Abstract
The hepatitis C virus NS5A protein is essential for RNA replication and virion assembly. NS5A is phosphorylated on multiple residues during infections, but these sites remain uncharacterized. Here we identify serine 222 of genotype 2a NS5A as a phosphorylation site that functions as a negative regulator of RNA replication. This site is a component of the hyperphosphorylated form of NS5A, which is in good agreement with previous observations that hyperphosphorylation negatively affects replication.
TEXT
Hepatitis C virus (HCV) is a human pathogen of global impact, with as many as 170 million chronically infected (1). Long-term infection results in progressive liver damage, including fibrosis, cirrhosis, and, oftentimes, hepatocellular carcinoma. HCV is a member of the Flaviviridae family of enveloped, positive-sense, single-stranded RNA viruses (reviewed in reference 2). The noncapped, nonpolyadenylated, 9.6-kb viral genome encodes a single internal ribosome entry site (IRES)-directed open reading frame flanked by highly structured 5′ and 3′ nontranslated regions. Translation yields a polyprotein of approximately 3,000 amino acids that undergoes a complex series of co- and posttranslational proteolytic events catalyzed by viral and host proteases, resulting in the production of 10 mature HCV proteins. These include the structural proteins that are involved in the production of progeny virions (C, E1, E2, p7) and the nonstructural replication proteins (NS2, NS3, NS4A, NS4B, NS5A, NS5B). There is considerable overlap in functionality between these groups, with a number of nonstructural proteins serving essential functions for both RNA replication and virion assembly, including the NS5A protein (3–11).
NS5A is a large, three-domain, hydrophilic phosphoprotein that exists in two distinct forms: a hypophosphorylated form of roughly 56 kDa and a hyperphosphorylated form of 58 kDa (12, 13). It is not known what residues in NS5A are phosphorylated or how various phosphorylation events contribute to the presence of the two phospho-forms (see reference 14 for a concise review). Evidence from mutagenesis suggests that hypophosphorylation primarily targets serine residues in domains II and III, whereas hyperphosphorylation sites cluster in and around domain I, LCSI (low complexity sequence block I), and domain II (12, 15–18). The function of NS5A phosphorylation in the viral life cycle is unknown, but a general theme has emerged in which hypophosphorylation is required for RNA replication, and hyperphosphorylation is a negative regulator of this process. Phosphorylation may also play a role in the function of NS5A in virion assembly, and it could regulate the interface between replication and assembly events (11). A number of phosphorylation sites have been identified in NS5A using different overexpression systems, but it is unknown in some cases if these modifications are actually used in the virus life cycle or what their function(s) might be (16–22).
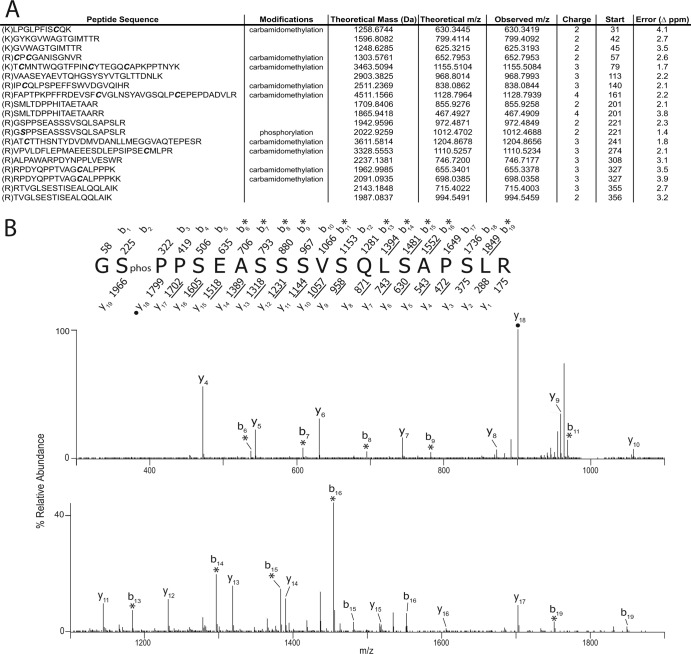
