Abstract
The NKG2D receptor is expressed on the surface of NK, T, and macrophage lineage cells and plays an important role in antiviral and antitumor immunity. To evade NKG2D recognition, herpesviruses block the expression of NKG2D ligands on the surface of infected cells using a diverse repertoire of sabotage methods. Cowpox and monkeypox viruses have taken an alternate approach by encoding a soluble NKG2D ligand, the orthopoxvirus major histocompatibility complex (MHC) class I-like protein (OMCP), which can block NKG2D-mediated cytotoxicity. This approach has the advantage of targeting a single conserved receptor instead of numerous host ligands that exhibit significant sequence diversity. Here, we show that OMCP binds the NKG2D homodimer as a monomer and competitively blocks host ligand engagement. We have also determined the 2.25-Å-resolution crystal structure of OMCP from the cowpox virus Brighton Red strain, revealing a truncated MHC class I-like platform domain consisting of a beta sheet flanked with two antiparallel alpha helices. OMCP is generally similar in structure to known host NKG2D ligands but has notable variations in regions typically used to engage NKG2D. Additionally, the determinants responsible for the 14-fold-higher affinity of OMCP for human than for murine NKG2D were mapped to a single loop in the NKG2D ligand-binding pocket.
INTRODUCTION
The ability to detect and remove abnormal or infected cells is a key function of the immune system. Many receptors are used to detect and respond to pathogens, and one of the most studied is the natural killer group 2, member D receptor (NKG2D), a homodimeric type II membrane protein found on NK cells, murine macrophages, and subsets of T cells (1). NKG2D is an activating receptor on NK cells and a costimulatory receptor on T cells (2–7). Humans and mice are known to express 8 and 9 sequence-diverse NKG2D ligands (NKG2DLs), respectively (8). Engagement of NKG2D by ligands presented on the surface of target cells initiates a signaling cascade mediated by either of the membrane adaptor proteins DAP10 and DAP12 that leads to release of cytolytic factors such as perforin and granzymes. Since cytolysis is potentially damaging to host tissues, expression of NKG2DLs is tightly regulated at both the transcriptional and posttranscriptional levels (9–11). This limits NKG2DL expression on the cell surface of normal cells. However, cellular stress induces the cell surface display of NKG2DLs, which target the abnormal cell for elimination by NKG2D-bearing lymphocytes (12, 13).
The importance of NKG2D surveillance in cancer immunity is well established. Tumor cells expressing high levels of NKG2DLs are more susceptible to immune clearance, and NKG2DLs have been implicated in immunoediting of tumor cells (14, 15). To avoid recognition by NKG2D-bearing cells, some human cancer cells shed soluble NKG2DLs from their surface, preventing NKG2D-mediated recognition and elimination (16–20).
Infection with many viruses induces the surface expression of NKG2DLs (21–26). Therefore, NKG2D surveillance is a frequent target of viral immune evasion strategies, with many viruses targeting the cell surface display of NKG2DLs. Herpesviruses encode multiple microRNAs (miRNAs) and proteins that target the expression and cell surface presentation of specific NKG2DLs (27–32). However, the considerable sequence divergence and large number of NKG2DLs limit the effectiveness of this strategy. Indeed, no viral protein or miRNA that can broadly target all NKG2DLs has been identified (8). For example, human cytomegalovirus (HCMV) expresses UL16 and UL142, which both sabotage NKG2DL surface expression. UL16 binds the human ligands ULBP1 to -3, ULBP6, and MICB and retains them intracellularly (9, 26, 33–38). However, despite binding MICB, UL16 does not bind the closely related ligand MICA. To compensate for this, UL142 can bind to MICA and retain it in the Golgi complex (39). Even this strategy is incomplete, because the MICA gene is extremely polymorphic and UL142 does not recognize all MICA alleles (39–41). Similarly, murine cytomegalovirus (MCMV) encodes four proteins that retain murine NKG2DLs inside the infected cell. H60 is retained by m155; MULT1 is retained by m145; MULT1, RAE-1ε, and H60 are retained by m138; and members of the RAE-1 family are retained by m152 (27–30, 42–44). As with UL142 recognition of MICA polymorphisms, some RAE-1 isoforms are more resistant to m152-induced retention (43). The large number of host NKG2DLs and viral NKG2DL-evasion proteins are thought to be a result of a continual cycle of evolution and adaptation between virus and host (45).
In addition to its protective role against herpesviruses, NKG2D is known to play an important role in the protection of mice from ectromelia virus, suggesting that NKG2D may also contribute to immunity to other poxviruses (46, 47). In support of this idea, Campbell et al. discovered a protein termed OMCP, uniquely encoded by cowpox and monkeypox viruses (CPXV and MPXV, respectively), which directly binds to the ectodomain of NKG2D (48). OMCP from the Brighton Red strain (CPXV018; GenBank sequence accession no. NP_619807) is a 171-residue protein that is abundantly secreted from infected cells and can block NKG2D-mediated target cell killing by NK cells in vitro (48). While other viral immune evasion strategies target the NKG2DLs, OMCP is the first viral protein shown to directly target NKG2D. This strategy has a significant advantage over targeting the NKG2DLs because while there are many NKG2DLs, the NKG2D receptor itself is monomorphic and is highly conserved across mammals.
While NKG2DLs are divergent in sequence, they are convergent in structure (49–53). The ectodomain of all known NKG2DLs contains a major histocompatibility complex class I-like (MHCI-like) platform domain composed of an antiparallel beta sheet flanked by two antiparallel alpha helices. This conserved structure allows the sequence-diverse NKG2DLs to interact with the NKG2D receptor using a conserved binding interface (54). Because OMCP is a viral antagonist of NKG2D, we tested whether OMCP efficiently competes for NKG2D binding with host NKG2DLs and show that OMCP is the strongest competitor for NKG2D binding. Since OMCP, like NKG2DLs, binds directly to NKG2D, we sought to solve the structure of OMCP to determine whether it made use of a similar overall structure. Here, we describe the 2.25-Å-resolution structure of OMCP from the Brighton Red strain of CPXV (OMCPBR) and show that it adopts a truncated MHCI-like fold related to but distinct from those of other structurally defined NKG2DLs. Evaluation of OMCP binding to NKG2D loop swap mutants, together with the competition of OMCP with host NKG2DLs for NKG2D binding, strongly suggests that OMCP binds the same groove of NKG2D used to recognize host NKG2DLs.
MATERIALS AND METHODS
Cloning, expression, and purification of OMCPBR.
The untagged, mature OMCPBR coding sequence (lacking its 19-residue N-terminal signal sequence) was amplified from genomic DNA and inserted between the XhoI and NdeI sites of the bacterial expression vector pET-21a. Recombinant protein was recovered as inclusion bodies and refolded as previously described (55). Refolded protein was purified by size exclusion chromatography on a Superdex 75 column (16/60; Amersham Biosciences) and eluted as a single monodisperse peak. Protein was further purified by hydrophobic interaction chromatography using a HiTrap Phenyl HP column (GE Healthcare Life Sciences). Fractions containing pure OMCP, verified by SDS-PAGE, were pooled, buffer exchanged to 25 mM HEPES (pH 7.4), and stored at 4°C. Inclusion bodies of selenomethionine-derivative OMCP (SeMet-OMCP) were refolded and purified identically to the native protein.
Protein expression and purification of NKG2DLs and NKG2Ds.
