 3), we tested whether MucR is required to form these bacterial clumps in B. melitensis 16M. The ΔmucR strain displayed a significant growth defect when cultured in 2YT medium containing 400 mM NaCl (see Fig. S2 in the supplemental material), suggesting that MucR is required for optimal growth under hypersaline conditions.
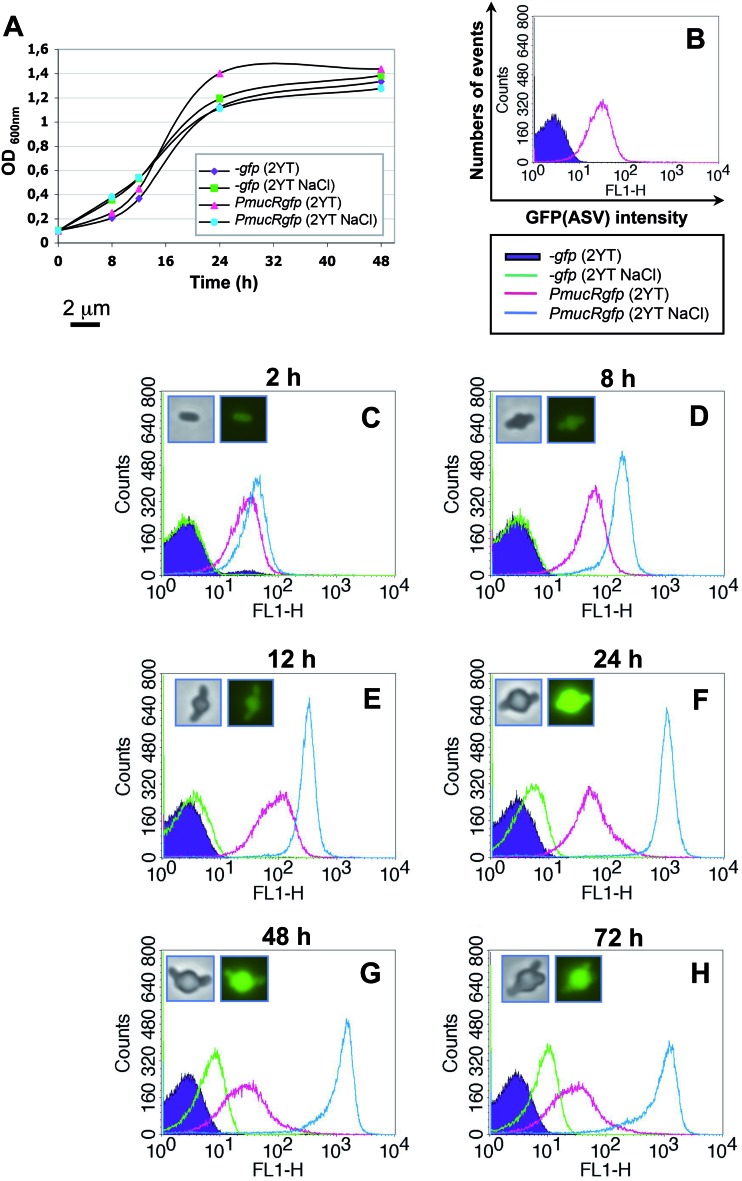
We used a B. melitensis strain carrying the reporter plasmid pBBRPmucRgfp(ASV) (see Materials and Methods) to monitor mucR promoter (PmucR) activity in B. melitensis at different culture times in unsupplemented 2YT medium or 2YT medium supplemented with 400 mM NaCl. The B. melitensis strain harboring the promoterless vector pBBR-gfp(ASV) was used as a negative control. These strains displayed similar growth curves under both conditions (Fig. 5A). Figure 5C to H represents the fluorescent signal intensities measured by flow cytometry in the reporter strain at different culture times in either 2YT medium or 2YT medium plus 400 mM NaCl. In 2YT medium, PmucR activity is constitutive and gives a mean fluorescent channel intensity (MFI) of approximately 40 to 50, independent of the culture time considered (Fig. 5). Under hypersaline conditions (blue curve), PmucR activity is induced beginning at 8 h p.i. (MFI = 200) and is then strongly enhanced as the culture time increases (MFI = 300, 1,000, and 1,500 for 12, 24, and 48 h p.i., respectively). These data were confirmed by fluorescence microscopy (Fig. 5, insets). The progressive and major morphological changes of the bacteria were most notable when grown at such salt concentrations (Fig. 5, insets). An equatorial bulge can be distinguished after only 8 h of growth in hypersaline 2YT (Fig. 5D). The subsequent increase in the bulge in the ongoing culture results in an increase in cell size, which is also evident in the small MFI shift for the negative control (Fig. 5, compare the green curve to the violet curve). This cell shape alteration seems to be salt specific, because it did not occur under iso-osmolal conditions with sucrose (see Fig. S1 in the supplemental material). At equivalent osmolalities, the NaCl-supplemented medium promotes cell shape alterations and strongly induces PmucR activity compared to cells grown under sucrose-supplemented conditions (see Fig. S1). However, the fluorescent signal for cells grown for 24 h in sucrose-supplemented 2YT medium seems to be slightly higher than the signal for cells grown in regular 2YT medium (see Fig. S1). Together, these data suggest that the mucR promoter is induced by osmotic stress but even more highly induced by ionic stress in B. melitensis.
Fig 5.
PmucR activity is induced in B. melitensis growing in 2YT medium containing 400 mM NaCl. (A) Growth curve of the reporter [B. melitensis WT expressing the transcriptional fusion PmucRgfp(ASV)] and the control [the WT harboring the plasmid pBBR-gfp(ASV)] strains in both 2YT medium (85.5 mM NaCl) and 2YT medium containing 400 mM NaCl. (B) Fluorescence intensity measured by flow cytometry (5 × 104 events acquired) of WT B. melitensis expressing the transcriptional fusion PmucRgfp(ASV) at the end of preculture in 2YT medium. The WT harboring the plasmid pBBR-gfp(ASV) was used as a negative control. (C to H) Fluorescence intensities measured in the reporter strains growing in unsupplemented 2YT medium or 2YT medium supplemented with 314.5 mM NaCl at 2 h (C), 8 h (D), 12 h (E), 24 h (F), 48 h (G), and 72 h (H) postinoculation. The insets are phase-contrast and corresponding fluorescence images of the major morphotype of the reporter strain in 2YT medium containing 400 mM NaCl at the indicated times. Scale bar, 2 μm.
MucR regulates cyclic β-glucan synthase mRNA levels.
The various susceptibilities reported above suggest that there is a major cell envelope alteration in B. melitensis mutants lacking mucR. Susceptibility to detergents (deoxycholic acid, SDS, and Zwittergent) has been described for a cgs mutant of Brucella abortus, which was unable to produce periplasmic cyclic β-glucans (cβG) (66). To determine whether MucR can transcriptionally regulate cβG levels, the relative levels of cgs (the gene encoding cβG synthase) and cgt (encoding the cβG transporter) mRNAs were evaluated by qRT-PCR on RNA purified from the different strains harvested during the exponential growth phase (OD600 = 0.5) in 2YT medium. Although a mucR deletion does not affect cgt transcript levels, we found a nearly 2-fold reduction in cgs transcripts in the ΔmucR mutant compared to the WT strain (Table 2). WT levels of cgs mRNA were restored upon complementation with the plasmid pBBRmucR (Table 2). Although additional studies are needed, these results suggest that MucR could be the first regulator of cβG identified in B. melitensis 16M.
