Abstract
In the human hepatoma cell line HepG2, retinoic acid, clofibric acid, and bile acid treatment can only modestly increase hepatitis B virus (HBV) biosynthesis. Utilizing the human embryonic kidney cell line 293T, it was possible to demonstrate that the retinoid X receptor α (RXRα) plus its ligand can support viral biosynthesis independently of additional nuclear receptors. In addition, RXRα/peroxisome proliferator-activated receptor α (PPARα) and RXRα/farnesoid X receptor α (FXRα) heterodimeric nuclear receptors can also mediate ligand-dependent HBV transcription and replication when activated by clofibric acid and bile acid, respectively, independently of a requirement for the ligand-dependent activation of RXRα. These observations indicate that there are at least three possible modes of ligand-mediated activation of HBV transcription and replication existing within hepatocytes, suggesting that multiple independent mechanisms control viral production in the livers of infected individuals.
INTRODUCTION
Hepatitis B virus (HBV) infection is primarily restricted to hepatocytes in the liver. This restriction is believed to occur at two distinct levels (1). The receptor(s) involved in viral entry is presumably present only on hepatocytes and governs species specificity (2). In addition, viral biosynthesis is restricted in a tissue- and cell-type-specific manner because HBV transcription is dependent on liver-enriched transcription factors (3, 4). A variety of nuclear receptors have been shown to regulate HBV pregenomic 3.5-kb RNA synthesis and hence viral replication (5–7). Three of these nuclear receptors, retinoid X receptor (RXR), peroxisome proliferator-activated receptor (PPAR), and farnesoid X receptor (FXR), are ligand-dependent transcription factors that are activated by retinoids, peroxisome proliferators, and bile acids, respectively (8, 9). Therefore, it is apparent that the ligands for these nuclear receptors might be critical determinants of viral biosynthesis under both normal and pathophysiological conditions within the livers of infected individuals (10, 11).
As the ligand-activated heterodimeric nuclear receptors RXRα/PPARα and RXRα/FXRα regulate HBV pregenomic RNA synthesis by the recruitment of coactivators, it was of interest to evaluate the relative contributions of the individual heterodimeric partners to the overall level of viral transcription and replication (5, 6, 12). Characterization of the relative roles of individual polypeptides in the transcriptional activity of various heterodimeric nuclear receptors has been evaluated (13–15). This approach indicated that one partner might play a dominant role in controlling promoter activity depending on the nuclear receptors involved (13–15). Therefore, it was of interest to evaluate the effects of retinoids, peroxisome proliferators, and bile acids, alone or in combination, on HBV transcription and replication.
In the current study, it is demonstrated that retinoids can activate HBV biosynthesis utilizing both RXRα-containing homodimers and heterodimers. Alternatively HBV transcription and replication can be supported by RXRα/PPARα and RXRα/FXRα in the absence of retinoids when these heterodimeric transcription factors are activated by peroxisome proliferators and bile acids, respectively. Additionally, it appears that the activation of both heterodimeric partners does not dramatically enhance the level of HBV transcription and replication compared to the level of viral biosynthesis observed with a single nuclear receptor ligand. These observations suggest that a single ligand can efficiently activate the heterodimeric nuclear receptors governing HBV biosynthesis independently of the ligand binding status of its partner. This indicates that the ligand-induced conformational changes occurring in one polypeptide that are necessary for coactivator recruitment can occur independently of its heterodimer partner or that the conformational change in the ligand-bound polypeptide is concurrently induced in the heterodimeric partner without its requirement to bind the ligand (15). If nuclear receptor antagonists are to be considered potential antiviral agents for the treatment of chronic HBV infections, it will be critical to distinguish between these different mechanisms of action so appropriate therapeutic modalities might be considered.
MATERIALS AND METHODS
Plasmid constructions.
The HBV DNA (4.1-kbp) construct that contains 1.3 copies of the HBV genome includes the viral sequence from nucleotide coordinates 1072 to 3182 plus 1 to 1990 (4).
The pRS-hRXRα, pCMV-rFXRα, pCMVPPARα-G, pCMX-hRARα, pCMX-mPXR.1, pCMX-mLXRα, and pCMX-hCAR vectors express RXRα, FXRα, PPARα-G, retinoic acid receptor α (RARα), pregnane X receptor 1 (PXR.1), liver (oxysterol) X receptor α (LXRα), and constitutive androstane receptor (CAR) polypeptides from the human RXRα, rat FXRα, mouse PPARα-G, human RARα, mouse PXR.1, mouse LXRα, and human CAR cDNAs, respectively, using the Rous sarcoma virus long terminal repeat (LTR) (pRS) or the cytomegalovirus (CMV) immediate-early promoter (pCMV and pCMX) (16–22). The PPARα-G polypeptide contains a mutation in the PPARα cDNA changing Glu282 to Gly, which may decrease the affinity of the receptor for the endogenous ligand. Consequently, this mutation increases the peroxisome proliferator-dependent (i.e., clofibric acid-dependent) activation of transcription from a peroxisome proliferator response element (PPRE)-containing promoter (21).
Cells and transfections.
The human hepatoma HepG2 cell line and human embryonic kidney 293T cell line were grown in RPMI 1640 medium and 10% fetal bovine serum at 37°C in air containing 5% CO2. Transfections for viral RNA and DNA analysis were performed as previously described (23) using 10-cm plates containing approximately 1 × 106 cells. DNA and RNA isolation was performed 3 days posttransfection. The transfected DNA mixture was composed of 5 μg of HBV DNA (4.1 kbp) plus 1.5 to 6.5 μg of the nuclear receptor expression vectors pRS-hRXRα, pCMV-rFXRα, pCMVPPARα-G, pCMX-hRARα, pCMX-mPXR.1, pCMX-mLXRα, and pCMX-hCAR (16–22). Controls were derived from cells transfected with HBV DNA and the expression vectors lacking a nuclear receptor cDNA insert (24). All-trans-retinoic acid, 9-cis-retinoic acid, chenodeoxycholic acid, and clofibric acid at 0.05 to 10 μM, 0.05 to 10 μM, 100 μM, and 1 mM, respectively, were used to activate the nuclear receptors RXRα, FXRα, and PPARα (4, 17, 25).
Characterization of HBV transcripts and viral replication intermediates.
