Abstract
Interstrain recombinants were observed in the progenies of the Cucumber mosaic virus (CMV) reassortant L1L2F3 containing RNAs 1 and 2 from LS-CMV and RNA 3 from Fny-CMV. We characterized these recombinants, and we found that their fixation was controlled by the nature of the replicating RNAs 1 and 2. We demonstrate that the 2b gene partially affects this fixation process, but only in the context of homologous RNAs 1 and 2.
TEXT
Single-stranded positive-sense RNA viruses are the most common described plant viruses, indicating their adaptability to a wide variety of hosts and environmental conditions. Several mechanisms underlying their extreme evolutionary capacities have been studied extensively, including processes involved in mutation, recombination, and reassortment. Comparatively less attention has been devoted to understanding the mechanisms underlying the fixation of newly generated variants in the viral population, even though this process constitutes a key aspect of evolutionary dynamics with important implications such as the emergence of drug resistance mutants in human diseases (AIDS, cancers, tuberculosis, etc.).
Cucumber mosaic virus (CMV; genus Cucumovirus, family Bromoviridae) is a well-established model for RNA virus evolution studies (1). CMV strains have been divided into subgroups IA, IB, and II (2, 3). CMV contains a single-stranded positive-sense RNA genome divided into three RNAs, designated RNAs 1, 2, and 3 (2, 4). RNA 1 encodes the 1a protein, which has methyltransferase and helicase domains and which is involved in virus replication (5, 6). RNA 2 encodes the 2a protein, which has a GDD box characteristic of RNA-dependent RNA polymerases and which is also involved in virus replication (5, 7), and the 2b protein, which is involved in virus movement, symptom expression, and suppression of RNA silencing (8, 9). RNA 3 encodes the 3a movement protein and the coat protein, which are required for virus movement and dissemination (10, 11).
Interviral recombinants have arisen from serial passages of a mixture of different strains of CMV in some host plants (12). Several such recombinants described in the literature involved the 3′ nontranslated region (3′ NTR) of RNA 3 of CMV (13–17). In this work, we describe recombinants in the progenies of a virus obtained by reassortment between CMV strains Fny-CMV (subgroup IA) and LS-CMV (subgroup II) and we studied their fixation process in viral populations developed in competition experiments in five host plants.
Plants and viruses are described by our accompanying article (18). Ten 3-week-old Nicotiana benthamiana plants were inoculated with reassortant/recombinant viruses containing recombinant RNA 3 and RNAs 1 and 2 from either Fny- or LS-CMV and, as a competitor, an equal concentration (100 μg/ml) of RNA 3 from either Fny- or LS-CMV. Control plants were inoculated with the buffer only (50 mM Na2HPO4, pH 9).
Total RNA extraction and high-fidelity (HiFi) reverse transcription-PCR (RT-PCR) were as described in our accompanying article (18), except that for the competition experiments, total RNA was isolated from systemically infected leaves at 7, 15, and 21 days postinoculation (dpi). The thermal cycling reactions were carried out as previously described (19) and were optimized to eliminate any recombination artifacts in vitro. Cloning and sequence and clone analyses were also as described in our accompanying article (18) except for the competition experiments; 10 white colonies (instead of 24) were selected from each plate and resuspended in 30 μl of water. Five microliters of the suspension was then used in thermal cycling reactions as previously described (19). PCR products were digested with restriction enzymes BglI and DdeI separately. Competitor RNAs 3 were distinguished by restriction fragment length polymorphism (RFLP).
Strains of Fny- and LS-CMV and the cDNA clones derived from these viruses that are capable of producing infectious RNA transcripts were previously described (20, 21). To obtain infectious clones of the interviral recombinants, total RNAs from plants where they occurred (Table 1) were selected and used for HiFi RT-PCR. After cloning and sequence analysis, the 3′ end recombined fragments of various interviral recombinants were subcloned into XhoI and EcoNI restriction sites of plasmid pFny 309 (22). An EcoNI site was engineered in the 3′ end of the recombined fragment by using primer TGGTCTCCTTTTGGAGGACC in the final thermal cycling reactions in lieu of primer 1. The RNA 2 mutant clones used here were as described elsewhere (18).
Table 1.
