Abstract
ADAR1, an interferon (IFN)-inducible double-stranded (ds) RNA-specific adenosine deaminase, downregulates host innate responses, including activation of the dsRNA-dependent protein kinase (PKR) and induction of IFN-β mRNA. Conversely, PKR amplifies IFN-β induction by measles virus (MV) and inhibits virus protein synthesis. Formation of stress granules (SGs), cytoplasmic aggregates of stalled translation complexes and RNA-binding proteins, is a host response to virus infection mediated by translation initiation factor eIF2α phosphorylation. We examined the roles of PKR and ADAR1 in SG formation using HeLa cells stably deficient in either PKR (PKRkd) or ADAR1 (ADAR1kd) compared to control (CONkd) cells. Infection with either wild-type (WT) MV or an isogenic mutant lacking C protein expression (Cko) comparably induced formation of SG in ADAR1kd cells, whereas only the Cko mutant was an efficient inducer in control cells. Both ADAR1 and PKR colocalized with SG following infection. MV-induced; SG formation was PKR dependent but impaired by ADAR1. Complementation of ADAR1kd cells by expression of either p150 WT isoform or the p150 Zα (Y177A) Z-DNA-binding mutant of ADAR1 restored suppression of host responses, including SG formation and PKR activation. In contrast, neither the p110 WT isoform nor the p150 catalytic (H910A, E912A) mutant of ADAR1 complemented the ADAR1kd phenotype. These results further establish ADAR1 as a suppressor of host innate responses, including activation of PKR and the subsequent SG response.
INTRODUCTION
Measles virus (MV) possesses a negative-sense single-stranded RNA genome of ∼15.9 kb and is a member of the Morbillivirus genus of the Paramyxoviridae family. From the six MV specified genes, N, P/V/C, M, F, H, and L genes, eight proteins are encoded; five of them are structural proteins from monocistronic genes, whereas the polycistronic P/V/C gene encodes the structural P protein, a polymerase cofactor, in addition to the V and C accessory proteins that suppress the host immune response (1). Infection with MV causes acute febrile illness, and despite the existence of an effective vaccine, MV remains a leading cause of morbidity and mortality worldwide (2). Persistent infection with MV, although rare, may lead to a serious and often fatal neurodegenerative disease, subacute sclerosing panencephalitis (3). The potential for use of attenuated MV strains as oncolytic agents for cancer therapy (4), coupled with the need for improved MV vaccines as well as increased adherence to vaccine regimens (1), has motivated research to further understand the molecular mechanisms of the host response to MV infection. MV is immunomodulatory, with the capacity to affect both the innate and adaptive immune responses in part through the functions of the V and C accessory proteins as revealed from studies using isogenic knockout mutant viruses defective for either V (Vko) or C (Cko) protein expression (5).
The interferon (IFN) system plays a central role in the innate antiviral immune response to virus infection. Pathogen-associated molecular patterns of viruses, including structural characteristics of viral RNAs that differentiate them from cellular RNAs, trigger the transcriptional activation of IFN through multiple signaling pathways. Among these are the cytoplasmic retinoic-acid-inducible protein (RIG-I) sensors that detect viral RNAs, including MV RNA, as foreign RNA (5–7). IFNs are produced and secreted and can act in autocrine and paracrine manners. Following binding to their cognate receptors, IFNs trigger signaling, including by the canonical JAK-STAT pathway that leads to the induction of several interferon-stimulated genes (ISGs), some of which encode proteins with antiviral activities (8). Among the ISGs are two enzymes that are double-stranded RNA (dsRNA)-binding proteins, the protein kinase PKR, regulated by dsRNA, and the adenosine deaminase ADAR1, which acts on dsRNA (5, 9–12).
The PKR kinase is an IFN-inducible kinase that is either activated or antagonized following binding of dsRNA or single-stranded RNA (ssRNA) with ds character. PKR typically displays proapoptotic and antiviral activities (8, 10, 11, 13). dsRNA bound to a repeated domain present in the N-terminal region of PKR can lead to activation by autophosphorylation, including at threonine 446, dimerization, and subsequent substrate phosphorylation catalyzed by the kinase subdomains present in the C-terminal region of the PKR protein (13, 14). The best-characterized PKR substrate, eIF2α (15), when phosphorylated, alters the translational pattern in cells, which can lead to apoptosis (8, 11).
The ADAR1 RNA deaminase, like PKR, is an IFN-inducible enzyme, but in the case of ADAR1, dsRNA is typically the substrate rather than an effector of activity as in the case of PKR (12, 13). ADAR1 catalyzes the C-6 deamination of adenosine (A) to inosine (I) in dsRNA, a process known as A-to-I editing (13, 16). ADAR1 editing activity is a physiologically relevant process that can cause changes in genetic decoding at the translational level and also lead to destabilization of RNA structures (12, 17). Dependent upon their structure, substrate RNAs may be selectively edited in a manner that results in altered translational decoding by amino acid substitutions such as “I” base pairs like “G” instead of like “A”. This occurs in RNAs exemplified by those encoding the neurotransmitter receptors GluR-B and 5HT-2cR and in the hepatitis delta virus antigenome RNA (16). RNAs also can undergo nonselective hyperediting as illustrated by MV and polyomavirus RNAs (12). In addition to effects on coding potential, A-to-I editing can destabilize dsRNA structures because product I:U base pairs generated by editing are less stable than substrate A·U base pairs (17–19). The ADAR1 protein is organized in three functional regions: (i) either one or two Z-DNA-binding domains are present at the N-terminal region; (ii) three dsRNA-binding domains similar to those found in PKR are present in the central region; and (iii) the C-terminal region includes the deaminase catalytic domain (9, 20, 21). ADAR1 is encoded by a single gene, but two different-sized isoforms are produced through the use of alternate promoters and alternative splicing (9, 22). A short form of ADAR1, known as p110, is constitutively expressed and predominantly, if not exclusively, localized in the nucleus; a long, IFN-inducible form, known as p150, is found in both the cytoplasm and the nucleus (9, 16, 22, 23). The human p150 protein possesses two Z-DNA-binding domains, Zα and Zβ (20), and a nuclear export signal (23) within an N-terminal 295-amino-acid extension compared to the constitutive p110 protein that initiates at methionine 296 and hence lacks Zα. Both p110 and p150 are active deaminases (9, 12, 17, 22, 24).
PKR and ADAR1 play opposing functional roles in the host innate immune response to MV infection (10–12, 25). PKR is antiviral and proapoptotic; PKR activation correlates with enhanced IFN induction (6), increased mitogen-activated protein (MAP) kinase pathway activation (6, 26), reduction in virus growth (27), and increased cytotoxicity and apoptosis of infected cells (27). ADAR1, conversely, generally acts in a proviral and antiapoptotic manner, at least in cell culture; ADAR1 deficiency leads to enhanced PKR activation (28, 29), increased IFN induction (30, 31), and enhanced IRF-3 phosphorylation and dimerization (31), whereas ADAR1 sufficiency or ectopic overexpression leads to increased virus growth and suppression of apoptosis (29, 31, 32).
The host responses to virus infection include one that involves translational arrest resulting from phosphorylation and inactivation of eukaryotic initiation factor 2α (eIF2α) by cellular kinases, most commonly PKR (15, 33). Formation of stress granules (SGs) is a mark of the response to infection that results from eIF2α inactivation (34) that can occur during virus-mediated PKR activation (35). SGs, whose formation is triggered when polysomes are disassembled, represent aggregates of translationally silent mRNA, stalled 48S initiation complexes, and multiple RNA-binding proteins (RBPs) including the SG markers T-cell restricted intracellular antigen-1 (TIA-1) and Ras GAP SH3-domain-binding protein 1 (G3BP1) as well as other RBPs (35, 36). Among the roles played by SGs is the storage of translationally inactive mRNAs under conditions of physiologic stress, RNAs that subsequently are either expressed when the stress is eliminated and translation is restored or alternatively triaged for degradation by processing bodies if the cells do not survive but undergo apoptosis (35–37). Several viruses including both positive-stranded and negative-stranded ssRNA viruses and dsDNA viruses are capable of inducing SG formation (38–42). In some instances, infection with wild-type (WT) viruses causes suppression of SG formation, whereas mutants can become efficient inducers of SG formation. For example, influenza virus A/Puerto Rico/8/34 suppresses SG formation; however, when the gene for the nonstructural protein NS1, an inhibitor of PKR, is deleted, the mutant virus is capable of SG induction (43). Similarly, WT vaccinia virus is a poor inducer of formation of SG-like structures termed “antiviral granules” compared to a mutant with deletion of the E3L protein (42); E3L, like NS1, is a PKR antagonist (5, 12). Another example is West Nile virus (WNV), in which SG formation is lineage dependent; lineage 1 viruses inhibited SGs, but lineage 2/1 chimeric viruses induced SGs (44, 45).
