Abstract
Bacteria within biofilms are protected from multiple stresses, including immune responses and antimicrobial agents. The biofilm-forming ability of bacterial pathogens has been associated with increased antibiotic resistance and chronic recurrent infections. Although biofilms have been well studied for several gut pathogens, little is known about biofilm formation by anaerobic gut species. The obligate anaerobe Clostridium difficile causes C. difficile infection (CDI), a major health care-associated problem primarily due to the high incidence of recurring infections. C. difficile colonizes the gut when the normal intestinal microflora is disrupted by antimicrobial agents; however, the factors or processes involved in gut colonization during infection remain unclear. We demonstrate that clinical C. difficile strains, i.e., strain 630 and the hypervirulent strain R20291, form structured biofilms in vitro, with R20291 accumulating substantially more biofilm. Microscopic and biochemical analyses show multiple layers of bacteria encased in a biofilm matrix containing proteins, DNA, and polysaccharide. Employing isogenic mutants, we show that virulence-associated proteins, Cwp84, flagella, and a putative quorum-sensing regulator, LuxS, are all required for maximal biofilm formation by C. difficile. Interestingly, a mutant in Spo0A, a transcription factor that controls spore formation, was defective for biofilm formation, indicating a possible link between sporulation and biofilm formation. Furthermore, we demonstrate that bacteria in clostridial biofilms are more resistant to high concentrations of vancomycin, a drug commonly used for treatment of CDI. Our data suggest that biofilm formation by C. difficile is a complex multifactorial process and may be a crucial mechanism for clostridial persistence in the host.
INTRODUCTION
Biofilms are sessile surface-associated microbial communities, encapsulated within self-produced polymeric matrices (1). Biofilms represent the predominant state of bacteria in nature; only a small fraction of bacteria in natural ecosystems are believed to exist planktonically (2). Bacteria in biofilms are known to be more resistant to different environmental stresses, including antibiotics (2). The human large intestine is a good example of an extremely complex ecosystem, home to numerous bacterial species of microflora, which play an important role in protection against gut diseases (3). Various gut pathogens, including enterotoxigenic Escherichia coli, Salmonella, Yersinia, etc., can alter the dynamics of the gut due to their highly adhesive and invasive properties (4) and thus establish infections. Biofilm formation by gut pathogens such as enteroaggregative E. coli has been well studied both in vitro and in vivo (5, 6). Several bacterial factors such as adhesins and pili, which mediate biofilm formation, have been implicated in bacterial colonization and virulence (7).
Biofilm formation has been characterized for very few individual bacterial gut species of the numerous anaerobic species that populate the gut. This could be attributed to difficult cultivation and genetic manipulation of such bacteria. Clostridium difficile is a spore-forming, Gram-positive anaerobic bacillus that can causes severe gastrointestinal infections in humans (8). C. difficile infection (CDI) is usually associated with antimicrobial therapy since it results in disruption of the normal microbiota. The clinical symptoms of CDI range from mild or severe diarrhea to serious inflammatory conditions, including pseudomembranous colitis (8). CDI is one of the predominant nosocomial infections worldwide today. The population with the largest risk for CDI is the elderly, although CDI in younger patients is on the rise (9). The transmission of clostridial disease occurs through spores, as demonstrated recently using a murine model for C. difficile infection (10, 11).
CDI is primarily a toxin-mediated disease. The two large clostridial toxins A (TcdA) and B (TcdB) are the best-characterized virulence factors of C. difficile (12–15). C. difficile toxins have been shown to have many effects, including disorganization of the cell actin cytoskeleton and tight junctions, induction of apoptosis, fluid accumulation, and destruction of the epithelium (14, 15). Although the toxins are key virulent factors, a role for bacterial colonization has been highlighted in recent years. Mice immunized with C. difficile surface proteins demonstrated reduced intestinal colonization with C. difficile (16). Recently, the fibronectin-binding adhesin protein A was shown to play a role in C. difficile colonization (17). The high- and low-molecular-weight surface layer proteins (SLPs) are predicted to be involved in adherence of C. difficile to host cells during the infection (18, 19). Cell wall proteins (CWPs) such as Cwp66 and Cwp84 have been shown to be important in the adherence and degradation of the extracellular matrix (20, 21).
Recurrent bacterial infections have been associated with the ability to form resilient biofilms for various pathogens (22). Persistent recurring C. difficile infections have presented a major hurdle in the treatment of CDI (23). The ability to form biofilms has been demonstrated recently for related clostridial species and for C. difficile in relation to other intestinal species (24, 25). However, biofilm formation by C. difficile, or the mechanisms involved in this process, has not yet been characterized. We describe biofilm formation by clinical C. difficile strains in vitro using multiple techniques. Using isogenic mutants of proteins associated with clostridial pathogenesis we have identified surface and regulatory proteins that are required for biofilm development by the hypervirulent strain R20291. We also report a role for C. difficile biofilms in resistance to antibiotics commonly used for the treatment of CDI.
MATERIALS AND METHODS
Bacterial strains and media.
Two C. difficile strains, 630 and strain B1/NAP1/027 R20291 (isolated from the Stoke Mandeville outbreak in 2004 and 2005), were used in our studies. C. difficile strains were grown at 37°C in an anaerobic workstation (Don Whitley, United Kingdom) in BHIS, brain heart infusion (Bacto, USA) supplemented with l-cysteine (0.1% (wt/vol), and yeast extract (5 mg/ml; Bacto, USA). Details about antibiotics are described in the supplemental material. Table S1 in the supplemental material lists the strains and plasmids used in the present study.
Biofilm formation assay.
For the generation of biofilms, overnight cultures of C. difficile were diluted 1:100 into fresh BHIS, containing 0.1 M glucose or 0.3 M NaCl, and incubated in the tissue culture treated 24-well polystyrene plates (Costar, USA), 1 ml per well, in anaerobic conditions at 37°C for 1 h to 120 h. Twenty-four-well cell culture plates were preincubated in anaerobic conditions for 48 h prior to use. To prevent evaporation of liquid, the plates were wrapped with parafilm.
Measurement of biofilm biomass.
Measurement of biofilm biomass with crystal violet (CV) was done as described in previously published methods (24). After the required incubation, wells of the 24-well plate were gently washed twice with sterile phosphate-buffered saline (PBS) and then allowed to dry for 10 min. The biofilm was stained with 1 ml of filter-sterilized 0.2% CV and incubated for 30 min at 37°C in anaerobic conditions. CV was removed from the wells, followed by two washes with sterile PBS. The dye was extracted by adding 1 ml of methanol to each well, followed by incubation for 30 min at room temperature in aerobic conditions. The methanol-extracted dye was diluted 1:1, 1:10, or 1:100 and A570 was measured with spectrophotometer Ultrospec 500pro (Amersham Biosciences).
For bacterial cell counts from biofilms, the planktonic phase was first removed and wells were washed twice with PBS. The adherent biofilms were then detached by scraping with a sterile pipette tip, washed in PBS, and plated on BHIS for determination of the number of CFU present in the biofilm.
Enzymatic inhibition of biofilms.
To study whether enzymes affect the biofilm formation, 0.1 mg of proteinase K/ml and 2 U of DNase I/ml was added to biofilms at time zero. To see whether these enzymes affect preformed biofilms, biofilms were allowed to form in 24-well plate as described above, washed twice with sterile PBS. Fresh BHIS plus 0.1 M Glc with 0.1 mg of proteinase K/ml or 2 U of DNase I/ml was added, and the biofilms were incubated for a further 24 h, followed by staining with 0.2% CV as described above.
Confocal microscopy analysis of biofilm formation.
C. difficile strains were grown in 4-well glass chamber slides (BD Falcon, USA) in BHIS plus 0.1 M glucose at 37°C in anaerobic conditions for 24 or 72 h and then stained with various staining reagents. For biofilm staining with the fluorescent BacLight Live/Dead stain mixture (Molecular Probes/Invitrogen), which contains the nucleic acid stains Syto 9 and propidium iodide, wells were gently washed twice with PBS–0.1% saponin to remove unattached cells, followed by incubation with dye for 15 min at 37°C. Before removal from the anaerobic chamber, the bacteria were fixed with 4% paraformaldehyde (PFA) for 15 min.
For immunofluorescent staining bacteria were fixed with 4% PFA for 15 min, washed three times with 2% bovine serum albumin (BSA)–PBS, and blocked with 2% BSA and 0.1% saponin in PBS for 10 min. The samples were then incubated with mouse serum against fixed R20291 strain or against a synthetic C. difficile PSII polysaccharide (synthetic phosphorylated hexasaccharide repeating unit) (26) diluted 1:1,000 for 1 h, followed by incubation with Alexa Fluor 568 goat anti-mouse antibodies diluted 1:500 for 1 h.
