Abstract
The replication mechanism of bluetongue virus (BTV) has been studied by an in vivo reverse genetics (RG) system identifying the importance of certain BTV proteins for primary replication of the virus. However, a unique in vitro cell-free virus assembly system was subsequently developed, showing that it did not require the same set of viral components, which is indicative of differences in these two systems. Here, we studied the in vivo primary replicase complex more in-depth to determine the minimum components of the complex. We showed that while NS2 is an essential component of the primary replication stage during BTV infection, NS1 is not an essential component but may play a role in enhancing BTV protein synthesis. Furthermore, we demonstrated that VP7, a major structural protein of the inner core, is not required for primary replication but appears to stabilize the replicase complex. In contrast, VP3, the other major structural core protein, is an essential component of the complex, together with the three minor enzymatic proteins (VP1, VP4, and VP6) of the core. In addition, our data have demonstrated that the smallest minor protein, VP6, which is known to possess an RNA-dependent helicase activity, may also act as an RNA translocator during assembly of the primary replicase complex.
INTRODUCTION
Bluetongue virus (BTV), the etiological agent of bluetongue disease of livestock, is a member of the Orbivirus genus of the Reoviridae family. BTV particles have three consecutive layers of proteins that are organized into two capsids, an outer capsid of two proteins (VP2 and VP5) and an inner icosahedral capsid (core) composed of two major proteins, VP7 and VP3, which encloses the three minor proteins VP1, VP4, and VP6, in addition to the viral genome. The viral genome consists of 10 linear double-stranded (dsRNA) molecules (S1 to S10), which encode 7 structural proteins (VP1, VP2, VP3 to VP7) and 4 nonstructural (NS1 to NS4) proteins (1–3).
The replication of members of the Reoviridae family, including BTV, occurs in two stages (4). In the first stage, the outer capsid is removed shortly after cell entry and the whole inner capsid or core particle is released from the host cell endosome. The single-stranded RNAs (ssRNAs) from the 10 genomic segments are then repeatedly transcribed by core-associated enzymes within the core compartment and released into the host cell cytoplasm to act as the templates for translation, as well as serve as the templates for negative-strand viral RNA synthesis enzymes (5–7). Newly synthesized transcripts, released from the cores, initiate the primary replication cycle of BTV, generating the replicase complex (8, 9). In our current model, during this early stage of replication, the three minor enzymatic proteins, VP1 (polymerase), VP4 (capping enzyme), and VP6 (helicase), together with the 10 ssRNAs, are assembled along with a major protein, VP3, forming the subcore (10–12). In the second stage, VP7 is added onto the VP3 layer to form the stable core particle, which subsequently acquires the two outer capsid proteins, VP2 and VP5, to form mature progeny virions prior to virus egress (13, 14).
In addition to the structural proteins, BTV also encodes three or four nonstructural (NS) proteins in infected cells, which are involved in virus replication, assembly, and morphogenesis (1, 15–21).
In the past few years, having successfully established a reverse genetics (RG) system for BTV (22, 23), we reported a mechanism of primary replication that enhances virus production (8, 9). Additionally, we demonstrated that subcore proteins (VP1, VP3, VP4, and VP6) and two nonstructural proteins, NS1 and NS2, are involved in primary replication in vivo (9). Recently, using a cell-free in vitro assembly (CFA) system, we have shown that three minor proteins, VP1, VP4, and VP6, are first assembled in the presence of a complete set of 10 BTV plus-strand ssRNAs (+ssRNAs), followed by assembly of a subcore-like complex upon VP3 addition, and that the packaged +ssRNAs within the VP3 layer could synthesize genomic dsRNAs in the presence of ribonucleoside triphosphates (rNTPs) (10). However, addition of VP7 was essential to protect the ssRNAs, which is in contrast to our in vivo primary replication assay system, where VP7 was shown to be not required (9). Similarly, neither NS1 nor NS2 was needed for the in vitro assembly of the BTV subcore. Thus, their involvement in primary replication needs to be reexamined.
Of the three minor catalytic core proteins, the polymerase activity of VP1 and the mRNA capping activity of VP4 have been confirmed by a range of in vitro studies (24–28). In addition, structural studies have revealed their close association as a complex, located at the 5-fold vertices of the VP3 subcore (29, 30). In contrast, although VP6 has been demonstrated to be an ATP hydrolysis-dependent RNA helicase protein by an in vitro assay (31, 32), to date it has not been possible to determine the precise role of the protein in the virus replication process or its precise location within the core. Further, although VP6 may unwind dsRNA either ahead of or behind the transcribing polymerase, it is also plausible that the VP6 helicase also plays an entirely different role in virus replication, for example, by assisting with the packaging of viral RNA into the capsid. Neither rotaviruses nor bacteriophages with dsRNA genomes have an equivalent protein with helicase activity inside the viral core, although a reovirus inner capsid protein, lambda1, which is the primary constituent of the core shell, has been reported to possess helicase activity (33). Thus, the exact function of BTV VP6 in BTV biology still remains to be addressed, in particular, deciphering whether it is an integral component of the transcription machinery or a packaging promoter for translocating RNA during the assembly of nascent core particle.
Recently, we constructed VP6-deficient BTV strains using an RG system and a complementary BSR-VP6 cell line, demonstrating that VP6-deficient mutant viruses were replication deficient in noncomplementary cells (9, 34). These VP6-deficient viruses could provide information about the role of the protein and may also reveal the mechanism of assembly of the primary replicase complex.
To understand the BTV assembly pathway in-depth, it is necessary to revisit the exact BTV protein requirement in in vivo primary replication. In particular, the role of VP6 should be examined further, as it is an internal protein of the core particle and believed to form the replicase complex with viral genomic RNAs, the viral polymerase (VP1) and capping enzyme (VP4), as an initial step of the assembly pathway. Thus, in this study, we extended our previous findings, and by using the RG system, we have determined the essential components of the primary replicase complex in vivo. We demonstrated that NS2, but not NS1, is an essential component for in vivo replicase assembly. Our previous study also indicated that the surface core protein VP7 is not required in this process; however, the VP7 (S7) transcript used in the second transfection was capped at the 5′ end, which facilitates protein synthesis; thus, it may have contributed to the enhancement of virus recovery (9). Therefore, to eliminate this possibility, in this study we have examined the role of VP7 using an uncapped S7 transcript. Most importantly, we provide evidence of the role of BTV VP6 as an essential element for the initiation of primary replication and its plausible role in RNA packaging.
Together, these data demonstrate that VP6-deficient viruses are not competent for replication due to their failure to assemble functional replicase complexes.
MATERIALS AND METHODS
Cell lines and virus.
BSR cells (BHK-21 subclone) were maintained in Dulbecco's modified Eagle's medium (DMEM; Sigma) supplemented with 4% (vol/vol) fetal bovine serum (FBS; Invitrogen). The stable cell lines BSR-VP6 and BSR-NS2 were grown in DMEM–4% FBS supplemented with 7.5 μg/ml of puromycin (Sigma).