Whether or not SGs are induced during MV infection is not known. We therefore wished to test whether MV induced the formation of SG and whether deletion of either the V or C accessory protein affected the SG response. We furthermore examined whether PKR affected the SG response as anticipated from prior work with other viruses and more importantly the effect of ADAR1 on the SG response. These studies were possible via combined genetic and biochemical approaches because of the availability of isogenic virus mutants deleted for either V or C protein expression and because of the availability of human cell clones stably deficient for either PKR or ADAR1 protein expression. Our results revealed that C mutant MV was a robust inducer of SG formation compared to either WT or V mutant MV, both of which were poor inducers; that SG formation was PKR dependent; and surprisingly, that ADAR1, while associated with SG, led to an impairment of SG formation. To gain insight into the functional requirements of ADAR1 for suppression of SG formation as well as ADAR1 roles in IFN-β induction, PKR activation, and enhancement of MV growth, a complementation analysis of ADAR1kd cells with WT and mutant forms of ADAR1 was carried out.
MATERIALS AND METHODS
Cells and viruses.
Parental HeLa cells and Vero cells were cultured in Dulbecco modified Eagle's medium (DMEM) supplemented with 5% (vol/vol) fetal bovine serum (HyClone), penicillin (100 μg/ml), and streptomycin (100 units/ml) (Gibco/Invitrogen) as previously described (46). HeLa cells made stably deficient in either ADAR1 (ADAR1kd) (29, 47) or PKR (PKRkd) (46, 48) by a short hairpin RNA interference silencing strategy and the drug-resistant pSUPER.retro.puro vector control cells (CONkd) were as previously described. Knockdown cells were cultured in DMEM maintenance media with puromycin (1 μg/ml) (Sigma). The recombinant parental Moraten measles virus vaccine strain, here designated WT, and isogenic V-deficient (Vko) and C-deficient (Cko) mutants were as described previously (27, 49); they were generously provided by Roberto Cattaneo (Mayo Clinic, Rochester, MN). The recombinant MV includes the gene encoding green fluorescent protein (GFP) inserted downstream of the viral H gene. Infections were carried out at a 50% tissue culture dose (TICD50) per cell multiplicity of infection (MOI) of 1 or 3 TICD50/cell as indicated (6, 27). Where indicated, arsenite (Sigma-Aldrich) treatment was at 0.5 mM for 1 h.
Western immunoblot analysis.
Cells were harvested 24 h after infection, and whole-cell extracts prepared as described previously (27). The Bradford assay method (Bio-Rad) was used to determine protein concentrations. Electrophoretic fractionation of extract protein (20 to 30 μg) was on either 8% or 10% sodium dodecyl sulfate-polyacrylamide gels. Proteins were transferred to nitrocellulose membranes and the membranes blocked with either 5% milk (wt/vol) in phosphate-buffered saline or 5% bovine serum albumin (BSA) (wt/vol) and 0.1% Tween 20 (vol/vol) in Tris-buffered saline for detection of phosphoproteins. Antibodies for the detection of proteins were rabbit polyclonal antibody against ADAR1 (9), MV H protein (27), PKR and GFP (Santa Cruz Biotechnology), goat polyclonal antibody against TIA-1 (Santa Cruz Biotechnology), mouse monoclonal antibody against G3BP1 and α-tubulin (Sigma), and rabbit monoclonal antibody against phospho-Thr446 PKR (Epitomics). An Odyssey infrared imaging system (Li-COR) was used for detection in immunoblots (6).
Immunofluorescence.
Cells were grown on 18-mm glass coverslips in 12-well plates and infected at an MOI of 1 TICD50/cell for 24 h. Fixation was with 10% (vol/vol) neutral buffered formalin (Sigma) at room temperature for 15 min. Blocking and permeabilization were done with Tris-buffered saline (50 mM Tris [pH 7.5], 150 mM NaCl) containing 2% (wt/vol) normal donkey serum (Jackson ImmunoResearch) and 0.2% Triton X-100 (CBP buffer) for 30 min at 37°C. Permeabilized cells then were incubated with primary antibody overnight at 4°C using the following antibody dilutions in CBP buffer: mouse monoclonal anti-G3BP1 antibody, 1:500 (Sigma); goat polyclonal anti-TIA-1 antibody, 1:50 (Santa Cruz Biotechnology); rabbit monoclonal anti-phospho-Thr446 PKR antibody, 1:100 (Epitomics); rabbit polyclonal anti-ADAR1 antibody, 1:300 (9), rabbit anti-MV N antibody, 1:500 (27); and chicken IgY anti-GFP antibody, 1:500 (Molecular Probes). GFP is encoded by the recombinant Moraten vaccine strain of virus used in these experiments. However, because the GFP signal was weakened following preparation of samples for immunofluorescence microscopy, antibody against GFP was used to enhance and stabilize the signal. Incubation of coverslips with donkey secondary antibodies diluted in CBP was at 37°C for 1 h at a 1:200 dilution with the following: Alexa Fluor 350 anti-rabbit or anti-mouse IgG (Molecular Probes); AffiniPure fluorescein isothiocyanate (FITC)-conjugated anti-chicken IgG (Jackson ImmunoResearch); DyLight 594 anti-rabbit IgG (Jackson ImmunoResearch); and Texas Red-conjugated anti-goat IgG (Jackson ImmunoResearch). Images were obtained using an Olympus IX71 microscope with Q-Capture PRO software (QImaging) or an Olympus IX81 microscope equipped with a Disc Spinning Unit (Olympus) and a 60× oil immersion objective using IPLab software (BD Biosciences). Individual channels were linearly adjusted and overlaid in Adobe Photoshop in RGB for creation of final merged images.
Stress granule quantitation.
To determine the number of SG-positive cells, a minimum of four wide-field 40× images were captured per experiment, and a total of ∼150 to 200 cells per experiment were counted. Cells displaying punctate immunofluorescent foci of the G3BP1 SG marker protein were recorded as SG positive. Percentages were determined as the number of SG-positive cells divided by the total number of cells, multiplied by 102.
Transient transfections and virus titrations.
For complementation of ADAR1kd cells, the indicated ADAR1 expression plasmids were transfected at 2 μg per well of a 6-well plate using Fugene HD reagent (Promega) according to the manufacturer's instructions and as described previously (26). At 24 h after transfection, cells were either infected with MV or left uninfected (UI) as indicated. MV yields were determined at 24 h after infection. The titers of virus were determined by a modification of the protocol as described previously (50). Briefly, cells were subjected to 3 cycles of freeze-thaw; dilutions of virus were then used to infect Vero cells seeded in 96-well plates. Titers were determined by counting GFP fluorescent foci at 48 h postinfection. Individually infected cells and syncytia were considered foci; titers are expressed as focus-forming units per ml.
Plasmid constructions.
WT and mutant ADAR1 proteins were expressed using the pcDNA6/V5HisA expression vector (Invitrogen). Constructs were prepared using the cDNA clone previously described (9) and generously provided by A. M. Toth, of this laboratory. Briefly, the construction of the plasmids was as follows. The construct expressing the p110 size isoform corresponding to the constitutively expressed form of ADAR1 starting from methionine 296 (M296) was prepared by subcloning the 2,946-bp BamHI-XhoI fragment from pcDNA3.1/HisC-M296-a, which was previously described (51). The resulting expression construct, pcDNA6/V5HisA-M296-a (here referred to as ADAR1 p110 WT), encodes the physiologically relevant “a” splice variant form of the p110 protein (52). The construct for the p150 size isoform of ADAR1 that is IFN inducible and initiates from methionine 1 (M1) was prepared by using the 3,678-bp BamHI-XhoI fragment from the pcDNA3.1/HisC-FL-b (53) plasmid, obtained by digestion with XhoI followed by partial digestion with BamHI. The resulting expression construct, pcDNA6/V5HisA-FL-b (here referred to as ADAR1 p150 WT), expresses the physiologically relevant “b” splice variant form of the p150 full-length protein (52) and also contains an M296L mutation to prevent expression of the p110 protein by leaky scanning. Catalytic and Zα-DNA-binding mutants of the p150 isoform are referred to here as p150 cat (H910Q, E912A) and p150 Zα (Y177A), respectively. For the catalytic mutant, the point mutations H910Q and E912A of the highly conserved C-terminal deaminase catalytic CHAE motif were as described previously (53). The p150 Zα (Y177A) mutant was based on the essential tyrosine 177 residue required for Z-DNA-binding activity identified by Schade et al. (54). The Y177A mutant was generated using the QuikChange II site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol using the following primer pairs (with the mutated bases underlined): (+), 5′-ACTCCGAAGAAAGAAATCAATCGAGTTTTAGCCTCCCTGGCAAAGAAG-3′, and (−), 5′-CTTCTTTGCCAGGGAGGCTAAAACTCGATTGATTTCTTTCTTCGGAGT-3′. Mutations were confirmed by sequence analysis.
qPCR.
