Abstract
Cell entry of rotaviruses is a complex process, which involves sequential interactions with several cell surface molecules. Among the molecules implicated are gangliosides, glycosphingolipids with one or more sialic acid (SA) residues. The role of gangliosides in rotavirus cell entry was studied by silencing the expression of two key enzymes involved in their biosynthesis—the UDP-glucose:ceramide glucosyltransferase (UGCG), which transfers a glucose molecule to ceramide to produce glucosylceramide GlcCer, and the lactosyl ceramide-α-2,3–sialyl transferase 5 (GM3-s), which adds the first SA to lactoceramide-producing ganglioside GM3. Silencing the expression of both enzymes resulted in decreased ganglioside levels (as judged by GM1a detection). Four rotavirus strains tested (human Wa, simian RRV, porcine TFR-41, and bovine UK) showed a decreased infectivity in cells with impaired ganglioside synthesis; however, their replication after bypassing the entry step was not affected, confirming the importance of gangliosides for cell entry of the viruses. Interestingly, viral binding to the cell surface was not affected in cells with inhibited ganglioside synthesis, but the infectivity of all strains tested was inhibited by preincubation of gangliosides with virus prior to infection. These data suggest that rotaviruses can attach to cell surface in the absence of gangliosides but require them for productive cell entry, confirming their functional role during rotavirus cell entry.
INTRODUCTION
Rotaviruses, the leading cause of severe dehydrating diarrhea, are members of the family Reoviridae. They are nonenveloped viruses formed by three concentric protein layers with a genome composed of 11 segments of double-stranded RNA (dsRNA). The outer capsid is formed by two proteins, VP4 and VP7, both of which have essential functions in receptor binding and cell penetration. Rotavirus cell entry seems to be mediated by a series of interactions that involve both viral surface proteins and several cell surface molecules (reviewed in reference 1). The susceptibility of some rotavirus strains to the treatment of cells with neuraminidase (NA) (an enzyme which cleaves terminal sialic acids [SA] in glycoconjugates) led to the description of NA-sensitive and NA-resistant strains (2). The SA-binding domain on the virus surface proteins has been located to the VP8 trypsin cleavage product of VP4 (3, 4). The crystal structures of the VP8 protein from two NA-resistant human rotavirus strains (Wa and DS-1) and two NA-sensitive animal viruses (RRV and CRW-8) (5–7) have been described. All structures share a galectin-like fold, but, while the VP8 of the NA-sensitive strains RRV and CRW-8 was shown to interact with SA in a shallow groove located between two β sheets (6, 8), it was predicted that the VP8 of NA-resistant human rotavirus strain DS-1 would not bind a carbohydrate at this location (7), and the VP8 of the NA-resistant rotavirus Wa did not bind SA (5). An alternative groove which is present in strain CRW-8 capable of interacting with carbohydrates, and which is conserved in rotavirus Wa, was recently identified; however, whether this site is involved in oligosaccharide binding remains to be determined (5). The recent finding that the human strain Wa was able to bind to subterminal SA through VP8 (9) supports the possibility that even NA-resistant rotavirus strains interact with SA. This observation is consistent with the fact that subterminal SAs are resistant to NA treatment (10).
Glycosphingolipids are a large and heterogeneous family of amphipathic lipids present on the extracellular leaflet of all mammalian plasma membranes. They are composed of a ceramide anchor linked to an oligosaccharide chain of variable size. Gangliosides are glycosphingolipids with one or more SA residues. Although not well understood, they play an important role in cell adhesion, cell-cell interactions, and signal transduction, and their cellular expression is tightly controlled (11–13). Besides these important biological functions they are also used as receptors for many viruses, including different members of the Polyomaviridae family (simian virus 40, murine polyomavirus, BK virus, JC virus, Merkel cell polyomavirus) (14, 15), paramyxoviruses (Newcastle disease virus and Sendai virus) (16), bovine adeno-associated virus (17), influenza virus (18), murine norovirus (19), and rotavirus (10, 20).
Ganglioside synthesis begins with the synthesis of ceramide in the endoplasmic reticulum (ER), which is then transported to the Golgi complex, where it is modified by the UDP-glucose:ceramide glucosyltransferase (UGCG), which transfers a glucose molecule to ceramide to produce glucosylceramide (GlcCer). GlcCer is then transformed by the addition of galactose by galactosyltransferase I (GalT1) to produce lactoceramide (LacCer), which after the addition of the first sialic acid, by action of the lactosyl ceramide-α-2,3–sialyl transferase 5 (GM3-synthase [GM3-s]), yields ganglioside GM3, which is the critical branch point in the synthesis of gangliosides (Fig. 1) (21). Once the synthesis of gangliosides is completed in the Golgi apparatus, they are delivered to the plasma membrane (22).
Fig 1.
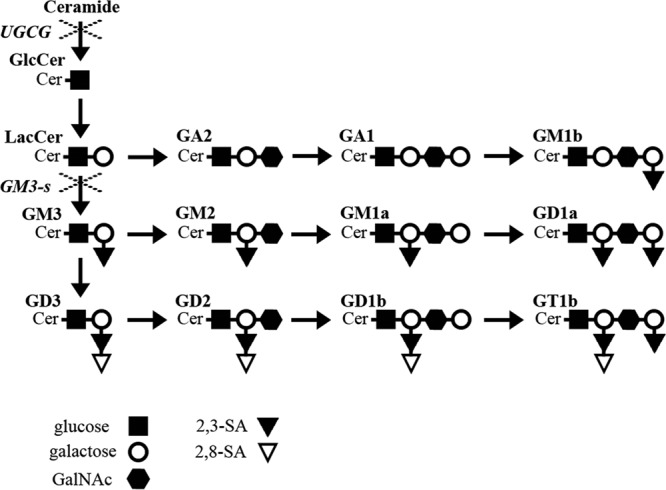
Ganglioside biosynthesis. Schematic representation of main ganglioside synthesis. × indicates silenced enzymes UGCG (UDP-glucose:ceramide glucosyltransferase) and GM3-s (lactosyl ceramide-α-2,3–sialyl transferase 5). The key code for saccharide units composing gangliosides is shown. Ganglioside nomenclature according to IUPAC-IUB (46) was used.