Incidence of the recombinants observed in L1L2F3 progenies in five hosts
| Host | Plant | No. of recombinants |
|||||
|---|---|---|---|---|---|---|---|
| Parental F3 | Rec-1 | Rec-2 | Rec-3 | Rec-4 | Rec-5 | ||
| N. benthamiana | 1 | 4 | 6 | 0 | 0 | 0 | 0 |
| 2 | 0 | 13 | 0 | 0 | 0 | 0 | |
| 3 | 0 | 21 | 0 | 0 | 0 | 0 | |
| Pepper | 1 | 0 | 21 | 0 | 0 | 0 | 0 |
| 2 | 0 | 22 | 0 | 0 | 0 | 0 | |
| 3 | 0 | 4 | 0 | 0 | 0 | 16 | |
| Tomato | 1 | 0 | 23 | 0 | 0 | 1 | 0 |
| 2 | 0 | 19 | 0 | 0 | 0 | 0 | |
| 3 | 0 | 6 | 0 | 0 | 17 | 0 | |
| Squash | 1 | 12 | 5 | 0 | 0 | 1 | 0 |
| 2 | 4 | 18 | 0 | 0 | 0 | 0 | |
| 3 | 0 | 23 | 0 | 0 | 0 | 0 | |
| Tobacco | 1 | 0 | 6 | 17 | 1 | 0 | 0 |
| Total | 20 | 187 | 17 | 1 | 19 | 16 | |
We were not surprised to see interviral recombinants in the populations of reassortant viruses, as these have been described previously (12–16, 23). However, recombinants appeared only in the progeny of one of the six reassortants, irrespective of the hosts tested. No recombinant was detected in the progenies of wild types (wt) F1F2F3 and L1L2L3 or in the progenies of reassortants F1L2L3, F1F2L3, F1L2F3, L1F2F3, and L1F2L3, while several recombinants were identified in the progeny of L1L2F3 in all hosts. For each recombinant, part of the 3′ NTR of L2 was inserted in F3.
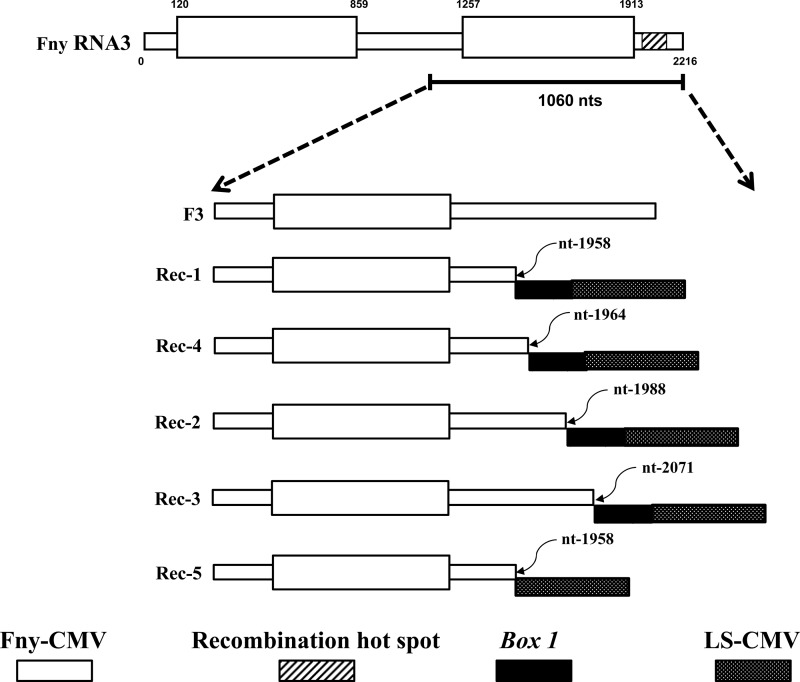
Based on the sizes of the inserted fragments and depending on the insertion sites, we could distinguish several different recombinants (Fig. 1), designated Rec-1 to Rec-5. Also, we noticed that in all five hosts, the interviral recombinants always outcompeted the parental F3, which was lost from the population with time (Table 1). For example, of the 10 clones sequenced from N. benthamiana plant 1, four were parental F3 and six were Rec-1 but the 21 clones sequenced from N. benthamiana plant 3 were all Rec-1, with no parental F3 (Table 1). This indicates that the interviral recombinants have a selective advantage over the parental F3. To further study the mechanisms underlying the fixation of the 3′ NTR recombinants in the viral population, we recloned the recombinant-containing progeny populations to obtain full-length infectious clones using cDNA from the viral RNAs as templates. In the cloning process we discovered some additional recombinants. Their nucleotide sequences were compared to those of the original recombinants, Rec-1 to Rec-5. They fall into four groups depending on their reporter sizes (a reflection of the insert size and position). The first group includes clones a, b, c, and d, of 1,111 nucleotides (nt), which were found to be identical to Rec-1. The second group includes clones h and i, of 1,141 nt, which were identical to Rec-2. Group 3 includes clones e and f, of 1,118 nt, identical to Rec-4. The last group contains clones g, j and k, of various sizes, which were not isolated previously. This suggests that there is a richer pool of interviral recombinants in L1L2F3 progenies. No recombinants identical to Rec-3 or Rec-5 were found in this experiment. Two important characteristics of the infectious clones are the sizes and the nucleotide sequences of the inserts corresponding to the 3′ termini. Except for Rec-5 and clone j, the size of the insert is 309 nt, with the sequence corresponding to the 3′ terminus identical to that of an important motif called Box 1 (17). Recombinants j and Rec-5 do not contain the Box 1 motif.