Transfected cells from a single plate were divided equally and used for the preparation of total cellular RNA and viral DNA replication intermediates as described previously (26) with minor modifications. RNA (Northern) and DNA (Southern) filter hybridization analyses were performed using 10 μg of total cellular RNA and 30 μl of viral DNA replication intermediates, respectively, as described previously (27). Filter hybridization analyses were quantified by phosphorimaging using a Packard Cyclone storage phosphor system.
RESULTS
Retinoids, peroxisome proliferators, and bile acids modulate HBV biosynthesis in human hepatoma HepG2 cells.
Transfection of the HBV DNA (4.1-kbp) construct into HepG2 cells supports HBV transcription and replication (Fig. 1A and B, lane 1). Treatment of HepG2 cells with retinoic acid, clofibric acid, and chenodeoxycholic acid modestly enhanced the level of HBV biosynthesis (Fig. 1A and B, lanes 2, 5, and 8), suggesting that these hepatoma cells express RXR (and/or RAR), PPAR, and FXR that are capable of being activated in the presence of an exogenously added ligand. Exogenous expression of RXRα, PPARα, and FXRα in the absence of added ligands had a limited effect on HBV biosynthesis (Fig. 1A and B, lanes 3, 6, and 9). Exogenous expression of RXRα, PPARα, and FXRα in the presence of added ligands resulted in a level of viral replication similar to that seen with ligand alone (Fig. 1B, lanes 4, 7, and 10). Although all of these effects in HepG2 cells are relatively modest, they indicate that HepG2 cells contain excess inactive RXR (and/or RAR), PPAR, and FXR that are capable of enhancing HBV biosynthesis if activated by the appropriate ligand. In addition, it appears that ligand-activated PPAR or FXR might be able to enhance HBV biosynthesis in the absence of ligand-activated RXR. However, due to the modest nature of the effects in HepG2 cells (Fig. 1C), it is not possible to conclusively determine the relative roles of the various nuclear receptors and their ligands in HBV biosynthesis without examining their effects in a more tractable system.
Fig 1.
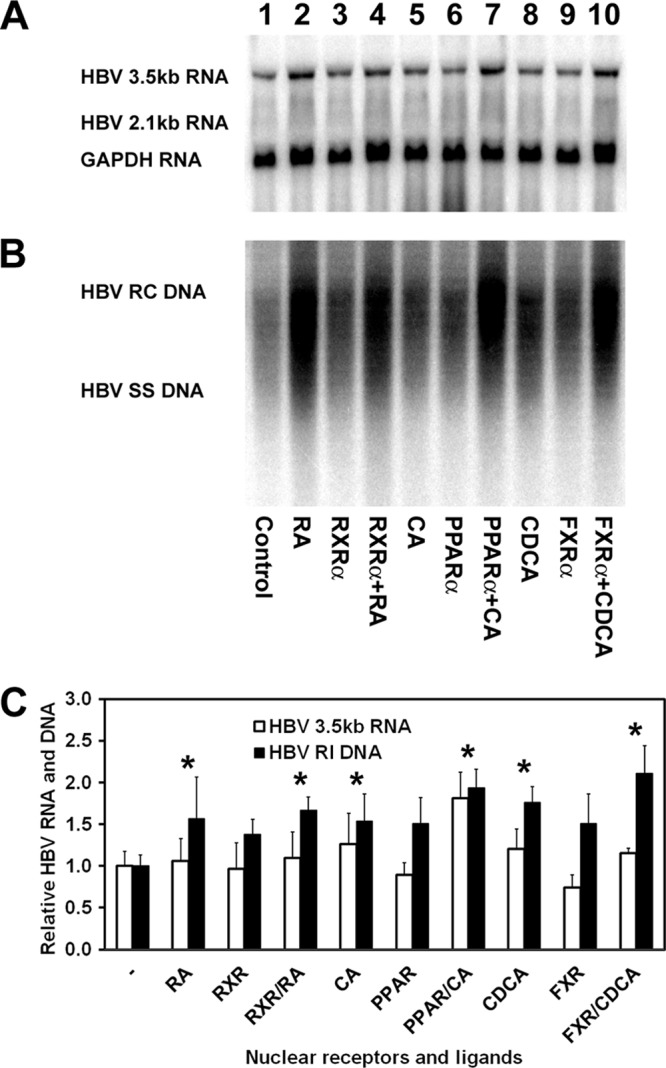
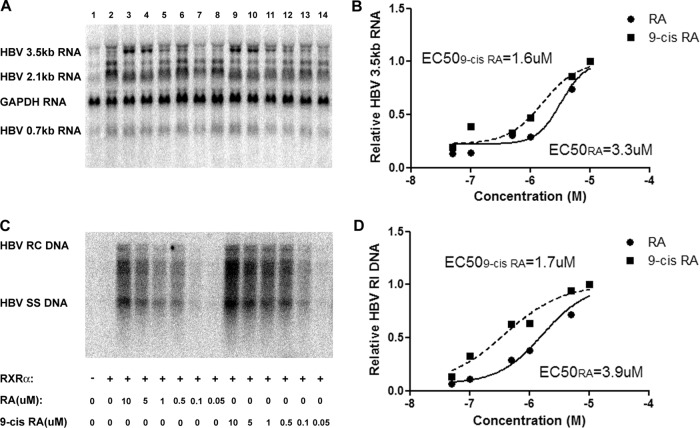
Effects of retinoids, peroxisome proliferators, and bile acids on HBV biosynthesis in the human hepatoma cell line HepG2. (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. Cells were transfected with the HBV DNA (4.1-kbp) construct alone (lane 1) or the HBV DNA (4.1-kbp) construct plus the RXRα (lanes 3 and 4), PPARα (lanes 6 and 7), and FXRα (lanes 9 and 10) expression vectors as indicated. In addition, cells were treated with 1 μM retinoic acid (RA; lanes 2 and 4), 1 mM clofibric acid (CA; lanes 5 and 7), and 100 mM chenodeoxycholic acid (CDCA; lanes 8 and 10) to activate the RXRα, PPARα, and FXRα nuclear receptors, respectively. (C) Quantitative analysis of the HBV 3.5-kb RNA and HBV DNA replication intermediates. The levels of the HBV 3.5-kb RNA and total HBV DNA replication intermediates (RI) relative to that of the HBV DNA (4.1-kbp) construct in the absence of nuclear receptor expression or ligand (left bars) are reported. The mean RNA and DNA levels plus standard deviations from four independent analyses are shown. The levels of the transcripts and replication intermediates in the nuclear receptor and/or ligand-treated cells which are statistically significantly higher than their levels in the corresponding untreated cells by Student's t test (P < 0.05) are indicated with asterisks.