The possible role of gangliosides in rotavirus cell entry has been investigated previously. Using a binding assay based on thin-layer chromatography, NA-sensitive rotavirus strains (simian SA11 and bovine NCDV) were shown to bind gangliosides with terminal SA, while the NA-resistant bovine strain UK recognized gangliosides with subterminal SA (23). In line with this observation, ganglioside GM1a, which contains a subterminal SA, was reported to be important for infectivity of the NA-resistant human strains KUN and MO (10), while GM3 (which contains a terminal SA residue) blocked the infection of the NA-sensitive porcine rotavirus strain OSU (20). Recently, it was determined that aceramido-GM1a binds to the VP8 protein of the NA-resistant strain Wa, while aceramido-GD1a (containing terminal and subterminal SA moieties) binds to the VP8 of the NA-sensitive strain CRW-8 (9). Of note, there was neither binding of aceramido-GM1a to the VP8 domain of CRW-8 nor binding of aceramido-GD1a to Wa VP8 (9).
It was recently described that the VP8 protein of human rotavirus strain HAL1166 interacts with A-type histo-blood group antigen (HBGA) at the same location where the VP8 of NA-sensitive RRV rotavirus interacts with SA (24). The infectivity of strain HAL1166 was increased in CHO cells expressing A-type HBGA and decreased by anti-A-type HBGA monoclonal antibody, suggesting the involvement of other oligosaccharides in rotavirus cell entry. Interestingly, the VP8 proteins of other human rotavirus strains were also found to interact with HBGA (25, 26); however, their role during the entry process has not been defined yet.
In this work, the functional relevance of gangliosides in rotavirus infection was studied by knocking down by RNA interference (RNAi) the expression of two key enzymes (UGCG and GM3-s) involved in the ganglioside biosynthetic pathway. Our results suggest that both NA-resistant and NA-sensitive rotaviruses use gangliosides with terminal or subterminal SAs at a step different from the initial attachment to the cell surface, during their productive entry into the cell.
MATERIALS AND METHODS
Cells, reagents, and viruses.
The monkey kidney epithelial cell line MA104 was grown in advanced Dulbecco modified Eagle medium (DMEM), supplemented with 3% fetal bovine serum (FBS). Rhesus rotavirus strain RRV and human strain Wa were obtained from H. B. Greenberg (Stanford University, Stanford, CA), bovine rotavirus UK was obtained from D. R. Snodgrass (Moredun Research Institute, Edinburgh, United Kingdom), and porcine strain TFR-41 was obtained from I. Holmes (University of Melbourne, Victoria, Australia). All rotavirus strains were amplified in MA104 cells and, when needed, purified as described previously (27). Rabbit polyclonal antibodies to integrin subunits α2 and β3, were from Chemicon (Temecula, CA), and the rabbit anti-vimentin serum was produced in our laboratory. Mouse monoclonal antibody to heat shock cognate protein (hsc70), clone B-6, and rabbit polyclonal antibody to caveoline (N-20) were from Santa Cruz Biotechnology (Santa Cruz, CA), and horseradish peroxidase-conjugated goat anti-rabbit polyclonal antibody was from PerkinElmer Life Sciences (Boston, MA). Horseradish peroxidase-conjugated anti-mouse, peroxidase-conjugated cholera toxin B (CTB) subunit, gangliosides GA1, GM1a, GM2, GM3, GD3, GD1a, and GD1b (isolated from bovine brain), ganglioside GM3 (isolated from canine blood), ganglioside GD3 (isolated from bovine milk), and neuraminidase from Arthrobacter ureafaciens were from Sigma (St. Louis, MO). Goat anti-rabbit IgG coupled to Alexa 568 and cholera toxin B subunit coupled to Alexa 488 were from Invitrogen.
Transfection of siRNAs.
The specific small interfering RNAs (siRNAs) or SMARTpool siRNAs were obtained from Dharmacon Research (Lafayette, CO) as annealed duplexes. The target sequences of the siRNAs are as follows: siUGCG (AAUCAACAACCUGGAAACAUU), siGM3-s (AAGAGAGCUCAGAAAUAUGCU), sigrp94 (AAGCCGAAGUUAACAGAAUGA). Control irrelevant siRNAs (siIrr) were specific for green fluorescent protein (AAC UUA CCC UGA AGU UCA UCU) or luciferase (AAG UGC GUU GCU AGU ACC AAC). Neither of the control siRNAs used affected viral protein synthesis or infectivity, and they are generically referred to as siIrrs. Two methods were used for transfection. (i) MA104 cells growing in exponential phase were trypsinized and seeded at 5 × 105cells/ml in Eagle's minimal essential medium (MEM) supplemented with 5% dialyzed FBS; 24 h later, when cells reached approximately 70% confluence, the siRNAs were introduced by transfection. The transfection mixture contained 3 μl of the Oligofectamine reagent (Invitrogen, Carlsbad, CA) and 60 pmol of siRNAs per 100 μl in MEM without FBS and antibiotics. The liposome complexes were added to cells after washing off the growth medium. After 14 h of incubation at 37°C in a 5% CO2 incubator, the mixture was replaced by fresh MEM supplemented with 5% dialyzed FBS, and the cells were allowed to grow for an additional 48 h, the time at which they reached confluence. (ii) A reverse transfection method was performed as previously described (28). Briefly, 22.5 μl Oligofectamine was diluted in 1.7 ml MEM and incubated for 10 min at room temperature. This mixture was then mixed with 120 pmol of siRNA diluted in MEM. After an incubation of 20 min at room temperature, 1 ml of a suspension of 1.8 × 106 cells in advanced DMEM was added to each tube and the tubes were incubated at 37°C for 1 h with gentle mixing. Then, transfected cells were plated in a 25-cm2 flask, advanced DMEM was added to a final volume of 5 ml, and cells were left to grow for 72 h to reach confluence. The cells were washed twice with MEM, and cultures were processed as indicated. Both transfection methods showed similar levels of RNA silencing, GM1a decrease, and rotavirus infectivity reduction.
Density gradient centrifugation.
MA104 cells transfected by the reverse transfection method described above were grown for 72 h to reach confluence. Cells were harvested and processed basically as described previously (29). As a control of purification, nontransfected cells (106) in 10 ml of MEM were incubated with 10 mM methyl-β-cyclodextrine (MβCD) (Sigma, St. Louis, MO) for 1 h at 37°C, with occasional mixing. After two washes with cold MEM, cells were transferred to an Eppendorf tube on ice. All following steps were performed on ice in a cold room. The pelleted cells were lysed for 20 min in 50 μl of TNC buffer (25 mM Tris, pH 7.5, 150 mM NaCl, 2 mM CaCl2) with 1% TX-100 (vol/vol) and a mixture of protease inhibitors (20 μg/ml aprotinin, 20 μg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride [PMSF]). Lysates were then brought to a final concentration of 40% iodixanol (Sigma, St. Louis, MO), placed at the bottom of a centrifuge tube (SW50.1; Beckman), and overlaid with 800 μl each of 35%, 30%, 25%, and 20% iodixanol in TNC, followed by 800 μl of TNC. Samples were centrifuged for 4 h at 200,000 × g at 4°C, and 10 fractions (430 μl each) were collected from the tops of the tubes.