Fig 1.
Schematic representation of the recombined RNA 3 mutants generated in planta. The white box represents the recombination hot spot in Fny-CMV RNA 3. The insertion site of the recombined fragment is indicated for each recombinant. Box 1 (black box) corresponds to a 21-nucleotide conserved sequence containing the initiation nucleotide for subgenomic RNA 5 of subgroup II strains of CMV lacking an open reading frame (ORF). The stippled boxes represent the LS-CMV RNA 2 3′ NTR.
The clones a, e, g, h, and j were tested for their infectivity (Table 2). In vitro transcripts of RNAs 1 and 2 of Fny-CMV or LS-CMV were mixed with transcripts of the various recombinant RNAs 3 and inoculated into N. benthamiana plants. All the clones tested were infectious except clone j, which is the only one lacking Box 1, suggesting that Box 1 may be indispensable for establishing an infection. The infectivity tests also showed that replacement of LS-CMV RNA 3 by any of the infectious recombinant RNAs increased the virulence of LS-CMV (Table 2): the symptoms appeared earlier and more plants were infected. This was not the case for Fny-CMV, which already displays greater virulence than LS-CMV. The selective advantage of the interviral recombinant RNAs 3 over the parental F3 in L1L2F3 progenies could be due to a better affinity of the replicase L1L2 for the recombined 3′ NTR, which seems to fit even better than the parental L3 3′ NTR.
Table 2.
Infectivity assays in Nicotiana benthamiana
| Inoculum | Box 1 in RNA-3a | Infectivityb | Initiation of symptoms (dpic) |
|---|---|---|---|
| F1F2F3 (parental) | − | 4/4b | 3 |
| F1F2 + a | + | 4/4 | 3 |
| F1F2 + e | + | 2/4 | 3 |
| F1F2 + g | + | 3/4 | 3 |
| F1F2 + h | + | 4/4 | 3 |
| F1F2 + j | − | 0/4 | Not infectious |
| L1L2L3 (parental) | + | 1/4 | 8 |
| L1L2 + a | + | 3/4 | 4 |
| L1L2 + e | + | 2/4 | 4 |
| L1L2 + g | + | 2/4 | 4 |
| L1L2 + h | + | 3/4 | 4 |
| L1L2 + j | − | 0/4 | Not infectious |
−, absent; +, present.
Number of plants infected/number inoculated.
dpi, days postinoculation.
Interviral recombinants were detected only in the progenies of one reassortant, L1L2F3. It is possible that 15 days was not enough time to generate recombinants, as previous studies have shown that this can be a slow process (12, 24), but the rapid generation of recombinants in L1L2F3 suggests that other factors are more important. In addition, the high reproducibility of this asymmetrical recombination event, coupled with the rapid fixation of the recombinants in the viral population, indicates a strong selective advantage of the recombinants in the context of the L1L2 replicase. Thompson et al. (17) showed that the uncapped and noncoding subgenomic RNA 5 that is produced in subgroup II strains of CMV (like LS-CMV), but not in subgroup I strains (like Fny-CMV), has its initiation nucleotide located in a 21-nucleotide conserved sequence named Box 1, which is absent in CMV subgroup I. Interestingly, the recombination event described here took place just one nucleotide upstream of this Box 1 and led to the acquisition of most of the LS-RNA 2 3′ NTR, including the Box 1 motif. Infectivity assays performed for several interviral recombinants (Table 2) also revealed that the recombinants have a slight advantage over the parental LS-CMV RNA 3 and indicate that the acquired Box 1 is required for their infectivity, as clone j, which lacks Box 1, is not infectious. Transcomplementation may explain the survival of the Box 1-negative clones j and Rec-5 in the viral progenies (Table 1).