Table 2.
MucR regulates the mRNA levels of the flagellar and cyclic β-glucan synthase genesa
| Gene | Locus tag | ΔmucR/WT ratio | ΔmucR pBBRmucR/WT ratio |
|---|---|---|---|
| fliC | BMEII0150 | 4.62 | 0.95 |
| flgE | BMEII0159 | 3.78 | 0.96 |
| ftcR | BMEII0158 | 4.45 | 1.35 |
| fliF | BMEII0151 | 6.81 | 1.69 |
| cgs | BMEI1837 | 0.55 | 0.96 |
The relative levels of cgs, fliC, flgE, ftcR, and fliF mRNAs were determined using quantitative real-time PCR on RNA isolated from bacteria harvested at mid-exponential growth phase in rich medium (2YT). Normalization was performed using 16S rRNA. ANOVA I on ΔCT values from biological triplicates was used for statistical analysis after testing the homogeneity of variance (Bartlett test). Scheffe's comparison test was performed, and statistical significance was obtained (at a P value of <0.05) between the ΔCT values of the ΔmucR and WT or complemented strains.
Restoration of EPS I production in an S. meliloti mucR mutant through constitutive expression of B. melitensis mucR.
The mucR gene is well conserved within Rhizobiales, especially in S. meliloti (61% amino acid identity), where its function has been well studied. In S. meliloti, MucR has been described as a regulator of both flagellar-gene expression and EPS synthesis (21). S. meliloti produces two different types of EPS: succinoglycan, which was first described as a calcofluor-binding acidic exopolysaccharide (EPS I), and galactoglucan (EPS II). Strains producing one versus the other type of EPS can be rapidly discriminated on agar medium containing calcofluor when placed under UV light (65). Under standard laboratory conditions, both wild-type strains, Rm1021 and Rm2011, produce EPS I and appear fluorescent on calcofluor-containing medium, whereas non-EPS I-producing strains do not (Rm1021 exoY:Tn5) (Fig. 6). An S. meliloti mucR mutant (Rm101) forms colonies that are more mucoid and lack the blue-green color characteristic of EPS I-producing strains (18, 65) (Fig. 6). We observed that the constitutive expression of mucRBm in an S. meliloti Rm101 background restored EPS I production (Fig. 6). These data suggest that the mucR gene from B. melitensis 16M encodes a fully functional protein that is at least able to regulate the expression of EPS biosynthesis genes in S. meliloti.
Fig 6.
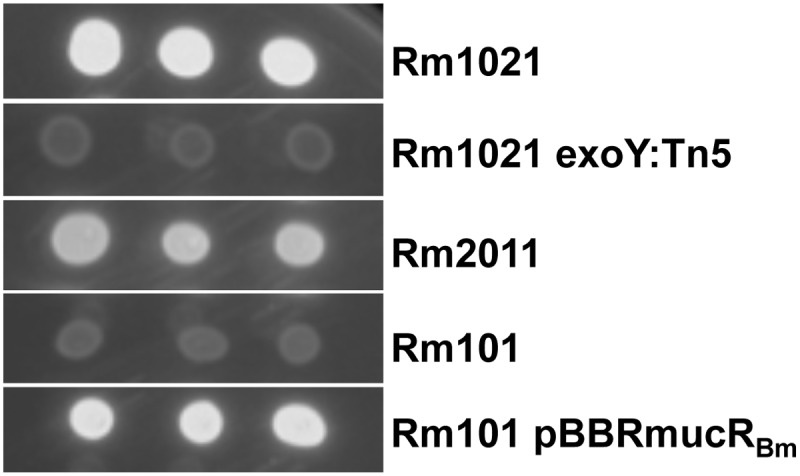
Heterospecific complementation of an S. meliloti mucR mutant with the mucR gene from B. melitensis on LB plates containing calcofluor. The strains Rm1021 (wild type), Rm1021 exoY:Tn5 (non-EPS I-producing strain), Rm2011 (wild type), Rm101 (mucR mutant), and Rm101 pBBRmucRBm (mucR mutant carrying the wild-type mucR gene of B. melitensis 16M) were spotted in triplicate on LB medium containing 0.02% calcofluor and incubated at 30°C for 2 days before being subjected to UV light.
MucR represses flagellar-gene expression by modulating mRNA levels.
In addition to regulating EPS production, MucR also regulates flagellar-gene expression in S. meliloti. Indeed, it has been shown that MucR inhibits expression of rem, the flagellar master regulator (67), and consequently, the expression of rem-regulated genes (21). Our laboratory has previously shown that the Brucella flagellar master regulator is orthologous to Rem (68). We therefore examined the putative impact of MucR on flagellar-gene regulation in B. melitensis 16M. Even though they are described as nonmotile, Brucella spp. possess flagellar genes that are expressed only in the early log phase of growth in rich medium (26). Using specific antibodies against FliC (flagellin), we examined FliC protein expression at different growth phases in the mucR mutant compared to the WT strain (Fig. 7A). We detected FliC both at the beginning of log-phase growth, as seen in the WT strain, and during stationary phase, when the protein is no longer present in the WT background. Similar results were obtained for FlgE, the hook monomer (data not shown). Complementation of mucR in trans restored the WT phenotype for both FlgE and FliC (Fig. 7B and C). This result indicates that MucR could also be a repressor of flagellar-gene expression in B. melitensis 16M.
Fig 7.