BTV serotype 1 (BTV-1) stock was obtained by infecting BSR cells at a low multiplicity of infection and harvested when a 100% cytopathic effect was evident. VP6-deficient BTV (S9E2) stocks were obtained from the complementary cell line BSR-VP6 as described previously and kept at a low passage number (less than 5) for all experiments (9, 34). Virus stocks were stored at 4°C.
Plasmids.
For the modified BTV RG system, 5 additional mammalian expression plasmids, pCAG-PBTV1VP1, pCAG-PBTV1VP3, pCAG-PBTV1VP4, pCAG-PBTV1VP7, and pCAG-PBTV1NS2, were generated, as described for VP6 expression plasmid pCAG-PBTV10VP6 (9). The coding region (CDR) of each of BTV-1 S1, S3, S4, S7, and S8, encoding VP1, VP3, VP4, VP7, and NS2, respectively, was inserted into the mammalian expression plasmid pCAG-PM (35). The sequence of each expression plasmid was confirmed. The expression of each protein was confirmed by immunoblotting analysis or immunofluorescence using standard methods.
Development of a BSR-NS2 cell line to express NS2 constitutively.
The complementary cell line expressing NS2, BSR-NS2, was generated as described previously (9). Briefly, BSR cells were transfected with the NS2 expression vector, pCAG-PBTV1NS2. After transfection, cells with integrated copies of the expression vector were selected in the presence of puromycin (Sigma). Surviving clones were tested for the expression level of NS2 by immunoblotting analysis, and the best-expressing clone was used.
Synthesis of BTV transcripts from cDNA plasmid clones.
Synthesis of capped and uncapped BTV transcripts was performed as described previously (9, 22). All capped T7 transcripts were synthesized using an mMESSAGE mMACHINE T7 ultrakit (Ambion). For synthesis of uncapped T7 transcripts, a RiboMAX large-scale RNA production system T7 (Promega) was used according to the manufacturer's protocols. The synthesized RNA transcripts were resuspended in nuclease-free water and stored at −80°C.
Primary replicase assay.
BSR or BSR-NS2 cells were transfected twice with BTV T7 transcripts using Lipofectamine 2000 reagent (Invitrogen) as described previously (9). For the modified RG system, the first transfection was performed with 100 ng of each expression plasmid (representing the coding regions of each genome segment), followed by a second transfection with 50 ng of each of the 10 T7 transcripts (representing exact copies of each segment) at 18 h after the first transfection. At 6 h after the second transfection, the culture medium was replaced with 1.5 ml overlay consisting of DMEM, 2% FBS, and 1.5% (wt/vol) agarose type VII (Sigma), and the plates were incubated at 35°C in 5% CO2 for 3 days to allow plaques to appear.
Detection of BTV genomic RNA (S10) in infected cells.
BTV genomic RNAs were detected by the reverse transcription-PCR (RT-PCR) amplification method. Briefly, BSR cells infected with VP6-deficient virus were lysed with the lysis buffer (50 mM sodium phosphate, pH 8.0, 10% [vol/vol] glycerol, 0.5% [vol/vol] NP-40). For detection of BTV mRNA, total RNA was extracted from the whole lysate using TRI reagent (Sigma). For detection of BTV dsRNA, the lysate was treated with 1 U/μl of RNase One RNase (Promega) at 37°C for 15 min prior to the RNA extraction. cDNA copies of the BTV S10 segment were amplified from extracted RNA in a sequence-dependent manner using SuperScript II (Invitrogen) as described in the manufacturer's protocol. For S10 mRNA and plus-strand ssRNA (+ssRNA) of S10 dsRNA, the primer BTVS10R (5′-GTAAGTGTGTAGTATCGCGC-3′) was used. For the minus-strand ssRNA (−ssRNA) of S10 dsRNA, the primer NS3BamHI (17) was used. cDNA copies of S10 mRNA were diluted 1 in 10 and used as a template for subsequent PCR, whereas cDNA copies for S10 +ssRNA and −ssRNA were used without dilution. The set of primers NS3BamHI and BTVS10R was used for subsequent PCR amplification. Note that PCR was run for 20 cycles to stop the reaction before saturation. Then, each band of amplified product was quantified using ImageJ software (NIH; http://rsb.info.nih.gov/ij/).
Purification of BTV core particles.
BTV core particles were semipurified as described previously (6, 23). Further purification of core particles by cesium chloride (CsCl) equilibrium centrifugation was performed using a modification of previous methods (35, 36). Briefly, CsCl was added to the semipurified core particles to reach a final concentration of 50% (wt/vol). The mixture was centrifuged at 150,000 × g for 18 h at 16°C. After centrifugation, 22 fractions (250 μl each) were collected from the top and pelleted through a 30% (wt/vol) sucrose cushion by centrifugation at 14,500 × g for 1.5 h at 4°C. The pellets were resuspended in 20 mM Tris-HCl (pH 8.0) and analyzed by SDS-PAGE.
Electron microscopy.
Aliquots of purified cores were absorbed onto Formvar/carbon support film copper 400-mesh grids (TAAB) for 1 min, washed with water, and negatively stained with 2% (wt/vol) uranyl acetate. The grids were examined under a JEOL 100CX electron microscope at 80 kV.
RESULTS
Roles of BTV nonstructural proteins NS1 and NS2 in primary replication.
In our previous report, we demonstrated the existence of a primary replication cycle in BTV infection using an RG system (9). BSR cells were first transfected with in vitro-synthesized T7 transcripts of only those BTV segments (S1/VP1, S3/VP3, S4/VP4, S9/VP6, S6/NS1, and S8/NS2) encoding products shown to be associated with primary BTV replication in a previous study. At 18 h after the first transfection, these transfected cells were then transfected for a second time with all 10 BTV T7 transcripts. The second set of transcripts provided the templates for BTV dsRNA genome synthesis. We showed that when the four subcore proteins (VP1, VP3, VP4, and VP6) together with two nonstructural proteins (NS1 and NS2) were expressed prior to the second transfection of all 10 BTV T7 transcripts, virus recovery was enhanced considerably (9). We interpreted that the primary transfection components allowed formation of a replicase complex, which then efficiently concluded the replication cycle when the complete set of BTV transcripts was provided. Moreover, it was suggested that these six proteins are important for the BTV primary replication and are probably responsible for the formation of a primary replicase complex. However, our recent CFA system showed that neither NS1 nor NS2 was needed for core assembly and ssRNA packaging (10). Thus, it was important to determine whether NS1 and NS2 are indeed required for in vivo primary replication.