Ligand-activated RXRα supports HBV biosynthesis in human embryonic kidney 293T cells.
Transfection of the HBV DNA (4.1-kbp) construct with the RXRα expression vector into 293T cells fails to support HBV transcription and replication in the absence of a ligand (Fig. 2A and C, lanes 1 and 2). Activation of RXRα by all-trans-retinoic acid, presumably by its isomerization to 9-cis-retinoic acid (17, 25), or 9-cis-retinoic acid treatment of the 293T cells resulted in a dose-dependent induction of both HBV RNA and DNA synthesis (Fig. 2). The half-maximal induction of HBV biosynthesis was observed at approximately 1.6 μM 9-cis-retinoic acid and 3.6 μM all-trans-retinoic acid (Fig. 2). The relatively high concentrations of retinoids required to activate RXRα suggest that RAR, which can be activated by both all-trans- and 9-cis-retinoic acid (28–31), is probably not the RXRα heterodimer partner involved in mediating HBV transcription and replication in 293T cells. Additionally the requirement for high concentrations of 9-cis-retinoic acid, the natural ligand for RXRα (17, 25), suggests that the intracellular concentration of this retinoic acid isomer must be relatively low, presumably due to the activity of endogenous isomerases within the 293T cells (see Discussion for details). Therefore, these observations suggest that RXRα activates HBV biosynthesis either as a homodimer or as a heterodimer with PPARα or FXRα, although the latter suggestion seems less likely given the embryonic origin of the 293T cell line (32).
Fig 2.
Effect of all-trans-retinoic acid and 9-cis-retinoic acid concentrations on HBV biosynthesis in the human embryonic kidney cell line 293T expressing RXRα. Cells were transfected with the HBV DNA (4.1-kbp) construct alone (lane 1) or the HBV DNA (4.1-kbp) construct plus the RXRα expression vector (lanes 2 to 14) as indicated. Cells were treated with various concentrations of all-trans-retinoic acid (0.05 to 10 μM RA; lanes 3 to 8) and 9-cis-retinoic acid (0.05 to 10 μM 9-cis-RA; lanes 9 to 14). (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B) Quantitative analysis of the HBV 3.5-kb RNA from two independent experiments. Trend lines were calculated using GraphPad Prism 5 software to determine the sigmoidal dose response (variable slope) curve plus 50% effective concentrations (EC50s). (C) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. (D) Quantitative analysis of the HBV replication intermediates from two independent experiments. Trend lines were calculated using GraphPad Prism 5 software to determine the sigmoidal dose response (variable slope) curve plus EC50s.
Selective effects of specific nuclear receptors on ligand-activated RXRα-mediated HBV biosynthesis in human embryonic kidney 293T cells.
Unlike the situation in HepG2 cells, transfection of the HBV DNA (4.1-kbp) construct into 293T cells in the presence of retinoic acid fails to support HBV transcription and replication (Fig. 3A and B, lanes 1 and 2). In addition, the exogenous expression of RXRα also failed to support HBV biosynthesis in the absence of retinoic acid in 293T cells (Fig. 3A and B, lane 3). However, the exogenous expression of RXRα in the presence of retinoic acid is sufficient to support robust HBV transcription and replication (Fig. 3A and B, lane 4). To address the potential role additional nuclear receptors might have in governing HBV biosynthesis in 293T cells, several potential heterodimer partners were overexpressed in the presence of a constant amount of coexpressed RXRα polypeptide (Fig. 3A and B, lanes 5 to 11). Overexpression of RXRα by transfecting approximately 4-fold more of the RXRα expression vector did not greatly modulate HBV biosynthesis (Fig. 3A and B, lanes 4 and 5). Comparable overexpression of FXRα, PPARα, LXRα, and CAR did not greatly modulate RXRα-mediated HBV biosynthesis (Fig. 3A and B, lanes 7, 8, 10, and 11), suggesting that in the absence of their ligands these nuclear receptors did not affect viral transcription and replication (Fig. 3C). Interesting, overexpression of RARα inhibited RXRα-mediated HBV biosynthesis (Fig. 3A and B, lane 6). This observation strongly suggests that the RXRα/RARα heterodimer does not activate HBV biosynthesis, and therefore it appears likely that RXRα can directly activate HBV transcription and replication in the absence of any additional ligand-activated nuclear receptors. The mechanism(s) of inhibition of HBV biosynthesis by RARα and PXR is unclear (Fig. 3A and B, lanes 6 and 9). It is possible that these nuclear receptors might shift the cellular equilibrium from RXRα homodimers capable of activating HBV transcription to RXRα/RARα or RXRα/PXR heterodimers that are incapable of binding to the nucleocapsid promoter to activate HBV transcription. This equilibrium shift would reduce HBV biosynthesis, but then it is difficult to rationalize why LXRα and CAR would not affect viral RNA and DNA synthesis similarly. In the case of the RXRα/RARα heterodimer, it is likely that binding to the direct repeat 1 (DR1) sequence comprising two copies of the AGGTCA-related sequence separated by a single nucleotide within the HBV nucleocapsid promoter mediates corepressor recruitment and hence transcriptional repression (24, 30, 31). In contrast, RXR/PXR does not bind to DR1 sequences (18, 33) but might sequester coactivators, such as peroxisome proliferator-activated receptor γ coactivator 1α (PGC1α), that can activate nuclear receptor-mediated HBV biosynthesis (5, 6, 34, 35).
Fig 3.