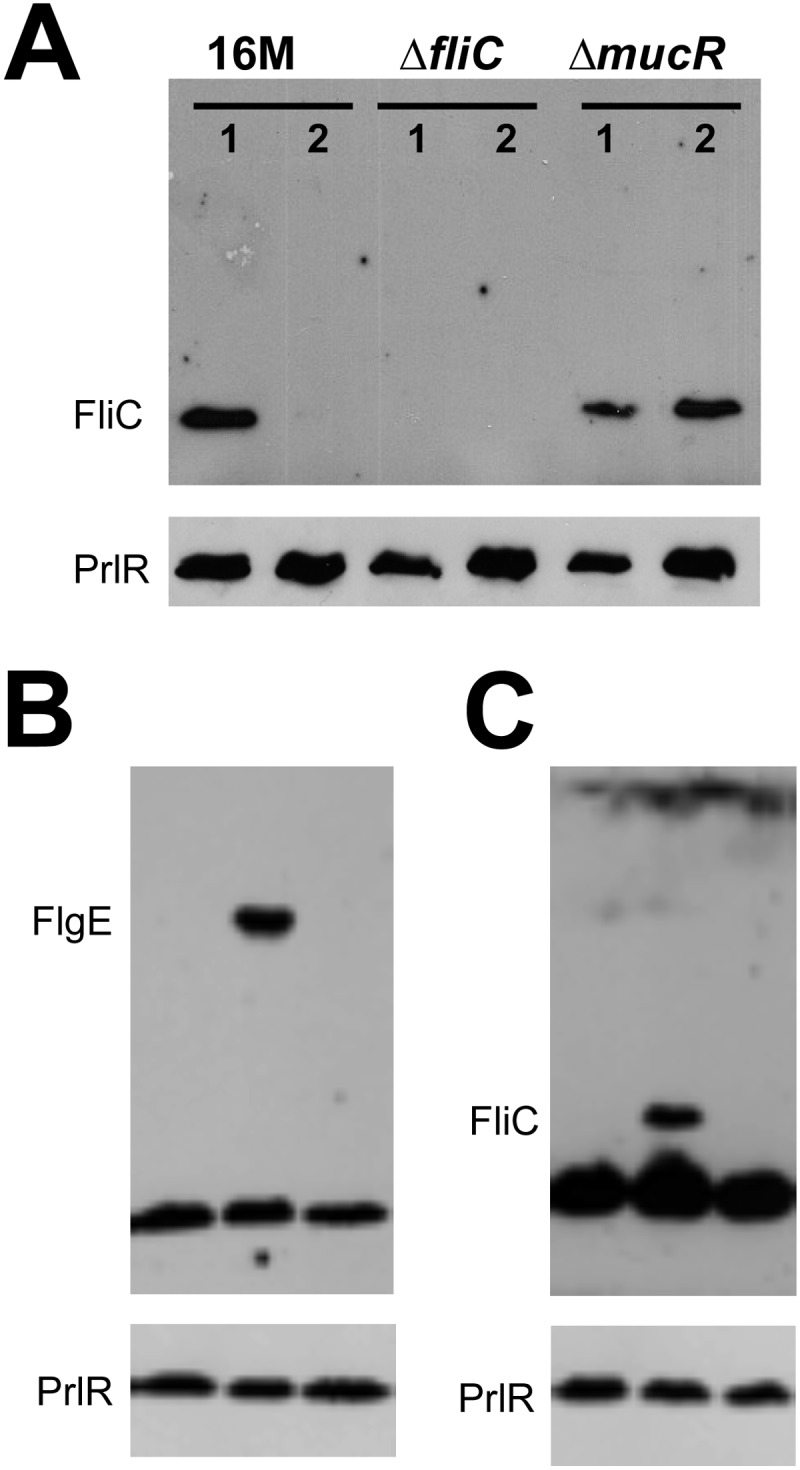
Western blot detection of the flagellar proteins FliC and FlgE. (A) Detection of flagellin (FliC) production in B. melitensis WT, ΔfliC, and ΔmucR strains in both early log phase and stationary phase. The strains were cultivated in 2YT broth, and extracts were prepared from samples harvested at the beginning of the exponential phase of growth (OD600 = 0.2) (lanes 1) and in the stationary phase (OD600 = 1.0) (lanes 2). (B and C) FlgE (B) or FliC (C) expression in the ΔmucR mutant in stationary phase. Complementation of mucR in trans restored the WT phenotype for both FlgE and FliC. The strains were cultivated in 2YT broth, and extracts were prepared from samples harvested in stationary phase (OD600 = 1.0). Total extracts were separated by electrophoresis, transferred to nitrocellulose membranes, and probed with FlgE-specific or FliC-specific antiserum. A polyclonal anti-PrlR antibody was used to probe PrlR as a loading control.
To confirm this hypothesis, the relative levels of fliC (flagellin), flgE (hook monomer), fliF (membrane and supramembrane [MS] ring monomer), and ftcR mRNAs were determined by qRT-PCR for RNA extracted from the different strains harvested during the exponential growth phase (OD600 = 0.5) in 2YT medium. We found that a mucR deletion resulted in a significant increase in all the mRNA levels tested compared to the WT stain, including the mRNA levels corresponding to the master regulator FtcR (Table 2). For each mRNA, wild-type levels were restored by complementing the ΔmucR deletion (Table 2). Together, these results clearly indicate that B. melitensis MucR, as in S. meliloti, is a regulator that acts upstream of the master flagellar regulator FtcR, the orthologue of Rem.
MucR negatively regulates its own transcription.
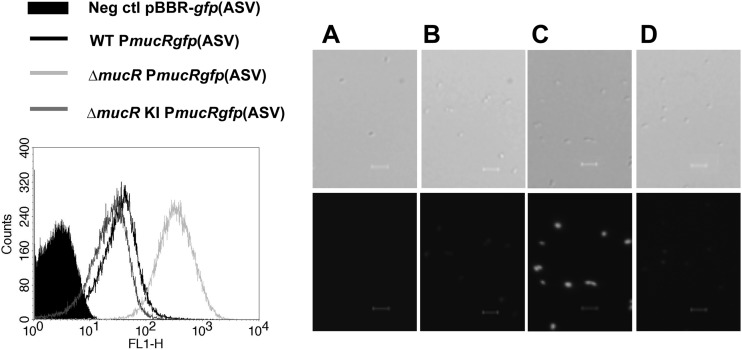
In S. meliloti, MucR negatively regulates its own transcription (18, 69). To pinpoint a potentially similar self-regulation, mucR promoter (PmucR) activity was monitored in B. melitensis WT, ΔmucR, and ΔmucRKI strains. All the strains harboring the plasmid pBBRPmucRgfp(ASV) were grown in 2YT, and the GFP(ASV) fluorescence intensity was measured by flow cytometry at different time points. Figure 8 shows the fluorescence signals measured by flow cytometry (left) and representative epifluorescence micrographs (right) of the different strains in mid-log growth phase. Similar results were obtained for each sampling time point (data not shown). The MFI of the ΔmucR mutant was higher (MFI = 350) than that of the WT strain (MFI = 45) or the corresponding complemented ΔmucRKI mutant (MFI = 30) (Fig. 8, left). The B. melitensis strain harboring the vector pBBR-gfp(ASV) was used as a negative control. PmucR is strongly induced when Brucella lacks the mucR gene, indicating that MucR negatively regulates its own transcription in B. melitensis 16M.
Fig 8.
MucR negatively autoregulates its own transcription. (Left) Fluorescence intensity measured by flow cytometry (5 × 104 events acquired) on B. melitensis WT, ΔmucR, and ΔmucRKI strains expressing the transcriptional fusion PmucRgfp(ASV) in mid-log-phase culture in 2YT medium. The WT harboring the plasmid pBBR-gfp(ASV) was used as a negative control. (Right) DIC (above) and corresponding fluorescence (bottom) images. (A) WT pBBR-gfp(ASV). (B) WT pBBRPmucRgfp(ASV). (C) ΔmucR pBBRPmucRgfp(ASV). (D) ΔmucRKI pBBRPmucRgfp(ASV). Scale bars, 3 μm.
DISCUSSION
In this study, we report the extensive characterization of an in-frame deletion mutant of B. melitensis mucR. This gene was previously identified as necessary for virulence in a transposon mutagenesis screen of B. melitensis but never characterized (40). Our study has confirmed that a ΔmucR mutant is attenuated in both cellular and murine models of infection. Our study has also shown that the most plausible explanation for its attenuation is a deficiency in intracellular survival, rather than a deficiency in cellular invasion.