Biofilm matrix was labeled by SYPRO Ruby biofilm matrix stain (Molecular Probes/Invitrogen), which stains matrices of biofilms (labels most classes of proteins, including glycoproteins, phosphoproteins, lipoproteins, calcium-binding proteins, and fibrillar proteins). After two washes with sterile PBS–0.1% saponin sample were incubated for 30 min with 1 ml of Ruby biofilm matrix stain at room temperature and then washed with distilled water.
All chamber slides were mounted with ProLong Gold Antifade reagent (Molecular Probes/Invitrogen) and analyzed with a Zeiss Observer LSM 710 confocal scanning microscope.
Construction of C. difficile mutants and complemented strains.
Mutations in the genes sleC, spo0A, fliC, and luxS were generated using the insertional inactivation system, ClosTron, as previously described (27–30). Construction of the fliC and luxS mutants is described in the supplemental material. The positions of in-frame deletions and ClosTron insertions are reported in Table S1 in the supplemental material. To generate the cwp84 mutant, the 3′ end of the cwp84 gene was deleted by using allelic exchange as described elsewhere (Y. K. Ng et al. unpublished data). Complementation studies are described in the supplemental material.
Measurement of C. difficile spores in biofilm.
C. difficile spores were measured as previously described (27). At day 1, 3, and 5 of biofilm formation samples were removed from the anaerobic chamber and heated at 80°C for 25 min to kill the vegetative cells but not the spores. Serially diluted were then plated onto BHI agar and BHI supplemented with 0.1% taurocholate (Sigma-Aldrich, USA). The plates were incubated for 24 to 48 h.
Biofilm resistance to antibiotics.
To determine the resistance capabilities of planktonic and biofilm cells, we used antibiotic vancomycin (Sigma-Aldrich) at 100 times the MIC (the MIC was determined to be 0.2 μg/ml for R20291 [data not shown]). Resistance was determined to both sessile and planktonic populations from the same C. difficile culture. The supernatant (planktonic fraction) from 1- or 3-day biofilm cultures in six-well plates was removed, washed once in sterile PBS, resuspended in BHIS–0.1 M glucose with 20 μg of vancomycin (Sigma-Aldrich)/ml, and incubated in fresh six-well plates. In parallel, to six-well plate where supernatants were removed, fresh BHIS–0.1 M glucose containing 20 μg of vancomycin/ml was added to the adherent biofilms (sessile part). Treatment with vancomycin was performed for 6 or 24 h. CFU from planktonic and adherent phases were determined by plating dilutions on BHIS agar. Biofilm cell counts were determined as described above. The percent survival was calculated by dividing the number of CFU posttreatment by the initial number of CFU.
To investigate whether the protection from vancomycin is a result of biofilm structure or a physiological attribute of the cells in the biofilm, we perform experiment as described above. The sessile part of biofilm was disrupted gently by pipetting and incubated with BHIS with 0.1 M Glc and 20 μg of vancomycin/ml. The planktonic growth from the same well was washed and incubated with BHIS with 0.1 M Glc and 20 μg of vancomycin/ml. CFU counts were determined after 6 and 24 h, and counts before and after different times of treatment were compared.
Statistical analyses.
All biofilm experiments were performed in triplicates and at least three independent experiments were performed. A paired Student t test was performed to determine whether the differences between two groups was significant. P values of <0.05 were considered statistically significant.
RESULTS
C. difficile forms time- and strain-dependent biofilms in vitro.
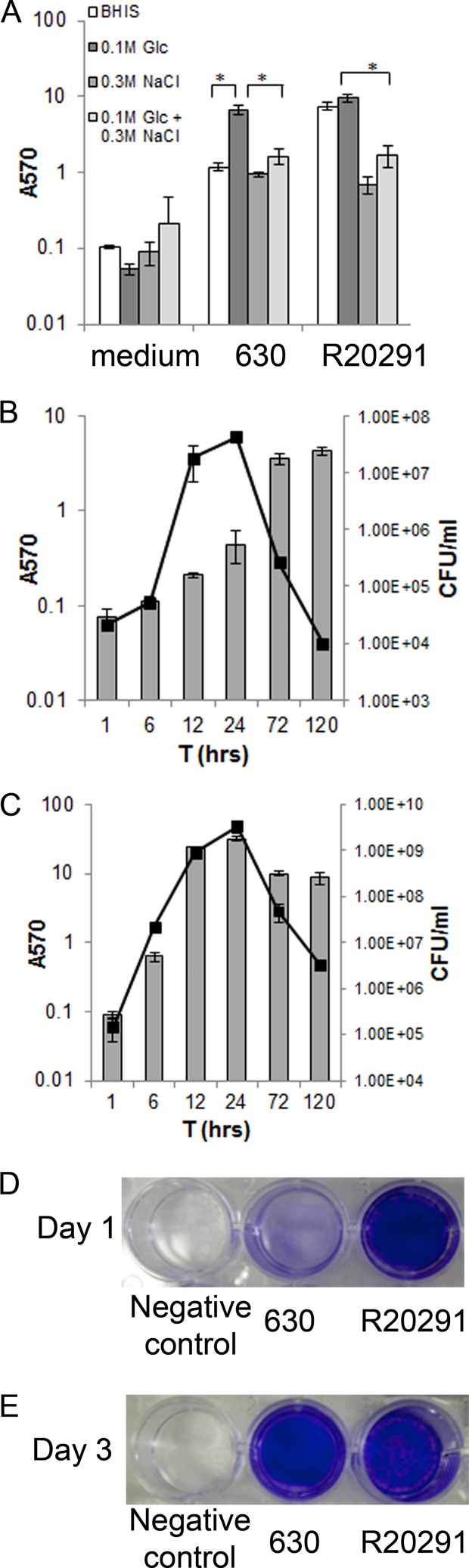
In order to investigate biofilm formation by C. difficile, we studied two strains, C. difficile 630, a commonly studied clinical strain, and the hypervirulent strain R20291. Sessile biomass (biofilm) was measured by staining the biofilms with CV, as described in Materials and Methods. We tested biofilm formation in several media (chemically defined medium, rich medium TYM, and BHIS) supplemented with combinations of sodium chloride and glucose (data not shown). Both strains form higher amounts of biofilm in rich medium BHIS supplemented with 0.1 M glucose. In the absence of glucose, strain 630 forms significantly lower amounts of biofilm compared to that seen in the presence of 0.1 M glucose. However, the presence of glucose does not significantly affect biofilm formation for the strain R20291. The presence of 0.3 M sodium chloride inhibits biofilm formation in both strains (Fig. 1A).
Fig 1.
C. difficile biofilm formation in vitro. (A) Biofilm formation by strain 630 and hypervirulent strain R20291, in BHIS supplemented with 0.1 M glucose or 0.3 M NaCl, for 3 days at 37°C in anaerobic conditions. Biofilm formation was measured by CV staining. (B and C) Time course for biofilm formation for strains 630 (B) and R20291 (C) measured by CV staining (bars) and colony counts (CFU/ml, line). The results are presented in log scale, and the error bars represent standard deviations (P < 0.05). The data are representative of at least three independent experiments, each performed in triplicates. (D and E) Photographs of biofilms formed on a 24-well plate for strains 630 and R20291 on day 1 (D) and day 3 (E) are shown.
Biofilms formed in all conditions tested were adherent in nature and were found to form only on the bottoms of the wells (of 24-well plates) and not on the surface of the culture medium. Biofilm formation in BHIS containing 0.1 M glucose was monitored over a time period of 1 to 120 h and quantitated by both CV staining and bacterial counts. C. difficile 630 biofilm accumulation appears to be maximal at day 5 (120 h) (Fig. 1B). Strain R20291 starts to form biofilm after 6 h of incubation, with maximum biofilm formation after 24 h, as measured by CV staining (Fig. 1C). Biofilm formed by the strain R20291 uniformly covers the entire surface area of the bottom of the wells (24-well plate) at 24 h (Fig. 1D), while at the same time point for strain 630 (Fig. 1D) we saw much less biofilm formation. At 72 h (Fig. 1E), we observed relatively more uniform biofilms for strain 630 and uneven biofilms for strain R20291 (Fig. 1E). Bacterial counts from biofilms indicated a similar trend of increasing bacterial numbers until 24 h and a decrease after this period for both strains. However, it is clear that R20291 biofilms have significantly higher number of bacteria as well as higher amounts of biofilms as quantitated by CV, compared to 630, under the conditions tested.