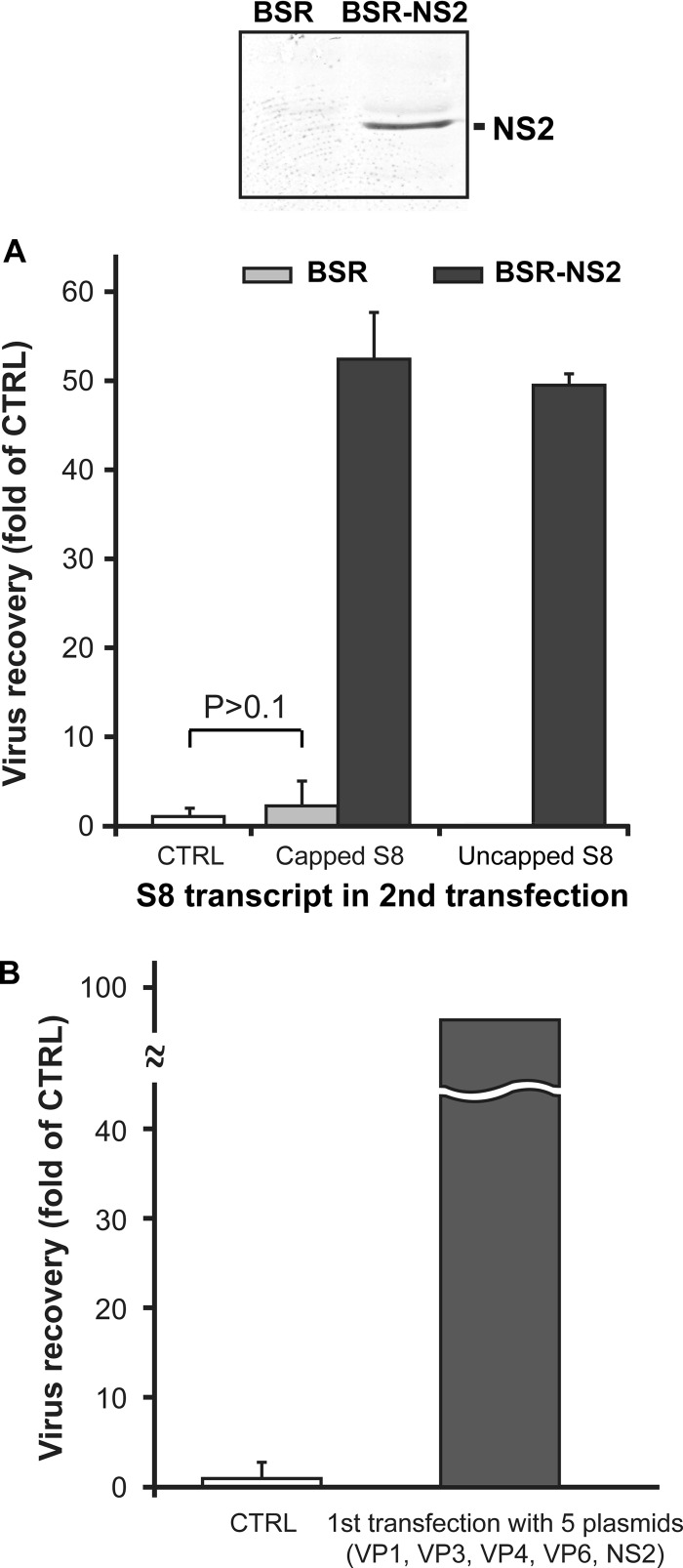
In virus-infected cells, NS2 forms the virus inclusion bodies and is known to interact with the core component (18, 19). Therefore, it is logical to hypothesize that NS2 is involved in BTV primary replication. To obtain direct evidence, a stable cell line that expresses NS2 constitutively was first generated by transfection of an NS2-expressing plasmid, pCAG-PBTV1NS2, as described in Materials and Methods. One clone (clone 1-2) was selected as a candidate for a complementary cell line expressing NS2 (BSR-NS2) by Western blotting (Fig. 1A, top). Further, the expression of NS2 protein was also confirmed by immunofluorescence analysis (data not shown).
Fig 1.
Influence of nonstructural proteins NS2 and NS1 in primary replication. (A) The complementary cell line BSR-NS2 was constructed, and detection of NS2 was performed using a polyclonal NS2 antibody (top). The recovery of BTV was compared between BSR cells (bottom, light gray columns) and BSR-NS2 cells (bottom, dark gray columns) transfected first with 5 capped transcripts, S1 (VP1), S3 (VP3), S4 (VP4), S6 (NS1), and S9 (VP6), and subsequently with capped or uncapped S8 (NS2) together with the remaining 9 capped transcripts. As a control (bottom, open column), BSR cells were transfected only once with 10 capped transcripts (S1 to S10). The recovery of virus was shown as a fold of that for the control (mean ± SD). (B) Virus recovery was compared in the presence and absence of the first transfection with 5 plasmids expressing VP1, VP3, VP4, VP6, and NS2 but not NS1. At 18 h after the first transfection with 5 expression plasmids, transfected BSR cells were transfected a second time with 10 capped transcripts (S1 to S10). As a control (open column), BSR cells were transfected only once with 10 capped transcripts (S1 to S10). The recovery of virus was shown as a fold of that for the control (mean ± SD).
Both BSR cells and BSR-NS2 cells were first transfected with 5 transcripts (encoding VP1, VP3, VP4, VP6, and NS1) lacking the S8 transcript that encodes NS2. Both types of transfected cells were subsequently transfected a second time at 18 h posttransfection with either capped or uncapped S8, together with the remaining 9 capped transcripts (S1 to S7, S9, and S10). Note that uncapped transcripts should not be translated efficiently, unless they are packaged into the assembling subcore. As a control, BSR cells were transfected only once with all 10 capped transcripts (S1 to S10). Virus rescue was measured by plaque assay at 3 days after the second transfection. The first transfection of normal BSR cells without S8 (NS2) did not enhance virus recovery at all (Fig. 1A, bottom, control [CTRL] column versus light gray column), while the presence of S8 in the first transfection significantly enhanced virus recovery (9). Moreover, the complete lack of NS2 abolished virus rescue from transfected BSR cells (Fig. 1A, bottom). However, when NS2 was supplemented by the complementary cell line, the virus recovery was enhanced at a considerably higher level (Fig. 1A, bottom, dark gray columns). These findings suggested that NS2 is important for the primary replication.
Since in virus-infected cells NS1, like NS2, is synthesized abundantly in an early stage and assembles into a tubule structure (37, 38), it may also be involved in primary replication. The role of the NS1 tubule structure has yet to be defined, although our study recently demonstrated that NS1 enhances translation of BTV proteins (39). To clarify the role of NS1 in primary replication, we have used a modified RG system based on plasmid expression of BTV proteins under the control of the CAG promoter, as described in Materials and Methods. The system appears to be efficient for BTV protein expression. BSR cells were therefore first transfected with 5 BTV plasmids, pCAG-PBTV1VP1, pCAG-PBTV1VP3, pCAG-PBTV1VP4, pCAG-PBTV10VP6, and pCAG-PBTV1NS2, expressing the four subcore proteins (VP1, VP3, VP4, and VP6) and NS2, respectively, but not NS1, followed by a second transfection with all 10 capped transcripts as described above, and the resulting plaques were rescued. The virus recovery was considerable (Fig. 1B) and equivalent to that shown in Fig. 1A and a previous report (9), in which S6 (NS1) was present in the first transfection. The result suggested that NS1 is not an essential component of the primary replicase complex. Thus, it can be postulated that as long as the other essential BTV proteins are expressed at a level that could initiate primary replication, NS1 is accessory.
The core protein VP7 is required after subcore protein assembly into the primary replicase complex.