In addition to reduced virulence, the ΔmucR strain exhibits pleiotropic complementable phenotypes, which we discuss here and tentatively correlate with the attenuation of the ΔmucR strain.
B. melitensis MucR is a functional orthologue of S. meliloti MucR.
The mucR gene is highly conserved in Rhizobiales (70) and encodes a protein predicted to contain a C2H2-type zinc finger motif. Zinc finger-containing proteins include DNA binding proteins that are able to bind a zinc ion via a conserved structure (e.g., through cysteine and histidine amino acids) (71). In all the bacteria in which MucR orthologues have been characterized, this regulator controls various cell envelope modifications with a common theme of exopolysaccharide synthesis and altered host-bacterial interaction (72–74). In S. meliloti, where MucR (61% identity with B. melitensis MucR) has been the most extensively studied, the regulator couples motility regulation with EPS production (21). The S. meliloti MucR protein appears to be highly specific for its own DNA recognition sequence, because it does not bind to sites recognized by Ros, the orthologous regulator in Agrobacterium tumefaciens (61% identity to B. melitensis MucR) (69, 75). In contrast, as shown on calcofluor plates (Fig. 6), the constitutive expression of the B. melitensis mucR gene in a mucR mutant of S. meliloti (Rm101) was able to restore EPS I (succinoglycan) production. This heterospecific-complementation experiment suggests that the mucR gene of B. melitensis 16M encodes a functional protein able to recognize MucR-specific promoter-targeting sequences, or at least the promoter(s) regulating the expression of EPS biosynthesis genes in S. meliloti.
B. melitensis MucR also controls flagellar-gene expression and the formation of the aggregative phenotype.
Our results clearly indicate that MucR of B. melitensis is also a repressor of flagellar-gene expression and likely acts upstream of FtcR, the flagellar master regulator orthologous to Rem (68) through which MucR of S. meliloti acts (21, 67). In B. melitensis 16M, flagellar-gene expression is very tightly regulated, given that the QS regulator VjbR (27), the sigma factors rpoEI (76, 77) and rpoH2 (76), and the cyclic-di-GMP phosphodiesterase BpdA (78) have already been shown to be involved in flagellar regulation. Here, we show that MucR is an additional actor within this complex regulatory network. According to the previous transcriptomic analyses, mucR expression is under the control of QS regulators (39). We confirmed these data by qRT-PCR using RNA extracts from ΔvjbR and WT strains at the end of log phase in rich medium (data not shown). This indicates that VjbR represses mucR expression and thus allows ftcR expression and subsequent flagellar-gene expression. Together, these results support the hypothesis that MucR plays an intermediate role between VjbR and the regulation of flagellar-gene expression.
It has previously been reported that B. melitensis QS mutants form bacterial aggregates in 2YT medium (29, 79). Moreover, we recently described a peculiar growth condition (2YT plus 400 mM NaCl for 72 h) in which WT B. melitensis also has a similar aggregative phenotype (57). This behavior could result from the production of EPS and/or a modification of envelope/surface properties. Because MucR is functionally conserved between S. meliloti and B. melitensis and because MucR in S. meliloti regulates EPS I biosynthesis (Fig. 6) (18, 65), we tested whether MucR is involved in the formation of bacterial clumps in B. melitensis 16M. Unfortunately, the ΔmucR strain displays a growth defect in hypersaline medium (see Fig. S2 in the supplemental material). Therefore, the absence of clumps in the culture could be due to poor growth or to the inability to properly respond to hypersaline stress. These data are consistent with the strong induction of the promoter PmucR due to ionic stress, which reinforces the idea that MucR is necessary to optimally respond to the hypersaline stress and could be required to promote aggregation. In this context, we showed that a mucR-overexpressing strain, in contrast to the WT strain, developed bacterial aggregates when grown in standard 2YT medium for 72 h (see Fig. S3 in the supplemental material), indicating that MucR could actually be involved in clump formation.
The aggregative phenotype of the mucR-overexpressing strain and the cell envelope modifications and flagellar-gene activation of the ΔmucR mutant appear to be coordinated phenotypes that are reminiscent of the paradigmatic transition from a “planktonic” to a “sessile” form of life, which often implies the inversely coordinated expression of surface polysaccharide components and the flagellar apparatus (11, 21, 80, 81).
Altogether, the negative autoregulation of MucR (Fig. 8), its negative impact on both flagellar mRNA and protein levels (Fig. 7 and Table 2), and the ability to form aggregates (Fig. 6; see Fig. S3 in the supplemental material) seem to be conserved between S. meliloti and B. melitensis. Moreover, in both species, MucR may possibly bind to a similar DNA binding site. This reinforces the idea that these bacterial species have evolved from a common ancestor and share molecular mechanisms for their interactions with their respective hosts (82, 83). Notably, the induction of PmucR activity by ionic stress and, to a lesser extent, osmotic stress (see Fig. S1 in the supplemental material) constitute the first factors identified that affect the expression of the autoregulated mucR gene in an alphaproteobacterium.
The mucR mutant has an altered growth phenotype and an exacerbated sensitivity to oxidative stress.
Despite having the same lag phase and a similar growth rate in a rich medium in vitro, the ΔmucR strain enters stationary phase much earlier (and thus at a lower OD600) than the WT strain. As previously demonstrated in a B. abortus hfq mutant, stationary-phase physiology plays an important role in the ability of brucellae to establish and maintain long-term intracellular residence in host macrophages (84) and probably also to cope with the stresses of the early, nonreplicative phase (the first 10 h) of intracellular infection (85). A common feature of all these conditions is exposure to oxidative stress either through the accumulation of endogenous oxygen radicals (86–88) or following the oxidative burst inside infected macrophages (89). Based on unpublished microarray analysis, the involvement of mucR in stress response mechanisms has been suggested for B. melitensis (41). Consistent with this hypothesis, we showed a strong increase in the sensitivity of the ΔmucR mutant to exogenous oxidative stress compared to the WT strain. This feature could partially explain the 2-log-unit decrease in survival in macrophages that this and other studies have described (Fig. 1) (40) and that we have also seen in HeLa cells.
ΔmucR sensitivity to detergents and polymyxin correlates with cell envelope alterations.
In addition to the sensitivity to oxidative stress, an increased susceptibility to detergents (SDS and Triton) and polymyxin B was also observed with the ΔmucR strain. These observations and the altered staining of the mutant colonies with Congo red suggest that there are major cell surface alterations in this B. melitensis mutant. A link between reduced virulence and sensitivity to detergents has also been observed in B. abortus cyclic β-glucan synthase (cgs) (66) and bvrR bvrS TCS mutants, which are also more sensitive to bactericidal polycationic substances (polymyxin B, melittin, and poly-l-lysine) (28). Mutants in the latter system have a severely altered cell envelope, including lipid A modifications (37, 38, 90).
Consistent with the detergent susceptibility assays, our qRT-PCR data suggest that MucR positively regulates B. melitensis cyclic glucan synthase (cgs). Unfortunately, we were unable to show modified Cgs protein expression by Western blotting using a previously described antibody (91) because the antibody generated a positive signal only in the cgs-overexpressing strain (D. Comerci, personal communication).