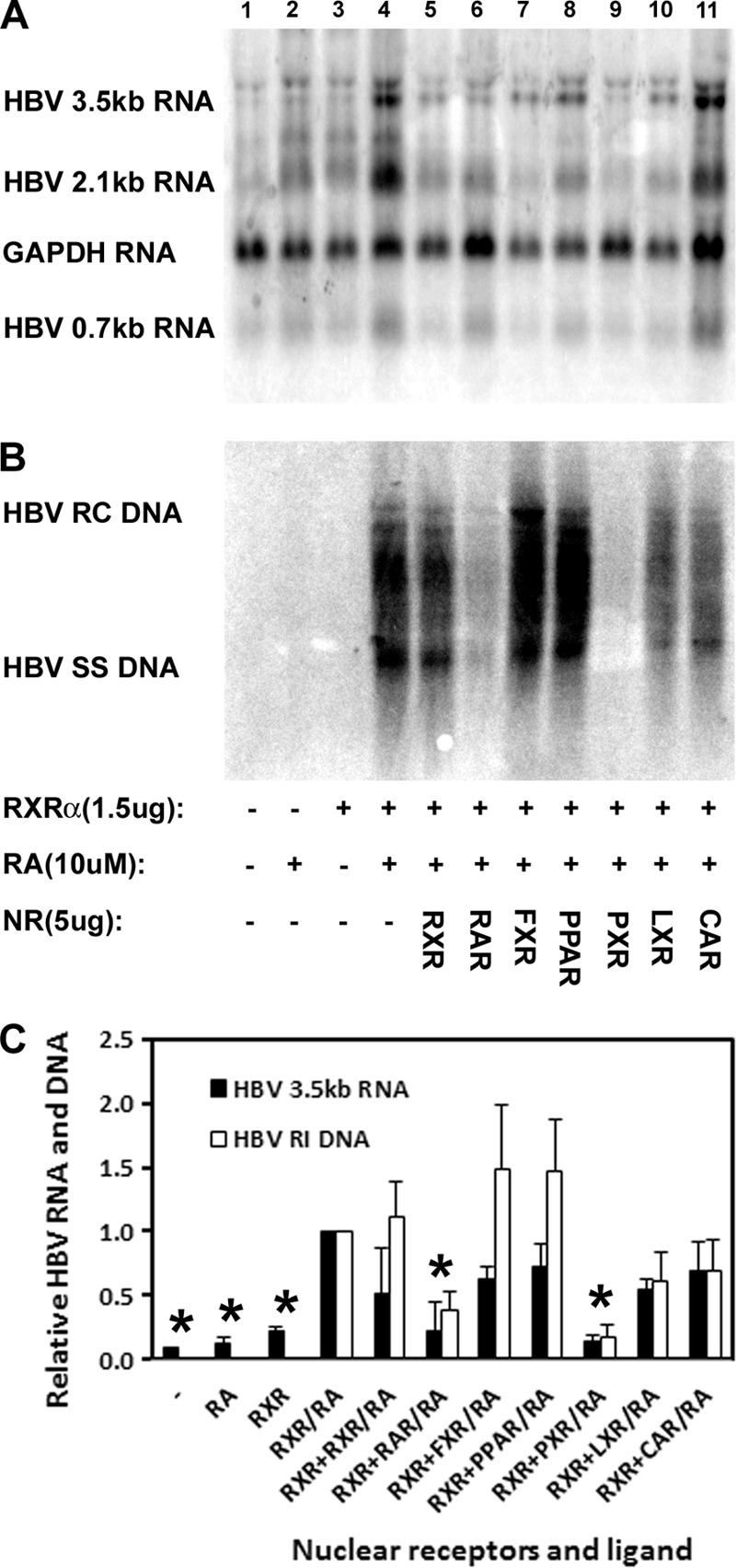
Effects of nuclear receptor expression on RXRα-mediated HBV biosynthesis in the human embryonic kidney cell line 293T. Cells were transfected with 5 μg of the HBV DNA (4.1-kbp) construct alone (lanes 1 and 2) or 5 μg of the HBV DNA (4.1-kbp) construct plus 1.5 μg of the RXRα expression vector (lanes 3 to 11) as indicated. In addition, cells were treated with 10 μM retinoic acid (RA; lanes 2 and 4 to 11) to activate RXRα. An additional 5 μg of nuclear receptor RXRα (lane 5), RARα (lane 6), FXRα (lane 7), PPARα (lane 8), PXR (lane 9), LXRα (lane 10), and CAR (lane 11) expression vectors was also included in the transfections as indicated. (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA; NR, nuclear receptor. (C) Quantitative analysis of the HBV 3.5-kb RNA and HBV DNA replication intermediates. Reported are the levels of the HBV 3.5-kb RNA and total HBV DNA replication intermediates relative to that of the HBV DNA (4.1-kbp) construct in the presence of RXRα expression and 10 μM all-trans-retinoic acid (RXR/RA bars), which are designated as having a relative activity of 1.0. The mean RNA and DNA levels plus standard deviations from two independent analyses are shown. The levels of the transcripts and replication intermediates which are statistically significantly lower than those observed in the all-trans-retinoic acid-treated RXR-expressing cells by Student's t test (P < 0.05) are indicated with asterisks.
Independent activation of HBV biosynthesis by retinoids, peroxisome proliferators, and bile acids in human embryonic kidney 293T cells.
Previously, it has been shown that, when both nuclear receptor partners are ligand activated, RXRα plus PPARα and RXRα plus FXRα can support HBV biosynthesis in nonhepatoma cells (4–7). The relative roles of the individual heterodimer polypeptides in nuclear receptor-dependent activation of HBV biosynthesis have not been evaluated. In 293T cells, transfection of the HBV DNA (4.1-kbp) construct in the presence of retinoic acid fails to support HBV transcription and replication (Fig. 4A and B, lane 16). In addition, the exogenous expression of RXRα also failed to support HBV biosynthesis in the absence of retinoic acid (Fig. 4A and B, lane 17). However, the exogenous expression of RXRα in the presence of retinoic acid is sufficient to support robust HBV transcription and replication (Fig. 4A and B, lane 18). These observations suggest that an RXRα homodimer is sufficient to support HBV biosynthesis in 293T cells. However, it is also possible that RXRα forms heterodimers with an endogenously expressed nuclear receptor and that this transcription factor complex is directing the expression of the HBV pregenomic 3.5-kb RNA. To evaluate this possibility, the roles of known functional RXRα partners were evaluated for their effects on HBV biosynthesis in the presence or absence of activating ligands. FXRα, its ligand alone, or both together failed to support robust HBV transcription or replication (Fig. 4A and B, lanes 2 to 4). RXRα plus FXRα in the absence of ligands also failed to support robust HBV biosynthesis (Fig. 4A and B, lane 5). In contrast, RXRα plus FXRα in the presence of ligands supported robust HBV biosynthesis (Fig. 4A and B, lanes 6 to 8). Activation of FXRα by its ligand, chenodeoxycholic acid, appeared to be somewhat more potent than retinoic acid alone, but the effect of combining both ligands was only somewhat greater than the effect observed with chenodeoxycholic acid alone. Similar observations were apparent when PPARα and clofibric acid were evaluated with RXRα and retinoic acid (Fig. 4A and B, lanes 9 to 15). Overall, these results indicate that ligand-activated RXRα can activate HBV biosynthesis in nonhepatoma cells. In the presence of FXRα or PPARα, RXRα/FXRα or RXRα/PPARα heterodimers can bind to the HBV nucleocapsid promoter and direct viral pregenomic RNA synthesis either through ligand-dependent activation of the common heterodimer partner, RXRα, or conversely through ligand-dependent activation of the unique heterodimer partner. HBV transcription does not appear to require the activation of both polypeptides in the heterodimer, although activation of the unique partner or both partners appears to induce slightly more-robust viral biosynthesis.