IFN-β transcripts were measured by quantitative real-time PCR (qPCR) as previously described (6). Briefly, 6-well cultures were infected with WT MV or left uninfected. Total RNA was isolated using TRIzol (Invitrogen) at 24 h after infection and from parallel uninfected control cells. Random-primed cDNA was prepared using ∼2 μg of RNA and SuperScript II (Invitrogen) according to the manufacturer's protocol. For the qPCR analyses, the primer pairs and cycle programs for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and IFN-β were as previously described (31). qPCRs were performed with reverse transcription templates by using IQ SYBR green Supermix (Bio-Rad) and a Bio-Rad MyIQ real-time qPCR instrument. The IFN-β values were normalized to GAPDH values.
RESULTS
Measles virus deficient in C-protein expression efficiently induces stress granule formation, but wild-type virus does not.
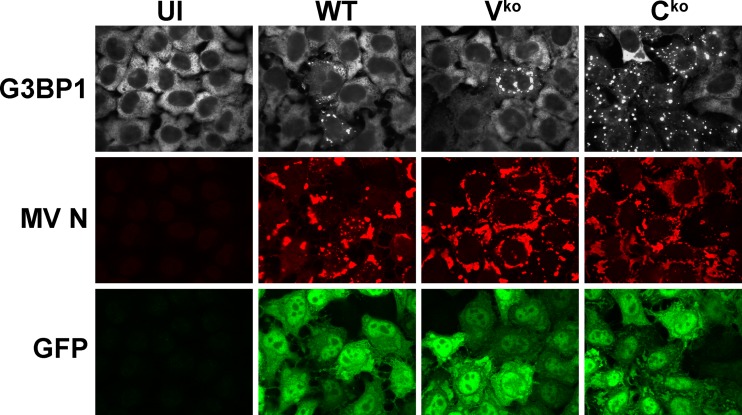
To test whether measles virus (MV) infection leads to stress granule (SG) formation as a mark of virus infection, we used the Moraten vaccine strain of virus because parental (WT) and isogenic mutants lacking the expression of either the V (Vko) or C (Cko) protein are available and entry of the Moraten strain is via the CD46 receptor present on HeLa cells (27, 55). G3BP1 is a well-established SG associated protein and serves as a marker of SG formation (28, 34, 56). To determine the expression pattern of G3BP1, HeLa parental cells were left uninfected or were infected with either WT, Vko, or Cko virus (Fig. 1). Immunofluorescence microscopy revealed punctate cytoplasmic G3BP1 foci that formed most efficiently in the Cko mutant-infected cells compared to cells infected with either WT or the Vko mutant virus (Fig. 1). In uninfected cells, the G3BP1 signal (white) was homogeneously dispersed throughout the cytoplasm and did not appear punctate. GFP reporter (green), engineered into the MV genome downstream of the H gene, and the MV N protein (red) were used to monitor MV infection (Fig. 1). These results establish that MV induces SG formation and suggest that the MV C protein plays a role during infection that results in downregulation of responses leading to SG formation.
Fig 1.
Measles virus deficient for C-protein expression is an efficient inducer of stress granule formation, but wild-type virus is not. Cells were either left uninfected (UI) or infected with WT, Vko, or Cko MV at a multiplicity of infection of 3 TICD50/cell. At 24 h after infection, cells were analyzed by immunofluorescence microscopy as described in Materials and Methods using antibodies to G3BP1 (white) as a marker for SG formation and N (red) and GFP (green) for detection of MV infection. Individual channels are shown.
Stress granule formation by measles virus is enhanced by PKR and impaired by ADAR1.
The dsRNA-dependent protein kinase PKR and the dsRNA-selective adenosine deaminase ADAR1 are established modulators of the host innate response to infection (12, 25, 30, 31), including during MV infection (6, 27, 29, 31, 31). Clonal HeLa cell lines made stably deficient in either PKR or ADAR1 (PKRkd and ADAR1kd, respectively) were used in loss of function analyses to assess the roles of PKR and ADAR1 in SG formation. The PKRkd cells express less than 5% of PKR compared to CONkd drug control or ADAR1kd cells, and the ADAR1kd cells express less than 15% of the p110 and p150 forms of ADAR1 compared to the CONkd or PKRkd cells (27, 29, 31, 48).
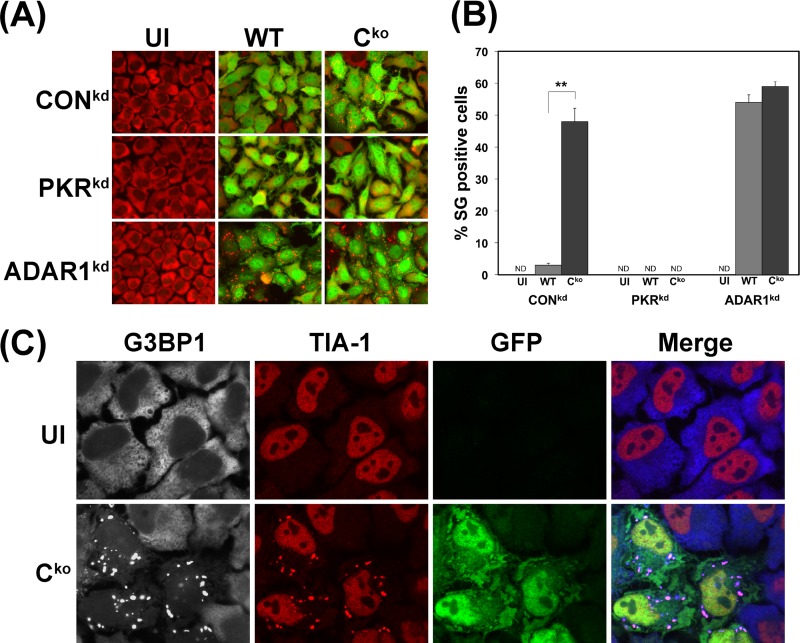
SG formation measured by the punctate G3BP1 staining pattern was more efficient in Cko mutant-infected than WT virus-infected CONkd HeLa cells (Fig. 2A), similar to the results obtained with parental HeLa cells (Fig. 1). Both the parental and the CONkd HeLa cells are PKR sufficient and ADAR1 sufficient. In contrast, SG formation was not detectable in the PKRkd cells infected with either Cko or WT virus or in uninfected PKRkd or CONkd cells, as shown in Fig. 2A. These results establish a dependency on PKR for formation of SG in MV-infected cells.
Fig 2.
Stress granule formation is impaired by PKR deficiency and amplified by ADAR1 deficiency following measles virus infection. (A) PKRkd, ADAR1kd, and CONkd cells were either left uninfected (UI) or infected with WT or Cko mutant MV as indicated. At 24 h after infection, cells were analyzed by immunofluorescence microscopy as described in Materials and Methods using antibodies to G3BP1 (red) as a marker for detection of SG formation and GFP (green) for detection of MV infection. Images shown are merged. (B) Quantitation of SG-positive cells. Wide-field 40× images were obtained, and a minimum of 150 cells were counted per experiment. Results shown are means and standard errors of independent experiments. **, P < 0.0005 by Student's t test comparing WT-infected to Cko-infected cells. (C) CONkd cells were infected with Cko MV or left uninfected (UI). At 24 h after infection, cells were analyzed by confocal microscopy using antibodies against G3BP1 (white), TIA-1 (red), and GFP (green). Individual channels and merged images are shown; G3BP1 appears blue in merged images.