Further investigations would be necessary to test whether there is a change in cyclic β-glucan production in the different strains. If so, MucR would be the first regulator of cgs identified in brucellae.
In conclusion, we propose that the conserved MucR regulator plays multiple roles in B. melitensis 16M, controlling growth in bacteriological medium, virulence in macrophages and mice, flagellar expression, aggregation, and cell envelope homeostasis. Interestingly, in B. abortus 2308, MucR (100% amino acid identity with B. melitensis MucR) was very recently found (C. Caswell and R. M. Roop, personal communication) to be involved in virulence (in macrophage and spleen colonization in mice) and growth in bacteriological medium, illustrating the similarities between the roles of MucR in these two Brucella strains. Moreover, the autoregulation of mucR also occurs in B. abortus, in which MucR as been shown by electrophoretic mobility shift assay (EMSA) to bind its own promoter (100% nucleotide identity with B. melitensis) (C. Caswell and R. M. Roop, personal communication). In B. abortus 2308, MucR controls the expression of several transcriptional regulators, such as babR and nolR (C. Caswell and R. M. Roop, personal communication), suggesting that it can coordinate the expression of several regulons, which would explain the pleiotropic phenotype of mucR mutants.
Interestingly, we have identified a probable alteration in the core lipid A structure of the B. melitensis ΔmucR mutant. In fact, the ΔmucR background has a modified Western blot staining pattern for anti-core MAbs compared to a WT background or the complemented mutants. Interestingly, recognition of the free (not linked to the O chain) core lipid A structure is far less altered than the recognition of the O-chain-linked core (Fig. 4B). This staining pattern is reminiscent of a staining pattern described recently for a mutant for a putative core glycosyl transferase (WadC) (92). wadC transcript levels were not altered in the mucR mutant background (data not shown). Surprisingly, the B. abortus ΔmucR mutant is rough (by crystal violet staining), and this phenotype is complementable (C. Caswell and R. M. Roop, personal communication). Conversely, the B. melitensis 16M ΔmucR mutant reported here remains smooth, illustrating that there are remarkable differences in MucR function between Brucella strains that would be worth investigating. These phenotypic differences could likely be linked to still unidentified differences in the core lipid A proteins of these closely related species (93). For example, differences in the negative charge of the core lipid A (64) could also account for the differences in polymyxin B resistance between B. abortus and B. melitensis (94). This impact of MucR on the core lipid A has never been reported in the alphaproteobacteria studied previously and could be a more general compensatory mechanism that allows the bacteria to adapt to an altered envelope homeostasis (6). Alternatively, because MucR function is suspected to be interwoven into the cell cycle regulatory circuitry of Caulobacter crescentus (P. Viollier, personal communication), its differential impact on the envelopes of the two sibling cells is worth investigating in other asymmetrically dividing alphaproteobacteria (95). This opens a new avenue into deciphering why, how (directly or indirectly through other induced membrane changes), and in which part of the bacterial life cycle the lipid A core structure is modulated.
Supplementary Material
ACKNOWLEDGMENTS
We thank A. Becker (CeBiTec) and G. Walker (MIT) for providing the S. meliloti strains and C. Didembourg for his helpful technical assistance.
Part of this work was funded by an ARC Convention from the French community of Belgium (no. 08/13-015) and by the Interuniversity Attraction Poles Programme initiated by the Belgian Science Policy Office. A.M. is the recipient of a specialization grant from FRIA (Fonds pour la Formation à la Recherche dans l'Industrie et l'Agriculture).
Footnotes
Published ahead of print 16 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01336-12.
REFERENCES
- 1. Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Janczarek M. 2011. Environmental signals and regulatory pathways that influence exopolysaccharide production in rhizobia. Int. J. Mol. Sci. 12:7898–7933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li Y, Powell DA, Shaffer SA, Rasko DA, Pelletier MR, Leszyk JD, Scott AJ, Masoudi A, Goodlett DR, Wang X, Raetz CR, Ernst RK. 2012. LPS remodeling is an evolved survival strategy for bacteria. Proc. Natl. Acad. Sci. U. S. A. 109:8716–8721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. La MV, Raoult D, Renesto P. 2008. Regulation of whole bacterial pathogen transcription within infected hosts. FEMS Microbiol. Rev. 32:440–460 [DOI] [PubMed] [Google Scholar]
- 5. Reuhs BL, Williams MN, Kim JS, Carlson RW, Cote F. 1995. Suppression of the Fix− phenotype of Rhizobium meliloti exoB mutants by lpsZ is correlated to a modified expression of the K polysaccharide. J. Bacteriol. 177:4289–4296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hozbor DF, Pich Otero AJ, Lodeiro AR, Del Papa MF, Pistorio M, Lagares A. 2004. The symbiotic defect in a Sinorhizobium meliloti lipopolysaccharide mutant can be overcome by expression of other surface polysaccharides. Res. Microbiol. 155:855–860 [DOI] [PubMed] [Google Scholar]
- 7. Glazebrook J, Walker G. 1989. A novel exopolysaccharide can function in place of the calcofluor-binding exopolysaccharide in nodulation of alfalfa by Rhizobium meliloti. Cell 56:661–672 [DOI] [PubMed] [Google Scholar]
- 8. Cheng HP, Walker GC. 1998. Succinoglycan production by Rhizobium meliloti is regulated through the ExoS-ChvI two-component regulatory system. J. Bacteriol. 180:20–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reed JW, Glazebrook J, Walker GC. 1991. The exoR gene of Rhizobium meliloti affects RNA levels of other exo genes but lacks homology to known transcriptional regulators. J. Bacteriol. 173:3789–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wells D, Chen E, Fisher R, Long S. 2007. ExoR is genetically coupled to the ExoS-ChvI two-component system and located in the periplasm of Sinorhizobium meliloti. Mol. Microbiol. 64:647–664 [DOI] [PubMed] [Google Scholar]
- 11. Yao SY, Luo L, Har KJ, Becker A, Ruberg S, Yu GQ, Zhu JB, Cheng HP. 2004. Sinorhizobium meliloti ExoR and ExoS proteins regulate both succinoglycan and flagellum production. J. Bacteriol. 186:6042–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ozga DA, Lara JC, Leig JA. 1994. The regulation of exopolysaccharide production is important at two levels of nodule development in Rhizobium meliloti. Mol. Plant Microbe Interact. 7:758–765 [DOI] [PubMed] [Google Scholar]