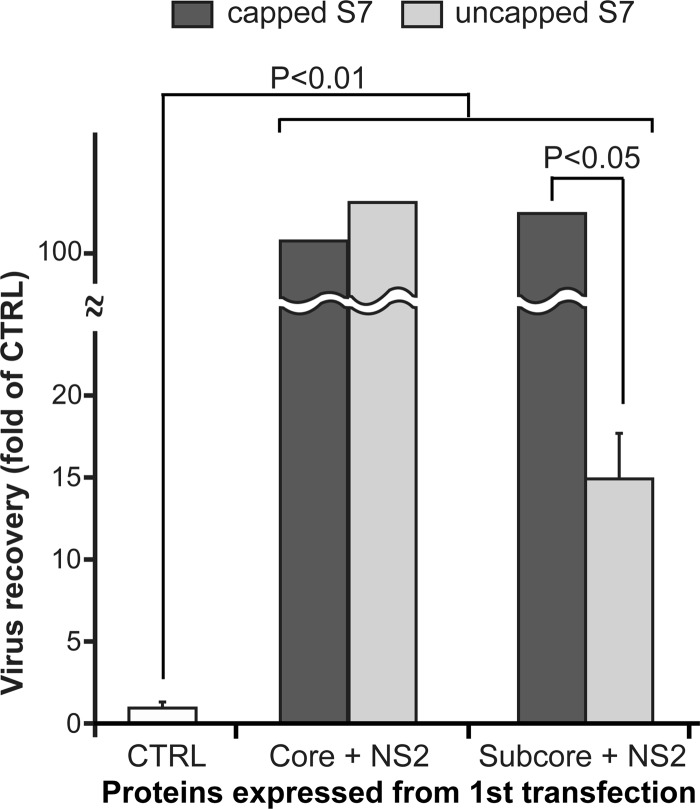
After entry into cells, the BTV core is released into the cytoplasm and extrudes newly synthesized transcripts. It is believed that only the intact core and not the subcore is transcriptionally active, so it may be involved in primary replication. Further, our recent in vitro CFA assay demonstrated that VP7 is essential to protecting the packaged ssRNA molecules and for stabilizing the assembled cores. Although our previous report showed that only 4 of 5 core proteins, VP1, VP3, VP4, and VP6, but not VP7, were required to enhance virus recovery (9), there was still some possibility that VP7, expressed from the capped S7 transcript in the second transfection, could be involved in this enhancement. To determine the role of VP7 in primary replication, instead of T7 RNA transcripts, we used DNA plasmids of BTV RNA segments for the primary transfection, as described above. BSR cells were first transfected with either 6 BTV plasmids expressing core proteins (VP1, VP3, VP4, VP6, VP7) and NS2 or 5 plasmids expressing only subcore proteins (VP1, VP3, VP4, VP6) and NS2 and not a VP7-expressing plasmid. All cells were then transfected for a second time with a mixture of an uncapped S7 (incapable of VP7 expression) or capped S7 (capable of VP7 expression) transcript, together with the remaining 9 capped transcripts (S1 to S6, S8 to S10). As a control, BSR cells were transfected only once with all 10 capped transcripts (S1 to S10). As shown in Fig. 2, although the virus recovery from BSR cells lacking VP7 expression was significantly less than that from BSR cells synthesizing VP7 in either the first or second transfection, it was still considerably higher than that of the single-transfection control. Moreover, VP7 expressed from the first transfection did not affect virus recovery, as long as the S7 transcript was capped in the second transfection (Fig. 2, dark gray columns). This result was consistent with our previous data, which showed that the addition of S7 transcripts to the first transfection did not affect virus recovery (9). Further, it is noteworthy that virus recovery was likely to be regulated by VP7 expressed from the second transfection and not that expressed from the first transfection (Fig. 2, right dark gray column versus right light gray column). All together, these data suggested that although VP7 is not essential for primary replication, it is most likely required once the subcore proteins have assembled into the primary replicase complex to stabilize the complex in order to continue synthesis of transcripts.
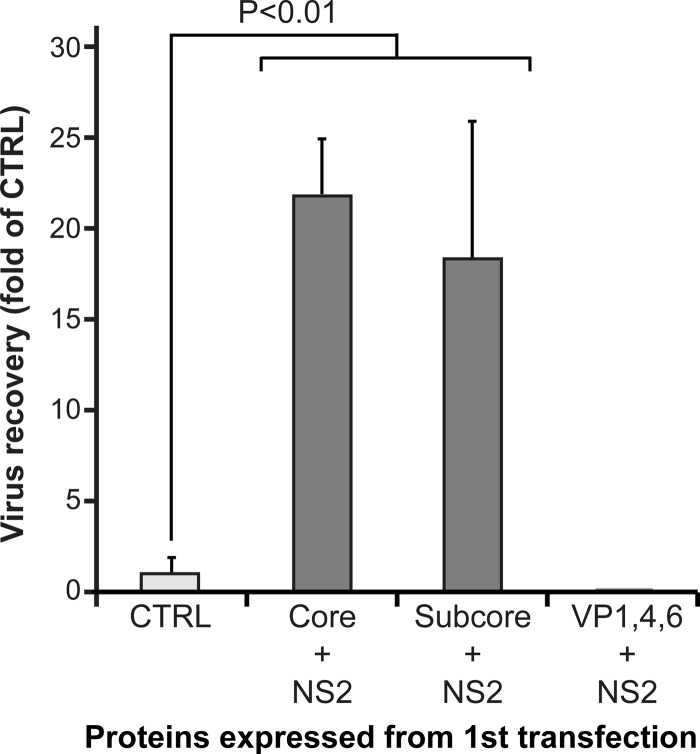
Fig 2.
Impact of the VP7 protein in primary replication. Total numbers of plaques were compared in the presence and absence of VP7. BSR cells were first transfected with 6 expression plasmids, VP1, VP3, VP4, VP6, VP7, and NS2 (Core + NS2), or with 5 expression plasmids, VP1, VP3, VP4, VP6, and NS2 (Subcore + NS2), followed by a second transfection with capped (dark gray columns) or uncapped (light gray columns) S7 transcript as well as the 9 remaining capped transcripts. As a control (open column), BSR cells were transfected only once with 10 capped transcripts (S1 to S10). The recovery of virus was shown as a fold of that for the control (mean ± SD).
Minimum requirement for the functional primary replicase complex.