C. difficile biofilms are multilayered and encased in a thick matrix.
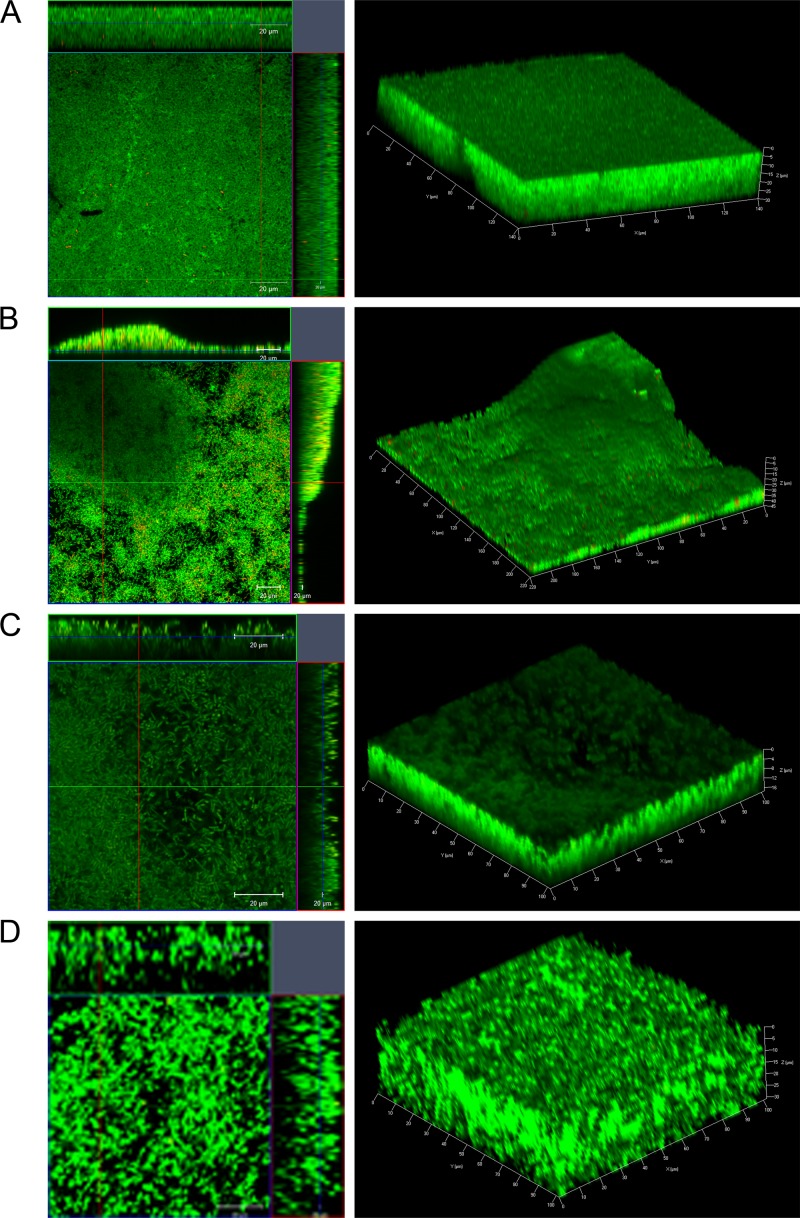
To examine biofilm formation by strains R20291 and 630 further, biofilms were allowed to form on glass slides in BHIS containing 0.1 M glucose for one or 3 days. Live/Dead staining (with Syto 9 and propidium iodide dyes staining live bacteria green and dead bacteria red, respectively) was used in order to evaluate the bacterial viability and biofilm thickness. We found that the majority of bacteria in both R20291 (Fig. 2A and B) and 630 (Fig. 2C and D) biofilms were alive with a minor number of dead cells after 1 day (Fig. 2A and C) and 3 days (Fig. 2B and D). Z-stack acquisitions revealed multiple layers of bacteria in the biofilm underneath a dense layer of material (Fig. 2A to D, right panels). Three-dimensional (3D) images show the presence of uniformly spread biofilms for R20291 on day 1 (Fig. 2A), with uneven secondary structures by day 3 (Fig. 2B), and with a thickness ranging from 30 to 45 μm. For strain 630, we observe that the Live/Dead staining of biofilms was not homogeneous as seen for R20291 after day 1 (Fig. 2C) or day 3 (Fig. 2D). The maximum thickness of the biofilm was 30 μm (72 h). Thus, microscopic analysis suggests that C. difficile biofilms are structured, with several layers of largely live bacteria encased within a dense matrix.
Fig 2.
Confocal microscopy analysis of biofilms formed by C. difficile. Live/Dead staining shows dead (red) and live (green) bacteria (propidium iodide and Syto 9, respectively) in strains R20291 (A and B) and 630 (C and D) biofilms after incubation for 1 day (A and C) and 3 days (B and D). 3D images of biofilms depicting biofilm thickness in μm are shown in the left panels.
Composition of the C. difficile biofilm matrix.
Since the R20291 biofilms were more robust and reproducible, we examined the R20291 biofilms further using multiple staining to evaluate the extracellular matrix. Interestingly, antibodies against whole bacteria were able to stain a complex biofilm matrix and some superficial individual bacteria (Fig. 3A). The formation of a thick mature biofilm was visualized by a biofilm matrix tracer Ruby stain, which labels a range of protein classes including glycoproteins, lipoproteins, and phosphoproteins (Fig. 3B). In addition, when using staining with an antibody against a synthetic C. difficile PSII polysaccharide (synthetic phosphorylated hexasaccharide repeating unit) (26), we observed staining of fiber-like structures on biofilms (Fig. 3C). To confirm whether proteins are part of biofilm matrix, we treated biofilms with 0.1 mg of proteinase K/ml. Although planktonic growth was not inhibited by proteinase K (see Fig. S1 in the supplemental material), incubation of the bacterial cultures with the enzyme resulted in a significant decrease in biofilm formation on days 1 and 3, with protease (Fig. 3E, dark-shaded bars). One-day-old biofilms were also disrupted when treated with proteinase K (Fig. 3E, light-shaded bars). We also performed similar experiments with DNase I treatment and found that DNase I also inhibited biofilm formation (Fig. 3F, dark-shaded bars) and is able to reduce preformed biofilms (Fig. 3F, light-shaded bars), although to a lesser degree compared to proteinase K. These results indicate that bacterial proteins, DNA, and surface polysaccharide are components of the biofilm matrix formed by C. difficile in vitro.
Fig 3.
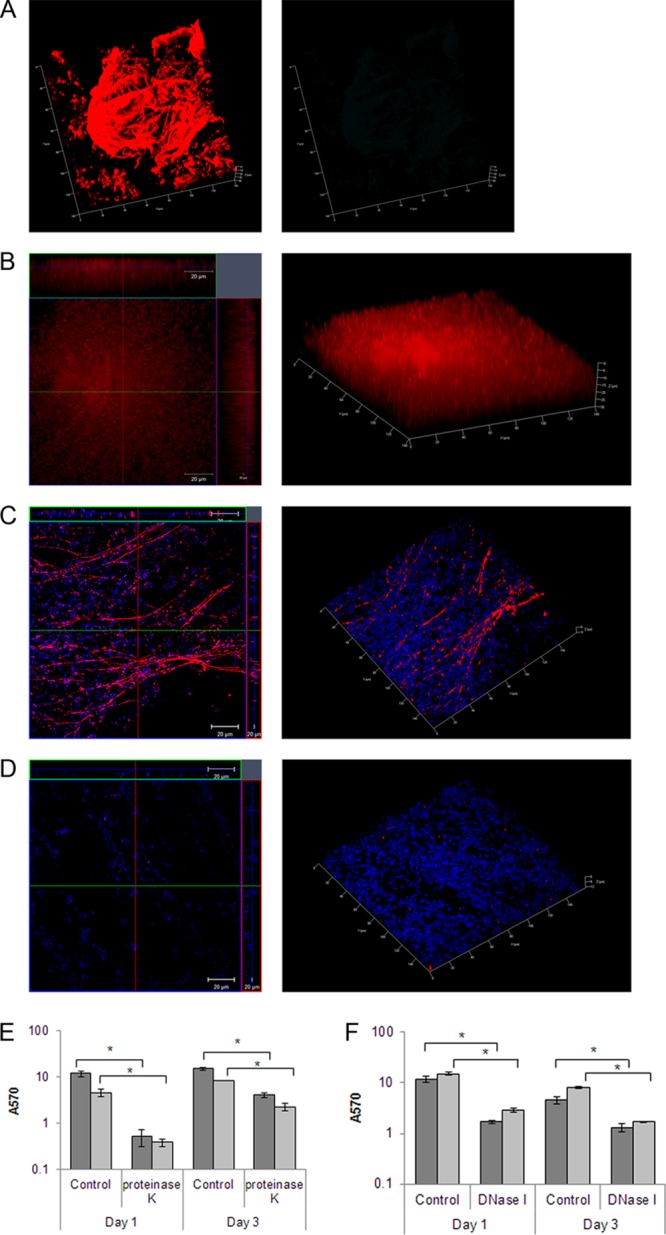
Characterization of C. difficile biofilm matrix. (A) shows 3D confocal microscopy images of staining of R20291 biofilms with murine anti-R20291 (left panel) and mouse preimmune serum (right panel) after 3 days of incubation. (B) Biofilms stained with Ruby matrix stain after 3 days of incubation. Biofilms were stained with antibodies to a synthetic C. difficile PSII polysaccharide (red) and DAPI (blue), which stains the bacterial DNA (C), or with the control mice preimmune serum and DAPI (D). Biofilms were incubated with proteinase K (E) or DNase I (F) as described in Materials and Methods. The dark gray bars represent data from treatment of either enzyme at the start of incubation (inhibition of biofilm formation), and the light gray bars represent data from incubating preformed 1-day-old biofilms with either enzyme (disruption of biofilms). The data shown are representative of at least two independent experiments performed in triplicates (P < 0.05).