The 3′ NTRs of CMV and related viruses have been studied extensively (10, 25–27), although the function(s) of RNA 5 is not known. Members of the Bromovirus and Cucumovirus genera have a tRNA-like structure at the 3′ end of their genomic RNAs that interacts with the replicase and is required for minus-strand synthesis (28, 29). Sivakumaran et al. (27) showed that the 3′ NTR of Fny-CMV contains a stem-loop structure similar to that of Brome mosaic virus, which is necessary for the binding of CMV replicase. Therefore, the 3′ NTR of CMV clearly contains the promoter for minus-strand synthesis of the viral RNA during replication, and its primary function involves interaction with the replicase. In addition, phylogenetic analyses have shown that RNA-specific functions (translation, encapsidation) may also be encoded in this region (26). Our data indicate that the 3′ NTR of Fny-CMV RNA 3 was exchanged for that of LS-CMV RNA 2 by recombination, resulting in increased fitness of the newly formed molecule (presumably because it better accommodates the replicase). Others have shown that cucumoviral recombinants outcompete the parental viruses (30). It was suggested that viruses of subgroup II of CMV (like LS-CMV) have obtained their 3′ NTRs from the related Cucumovirus Tomato aspermy virus (TAV), by means of recombination (23). It seems clear that the location of the recombination event that generated the interviral recombinants described here, one nucleotide upstream of Box 1, is of biological significance. De Wispelaere and Rao (31) showed that the first nucleotide of RNA 5 of Q-CMV RNAs 1, 2, and 3 is located two nucleotides upstream of Box 1. We are confident that RNA 2 and not RNA 1 is the donor of the 3′ NTR in the recombination event described here, for the following reasons: (i) the sequence of the 309 nt exchanged is 100% identical to the corresponding fragment in RNA 2 and only 97.7% identical to the corresponding fragment in RNA 1; (ii) the sequence of LS-RNA 1 does not contain the 21-nt conserved motif (Box 1). However, according to de Wispelaere and Rao (31), RNA 5 and not RNA 2 per se is the preferred template for this type of recombination. There is evidence that RNA 5 is recognized by CMV replicase (24, 32). Hence, our system is offering new tools to further study the mechanisms involved in this type of recombination. The lack of RNA 5 in subgroup I strains may explain the lack of recombinants in reassortants with Fny RNA 2, but it cannot explain why strains such as F1L2F3 did not generate recombinants or the differential selection of recombinants in infections with mixtures of RNA 3.
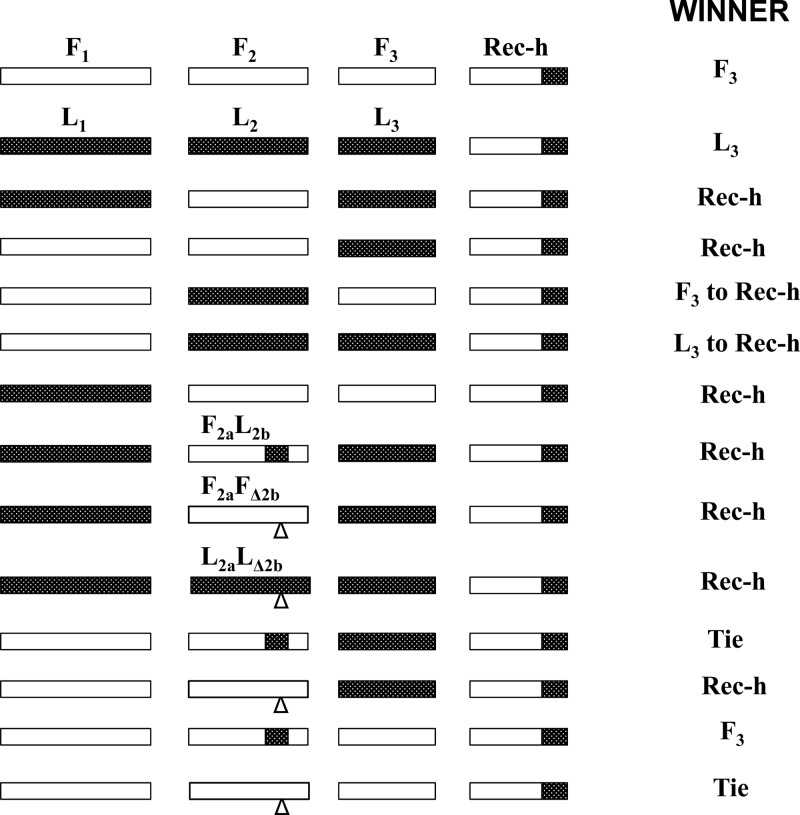
To uncover the mechanisms underlying the fixation phenomenon described here, we conducted experiments involving competition between the interviral recombinant RNA 3 (h) and the parental RNA 3 (L3 or F3) in planta, with various reassortments of RNAs 1 and 2 of Fny- and LS-CMV (Table 3; Fig. 2). Because of its size, clone h is easily distinguishable from the parental F3 or L3.