Fig 4.
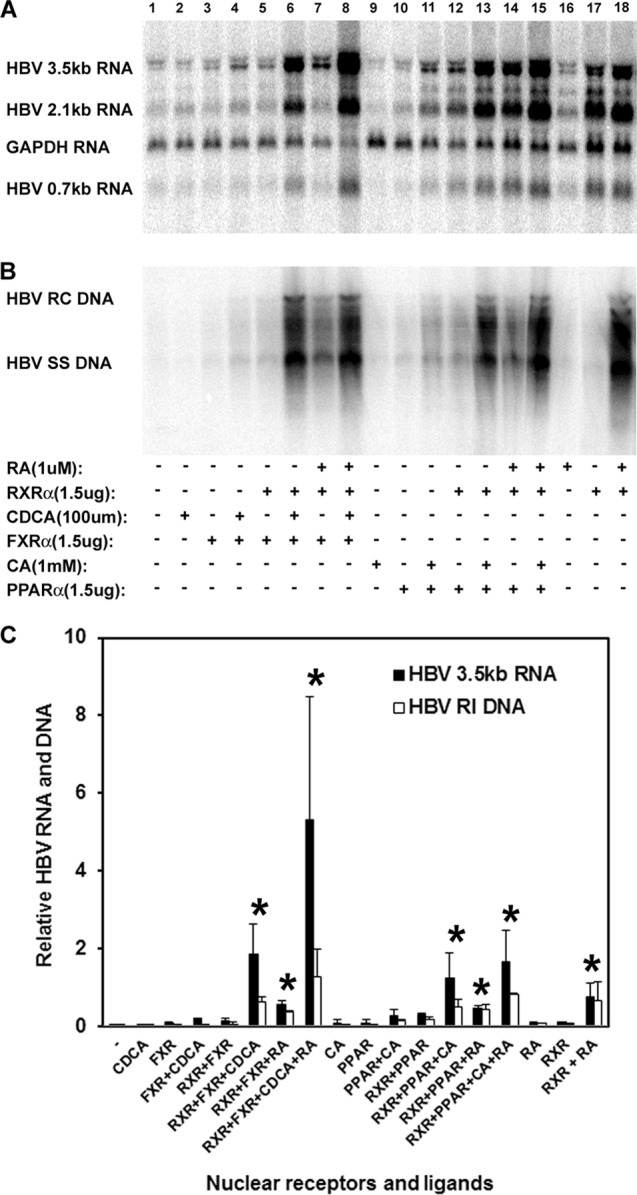
Independent activation of HBV biosynthesis by retinoids, peroxisome proliferators, and bile acids in the human embryonic kidney cell line 293T. Cells were transfected with the HBV DNA (4.1-kbp) construct alone (lanes 1, 2, 9, and 16), the HBV DNA (4.1-kbp) construct plus the RXRα expression vector (lanes 5 to 8, 12 to 15, 17, and 18), the HBV DNA (4.1-kbp) construct plus the FXRα expression vector (lanes 3 to 8), or the HBV DNA (4.1-kbp) construct plus the PPARα expression vector (lanes 10 to 15) as indicated. In addition, cells were treated with 1 μM retinoic acid (RA; lanes 7 and 8, 14 to 16, and 18) to activate RXRα, 100 μM chenodeoxycholic acid (CDCA; lanes 2, 4, 6, and 8) to activate FXRα, and 1 mM clofibric acid (CA; lanes 9, 11, 13, and 15) to activate PPARα as indicated. (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. (C) Quantitative analysis of the HBV 3.5-kb RNA and HBV DNA replication intermediates. The levels of the HBV 3.5-kb RNA and total HBV DNA replication intermediates relative to the HBV DNA (4.1-kbp) construct in the presence of RXRα expression and 1 μM all-trans retinoic acid (RXR + RA bars) are reported. The mean RNA and DNA levels plus standard deviations from two independent analyses are shown. The levels of the transcripts and replication intermediates in the ligand-treated cells which are statistically significantly higher than their levels in the corresponding untreated cells by Student's t test (P < 0.05) are indicated with asterisks.
DISCUSSION
Nuclear receptors are a major determinant of HBV tropism because they contribute to the liver-specific expression of the viral 3.5-kb pregenomic RNA (4–6). This HBV transcript encodes the viral polymerase and core polypeptides and is reverse transcribed into the 3.2-kbp partially double-stranded genomic DNA present in the virion (36). Nuclear receptors are ligand-dependent transcription factors, and hence their activities are governed by the availability of their specific ligands (8, 9). Consequently, nuclear receptors represent potential targets for drug development because small-molecular-weight compounds can act as agonists or antagonists to modulate their activities and hence alter disease progression. Indeed, the fibrate class of compounds has been used to modulate PPARα activity and treat hypertriglyceridemia and hypercholesterolemia (37). Likewise, the thiazolidinedione class of compounds, which act on PPARγ, represent a group of antidiabetic drugs, and tamoxifen is an antiestrogen compound used in the treatment of breast cancer (38, 39). Therefore, understanding the mechanism of action of ligands capable of modulating the activities of the nuclear receptors governing HBV biosynthesis may be informative in the development of strategies aimed at treating chronic HBV infections.