An intact S layer and flagella are important in C. difficile biofilm formation.
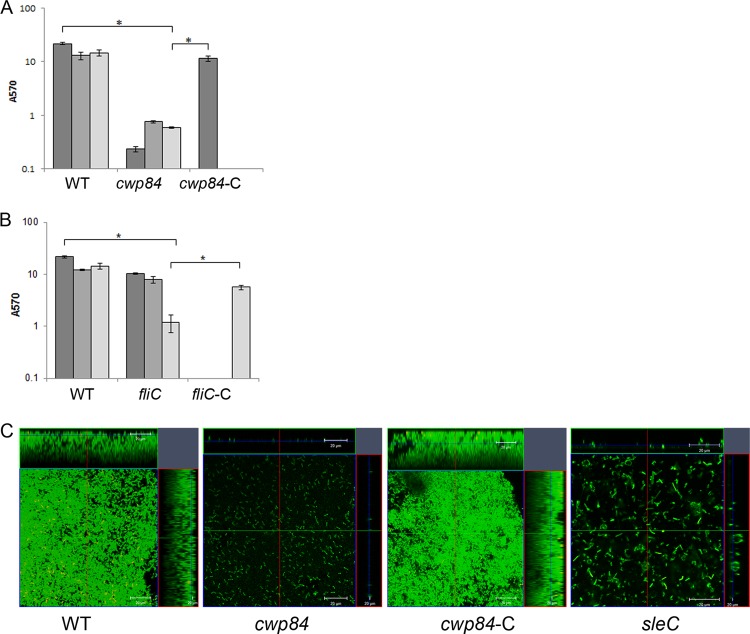
Numerous cell surface proteins and surface structures such as flagella and pili have been shown to be important for biofilm formation in Gram-positive bacteria. Cwp84 is a key surface protease involved in maturation of the S layer of C. difficile. We compared biofilm formation of a strain with a deletion of the cwp84 gene, CdiR20291Δcwp84 (Δcwp84), with the wild type (WT) using multiple methods. Although there was no defect in planktonic growth (see Fig. S2 in the supplemental material), we saw a dramatic decrease in biofilm accumulation for cwp84 strain, as measured by CV staining (Fig. 4A). The cwp84 mutant showed a more dramatic defect in biofilm formation on day 1 compared to days 3 and 5. Microscopic analysis showed a single layer of bacteria for cwp84 strain (Fig. 4C, panel 2) compared to the WT (Fig. 4C, panel 1). The biofilm defect for the cwp84 strain was fully complemented by restoring the WT gene on the chromosome CdiR20291Δcwp8-C (referred to here as cwp84-C, where the suffix “-C” indicates that the gene is complemented).
Fig 4.
Role of S layer and flagella in biofilm formation. (A) Biofilm formation by WT R20291, a cwp84 mutant (Δcwp84) for days 1, 3, and 5 and a complemented strains (cwp84-C) for day 1 in vitro as measured by CV staining. (B) Biofilm formation by WT R20291, fliC mutant (fliC) for days 1, 3, and 5 and complemented strains fliC (fliC-C) for day 5 in vitro as measured by CV staining. (C) Confocal microscopy images of Live/Dead staining of biofilms formed by the WT, Δcwp84, cwp84-C, and fliC. The results are presented in log scale, and the error bars represent standard deviations (P < 0.05). Biofilm assays were performed in triplicates, and data are representative of at least three independent experiments.
To examine the role of flagella, a mutant in the flagellin gene, fliC, CRG3351 (fliC), was tested for biofilm formation. We observed a significant decrease in biofilm accumulation for the fliC mutant compared to the WT on day 5 but not at earlier times using CV staining (Fig. 4B) and microscopic analysis (Fig. 4C, panel 4). This was reversed upon expressing the FliC protein episomally from its native promoter (fliC-C) (Fig. 4B). No differences in planktonic growth were observed between these strains (data not shown). These data may indicate that while a mature S layer is important in early biofilm stages such as adhesion, flagella may be more important for later stages in biofilm formation.
A putative role for quorum sensing in biofilm formation.
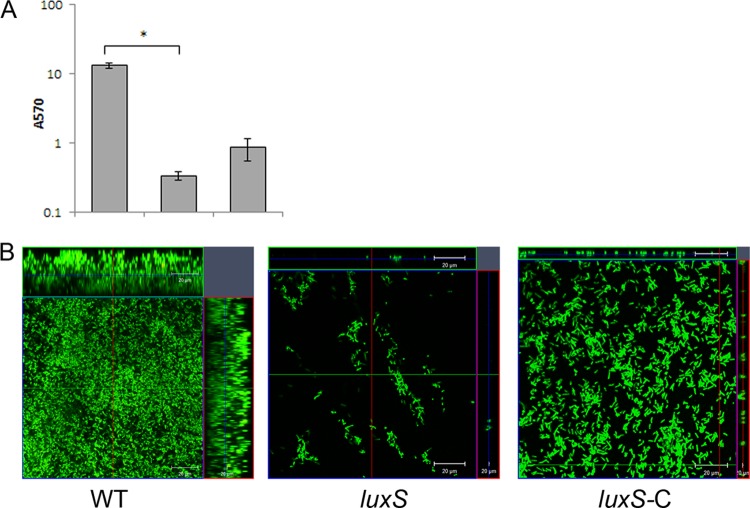
Quorum sensing plays a vital part in biofilm formation, and the involvement of quorum sensing regulators such as luxS has been demonstrated for various other bacteria (31–33). We tested a mutant of a luxS, homologue in C. difficile, by homology to other Gram-positive bacteria. A dramatic defect in biofilm formation was observed for the luxS mutant, CRG1183 (luxS), both by CV staining (Fig. 5A) and by microscopy (Fig. 5B, center panel), with no significant differences at different times after incubation (data not shown). Examination of the luxS mutant showed that it is unable to form even a bacterial monolayer on glass surface. Biofilm defects of this mutant were complemented by episomal expression of the full-length gene under the control of the native promoter (luxS-C). Although the complementation did not completely restore the WT phenotype, we were able to detect several layers of bacteria for the complemented strain (Fig. 5B, right panel). Although the growth curves for mutant and complemented strains were similar (see Fig. S3 in the supplemental material), it is possible that expressing luxS episomally may be toxic for the bacteria, since we observed a smaller colony size for this strain on plates (data not shown).
Fig 5.
Potential role for quorum sensing in C. difficile biofilm formation. (A) Biofilm formation by WT R20291, putative quorum-sensing gene luxS mutant (luxS), and complemented strains (luxS-C) as measured by CV staining. (B) Confocal microscopy analysis of WT, luxS, and complemented strain luxS-C. The results are presented in log scale, and the error bars represent the standard deviations (P < 0.05). The data from biofilm assays are representative of at least three independent experiments performed in triplicates.
Sporulation/germination pathway mutants are defective in biofilm formation.