While SG formation was not readily detectable in CONkd cells infected with WT virus, surprisingly, WT virus became a potent inducer of SG formation in ADAR1kd cells (Fig. 2A). Quantitation revealed that less than 3 to 5% of the CONkd cells were SG positive following WT virus infection, but ∼50% of the CONkd cells were SG positive following infection with the Cko mutant (Fig. 2B). In contrast, a comparably high percentage (∼50 to 60%) of the ADAR1-deficient (ADAR1kd) cells were SG positive at 24 h after infection with either WT or Cko virus (Fig. 2B). SGs were undetectable in uninfected CONkd, PKRkd, and ADAR1kd cells, or in infected PKRkd cells (Fig. 2B). Results obtained with G3BP1 were confirmed with TIA-1 as a second marker of the SG. The TIA-1 protein in uninfected cells is largely nuclear, in contrast to the cytoplasmic G3BP1 protein; however, TIA-1 translocated to the cytoplasm and colocalized with the G3BP1 foci as shown by Cko infection of CONkd cells (Fig. 2C). These results suggest that ADAR1 acts to impair SG formation following MV infection; the findings also further suggest that the viral C protein plays a role in modulating the efficiency of SG formation dependent upon the level of ADAR1 protein in PKR-sufficient cells.
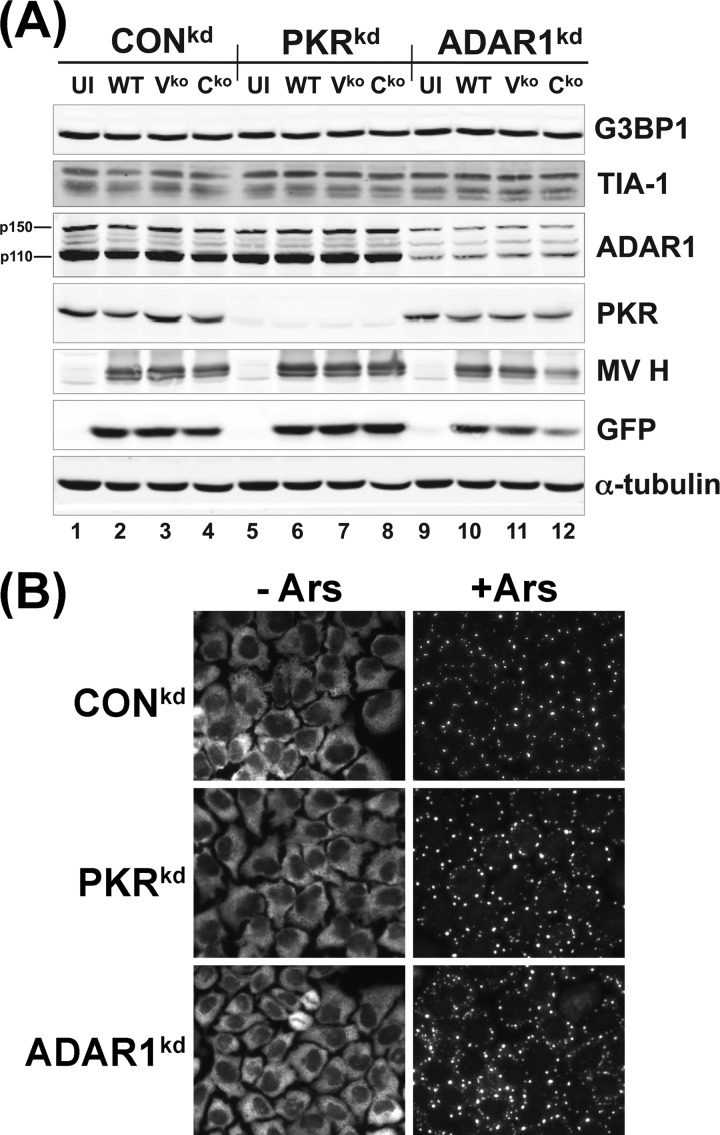
While the localization of G3BP1 and TIA-1 to foci characteristic of SG was induced by MV infection (Fig. 1 and 2), the steady-state expression levels of G3BP1 and TIA-1 were not detectably altered following infection. The three cell types, CONkd, PKRkd, and ADARkd, expressed similar levels of both G3BP1 and TIA-1 as assayed by Western immunoblot analysis (Fig. 3A). Infection with MV, either WT, Vko, or Cko virus, did not affect the steady-state level of either G3BP1 or TIA-1. In contrast to the enhanced induction of SG in ADAR1-deficient ADAR1kd cells and virtual absence of SGs in PKR-deficient PKRkd cells following MV infection (Fig. 2), arsenite treatment comparably induced the formation of SGs in uninfected CONkd, PKRkd, and ADAR1kd cells (Fig. 3B).
Fig 3.
Stress granule marker proteins G3BP1 and TIA-1 are comparably expressed in uninfected and measles virus-infected CONkd, PKRkd, and ADAR1kd cells. (A) Cells were either left uninfected (UI) or infected with WT, Vko, or Cko MV at a multiplicity of infection of 3 TICD50/cell. At 24 h after infection, whole-cell extracts were prepared and analyzed by Western immunoblot assay with antibodies against ADAR1, PKR, G3BP1, TIA-1, MV H, GFP, and α-tubulin as described in Materials and Methods. (B) Uninfected cells treated with arsenite (+Ars) or not (−Ars) as described in Materials and Methods were analyzed by immunofluorescence microscopy using antibody against G3BP1.
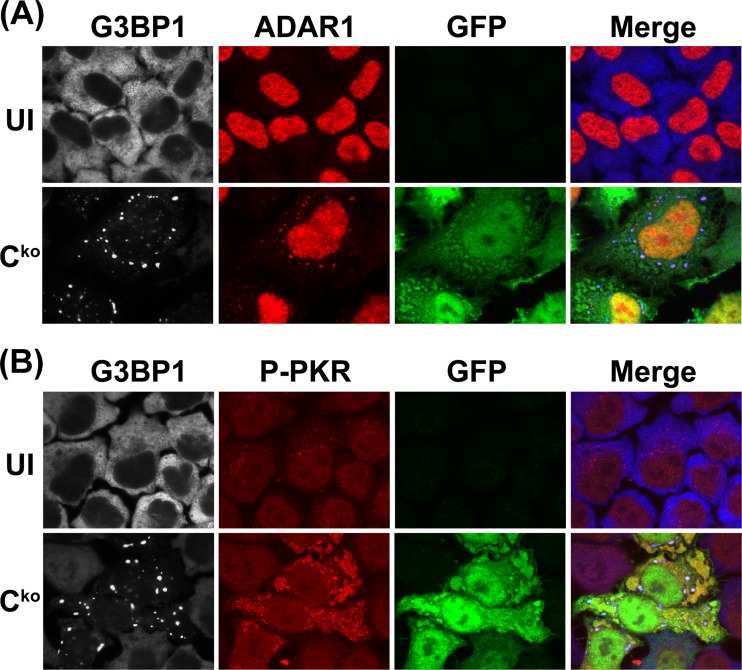
ADAR1 and phospho-PKR colocalize with MV-induced stress granules.
Multiple RNA-binding proteins (RBPs) colocalize with SG markers (34, 57–59). Among these, ADAR1, when either ectopically expressed as p150 or endogenously expressed, is described as a component of HeLa cell SG following arsenite treatment or transfection with polyinosinic-poly(C) (pIpC) (60). We wished to test whether a physiologically natural stressor, virus infection, also would cause colocalization of ADAR1 with the induced SG. This possibility was examined with CONkd cells infected with Cko virus, because this cell-virus combination possesses endogenous ADAR1 and SGs are abundantly formed following infection (Fig. 2). At 24 h after infection, cells were analyzed by confocal microscopy using antibodies to G3BP1 (white; blue in merged images), ADAR1 (red), and GFP (green). Individual channels and merged images are shown in Fig. 4. The ADAR1 signal detected in uninfected cells not treated with IFN was largely nuclear, as anticipated (9), because the ADAR1 p110 isoform is constitutively expressed and is known to display predominantly, if not exclusively, nuclear localization (12). Upon infection with Cko virus, the ADAR1 signal became punctate in the cytoplasm and colocalized with the G3BP1 marker of SG (Fig. 4A). This colocalization of ADAR1 and G3PB1 was observed in <10% of cells (data not shown). Our finding that ADAR1 localizes to the MV-induced SG (Fig. 4A) is in agreement with the recent reports using either the nonphysiologic pIpC inducer of stress (60) or hepatitis C virus infection (41). However, the presence of ADAR1 protein with SG does not appear to be an obligatory requirement for maintenance of SGs but, rather unexpectedly, the converse; the presence of SG structures was substantially increased in ADAR1-deficient cells following MV infection compared to ADAR1-sufficient cells (Fig. 2).
Fig 4.