To determine further the minimum components of the primary replicase complex, we utilized our modified plasmid-based RG system with uncapped transcripts for the second transfection to provide only the template RNAs. BSR cells were first transfected with 5 BTV plasmids (VP1, VP3, VP4, VP6, NS2), followed by the second transfection with 10 uncapped transcripts. As a control, BSR cells were transfected only once with all 10 capped transcripts. As long as core (VP1, VP3, VP4, VP6, and VP7) or subcore (VP1, VP3, VP4, and VP6) proteins together with NS2 were expressed in the first transfection, the virus was recovered from BSR cells transfected with uncapped transcripts in the second transfection (Fig. 3, Core + NS2 and Subcore + NS2 columns). In addition, the amount of virus recovery was still higher than that for the control cells that were transfected only once with all 10 capped transcripts. These results suggested that 10 uncapped transcripts were packaged into the primary replicase complex and capped transcripts were newly synthesized from the complex. However, when VP3 was not expressed prior to the second transfection, the virus was not recovered at all when the transcripts in the second transfection were uncapped (Fig. 3, VP1, 4, 6 + NS2 column). The data also indicate that VP4, synthesized by the first transfection of plasmid pCAG-PBTV1VP4 in BSR cells, was incapable of supporting translation of BTV proteins at a level sufficient to lead to formation of virus particles. Moreover, these data suggested that the presence of NS2 and only the 3 enzymatic proteins, VP1, VP4, and VP6, with 10 +ssRNAs is insufficient for initiating primary replication and that their association with VP3 is essential. The CFA system demonstrated that 3 minor proteins, VP1, VP4, and VP6, first assembled in the presence of the +ssRNAs of all 10 segments, followed by assembly of a subcore-like complex upon VP3 addition (10). Together, these results strongly suggest that all four subcore proteins and BTV ssRNAs comprise the minimum components of the primary replicase complex.
Fig 3.
Minimum requirements for primary replication. The total numbers of plaques in each experiment were counted when BSR cells were first transfected with 6 expression plasmids, VP1, VP3, VP4, VP6, VP7, and NS2 (Core + NS2), or with 5 expression plasmids, VP1, VP3, VP4, VP6, and NS2 (Subcore + NS2), or 4 expression plasmids, VP1, VP4, VP6, and NS2 (VP1, 4, 6 + NS2). The transfected cells were transfected a second time with all 10 uncapped transcripts (S1 to S10). The recovery of BTV was compared with that for the control (open column), in which BSR cells were transfected only once with the 10 capped transcripts (S1 to S10). The recovery of virus was shown as a fold of that for the control (mean ± SD).
VP6-deficient virus is replication deficient due its inability to incorporate viral RNAs.
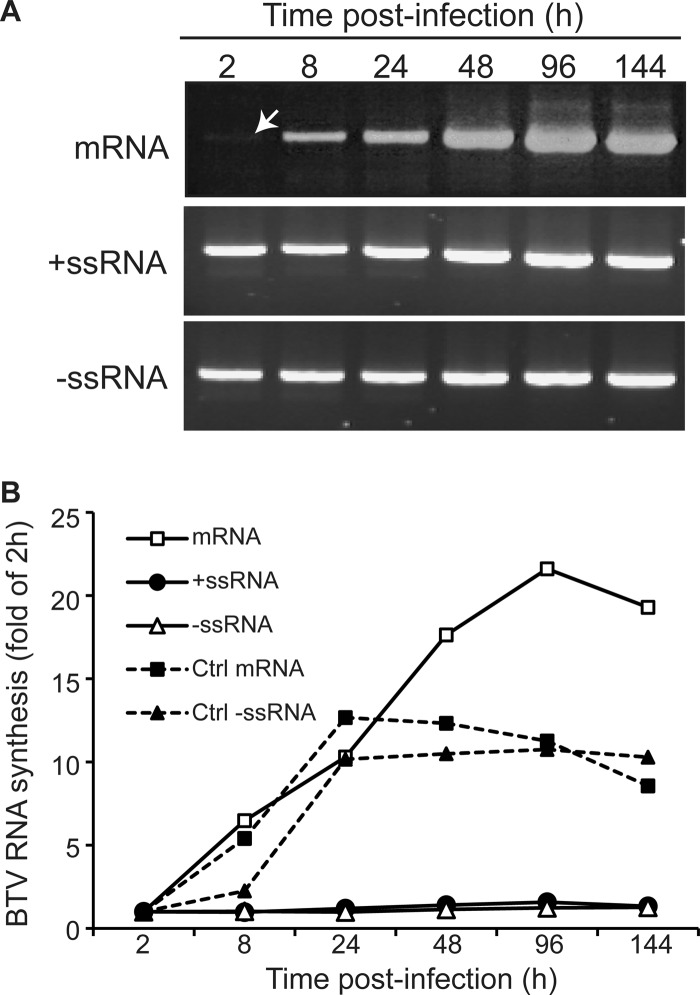
Since VP6 is an internal subcore protein and its role in vivo has not yet been clarified, we wanted to examine if it has any specific role in primary replication. Our previous data demonstrated that although VP6-deficient BTV could synthesize BTV proteins as well as chimeric S9EGFP proteins at 2 days postinfection, replication was not possible in normal BSR cells, suggesting that VP6-deficient BTV is replication deficient (9, 34). However, it was unclear why VP6-deficient mutant viruses were incapable of replication in BSR cells. In particular, at which step was BTV replication hampered by VP6 deficiency: unwinding of dsRNA or another function during assembly? To address this, BSR cells were infected with one of the previously recovered VP6-deficient viruses, BTV S9E2 (Fig. 4) and total RNA was extracted from the infected cells at various time points (2, 8, 24, 48, 96, and 144 h) postinfection. As a control, BSR cells were infected with wild-type (WT) BTV. Viral mRNAs were detected as described in Materials and Methods. For detection of dsRNA, BSR cell lysate was treated with RNase before RNA extraction, and then +ssRNAs and −ssRNAs were detected as described in Materials and Methods. As shown in Fig. 5, S10 mRNA was detected at as early as 2 h postinfection and continuously increased (Fig. 5A, top). Similar results were obtained with the control (Fig. 5B, Ctrl mRNA). In contrast, the level of both positive- and negative-sense RNA detected from RNase-treated cell lysate did not alter and remained the same throughout the infection (Fig. 5A, middle and bottom), unlike that of the control (Fig. 5B, Ctrl −ssRNA), indicating that dsRNA synthesis was aborted. As the level of dsRNA, which was supposed to increase during assembly of a new primary replicase complex, did not increase (Fig. 5A and B, +ssRNA and −ssRNA), the increase in the mRNA level detected in the infected BSR cells (Fig. 5A and B, mRNA) was evidently due to transcription from the inoculated residual core particles. It has been reported that VP6 is not necessary for dsRNA synthesis, at least in vitro (24). Indeed, our CFA system demonstrated that dsRNAs were synthesized after the positive-sense viral ssRNAs were assembled with other integral components of the primary replicase complex (10). Therefore, it is likely that the assembly of the replicase complex itself and not transcription from newly assembled complex failed in VP6-deficient virus.
Fig 4.
Schematic representation of the changes introduced in the S9 (VP6) segment of BTV. The name of the mutation is indicated on the left. Numbers indicate nucleotide positions in S9 (VP6) where deletions were introduced. UTR, untranslated region.
Fig 5.
Detection of genomic RNA in BSR cells infected with VP6-deficient BTV. RT-PCR amplification was performed using primers specific for S10 mRNA, +ssRNA, and −ssRNA, as described in Materials and Methods. Bands of amplified PCR products were detected by ethidium bromide staining (A), quantified using ImageJ software (NIH: http://rsb.info.nih.gov/ij/), and plotted (B). The RNA synthesis of each time point was shown as a fold of that at 2 h.