Table 3.
Competition experiment in Nicotiana benthamiana with various reassortants of Fny- and LS-CMV and with viruses containing deleted or exchanged 2b
| Inoculum | Plant | No. of clones of indicated reassortant/no. of clones identifieda at: |
|||||
|---|---|---|---|---|---|---|---|
| 7 dpi |
15 dpi |
21 dpi |
|||||
| F3 or L3 | h | F3 or L3 | h | F3 or L3 | h | ||
| L1L2L3 + h | 1 | NIb | NI | NI | NI | NI | NI |
| 2 | −c | − | 6/9 | 3/9 | 10/10 | 0/10 | |
| L1F2L3 + h | 1 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 |
| 2 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 | |
| F1F2L3 + h | 1 | 0/9 | 9/9 | 0/9 | 9/9 | 0/10 | 10/10 |
| 2 | 0/10 | 10/10 | 0/8 | 8/8 | 0/9 | 9/9 | |
| F1F2F3 + h | 1 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 |
| 2 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 | |
| F1L2F3 + h | 1 | 8/8 | 0/8 | 10/10 | 0/10 | 2/9 | 7/9 |
| 2 | NI | NI | NI | NI | NI | NI | |
| F1L2L3 + h | 1 | NI | NI | NI | NI | NI | NI |
| 2 | 9/10 | 1/10 | 0/8 | 8/8 | 2/6 | 4/6 | |
| L1F2F3 + h | 1 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 |
| 2 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 | |
| L1F2F3L3 + h | 1 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 |
| 2 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 | |
| F1F2L3F3 + h | 1 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 |
| 2 | 10/10d | 0/10 | 10/10d | 0/10 | 10/10d | 0/10 | |
| L1F2aL2bL3 + h | 1 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 |
| 2 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 | |
| L1F2aFΔ2bL3 + h | 1 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 |
| 2 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 | |
| L1L2aLΔ2bL3 + h | 1 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 |
| 2 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 | |
| F1F2aL2bL3 + h | 1 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 |
| 2 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 | |
| F1F2aFΔ2bL3 + h | 1 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 |
| 2 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 | |
| F1F2aL2bF3 + h | 1 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 |
| 2 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 | |
| F1F2aFΔ2bF3 + h | 1 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 |
| 2 | 0/10 | 10/10 | 0/10 | 10/10 | 0/10 | 10/10 | |
The “winner” is highlighted in grey.
NI, not infected.
−, the RT-PCR is negative.
The RNA 3 identified is F3.
Fig 2.
Schematic representation of the competition experiments. The winner among the competitor RNAs 3 is indicated.