Recombinant proteins for the ectodomains of MULT1, H60-birA, RAE-1ε, RAE-1ε-birA, mNKG2D-birA, mNKhL2-birA, mNKhL3-birA, and mNKHL2+3-birA were expressed in bacteria, recovered as inclusion bodies, and refolded as previously described (55). Briefly, inclusion bodies were denatured in 6 M guanidine-HCl and refolded against 400 mM arginine, 100 mM Trizma, 2 mM EDTA, 5/0.5 mM reduced/oxidized glutathione, and 0.2 mM phenylmethylsulfonyl fluoride (PMSF). Refolded protein was purified by size exclusion chromatography on a Superdex 75 column (16/60; Amersham Biosciences). Recombinant hNKG2D-birA was prepared as described above with an additional purification step over a Q-Sepharose column (Pharmacia) by gradient elution in 30 mM Tris-HCl (pH 8.5), 0 to 500 mM NaCl. Purified hNKG2D-birA eluted from the column at ∼150 mM NaCl.
Crystallization, data collection, and processing.
Native protein crystals were grown by hanging drop vapor diffusion at 20°C from a mixture of 0.5 μl OMCP protein solution at 30 mg/ml in 25 mM HEPES, pH 7.4, with well solution containing 0.9 M ammonium phosphate, pH 5.5. Crystals were cryoprotected with well solution containing 15% ethylene glycol and 1.5 M sodium malonate before being flash cooled directly in a liquid nitrogen bath. SeMet-OMCP crystals were obtained similarly, with a well solution containing 0.75 M ammonium phosphate (pH 5.1), 10 mM trimethylamine HCl, and 100 mM CaCl2 and flash cooled with cryoprotectant containing 333 mM ammonium phosphate (pH 5.1) and 2.8 M sodium malonate (pH 5.5). Diffraction data were collected at the Advanced Light Source synchrotron (beamline 4.2.2). Native OMCP crystal diffraction data were collected at a single wavelength of 0.9790 Å, while SeMet-OMCP crystal diffraction was collected at wavelengths of 0.9792 Å and 0.9641 Å (Table 1). Data processing with HKL2000 showed that the crystals belonged to the primitive hexagonal space group P3221 (space group 154) with unit cell dimensions of a = b = 105.14 Å and c = 108.61 Å (56). These crystals have a solvent content of 62% with three OMCP monomers in the asymmetric unit.
Table 1.
Data collection, phasing, and refinement statistics for OMCP
| Statisticd | Native | High remote | Inflection |
|---|---|---|---|
| Data collection | |||
| Wavelength (Å) | 0.9790 | 0.9641 | 0.9792 |
| Resolution range (Å) | 50–2.25 (2.33–2.25) | 50–2.9 (3.00–2.90) | 50–2.65 (2.74–2.65) |
| Space group | P3221 | P3221 | P3221 |
| Unit cell (Å) | a = b = 105.17, c = 108.66 | a = b = 105.31, c = 109.51 | a = b = 105.35, c = 108.48 |
| Total reflections | 251,334 | 168,403 | 162,234 |
| Unique reflections | 32,729 | 29,903 | 39,189 |
| Redundancy | 7.7 (5.3) | 5.6 (5.1) | 4.1 (3.5) |
| Completeness (%) | 97.60 (88.8) | 100.00 (100.0) | 99.90 (99.5) |
| I/σ (I)a | 30.86 (2.73) | 19.76 (3.51) | 19.09 (2.53) |
| R-mergea | 0.050 (0.486) | 0.088 (0.496) | 0.069 (0.504) |
| Phasing | |||
| Heavy atom sites | 12 | ||
| Phasing power (ano/iso) | 1.1 | 1.4/2.0 | |
| FOM | 0.56 | ||
| Refinement | |||
| Resolution range (Å) | 47.35–2.25 (2.31–2.25) | ||
| R-freeb | 0.251 (0.387) | ||
| No. of atoms | 3,959 | ||
| Protein residues | 450 | ||
| Water molecules | 164 | ||
| RMS (bonds)b | 0.002 | ||
| RMS (angles)b | 0.662 | ||
| Ramachandran favored (%)c | 98 | ||
| Ramachandran outliers (%)c | 0 | ||
| Clash score, all atomsc | 6.78 | ||
| Average B-factor | 50.2 | ||
| Wilson B-factor | 40.7 | ||
| ML-est. coordinate error (Å)b | 0.35 |
Model building and refinement.
An initial phase solution was obtained by multiwavelength anomalous dispersion (MAD) through the autoSHARP server using both wavelengths of SeMet-OMCP diffraction data (59). This initial model was used to provide a solution for the higher-resolution native data set by molecular replacement through Phenix AutoMR (57). Subsequent use of Phenix AutoBuild produced a much-improved model, suitable for manual building. Reiterative refinement and manual rebuilding were performed using phenix.refine and Coot, respectively (57, 60). Both 2Fo-Fc and Fo-Fc maps were used for manual building and to place solvent molecules. The final model yielded an Rfree of 25.1 and an Rwork of 21.4 with 6% of all reflections set aside for free R factor cross-validation. Additional refinement statistics are summarized in Table 1. The final model contains residues His1 to Arg149 (mature protein numbering, after signal sequence removal), with an additional N-terminal glycine included in all copies as part of the bacterial expression construct (Gly0). No density was observed for the last three C-terminal residues (Asn150-Thr151-Gly152). Molecular images were produced using the program PyMOL (61).
MALS.
Multiple-angle light scattering (MALS) experiments with OMCP and OMCP/hNKG2D were performed as previously described (62). Briefly, 100 μg (1 mg/ml) of OMCP or OMCP/hNKG2D was loaded into a gel filtration column in series with a Dawn Helios II multiangle light scattering detector (Wyatt), Optilab rEX (Wyatt), differential refractive index detector, and photodiode array detector 996 (Waters). The data were analyzed with the Astra V macromolecular characterization software (Wyatt).
Surface plasmon resonance (SPR).
A Biacore T100 instrument (GE Health Sciences) was used to assess the competition between OMCP/NKG2DLs and to determine the kinetics of OMCP interaction with mNKG2D/hNKG2D loop swap mutants. All experiments were carried out at a flow rate of 50 μl/min, 25°C, and in running buffer containing 10 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% Triton X-100. CM5 chips with immobilized neutravidin were made as previously described (63).
For competition experiments, 10 nM mNKG2D was preincubated with a titration of concentrations of OMCP, RAE-1ε, or MULT1. The concentration of the three ligands was titrated in 2-fold dilutions from 1.25 to 40 nM. As a negative control for nonspecific inhibition, 10 nM mNKG2D was also preincubated with 80 nM m144, an MHCI-like protein encoded by MCMV that does not bind NKG2D (Protein Data Bank [PDB] code 1PQZ). The maximum binding response for mNKG2D at each concentration was normalized to the mNKG2D binding seen with m144 and then averaged across three separate experiments.
To assess the binding kinetics of the mNKG2D/hNKG2D loop swap mutants, each loop swap mutant was site specifically biotinylated and immobilized on a neutravidin-CM5 chip. OMCP was then injected across different flow cells containing the respective loop swap mutants. Curves were fitted with a 1:1 binding model. SPR experiments with the mNKhL2 and mNKhL2+3 variants indicate apparent mass transport limitations, and therefore, quantitative assessment of disassociation rates is potentially unreliable.
Atomic coordinate accession number.
The atomic coordinates (accession code 4FFE) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics (Rutgers University, New Brunswick, NJ).
RESULTS
Competition of OMCP with host NKG2DLs.