As no phosphorylation sites have yet been definitively mapped in NS5A in the context of the protein functioning in authentic RNA replication, we developed a method to extract NS5A from cells harboring subgenomic replicons and enrich for the protein by immunoprecipitation. Enriched fractions were then subjected to SDS-PAGE gel separation, proteolytic digestion and peptide extraction, and mass spectrometry. Briefly, 7.5 × 107 Huh-7.5 cells containing the genotype 2a replicon pSGR-JFH-1 were lysed in NETN buffer containing protease and phosphatase inhibitors. The NS5A protein was extracted by immunoprecipitation using the 9E10 anti-NS5A monoclonal antibody covalently coupled to agarose beads (21). Following extensive washing in NETN, NS5A was eluted from the beads by the addition of 2× SDS-PAGE nonreducing sample buffer and boiling for 5 min. Extracts were then resolved on 10% acrylamide SDS-PAGE gels, and the region of the gel corresponding to the migration point of NS5A was excised and subjected to in-gel reduction, alkylation, and trypsin digestion following the standard methods of the Scripps Proteomic Core facility. Eluted peptides were then loaded onto a fused silica (outer diameter [OD], 360 μm; inner diameter [ID], 75 μm) capillary C18 (ODS-AQ, 20 μm) precolumn and washed with 0.1% acetic acid. Once salts were removed, the precolumn was attached to a fused silica (OD, 360 μm; ID, 50 μm) C18 column (ODS-AQ; 5 μm) attached to a fused silica emitter tip. Peptides were high-performance liquid chromatography (HPLC) gradient eluted and analyzed by a Thermo Electron LTQ Orbitrap ion trap mass spectrometer set to data-dependent mode to acquire tandem mass spectrometric (MS-MS) spectra on the five most abundant m/z peaks in each MS scan. All MS-MS spectra were searched using Sequest (Thermo Electron) and Mascot (Matrix Science) software. Potential posttranslational modifications were searched (STY = 80 [phosphorylation]) as well as static modifications for carbamidomethylation (C = 57). Search results were manually confirmed to ensure correct sequence identifications. This methodology produced 61% sequence coverage of the genotype 2a JFH-1 isolate of NS5A and identified a single phosphorylation posttranslational modification at serine 222 (Fig. 1). To provide context to the more widely studied Con1 1b NS5A protein, this serine residue would reside at position 222 in that protein and position 2194 in the Con1 1b polyprotein. It should be noted that a number of serine-rich regions of NS5A were not identified in this analysis, and these may contain additional phosphorylation sites. Nonetheless, the data definitively identify serine 222 as a phosphoacceptor used during the course of authentic HCV RNA replication.
Fig 1.
Identification of serine 222 as a phosphorylation acceptor. (A) A comprehensive list of NS5A peptides identified following LC-MS-MS analysis. Peptide sequences are listed along with their theoretical mass, theoretical and observed m/z values, and associated mass error (ppm). The charge state, start residue of each peptide, and identified modifications are also provided. Modifications observed are indicated, including the phosphorylation of serine 222. (B) MS-MS spectrum of phosphorylated NS5A peptide, residues 221 to 240. The [M + 2H]+2 ion of the phosphorylated NS5A peptide (m/z 1012.47) was selected for dissociation, and the amino acid sequence of the peptide is shown above the spectrum. The masses above and below the sequence correspond to the theoretical, singly protonated b- and y-type product ions, respectively. The observed singly protonated product ions are underlined, and doubly protonated ions are denoted with filled circles. Asterisks indicate product ions that result from neutral loss of H3PO4. As indicated in the sequence, the phosphorylated residue was identified as serine residue 222.