Comparison of L1F2L3 plus h and F1F2L3 plus h, where the recombinant “won” in both cases, indicates that the nature of RNA 1 alone does not affect the selection process and Fny RNA 2 may have better affinity with the interviral recombinant than with L3. However, comparison of L1F2F3 plus h, where the recombinant “wins,” and F1F2F3 plus h, where the wt F3 “wins,” indicates that the source of RNA 1 does control the fixation of the recombinant. Comparison of L1F2F3 plus h and L1F2L3 plus h, where the recombinant “wins” in both cases, indicates that the nature of the competing RNA 3 may not matter in the presence of heterologous RNAs 1 and 2 (L1 and F2). However, F1F2L3 plus h and F1F2F3 plus h indicate the contrary, i.e., that the nature of the competing RNA 3 may influence the selection of the recombinant. In the presence of homologous RNAs 1 and 2 from Fny, F3 is selected over the recombinant RNA 3 while the recombinant is selected over L3. F1F2L3F3 plus h (Table 3) confirms this, as F3 is selected from among the three choices. In general, these sets of competition experiments indicate that the selection/fixation process is controlled by the interactions between the proteins encoded by RNAs 1 and 2 (i.e., the 1a, 2a, and 2b proteins) or between the RNAs 3 themselves.
On the other hand, comparison of L1L2L3 plus h and L1F2L3 plus h indicates that Fny RNA 2 may promote the selection of the interviral recombinant. Also, comparison of F1F2F3 plus h and F1L2F3 plus h corroborates the role of RNA 2 in the selection process: when Fny RNA 2 is replaced by that of LS-CMV, the parental F3 is gradually replaced by the recombinant over time. This is further confirmed by F1L2F3 plus h and F1L2L3 plus h and by F1L2L3 plus h and F1F2L3 plus h: the replacement of F2 by L2 leads to the gradual fixation of the recombinant, irrespective of the wt source of RNA 3. This indicates that the selection process is mainly controlled by the interaction between the replicase complex and the replicating RNA 3. In the presence of the homologous RNAs 1 and 2, L1L2 or F1F2, the cognate RNA 3, L3, or F3 is selected over the recombinant, although with L1L2 the recombinant and L3 seem to be in a tie until late in infection (Table 3). In the presence of the heterologous RNAs 1 and 2, L1F2, or F1L2, the recombinant outcompetes the parental RNA 3. Consequently, we investigated the putative role of the 2b gene in the fixation process described here with various reassortant viruses in which the 2b genes were deleted or exchanged between Fny- and LS-CMV. Comparison of L1F2aL2bL3 plus h, L1F2aF2bL3 plus h, and L1F2aFΔ2bL3 plus h, where the recombinant wins in all cases (Table 3 and Fig. 2) indicates that, in the presence of the heterologous RNAs 1 and 2 (L1F2), the source or even the presence of 2b is not critical for the fixation of the interviral recombinant. However, this is not the case in the presence of homologous RNAs 1 and 2 of L1L2 (L1L2aLΔ2bL3 plus h and L1L2aL2bL3 plus h), where the recombinant won in the presence of L2b and the wt L3 won in the absence of L2b, or F1F2 (F1F2aL2bL3 plus h and F1F2aF2bL3 plus h; F1F2aFΔ2bF3 plus h and F1F2aF2bF3 plus h; F1F2aL2bF3 plus h and F1F2aFΔ2bF3 plus h), where the source or the absence of 2b only partially influences the fixation process. Comparison of F1F2aF2bL3 plus h and F1F2aFΔ2bL3 plus h, where the recombinant wins in both cases, shows that the absence of Fny 2b does not affect the fixation process in the context of RNAs 1 and 2 from Fny-CMV and RNA 3 of LS-CMV. These findings are different from those published by Shi et al. (32). These authors used a similar system in which an interviral recombinant emerged from a reassortant virus with RNAs 1 and 2 from Q-CMV and RNA 3 from TAV, with the 2b gene of TAV (or WAII-CMV) replacing that of Q-CMV. They indicated that the 2b gene controlled the generation and the selection of the interviral recombinants. It is possible that two different mechanisms control the fixation process in these two systems because in our case, a cognate 2b gene (L1L2aL2bF3) is sufficient to generate the recombinants and the fixation is a fairly rapid process, whereas this was not the case in the system described by Shi et al. (32). Indeed, our data clearly show an effect of the 2b gene in the fixation process, but only in the context of homologous RNAs 1 and 2, which was the only context tested by Shi et al. (32). Their findings could have been different had they tested the role of the 2b in a different context with Q1T2 or T1Q2, but these combinations are not viable. The role of 2b in the fixation process described here (in the context of a homologous interaction of RNAs 1 and 2) may be explained by the 2b protein preferentially binding to the cognate parental RNA 3 for cell-to-cell movement.