Initially the human hepatoma cell line HepG2 was treated with retinoid, a peroxisome proliferator, and a bile acid to examine the role of the nuclear receptors RXR (and/or RAR), PPAR, and FXR in the regulation of HBV transcription and replication (Fig. 1). Treatment with retinoic acid, clofibric acid, and chenodeoxycholic acid enhanced HBV biosynthesis, indicating that HepG2 cells express RXR (and/or RAR), PPAR, and FXR capable of being activated by the appropriate ligand. As PPARα and FXRα bind to their recognition sequences in the HBV nucleocapsid promoter as RXR/PPAR and RXR/FXR heterodimers (4, 7, 24), these observations suggest that HepG2 cells contain RXR capable of forming transcriptionally active complexes with their activated heterodimer partners. Consistent with this assumption is the observation that retinoic acid can activate HBV biosynthesis in HepG2 cells without the exogenous expression of any additional nuclear receptors. Therefore, it appears that HepG2 cells contain inactive RXR, which can be activated by relatively high concentrations of retinoic acid when it is converted into 9-cis-retinoic acid, the natural ligand for RXR (17, 25). Alternatively, the RXR homodimer may be capable of supporting HBV biosynthesis when activated by retinoids without the requirement for any interaction with additional nuclear receptors. However, due to the high constitutive level of HBV biosynthesis mediated by endogenous transcription factors and the relatively modest induction of viral RNA and DNA synthesis by ligand-activated nuclear receptors in HepG2 cells, a definitive characterization of the relative roles of ligands and their corresponding nuclear receptors in governing HBV biosynthesis was not possible in this system. To address this limitation, the nonhepatoma HBV replication system, where viral transcription is completely dependent on the activation of exogenously expressed nuclear receptors by their cognate ligands, was exploited (Fig. 2 to 4).
HBV transcription and replication in the human embryonic kidney cell line 293T require the ligand-dependent activation of exogenously expressed RXRα (Fig. 2). 9-cis-Retinoic acid and all-trans-retinoic acid treatment of 293T cells displayed dose-dependent increases in HBV RNA and DNA synthesis, with half-maximal levels being observed at concentrations of approximately 1.6 μM and 3.6 μM, respectively. The modest preference of RXRα for its cognate ligand, 9-cis-retinoic acid, and the relatively high concentrations of retinoids required for this level of viral biosynthesis were initially unexpected. In the monkey kidney cell line CV-1, half-maximal levels of transcription from RXR-responsive promoters are observed at approximately 50 nM 9-cis-retinoic acid and 2 μM all-trans-retinoic acid (17, 25). However, the reason the 9-cis-retinoic acid was relatively ineffective at activating HBV biosynthesis might be the 64-h treatment with retinoids required for the evaluation of viral RNA and DNA production. It is possible that during this extended incubation period both the 9-cis-retinoic acid and the all-trans-retinoic acid are rapidly isomerized to a similar pool of retinoic acid isomers, resulting in similar effective intracellular concentrations of the RXRα ligand, 9-cis-retinoic acid (17, 25). Consequently, there is only a minor shift in the dose-response curves when HBV biosynthesis is evaluated and a relatively high concentration of retinoid is required because only a minor fraction is the appropriate isomer (17, 25). Regardless of the reason for these observations, it is apparent that ligand-activated RXRα directly activates viral transcription, presumably by binding to the HBV nucleocapsid promoter DR1 sequence as a homodimer (4, 7, 24).
Previously, RXRα/PPARα and RXRα/FXRα heterodimers have been shown to activate HBV transcription and replication (4–7, 24, 40, 41). The relative importance of the different polypeptides in these heterodimers and the role of their ligands in the activation of HBV biosynthesis have not been established. The observation that ligand-activated RXRα alone is sufficient to support HBV RNA and DNA synthesis raised the issue of the role of additional RXRα heterodimer partners in governing the level of HBV biosynthesis. Initially, additional nuclear receptors capable of heterodimerizing with RXRα were evaluated for their effects on retinoic acid-activated RXRα-mediated HBV RNA and DNA synthesis (Fig. 3). Interestingly, FXRα, PPARα, LXRα, and CAR failed to modulate ligand-activated RXRα-mediated HBV transcription and replication (Fig. 3). In contrast, RARα and PXR inhibited ligand-activated RXRα-mediated HBV biosynthesis (Fig. 3). The observation that RARα inhibited retinoic acid-activated RXRα-mediated HBV biosynthesis is consistent with the suggestion that RXRα/RARα heterodimers activate transcription when bound to direct repeat 5 (DR5) transcriptional regulatory elements but repress transcription when bound to DR1 transcriptional regulatory elements (30, 31). As the HBV nucleocapsid promoter contains a functional DR1 transcriptional regulatory element that binds various nuclear receptors but lacks a known functional DR5 transcriptional regulatory element (4, 7, 24), it appears that the binding of an RXRα homodimer to this DR1 element activates viral transcription, whereas the binding of an RXRα/RARα heterodimer represses HBV biosynthesis (Fig. 3). The observation that the expression of CAR, which binds to DR5 transcriptional regulatory elements as an RXR/CAR heterodimer (16, 42), did not affect ligand-activated RXRα-mediated HBV biosynthesis is also consistent with this suggestion (Fig. 3). Together these observations suggest that enhanced RAR activity may inhibit HBV biosynthesis under some conditions and therefore may have some therapeutic benefits for chronic HBV carriers. Interestingly, the nuclear receptor PXR also inhibited retinoic acid-activated RXRα-mediated HBV biosynthesis. PXR binds to DR3, DR4, and inverted repeat 6 (IR6) transcriptional regulatory elements as an RXR/PXR heterodimer and is activated by various steroid hormones, xenobiotic drugs, including rifampin, and dietary compounds such as phytoestrogens (18, 33, 43). The absence of detectable PXR response elements within the HBV transcriptional regulatory sequence elements suggests that PXR might inhibit ligand-activated RXRα-mediated HBV biosynthesis by sequestering RXRα as RXRα/PXR heterodimers apart from the viral nucleocapsid promoter. However, if PXR can inhibit HBV transcription in this manner, it is unclear why LXRα and CAR cannot also prevent viral RNA synthesis by a similar mechanism. Alternatively, PXR might act in a manner similar to RAR or by preventing coactivator-mediated activation of RXRα (30, 31, 34, 35). Further analysis will be required to determine the mechanism of PXR-mediated inhibition of ligand-activated RXRα-mediated HBV biosynthesis, but these observations suggest that activation of PXR by small-molecular-weight compounds might represent another possible approach for the treatment of chronic HBV infections.