With S222 identified, we proceeded to determine what role the posttranslational modification of this residue might have in HCV biology. We generated a series of phosphomimetic mutants at this residue, using methods described previously (11), including an aspartic acid substitution to mimic phosphorylation and an alanine residue to generate a site incapable of phosphorylation. These were built into the genotype 2a subgenomic replicon, pSGR-JFH1, and the RNA replication kinetics of RNAs harboring these mutations were assessed relative to a wild-type pSGR-JFH1 RNA and a nonreplicative pSGR-JFH1 RNA containing a lethal lesion in the RNA-dependent RNA polymerase (Pol−) (23). It is important to note that the pSGR-JFH1 construct, unlike many other replicons of different HCV genotypes, replicated efficiently without adaptive mutations, so the serine 222 mutations are analyzed in the context of a wild-type NS5A sequence. Figure 2A shows the results of the delivery of 1 μg of these RNAs to Huh-7.5 cells via electroporation and the time course analysis of total HCV RNA accumulation inside cells over a 48-h period via real-time reverse transcriptase PCR analysis as described previously (11, 24). In this analysis the replication of pSGR-JFH1 with the serine 222-to-alanine (S222A) mutation is indistinguishable from the wild-type pSGR-JFH1 construct, suggesting that under these conditions, the inability to phosphorylate serine 222 is phenotypically null. Analysis of the serine 222-to-aspartic acid mutation is more interesting, as pSGR-JFH1 genomes containing this mutation (S222D) are impaired in RNA replication approximately 2- to 3-fold compared to the parental pSGR-JFH1 construct at all data points after 24 h postelectroporation. Although this change is relatively small, it is statistically significant and reproducible. The data presented are normalized to total RNA, but it should be noted that further normalization of the data to the 4-h time point values did not alter the observed differences or their statistical validity. It is important to note that the S222D mutant containing replicons do replicate RNA, just at a lower efficiency, as can be seen by comparison to nonreplicating (Pol−) pSGR-JFH1 RNA. To investigate the impact of serine 222 modification on replication of an infectious viral genome, we built the same mutations into the JC1 HCV infectious clone and performed a similar electroporation-based RNA accumulation assay to that performed with the replicon (Fig. 2B). Similarly to what was observed in the replicon, the serine 222A mutation was identical in RNA replication to the JC1 parental viral genome, and the serine 222D was again attenuated in RNA replication. The data presented are normalized to total RNA, but it should be noted that further normalization of the data to the 4-h time point values did not alter the observed differences or their statistical validity. As NS5A is known to be an important factor in infectious virus production and as this may be linked to phosphorylation, we harvested supernatants of cells 48 h after the delivery of our mutant RNAs and determined the titers of the amount of infectious virus in these samples by the limiting dilution immunohistochemical assay described previously (21). Despite the clear difference in RNA replication efficiency of the various serine phosphomimetic mutants, we observed no statistically significant difference in infectivity, although a slight (about 1.8-fold) downward trend was seen in the S222D mutant-bearing virus. Normalization of titer data to RNA replication level eliminated this difference, indicating that the S222D mutant does not impact virus production. As our mutations alter the sequence of the NS5A protein, we felt it was prudent to monitor the stability and processing of NS5A over the course of these experiments. To avoid differences in levels of NS5A due to different levels of RNA replication for the various mutations and other complications, we opted to express NS5A in the context of the full-length viral JC1 genome using the VTF7-3 vaccinia T7 system (25). Figure 2D shows the results of this analysis 24 h after transfection of 1 μg of each plasmid via transfection as described previously (24) and infection of Huh-7.5 cells with VTF7-3 at a multiplicity of 1, prior to cell lysate preparation and SDS-PAGE and Western blot analysis with the 9E10 NS5A-specific antibody and an actin control as described previously, and the results indicate that there is no gross defect in NS5A stability or polyprotein processing. Analysis of the phospho-form distribution of NS5A suggests that S222A bearing NS5A produces a 44.9% reduction in hyperphosphorylation compared to the wild-type construct while exhibiting only a 4.9% reduction in hypophosphorylated NS5A relative to the control. S222D hyperphosphorylation, in contrast, is reduced only 9.8% relative to the wild type, with the hypophosphorylated form reduced 19.3% compared to the control. This suggests that S222 might control multiple phosphorylation sites when modified. Collectively, these results indicate that serine 222 of genotype 2A NS5A is phosphorylated in the context of HCV replication, and this modification is a negative regulator of replication, albeit a minor one.