OMCP is an immunomodulatory protein expressed by CPXV and MPXV that directly binds to NKG2D (48). OMCP is highly secreted from infected cells, and previous work has shown that OMCP can inhibit the lysis of NKG2DL-expressing target cells by NK cells (48). However, OMCP also binds to at least one other cellular receptor, FcRL5, in addition to NKG2D (64). We therefore wanted to test the ability of OMCP to compete with host NKG2DLs for NKG2D binding in a cell-free system, excluding the possibility of other receptors cooperating in this interaction.
OMCP binds to both the human and murine NKG2Ds with affinities that are equivalent to or greater than those of all known host NKG2DLs. The reported affinities for hNKG2D with human NKG2DLs MICA, MICB, and ULBP3 are ∼1 μM, while the affinity of OMCP for hNKG2D is ∼0.2 nM, an ∼5,000-fold difference (12, 51). In contrast, murine NKG2DLs bind mNKG2D with affinities ranging from 350 to 730 nM for RAE-1α-δ, 20 to 80 nM for RAE-1ε, and 6 to 20 nM for MULT1 and H60 (12, 63, 65, 66). Like H60 and MULT1, OMCP has an affinity in the low-nanomolar range (48, 65, 67). To stringently test the competition between host NKG2DLs and OMCP, we used high-affinity murine NKG2DLs.
The extracellular domains of birA-tagged H60 and RAE-1ε were refolded from bacterial inclusion bodies. Their birA tags were then site specifically biotinylated with birA ligase. H60 and RAE-1ε were then immobilized on SPR chips coated with neutravidin. To test the ability of OMCP to compete with surface-immobilized NKG2DLs, a constant amount of the mNKG2D extracellular domain was injected across the surface of the SPR chip with increasing amounts of OMCP. As a positive control for NKG2D binding competition, the extracellular domains of RAE-1ε and MULT1 were also tested. As a negative control for NKG2D binding competition, mNKG2D was also injected with an 80 nM concentration of an irrelevant protein, m144. Each injection contained 10 nM mNKG2D with a different concentration of OMCP, MULT1, or RAE-1ε. The concentrations of NKG2L or OMCP ranged from 1.25 nM to 40 nM in 2-fold increments (Fig. 1).
Fig 1.
Competition with immobilized host NKG2DLs for mNKG2D binding. Murine NKG2D preincubated with each concentration of OMCP, MULT1, or RAE-1ε was injected over an SPR chip containing immobilized biotinylated NKG2DLs (H60 or RAE-1ε). Binding was normalized to the amount of binding seen with mNKG2D in the presence of an irrelevant protein. Each data point is the average of 3 experiments, with error bars indicating the standard errors.
As expected, MULT1 and RAE-1ε inhibited mNKG2D binding to the surface-immobilized NKG2DLs. Between the two host ligands, MULT1 (50% inhibitory concentration [IC50] of 10 nM) competed for NKG2D binding more effectively than did RAE-1ε (IC50 of 20 nM), consistent with the higher affinity of MULT1. OMCP also inhibited the binding of mNKG2D to surface-immobilized NKG2DLs. OMCP and MULT1 inhibited mNKG2D with very similar dose responses (IC50s of 8 nM and 10 nM, respectively), demonstrating that OMCP can effectively compete for mNKG2D with the highest-affinity host ligands in the absence of other cellular receptors.
OMCP structure and oligomeric state.
The structures of several low-affinity host NKG2DLs have been solved, but no structure of a high-affinity NKG2DL has been reported. Additionally, OMCP has the unique role of being a secreted viral decoy NKG2DL. To better understand these unique properties, we determined the structure of OMCP. OMCP protein from the Brighton Red strain of CPXV was obtained by refolding bacterial inclusion bodies and crystallized. Diffraction data for native OMCP crystals were collected to 2.25-Å resolution. The phase problem was solved by multiple-wavelength anomalous dispersion (MAD) methods using selenomethionine-labeled OMCP. OMCP crystallized with three molecules in the asymmetric unit. Within the asymmetric unit, chain X contained the lowest overall B factors, and this molecule is displayed throughout this report (Fig. 2A). Continuous electron density was observed in all regions, with the exception of Gln108 in all three monomers, where the side chain protrudes into a crystal solvent channel (Fig. 3A). The final model has excellent geometry and R factors of 21.4% and 25.1% for Rwork and Rfree, respectively (Table 1).
Fig 2.
OMCP structure. (A) Ribbon diagram of CPXV OMCP. Secondary structure elements are noted, S for beta strands and H for helix. The α1/α2 portions of the platform domain are indicated in cyan and magenta, respectively. (B) Ribbon diagram of the α1/α2 domain of MICA (PDB identifier 1HYR), with the α3 domain removed for clarity. Residues that contact NKG2D are colored yellow. (C) Structure alignment of OMCP with NKG2DLs. The mature sequences of OMCPBR (CPXV-BR-018; GenBank accession number NP_619807; PDB identifier 4FFE) and OMCPMPX (MPXV-ZAR_1979_005-198; N3R; GenBank accession number AAY97396) are aligned with the ectodomain sequences of MICA (1HYR), MICB (1JE6), ULBP3 (1KCG), and RAE-1β (1JFM). Known NKG2D contact residues for NKG2DLs are indicated in yellow. Asn residues likely to be glycosylated are noted by black boxes in panel C and as black side chains in panels A and B.
Fig 3.
Assessment of OMCP oligomerization and NKG2D binding stoichiometry. (A) Packing of OMCP monomers within the crystal. OMCP crystallized with three monomers related by 3-fold noncrystallographic symmetry (colored cyan, magenta, and yellow) in the asymmetric unit of the P3221 space group. For the displayed cyan monomer, trimer contacts bury 520 Å2 and 510 Å2 of surface area (interfaces labeled 1 and 2, respectively), while crystal symmetry mate contacts bury 856 Å2 and 737 Å2 (interface 3 and data not shown). The location of Gln108, which lacked continuous electron density, is indicated in the crystal solvent channel. (B) MALS molecular mass assessment of OMCP alone and OMCP bound to hNKG2D. OMCP or OMCP/hNKG2D was first passed through a gel filtration column before assessment by multiple-angle light scattering, which allows for unambiguous mass determination for proteins eluting as monodisperse peaks. Calc, calculated; Exp, experimental; amu, atomic mass units.
The structure of OMCP consists of an MHCI-like α1/α2 platform domain (Fig. 2A). The helix of the OMCP α1 domain (H1) is continuous, while the helix of the α2 domain is broken into two regions (H2a and H2b). The helices flank a six-stranded beta sheet and together form the characteristic platform that defines MHC proteins. Like other NKG2DLs (Fig. 2B), the alpha helices of OMCP are close together and thus have no groove for binding peptides or other ligands like antigen-presenting MHC platform domains. OMCP contains one disulfide bond between S5 and H2b, and this disulfide bond is conserved in most NKG2DLs (Fig. 2C). OMCP has two predicted N-linked glycosylation sites, one in the S1-S2 loop and one in the S5-S6 loop (Fig. 2A and C). Both Asn side chains are exposed to solvent and are likely to be glycosylated when expressed in mammalian cells.
OMCP is significantly divergent in sequence from MHCI-like proteins, with the Basic Local Alignment Search Tool (BLAST) failing to align OMCP with any MHCI molecules (48). However, three-dimensional (3D) superimposition analysis of OMCP using the DALI server showed high similarity of OMCP with both classical and nonclassical MHCI platform domains (68). The top hit was with the human neonatal FcR (3m17), where 136/150 OMCP residues were aligned with 13% sequence identity and a root mean square deviation (RMSD) of 2.7 Å. DALI identified all host NKG2DLs as being related to OMCP, with MICB (2wy3) scoring highest with statistics very similar to those of FcRn, aligning 136/150 OMCP residues with 15% sequence identity and an RMSD of 3.1.