The observation that FXRα and PPARα failed to enhance retinoic acid-activated RXRα-mediated HBV biosynthesis in the absence of their cognate ligands (Fig. 3) questioned the importance of their role in HBV biosynthesis. However, in the absence of retinoic acid, bile acids and peroxisome proliferators activated HBV biosynthesis, demonstrating that these ligands were critical determinants of viral pregenomic RNA synthesis as part of RXRα/FXRα and RXRα/PPARα heterodimer complexes, respectively (Fig. 4). Inclusion of both ligands for either nuclear receptor heterodimer combination did not synergistically enhance HBV RNA and DNA synthesis, suggesting that activation of any one partner in the heterodimer was sufficient to induce the majority of the potential transcriptional activity associated with the two nuclear receptor polypeptides. A further modest enhancement of transcriptional activity was apparent when the second ligand was included, but overall these results suggest that the activation of a single nuclear receptor by ligand binding is sufficient to support most of the observed RNA synthesis from the HBV nucleocapsid promoter. These observations suggest that either a single nuclear receptor within the heterodimer is capable of recruiting the majority of the transcriptional machinery necessary for maximal promoter activity or the activation of one partner by ligand binding leads to the activation of the other partner, presumably by appropriate allosteric interactions (13–15, 44).
The observation that peroxisome proliferators activate viral transcription and replication in vivo in the HBV transgenic mouse model of chronic infection demonstrates that activation of PPARα is functionally important in this animal system (10). Similarly, bile acids can activate HBV biosynthesis to a limited extent in vivo under certain circumstances (11). Together these findings suggest that ligand-activated nuclear receptor-mediated HBV biosynthesis may represent a proportion of the virus production occurring during natural infection, whereas the remainder of the HBV RNA and DNA synthesis is probably mediated by orphan nuclear receptors (11) plus additional classes of liver-enriched and ubiquitous transcription factors (3). Consequently therapeutic approaches aimed at the reduction or elimination of HBV transcription will probably benefit from selective targeting of both ligand-dependent and ligand-independent modes of viral RNA synthesis if this step in the HBV life cycle is going to be efficiently inhibited.
ACKNOWLEDGMENTS
We are grateful to Ronald M. Evans (The Salk Institute, La Jolla, CA) for plasmids pRS-hRXRα and pCMV-rFXRα, Eric F. Johnson (The Scripps Research Institute, La Jolla, CA) for plasmid pCMVPPARα-G, and David Mangelsdorf (Southwestern Medical Center, Dallas, TX) for plasmids pCMX-hRARα, pCMX-mPXR.1, pCMX-mLXRα, and pCMX-hCAR.
This work was supported by Public Health Service grant AI30070 and Ruth L. Kirschstein National Research Service Award AI081451 from the National Institutes of Health.
Footnotes
Published ahead of print 7 November 2012
REFERENCES
- 1. Raney AK, McLachlan A. 1991. The biology of hepatitis B virus, p 1–37 In McLachlan A. (ed), Molecular biology of the hepatitis B virus. CRC Press, Boca Raton, FL [Google Scholar]
- 2. Glebe D, Urban S. 2007. Viral and cellular determinants involved in hepadnaviral entry. World J. Gastroenterol. 13:22–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tang H, Banks KE, Anderson AL, McLachlan A. 2001. Hepatitis B virus transcription and replication. Drug News Perspect. 14:325–334 [PubMed] [Google Scholar]
- 4. Tang H, McLachlan A. 2001. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc. Natl. Acad. Sci. U. S. A. 98:1841–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ondracek CR, Reese VC, Rushing CN, Oropeza CE, McLachlan A. 2009. Distinct regulation of hepatitis B virus biosynthesis by peroxisome proliferator-activated receptor γ coactivator 1α and small heterodimer partner in human hepatoma cell lines. J. Virol. 83:12545–12551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ondracek CR, Rushing CN, Reese VC, Oropeza CE, McLachlan A. 2009. Peroxisome proliferator-activated receptor γ coactivator 1α and small heterodimer partner differentially regulate nuclear receptor-dependent hepatitis B virus biosynthesis. J. Virol. 83:12535–12544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reese VC, Ondracek CR, Rushing CN, Li L, Oropeza CE, McLachlan A. 2011. Multiple nuclear receptors may regulate hepatitis B virus biosynthesis during development. Int. J. Biochem. Cell Biol. 43:230–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mangelsdorf DJ, Evans RM. 1995. The RXR heterodimers and orphan receptors. Cell 83:841–850 [DOI] [PubMed] [Google Scholar]
- 9. Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Guidotti LG, Eggers CM, Raney AK, Chi SY, Peters JM, Gonzalez FJ, McLachlan A. 1999. In vivo regulation of hepatitis B virus replication by peroxisome proliferators. J. Virol. 73:10377–10386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reese VC, Moore DD, McLachlan A. 2012. Limited effects of bile acids and small heterodimer partner on hepatitis B virus biosynthesis in vivo. J. Virol. 86:2760–2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu L, Glass CK, Rosenfeld MG. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 9:140–147 [DOI] [PubMed] [Google Scholar]
- 13. Forman BM, Umesono K, Chen J, Evans RM. 1995. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell 81:541–550 [DOI] [PubMed] [Google Scholar]
- 14. Germain P, Iyer J, Zechel C, Gronemeyer H. 2002. Co-regulator recruitment and the mechanism of retinoic acid receptor synergy. Nature 415:187–192 [DOI] [PubMed] [Google Scholar]
- 15. Schulman IG, Li C, Schwabe JWR, Evans RM. 1997. The phantom ligand effect: allosteric control of transcription by the retinoid X receptor. Genes Dev. 11:299–308 [DOI] [PubMed] [Google Scholar]
- 16. Choi HS, Chung M, Tzameli I, Simha D, Lee YK, Seol W, Moore DD. 1997. Differential transactivation by two isoforms of the orphan nuclear hormone receptor CAR. J. Biol. Chem. 272:23565–23571 [DOI] [PubMed] [Google Scholar]