Infectivity assays.
MA104 cells grown in 48-well plates and transfected as described above were treated or not with neuraminidase from A. ureafaciens (40 mU/ml) for 1 h at 37°C. Cultures were washed and infected with different rotavirus strains (2,000 PFU/well). Infection was left to proceed for 14 h, after which the cells were fixed and stained with polyclonal anti-rotavirus sera. The number of infected cells was determined as described previously (30).
Blocking assays.
Rotaviruses (2,000 PFU/well) were mixed with different ganglioside dilutions in MEM and incubated for 1 h at 37°C with gentle rocking. Afterward, the mixture was added to MA104 cells grown in 48-well plates and incubated for 1 h at 37°C. Cultures were washed twice with MEM without serum and processed for an infectivity assay as described above.
Virus binding.
MA104 cells grown in 48-well plates, transfected as described above and treated or not with 40 mU of NA, were washed twice with MEM and incubated in MEM without FBS for 30 min at 37°C. Medium was removed, and cells were blocked with blocking buffer (phosphate-buffered saline [PBS]-1% bovine serum albumin [BSA]) for 1 h at 37°C. After washing twice with PBS-0.5% BSA, the cells were placed on ice, 2 μg of purified triple-layered particles (TLPs) in blocking buffer was added per well, and the wells were incubated for 1 h at 4°C. Unbound virus was removed, and cells were washed four times with MEM and then lysed with lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Triton X-100). Lysates were freeze-thawed twice, and the amount of viral protein present in the lysates was determined by enzyme-linked immunosorbent assay (ELISA) as previously described (27).
Real-time RT-PCR.
MA104 cells grown in 48-well plates and transfected as described above were lysed with TRIzol (Invitrogen) and total RNA was purified according to the manufacturer's protocol. Total RNA was treated with RNase-free DNase (Roche, Mannheim, Germany), and the levels of mRNAs of interest were determined by one-step real-time reverse transcription-PCR (RT-PCR) using the adequate primers (Table 1). Each reaction tube contained 100 ng of total RNA, 12.5 μl of SYBR green master mix (2×) (Applied Biosystems), 0.125 μl of reverse transcriptase (50 U/μl) (Applied Biosystems), 0.25 μl of RNase inhibitor (20 U/μl) (Applied Biosystems), and 1 μl of each primer (2.5 pmol/μl) in a total volume of 20 μl. Amplification was carried out in an ABI Prism 7500 sequence detector system (Applied Biosystems). The results were normalized to the level of GAPDH mRNA detected in each sample (31). The level of each mRNA was calculated by the 2−ΔΔCT method, where CT is the threshold cycle (32).
Table 1.
Primers used in real-time RT-PCR
| Primer | Direction | Sequence |
|---|---|---|
| GM3s | Forward | 5′ GGTCAGGGTCCACATAATGC 3′ |
| Reverse | 5′ GCTTGTGTTTGGAGTGTGGA 3′ | |
| UGCG | Forward | 5′ AGACACCTGGGAGCTTGCTA 3′ |
| Reverse | 5′ TTCGTCCTCTTCTTGGTGCT 3′ | |
| grp94 | Forward | 5′ TCCGCCTTCCTTGTAGCAGATA 3′ |
| Reverse | 5′ TTGTCGTTCCCCGTCCTAGA 3′ | |
| GAPDH | Forward | 5′ ACCTGACCTGCCGTCTAGAAA 3′ |
| Reverse | 5′ CCTGCTTCACCACCTTCTTGAT 3′ |
Immunofluorescence.
MA104 cells were grown on coverslips and transfected as described above. After transfection, cells were washed with MEM and infected with different rotavirus strains at a multiplicity of infection (MOI) of 3. Eight hours postinfection (hpi), cells were fixed with 2% paraformaldehyde, permeabilized with Triton X-100, and stained with rabbit polyclonal sera to NSP2 protein, followed by goat anti-rabbit IgG coupled to Alexa 568, as previously described (33), and with cholera toxin B subunit coupled to Alexa 488. Cell nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole). Coverslips were mounted on glass slides with Fluoprep (bioMérieux, Inc. Durham, NC). Slides were analyzed with a Nikon E600 epifluorescence microscope coupled to a Nikon DXM1200 digital still camera. Images were captured digitally and processed with Adobe Photoshop CS2 as previously described (34). In each experiment, two persons analyzed at least 100 cells independently for GM1a expression and virus infection, and their results were compared.
Immunoblots.
MA104 cells grown in 48-well plates were transfected with siRNA and, 48 h after transfection, were washed and harvested with Triton X-100 buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% Triton X-100) or with Laemmli sample buffer. For detection of ganglioside GM1a, the protein content of Triton X-100-extracted samples was determined and 250 ng of samples was blotted onto nitrocellulose membrane (Millipore, Bedford, MA), using Bio-Dot (Bio-Rad) following the manufacturer's description. The membrane was blocked with 5% nonfat dry milk, and GM1a was detected with peroxidase-conjugated cholera toxin B subunit. To detect other cellular proteins, samples harvested in Laemmli sample buffer were separated by sodium dodecyl sulfate-7.5% polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to nitrocellulose membranes. To analyze cellular proteins after density gradient centrifugation, the same amount of gradient fraction was precipitated by methanol-chloroform-distilled water and proteins were separated by 15% SDS-PAGE and transferred to nitrocellulose membranes. All membranes were blocked with 5% nonfat dry milk and developed with primary and peroxidase-conjugated secondary antibodies. The peroxidase activity was developed using the Western Lightning chemiluminescence kit following the manufacturer's instructions (PerkinElmer Life Sciences).
Statistical analysis.
Statistical analysis included the Student t test, one-way analysis of variance (ANOVA), and a Tukey posttest. Tests were performed using GraphPad Prism 5.0.
RESULTS
RNA interference of the ganglioside biosynthesis pathway in MA104 cells.