Fig 2.
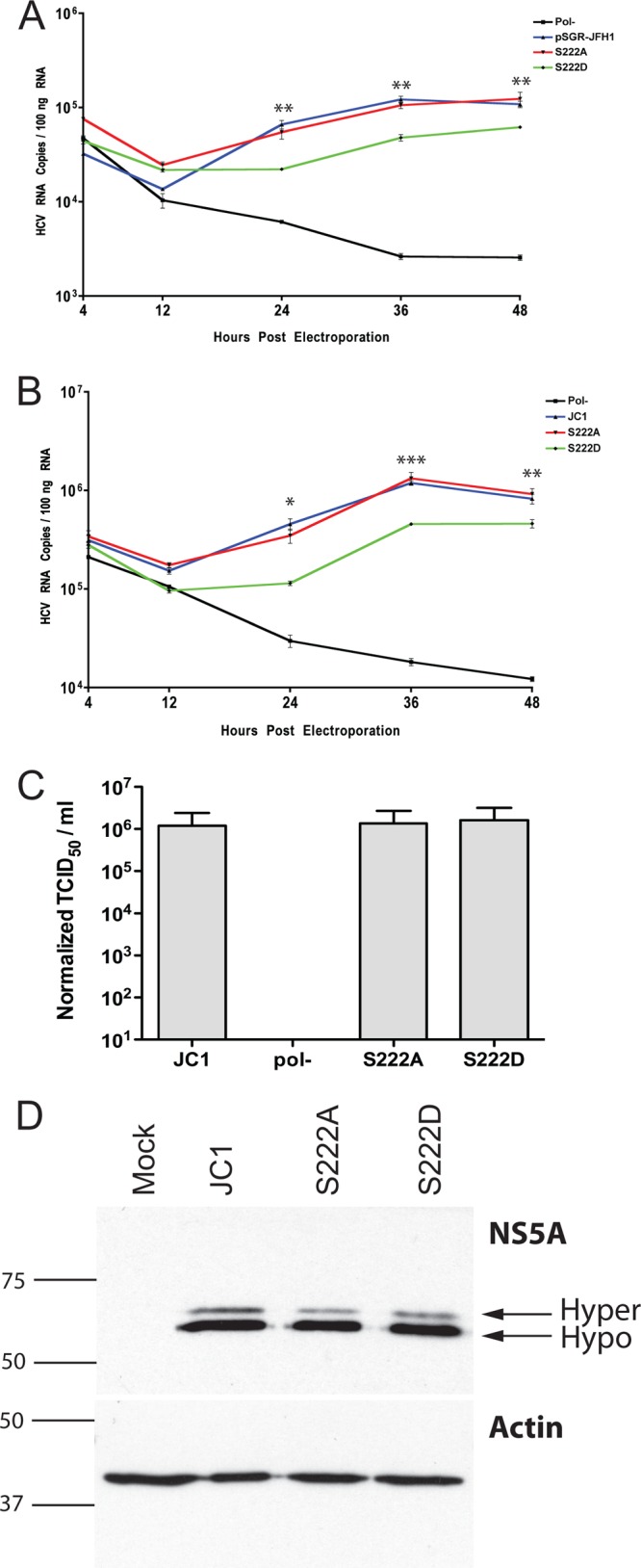
Replication analysis of phosphomimetic mutations of serine 222. (A) Replication of serine 222 mutants in the context of the pSGR-JFH1 subgenomic replicon. HCV RNA copies per 100 nanograms of total RNA as determined via real-time reverse transcriptase PCR analysis of cells electroporated with wild-type pSGR-JFH1 RNA (pSGR-JFH1), a pSGR-JFH1 replicon harboring the serine 222-to-alanine mutation (S222A), a psGR-JFH1 replicon harboring the serine 222-to-aspartic acid mutation (S222D), and a pSGR-JFH1 replicon harboring a lethal lesion in the RNA-dependent RNA polymerase (Pol−). RNAs were harvested at indicated times postelectroporation from triplicate samples for calculation of statistical significance using a Student t test for each indicated data point. (B) Samples as described for panel A but with mutations built into the full-length infectious JC1 virus clone (JC1). (C) Titer release from cells in panel B 48 h after electroporation. Values are in normalized tissue culture infectious doses, 50% value, per milliliter as determined by limiting dilution assay. Values shown are the averages of three independent electroporations of viral RNA. Samples are designated as in the previous panel. Data for this experiment were normalized using the RNA replication data from panel B. (D) NS5A expressed from VTF7-3 transfection/infection experiments separated on a 12% SDS-PAGE gel to monitor polyprotein processing and stability. The single band represents the NS5A protein. The samples shown are from wild-type JC1 virus encoding a genomic infectious clone plasmid (JC1), the JC1 infectious clone plasmid containing the serine 222-to-alanine mutation (S222A), and the JC1 infection clone plasmid containing the serine 222-to-aspartic acid mutation (S222D). NS5A was detected by Western blotting using the 9E10 anti-NS5A monoclonal antibody. An actin Western blot loading control is shown as the lower panel in this composite figure. Arrows indicate the migration distances of the hypophosphorylated (hypo) and hyperphosphorylated (hyper) forms of NS5A. In both panels, the numbers and lines to the left of the image indicate the sizes (in kilodaltons) and mobilities of molecular weight markers.