ADAR1 and PKR colocalize with stress granules induced by infection with measles virus. CONkd cells were infected with Cko mutant MV and analyzed at 24 h postinfection by confocal microscopy using antibodies against G3BP1 (white), GFP (green), and either ADAR1 (red) (A) or phospho-Thr446-PKR (P-PKR) (red) (B). Individual channels and merged images are shown; G3BP1 appears blue in merged images.
We also examined whether the dsRNA-dependent kinase PKR, following activation by phosphorylation, colocalized with MV-induced SG. At 24 h after Cko infection of CONkd cells, confocal microscopy was carried out using antibodies for G3BP1 (white; blue in merged images), phospho-Thr446-PKR (red), and GFP (green). The phospho-PKR signal was weak in uninfected cells, but following MV infection the signal was greatly increased within the cytoplasm, characteristic of activation of PKR (Fig. 4B). Furthermore, the staining pattern of phospho-PKR appeared punctate in part, and the punctate signal colocalized with the G3BP1 marker of SG.
Deaminase catalytic activity of the p150 ADAR1 protein is required for suppression of MV-induced host responses, including stress granule formation.
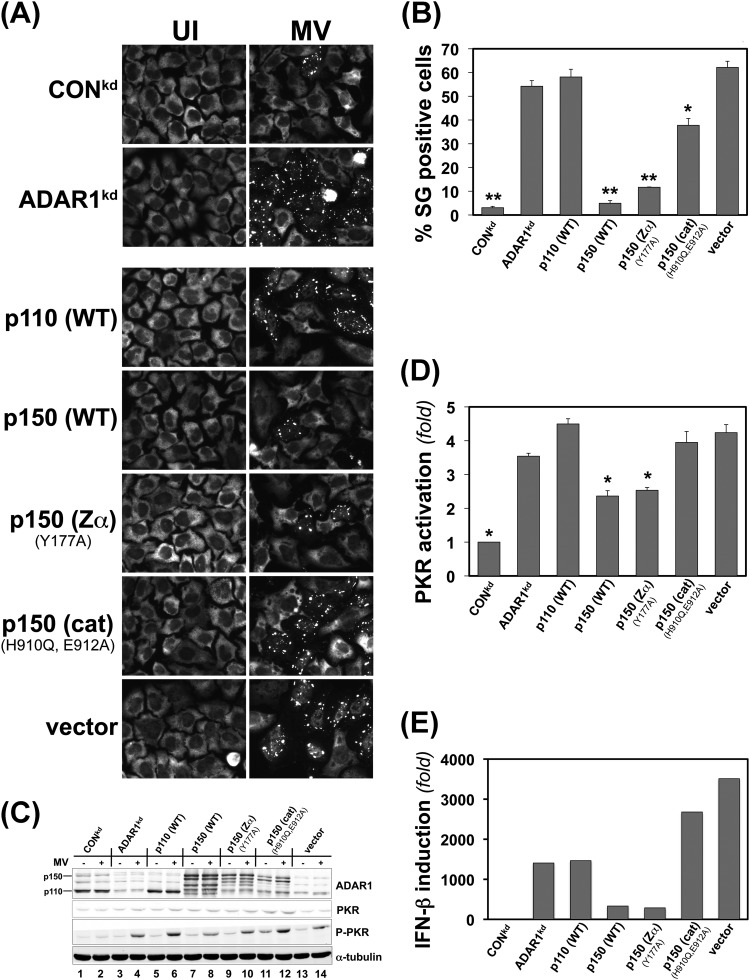
We earlier found that ADAR1 acts as a suppressor of PKR activation during MV infection (29, 31). In ADAR1-deficient cells, PKR activation is enhanced compared to ADAR1-sufficient cells. As was shown in Fig. 2, ADAR1-deficient cells displayed enhanced SG formation after infection with WT MV. To probe the mechanistic basis of the suppression by ADAR1 of SG formation (Fig. 2) and also PKR activation and IFN-β induction (29, 31), we used a complementation strategy. We tested the functional effects of ectopic expression of different forms of ADAR1 that included the WT p110 and WT p150 ADAR1 proteins and mutant p150 proteins defective in either the deaminase catalytic or the Z-DNA-binding activities. SG formation and PKR activation as well as IFN-β transcript production were examined in complemented ADAR1kd cells, both uninfected and MV infected (Fig. 5).
Fig 5.
ADAR1 deaminase activity of the p150 protein isoform is required for suppression of stress granule formation as well as suppression of PKR activation and IFN-β induction following measles virus infection. ADAR1kd cells deficient in p150 and p110 were complemented by transfection with ADAR1 constructs or the vector plasmid as indicated, or ADAR1kd and CONkd cells were also left untransfected as controls. At 24 h after transfection, cells were either infected with WT measles virus (MV) or left uninfected (UI). Infection was with a multiplicity of infection of 1 TICD50/cell for microscopy experiments or 3 TICD50/cell for Western immunoblot assays, IFN-β mRNA quantitation, and yield determinations. ADAR1kd cells complemented with ADAR1 proteins are designated p110 (WT) and p150 (WT) for wild-type protein isoforms and p150 Zα (Y177A) and p150 (cat) (H910Q, E912A) for p150 proteins with mutated domains. Vector, empty vector. (A) At 24 h after infection, cells were analyzed by immunofluorescence microscopy using G3BP1 antibody as a marker for SG formation. (B) Quantitation of SG-positive cells. Wide-field 40× images were obtained, and a minimum of 150 cells were analyzed per experiment for the presence of stress granules as shown in panel A. *, P < 0.001; **, P ≤ 3 × 10−5 by Student's t test comparing uninfected ADAR1kd cells to infected cells. The results shown are means and standard errors from three independent experiments. (C) Western immunoblot analysis of ectopic ADAR1 expression. At 24 h postinfection, whole-cell extracts were prepared and analyzed using antibodies against ADAR1, PKR, phospho-Thr446-PKR, and α-tubulin as indicated. (D) Quantitation of PKR activation (n-fold) as measured by the level of phospho-Thr446-PKR to total PKR protein determined by Western immunoblot analysis as shown in panel C. *, P < 0.05 by Student's t test to compare the level of P-PKR in infected ADAR1kd cells to infected cells complemented as indicated or to CONkd cells. The results shown are means and standard errors from three independent experiments. (E) IFN-β mRNA levels. At 24 h after infection, total RNA was isolated and IFN-β transcript levels were determined by qPCR and normalized to GAPDH. Results shown are representative of 4 independent experiments.
Uninfected (UI) ADAR1kd cells, either complemented or not by ectopic expression, did not show detectable SGs measured by G3BP1 staining (Fig. 5A, left). However, MV-infected ADAR1kd cells (Fig. 5A, right) showed robust SG formation either when they had not been transfected (ADAR1kd) or when they had been complemented by expression of the p110 (WT) ADAR1 protein or transfected with the empty vector as a control. However, cells complemented with either the p150 (WT) protein or the p150 Z-DNA-binding defective mutant Y177A showed low levels of SG formation following infection (Fig. 5A, B). In contrast, the p150 catalytic mutant H910Q, E912A showed high levels of SG formation (Fig. 5A, B). These results suggest that the p150 size isoform of ADAR1 and, furthermore, catalytic activity of the p150 protein are required to suppress SG formation following infection with WT MV.
Because ADAR1 is known to function as a suppressor of PKR activation (12, 25, 29), PKR phosphorylation at Thr446 as a measure of PKR activation was determined in ADAR1-complemented ADAR1kd cells (Fig. 5C). Only the p150 (WT) and p150 Zα mutant constructs were able to significantly suppress PKR activation (Fig. 5C and D). Because ADAR1 also is implicated as a suppressor of IFN induction (30, 31), induction levels of IFN-β transcripts were determined. As shown by the representative data set in Fig. 5E, ADAR1kd cells expressing either p150 (WT) or the p150 Zα mutant suppressed IFN-β RNA expression to levels approaching that seen with CONkd cells (Fig. 5E). In contrast, complementation of ADAR1kd cells with either p110 (WT) or the p150 catalytic mutant did not decrease the level of either phospho-PKR (Fig. 5D) or IFN-β RNA (Fig. 5E) from that seen in ADAR1kd cells either not complemented or transfected with empty vector. These results suggest that deaminase catalytic activity of the ADAR1 p150 protein is required for suppression of both PKR activation and IFN-β induction. Because PKR is the eIF2α kinase activated following MV infection, ADAR1 suppression of PKR activation emerges as a likely mechanism by which ADAR1 deficiency contributes to enhanced SG formation, in part because of enhanced PKR activation, just as we recently established for induction of IFN-β (31).