- 13. Keating DH. 2007. The Sinorhizobium meliloti ExoR protein is required for the downregulation of lpsS transcription and succinoglycan biosynthesis in response to divalent cations. FEMS Microbiol. Lett. 267:23–29 [DOI] [PubMed] [Google Scholar]
- 14. Bélanger L, Dimmick KA, Fleming JS, Charles TC. 2009. Null mutations in Sinorhizobium meliloti exoS and chvI demonstrate the importance of this two-component regulatory system for symbiosis. Mol. Microbiol. 74:1223–1237 [DOI] [PubMed] [Google Scholar]
- 15. Doherty D, Leigh JA, Glazebrook J, Walker GC. 1988. Rhizobium meliloti mutants that overproduce the R. meliloti acidic calcofluor-binding exopolysaccharide. J. Bacteriol. 170:4249–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gibson KE, Campbell GR, Lloret J, Walker GC. 2006. CbrA is a stationary-phase regulator of cell surface physiology and legume symbiosis in Sinorhizobium meliloti. J. Bacteriol. 188:4508–4521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bertram-Drogatz PA, Quester I, Becker A, Puhler A. 1998. The Sinorhizobium meliloti MucR protein, which is essential for the production of high-molecular-weight succinoglycan exopolysaccharide, binds to short DNA regions upstream of exoH and exoY. Mol. Gen. Genet. 257:433–441 [DOI] [PubMed] [Google Scholar]
- 18. Keller M, Roxlau A, Weng WM, Schmidt M, Quandt J, Niehaus K, Jording D, Arnold W, Puhler A. 1995. Molecular analysis of the Rhizobium meliloti mucR gene regulating the biosynthesis of the exopolysaccharides succinoglycan and galactoglucan. Mol. Plant Microbe Interact. 8:267–277 [DOI] [PubMed] [Google Scholar]
- 19. McIntosh M, Krol E, Becker A. 2008. Competitive and cooperative effects in quorum-sensing-regulated galactoglucan biosynthesis in Sinorhizobium meliloti. J. Bacteriol. 190:5308–5317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rüberg S, Pühler A, Becker A. 1999. Biosynthesis of the exopolysaccharide galactoglucan in Sinorhizobium meliloti is subject to a complex control by the phosphate-dependent regulator PhoB and the proteins ExpG and MucR. Microbiology 145:603–611 [DOI] [PubMed] [Google Scholar]
- 21. Bahlawane C, McIntosh M, Krol E, Becker A. 2008. Sinorhizobium meliloti regulator MucR couples exopolysaccharide synthesis and motility. Mol. Plant Microbe Interact. 21:1498–1509 [DOI] [PubMed] [Google Scholar]
- 22. Mueller K, Gonzalez JE. 2011. Complex regulation of symbiotic functions is coordinated by MucR and quorum sensing in Sinorhizobium meliloti. J. Bacteriol. 193:485–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moreno E, Stackebrandt E, Dorsch M, Wolters J, Busch M, Mayer H. 1990. Brucella abortus 16S rRNA and lipid A reveal a phylogenetic relationship with members of the alpha-2 subdivision of the class Proteobacteria. J. Bacteriol. 172:3569–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pappas G, Papadimitriou P, Akritidis N, Christou L, Tsianos EV. 2006. The new global map of human brucellosis. Lancet Infect. Dis. 6:91–99 [DOI] [PubMed] [Google Scholar]
- 25. O'Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli ML, Bourg G, Foulongne V, Frutos P, Kulakov Y, Ramuz M. 1999. A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol. Microbiol. 33:1210–1220 [DOI] [PubMed] [Google Scholar]
- 26. Fretin D, Fauconnier A, Kohler S, Halling S, Leonard S, Nijskens C, Ferooz J, Lestrate P, Delrue RM, Danese I, Vandenhaute J, Tibor A, DeBolle X, Letesson JJ. 2005. The sheathed flagellum of Brucella melitensis is involved in persistence in a murine model of infection. Cell Microbiol. 7:687–698 [DOI] [PubMed] [Google Scholar]
- 27. Delrue RM, Deschamps C, Leonard S, Nijskens C, Danese I, Schaus JM, Bonnot S, Ferooz J, Tibor A, De Bolle X, Letesson JJ. 2005. A quorum-sensing regulator controls expression of both the type IV secretion system and the flagellar apparatus of Brucella melitensis. Cell Microbiol. 7:1151–1161 [DOI] [PubMed] [Google Scholar]
- 28. Sola-Landa A, Pizarro-Cerda J, Grillo MJ, Moreno E, Moriyon I, Blasco JM, Gorvel JP, Lopez-Goni I. 1998. A two-component regulatory system playing a critical role in plant pathogens and endosymbionts is present in Brucella abortus and controls cell invasion and virulence. Mol. Microbiol. 29:125–138 [DOI] [PubMed] [Google Scholar]
- 29. Uzureau S, Godefroid M, Deschamps C, Lemaire J, De Bolle X, Letesson JJ. 2007. Mutations of the quorum sensing-dependent regulator VjbR lead to drastic surface modifications in Brucella melitensis. J. Bacteriol. 189:6035–6047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ugalde R. 1999. Intracellular lifestyle of Brucella spp. Common genes with other animal pathogens, plant pathogens, and endosymbionts. Microbes Infect. 1:1211–1219 [DOI] [PubMed] [Google Scholar]
- 31. Lestrate P, Dricot A, Delrue R, Lambert C, Martinelli V, De Bolle X, Letesson J, Tibor A. 2003. Attenuated signature-tagged mutagenesis mutants of Brucella melitensis identified during the acute phase of infection in mice. Infect. Immun. 71:7053–7060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fernandez-Prada CM, Nikolich M, Vemulapalli R, Sriranganathan N, Boyle SM, Schurig GG, Hadfield TL, Hoover DL. 2001. Deletion of wboA enhances activation of the lectin pathway of complement in Brucella abortus and Brucella melitensis. Infect. Immun. 69:4407–4416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Porte F, Naroeni A, Ouahrani-Bettache S, Liautard JP. 2003. Role of the Brucella suis lipopolysaccharide O antigen in phagosomal genesis and in inhibition of phagosome-lysosome fusion in murine macrophages. Infect. Immun. 71:1481–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Fernandez-Prada CM, Zelazowska EB, Nikolich M, Hadfield TL, Roop RM, II, Robertson GL, Hoover DL. 2003. Interactions between Brucella melitensis and human phagocytes: bacterial surface O-polysaccharide inhibits phagocytosis, bacterial killing, and subsequent host cell apoptosis. Infect. Immun. 71:2110–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Arellano-Reynoso B, Lapaque N, Salcedo S, Briones G, Ciocchini AE, Ugalde R, Moreno E, Moriyón I, Gorvel J-P. 2005. Cyclic β-1,2-glucan is a Brucella virulence factor required for intracellular survival. Nat. Immunol. 6:618–625 [DOI] [PubMed] [Google Scholar]