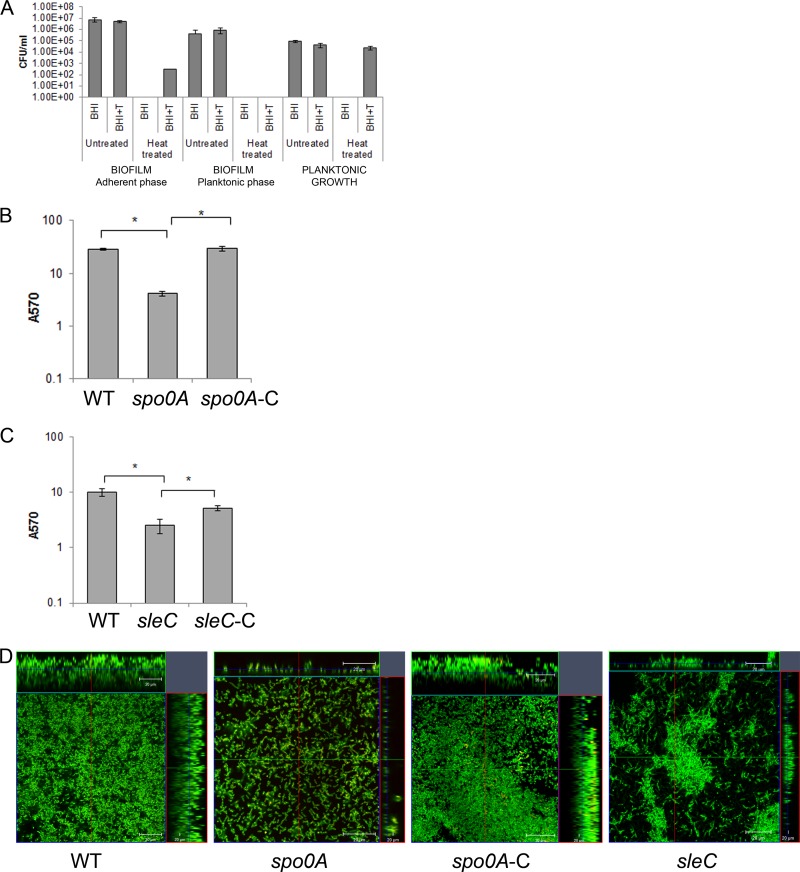
Sporulation is a key pathway that is initiated by C. difficile under conditions of stress. Regulators of sporulation such as Spo0A are also involved in formation of biofilms in other Gram-positive bacteria (34). We first studied whether spore formation occurs in C. difficile biofilms in our growth conditions, for adherent biofilms and planktonic phase from the same well of the 24-well plate (Fig. 6A). We found that there are very few spores in the biofilm (0.0001%) and in planktonic phases (none detectable) on day 3 and day 5 (0.0001%, data not shown). However, in the control, which was bacteria cultured in a tube (where biofilm formation did not occur), spores were formed by day 3 (40 to 50%) (Fig. 6A). Nevertheless, a spo0A mutant CRG1375 (spo0A), which is defective in sporulation (see Fig. S5A in the supplemental material), demonstrated significantly lesser biofilm formation compared to WT both by CV staining (Fig. 6B) and microscopy (Fig. 6D, panel 2) at day 1 and days 3 and 5 (data not shown). We also tested a mutant for a protein involved in C. difficile germination, CRG1166 (sleC) (Fig. S5B). The sleC mutant is able to form biofilm-like structures (Fig. 6C), but the biofilm is uneven and the thickness of biofilm produced by this strain is never more than 20 μm (Fig. 6D, panel 4). The cellular morphology of the bacteria in these biofilms is different to wild-type, and appears to form filamentous structures. Biofilm defects of both mutants were complemented by episomal expression of genes spo0A or sleC (spoA-C and sleC-C) under the control of their respective native promoters (Fig. 6B to D).
Fig 6.
Sporulation/germination proteins affect C. difficile biofilm formation. (A) Quantitation of the number of spores present in the adherent, planktonic phases of biofilm and in planktonic tube culture, in brain heart infusion media (BHI) with sodium taurocholate (BHI+T) and heat treatment (80°C), as described in Materials and Methods. (B and C) Biofilm formation by WT R20291, sporulation transcription factor spo0A mutant (spo0A) and complemented spo0A mutant (spo0A-C) after 1 day (B) and WT R20291, sleC mutant (sleC) and complemented sleC mutant (sleC-C) after 3 days (C). (D) Confocal microscopy analysis of WT and mutants spo0A, complemented spo0A mutant (spo0A-C), and sleC. The results are presented in log scale, and the error bars represent standard deviations (P < 0.05). Both biofilm and spore quantitation experiments were performed in triplicates, and the data shown are representative of at least three independent experiments.
In addition to the ClosTron mutants described above (fliC, luxS, spo0A, and sleC), other mutants in the binary toxins, cdtA and cdtB, or germination-specific N-acetylmuramoyl-l-alanine amidase, cwlD (generated using ClosTron) were also tested, but did not display significant defects in biofilm formation (data not shown).
Effects of vancomycin on C. difficile biofilms.
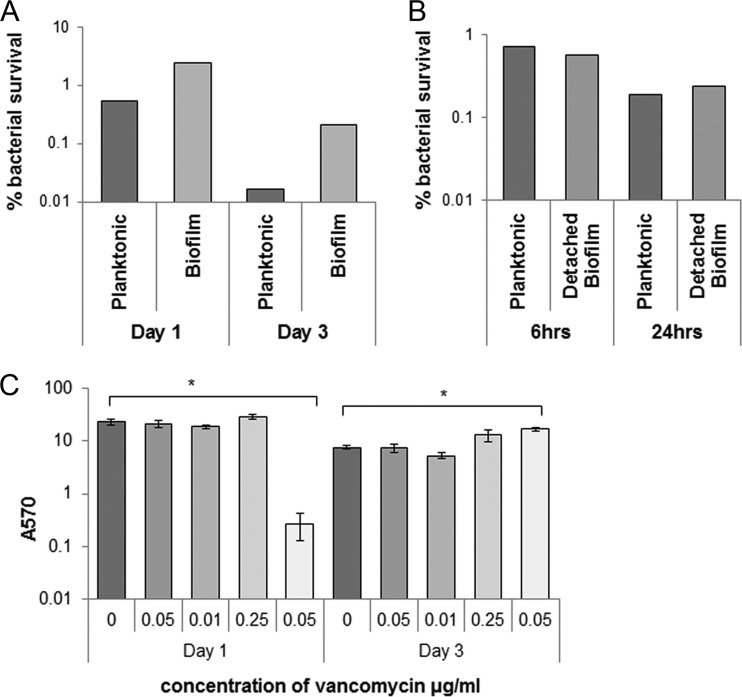
Since biofilms are known to be a means by which bacteria protect themselves from antibiotics, we studied whether C. difficile biofilms are important in mediating antibiotic resistance. We tested the resistance of bacteria in biofilms to the antibiotic vancomycin, which is commonly used for the treatment of CDI. In vitro, vancomycin, has excellent activity against C. difficile; an MIC of 0.75 to 2.0 μg/ml is sufficient to inhibit 90% of strains (23). Both sessile and planktonic phases, from the same C. difficile biofilm culture, 1-day-old (see Fig. S6A in the supplemental material) and 3-day-old biofilms (see Fig. S6B in the supplemental material) were exposed to 20 μg of vancomycin/ml (100 times the MIC of strain R20291) for 6 and 24 h as described in Materials and Methods. The percentages of surviving bacteria after treatment with antibiotics for 24 h for 1-day-old and 3-day-old biofilms are presented in Fig. 7A. Bacteria in 1-day-old and 3-day-old biofilms survived 5- and 12-fold more, respectively, than planktonic bacteria. These data support a role for C. difficile biofilms in resisting antibiotics.
Fig 7.
Effect of antibiotics on C. difficile biofilms. (A) Clostridium difficile 1- to 3-day-old biofilms and the corresponding planktonic growth were exposed to 20 μg of vancomycin/ml (100 times the MIC) for 24 h. The data are presented as percentage of surviving bacteria after treatment with antibiotics for 24 h for 1- and 3-day-old biofilms. (B) Bacterial counts from disrupted 1-day-old biofilms were incubated with 20 μg of vancomycin/ml for 6 h or 24 h. (C) Biofilm formation measured by CV staining at days 1 and 3 after treatment with subinhibitory and inhibitory concentration of antibiotic vancomycin (MIC for R20291 was 0.2 μg/ml). The results are presented in log scale, and the error bars represent standard deviations (P < 0.05). An asterisk (*) denotes significant differences compared to biofilm formation in the absence of vancomycin (0 μg/ml).