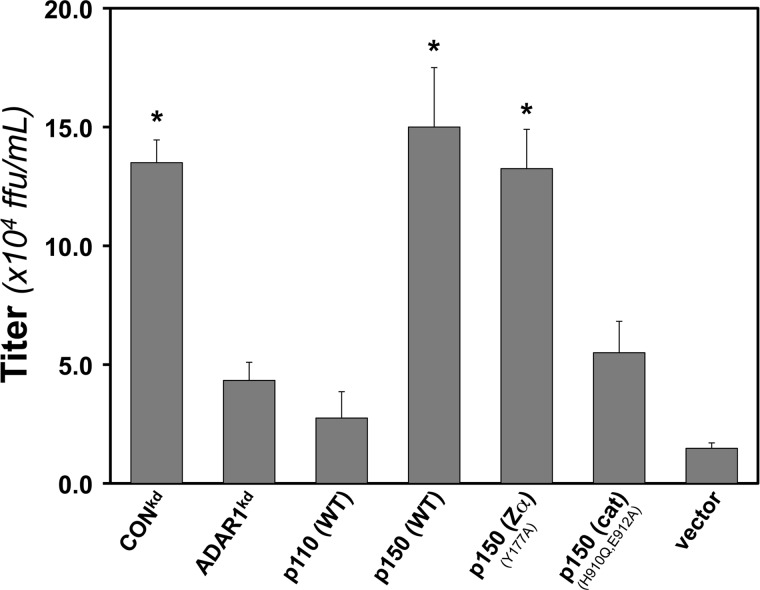
Wild-type p150 ADAR1 but not the catalytic mutant enhances MV growth.
ADAR1 displays a proviral effect with a range of RNA viruses (12, 25). The mechanism remains unclear. To gain insight into the functional domains of ADAR1 required and to determine whether the enhancement of MV growth by ADAR1 correlates with the suppression of SG formation, we again employed the complementation strategy. The results are shown in Fig. 6. Enhancement of WT MV growth in ADAR1kd cells was observed only in the cells complemented to express either the p150 (WT) or the p150 Zα mutant (Fig. 6). Virus yields in the ADAR1kd cells expressing either the p110 (WT) or the p150 catalytic mutant were low and comparable to those in ADAR1kd cells that either had not been complemented or had been transfected with empty vector (Fig. 6).
Fig 6.
ADAR1 p150 wild-type but not catalytic mutant protein enhances measles virus growth. ADAR1kd cells were complemented as described in the legend for Fig. 5. The yield of WT virus was measured at 24 h postinfection as described in Materials and Methods; titers are expressed as fluorescent focus units per ml (ffu/ml) on Vero cells. *, P < 0.02 by Student's t test. Results shown are means and standard errors from four independent experiments.
DISCUSSION
Our studies were undertaken with the aim of testing the roles of PKR and ADAR1, two dsRNA-binding proteins, in virus-induced stress granule formation following MV infection and of determining whether the viral C and V accessory proteins affect the SG response. By using cells stably deficient either in PKR (PKRkd) or ADAR1 (ADAR1kd), we established that SG formation was induced by MV infection in a PKR-dependent manner. But conversely, virus-induced SG formation was impaired by ADAR1. Furthermore, MV deficient in C protein expression was a more efficient inducer of SG formation than either WT or the isogenic V-deficient mutant. Complementation analyses provided mechanistic insight. Ectopic expression of ADAR1 in cells deficient in both p110 and p150 ADAR1 proteins established that the deaminase catalytic activity of the ADAR1 p150 size isoform was necessary for suppression of SG formation. p150 ADAR1 catalytic activity was also necessary for suppression of PKR activation and IFN-β induction. Several important points emerge from these findings, which extend prior observations.
First, we found that following infection with either WT or Vko virus, SGs were not efficiently induced in PKR-sufficient, ADAR1-sufficient HeLa cells, either parental cells or CONkd control cells (<5% SG positive). However, induction of SG in these cells after infection with Cko virus was striking; ∼50 to 60% of the control cells became SG positive following infection with the mutant virus. The finding that SG-positive PKRkd cells were virtually undetectable following infection with measles virus is consistent with recent observations for West Nile virus, hepatitis C virus, influenza virus, and respiratory syncytial virus (39, 41, 43, 44), whereby SG formation induced by these viruses also was PKR dependent. Because SG formation is dependent on eIF2α phosphorylation (34, 58), these findings further suggest that PKR is the predominant eIF2α kinase responsible for SG formation by these viruses rather than PERK, activated by endoplasmic reticulum stress, HRI, activated by oxidative stress and heavy metals, or GCN2, activated by nutritional stress, all of which also are eIF2α kinases (33, 61).
In contrast to the PKR dependency for SG formation with MV, we observed in cells deficient for ADAR1 that SG formation was far more efficient than in ADAR1-sufficient cells infected with WT virus. Furthermore, the level of SG formation seen with WT and Cko virus in the ADAR1kd cells was comparable to the same high level seen in CONkd cells infected with Cko virus. The impairment of SG formation by ADAR1 in WT virus-infected CONkd cells was striking. These results suggest that ADAR1 was capable of suppression of SG formation induced by WT virus infection but not with Cko infection. What then is the mechanistic basis of these novel effects? Does ADAR1 act as an enzyme, or as an RNA-binding protein similar to G3BP1, TIA-1, or Staufen (62, 63), or possibly as both to mediate the suppression of SG formation?
To begin to address the question of the functional activity and size isoform of ADAR1 required to mediate the suppressing effects, we employed a complementation strategy by expressing WT and mutant forms of ADAR1 in the ADAR1kd cells.
The results established that deaminase catalytic activity of the p150 size isoform was required; the p110 size isoform of ADAR1 was unable to suppress either SG formation or activation of PKR following MV infection. The p150 size isoform is IFN inducible and is the only known ADAR protein found in the cytoplasm, whereas the p110 size isoform is constitutively expressed and predominantly if not exclusively nuclear (9, 64). Both p150 and p110 are efficient dsRNA-binding proteins that possess three copies of the prototypical dsRNA-binding domain first identified in PKR (9, 12, 53). Because p110 is unable to rescue the ADAR1kd phenotype, the RNA-binding activity of ADAR1 alone appears insufficient for rescue because the three RNA-binding domain copies are identical between p110 and p150. In addition to the p150 WT protein, the p150 Zα mutant also rescued the SG phenotype. The Zα domain of ADAR1 p150 was mutated for two reasons: first, Zα is unique to p150 and is not present in the p110 protein (65, 66); and second, another Z-DNA-binding protein, ZBP1, which has two Zα domains, is reported to localize to SGs upon heat shock and arsenite treatment (67). Our results indicate that ADAR1 suppression of SG formation requires A-to-I editing activity but not Z-DNA-binding activity of Zα.
While MV infection induced SG formation as measured by the appearance of punctate cytosolic staining patterns of two beacons of SG, the G3BP1 and TIA-1 RNA-binding proteins, virus infection did not alter the steady-state level of either G3BP1 or TIA-1, only the aggregation of them in the cytoplasm. The levels of TIA-1 and G3BP1 were comparable in the three engineered cell lines, PKRkd, ADAR1kd, and CONkd cells, both uninfected and infected for 24 h with WT virus or Vko or Cko mutant virus. MV-induced SGs characterized by the TIA-1 and G3BP1 markers also contained ADAR1 and phospho-PKR. However, the SGs did not contain MV proteins (N or P) to a detectable level (Fig. 1 and data not shown). Our finding that ADAR1 was associated with SG following MV infection is in agreement with recent reports that likewise showed endogenous ADAR1 associated with SG, either following infection with HCV as the stress inducer (41) or ectopically expressed ADAR1 p150 following treatment with arsenite or transfection with synthetic pIpC dsRNA as the stress inducer (60). Curiously, the cellular protein Staufen displays some characteristics similar to those of ADAR1. Staufen is a dsRNA-binding protein (62) and is SG associated (68–70). Overexpression of Staufen leads to decreased SG formation, while transient knockdown of Staufen leads to enhanced SG formation (70). Likewise, overexpression of p150 (WT) ADAR1 led to decreased SG formation, whereas stable knockdown of ADAR1 enhanced SG formation following infection. These results suggest that both ADAR1 and Staufen, while not essential components of induced SGs, yet modulate the efficiency of SG formation.