As our method of isolation of NS5A does not discriminate between hyper- and hypophosphorylated forms, we wanted to determine if serine 222 was a discrete component of one form over the other or a component of both forms. We therefore metabolically labeled cells harboring the pSGR-JFH1 replicon and a replicon harboring S222A using radioactive orthophosphate as described previously (26). Cell lysates were then prepared, NS5A was immunoprecipitated, and precipitates were analyzed by scintillation counting. Samples containing Huh-7.5 cell lysates devoid of HCV and samples precipitated with isotype-matched control antibodies contained background levels of radioactivity after washing and were not analyzed further. Samples containing NS5A were then separated by SDS-PAGE as described in the aforementioned reference, with the substitution of the 9E10 NS5A antibody for precipitations. Gels were then exposed to X-ray film to determine incorporation of radioactive phosphate into NS5A. Figure 3A presents these data, with the electrophoretic migration of hyper- and hypophosphorylated NS5A indicated for both the wild-type and mutant replicon extracts. Measurement of radioactivity in the various phospho-forms indicated that although the hypophosphorylated form of NS5A differed by only 3% between wild type and mutant, the hyperphosphorylated form differed by 19.1% between the two samples, suggesting that phosphorylated serine 222 is predominantly a component of the hyperphosphorylated form of NS5A. We also investigated phospho-form distribution by Western blotting of NS5A from cells infected with J6/JFH1 viral genomes containing our serine 222 mutants (Fig. 3B). Densitometric analysis of these blots shows that NS5A from S222A viruses has 36% less hyperphosphorylation than a wild-type construct, while reduction in hypophosphorylation was only 6%. Similarly, S222D virus showed a reduction of 23.4% hyperphosphorylation compared to the wild type, with a 7% reduction in hypophosphorylation. The reduction in hyperphosphorylation difference between these two mutants is quite interesting, as neither is capable of being phosphorylated, and this could be interpreted as an indication that S222D controls the use of other phosphorylation sites in NS5A. The location of this residue in relation to the domain model of NS5A, with serine 222 in the LCSI region between domain I and II, is in good agreement with previous data suggesting that hyperphosphorylation sites map in this area of NS5A.
Fig 3.