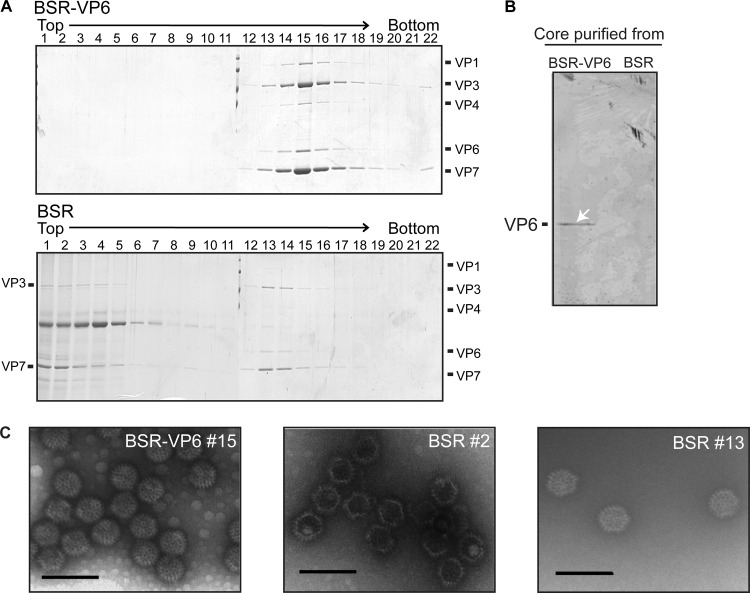
To further substantiate this, normal BSR cells and BSR-VP6 cells were infected with BTV S9E2 and core particles were purified at 3 days postinfection, first by a sucrose-ultracentrifugation method and then by cesium chloride equilibrium centrifugation, as described in Materials and Methods. Core particles produced in BSR-VP6 cells showed a single peak (predominantly in fraction 15), which contains VP6 (Fig. 6A, top). In contrast, core particles produced in BSR cells showed two peaks; one was with VP6 (fraction 13; Fig. 6A, bottom) and the other was without VP6 (fraction 2; Fig. 6A, bottom). The absence of VP6 in the core particles purified from normal BSR cells was also confirmed by Western blotting (Fig. 6B). The visualization by electron microscopy clearly showed that all core particles in the peak lacking VP6 were empty particles (Fig. 6C, middle), whereas all core particles of BSR-VP6 in fraction 15 were complete (Fig. 6C, left). Further, neither VP1 nor VP4 was detected in the fractions containing empty core particles (Fig. 6A, bottom), indicating that not only the genomic RNAs but also VP1 and VP4 were also not recruited. The second peak (fraction 13) of the BSR core particle, which was slightly lighter than that of the BSR-VP6 core particles, contained particles visually similar to BSR-VP6 core particles (Fig. 6C, right). These particles are most likely the remaining cores of the inoculated virus which transcribed mRNAs for a limited period only (Fig. 5) in the noncomplementary cells and eventually degraded.
Fig 6.
Analysis of VP6-deficient BTV core particles purified from BSR and BSR-VP6 cells by CsCl gradient centrifugation. (A) Supernatants from BSR-VP6 (top) or BSR (bottom) infected cell lysates were spun down over a 30% (wt/vol) sucrose cushion and subjected to CsCl equilibrium centrifugation. Twenty-two fractions were collected from the top and then analyzed by SDS-PAGE. The gels were stained by Coomassie brilliant blue. (B) BTV core particles purified from BSR-VP6 and BSR cells were resolved by SDS-PAGE, and VP6 was detected by Western blotting using VP6-specific antibody. (C) Electron micrograph of core particles of peak fractions. Bars, 100 nm.
Together, these data demonstrate that VP6-deficient viruses were not competent for replication due to their failure to assemble functional replicase complexes. The only difference between the normal BSR cells and the BSR-VP6 cells infected with VP6-deficient virus was the absence or presence of VP6. All other BTV proteins, including the essential protein NS2, were expressed. Thus, our results suggest that VP6 is an essential protein for the assembly of the primary replicase complex. Further, since our previous CFS system demonstrated that 10 BTV ssRNAs initiate the in vitro replicase complex and since VP6 is an ssRNA binding protein, VP6 and, by default, VP1 and VP4 may be involved in recruitment of ssRNAs in vivo during assembly.
DISCUSSION
The development of RG systems has revolutionized dsRNA virus research. The RG system for orbiviruses has previously allowed us to demonstrate that in order to enhance virus production, orbiviruses are likely to first assemble a primary replicase complex, which then increases the level of transcripts synthesized from all 10 genomic segments (8, 9). Together with the recently developed CFA system of the BTV core (10), we were able to understand BTV primary replication in more detail. We present here further evidence of the existence of primary replication and the roles of the essential proteins required in primary replication. In particular, we demonstrated that the smallest core protein, VP6, plays an essential role in primary replication and also probably in RNA packaging during core assembly.
Our previous in vivo and in vitro assembly studies (10, 11, 34, 37) indicated that BTV cores may assemble in separate stages, starting from the assembly of the minor proteins VP1, VP4, and VP6 with 10 +ssRNAs, which is then followed by the assembly of VP3 decamers. This then triggers the polymerase protein VP1 to synthesize all BTV dsRNAs and the formation of subcore particles. The major core protein VP7, which forms 260 trimers, is then attached to the VP3 surface layer in a highly ordered fashion to form a stable core particle. Our RG data in this report strongly suggested that a newly assembled complex, which is composed of the same components as subcore particles, initiates transcription activity. Thus, the assembly pathway of the primary replicase complex should be same as that for core assembly.
Although the CFA system did not show how nonstructural proteins, especially NS2, affect the assembly of the primary replicase complex (10), the importance of nonstructural proteins in BTV replication has already been reported (15–19). NS2 is the main component of viral inclusion bodies (VIBs) in virus-infected cells where assembly of core particle takes place (18, 19, 37, 40, 41). The data obtained from this study showed that NS2 is active in primary replication. Thus, although NS2 is not the main component of the primary replicase complex in vitro, NS2 is essential in vivo for the formation of VIBs, which could have a role as a concentrator of components of the complex as well as for protection of newly synthesized ssRNAs from degradation, prior to their assembly into the complex. The other nonstructural protein, NS1, which has been shown to be responsible for enhancing BTV protein synthesis (39), was not necessary in primary replication, as long as subcore proteins and NS2 were expressed well. Thus, NS1 is probably required as a translation enhancer during primary replication.
An RNase protection assay using in vitro assembly systems showed that the presence of VP7 was necessary to protect +ssRNAs in the complex against RNase A treatment (10). Additionally, the polymerase reaction was successfully achieved after the assembly of subcore proteins and ssRNA (10). These data suggest that an in vitro-assembled subcore-like complex is unstable, which is probably due to the weak interactions between the VP3 decamers. Interestingly, RG data showed that the assembled subcore-like complex was transcriptionally active but that VP7 was required to stabilize the assembled complex.
VP7 has been reported to colocalize with NS2 only in the presence of VP3 (18). Additionally, core particles are commonly observed in the cytoplasm as well as VIBs (16, 18, 34). Although further investigation is required, it is possible that addition of VP7 onto the subcore may be necessary prior to the trafficking of the assembled complex out of VIBs and that the lack of VP7 hampered the transfer of the complex from VIBs, resulting in a reduction of protein synthesis by the new BTV.