Different lines of evidence suggest that gangliosides are involved in rotavirus cell entry (9, 10, 20, 30). It was previously reported that ganglioside synthesis could be inhibited by silencing the expression of enzymes involved in ganglioside biosynthesis by RNAi (35). To characterize if gangliosides are important for rotavirus cell entry, we first tested if it was possible to knock down the expression of two key enzymes involved in their biosynthesis (UGCG and GM3-s) in MA104 cells. Given the lack of specific antibodies to these enzymes, the effectiveness of RNA interference was assessed by quantification of their mRNA levels by real-time PCR. As a negative control, the expression of the ER chaperone grp94 was silenced, since it has been previously shown that its knockdown by RNAi does not affect rotavirus infectivity (37). The siRNAs directed to UGCG and GM3-s decreased the level of the cognate, target mRNA, while they did not affect grp94 mRNA (Fig. 2A). Similarly, silencing the expression of grp94 had no effect on the levels of UGCG and GM3-s mRNAs (Fig. 2A).
Fig 2.
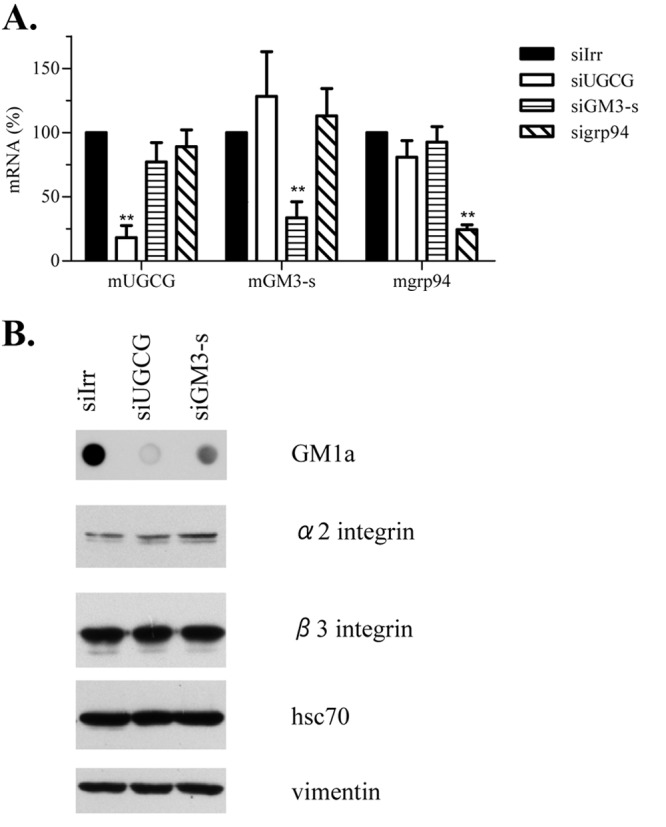
siRNAs directed to enzymes involved in ganglioside biosynthesis decrease ganglioside levels, without affecting expression of other rotavirus receptors. MA104 cells were transfected with siRNAs against UGCG and GM3-s, and 72 h posttransfection parallel wells were harvested for mRNA quantification and for immunoblotting. (A) Total RNA was extracted and levels of mRNA were determined by one-step RT-PCR as described in Materials and Methods. Results are expressed as percentages relative to mRNA values obtained in cells lipofected with control siRNA (siIrr). As a negative control, the expression of endoplasmic reticulum chaperone grp94 was silenced. Means and standard deviations for results of at least three independent experiments are shown. (B) Harvested cellular proteins were resolved by SDS-PAGE, and proteins associated with rotavirus cell entry (integrin subunits α2 and β3 and hsc70 protein) were detected by immunostaining. Vimentin was used as a loading control. The result of one representative experiment of three is shown. To detect GM1a, 250 ng of total protein was applied to nitrocellulose membranes and stained using cholera toxin B subunit. Statistical analysis was done using a Student t test, and statistically significant values are shown (**, P < 0.01).
As the next step, the effect of silencing the expression of UGCG and GM3-s on the level of ganglioside GM1a was analyzed by dot blot. The UGCG catalyzes the addition of the first glucose to ceramide; therefore, it was expected to cause a general inhibition of ganglioside synthesis, while GM3-s is responsible for addition of the first SA to LacCer to yield ganglioside GM3; its inhibition should then prevent the synthesis of most gangliosides, with only LacCer, GA2, GA1, and GM1b (a monosialylated glycosphingolipid) being synthesized (Fig. 1). In both cases the synthesis of ganglioside GM1a, the target of cholera toxin (38, 39), is expected to be inhibited. The decreased expression of both enzymes resulted in decreased levels of ganglioside GM1a by 82% (UGCG) and 62% (GM3-s), respectively (Fig. 2B), suggesting that the general biosynthesis of gangliosides was impaired. We also analyzed whether silencing the expression of UGCG and GM3-s affected the expression of integrin subunits α2 and β3 and hsc70 protein, molecules that have been proposed to be involved in rotavirus infection. Western blot analysis of these molecules in cells that had been transfected with the siRNAs to either UGCG or GM3-s showed that knocking down the ganglioside biosynthetic pathway did not affect the cellular abundance of gangliosides (Fig. 2B).
It was previously shown that detergent-resistant membranes (DRMs) are important for rotavirus infectivity (29). To test if decreased ganglioside levels affected the formation of DRMs, these membranes were purified from cells in which the expression of UGCG was silenced. As can be seen in Fig. 3, the knockdown of ganglioside biosynthetic pathway did not affect the physiology of the cell membrane, since DRMs were present at the same gradient fractions (as observed by staining caveolin 1 as a DRM marker), compared to cells that were transfected with an irrelevant siRNA, despite the low level of gangliosides. Of interest, the remaining GM1a was also observed in DRMs fractions (Fig. 3). Cholesterol removal by MβCD resulted in DRM destabilization and in the shift of caveolin 1 and GM1a to higher-density fractions, where soluble proteins locate (Fig. 3).
Fig 3.