- 36. Guzman-Verri C, Manterola L, Sola-Landa A, Parra A, Cloeckaert A, Garin J, Gorvel Moriyon J-PI, Moreno E, Lopez-Goni I. 2002. The two-component system BvrR/BvrS essential for Brucella abortus virulence regulates the expression of outer membrane proteins with counterparts in members of the Rhizobiaceae. Proc. Natl. Acad. Sci. U. S. A. 99:12375–12380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Manterola L, Moriyon I, Moreno E, Sola-Landa A, Weiss D, Koch M, Howe J, Brandenburg K, Lopez-Goni I. 2005. The lipopolysaccharide of Brucella abortus BvrS/BvrR mutants contains lipid A modifications and has higher affinity for bactericidal cationic peptides. J. Bacteriol. 187:5631–5639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Viadas C, Rodríguez MC, Sangari FJ, Gorvel García-Lobo J-PJM, López-Goñi I. 2010. Transcriptome analysis of the Brucella abortus BvrR/BvrS two-component regulatory system. PLoS One 5:e10216 doi:10.1371/journal.pone.0010216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Uzureau S, Lemaire J, Delaive E, Dieu M, Gaigneaux A, Raes M, De Bolle X, Letesson JJ. 2010. Global analysis of quorum sensing targets in the intracellular pathogen Brucella melitensis 16M. J. Proteome Res. 9:3200–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu Q, Pei J, Turse C, Ficht TA. 2006. Mariner mutagenesis of Brucella melitensis reveals genes with previously uncharacterized roles in virulence and survival. BMC Microbiol. 6:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Arenas-Gamboa AM, Rice-Ficht AC, Kahl-McDonagh MM, Ficht TA. 2011. Protective efficacy and safety of Brucella melitensis 16MDeltamucR against intraperitoneal and aerosol challenge in BALB/c mice. Infect. Immun. 79:3653–3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Meade HM, Long SR, Ruvkun GB, Brown SE, Ausubel FM. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leigh JA, Signer ER, Walker GC. 1985. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. U. S. A. 82:6231–6235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Casse F, Boucher C, Julliot JS, Michel M, Dénarié J. 1979. Identification and characterization of large plasmid in Rhizobium meliloti using agarose gel electrophoresis. Microbiology 113:229–242 [Google Scholar]
- 45. Becker A, Ruberg S, Kuster H, Roxlau AA, Keller M, Ivashina T, Cheng HP, Walker GC, Puhler A. 1997. The 32-kilobase exp gene cluster of Rhizobium meliloti directing the biosynthesis of galactoglucan: genetic organization and properties of the encoded gene products. J. Bacteriol. 179:1375–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Simon RPU, Pühler A. 1983. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Nat. Biotechnol. 1:784–791 [Google Scholar]
- 47. Kovach ME, Phillips RW, Elzer PH, Roop RM, II, Peterson KM. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800–802 [PubMed] [Google Scholar]
- 48. Donnenberg MS, Kaper JB. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Andersen JB, Sternberg C, Poulsen LK, Bjorn SP, Givskov M, Molin S. 1998. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64:2240–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ausubel FM, Brent R, Kingston RE, Moore DE, Seidmann JG, Smith JA, Struhl K. 1991. Current protocols in molecular biology. Wiley-Interscience, New York, NY [Google Scholar]
- 51. Mignolet J, Van der Henst C, Nicolas C, Deghelt M, Dotreppe D, Letesson JJ, De Bolle X. 2010. PdhS, an old-pole-localized histidine kinase, recruits the fumarase FumC in Brucella abortus. J. Bacteriol. 192:3235–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Delrue RM, Martinez-Lorenzo M, Lestrate P, Danese I, Bielarz V, Mertens P, De Bolle X, Tibor A, Gorvel JP, Letesson JJ. 2001. Identification of Brucella spp. genes involved in intracellular trafficking. Cell Microbiol. 3:487–497 [DOI] [PubMed] [Google Scholar]
- 53. Zhang X, Ren J, Li N, Liu W, Wu Q. 2009. Disruption of the BMEI0066 gene attenuates the virulence of Brucella melitensis and decreases its stress tolerance. Int. J. Biol. Sci. 5:570–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Garin-Bastuji B, Bowden RA, Dubray G, Limet JN. 1990. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting analysis of smooth-lipopolysaccharide heterogeneity among Brucella biovars related to A and M specificities. J. Clin. Microbiol. 28:2169–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cloeckaert A, Zygmunt MS, Nicolle JC, Dubray G, Limet JN. 1992. O-chain expression in the rough Brucella melitensis strain B115: induction of O-polysaccharide-specific monoclonal antibodies and intracellular localization demonstrated by immunoelectron microscopy. J. Gen. Microbiol. 138:1211–1219 [DOI] [PubMed] [Google Scholar]
- 56. Bowden RA, Cloeckaert A, Zygmunt MS, Bernard S, Dubray G. 1995. Surface exposure of outer membrane protein and lipopolysaccharide epitopes in Brucella species studied by enzyme-linked immunosorbent assay and flow cytometry. Infect. Immun. 63:3945–3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mirabella A, Yanez Villanueva RM, Delrue RM, Uzureau S, Zygmunt MS, Cloeckaert A, De Bolle X, Letesson JJ. 2012. The two-component system PrlS/PrlR of Brucella melitensis is required for persistence in mice and appears to respond to ionic strength. Microbiology 158:2642–2651 [DOI] [PubMed] [Google Scholar]
- 58. Jacobs C, Domian IJ, Maddock JR, Shapiro L. 1999. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell 97:111–120 [DOI] [PubMed] [Google Scholar]
- 59. Jubier-Maurin V, Rodrigue A, Ouahrani-Bettache S, Layssac M, Mandrand-Berthelot MA, Kohler S, Liautard JP. 2001. Identification of the nik gene cluster of Brucella suis: regulation and contribution to urease activity. J. Bacteriol. 183:426–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Roop RM, II, Gee JM, Robertson GT, Richardson JM, Ng WL, Winkler ME. 2003. Brucella stationary-phase gene expression and virulence. Annu. Rev. Microbiol. 57:57–76 [DOI] [PubMed] [Google Scholar]
- 61. Jenkins DE, Schultz JE, Matin A. 1988. Starvation-induced cross protection against heat or H2O2 challenge in Escherichia coli. J. Bacteriol. 170:3910–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lapaque N, Moriyon I, Moreno E, Gorvel JP. 2005. Brucella lipopolysaccharide acts as a virulence factor. Curr. Opin. Microbiol. 8:60–66 [DOI] [PubMed] [Google Scholar]
- 64. Velasco J, Bengoechea JA, Brandenburg K, Lindner B, Seydel U, Gonzalez D, Zahringer U, Moreno E, Moriyon I. 2000. Brucella abortus and its closest phylogenetic relative, Ochrobactrum spp., differ in outer membrane permeability and cationic peptide resistance. Infect. Immun. 68:3210–3218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhan HJ, Levery SB, Lee CC, Leigh JA. 1989. A second exopolysaccharide of Rhizobium meliloti strain SU47 that can function in root nodule invasion. Proc. Natl. Acad. Sci. U. S. A. 86:3055–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Briones G, Inon de Iannino N, Roset M, Vigliocco A, Paulo PS, Ugalde RA. 2001. Brucella abortus cyclic beta-1,2-glucan mutants have reduced virulence in mice and are defective in intracellular replication in HeLa cells. Infect. Immun. 69:4528–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Rotter C, Muhlbacher S, Salamon D, Schmitt R, Scharf B. 2006. Rem, a new transcriptional activator of motility and chemotaxis in Sinorhizobium meliloti. J. Bacteriol. 188:6932–6942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Leonard S, Ferooz J, Haine V, Danese I, Fretin D, Tibor A, de Walque S, De Bolle X, Letesson JJ. 2007. FtcR is a new master regulator of the flagellar system of Brucella melitensis 16M with homologs in Rhizobiaceae. J. Bacteriol. 189:131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bertram-Drogatz PA, Ruberg S, Becker A, Puhler A. 1997. The regulatory protein MucR binds to a short DNA region located upstream of the mucR coding region in Rhizobium meliloti. Mol. Gen. Genet. 254:529–538 [DOI] [PubMed] [Google Scholar]