To try and understand whether resistance to vancomycin is due to protection conferred by biofilm matrix structure or an inherent property of the bacteria in biofilm, we studied the effect of vancomycin on adherent biofilms that were disrupted by pipetting (Fig. 7B). Disrupted sessile biofilm and the planktonic phase from 1-day-old biofilm were incubated for 6 and 24 h with 20 μg of vancomycin/ml. Bacteria from the disrupted adherent biofilms were not more resistant to high concentrations of antibiotics of biofilm compared to bacteria from the planktonic phase. Although the bacteria from disrupted biofilms do not form new adherent biofilms after the incubation with antibiotics even after 48 h of incubation (data not shown), we observe unstable, thread-like structures in the wells (which were disrupted by pipetting before performing CFU counts). The lack of antibiotic resistance by disrupted biofilm bacteria may indicate a lack of genetic changes in the bacteria within resistant biofilms and suggest that the biofilm matrix and/or other epigenetic mechanisms are involved in mediating vancomycin resistance.
Biofilm formation, on the other hand, has been reported to be stimulated by subinhibitory concentrations of antibiotics (35). To study whether vancomycin stimulates the biofilm formation in vitro, bacteria were treated with a range of concentrations of vancomycin (0 to 0.5 μg/ml), both lower and higher than the tube growth MIC (0.2 μg/ml), and biofilm formation was measured at day 1 and day 3. No significant induction of biofilm was observed for any of the vancomycin concentrations after 1 day. Inhibition of biofilm formation was evident for concentrations of vancomycin of ≥0.5 μg/ml (Fig. 7C). Interestingly, after 3 days incubation, a significant induction of biofilms was observed with 0.5 μg of vancomycin/ml and to a lesser extent with a subinhibitory concentration (0.25 μg/ml) of vancomycin (Fig. 7C). These results suggest that exposure to inhibitory vancomycin concentrations can stimulate biofilm formation in vitro.
DISCUSSION
Biofilms are the most representative form of growth of bacteria in the large intestine (4). The ability to form biofilms is known to influence the virulence and also the transmission of intestinal pathogenic bacteria such as Vibrio cholerae (36–38). In the present study, we report for the first time the ability of two clinically relevant Clostridium difficile strains, “nonepidemic” strain 630 and “epidemic” hypervirulent strain R20291, to form structured biofilms in vitro. Furthermore, we identify several genes that may influence C. difficile biofilm development.
The ability to adhere and form biofilms influences the ability of pathogens to colonize and establish an infection (39, 40). Our data show that R20291 forms more biofilm in all tested conditions in vitro. It has been reported that strain R20291, a hypervirulent strain, produces higher levels of toxin in vitro compared to 630 (41). Although colonization of these strains has yet to be examined carefully in vivo, higher biofilm formation by this strain could indicate better colonization in vivo. While for the strain R20291 addition of glucose does not increase the levels of biofilm formation, it is an important factor for biofilm formation for strain 630. It is well known that different carbohydrates can modulate biofilm formation. Carbohydrates induce biofilm formation in Streptococcus gordonii (42), while in Bacillus subtilis the CcpA protein represses formation of biofilm in medium with high levels of glucose (43). It is possible that in vivo the nutritional environment in the gut modulates colonization of C. difficile.
The biofilm matrix is known to protect bacteria during infections by providing an enclosed environment to escape immune responses (5, 36). Similar to many clinically relevant biofilm formers such as Pseudomonas aeruginosa and Staphylococcus aureus, C. difficile also appears to form a complex matrix comprising of proteins, DNA and polysaccharide (44–46). Staining with antibodies against C. difficile suggests that this compact biofilm may comprise surface-associated or secreted bacterial components and may be impenetrable to antibodies. Furthermore, it is interesting that we observe biofilm matrix staining for a synthetic derivative of the C. difficile surface PSII polysaccharide, which was recently reported to be immunogenic (26). The presence of a complex and perhaps impermeable matrix may protect C. difficile from unfavorable agents in vivo in the gut.
Regulation of biofilm formation in Gram-positive bacteria involves multiple factors including adhesins, surface structures such as flagella and pili (47). The C. difficile S layer is composed of S-layer proteins (SLPs) that are present as heterodimeric complex (48). The signal peptide of protein precursor SlpA is removed by proteolytic cleavage. Additional cleavage is essential for maturation of SLPs, which is mediated by the cysteine protease Cwp84 (49, 50). A mutant in cwp84 has previously been shown to be defective in the S-layer synthesis (50). This protease is also important for the degradation of extracellular matrix proteins such as fibronectin, laminin, and vitronectin (21). Our data prove that a mature S layer is essential for C. difficile biofilm formation, perhaps due to the fact that this layer may be essential for anchoring various cell wall associated proteins, which may be required for the early steps such as adhesion during biofilm formation. A role for specific CWPs in biofilms remains to be investigated. Furthermore, the observation that antibodies raised against fixed whole bacteria recognize complex structures on biofilms may indicate that surface components such as CWPs may compose the biofilm matrix.
Bacterial flagella are known to modulate attachment, the first step in biofilm formation in motile bacteria, however, the precise role varies between species. For Gram-positive bacteria such as B. subtilis the presence of flagella is important but not essential for formation of biofilms (51), while for Listeria monocytogenes, motility is essential for mature biofilm formation (52). In our experiments a mutant in flagellin, a principal component of flagella, clearly affects biofilms in vitro, indicating that motility of C. difficile is key in formation of biofilms. Recently, strain 630 C. difficile flagellar mutants were reported to have better adherence in an in vitro model (53). Our data further suggest that flagella are important at later biofilm stages, as the mutant does not display defects in our in vitro assays at earlier time points.
Cell-cell communication is crucial in a complex structure like biofilms where bacteria are in strict contact with one another. Quorum sensing has an important role in bacterial biofilm formation (33, 54). The enzyme LuxS, that synthesizes autoinducer-2 (AI-2), is one of the major modulators of QS and is largely conserved across bacterial species (55). It was demonstrated that C. difficile genome carries a 453-bp gene that encodes a protein which shares 40% identity to the V. harveyi LuxS protein and is responsible for AI-2 production (56). The role of luxS in toxin production is not clear as there are conflicting reports in the literature (56, 57). While precise mechanisms by which luxS functions in C. difficile is unclear at present, our data suggest a role for putative luxS-encoded molecules in formation of biofilms in vitro and may indicate that a luxS-mediated quorum-sensing system exists in C. difficile.
Sporulation is a critical pathway in bacterial responses to environmental stresses. Spo0A is the main response regulator which controls entry into sporulation and is well conserved in Bacillus and Clostridium species (58, 59). Spo0A is also known to regulate a range of regulatory factors, thus affecting pathways unrelated to sporulation (60). C. difficile spo0A mutants have been reported to modulate toxin production, although there are contradictory data about the nature of this regulation (11, 59). Although we do not know yet if there is a link between toxin production and biofilms, clearly C. difficile spo0A is involved in the formation of biofilms in vitro. Biofilm environments have been shown to be optimal for spore formation and spores are part of biofilms for many spore-forming bacteria under nutrient-starved conditions (47). However, we find extremely low numbers of spores in biofilms (adherent and planktonic phases of biofilms) under our conditions. Previously, Hamon et al. demonstrated that a B. subtilis mutant of spo0A was defective for biofilm formation, and similar to our observation, they did not detect spores in Bacillus biofilms in vitro (34). We hypothesize that spo0A in C. difficile acts as a switch between different stress-related pathways such as sporulation, biofilm formation, and toxin production. Our observation that bacteria sporulate during tube culture, but not in biofilms, when incubated under similar conditions, may support this hypothesis. Recently, spo0A mutants were shown to be defective in persistence and transmission in a murine infection model, primarily due to the inability of the spo0A mutant strain to form spores (11). In addition to production of spores, the formation of biofilms in vivo may also account for the persistence defects observed for the spo0A strain. Indeed, under other in vitro conditions or in vivo, spores may also form part of biofilms.
Interestingly, we find that a mutant lacking SleC, a protein recently reported to be specifically involved in germination of C. difficile spores (27), is defective for biofilm formation. A role for spore germination in biofilms has not been well studied; however, in our conditions given the lack of spores, it is unlikely that SleC is involved in germination of spores. Given that the sleC mutant biofilms shows strikingly different cellular morphologies, it is possible that SleC has other functions such as in hydrolysis of vegetative cell peptidoglycans.