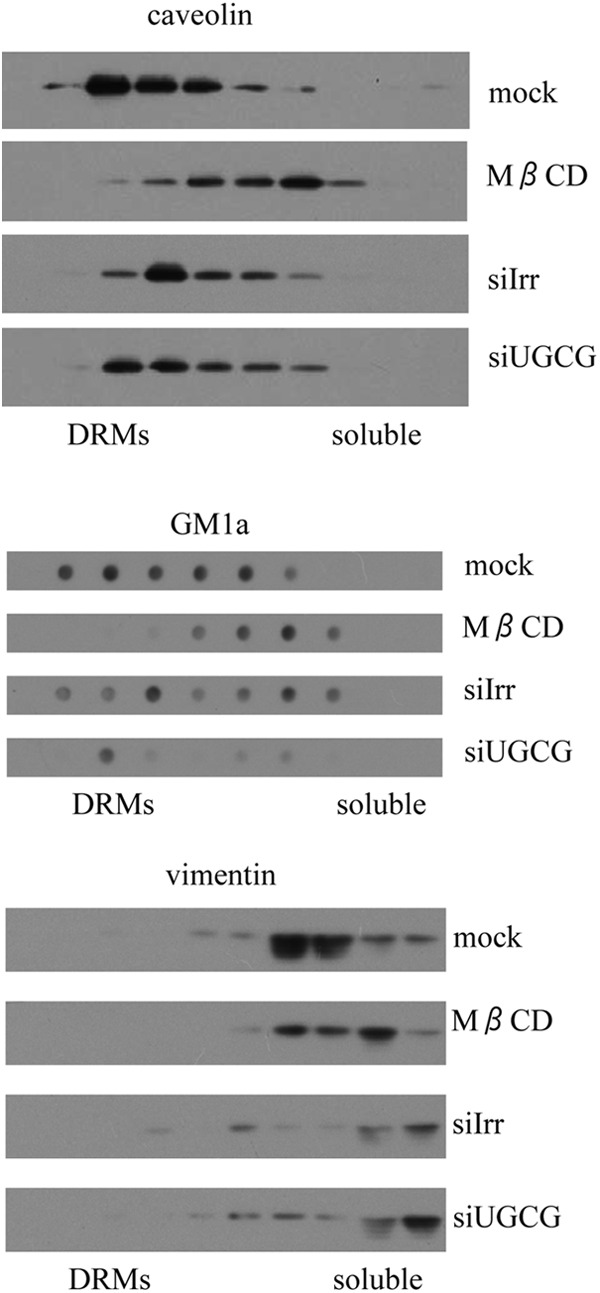
The formation of detergent-resistant membrane domains is unaffected in cells with decreased ganglioside synthesis. MA104 cells were transfected with siRNAs (siIrr and siUGCG) in 25-cm2 flasks, and 72 h posttransfection they were brought to single cell suspension and incubated or not with 10 mM MβCD. Then cells were lysed with 1% Triton X-100 at 4°C and subjected to density gradient centrifugation in discontinuous iodixanol gradients, as described in Materials and Methods. The proteins from equal volumes of individual fractions were separated by 15% SDS-PAGE, transferred to a nitrocellulose membrane, and probed with rabbit antibodies to caveolin 1 and vimentin, followed by a peroxidase-labeled secondary antibody. For GM1 detection, the samples were applied to nitrocellulose membranes with a dot blotter and developed using a horseradish peroxidase-conjugated cholera toxin B subunit. DRMs localize in the top fractions of gradient, while soluble proteins are in the bottom high-density fractions.
Reduced expression of UGCG and GM3-s decreases virus entry but not postentry virus replication steps.
The infectivity of four rotavirus strains that differ in their sensitivity to NA treatment of the cells was assayed in MA104 cells previously transfected with siRNAs to UGCG or GM3-s. The infectivity of both NA-sensitive (RRV and TFR-41) and NA-resistant (Wa and UK) rotaviruses showed a significant decrease (Fig. 4A), resulting in a 45 to 65% or 30 to 55% reduction for NA-sensitive and NA-resistant strains, respectively.
Fig 4.
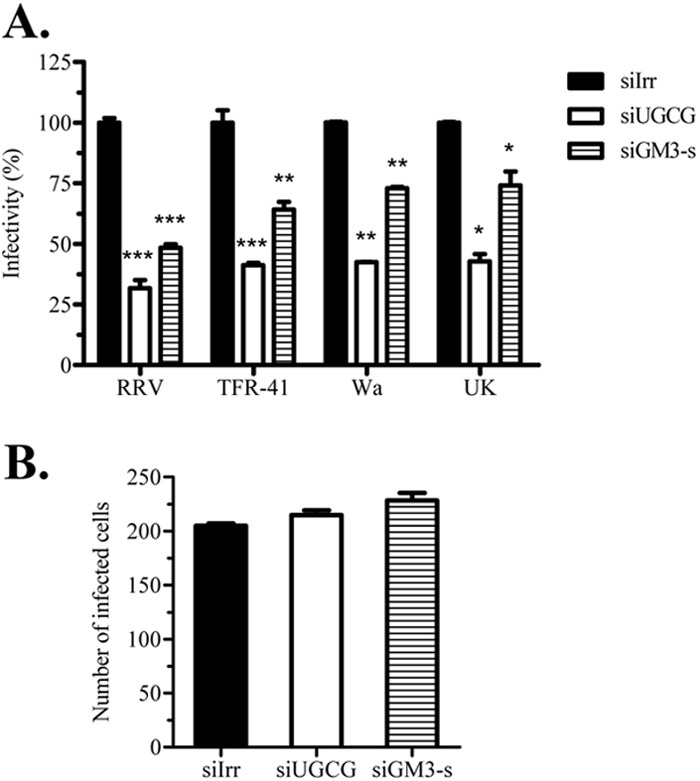
Inhibition of ganglioside synthesis decreases rotavirus cell entry. MA104 cells grown in 48-well plates were transfected with siRNAs (siIrr, siUGCG, and siGM3-s) and 72 h posttransfection were infected with rotavirus. In each experiment, parallel wells were harvested and ganglioside inhibition analyzed by GM1a immunoblotting as described in Materials and Methods. (A) Four different rotavirus strains (RRV, TFR-41, Wa, and UK) were inoculated for 1 h, washed, and left for 14 h for infection to proceed. Then infected cells were detected using peroxidase immunostaining with anti-rotavirus polyclonal antibodies. Results are expressed as percentages relative to irrelevant control treatment (siIrr) for each rotavirus strain. Means and standard deviations for results of at least three independent experiments are shown. (B) RRV DLPs were lipofected in ganglioside-decreased cells, and at 14 h posttransfection focus-forming units (FFUs) were detected as described previously. As a negative control, DLPs without lipofectant were inoculated (data not shown), and no FFUs were detected. Results are expressed as the number of FFUs obtained in each treatment. Means and standard deviations for results of at least three independent experiments are shown. Statistical analysis was done using a Student t test, and statistically significant values are shown as follows: ***, P < 0.001, **; P < 0.01; *, P < 0.05.