- 70. Bouhouche N, Syvanen M, Kado CI. 2000. The origin of prokaryotic C2H2 zinc finger regulators. Trends Microbiol. 8:77–81 [DOI] [PubMed] [Google Scholar]
- 71. Berg JM. 1990. Zinc fingers and other metal-binding domains. Elements for interactions between macromolecules. J. Biol. Chem. 265:6513–6516 [PubMed] [Google Scholar]
- 72. Bittinger MA, Handelsman J. 2000. Identification of genes in the RosR regulon of Rhizobium etli. J. Bacteriol. 182:1706–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Janczarek M, Kutkowska J, Piersiak T, Skorupska A. 2010. Rhizobium leguminosarum bv. trifolii rosR is required for interaction with clover, biofilm formation and adaptation to the environment. BMC Microbiol. 10:284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Janczarek M, Skorupska A. 2007. The Rhizobium leguminosarum bv. trifolii RosR: transcriptional regulator involved in exopolysaccharide production. Mol. Plant Microbe Interact. 20:867–881 [DOI] [PubMed] [Google Scholar]
- 75. Close TJ, Tait RC, Kado CI. 1985. Regulation of Ti plasmid virulence genes by a chromosomal locus of Agrobacterium tumefaciens. J. Bacteriol. 164:774–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Delory M, Hallez R, Letesson J, De Bolle X. 2006. An RpoH-like heat shock sigma factor is involved in stress response and virulence in Brucella melitensis 16M. J. Bacteriol. 188:7707–7710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ferooz J, Lemaire J, Delory M, De Bolle X, Letesson JJ. 2011. RpoE1, an extracytoplasmic function sigma factor, is a repressor of the flagellar system in Brucella melitensis. Microbiology 157:1263–1268 [DOI] [PubMed] [Google Scholar]
- 78. Petersen E, Chaudhuri P, Gourley C, Harms J, Splitter G. 2011. Brucella melitensis cyclic di-GMP phosphodiesterase BpdA controls expression of flagellar genes. J. Bacteriol. 193:5683–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Godefroid M, Svensson MV, Cambier P, Uzureau S, Mirabella A, De Bolle X, Van Cutsem P, Widmalm G, Letesson J-J. 2010. Brucella melitensis 16M produces a mannan and other extracellular matrix components typical of a biofilm. FEMS Immunol. Med. Microbiol. 59:364–377 [DOI] [PubMed] [Google Scholar]
- 80. Merritt JH, Brothers KM, Kuchma SL, O'Toole GA. 2007. SadC reciprocally influences biofilm formation and swarming motility via modulation of exopolysaccharide production and flagellar function. J. Bacteriol. 189:8154–8164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yeung AT, Torfs EC, Jamshidi F, Bains M, Wiegand I, Hancock RE, Overhage J. 2009. Swarming of Pseudomonas aeruginosa is controlled by a broad spectrum of transcriptional regulators, including MetR. J. Bacteriol. 191:5592–5602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Batut J, Andersson SGE, O'Callaghan D. 2004. The evolution of chronic infection strategies in the α-proteobacteria. Nat. Rev. Microbiol. 2:933–945 [DOI] [PubMed] [Google Scholar]
- 83. Bellefontaine AF, Pierreux CE, Mertens P, Vandenhaute J, Letesson JJ, De Bolle X. 2002. Plasticity of a transcriptional regulation network among alpha-proteobacteria is supported by the identification of CtrA targets in Brucella abortus. Mol. Microbiol. 43:945–960 [DOI] [PubMed] [Google Scholar]
- 84. Robertson G, Roop R. 1999. The Brucella abortus host factor I (HF-I) protein contributes to stress resistance during stationary phase and is a major determinant of virulence in mice. Mol. Microbiol. 34:690–700 [DOI] [PubMed] [Google Scholar]
- 85. Barbier T, Nicolas C, Letesson JJ. 2011. Brucella adaptation and survival at the crossroad of metabolism and virulence. FEBS Lett. 585:2929–2934 [DOI] [PubMed] [Google Scholar]
- 86. Dukan S, Nystrom T. 1999. Oxidative stress defense and deterioration of growth-arrested Escherichia coli cells. J. Biol. Chem. 274:26027–26032 [DOI] [PubMed] [Google Scholar]
- 87. Gonzalez-Flecha B, Demple B. 1995. Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli. J. Biol. Chem. 270:13681–13687 [DOI] [PubMed] [Google Scholar]
- 88. Steele KH, Baumgartner JE, Valderas MW, Roop RM., II 2010. Comparative study of the roles of AhpC and KatE as respiratory antioxidants in Brucella abortus 2308. J. Bacteriol. 192:4912–4922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jiang X, Leonard B, Benson R, Baldwin CL. 1993. Macrophage control of Brucella abortus: role of reactive oxygen intermediates and nitric oxide. Cell Immunol. 151:309–319 [DOI] [PubMed] [Google Scholar]
- 90. Lamontagne J, Butler H, Chaves-Olarte E, Hunter J, Schirm M, Paquet C, Tian M, Kearney P, Hamaidi L, Chelsky D, Moriyón I, Moreno E, Paramithiotis E. 2007. Extensive cell envelope modulation is associated with virulence in Brucella abortus. J. Proteome Res. 6:1519–1529 [DOI] [PubMed] [Google Scholar]
- 91. Ciocchini AE, Roset MS, Inon de Iannino N, Ugalde RA. 2004. Membrane topology analysis of cyclic glucan synthase, a virulence determinant of Brucella abortus. J. Bacteriol. 186:7205–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Conde-Alvarez R, Arce-Gorvel V, Iriarte M, Mancek-Keber M, Barquero-Calvo E, Palacios-Chaves L, Chacon-Diaz C, Chaves-Olarte E, Martirosyan A, von Bargen K, Grillo MJ, Jerala R, Brandenburg K, Llobet E, Bengoechea JA, Moreno E, Moriyon I, Gorvel JP. 2012. The lipopolysaccharide core of Brucella abortus acts as a shield against innate immunity recognition. PLoS Pathog. 8:e1002675 doi:10.1371/journal.ppat.1002675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. González D, Grilló M-J, De Miguel Ali M-JT, Arce-Gorvel V, Delrue R-M, Conde-Álvarez R, Muñoz P, López-Goñi I, Iriarte M, Marín Weintraub C-MA, Widmalm G, Zygmunt M, Letesson J-J, Gorvel J- P, Blasco Moriyón J-MI. 2008. Brucellosis vaccines: assessment of Brucella melitensis lipopolysaccharide rough mutants defective in core and O-polysaccharide synthesis and export. PLoS One 3:e2760 doi:10.1371/journal.pone.0002760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Farrell ID, Robertson L. 1967. The sensitivity of the biotypes of Brucella abortus to three antibiotics used in selective media, and the description of a new biotype. J. Hyg. (London) 65:165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hallez R, Bellefontaine AF, Letesson JJ, De Bolle X. 2004. Morphological and functional asymmetry in alpha-proteobacteria. Trends Microbiol. 12:361–365 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.