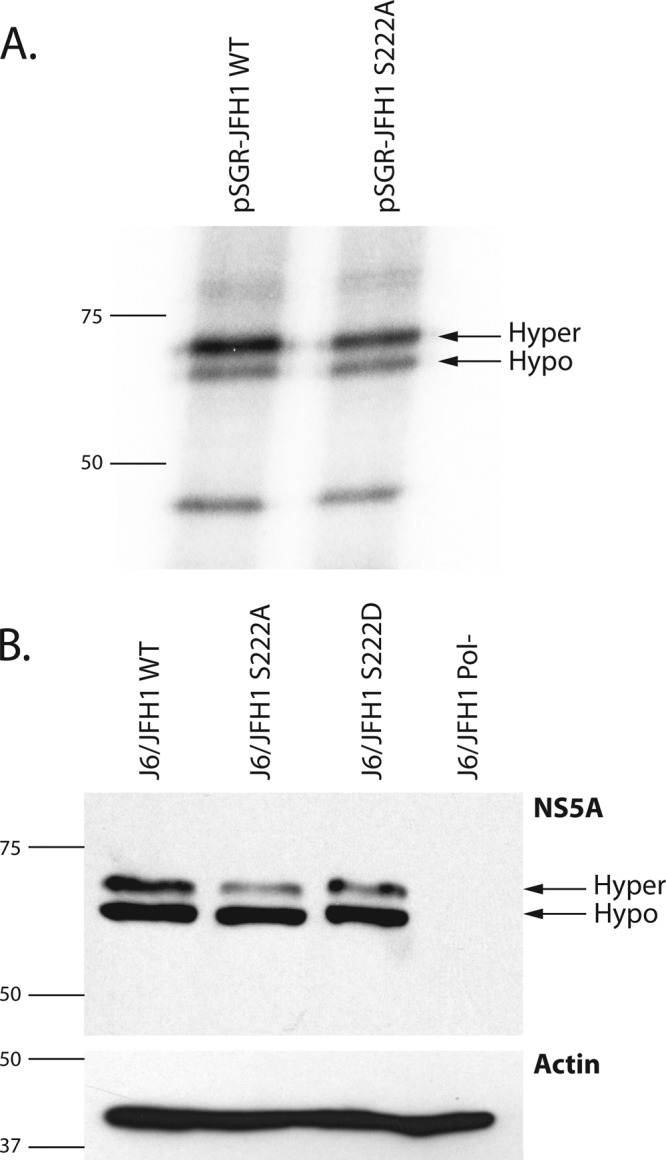
Metabolic labeling of replicon containing cell lines. (A) Film from 10% SDS-PAGE separation of 32Pi-labeled Huh-7.5 cell lysates from cells harboring wild-type pSGR-JFH1 (pSGR-FJH1 WT) or the pSGR-JFH1 replicon harboring the serine 222-to-alanine mutation (pSGR-JFH1 S222A). Arrows indicate the migration distances of the hypophosphorylated (hypo) and hyperphosphorylated (hyper) forms of NS5A. Numbers and lines to the left of the image indicate the sizes (in kilodaltons) and mobilities of molecular weight markers. (B) Upper panel, Western blot analysis detecting NS5A protein from cells electroporated with HCV J6/JFH1 viral genomic RNA containing S222A, S222D, wild-type, and Pol− RNAs. Arrows indicate the migration distances of the hypophosphorylated (hypo) and hyperphosphorylated (hyper) forms of NS5A. Lower panel, a Western blot of lysates identical to those in the upper panel but probed for the cellular actin protein as a loading control. In both panels, the numbers and lines to the left of the image indicate the sizes (in kilodaltons) and mobilities of molecular weight markers.