The results presented here provide insight into the viral factors controlling the fixation of interstrain recombinants in viral populations in planta. We have shown that the increase in relative fitness of the interviral recombinants is associated with a better adaptation to the replicase complex. This could be a mechanism used by interviral recombinants to evolve into new species in viruses with segmented genomes, a process that is probably a major driving force in deep evolution of RNA viruses (33).
ACKNOWLEDGMENTS
This work was supported by the Samuel Roberts Noble Foundation and by the Pennsylvania State University College of Agricultural Sciences.
We thank Luis Márquez and Xiaodong Bao for careful reading of the manuscript.
Footnotes
Published ahead of print 31 October 2012
REFERENCES
- 1. Roossinck MJ. 2001. Cucumber mosaic virus, a model for RNA virus evolution. Mol. Plant Pathol. 2:59–63 [DOI] [PubMed] [Google Scholar]
- 2. Palukaitis P, García-Arenal F. 2003. Cucumoviruses. Adv. Virus Res. 62:241–323 [DOI] [PubMed] [Google Scholar]
- 3. Roossinck MJ, Zhang L, Hellwald K-H. 1999. Rearrangements in the 5′ nontranslated region and phylogenetic analyses of cucumber mosaic virus RNA 3 indicate radial evolution of three subgroups. J. Virol. 73:6752–6758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Palukaitis P, Roossinck MJ, Dietzgen RG, Francki RIB. 1992. Cucumber mosaic virus. Adv. Virus Res. 41:281–348 [DOI] [PubMed] [Google Scholar]
- 5. Hayes RJ, Buck KW. 1990. Complete replication of a eukaryotic virus RNA in vitro by a purified RNA-dependent RNA polymerase. Cell 63:363–368 [DOI] [PubMed] [Google Scholar]
- 6. Nitta N, Takanami Y, Kuwata S, Kubo S. 1988. Inoculation with RNAs 1 and 2 of cucumber mosaic virus induces viral RNA replicase activity in tobacco mesophyll protoplasts. J. Gen. Virol. 69:2695–2700 [Google Scholar]
- 7. Nitta N, Masuta C, Kuwata S, Takanami Y. 1988. Comparative studies on the nucleotide sequence of cucumber mosaic virus RNA3 between Y strain and Q. strain. Ann. Phytopathol. Soc. Jpn. 54:516–522 [Google Scholar]
- 8. Brigneti G, Voinnet O, Li W-X, Ji L-H, Ding S-W, Baulcombe DC. 1998. Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J. 17:6739–6746 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9. Shi B-J, Miller J, Symons RH, Palukaitis P. 2003. The 2b protein of cucumoviruses has a role in promoting the cell-to-cell movement of pseudorecombinant viruses. Mol. Plant Microbe Inteact. 16:261–267 [DOI] [PubMed] [Google Scholar]
- 10. Boccard F, Baulcombe D. 1993. Mutational analysis of cis-acting sequences and gene function in RNA3 of cucumber mosaic virus. Virology 193:563–578 [DOI] [PubMed] [Google Scholar]
- 11. Suzuki M, Kuwata S, Kataoka J, Masuta C, Nitta N, Takanami Y. 1991. Functional analysis of deletion mutants of cucumber mosaic virus RNA3 using an in vitro transcription system. Virology 183:106–113 [DOI] [PubMed] [Google Scholar]
- 12. Masuta C, Ueda S, Suzuki M, Uyeda I. 1998. Evolution of a quadripartite hybrid virus by interspecific exchange and recombination between replicase components of two related tripartite RNA viruses. Proc. Natl. Acad. Sci. U. S. A. 95:10487–10492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bonnet J, Fraile A, Sacristán S, Malpica JM, GarcíA-Arenal F. 2005. Role of recombination in the evolution of natural populations of Cucumber mosaic virus, a tripartite RNA plant virus. Virology 332:359–368 [DOI] [PubMed] [Google Scholar]
- 14. Canto T, Choi SK, Palukaitis P. 2001. A subpopulation of RNA 1 of Cucumber mosaic virus contains 3′ termini originating from RNAs 2 or 3. J. Gen. Virol. 82:941–945 [DOI] [PubMed] [Google Scholar]