OMCP formed a trimer in the asymmetric unit of the crystal with a total buried surface area of 1,550 Å2 (Fig. 3A). The beta sheet of one OMCP monomer formed the primary contact on one side of the interface, with every beta strand but strand 4 interacting with an adjacent OMCP monomer, burying 520 Å2 (interface 1 in Fig. 3A). The other side of the interface consisted of strand 4 and the loop between strand 4 and H1 and buried 510 Å2 (interface 2 in Fig. 3A). The largely hydrophobic interface suggested that OMCP might be secreted from infected cells as a trimer, although it is important to point out that each monomer buries more surface area in adjacent lattice contacts (856 Å2 and 737 Å2) than in trimer contacts.
While multimeric OMCP could have a higher avidity for NKG2D and thus compete more effectively with host NKG2DLs for NKG2D, crystal lattice contacts provide limited support for such a hypothesis. Therefore, we assessed the solution oligomeric state of OMCP, both alone and in complex with hNKG2D, using multiple-angle light scattering (MALS) (Fig. 3B). Both OMCP alone and OMCP/hNKG2D were resolved as monodisperse peaks, indicating that only one species exists in solution. The experimental molecular masses for OMCP and OMCP/hNKG2D were 20.5 kDa and 51.6 kDa, respectively. These experimental molecular masses are highly consistent with the calculated molecular masses of monomeric OMCP (18.3 kDa) and with a 1:1 binding between OMCP and a NKG2D homodimer (47.0 kDa). Native gel shifts with OMCP titrations also support this binding stoichiometry (data not shown). Together, these data demonstrate that OMCP is a monomer in solution and that a single OMCP binds to a single NKG2D homodimer.
Comparison of OMCP to host NKG2D ligands.
The structures of four host NKG2DLs (MICA, MICB, ULBP3, and RAE-1β) have been determined, either unligated, bound to NKG2D, or both, and OMCP differs prominently in several ways from all of these NKG2DL structures (Fig. 2C) (49–53). The beta sheet of the platform for OMCP has been truncated by two beta strands in the α2 domain, resulting in a six-stranded beta sheet (Fig. 2B). Deviations from an eight-stranded beta sheet are rare for MHCI-like molecules, with the γδ T-cell receptor ligand T22 and MCMV protein m144 being notable examples (69–71). The alpha helices for OMCP have also been truncated compared to those for other NKG2DLs (Fig. 2). Host NKG2DLs have between 6 and 7 turns in H1, 4 and 5 turns in H2a, and 6 and 8 turns in H2b. In contrast, H1, H2a, and H2b of OMCP all have only 4 turns.
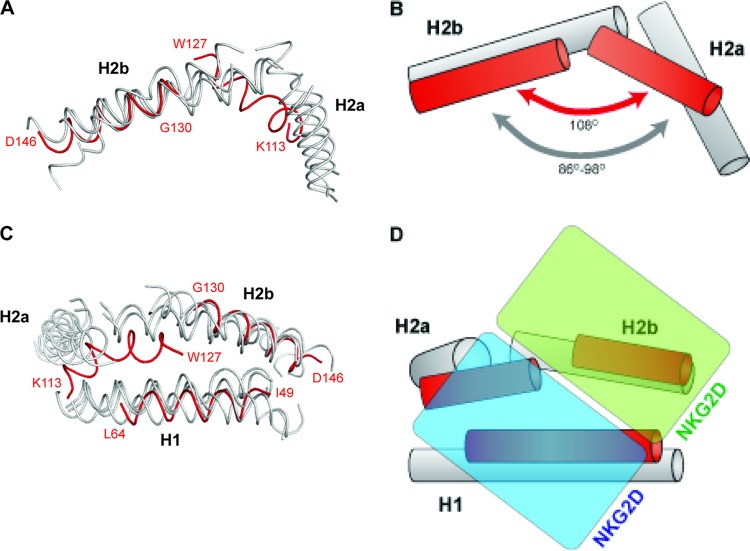
The position of the H2a helix relative to the beta sheet is also unique in OMCP. The α2 helix of NKG2DLs is discontinuous, with H2a and H2b hinged relative to each other. For host ligands, this results in H2a being oriented away from the binding pocket of NKG2D. As a result, H2a is not a part of the binding interface. The angle between H2a and H2b in host NKG2DLs ranges from 86 to 98°. In contrast, the angle between H2a and H2b in OMCP is 108°. This orientation makes the H2a and H2b helices flatter against the beta sheet (Fig. 4A and B). The break between H2a and H2b of OMCP hinges less than other NKG2DLs and instead translates H2a closer to H1 (Fig. 4C and D). Thus, the position of H2a is significantly differently in OMCP than in host NKG2DLs. The combination of shorter helices, a translated H2a helix, and a larger hinge angle between H2a and H2b in OMCP makes the putative binding surface of OMCP for NKG2D overall more compact and flatter than those of host NKG2DLs.
Fig 4.
Comparison of H1, H2a, and H2b between OMCP and host NKG2DLs. (A) Side view of OMCP helices (red) overlaid with host NKG2DL helices (MICA [1HYR[, MICB [1JE6], ULBP3 [1KCG], and RAE-1β [1JFM]) (gray). (B) Cartoon model showing the distinct helical angle for OMCP. (C) Top view of OMCP helices (red) overlaid with host NKG2DL helices (gray). (D) Cartoon model with each half of the NKG2D dimer (cyan and green) shown in the same orientation used to bind host NKG2DLs.
The structures of MICA and ULBP3 bound to human NKG2D and of RAE-1β bound to murine NKG2D have been solved (50, 51, 53). Comparison of the three structures reveals a common paradigm for interaction of NKG2DLs with NKG2D (Fig. 4D). All three ligands primarily contact NKG2D using three elements: the H1 helix, the H2b helix, and the loop between S1 and S2. An alignment of OMCP secondary structure with the host NKG2DLs reveals that OMCP diverges from host ligands in all three of these NKG2D-contacting regions (Fig. 2C).
The S1-S2 loop of OMCP has been reduced to a two-residue turn, likely preventing it from being close enough to interact with NKG2D. Like the S1-S2 loop, the H1 and H2b helices of OMCP have also been truncated. Based upon the alignments, the regions of H1 and H2b in host ligands that make up the majority of NKG2D contacts are missing from OMCP. Of the remaining regions of H1 and H2b of OMCP, the only residue that appears to be similar to a host NKG2DL contact residue is Asp132. An acidic residue is found at the analogous position in the other NKG2DLs, and this residue makes a contact with Tyr199 of NKG2D, one of three critical binding residues of NKG2D (72). The high affinity of OMCP for NKG2D, despite the loss of these regions, suggests that OMCP has found a way to compensate. The reorientation of H2a into a position that could contact NKG2D suggests that H2a may have a role in interacting with NKG2D, unlike in host NKG2DLs (Fig. 4D).
Mapping NKG2D regions required for high-affinity OMCP binding.
CPXV and MPXV are capable of infecting many species and are generally thought to have small-rodent reservoirs (77). Interestingly, despite the NKG2Ds being 70% identical, there is a 14-fold difference between the affinity of OMCP for human NKG2D and that for murine NKG2D (0.20 ± 0.06 nM versus 2.87 ± 0.88 nM). The difference in OMCP affinity between the two NKG2Ds is associated with the 2.3-fold-higher on-rate (4.67 × 106 ± 1.7 × 106 M−1 s−1 versus 10.6 × 106 ± 1.7 × 106 M−1 s−1) and 6.4-fold-shorter half-life (5.8 ± 1.4 min versus 0.9 ± 0.28 min) of OMCP for hNKG2D. Previous experiments with a different Biacore instrument gave very similar results, with the only difference being a lower affinity of OMCP for mNKG2D than that in our experiments (48). To understand the region of NKG2D responsible for this difference in affinity, we designed loop swap mutants between the two NKG2Ds.