Here we have identified serine 222 of the genotype 2a NS5A protein as a site of phosphorylation used in cells harboring a functional replicon, the first site to be found in the context of RNA replication. Our findings are in good agreement with the concept that hyperphosphorylation is a negative regulator of replication, but the effects we observe are small, and it is therefore unlikely that serine 222 is a major regulator of RNA replication. Mutation of this residue to aspartic acid produces a different phospho-form distribution than when it is changed to alanine (or serine), and these data might be interpreted to indicate that S222 modification directs the phosphorylation of other sites in NS5A that are components of the hyperphosphorylated isoform. Unlike serine residues in domain III of NS5A, serine 222 appears to have no impact on virus production, suggesting that different serine phosphorylation events can impact different aspects of the HCV life cycle, even when those serine residues are both components of the same NS5A phospho-form. The data suggest that a more complex regulation than explained in current models probably exists. Serine 222 is an absolutely conserved residue among divergent HCV genotypes, and indeed, this residue has been identified as a phosphoacceptor in an early NS5A overexpression phosphopeptide-mapping experiment in genotype 1a (designated S2194 in that work) (16). Phosphomimetic mutation of this position in the 1b replicon does not produce dramatic replication or phospho-form distribution phenotypes in the context of one set of adaptive mutations (15), but it does produce these effects in another adaptive genetic context (27). Adaptive mutations in genotype 1b replicons increase RNA replication and tend to reduce NS5A hyperphosphorylation (19), and one can directly manipulate hyperphosphorylation to bypass the need for adaptive mutations (15, 17, 28, 29), but the mechanistic details of these observations are not understood. Perhaps different adaptive mutations produce different patterns of phosphorylation site usage on NS5A, and in the example of S222, this may be important in one context but not in another. The genotype 2a NS5A used in this work requires no adaptive mutations for efficient replication, eliminating complexities of genetic background from the analysis and simplifying the identification of the phosphorylation of this residue as a minor negative regulator of replication. Perhaps the overwhelming questions suggest why this virus utilizes such a complex mechanism to fine-tune RNA replication efficiency and what precisely NS5A phosphorylation actually regulates. It will likely be some time before we develop a comprehensive view of the role of NS5A phosphorylation, but the identification of S222 as an authentic phosphorylation site is an important step in this process.
ACKNOWLEDGMENTS
These experiments were funded, in part, by the Burroughs Wellcome Fund Investigators in the Pathogenesis of Infectious Diseases program (T.L.T.). Additional funds supporting this work came from the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, and the National Institute of Allergy and Infectious Disease (T.L.T.).
We thank Marilena Fernandez, Sarae Copeland, and Melissa Tellinghuisen for assistance in the preparation of the manuscript and the members of the Scripps Proteomics Core facility for technical assistance. We thank Bernard Moss for the vaccinia T7 system reagents and Charles Rice for the Huh-7.5 cells and 9E10 monoclonal antibody.
Footnotes
Published ahead of print 31 October 2012
This article is number 21903 from the Scripps Research Institute.
REFERENCES
- 1. Anonymous 1997. World Health Organization—Hepatitis C: global prevalence. Wkly. Epidemiol. Rec. 72:341–344 [PubMed] [Google Scholar]
- 2. Lindenbach BD, Thiel HJ, Rice CM. 2007. Flaviviridae: the viruses and their replication, p 1101–1152 In Knipe DM, Howley PM. (ed), Fields virology, fifth ed, vol 1 Lippincott-Raven Publishers, Philadelphia, PA [Google Scholar]
- 3. Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, Kallis S, Engel U, Bartenschlager R. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035 doi:10.1371/journal.ppat.1000035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hughes M, Gretton S, Shelton H, Brown DD, McCormick CJ, Angus AG, Patel AH, Griffin S, Harris M. 2009. A conserved proline between domains II and III of hepatitis C virus NS5A influences both RNA replication and virus assembly. J. Virol. 83:10788–10796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hughes M, Griffin S, Harris M. 2009. Domain III of NS5A contributes to both RNA replication and assembly of hepatitis C virus particles. J. Gen. Virol. 90:1329–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jones CT, Murray CL, Eastman DK, Tassello J, Rice CM. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 81:8374–8383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jones DM, Patel AH, Targett-Adams P, McLauchlan J. 2009. The hepatitis C virus NS4B protein can trans-complement viral RNA replication and modulates production of infectious virus. J. Virol. 83:2163–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ma Y, Yates J, Liang Y, Lemon SM, Yi M. 2008. NS3 helicase domains involved in infectious intracellular hepatitis C virus particle assembly. J. Virol. 82:7624–7639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Masaki T, Suzuki R, Murakami K, Aizaki H, Ishii K, Murayama A, Date T, Matsuura Y, Miyamura T, Wakita T, Suzuki T. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 82:7964–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, Steinmann E, Abid K, Negro F, Dreux M, Cosset FL, Bartenschlager R. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. U. S. A. 103:7408–7413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tellinghuisen TL, Foss KL, Treadaway J. 2008. Regulation of hepatitis C virion production via phosphorylation of the NS5A protein. PLoS Pathog. 4:e1000032 doi:10.1371/journal.ppat.1000032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tanji Y, Kaneko T, Satoh S, Shimotohno K. 1995. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J. Virol. 69:3980–3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tellinghuisen TL, Marcotrigiano J, Gorbalenya AE, Rice CM. 2004. The NS5A protein of hepatitis C virus is a zinc metalloprotein. J. Biol. Chem. 279:48576–48587 [DOI] [PubMed] [Google Scholar]
- 14. Huang Y, Staschke K, De Francesco R, Tan SL. 2007. Phosphorylation of hepatitis C virus NS5A nonstructural protein: a new paradigm for phosphorylation-dependent viral RNA replication? Virology 364:1–9 [DOI] [PubMed] [Google Scholar]
- 15. Appel N, Pietschmann T, Bartenschlager R. 2005. Mutational analysis of hepatitis C virus nonstructural protein 5A: potential role of differential phosphorylation in RNA replication and identification of a genetically flexible domain. J. Virol. 79:3187–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Katze MG, Kwieciszewski B, Goodlett DR, Blakely CM, Neddermann P, Tan SL, Aebersold R. 2000. Ser(2194) is a highly conserved major phosphorylation site of the hepatitis C virus nonstructural protein NS5A. Virology 278:501–513 [DOI] [PubMed] [Google Scholar]
- 17. Neddermann P, Quintavalle M, Di Pietro C, Clementi A, Cerretani M, Altamura S, Bartholomew L, De Francesco R. 2004. Reduction of hepatitis C virus NS5A hyperphosphorylation by selective inhibition of cellular kinases activates viral RNA replication in cell culture. J. Virol. 78:13306–13314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reed KE, Rice CM. 1999. Identification of the major phosphorylation site of the hepatitis C virus H strain NS5A protein as serine 2321. J. Biol. Chem. 274:28011–28018 [DOI] [PubMed] [Google Scholar]
- 19. Blight KJ, Kolykhalov AA, Rice CM. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972–1974 [DOI] [PubMed] [Google Scholar]
- 20. Evans MJ, Rice CM, Goff SP. 2004. Phosphorylation of hepatitis C virus nonstructural protein 5A modulates its protein interactions and viral RNA replication. Proc. Natl. Acad. Sci. U. S. A. 101:13038–13043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 22. Nordle Gilliver A, Griffin S, Harris M. 2010. Identification of a novel phosphorylation site in hepatitis C virus NS5A. J. Gen. Virol. 91:2428–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tellinghuisen TL, Foss KL, Treadaway JC, Rice CM. 2008. Identification of residues required for RNA replication in domains II and III of the hepatitis C virus NS5A protein. J. Virol. 82:1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fuerst TR, Niles EG, Studier FW, Moss B. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 83:8122–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Neddermann P. 2009. NS5A phosphorylation and hyperphosphorylation. Methods Mol. Biol. 510:95–110 [DOI] [PubMed] [Google Scholar]
- 27. Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for hepatitis C virus genomic and subgenomic RNA replication. J. Virol. 76:13001–13014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Quintavalle M, Sambucini S, Di Pietro C, De Francesco R, Neddermann P. 2006. The alpha isoform of protein kinase CKI is responsible for hepatitis C virus NS5A hyperphosphorylation. J. Virol. 80:11305–11312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Quintavalle M, Sambucini S, Summa V, Orsatti L, Talamo F, De Francesco R, Neddermann P. 2007. Hepatitis C virus NS5A is a direct substrate of casein kinase I-alpha, a cellular kinase identified by inhibitor affinity chromatography using specific NS5A hyperphosphorylation inhibitors. J. Biol. Chem. 282:5536–5544 [DOI] [PubMed] [Google Scholar]