Enhanced PKR activation and induction of IFN-β also are observed in ADAR1kd cells infected with MV (29, 31). Enhanced levels of PKR activation likewise are observed in vesicular stomatitis virus-infected ADAR1-deficient cells compared to ADAR1-sufficient cells in culture (28, 47), and genetic disruption of ADAR1 in mice results in upregulation of IFN expression in hematopoietic stem cells (30). The robustness of SG formation that we observed here correlated with the extent of PKR activation and also IFN-β induction. While it is tempting to speculate that formation of SG may play a role in antiviral innate signaling triggered by MV, including IFN-β induction, further analyses are necessary to establish more definitively or eliminate this possibility. ADAR1-suppressed virus-induced host responses seen with WT but not Cko mutant virus include, in addition to suppression of PKR activation (29, 31) and IFN-β induction (29, 31), also the suppression of SG formation as shown here. ADAR is known to destabilize dsRNA structures (17), and dsRNA is a potent activator of PKR and inducer of SG formation (8, 35, 58, 71). The requirement for ADAR1 catalytic activity for rescue of the ADAR1kd phenotype is consistent with a role of ADAR1 as a dsRNA destablizer, as I:U mismatch base pairs are less stable than A·U pairs (17).
The difference between the capability of WT and that of the Cko mutant to induce SG formation was striking. Among the possible explanations, perhaps the C protein of WT MV modulates viral RNA synthesis in a quality control manner to minimize aberrant RNA formation such as that seen with human parainfluenza virus type 1 (72). In the absence of C protein, aberrant viral RNAs with sufficient structure to activate PKR and trigger SG formation might accumulate to such an elevated concentration that the endogenous ADAR1 is unable to destabilize them. Thus, the ability of C protein expressed by WT virus to effectively downregulate innate host responses may be an indirect consequence during virus replication. With viruses other than WT measles virus, SG formation is sometimes impaired and multiple mechanisms may be operative (35). These range from direct antagonism of PKR and eIF2α phosphorylation by viral gene products, exemplified by the NS1 protein of influenza virus (43) and E3L protein of vaccinia virus (42), to the cleavage or sequestration of RNA-binding proteins that drive SG formation, exemplified by cleavage of G3BP1 by the 3C protease of poliovirus (73) or depletion of Staufen by the HIV gag protein through direct interaction (68).
Taken together, our results establish that SG formation induced by MV infection is modulated in an opposing manner by ADAR1 and PKR. SG formation is impaired by ADAR1, just as the activation of PKR and induction of IFN-β likewise are impaired by ADAR1 (29, 31). But MV-induced SG formation is dependent upon PKR. The opposing roles of ADAR1 and PKR suggest that these proteins may likely contribute to the dynamic oscillation of SG formation characteristic of the cellular response to virus infection (41, 74, 75). It is now of utmost importance to establish the qualitative and quantitative nature of the RNA produced in MV-infected cells that is responsible for PKR-dependent SG formation and whether SG formation per se is necessary for triggering an antiviral innate immune response characterized by IFN induction and action. The data presented here further illustrate the delicate balance between PKR and ADAR1 and add to a growing body of evidence that despite being an IFN-stimulated gene, ADAR1 in some instances both suppresses host responses characteristic of virus infection and enhances virus growth (12, 25).
ACKNOWLEDGMENT
This work was supported in part by research grants AI-12520 and AI-20611 from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, U.S. Public Health Service.
Footnotes
Published ahead of print 31 October 2012
REFERENCES
- 1. Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed). 2007. Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2. Griffin DE, Lin W-H, Pan C-H. 2012. Measles virus, immune control, and persistence. FEMS Microbiol. Rev. 36:649–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Oldstone MBA. 2009. Modeling subacute sclerosing panencephalitis in a transgenic mouse system: uncoding pathogenesis of disease and illuminating components of immune control. Curr. Top. Microbiol. Immunol. 330:31–54 [DOI] [PubMed] [Google Scholar]
- 4. Cattaneo R, Miest T, Shashkova EV, Barry MA. 2008. Reprogrammed viruses as cancer therapeutics: targeted, armed and shielded. Nat. Rev. Microbiol. 6:529–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Randall RE, Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1–47 [DOI] [PubMed] [Google Scholar]
- 6. McAllister CS, Toth AM, Zhang P, Devaux P, Cattaneo R, Samuel CE. 2010. Mechanisms of protein kinase PKR-mediated amplification of beta interferon induction by C protein-deficient measles virus. J. Virol. 84:380–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yoneyama M, Fujita T. 2010. Recognition of viral nucleic acids in innate immunity. Rev. Med. Virol. 20:4–22 [DOI] [PubMed] [Google Scholar]
- 8. Samuel CE. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Patterson JB, Samuel CE. 1995. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol. Cell. Biol. 15:5376–5388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pfaller CK, Li Z, George CX, Samuel CE. 2011. Protein kinase PKR and RNA adenosine deaminase ADAR1: new roles for old players as modulators of the interferon response. Curr. Opin. Immunol. 23:573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sadler AJ, Williams BRG. 2007. Structure and function of the protein kinase R. Curr. Top. Microbiol. Immunol. 316:253–292 [DOI] [PubMed] [Google Scholar]
- 12. Samuel CE. 2011. Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology 411:180–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Toth AM, Zhang P, Das S, George CX, Samuel CE. 2006. Interferon action and the double-stranded RNA-dependent enzymes ADAR1 adenosine deaminase and PKR protein kinase. Prog. Nucleic Acid Res. Mol. Biol. 81:369–434 [DOI] [PubMed] [Google Scholar]
- 14. McCormack SJ, Thomis DC, Samuel CE. 1992. Mechanism of interferon action: identification of a RNA binding domain within the N-terminal region of the human RNA-dependent P1/eIF-2 alpha protein kinase. Virology 188:47–56 [DOI] [PubMed] [Google Scholar]
- 15. Samuel CE. 1979. Mechanism of interferon action: phosphorylation of protein synthesis initiation factor eIF-2 in interferon-treated human cells by a ribosome-associated kinase processing site specificity similar to hemin-regulated rabbit reticulocyte kinase. Proc. Natl. Acad. Sci. U. S. A. 76:600–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. George CX, Gan Z, Liu Y, Samuel CE. 2011. Adenosine deaminases acting on RNA, RNA editing, and interferon action. J. Interferon Cytokine Res. 31:99–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bass BL. 2002. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71:817–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bass BL, Weintraub H. 1988. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55:1089–1098 [DOI] [PubMed] [Google Scholar]
- 19. Wagner RW, Smith JE, Cooperman BS, Nishikura K. 1989. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc. Natl. Acad. Sci. U. S. A. 86:2647–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Herbert A, Alfken J, Kim YG, Mian IS, Nishikura K, Rich A. 1997. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc. Natl. Acad. Sci. U. S. A. 94:8421–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. 1994. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc. Natl. Acad. Sci. U. S. A. 91:11457–11461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. George CX, Samuel CE. 1999. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc. Natl. Acad. Sci. U. S. A. 96:4621–4626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Poulsen H, Nilsson J, Damgaard CK, Egebjerg J, Kjems J. 2001. CRM1 mediates the export of ADAR1 through a nuclear export signal within the Z-DNA binding domain. Mol. Cell. Biol. 21:7862–7871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu Y, Herbert A, Rich A, Samuel CE. 1998. Double-stranded RNA-specific adenosine deaminase: nucleic acid binding properties. Methods 15:199–205 [DOI] [PubMed] [Google Scholar]
- 25. Gélinas J-F, Clerzius G, Shaw E, Gatignol A. 2011. Enhancement of replication of RNA viruses by ADAR1 via RNA editing and inhibition of RNA-activated protein kinase. J. Virol. 85:8460–8466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Taghavi N, Samuel CE. 2012. Protein kinase PKR catalytic activity is required for the PKR-dependent activation of mitogen-activated protein kinases and amplification of interferon beta induction following virus infection. Virology 427:208–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Toth AM, Devaux P, Cattaneo R, Samuel CE. 2009. Protein kinase PKR mediates the apoptosis induction and growth restriction phenotypes of C protein-deficient measles virus. J. Virol. 83:961–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nie Y, Hammond GL, Yang J-H. 2007. Double-stranded RNA deaminase ADAR1 increases host susceptibility to virus infection. J. Virol. 81:917–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Toth AM, Li Z, Cattaneo R, Samuel CE. 2009. RNA-specific adenosine deaminase ADAR1 suppresses measles virus-induced apoptosis and activation of protein kinase PKR. J. Biol. Chem. 284:29350–29356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hartner JC, Walkley CR, Lu J, Orkin SH. 2009. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat. Immunol. 10:109–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Z, Okonski KM, Samuel CE. 2012. Adenosine deaminase acting on RNA (ADAR1) suppresses the induction of interferon by measles virus. J. Virol. 86:3787–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Samuel CE. 1993. The eIF-2 alpha protein kinases, regulators of translation in eukaryotes from yeasts to humans. J. Biol. Chem. 268:7603–7606 [PubMed] [Google Scholar]