While the binding site on NKG2D for OMCP is not known, the similar structure of OMCP compared to those of other NKG2DLs and the competition for binding suggest that OMCP binds to the same surface on NKG2D as do host NKG2DLs. Therefore, we analyzed the cocrystal structures of both hNKG2D and mNKG2D bound to host NKG2DLs. NKG2DLs utilize a discrete surface of NKG2D for binding (Fig. 5A). We therefore assessed all surface-exposed side chains of NKG2D within 4 Å of NKG2DLs and compared the sequences of hNKG2D and mNKG2D at those positions (Fig. 5B to D). NKG2D primarily contacts NKG2DLs using three loops (abbreviated L1, L2, and L3 here), and the sequences of two of the loops differed between hNKG2D and mNKG2D. L2 (183 to 188, hNKG2D numbering) differs in both sequence and position, while L3 (202 to 206) contains sequence differences but limited structural differences (Fig. 5D). To test whether L2 and L3 play a role in the affinity difference between NKG2Ds, the divergent hNKG2D residues were substituted into an mNKG2D backbone (Fig. 5C and D). We chose this combination by reasoning that a reduction in the affinity of hNKG2D could be due to unpredicted problems with the mutations, while a gain in affinity for the lower-affinity mNKG2D would more rigorously test the role that these loops play in OMCP binding.
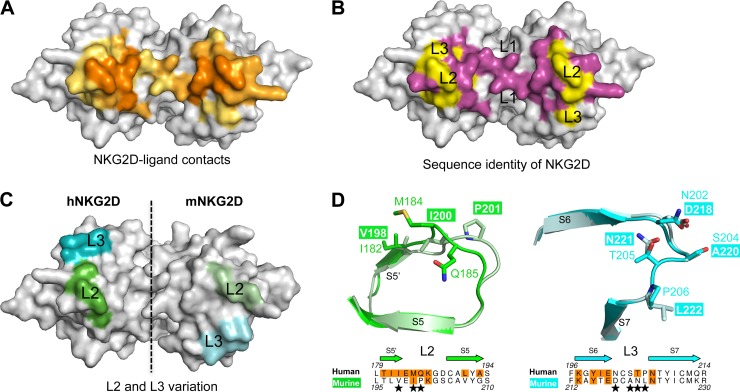
Fig 5.
Generation of NKG2D loop swap mutants. (A) View into the hNKG2D ligand-binding groove. NKG2D contact residues for the three NKG2DLs are colored according to frequency of use. Contacts in pale orange are used by a single NKG2DL, those in medium orange are used by two NKG2DLs, and those in dark orange are used by all NKG2DLs. (B) Conservation of NKG2D contact residues between hNKG2D and mNKG2D. Identical residues are colored magenta, and divergent residues are colored yellow. hNKG2D was used as the model. (C) Space-filling representation of NKG2D with L2 and L3 mutations colored green and cyan, respectively. To facilitate comparison, the left monomer is from hNKG2D and the right monomer is from mNKG2D. (D) Overlays of L2 and L3 between human (dark green/cyan) and murine (light green/cyan) NKG2Ds. The side chains of divergent amino acids are shown. Sequence comparison of mNKG2D and hNKG2D, with NKG2D ligand contact residues in orange and L2/L3 mutations noted by stars.
The affinity of OMCP for hNKG2D, mNKG2D, and the three loop swap mutants (designated mNKhL2 for L2, mNKhL3 for L3, and mNKhL2+3 for substitutions in both loops) was determined by SPR kinetic analysis. Each NKG2D contained a birA tag, which allowed for site-specific biotinylation. OMCP was then injected across each surface, and the on-rates and off-rates were compared (Fig. 6). The binding of OMCP to mNKhL3 had affinity and kinetics equivalent to those of parental mNKG2D. However, mNKhL2 and mNKhL2+3 displayed a low off-rate that is characteristic of hNKG2D, with an on-rate that was not significantly different from that of wild-type mNKG2D. Taken together, L3 does not appear to contribute significantly to the increase in OMCP affinity, while L2 alone is able to increase the off-rate of the OMCP-NKG2D interaction. Additionally, the effect of the loop swap mutations on OMCP binding implicates the NKG2D-host ligand-binding groove as the likely interface for OMCP binding.
Fig 6.
Binding curves of OMCP for NKG2D loop swap mutants. Biotinylated human, murine, and loop swap mutant NKG2Ds were immobilized to a neutravidin-coated SPR chip. OMCP concentrations of 0.16 to 20 nM were injected across each NKG2D. Curves are shown for a representative experiment. Two different batches of OMCP and two independent SPR chip surfaces were used to determine kinetic and affinity constants. Values are the averages of three experiments. *, SPR experiments with the mNKhL2 and mNKhL2+3 variants indicate apparent mass transport limitations, and therefore, quantitative assessment of disassociation rates is potentially unreliable.
DISCUSSION
In this study, we have demonstrated competitive binding of OMCP to NKG2D, defined receptor-ligand binding stoichiometry, and determined the structure of OMCP to 2.25-Å resolution. All known host NKG2DLs contain an MHCI-like platform domain, which is used to bind NKG2D (49–53). We show here that OMCP also is composed of an MHCI-like platform domain, despite low sequence identity with host NKG2DLs and other MHCI molecules. While the types of interactions between individual host NKG2DLs and NKG2D vary, the orientations of NKG2DL platform domains with NKG2D are similar (74). The alpha helices of NKG2DLs are oriented diagonally across the NKG2D binding pocket, creating an asymmetric interaction with the symmetric halves of the NKG2D homodimer (54). H1 and the S1-S2 loop of host NKG2DLs contact one NKG2D monomer, while H2b contacts the second NKG2D monomer. While OMCP is similar to other NKG2DLs in overall organization of its platform domain, it diverges in the specific elements used by NKG2DLs to bind NKG2D.
Each contact site in host ligands (H1, H2b, and the S1-S2 loop) has been shortened in OMCP. Additionally, the position of the H2a helix is dramatically different than that of the H2a helix of host NKG2DLs. The larger hinge angle between H2a and H2b makes the H2a helix flatter relative to the beta sheet than that in host NKG2DLs. This orients H2a into a position that could potentially interact with NKG2D (Fig. 4D). These changes would generate a unique surface to interact with NKG2D.
The most likely interaction orientation between OMCP and NKG2D would be one similar to canonical NKG2DL-NKG2D interactions. However, the truncations in OMCP in the regions analogous to host ligand binding suggest that NKG2D engagement may involve unique determinants. The diagonal orientation of host NKG2DLs across the NKG2D binding groove is thought to be forced by the hinge angle between H2a and H2b (54). The flattening of the helices caused by the increased hinge angle between H2a and H2b in OMCP may allow OMCP to rotate to a greater extent than that of other NKG2DLs. How much OMCP may rotate is unclear, but a change in orientation could bring H2a into contact with NKG2D. This would also potentially increase the buried surface area of the OMCP-NKG2D interaction, which could explain the high affinity of OMCP for NKG2D. Interestingly, Campbell et al. predicted that OMCP would adopt an MHCI-like fold using hidden Markov and structure threading methods (48). While modeling accurately predicted an OMCP deletion located in the S7-S8 region of the beta sheet, the unique helical elements were missed.