To determine if the decreased rotavirus infectivity was due to inhibition of viral entry, or if it was caused by a nonspecific postentry effect caused by the reduction in ganglioside synthesis, transcriptionally active double-layered particles (DLPs) of the four rotavirus strains were lipofected into cells in which the expression of UGCG and GM3-s had been silenced. The number of infected cells after DLP lipofection did not change in cells where either of the two enzymes was silenced (Fig. 4B, and results not shown), suggesting that the inhibition of rotavirus infection was due to a block in cell entry.
Since transfection efficiency with siRNAs in MA104 cells is only about 80% (40), the correlation of viral infectivity and the expression of gangliosides, as judged by the presence of GM1a, was determined at the individual cell level by immunofluorescence microscopy. MA104 cells transfected with the siRNA to UGCG were infected at an MOI of 3 and then fixed with paraformaldehyde at 8 hpi and labeled with the cholera toxin B subunit conjugated to Alexa 488 to detect GM1a and with a polyclonal antibody to the rotavirus nonstructural protein NSP2, followed by incubation with Alexa 568-conjugated anti-rabbit antibody, to detect virus-infected cells. While in cells transfected with the irrelevant siRNA the CTB signal appeared uniformly in all cells, and 93 to 98% of the cells became infected, cells transfected with the siRNA to UGCG showed a clear decrease of the CTB signal that corresponded to a significant decrease of rotavirus infection (Fig. 5). Depending on the virus strain, only between 15 and 76% of the cells with diminished levels of expression of GM1a became infected (Fig. 5).
Fig 5.
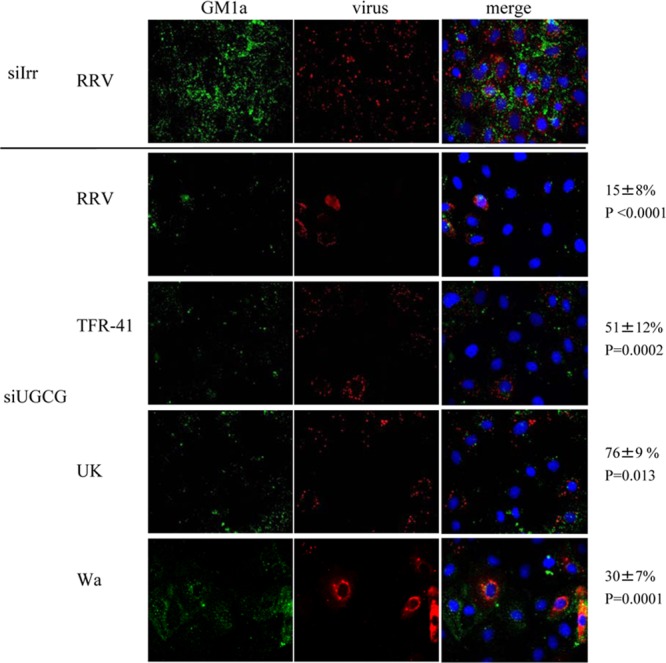
Cells with low ganglioside levels are less susceptible to rotavirus infection. MA104 cells grown in coverslips were transfected with the indicated siRNAs as described in Materials and Methods. Seventy-two hours later, cells were infected with the indicated rotavirus strain (MOI, 3), and eight hours postinfection they were fixed and processed as described in Materials and Methods. Cell membrane ganglioside GM1a was detected with Alexa 488-cholera toxin B subunit (in green); virus-infected cells were detected with polyclonal anti-NSP2 antibody and Alexa 568 anti-rabbit antibody (in red); nuclei were stained with DAPI (in blue). Images were scored independently by two persons, and scores were averaged. In the upper panel a representative staining of cells transfected with siIrr and infected with strain RRV is shown. The lower panel shows representative staining of cells transfected with siUGCG, infected with four different strains. Numbers at the right side correspond to the percentages of infected cells with low levels of ganglioside. Images acquired from at least 3 different experiments were analyzed and counted and represent at least 100 individual cells. Statistical analysis was done using a Student t test, and the values of statistical difference (P) for each virus are shown.
Rotavirus binds efficiently to cells with knocked-down ganglioside synthesis.
To test if gangliosides are involved in rotavirus binding to the cell surface, purified infectious virus particles were added to cells that had been previously transfected with siRNAs and treated or not with NA and were allowed to bind for 1 h at 4°C. After thoroughly washing the cells, the viral particles that remained bound to the cell surface were detected by an ELISA, as described previously (27). The cell attachment of the rotavirus strains tested was not affected by silencing the expression of either UGCG or GM3-s (Fig. 6). The binding of NA-sensitive strains RRV and TFR-41 was decreased after NA treatment, while binding of NA-resistant strains Wa and UK to cells after NA treatment was not affected (Fig. 6). These observations suggest that all rotavirus strains tested could bind to other cell surface molecules besides gangliosides and that at least the NA-sensitive strains need terminal sialic acids, which are cleaved by sialidases. It is not clear what cell molecules could be involved in the interaction of NA-resistant strains Wa and UK with cell membrane; however, the fact that a decreased expression of gangliosides reduces their infectivity suggests that this initial cell binding is not sufficient for viral entry.
Fig 6.
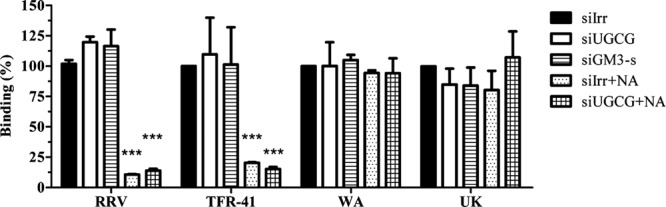
Neuraminidase treatment but not inhibition of ganglioside synthesis decreases cell surface binding of rotavirus strains RRV and TFR-41. MA104 cells were transfected with the indicated siRNAs, and 72 h posttransfection, the cell monolayers were incubated or not with 40 mU of neuraminidase for 1 h at 37°C. After this time, cells were blocked with BSA, and purified rotavirus TLPs were added for 1 h at 4°C, as described in Materials and Methods. Virus bound to cells was determined by an ELISA. Data are expressed as the percentage of virus bound to the cells relative to virus detected in siIrr control treatment cells. The means and standard errors of results from three independent experiments are shown. Statistical analysis was done using a Student t test, and statistically significant values are shown as follows: ***, P < 0.001; **, P < 0.01; *, P < 0.05.