The relevance of biofilm formation in the context of infection and treatment has been widely studied. The role of biofilms in mediating resistance to antibiotics had been well studied for several bacteria, e.g., methicillin-resistant S. aureus (61). Resistance to antibiotics in biofilm can increase from 10- to 1,000-fold more compared to planktonic bacteria (62). The rising incidence of resistance to antibiotic treatments for nosocomial pathogens such as S. aureus and C. difficile has been well documented in recent years (63). Highly antibiotic-resistant C. difficile strains and treatment of recurring clostridial infections have been the major challenges for managing CDI (23). Our findings indicate that C. difficile biofilms are more resistant to the antibiotic vancomycin, which is commonly used for treatment of CDI. Furthermore, our initial studies on the mechanisms involved in antibiotic resistance show that the bacteria from resistant biofilms do not appear to carry inheritable changes and may suggest a role for the C. difficile biofilm matrix structure in antibiotic penetration, as seen for other bacteria (62). It is also possible that the physiological state of bacteria within biofilms is important in mediating resistance (64). Interestingly, lower concentrations of vancomycin induce biofilm formation. A role for antibiotics in stimulating biofilm formation has been examined previously for other bacteria and second messenger signaling involving c-di-GMP has been implicated in this process (35, 65). It is believed that such an induction of biofilms could be clinically relevant when there is exposure to low doses of antibiotics, e.g., the beginning and end of antibiotic therapy, and could perhaps explain ineffective treatment (65). In CDI, the establishment of persistent biofilms in vivo, in addition to the formation of spores, could potentially explain the occurrence of recurrent infections.
In conclusion, we demonstrate that clinically relevant strains of the anaerobic gut pathogen C. difficile are able to form complex biofilms in vitro. C. difficile biofilm formation appears to be a multifactorial process with a role for proteins that are important in different aspects of bacterial physiology. Indeed, the details of the precise roles of each of these proteins/pathways and their regulation remain to be studied. A possible model of infection is that C. difficile colonizes the colon via formation of microcolonies or biofilms, followed by toxin production. Formation of biofilms in vivo perhaps provides the bacterium with a mechanism to protect itself from the cellular immune responses invoked by the toxins, in addition to a mechanism of persistence in the presence of antibiotics. Investigation of C. difficile biofilm development during infection and factors controlling it could give us a better insight into their role in C. difficile pathogenesis.
Supplementary Material
ACKNOWLEDGMENTS
The research leading to these results received funding from the European Community's Seventh Framework Programmes CLOSTNET (PEOPLE-ITN-2008-237942). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 23 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01980-12.
REFERENCES
- 1.Branda SS, Vik S, Friedman L, Kolter R. 2005. Biofilms: the matrix revisited. Trends. Microbiol. 13:20–26 [DOI] [PubMed] [Google Scholar]
- 2.Davey ME, O'Toole AG. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cummings JH, Antoine JM, Azpiroz F, Bourdet-Sicard R, Brandtzaeg P, Calder PC, Gibson GR, Guarner F, Isolauri E, Pannemans D, Shortt C, Tuijtelaars S, Watzl B. 2004. PASSCLAIM: gut health and immunity. Eur. J. Nutr. 43(Suppl 2):II118–II173 [DOI] [PubMed] [Google Scholar]
- 4.Macfarlane S, Dillon JF. 2007. Microbial biofilms in the human gastrointestinal tract. J. Appl. Microbiol. 102:1187–1196 [DOI] [PubMed] [Google Scholar]
- 5.Beloin C, Roux A, Ghigo JM. 2008. Escherichia coli biofilms. Curr. Top. Microbiol. Immunol. 322:249–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tzipori S, Montanaro J, Robins-Browne RM, Vial P, Gibson R, Levine MM. 1992. Studies with enteroaggregative Escherichia coli in the gnotobiotic piglet gastroenteritis model. Infect. Immun. 60:5302–5306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kline KA, Falker S, Dahlberg S, Normark S, Henriques-Normark B. 2009. Bacterial adhesins in host-microbe interactions. Cell. Host Microbe 5:580–592 [DOI] [PubMed] [Google Scholar]
- 8.Rupnik M, Wilcox MH, Gerding DN. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7:526–536 [DOI] [PubMed] [Google Scholar]
- 9.Freeman J, Bauer MP, Baines SD, Corver J, Fawley WN, Goorhuis B, Kuijper EJ, Wilcox MH. 2010. The changing epidemiology of Clostridium difficile infections. Clin. Microbiol. Rev. 23:529–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawley TD, Clare S, Walker AW, Goulding D, Stabler RA, Croucher N, Mastroeni P, Scott P, Raisen C, Mottram L, Fairweather NF, Wren BW, Parkhill J, Dougan G. 2009. Antibiotic treatment of clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect. Immun. 77:3661–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, Wren BW, Fairweather NF, Dougan G, Lawley TD. 2012. Clostridium difficile spo0A gene is a persistence and transmission factor. Infect. Immun. 80:2704–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. 2010. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467:711–713 [DOI] [PubMed] [Google Scholar]
- 13.Lyras D, O'Connor JR, Howarth PM, Sambol SP, Carter GP, Phumoonna T, Poon R, Adams V, Vedantam G, Johnson S, Gerding DN, Rood JI. 2009. Toxin B is essential for virulence of Clostridium difficile. Nature 458:1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voth DE, Ballard JD. 2005. Clostridium difficile toxins: mechanism of action and role in disease. Clin. Microbiol. Rev. 18:247–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen A. 2012. Clostridium difficile toxins: mediators of inflammation. J. Innate Immun. 4:149–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pechine S, Janoir C, Boureau H, Gleizes A, Tsapis N, Hoys S, Fattal E, Collignon A. 2007. Diminished intestinal colonization by Clostridium difficile and immune response in mice after mucosal immunization with surface proteins of Clostridium difficile. Vaccine 25:3946–3954 [DOI] [PubMed] [Google Scholar]
- 17.Barketi-Klai A, Hoys S, Lambert-Bordes S, Collignon A, Kansau I. 2011. Role of fibronectin-binding protein A in Clostridium difficile intestinal colonization. J. Med. Microbiol. 60:1155–1161 [DOI] [PubMed] [Google Scholar]
- 18.Calabi E, Calabi F, Phillips AD, Fairweather NF. 2002. Binding of Clostridium difficile surface layer proteins to gastrointestinal tissues. Infect. Immun. 70:5770–5778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karjalainen T, Waligora-Dupriet AJ, Cerquetti M, Spigaglia P, Maggioni A, Mauri P, Mastrantonio P. 2001. Molecular and genomic analysis of genes encoding surface-anchored proteins from Clostridium difficile. Infect. Immun. 69:3442–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waligora AJ, Hennequin C, Mullany P, Bourlioux P, Collignon A, Karjalainen T. 2001. Characterization of a cell surface protein of Clostridium difficile with adhesive properties. Infect. Immun. 69:2144–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Janoir C, Pechine S, Grosdidier C, Collignon A. 2007. Cwp84, a surface-associated protein of Clostridium difficile, is a cysteine protease with degrading activity on extracellular matrix proteins. J. Bacteriol. 189:7174–7180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall-Stoodley L, Stoodley P. 2009. Evolving concepts in biofilm infections. Cell. Microbiol. 11:1034–1043 [DOI] [PubMed] [Google Scholar]
- 23.Surawicz CM, Alexander J. 2011. Treatment of refractory and recurrent Clostridium difficile infection. Nat. Rev. Gastroenterol. Hepatol. 8:330–339 [DOI] [PubMed] [Google Scholar]
- 24.Varga JJ, Therit B, Melville SB. 2008. Type IV pili and the CcpA protein are needed for maximal biofilm formation by the gram-positive anaerobic pathogen Clostridium perfringens. Infect. Immun. 76:4944–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donelli G, Vuotto C, Cardines R, Mastrantonio P. 2012. Biofilm-growing intestinal anaerobic bacteria. FEMS Immunol. Med. Microbiol. 65:318–325 [DOI] [PubMed] [Google Scholar]