In this study, we have structurally characterized OMCP from the Brighton Red strain of CPXV, which exhibits less than 60% sequence identity to the highly conserved sequences (>97% identity) found in 17 other CPXV and MPXV strains (73). Comparing the mature sequences of OMCPBR and of OMCP encoded by MPXV strain Zaire_1979, which also binds NKG2D, indicates that 14/59 sequence differences are localized to the helical regions (H1, H2a, and H2b), with only 4 residues appreciably surface exposed (Ile49Leu, Met115Ser, Thr118Ile, and Met135Ile). Of these four substitutions, only Met135Ile is positioned at the structurally aligned NKG2D binding interface observed for host NKG2DLs (Fig. 2C). Thus, despite significant global sequence differences between OMCPBR and other strains, the conservation of exposed helical residues supports the hypothesis that these regions serve as determinants for NKG2D binding.
The location of the N-linked glycosylation sites in OMCP also supports the helices as the likely interface with NKG2D. Asn12 in the S1-S2 loop is positioned under the platform and is distal from the helices. This glycan site appears conserved in other OMCP variants and in host NKG2DLs, with most having a glycan under the platform domain and near S1. Unlike Asn12, Asn88 does not appear to be conserved with other OMCP variants and NKG2DLs, with the exception of MICA. Asn88 is oriented such that the glycan would be pointed to the side of the platform and away from the proposed interaction site with NKG2D.
The interaction of host ligands with NKG2D is centered on three key NKG2D residues: Tyr152, Met184, and Tyr199 (hNKG2D numbering). The two tyrosine residues are conserved between human and murine NKG2Ds, but Met184 in human NKG2D is changed to Ile200 in murine NKG2D. In addition, substitutions also exist in residues 181, 182, and 185, all of which are contact residues for at least one NKG2DL. Furthermore, the position of these residues is slightly different between the NKG2D homologues, with the backbone of murine NKG2D splaying farther away from the binding groove. This region of NKG2D was abbreviated as L2 in this report, and differences in OMCP affinity between NKG2D homologues map to this region. Together, these data suggest that OMCP makes use of the same core contact residues as do host ligands and that the differences in OMCP affinity for NKG2D homologues likely are due to altered contacts in the L2 loop.
The difference in OMCP affinity for NKG2D homologues is interesting in the context of zoonotic infection. While CPXV and MPXV are thought to have small-rodent reservoirs, it is unclear if the host NKG2D targeted by OMCP is a “mouse-like” or “human-like” NKG2D (75). However, CPXV and MPXV do infect humans, and the incidence of infection with MPXV has risen since the eradication of smallpox (75, 76). Therefore, it is important to understand what the contribution of high-affinity OMCP-hNKG2D binding would be to the pathogenesis of these viruses. The effect of OMCP on human disease is unclear, because the two poxviruses most commonly associated with human infections, variola virus and vaccinia virus, do not encode an OMCP-like protein (48). The role of OMCP in modulating the immune response has yet to be assessed in any in vivo pathogenesis models.
A further complication for understanding the effect of OMCP in pathogenesis arises because OMCP can bind FcRL5, which is expressed by marginal zone B cells, as well as an unidentified binding partner expressed on macrophages (64). Because of the diversity of receptors engaged by OMCP, further research will be required to identify the effect of OMCP binding on each receptor individually and in combination with each other.
ACKNOWLEDGMENTS
We thank Leon N. Carayannopoulos, Jessica A. Campbell, Michel Sun, Kelly Smith, and Anthony R. French for helpful discussions and experimental suggestions.
This work was supported in part by NIH/NIAID R01 AI073552 and U54 AI057160 (MRCE) and the Center for Structural Genomics of Infectious Diseases (CSGID) contract numbers HHSN272200700058C and HHSN272201200026C.
Footnotes
Published ahead of print 31 October 2012
REFERENCES
- 1. Obeidy P, Sharland AF. 2009. NKG2D and its ligands. Int. J. Biochem. Cell Biol. 41:2364–2367 [DOI] [PubMed] [Google Scholar]
- 2. Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, Spies T. 1999. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 285:727–729 [DOI] [PubMed] [Google Scholar]
- 3. Bottino C, Castriconi R, Moretta L, Moretta A. 2005. Cellular ligands of activating NK receptors. Trends Immunol. 26:221–226 [DOI] [PubMed] [Google Scholar]
- 4. Coudert JD, Held W. 2006. The role of the NKG2D receptor for tumor immunity. Semin. Cancer Biol. 16:333–343 [DOI] [PubMed] [Google Scholar]
- 5. Groh V, Rhinehart R, Randolph-Habecker J, Topp MS, Riddell SR, Spies T. 2001. Costimulation of CD8alphabeta T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nat. Immunol. 2:255–260 [DOI] [PubMed] [Google Scholar]
- 6. Lanier LL. 2008. Up on the tightrope: natural killer cell activation and inhibition. Nat. Immunol. 9:495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lodoen MB, Lanier LL. 2006. Natural killer cells as an initial defense against pathogens. Curr. Opin. Immunol. 18:391–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Champsaur M, Lanier LL. 2010. Effect of NKG2D ligand expression on host immune responses. Immunol. Rev. 235:267–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cosman D, Mullberg J, Sutherland CL, Chin W, Armitage R, Fanslow W, Kubin M, Chalupny NJ. 2001. ULBPs, novel MHC class I-related molecules, bind to CMV glycoprotein UL16 and stimulate NK cytotoxicity through the NKG2D receptor. Immunity 14:123–133 [DOI] [PubMed] [Google Scholar]
- 10. Radosavljevic M, Cuillerier B, Wilson MJ, Clement O, Wicker S, Gilfillan S, Beck S, Trowsdale J, Bahram S. 2002. A cluster of ten novel MHC class I related genes on human chromosome 6q24.2-q25.3. Genomics 79:114–123 [DOI] [PubMed] [Google Scholar]
- 11. Schrambach S, Ardizzone M, Leymarie V, Sibilia J, Bahram S. 2007. In vivo expression pattern of MICA and MICB and its relevance to auto-immunity and cancer. PLoS One 2:e518 doi:10.1371/journal.pone.0000518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mistry AR, O'Callaghan CA. 2007. Regulation of ligands for the activating receptor NKG2D. Immunology 121:439–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Raulet DH. 2003. Roles of the NKG2D immunoreceptor and its ligands. Nat. Rev. Immunol. 3:781–790 [DOI] [PubMed] [Google Scholar]