BTV, a member of the Orbivirus genus, is unique among the Reoviridae family in encoding VP6, a protein with nucleoside triphosphatase, RNA binding, and helicase activity (31, 32, 42). We found that VP6-deficient BTV was replication deficient because the BTV genome could not be packaged in the assembled particle. Common features of RNA translocators are that these proteins possess ATPase and helicase activity (43–46). For example, the dsRNA bacteriophage (ϕ6) packaging motor P4 has been shown to form a hexamer, like BTV VP6 (31), although the assembly mechanism of ϕ6 is quite different from that of BTV (45, 47–49). Thus, it is reasonable to hypothesize that VP6 also plays a role in RNA packaging, in addition to its role as an RNA helicase.
A series of BTV studies using the recombinant baculovirus expression system demonstrated that the two inner capsid proteins VP3 and VP7 expressed in insect cells could assemble into core-like particles (CLPs) and that enzymatic proteins VP1 and VP4 formed a complex and were recruited into CLPs when those proteins were expressed together with VP3 and VP7 (30, 50, 51). The CFA system clearly showed that 3 minor proteins, VP1, VP4, and VP6, first made a complex with ssRNAs of all 10 segments and that VP1 and VP4 also made a complex in the presence of BTV ssRNAs (10). Together with the fact that the VP1/VP4/VP6 complex has never been observed in the absence of BTV ssRNAs in any expression system, it is suggested that VP6 would indirectly interact with the VP1/VP4 complex via ssRNAs. In addition, VP6 is known to possess RNA binding activity (31, 32). Although further study is necessary, it is possible that VP1/VP4/ssRNA or VP3/VP1/VP4/ssRNA cannot be assembled into complexes in the absence of VP6, although VP3 and VP7, the two major proteins, may assemble as core-like particles in BTV-infected cells, similar to the case in a recombinant baculovirus expression system.
Overall, the data presented in this report demonstrate the assembly mechanisms of the primary replicase complex in vivo, particularly the importance of VP6 at an early stage. Together with our CFA system and in vivo RG system, it should be possible to define precisely the mechanisms of primary replication at a finer level. In addition, our data will contribute to clarification of the similarity and differences between orbiviruses and other viruses of the family Reoviridae in virus replication.
ACKNOWLEDGMENTS
We thank Maria McCrossan (LSHTM) for technical help with electron microscopy experiments.
This work is supported jointly by the Biotechnology and Biological Sciences Research Council (United Kingdom) and the National Institute of Allergy and Infectious Diseases (United States).
Footnotes
Published ahead of print 31 October 2012
REFERENCES
- 1. Ratinier M, Caporale M, Golder M, Franzoni G, Allan K, Nunes SF, Armezzani A, Bayoumy A, Rixon F, Shaw A, Palmarini M. 2011. Identification and characterization of a novel non-structural protein of bluetongue virus. PLoS Pathog. 7:e1002477 doi:10.1371/journal.ppat.1002477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Verwoerd DW. 1969. Purification and characterization of bluetongue virus. Virology 38:203–212 [DOI] [PubMed] [Google Scholar]
- 3. Verwoerd DW, Huismans H. 1972. Studies on the in vitro and the in vivo transcription of the bluetongue virus genome. Onderstepoort J. Vet. Res. 39:185–191 [PubMed] [Google Scholar]
- 4. Zarbl H, Millward S. 1983. The reovirus multiplication cycle. In Joklik W. (ed), The Reoviridae. Springer, New York, NY [Google Scholar]
- 5. Huismans H, van Dijk AA, Els HJ. 1987. Uncoating of parental bluetongue virus to core and subcore particles in infected L cells. Virology 157:180–188 [DOI] [PubMed] [Google Scholar]
- 6. Mertens PP, Burroughs JN, Anderson J. 1987. Purification and properties of virus particles, infectious subviral particles, and cores of bluetongue virus serotypes 1 and 4. Virology 157:375–386 [DOI] [PubMed] [Google Scholar]
- 7. Van Dijk AA, Huismans H. 1988. In vitro transcription and translation of bluetongue virus mRNA. J. Gen. Virol. 69:573–581 [DOI] [PubMed] [Google Scholar]
- 8. Matsuo E, Celma CC, Roy P. 2010. A reverse genetics system of African horse sickness virus reveals existence of primary replication. FEBS Lett. 584:3386–3391 [DOI] [PubMed] [Google Scholar]
- 9. Matsuo E, Roy P. 2009. Bluetongue virus VP6 acts early in the replication cycle and can form the basis of chimeric virus formation. J. Virol. 83:8842–8848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lourenco S, Roy P. 2011. In vitro reconstitution of Bluetongue virus infectious cores. Proc. Natl. Acad. Sci. U. S. A. 108:13746–13751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Roy P. 2008. Bluetongue virus: dissection of the polymerase complex. J. Gen. Virol. 89:1789–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roy P. 2008. Functional mapping of bluetongue virus proteins and their interactions with host proteins during virus replication. Cell Biochem. Biophys. 50:143–157 [DOI] [PubMed] [Google Scholar]
- 13. Bhattacharya B, Noad RJ, Roy P. 2007. Interaction between bluetongue virus outer capsid protein VP2 and vimentin is necessary for virus egress. Virol. J. 4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roy P. 1996. Orbivirus structure and assembly. Virology 216:1–11 [DOI] [PubMed] [Google Scholar]
- 15. Beaton AR, Rodriguez J, Reddy YK, Roy P. 2002. The membrane trafficking protein calpactin forms a complex with bluetongue virus protein NS3 and mediates virus release. Proc. Natl. Acad. Sci. U. S. A. 99:13154–13159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Celma CC, Roy P. 2011. Interaction of calpactin light chain (S100A10/p11) and a viral NS protein is essential for intracellular trafficking of nonenveloped bluetongue virus. J. Virol. 85:4783–4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Celma CC, Roy P. 2009. A viral nonstructural protein regulates bluetongue virus trafficking and release. J. Virol. 83:6806–6816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kar AK, Bhattacharya B, Roy P. 2007. Bluetongue virus RNA binding protein NS2 is a modulator of viral replication and assembly. BMC Mol. Biol. 8:4 doi:10.1186/1471-2199-8-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Modrof J, Lymperopoulos K, Roy P. 2005. Phosphorylation of bluetongue virus nonstructural protein 2 is essential for formation of viral inclusion bodies. J. Virol. 79:10023–10031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Owens RJ, Limn C, Roy P. 2004. Role of an arbovirus nonstructural protein in cellular pathogenesis and virus release. J. Virol. 78:6649–6656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wirblich C, Bhattacharya B, Roy P. 2006. Nonstructural protein 3 of bluetongue virus assists virus release by recruiting ESCRT-I protein Tsg101. J. Virol. 