- 26.Adamo R, Romano MR, Berti F, Leuzzi R, Tontini M, Danieli E, Cappelletti E, Cakici OS, Swennen E, Pinto V, Brogioni B, Proietti D, Galeotti CL, Lay L, Monteiro MA, Scarselli M, Costantino P. 2012. Phosphorylation of the synthetic hexasaccharide repeating unit is essential for the induction of antibodies to Clostridium difficile PSII cell wall polysaccharide. ACS Chem. Biol. 7:1420–1428 [DOI] [PubMed] [Google Scholar]
- 27.Burns DA, Heap JT, Minton NP. 2010. SleC is essential for germination of Clostridium difficile spores in nutrient-rich medium supplemented with the bile salt taurocholate. J. Bacteriol. 192:657–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heap JT, Pennington OJ, Cartman ST, Carter GP, Minton NP. 2007. The ClosTron: a universal gene knockout system for the genus Clostridium. J. Microbiol. Methods 70:452–464 [DOI] [PubMed] [Google Scholar]
- 29.Kuehne SA, Heap JT, Cooksley CM, Cartman ST, Minton NP. 2011. ClosTron-mediated engineering of Clostridium. Methods Mol. Biol. 765:389–407 [DOI] [PubMed] [Google Scholar]
- 30.Heap JT, Kuehne SA, Ehsaan M, Cartman ST, Cooksley CM, Scott JC, Minton NP. 2010. The ClosTron: mutagenesis in Clostridium refined and streamlined. J. Microbiol. Methods 80:49–55 [DOI] [PubMed] [Google Scholar]
- 31.Lombardia E, Rovetto AJ, Arabolaza AL, Grau RR. 2006. A LuxS-dependent cell-to-cell language regulates social behavior and development in Bacillus subtilis. J. Bacteriol. 188:4442–4452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohtani K, Hayashi H, Shimizu T. 2002. The luxS gene is involved in cell-cell signalling for toxin production in Clostridium perfringens. Mol. Microbiol. 44:171–179 [DOI] [PubMed] [Google Scholar]
- 33.Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR. 2005. Making ‘sense’ of metabolism: autoinducer-2, LuxS, and pathogenic bacteria. Nat. Rev. Microbiol. 3:383–396 [DOI] [PubMed] [Google Scholar]
- 34.Hamon MA, Lazazzera BA. 2001. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol. Microbiol. 42:1199–1209 [DOI] [PubMed] [Google Scholar]
- 35.Hoffman LR, D'Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171–1175 [DOI] [PubMed] [Google Scholar]
- 36.Watnick PI, Lauriano CM, Klose KE, Croal L, Kolter R. 2001. The absence of a flagellum leads to altered colony morphology, biofilm development and virulence in Vibrio cholerae O139. Mol. Microbiol. 39:223–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faruque SM, Biswas K, Udden SM, Ahmad QS, Sack DA, Nair GB, Mekalanos JJ. 2006. Transmissibility of cholera: in vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc. Natl. Acad. Sci. U. S. A. 103:6350–6355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu J, Mekalanos JJ. 2003. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 5:647–656 [DOI] [PubMed] [Google Scholar]
- 39.Nobbs AH, Lamont RJ, Jenkinson HF. 2009. Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73:407–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allsopp LP, Totsika M, Tree JJ, Ulett GC, Mabbett AN, Wells TJ, Kobe B, Beatson SA, Schembri MA. 2010. UpaH is a newly identified autotransporter protein that contributes to biofilm formation and bladder colonization by uropathogenic Escherichia coli CFT073. Infect. Immun. 78:1659–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, Frost E, McDonald LC. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079–1084 [DOI] [PubMed] [Google Scholar]
- 42.Gilmore KS, Srinivas P, Akins DR, Hatter KL, Gilmore MS. 2003. Growth, development, and gene expression in a persistent Streptococcus gordonii biofilm. Infect. Immun. 71:4759–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stanley NR, Britton RA, Grossman AD, Lazazzera BA. 2003. Identification of catabolite repression as a physiological regulator of biofilm formation by Bacillus subtilis by use of DNA microarrays. J. Bacteriol. 185:1951–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mann EE, Wozniak DJ. 2012. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol. Rev. 36:893–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. 2009. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One 4:e5822 doi:10.1371/journal.pone.0005822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Colvin KM, Gordon VD, Murakami K, Borlee BR, Wozniak DJ, Wong GC, Parsek MR. 2011. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 7:e1001264 doi:10.1371/journal.ppat.1001264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abee T, Kovacs AT, Kuipers OP, van der Veen S. 2011. Biofilm formation and dispersal in Gram-positive bacteria. Curr. Opin. Biotechnol. 22:172–179 [DOI] [PubMed] [Google Scholar]
- 48.Fagan RP, Albesa-Jove D, Qazi O, Svergun DI, Brown KA, Fairweather NF. 2009. Structural insights into the molecular organization of the S-layer from Clostridium difficile. Mol. Microbiol. 71:1308–1322 [DOI] [PubMed] [Google Scholar]
- 49.Calabi E, Ward S, Wren B, Paxton T, Panico M, Morris H, Dell A, Dougan G, Fairweather N. 2001. Molecular characterization of the surface layer proteins from Clostridium difficile. Mol. Microbiol. 40:1187–1199 [DOI] [PubMed] [Google Scholar]
- 50.Kirby JM, Ahern H, Roberts AK, Kumar V, Freeman Z, Acharya KR, Shone CC. 2009. Cwp84, a surface-associated cysteine protease, plays a role in the maturation of the surface layer of Clostridium difficile. J. Biol. Chem. 284:34666–34673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kobayashi K. 2007. Gradual activation of the response regulator DegU controls serial expression of genes for flagellum formation and biofilm formation in Bacillus subtilis. Mol. Microbiol. 66:395–409 [DOI] [PubMed] [Google Scholar]
- 52.Lemon KP, Freitag NE, Kolter R. 2010. The virulence regulator PrfA promotes biofilm formation by Listeria monocytogenes. J. Bacteriol. 192:3969–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dingle TC, Mulvey GL, Armstrong GD. 2011. Mutagenic analysis of the Clostridium difficile flagellar proteins, FliC and FliD, and their contribution to virulence in hamsters. Infect. Immun. 79:4061–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hammer BK, Bassler BL. 2003. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol. Microbiol. 50:101–104 [DOI] [PubMed] [Google Scholar]
- 55.Hardie KR, Heurlier K. 2008. Establishing bacterial communities by ‘word of mouth’: LuxS and autoinducer 2 in biofilm development. Nat. Rev. Microbiol. 6:635–643 [DOI] [PubMed] [Google Scholar]
- 56.Carter GP, Purdy D, Williams P, Minton NP. 2005. Quorum sensing in Clostridium difficile: analysis of a luxS-type signaling system. J. Med. Microbiol. 54:119–127 [DOI] [PubMed] [Google Scholar]
- 57.Lee AS, Song KP. 2005. LuxS/autoinducer-2 quorum sensing molecule regulates transcriptional virulence gene expression in Clostridium difficile. Biochem. Biophys. Res. Commun. 335:659–666 [DOI] [PubMed] [Google Scholar]
- 58.Lopez D, Kolter R. 2010. Extracellular signals that define distinct and coexisting cell fates in Bacillus subtilis. FEMS Microbiol. Rev. 34:134–149 [DOI] [PubMed] [Google Scholar]
- 59.Underwood S, Guan S, Vijayasubhash V, Baines SD, Graham L, Lewis RJ, Wilcox MH, Stephenson K. 2009. Characterization of the sporulation initiation pathway of Clostridium difficile and its role in toxin production. J. Bacteriol. 191:7296–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Molle V, Fujita M, Jensen ST, Eichenberger P, Gonzalez-Pastor JE, Liu JS, Losick R. 2003. The Spo0A regulon of Bacillus subtilis. Mol. Microbiol. 50:1683–1701 [DOI] [PubMed] [Google Scholar]
- 61.Olson KM, Starks CM, Williams RB, O'Neil-Johnson M, Huang Z, Ellis M, Reilly JE, Eldridge GR. 2011. Novel pentadecenyl tetrazole enhances susceptibility of methicillin-resistant Staphylococcus aureus biofilms to gentamicin. Antimicrob. Agents Chemother. 55:3691–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mah TF, O'Toole GA. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends. Microbiol. 9:34–39 [DOI] [PubMed] [Google Scholar]
- 63.French GL. 2010. The continuing crisis in antibiotic resistance. Int. J. Antimicrob. Agents 36(Suppl 3):S3–S7 [DOI] [PubMed] [Google Scholar]
- 64.Lewis K. 2008. Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 322:107–131 [DOI] [PubMed] [Google Scholar]
- 65.Boehm A, Steiner S, Zaehringer F, Casanova A, Hamburger F, Ritz D, Keck W, Ackermann M, Schirmer T, Jenal U. 2009. Second messenger signaling governs Escherichia coli biofilm induction upon ribosomal stress. Mol. Microbiol. 72:1500–1516 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.