The infectivity of rotavirus is differentially affected by gangliosides, depending on the type of sialic acid position.
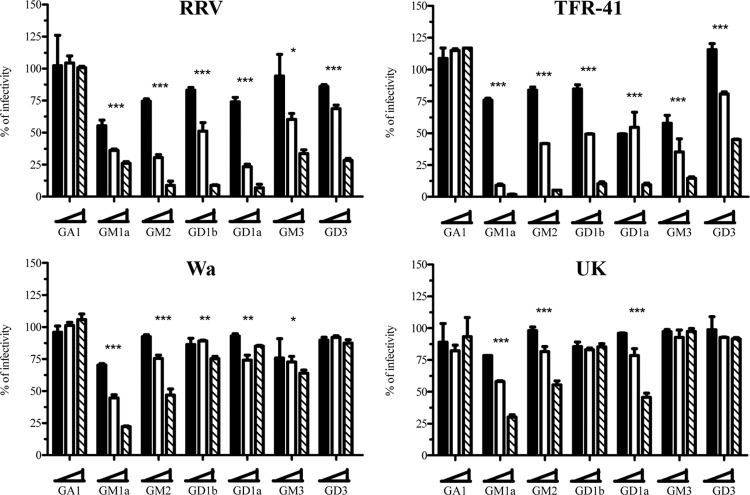
To analyze if the four rotavirus strains tested interact differentially with gangliosides, virus lysates were preincubated with various concentrations of gangliosides for 1 h at 37°C, and the virus-ganglioside mixture was used to infect MA104 cell monolayers. The glycosphingolipids tested could be classified into four groups, based on the presence or absence of SA and/or on the position of their SA residue (Fig. 1): (i) asialoglycosphingolipids (GlcCer, LacCer, and GA1), (ii) gangliosides with one (GM1a and GM2) or two (GD1b) subterminal SA residues, (iii) gangliosides with one (GM3) or two (GD3) terminal SA residues, and (iv) gangliosides with both terminal and subterminal SA (GD1a). None of the asialoglycosphingolipids affected the infectivity of the virus strains tested (shown in Fig. 7 for GA1), suggesting that SAs are key components in the rotavirus-ganglioside interaction. On the other hand, differential inhibition of rotavirus infectivity was observed when gangliosides with an SA moiety in different positions were used. The infectivity of NA-sensitive rotavirus strains RRV and TFR-41 was inhibited by preincubation with all sialic acid-containing gangliosides, with the greatest effect observed (P < 0.001 for the highest ganglioside concentration tested) after preincubation with GM1a, GM2, GD1b, GD1a, and GD3 (strain RRV) or with all gangliosides tested for rotavirus TFR-41 (Fig. 7). With regard to the effect on the infectivity of NA-resistant strains, the highest inhibition in the case of the human Wa virus was observed with GM1a and GM2 (between 50% and 70% decrease), followed by GD1a, GD1b, and GM3 (about 25% decrease), with GD3 having no effect (Fig. 7). In the case of the bovine UK strain, the highest inhibition was also observed with GM1a, GM2, and GD1a (between 50% and 70% decrease) (Fig. 7). Preincubation of UK virus with GM3 or GD3 had no effect on its infectivity, suggesting that these gangliosides do not bind the viral particle. These observations support the differential interaction of NA-sensitive and NA-resistant rotavirus strains with gangliosides, with NA-resistant strains interacting preferentially with gangliosides containing subterminal SA residues.
Fig 7.
Preincubation with gangliosides affects rotavirus infectivity. Four different rotavirus strains (RRV, TFR-41, Wa, and UK) were preincubated with the indicated gangliosides (GA1, GM1a, GM2, GD1b, GD1a, GM3, GD3) in three different concentrations (1.5 [black bars], 12.5 [white bars], and 50 [striped bars] μg/ml). After preincubation, the virus-ganglioside mixture was used to infect MA104 cells. Fourteen hours postinfection, cells were fixed and stained as described in Materials and Methods. The results are expressed as the percentages of infectivity relative to control (preincubation with 0 μg/ml) treatment. The means and standard errors from results of at least three independent experiments are shown. Statistical analysis was done using a Student one-way ANOVA test and a Tukey posttest, and statistically significant values for highest ganglioside concentration are shown as follows: ***, P < 0.001; **, P < 0.01; *, P < 0.05.
DISCUSSION
In this study, we showed that inhibition of ganglioside synthesis by RNAi results in a decreased infectivity of NA-sensitive and NA-resistant rotavirus strains, supporting the observation that gangliosides play a role during rotavirus cell entry. These results are in agreement with the previous observation that inhibition of the synthesis of glucosylceramide (a ganglioside precursor) by d,l-threo-1-phenyl-2-decanoylamino-3-morpholino-1-propanol (PDMP) decreased the infectivity of NA-sensitive and NA-resistant rotavirus strains (30). Interestingly, decreased levels of gangliosides did not affect cell binding of any of the viruses tested. This observation can be explained by the ability of rotaviruses to bind to SA present in other cell surface molecules, like glycoproteins, or they can interact with other cell surface molecules, like the recently described interaction with histo-blood group antigens (24, 25). Also, it cannot be discarded that the viruses could interact with ganglioside GM1b, which has terminal SA and whose synthesis would not be inhibited by the siRNA directed to GM3-s (Fig. 1), or to gangliosides present in nontransfected cells. However, the initial interaction with the cell surface does not seem to be sufficient to promote rotavirus cell entry, which, in this work, we showed requires the presence of gangliosides. Cell binding of NA-sensitive strains was inhibited by NA treatment, confirming that these viruses require terminal SA for attachment to the cell surface. The apparent higher level of inhibition of ganglioside synthesis by the UGCG knockdown (as judged by the detection of GM1a) probably explains the more efficient inhibition of rotavirus infectivity by the siRNA to this enzyme compared to that of the siRNA to GM3-s.