- 14. McGilvray RW, Eagle RA, Watson NF, Al-Attar A, Ball G, Jafferji I, Trowsdale J, Durrant LG. 2009. NKG2D ligand expression in human colorectal cancer reveals associations with prognosis and evidence for immunoediting. Clin. Cancer Res. 15:6993–7002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. O'Sullivan T, Dunn GP, Lacoursiere DY, Schreiber RD, Bui JD. 2011. Cancer immunoediting of the NK group 2D ligand H60a. J. Immunol. 187:3538–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fernandez-Messina L, Ashiru O, Boutet P, Aguera-Gonzalez S, Skepper JN, Reyburn HT, Vales-Gomez M. 2010. Differential mechanisms of shedding of the glycosylphosphatidylinositol (GPI)-anchored NKG2D ligands. J. Biol. Chem. 285:8543–8551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Groh V, Wu J, Yee C, Spies T. 2002. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature 419:734–738 [DOI] [PubMed] [Google Scholar]
- 18. Salih HR, Goehlsdorf D, Steinle A. 2006. Release of MICB molecules by tumor cells: mechanism and soluble MICB in sera of cancer patients. Hum. Immunol. 67:188–195 [DOI] [PubMed] [Google Scholar]
- 19. Salih HR, Rammensee HG, Steinle A. 2002. Cutting edge: down-regulation of MICA on human tumors by proteolytic shedding. J. Immunol. 169:4098–4102 [DOI] [PubMed] [Google Scholar]
- 20. Waldhauer I, Steinle A. 2006. Proteolytic release of soluble UL16-binding protein 2 from tumor cells. Cancer Res. 66:2520–2526 [DOI] [PubMed] [Google Scholar]
- 21. Draghi M, Pashine A, Sanjanwala B, Gendzekhadze K, Cantoni C, Cosman D, Moretta A, Valiante NM, Parham P. 2007. NKp46 and NKG2D recognition of infected dendritic cells is necessary for NK cell activation in the human response to influenza infection. J. Immunol. 178:2688–2698 [DOI] [PubMed] [Google Scholar]
- 22. Dupuy S, Lambert M, Zucman D, Choukem SP, Tognarelli S, Pages C, Lebbe C, Caillat-Zucman S. 2012. Human herpesvirus 8 (HHV8) sequentially shapes the NK cell repertoire during the course of asymptomatic infection and Kaposi sarcoma. PLoS Pathog. 8:e1002486 doi:10.1371/journal.ppat.1002486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pappworth IY, Wang EC, Rowe M. 2007. The switch from latent to productive infection in Epstein-Barr virus-infected B cells is associated with sensitization to NK cell killing. J. Virol. 81:474–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tokuyama M, Lorin C, Delebecque F, Jung H, Raulet DH, Coscoy L. 2011. Expression of the RAE-1 family of stimulatory NK-cell ligands requires activation of the PI3K pathway during viral infection and transformation. PLoS Pathog. 7:e1002265 doi:10.1371/journal.ppat.1002265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ward J, Bonaparte M, Sacks J, Guterman J, Fogli M, Mavilio D, Barker E. 2007. HIV modulates the expression of ligands important in triggering natural killer cell cytotoxic responses on infected primary T-cell blasts. Blood 110:1207–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Welte SA, Sinzger C, Lutz SZ, Singh-Jasuja H, Sampaio KL, Eknigk U, Rammensee HG, Steinle A. 2003. Selective intracellular retention of virally induced NKG2D ligands by the human cytomegalovirus UL16 glycoprotein. Eur. J. Immunol. 33:194–203 [DOI] [PubMed] [Google Scholar]
- 27. Krmpotic A, Hasan M, Loewendorf A, Saulig T, Halenius A, Lenac T, Polic B, Bubic I, Kriegeskorte A, Pernjak-Pugel E, Messerle M, Hengel H, Busch DH, Koszinowski UH, Jonjic S. 2005. NK cell activation through the NKG2D ligand MULT-1 is selectively prevented by the glycoprotein encoded by mouse cytomegalovirus gene m145. J. Exp. Med. 201:211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lenac T, Budt M, Arapovic J, Hasan M, Zimmermann A, Simic H, Krmpotic A, Messerle M, Ruzsics Z, Koszinowski UH, Hengel H, Jonjic S. 2006. The herpesviral Fc receptor fcr-1 down-regulates the NKG2D ligands MULT-1 and H60. J. Exp. Med. 203:1843–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lodoen M, Ogasawara K, Hamerman JA, Arase H, Houchins JP, Mocarski ES, Lanier LL. 2003. NKG2D-mediated natural killer cell protection against cytomegalovirus is impaired by viral gp40 modulation of retinoic acid early inducible 1 gene molecules. J. Exp. Med. 197:1245–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lodoen MB, Abenes G, Umamoto S, Houchins JP, Liu F, Lanier LL. 2004. The cytomegalovirus m155 gene product subverts natural killer cell antiviral protection by disruption of H60-NKG2D interactions. J. Exp. Med. 200:1075–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, Goldman-Wohl D, Greenfield C, Yagel S, Hengel H, Altuvia Y, Margalit H, Mandelboim O. 2007. Host immune system gene targeting by a viral miRNA. Science 317:376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thomas M, Boname JM, Field S, Nejentsev S, Salio M, Cerundolo V, Wills M, Lehner PJ. 2008. Down-regulation of NKG2D and NKp80 ligands by Kaposi's sarcoma-associated herpesvirus K5 protects against NK cell cytotoxicity. Proc. Natl. Acad. Sci. U. S. A. 105:1656–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Byun M, Verweij MC, Pickup DJ, Wiertz EJ, Hansen TH, Yokoyama WM. 2009. Two mechanistically distinct immune evasion proteins of cowpox virus combine to avoid antiviral CD8 T cells. Cell Host Microbe 6:422–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Byun M, Wang X, Pak M, Hansen TH, Yokoyama WM. 2007. Cowpox virus exploits the endoplasmic reticulum retention pathway to inhibit MHC class I transport to the cell surface. Cell Host Microbe 2:306–315 [DOI] [PubMed] [Google Scholar]
- 35. Dunn C, Chalupny NJ, Sutherland CL, Dosch S, Sivakumar PV, Johnson DC, Cosman D. 2003. Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2D ligands, protecting against natural killer cell cytotoxicity. J. Exp. Med. 197:1427–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rolle A, Mousavi-Jazi M, Eriksson M, Odeberg J, Soderberg-Naucler C, Cosman D, Karre K, Cerboni C. 2003. Effects of human cytomegalovirus infection on ligands for the activating NKG2D receptor of NK cells: up-regulation of UL16-binding protein (ULBP)1 and ULBP2 is counteracted by the viral UL16 protein. J. Immunol. 171:902–908 [DOI] [PubMed] [Google Scholar]
- 37. Vales-Gomez M, Browne H, Reyburn HT. 2003. Expression of the UL16 glycoprotein of human cytomegalovirus protects the virus-infected cell from attack by natural killer cells. BMC Immunol. 4:4 doi:10.1186/1471-2172-4-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu J, Chalupny NJ, Manley TJ, Riddell SR, Cosman D, Spies T. 2003. Intracellular retention of the MHC class I-related chain B ligand of NKG2D by the human cytomegalovirus UL16 glycoprotein. J. Immunol. 170:4196–4200 [DOI] [PubMed] [Google Scholar]
- 39. Chalupny NJ, Rein-Weston A, Dosch S, Cosman D. 2006. Down-regulation of the NKG2D ligand MICA by the human cytomegalovirus glycoprotein UL142. Biochem. Biophys. Res. Commun. 346:175–181 [DOI] [PubMed] [Google Scholar]
- 40. Perez-Rodriguez M, Arguello JR, Fischer G, Corell A, Cox ST, Robinson J, Hossain E, McWhinnie A, Travers PJ, Marsh SG, Madrigal JA. 2002. Further polymorphism of the MICA gene. Eur. J. Immunogenet. 29:35–46 [DOI] [PubMed] [Google Scholar]
- 41. Zou Y, Bresnahan W, Taylor RT, Stastny P. 2005. Effect of human cytomegalovirus on expression of MHC class I-related chains A. J. Immunol. 174:3098–3104 [DOI] [PubMed] [Google Scholar]