- 17. Heyman RA, Mangelsdorf DJ, Dyck JA, Stein RB, Eichele G, Evans RM, Thaller C. 1992. 9-cis-Retinoic acid is a high affinity ligand for the retinoid X receptor. Cell 68:397–406 [DOI] [PubMed] [Google Scholar]
- 18. Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterstrom RH, Perlmann T, Lehmann JM. 1998. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92:73–82 [DOI] [PubMed] [Google Scholar]
- 19. Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. 2000. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol. Cell 6:507–515 [DOI] [PubMed] [Google Scholar]
- 20. Mangelsdorf DJ, Ong ES, Dyck JA, Evans RM. 1990. Nuclear receptor that identifies a novel retinoic acid response pathway. Nature 345:224–229 [DOI] [PubMed] [Google Scholar]
- 21. Muerhoff AS, Griffin KJ, Johnson EF. 1992. The peroxisome profilerator-activated receptor mediates the induction of CYP4A6, a cytochrome P450 fatty acid omega-hydroxylase, by clofibric acid. J. Biol. Chem. 267:19051–19053 [PubMed] [Google Scholar]
- 22. Willy PJ, Umesono K, Ong ES, Evans RM, Heyman RA, Mangelsdorf DJ. 1995. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev. 9:1033–1045 [DOI] [PubMed] [Google Scholar]
- 23. McLachlan A, Milich DR, Raney AK, Riggs MG, Hughes JL, Sorge J, Chisari FV. 1987. Expression of hepatitis B virus surface and core antigens: Influences of pre-s and precore sequences. J. Virol. 61:683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Raney AK, Johnson JL, Palmer CNA, McLachlan A. 1997. Members of the nuclear receptor superfamily regulate transcription from the hepatitis B virus nucleocapsid promoter. J. Virol. 71:1058–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Levin AA, Sturzenbecker LJ, Kazmer S, Bosakowski T, Huselton C, Allenby G, Speck J, Kratzeisen C, Rosenberger M, Lovey A, Grippo JF. 1992. 9-cis-Retinoic acid stereoisomer binds and activates the nuclear receptor RXRα. Nature 355:359–361 [DOI] [PubMed] [Google Scholar]
- 26. Summers J, Smith PM, Huang M, Yu M. 1991. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J. Virol. 65:1310–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 28. Chambon P. 1996. A decade of molecular biology of retinoic acid receptors. FASEB J. 10:940–954 [PubMed] [Google Scholar]
- 29. Durand B, Saunders M, Leroy P, Leid M, Chambon P. 1992. All-trans and 9-cis retinoic acid induction of CRABPII transcription is mediated by RAR-RXR heterodimers bound to DR1 and DR2 repeated motifs. Cell 71:73–85 [DOI] [PubMed] [Google Scholar]
- 30. Kurokawa R, Söderström M, Hörlein A, Halachmi S, Brown M, Rosenfeld MG, Glass CK. 1995. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature 377:451–454 [DOI] [PubMed] [Google Scholar]
- 31. Kurokawa R, DiRenzo J, Boehm M, Sugarman J, Gloss B, Rosenfeld MG, Heyman RA, Glass CK. 1994. Regulation of retinoid signalling by receptor polarity and allosteric control of ligand binding. Nature 371:528–531 [DOI] [PubMed] [Google Scholar]
- 32. Kyrmizi I, Hatzis P, Katrakili N, Tronche F, Gonzalez FJ, Talianidis I. 2006. Plasticity and expanding complexity of the hepatic transcription factor network during liver development. Genes Dev. 20:2293–2305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Blumberg B, Sabbagh W, Juguilon H, Bolado J, Jr, Van Meter CM, Ono ES, Evans RM. 1998. SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev. 12:3195–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bhalla S, Ozalp C, Fang SS, Xiang LJ, Kemper K. 2004. Ligand-activated pregnane X receptor interferes with HNF-4 signaling by targeting a common coactivator PGC-1α. Functional implications in hepatic cholesterol and glucose metabolism. J. Biol. Chem. 279:45139–45147 [DOI] [PubMed] [Google Scholar]
- 35. Li T, Chiang JYL. 2005. Mechanism of rifampicin and pregnane X receptor inhibition of human cholesterol 7α-hydroxylase gene transcription. Am. J. Physiol. Gastrointest. Liver Physiol. 288:G74–G84 [DOI] [PubMed] [Google Scholar]
- 36. Will H, Reiser W, Weimer T, Pfaff E, Buscher M, Sprengle R, Cattaneo R, Schaller H. 1987. Replication strategy of human hepatitis B virus. J. Virol. 61:904–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Winkler K, Weltzien P, Friedrich I, Schmitz H, Nickell HH, Hoffmann MM, Baumstark MW, Wieland H, März W. 2004. Qualitative effect of fenofibrate and quantitative effect of atorvastatin on LDL profile in combined hyperlipidemia with dense LDL. Exp. Clin. Endocrinol. Diabetes 112:241–247 [DOI] [PubMed] [Google Scholar]
- 38. Lehmann JM, Moore LB, Smith-Oliver TA, Wilkison WO, Willson TM, Kliewer SA. 1995. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor gamma (PPARgamma). J. Biol. Chem. 270:12953–12956 [DOI] [PubMed] [Google Scholar]
- 39. Riggs BL, Hartmann LC. 2003. Selective estrogen-receptor modulators—mechanisms of action and application to clinical practice. N. Engl. J. Med. 348:618–629 [DOI] [PubMed] [Google Scholar]
- 40. Huan B, Kosovsky MJ, Siddiqui A. 1995. Retinoid X receptor α transactivates the hepatitis B virus enhancer 1 element by forming a heterodimeric complex with the peroxisome proliferator-activated receptor. J. Virol. 69:547–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ramiere C, Scholtes C, Diaz O, Icard V, Perrin-Cocon L, Trabaud MA, Lotteau V, Andre P. 2008. Transactivation of the hepatitis B virus core promoter by the nuclear receptor FXRα. J. Virol. 82:10832–10840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Baes M, Gulick T, Choi HS, Martinoli MG, Simha D, Moore DD. 1994. A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol. Cell. Biol. 14:1544–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bertilsson G, Heidrich J, Svensson K, Asman M, Jendeberg L, Sydow-Backman M, Ohlsson R, Postlind H, Blomquist P, Berkenstam A. 1998. Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. Proc. Natl. Acad. Sci. U. S. A. 95:12208–12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. DiRenzo J, Söderström M, Kurokawa R, Ogliastro MH, Ricote M, Ingrey S, Horlein A, Rosenfeld MG, Glass CK. 1997. Peroxisome proliferator-activated receptors and retinoic acid receptors differentially control the interactions of retinoid X receptor heterodimers with ligands, coactivators, and corepressors. Mol. Cell. Biol. 17:2166–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]