80:460–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Boyce M, Celma CC, Roy P. 2008. Development of reverse genetics systems for bluetongue virus: recovery of infectious virus from synthetic RNA transcripts. J. Virol. 82:8339–8348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Boyce M, Roy P. 2007. Recovery of infectious bluetongue virus from RNA. J. Virol. 81:2179–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Boyce M, Wehrfritz J, Noad R, Roy P. 2004. Purified recombinant bluetongue virus VP1 exhibits RNA replicase activity. J. Virol. 78:3994–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matsuo E, Roy P. 2011. Bluetongue virus VP1 polymerase activity in vitro: template dependency, dinucleotide priming and cap dependency. PLoS One 6:e27702 doi:10.1371/journal.pone.0027702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ramadevi N, Burroughs NJ, Mertens PPC, Jones IM, Roy P. 1998. Capping and methylation of mRNA by purified recombinant VP4 protein of bluetongue virus. Proc. Natl. Acad. Sci. U. S. A. 95:13537–13542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sutton G, Grimes JM, Stuart DI, Roy P. 2007. Bluetongue virus VP4 is an RNA-capping assembly line. Nat. Struct. Mol. Biol. 14:449–451 [DOI] [PubMed] [Google Scholar]
- 28. Wehrfritz JM, Boyce M, Mirza S, Roy P. 2007. Reconstitution of bluetongue virus polymerase activity from isolated domains based on a three-dimensional structural model. Biopolymers 86:83–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grimes JM, Burroughs JN, Gouet P, Diprose JM, Malby R, Zientara S, Mertens PP, Stuart DI. 1998. The atomic structure of the bluetongue virus core. Nature 395:470–478 [DOI] [PubMed] [Google Scholar]
- 30. Nason EL, Rothagel R, Mukherjee SK, Kar AK, Forzan M, Prasad BV, Roy P. 2004. Interactions between the inner and outer capsids of bluetongue virus. J. Virol. 78:8059–8067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kar AK, Roy P. 2003. Defining the structure-function relationships of bluetongue virus helicase protein VP6. J. Virol. 77:11347–11356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stauber N, Martinez-Costas J, Sutton G, Monastyrskaya K, Roy P. 1997. Bluetongue virus VP6 protein binds ATP and exhibits an RNA-dependent ATPase function and a helicase activity that catalyze the unwinding of double-stranded RNA substrates. J. Virol. 71:7220–7226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bisaillon M, Bergeron J, Lemay G. 1997. Characterization of the nucleoside triphosphate phosphohydrolase and helicase activities of the reovirus lambda1 protein. J. Biol. Chem. 272:18298–18303 [DOI] [PubMed] [Google Scholar]
- 34. Matsuo E, Celma CC, Boyce M, Viarouge C, Sailleau C, Dubois E, Breard E, Thiery R, Zientara S, Roy P. 2011. Generation of replication-defective virus-based vaccines that confer full protection in sheep against virulent bluetongue virus challenge. J. Virol. 85:10213–10221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Matsuo E, Tani H, Lim C, Komoda Y, Okamoto T, Miyamoto H, Moriishi K, Yagi S, Patel AH, Miyamura T, Matsuura Y. 2006. Characterization of HCV-like particles produced in a human hepatoma cell line by a recombinant baculovirus. Biochem. Biophys. Res. Commun. 340:200–208 [DOI] [PubMed] [Google Scholar]
- 36. Reimerink JH, Boshuizen JA, Einerhand AW, Duizer E, van Amerongen G, Schmidt N, Koopmans MP. 2007. Systemic immune response after rotavirus inoculation of neonatal mice depends on source and level of purification of the virus: implications for the use of heterologous vaccine candidates. J. Gen. Virol. 88:604–612 [DOI] [PubMed] [Google Scholar]
- 37. Roy P. 2007. Orbiviruses and their replication, p 1975–1997 In Knipe DM, et al. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 38. Urakawa T, Roy P. 1988. Bluetongue virus tubules made in insect cells by recombinant baculoviruses: expression of the NS1 gene of bluetongue virus serotype 10. J. Virol. 62:3919–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boyce M, Celma CC, Roy P. 2012. Bluetongue virus non-structural protein 1 is a positive regulator of viral protein synthesis. Virol. J. 9:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lymperopoulos K, Noad R, Tosi S, Nethisinghe S, Brierley I, Roy P. 2006. Specific binding of bluetongue virus NS2 to different viral plus-strand RNAs. Virology 353:17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lymperopoulos K, Wirblich C, Brierley I, Roy P. 2003. Sequence specificity in the interaction of bluetongue virus non-structural protein 2 (NS2) with viral RNA. J. Biol. Chem. 278:31722–31730 [DOI] [PubMed] [Google Scholar]
- 42. Luking A, Stahl U, Schmidt U. 1998. The protein family of RNA helicases. Crit. Rev. Biochem. Mol. Biol. 33:259–296 [DOI] [PubMed] [Google Scholar]
- 43. Beran RK, Serebrov V, Pyle AM. 2007. The serine protease domain of hepatitis C viral NS3 activates RNA helicase activity by promoting the binding of RNA substrate. J. Biol. Chem. 282:34913–34920 [DOI] [PubMed] [Google Scholar]
- 44. Carpio RV, Gonzalez-Nilo FD, Jayaram H, Spencer E, Prasad BV, Patton JT, Taraporewala ZF. 2004. Role of the histidine triad-like motif in nucleotide hydrolysis by the rotavirus RNA-packaging protein NSP2. J. Biol. Chem. 279:10624–10633 [DOI] [PubMed] [Google Scholar]
- 45. Kainov DE, Pirttimaa M, Tuma R, Butcher SJ, Thomas GJ, Jr, Bamford DH, Makeyev EV. 2003. RNA packaging device of double-stranded RNA bacteriophages, possibly as simple as hexamer of P4 protein. J. Biol. Chem. 278:48084–48091 [DOI] [PubMed] [Google Scholar]
- 46. Taraporewala ZF, Patton JT. 2001. Identification and characterization of the helix-destabilizing activity of rotavirus nonstructural protein NSP2. J. Virol. 75:4519–4527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huiskonen JT, Jaalinoja HT, Briggs JA, Fuller SD, Butcher SJ. 2007. Structure of a hexameric RNA packaging motor in a viral polymerase complex. J. Struct. Biol. 158:156–164 [DOI] [PubMed] [Google Scholar]
- 48. Lisal J, Kainov DE, Lam TT, Emmett MR, Wei H, Gottlieb P, Marshall AG, Tuma R. 2006. Interaction of packaging motor with the polymerase complex of dsRNA bacteriophage. Virology 351:73–79 [DOI] [PubMed] [Google Scholar]
- 49. Lisal J, Lam TT, Kainov DE, Emmett MR, Marshall AG, Tuma R. 2005. Functional visualization of viral molecular motor by hydrogen-deuterium exchange reveals transient states. Nat. Struct. Mol. Biol. 12:460–466 [DOI] [PubMed] [Google Scholar]
- 50. French TJ, Roy P. 1990. Synthesis of bluetongue virus (BTV) corelike particles by a recombinant baculovirus expressing the two major structural core proteins of BTV. J. Virol. 64:1530–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Limn CK, Staeuber N, Monastyrskaya K, Gouet P, Roy P. 2000. Functional dissection of the major structural protein of bluetongue virus: identification of key residues within VP7 essential for capsid assembly. J. Virol. 74:8658–8669 [DOI] [PMC free article] [PubMed] [Google Scholar]