- 34. Kedersha NL, Gupta M, Li W, Miller I, Anderson P. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147:1431–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. White JP, Lloyd RE. 2012. Regulation of stress granules in virus systems. Trends Microbiol. 20:175–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Buchan JR, Parker R. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36:932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lindquist ME, Lifland AW, Utley TJ, Santangelo PJ, Crowe JE. 2010. Respiratory syncytial virus induces host RNA stress granules to facilitate viral replication. J. Virol. 84:12274–12284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lindquist ME, Mainou BA, Dermody TS, Crowe JE. 2011. Activation of protein kinase R is required for induction of stress granules by respiratory syncytial virus but dispensable for viral replication. Virology 413:103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McInerney GM, Kedersha NL, Kaufman RJ, Anderson P, Liljeström P. 2005. Importance of eIF2alpha phosphorylation and stress granule assembly in alphavirus translation regulation. Mol. Biol. Cell 16:3753–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ruggieri A, Dazert E, Metz P, Hofmann S, Bergeest Mazur J-PJ, Bankhead P, Hiet Kallis M-SS, Alvisi G, Samuel CE, Lohmann V, Kaderali L, Rohr K, Frese M, Stoecklin G, Bartenschlager R. 2012. Dynamic oscillation of translation and stress granule formation mark the cellular response to virus infection. Cell Host Microbe 12:71–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Simpson-Holley M, Kedersha N, Dower K, Rubins KH, Anderson P, Hensley LE, Connor JH. 2011. Formation of antiviral cytoplasmic granules during orthopoxvirus infection. J. Virol. 85:1581–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Khaperskyy DA, Hatchette TF, McCormick C. 2012. Influenza A virus inhibits cytoplasmic stress granule formation. FASEB J. 26:1629–1639 [DOI] [PubMed] [Google Scholar]
- 44. Courtney SC, Scherbik SV, Stockman BM, Brinton MA. 2012. West Nile virus infections suppress early viral RNA synthesis and avoid inducing the cell stress granule response. J. Virol. 86:3647–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Emara MM, Brinton MA. 2007. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl. Acad. Sci. U. S. A. 104:9041–9046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang P, Samuel CE. 2007. Protein kinase PKR plays a stimulus- and virus-dependent role in apoptotic death and virus multiplication in human cells. J. Virol. 81:8192–8200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li Z, Wolff KC, Samuel CE. 2010. RNA adenosine deaminase ADAR1 deficiency leads to increased activation of protein kinase PKR and reduced vesicular stomatitis virus growth following interferon treatment. Virology 396:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang P, Jacobs BL, Samuel CE. 2008. Loss of protein kinase PKR expression in human HeLa cells complements the vaccinia virus E3L deletion mutant phenotype by restoration of viral protein synthesis. J. Virol. 82:840–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Devaux P, Messling von, Songsungthong VW, Springfeld C, Cattaneo R. 2007. Tyrosine 110 in the measles virus phosphoprotein is required to block STAT1 phosphorylation. Virology 360:72–83 [DOI] [PubMed] [Google Scholar]
- 50. Bossert B, Conzelmann K-K. 2002. Respiratory syncytial virus (RSV) nonstructural (NS) proteins as host range determinants: a chimeric bovine RSV with NS genes from human RSV is attenuated in interferon-competent bovine cells. J. Virol. 76:4287–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu Y, Wolff KC, Jacobs BL, Samuel CE. 2001. Vaccinia virus E3L interferon resistance protein inhibits the interferon-induced adenosine deaminase A-to-I editing activity. Virology 289:378–387 [DOI] [PubMed] [Google Scholar]
- 52. George CX, Wagner MV, Samuel CE. 2005. Expression of interferon-inducible RNA adenosine deaminase ADAR1 during pathogen infection and mouse embryo development involves tissue-selective promoter utilization and alternative splicing. J. Biol. Chem. 280:15020–15028 [DOI] [PubMed] [Google Scholar]
- 53. Liu Y, Samuel CE. 1996. Mechanism of interferon action: functionally distinct RNA-binding and catalytic domains in the interferon-inducible, double-stranded RNA-specific adenosine deaminase. J. Virol. 70:1961–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Schade M, Turner CJ, Kühne R, Schmieder P, Lowenhaupt K, Herbert A, Rich A, Oschkinat H. 1999. The solution structure of the Zalpha domain of the human RNA editing enzyme ADAR1 reveals a prepositioned binding surface for Z-DNA. Proc. Natl. Acad. Sci. U. S. A. 96:12465–12470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Navaratnarajah CK, Leonard VHJ, Cattaneo R. 2009. Measles virus glycoprotein complex assembly, receptor attachment, and cell entry. Curr. Top. Microbiol. Immunol. 329:59–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tourrière H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160:823–831 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 57. Anderson P, Kedersha N. 2009. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10:430–436 [DOI] [PubMed] [Google Scholar]
- 58. Anderson P, Kedersha N. 2008. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33:141–150 [DOI] [PubMed] [Google Scholar]
- 59. Beckham CJ, Parker R. 2008. P bodies, stress granules, and viral life cycles. Cell Host Microbe 3:206–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Weissbach R, Scadden ADJ. 2012. Tudor-SN and ADAR1 are components of cytoplasmic stress granules. RNA 18:462–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kedersha N, Tisdale S, Hickman T, Anderson P. 2008. Real-time and quantitative imaging of mammalian stress granules and processing bodies. Methods Enzymol. 448:521–552 [DOI] [PubMed] [Google Scholar]
- 62. St Johnston D, Brown NH, Gall JG, Jantsch M. 1992. A conserved double-stranded RNA-binding domain. Proc. Natl. Acad. Sci. U. S. A. 89:10979–10983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wickham L, Duchaîne T, Luo M, Nabi IR, DesGroseillers L. 1999. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol. Cell. Biol. 19:2220–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Samuel CE. 2012. ADARs: Viruses and innate immunity. Curr. Top. Microbiol. Immunol. 353:163–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Athanasiadis A, Placido D, Maas S, Brown BA, Lowenhaupt K, Rich A. 2005. The crystal structure of the Zbeta domain of the RNA-editing enzyme ADAR1 reveals distinct conserved surfaces among Z-domains. J. Mol. Biol. 351:496–507 [DOI] [PubMed] [Google Scholar]
- 66. Barraud P, Allain FH-T. 2012. ADAR proteins: double-stranded RNA and Z-DNA binding domains. Curr. Top. Microbiol. Immunol. 353:35–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Deigendesch N, Koch-Nolte F, Rothenburg S. 2006. ZBP1 subcellular localization and association with stress granules is controlled by its Z-DNA binding domains. Nucleic Acids Res. 34:5007–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Abrahamyan LG, Chatel-Chaix L, Ajamian L, Milev MP, Monette A, Clément Song J-FR, Lehmann M, DesGroseillers L, Laughrea M, Boccaccio G, Mouland AJ. 2010. Novel Staufen1 ribonucleoproteins prevent formation of stress granules but favour encapsidation of HIV-1 genomic RNA. J. Cell Sci. 123:369–383 [DOI] [PubMed] [Google Scholar]
- 69. Thomas MG, Martinez Tosar LJ, Loschi M, Pasquini JM, Correale J, Kindler S, Boccaccio GL. 2005. Staufen recruitment into stress granules does not affect early mRNA transport in oligodendrocytes. Mol. Biol. Cell 16:405–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Thomas MG, Martinez Tosar LJ, Desbats MA, Leishman CC, Boccaccio GL. 2009. Mammalian Staufen 1 is recruited to stress granules and impairs their assembly. J. Cell Sci. 122:563–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Anderson P, Kedersha N. 2002. Visibly stressed: the role of eIF2, TIA-1, and stress granules in protein translation. Cell Stress Chaperones 7:213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Boonyaratanakornkit J, Bartlett E, Schomacker H, Surman S, Akira S, Bae Collins Y-SP, Murphy B, Schmidt A. 2011. The C proteins of human parainfluenza virus type 1 limit double-stranded RNA accumulation that would otherwise trigger activation of MDA5 and protein kinase R. J. Virol. 85:1495–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. White JP, Cardenas AM, Marissen WE, Lloyd RE. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2:295–305 [DOI] [PubMed] [Google Scholar]
- 74. Anderson P, Kedersha N. 2006. RNA granules. J. Cell Biol. 172:803–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151:1257–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]