- 15. Chen Y-K, Goldbach R, Prins M. 2002. Inter- and intramolecular recombinations in the Cucumber mosaic virus genome related to adaptation to alstroemeria. J. Virol. 76:4119–4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morroni M, Thompson JR, Tepfer M. 2009. Analysis of recombination between viral RNAs and transgene mRNA under conditions of high selection pressure favour recombinants. J. Gen. Virol. 90:2798–2807 [DOI] [PubMed] [Google Scholar]
- 17. Thompson JR, Buratti E, Wispelaere M, Tepfer M. 2008. Structural and functional characterization of the 5′ region of subgenomic RNA5 of cucumber mosaic virus. J. Gen. Virol. 89:1729–1738 [DOI] [PubMed] [Google Scholar]
- 18. Pita JS, Roossinck MJ. 2013. Mapping viral functional domains for genetic diversity in plants. J. Virol. 87:790–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pita JS, deMiranda JR, Schneider WL, Roossinck MJ. 2007. Environment determines fidelity for an RNA virus replicase. J. Virol. 81:9072–9077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rizzo TM, Palukaitis P. 1990. Construction of full-length cDNA clones of cucumber mosaic virus RNAs 1,2 and 3: generation of infectious RNA transcripts. Mol. Gen. Genet. 222:249–256 [DOI] [PubMed] [Google Scholar]
- 21. Zhang L, Hanada K, Palukaitis P. 1994. Mapping local and systemic symptom determinants of cucumber mosaic cucumovirus in tobacco. J. Gen. Virol. 75:3185–3191 [DOI] [PubMed] [Google Scholar]
- 22. Roossinck MJ, Kaplan I, Palukaitis P. 1997. Support of a cucumber mosaic virus satellite RNA maps to a single amino acid proximal to the helicase domain of the helper virus. J. Virol. 71:608–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thompson JR, Tepfer M. 2009. The 3′ untranslated region of cucumber mosaic virus (CMV) subgroup II RNA3 arose by interspecific recombination between CMV and tomato aspermy virus. J. Gen. Virol. 90:2293–2298 [DOI] [PubMed] [Google Scholar]
- 24. Suzuki M, Hiba T, Masuta C. 2003. RNA recombination between cucumoviruses: possible role of predicted stem-loop strutures and an internal subgenomic promoter-like motif. Virology 306:77–86 [DOI] [PubMed] [Google Scholar]
- 25. Rao ALN, Grantham GL. 1994. Amplification in vivo of brome mosaic virus RNAs bearing 3′ noncoding region from cucumber mosaic virus. Virology 204:478–481 [DOI] [PubMed] [Google Scholar]
- 26. Roossinck MJ. 2002. Evolutionary history of Cucumber mosaic virus deduced by phylogenetic analyses. J. Virol. 76:3382–3387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sivakumaran K, Bao Y, Roossinck MJ, Kao CC. 2000. Recognition of the core RNA promoter for minus-strand RNA synthesis by the replicase of Brome mosaic virus and Cucumber mosaic virus. J. Virol. 74:10323–10331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ahlquist P, Dasgupta R, Kaesberg P. 1981. Near identity of the 3′ RNA secondary structure in bromoviruses and cucumber mosaic virus. Cell 23:183–189 [DOI] [PubMed] [Google Scholar]
- 29. Chapman MR, Kao CC. 1999. A minimal RNA promoter for minus-strand RNA synthesis by the brome mosaic virus polymerase. J. Mol. Biol. 286:709–720 [DOI] [PubMed] [Google Scholar]
- 30. Fernández-Cuartero B, Burgyán J, Aranda MA, Salánki K, Moriones E, García-Arenal F. 1994. Increase in the relative fitness of a plant virus RNA associated with its recombinant nature. Virology 203:373–377 [DOI] [PubMed] [Google Scholar]
- 31. de Wispelaere M, Rao ALN. 2009. Production of cucumber mosaic virus RNA5 and its role in recombination. Virology 384:179–191 [DOI] [PubMed] [Google Scholar]
- 32. Shi B-J, Symons RH, Palukaitis P. 2008. The cucumovirus 2b gene drives selection of inter-viral recombinants affecting the crossover site, the acceptor RNA and the rate of selection. Nuc Acids Res. 36:1057–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roossinck MJ. 2005. Symbiosis versus competition in the evolution of plant RNA viruses. Nat. Rev. Microbiol. 3:917–924 [DOI] [PubMed] [Google Scholar]