- 42. Karre K, Ljunggren HG, Piontek G, Kiessling R. 1986. Selective rejection of H-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature 319:675–678 [DOI] [PubMed] [Google Scholar]
- 43. Lanier LL. 2009. DAP10- and DAP12-associated receptors in innate immunity. Immunol. Rev. 227:150–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McFarland BJ, Kortemme T, Yu SF, Baker D, Strong RK. 2003. Symmetry recognizing asymmetry: analysis of the interactions between the C-type lectin-like immunoreceptor NKG2D and MHC class I-like ligands. Structure 11:411–422 [DOI] [PubMed] [Google Scholar]
- 45. Eagle RA, Trowsdale J. 2007. Promiscuity and the single receptor: NKG2D. Nat. Rev. Immunol. 7:737–744 [DOI] [PubMed] [Google Scholar]
- 46. Fang M, Lanier LL, Sigal LJ. 2008. A role for NKG2D in NK cell-mediated resistance to poxvirus disease. PLoS Pathog. 4:e30 doi:10.1371/journal.ppat.0040030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jacoby RO, Bhatt PN, Brownstein DG. 1989. Evidence that NK cells and interferon are required for genetic resistance to lethal infection with ectromelia virus. Arch. Virol. 108:49–58 [DOI] [PubMed] [Google Scholar]
- 48. Campbell JA, Trossman DS, Yokoyama WM, Carayannopoulos LN. 2007. Zoonotic orthopoxviruses encode a high-affinity antagonist of NKG2D. J. Exp. Med. 204:1311–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Holmes MA, Li P, Petersdorf EW, Strong RK. 2002. Structural studies of allelic diversity of the MHC class I homolog MIC-B, a stress-inducible ligand for the activating immunoreceptor NKG2D. J. Immunol. 169:1395–1400 [DOI] [PubMed] [Google Scholar]
- 50. Li P, McDermott G, Strong RK. 2002. Crystal structures of RAE-1beta and its complex with the activating immunoreceptor NKG2D. Immunity 16:77–86 [DOI] [PubMed] [Google Scholar]
- 51. Li P, Morris DL, Willcox BE, Steinle A, Spies T, Strong RK. 2001. Complex structure of the activating immunoreceptor NKG2D and its MHC class I-like ligand MICA. Nat. Immunol. 2:443–451 [DOI] [PubMed] [Google Scholar]
- 52. Li P, Willie ST, Bauer S, Morris DL, Spies T, Strong RK. 1999. Crystal structure of the MHC class I homolog MIC-A, a gammadelta T cell ligand. Immunity 10:577–584 [DOI] [PubMed] [Google Scholar]
- 53. Radaev S, Rostro B, Brooks AG, Colonna M, Sun PD. 2001. Conformational plasticity revealed by the cocrystal structure of NKG2D and its class I MHC-like ligand ULBP3. Immunity 15:1039–1049 [DOI] [PubMed] [Google Scholar]
- 54. Strong RK. 2002. Asymmetric ligand recognition by the activating natural killer cell receptor NKG2D, a symmetric homodimer. Mol. Immunol. 38:1029–1037 [DOI] [PubMed] [Google Scholar]
- 55. Nelson CA, Fremont MD, Sedy JR, Norris PS, Ware CF, Murphy KM, Fremont DH. 2008. Structural determinants of herpesvirus entry mediator recognition by murine B and T lymphocyte attenuator. J. Immunol. 180:940–947 [DOI] [PubMed] [Google Scholar]
- 56. Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Macromol. Crystallogr. A 276:307–326 [DOI] [PubMed] [Google Scholar]
- 57. Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948–1954 [DOI] [PubMed] [Google Scholar]
- 58. Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66:12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vonrhein C, Blanc E, Roversi P, Bricogne G. 2007. Automated structure solution with autoSHARP. Methods Mol. Biol. 364:215–230 [DOI] [PubMed] [Google Scholar]
- 60. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 61. Schrodinger LLC. 2010. The PyMOL molecular graphics system, version 1.3r1. Schrodinger LLC, Portland, OR [Google Scholar]
- 62. Luca VC, AbiMansour J, Nelson CA, Fremont DH. 2012. Crystal structure of the Japanese encephalitis virus envelope protein. J. Virol. 86:2337–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Carayannopoulos LN, Naidenko OV, Kinder J, Ho EL, Fremont DH, Yokoyama WM. 2002. Ligands for murine NKG2D display heterogeneous binding behavior. Eur. J. Immunol. 32:597–605 [DOI] [PubMed] [Google Scholar]
- 64. Campbell JA, Davis RS, Lilly LM, Fremont DH, French AR, Carayannopoulos LN. 2010. Cutting edge: FcR-like 5 on innate B cells is targeted by a poxvirus MHC class I-like immunoevasin. J. Immunol. 185:28–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. O'Callaghan CA, Cerwenka A, Willcox BE, Lanier LL, Bjorkman PJ. 2001. Molecular competition for NKG2D: H60 and RAE1 compete unequally for NKG2D with dominance of H60. Immunity 15:201–211 [DOI] [PubMed] [Google Scholar]
- 66. Schepis D, D'Amato M, Studahl M, Bergstrom T, Karre K, Berg L. 2009. Herpes simplex virus infection downmodulates NKG2D ligand expression. Scand. J. Immunol. 69:429–436 [DOI] [PubMed] [Google Scholar]
- 67. Carayannopoulos LN, Naidenko OV, Fremont DH, Yokoyama WM. 2002. Cutting edge: murine UL16-binding protein-like transcript 1: a newly described transcript encoding a high-affinity ligand for murine NKG2D. J. Immunol. 169:4079–4083 [DOI] [PubMed] [Google Scholar]
- 68. Holm L, Rosenstrom P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38:W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Adams EJ, Chien YH, Garcia KC. 2005. Structure of a gammadelta T cell receptor in complex with the nonclassical MHC T22. Science 308:227–231 [DOI] [PubMed] [Google Scholar]
- 70. Natarajan K, Hicks A, Mans J, Robinson H, Guan R, Mariuzza RA, Margulies DH. 2006. Crystal structure of the murine cytomegalovirus MHC-I homolog m144. J. Mol. Biol. 358:157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wingren C, Crowley MP, Degano M, Chien Y, Wilson IA. 2000. Crystal structure of a gammadelta T cell receptor ligand T22: a truncated MHC-like fold. Science 287:310–314 [DOI] [PubMed] [Google Scholar]
- 72. Hansen TH, Bouvier M. 2009. MHC class I antigen presentation: learning from viral evasion strategies. Nat. Rev. Immunol. 9:503–513 [DOI] [PubMed] [Google Scholar]
- 73. Lefkowitz EJ, Upton C, Changayil SS, Buck C, Traktman P, Buller RM. 2005. Poxvirus Bioinformatics Resource Center: a comprehensive Poxviridae informational and analytical resource. Nucleic Acids Res. 33:D311–D316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Strong RK, McFarland BJ. 2004. NKG2D and related immunoreceptors. Adv. Protein Chem. 68:281–312 [DOI] [PubMed] [Google Scholar]
- 75. Lewis-Jones S. 2004. Zoonotic poxvirus infections in humans. Curr. Opin. Infect. Dis. 17:81–89 [DOI] [PubMed] [Google Scholar]
- 76. Rimoin AW, Mulembakani PM, Johnston SC, Lloyd Smith JO, Kisalu NK, Kinkela TL, Blumberg S, Thomassen HA, Pike BL, Fair JN, Wolfe ND, Shongo RL, Graham BS, Formenty P, Okitolonda E, Hensley LE, Meyer H, Wright LL, Muyembe JJ. 2010. Major increase in human monkeypox incidence 30 years after smallpox vaccination campaigns cease in the Democratic Republic of Congo. Proc. Natl. Acad. Sci. U. S. A. 107:16262–16267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Essbauer S, Pfeffer M, Meyer H. 2010. Zoonotic poxviruses. Vet. Microbiol. 140:229–236 [DOI] [PMC free article] [PubMed] [Google Scholar]