Differences were observed in the ability of different gangliosides to block rotavirus infectivity when preincubated with the virus prior to infection. While the infectivity of NA-sensitive strains was efficiently inhibited by a variety of gangliosides (containing both terminal and subterminal SA residues), a marked inhibition (P < 0.001) of NA-resistant strains was caused only by gangliosides that contain one subterminal SA and, in the case of UK, also by GD1a, which contains an additional terminal SA. In agreement with these observations, Rolsma et al. (20) showed that the infectivity of the porcine, NA-sensitive strain OSU is inhibited by GM3, which is the predominant ganglioside species in the piglet intestine; the interaction of GM3 with the porcine NA-sensitive strain CRW-8 has also been reported (8). Likewise, RRV has been shown to interact with GM3, GM2, and GD1a gangliosides (23), all of which were shown in this work to block the infectivity of this virus (Fig. 7). Of interest, these three gangliosides, which contain both terminal and subterminal SA, seem to be the predominant species in the intestine of Macaca fascicularis (41), suggesting a potential role for rotavirus infection in vivo.
As stated above, gangliosides with a subterminal SA moiety, GM1a, GM2, and GD1b, which are resistant to neuraminidase treatment, blocked the infectivity of the two NA-sensitive strains tested. The interaction of NA-sensitive rotaviruses with gangliosides containing subterminal SA residues has been shown to occur previously; Superti and Donelli observed that the infectivity of the NA-sensitive strain SA11 was blocked by preincubation with GM1a ganglioside (42). Also, two NA-sensitive viruses (NCDV and SA11) were capable of binding to GM2 separated by thin-layer chromatography (23), and cell surface binding of rotavirus OSU was inhibited to different levels by preincubation with GM3, GM2, GM1a, and GD1a (43). The observation that NA-sensitive rotavirus strains interact with gangliosides containing subterminal SA and that this interaction blocks their infectivity contrasts with the fact that NA treatment of cells that removes terminal but not subterminal SA from gangliosides inhibits their infectivity. In this regard, it is important to stress that the in vitro interaction between gangliosides in solution and viruses that leads to the inhibition of virus infectivity does not necessarily imply that the same interaction takes place in the cell membrane during the infection.
The knockdown of ganglioside synthesis decreased the infection of the two NA-resistant strains tested. Furthermore, preincubation of these viruses with gangliosides containing subterminal SAs (GM1a, GM2, and GD1a, resistant to NA treatment) showed the highest efficiency to block viral infectivity. Although originally it was predicted that the NA-resistant human rotavirus strain DS-1 would not interact with carbohydrate ligands in the region that corresponds to the RRV SA binding site (7), a recent report showed an interaction between aceramido-GM1a and the VP8 protein of the NA-resistant strain Wa (9). In addition, GM1a was also shown to inhibit rotavirus KUN, another NA-resistant human strain (10), and bovine strain UK interacted specifically with GM1a, GM2 and GD1a separated by thin-layer chromatography (23). It is also of interest that GM1a is one of the predominant ganglioside species in the human gut (44), making this ganglioside a prime candidate to serve as a human rotavirus receptor in a natural infection. Alternative molecules (A-type HBGA and Lewis b and H type 1 HBGA) have been recently proposed that might be used by different human rotaviruses to interact with the cell surface (24, 25); however, the role of these interactions, particularly with Lewis b and H type 1 HBGA carbohydrates during rotavirus infection, remains to be determined.
We found in this work that the infectivity but not the cell binding of both NA-sensitive and NA-resistant rotavirus strains is affected after silencing the expression of two ganglioside biosynthetic enzymes. This observation suggests that gangliosides are important for rotavirus cell entry and do not simply serve as anchor molecules for virus attachment. Recently, it was proposed that ganglioside GD1a could be responsible for the intracellular sorting of murine polyomavirus to the endoplasmic reticulum rather than to facilitate the entry of the virus (45). In this regard, gangliosides could have different roles in the infection of different rotavirus strains, including the possibility of directing the intracellular traffic of the viruses. More experimental data are needed to elucidate the role of gangliosides in rotavirus infection; however, the present data clearly demonstrate gangliosides have a functional role during rotavirus cell entry.
ACKNOWLEDGMENTS
This work was supported by grants 55005515 from the Howard Hughes Medical Institute, IN210807 from DGAPA-UNAM, and 60025 and 102823 from CONACyT. M.A.M. is a recipient of a scholarship from CONACyT.
Footnotes
Published ahead of print 7 November 2012
REFERENCES
- 1. Lopez S, Arias CF. 2004. Multistep entry of rotavirus into cells: a Versaillesque dance. Trends Microbiol. 12:271–278 [DOI] [PubMed] [Google Scholar]
- 2. Isa P, Arias CF, Lopez S. 2006. Role of sialic acids in rotavirus infection. Glycoconj. J. 23:27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fuentes-Panana EM, Lopez S, Gorziglia M, Arias CF. 1995. Mapping the hemagglutination domain of rotaviruses. J. Virol. 69:2629–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Isa P, Lopez S, Segovia L, Arias CF. 1997. Functional and structural analysis of the sialic acid-binding domain of rotaviruses. J. Virol. 71:6749–6756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blanchard H, Yu X, Coulson BS, von Itzstein M. 2007. Insight into host cell carbohydrate-recognition by human and porcine rotavirus from crystal structures of the virion spike associated carbohydrate-binding domain (VP8*). J. Mol. Biol. 367:1215–1226 [DOI] [PubMed] [Google Scholar]
- 6. Dormitzer PR, Sun ZY, Wagner G, Harrison SC. 2002. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 21:885–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Monnier N, Higo-Moriguchi K, Sun ZY, Prasad BV, Taniguchi K, Dormitzer PR. 2006. High-resolution molecular and antigen structure of the VP8* core of a sialic acid-independent human rotavirus strain. J. Virol. 80:1513–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yu X, Coulson BS, Fleming FE, Dyason JC, von Itzstein M, Blanchard H. 2011. Novel structural insights into rotavirus recognition of ganglioside glycan receptors. J. Mol. Biol. 413:929–939 [DOI] [PubMed] [Google Scholar]
- 9. Haselhorst T, Fleming FE, Dyason JC, Hartnell RD, Yu X, Holloway G, Santegoets K, Kiefel MJ, Blanchard H, Coulson BS, von Itzstein M. 2009. Sialic acid dependence in rotavirus host cell invasion. Nat. Chem. Biol. 5:91–93 [DOI] [PubMed] [Google Scholar]
- 10. Guo CT, Nakagomi O, Mochizuki M, Ishida H, Kiso M, Ohta Y, Suzuki T, Miyamoto D, Hidari KI, Suzuki Y. 1999. Ganglioside GM(1a) on the cell surface is involved in the infection by human rotavirus KUN and MO strains. J. Biochem. 126:683–688 [DOI] [PubMed] [Google Scholar]
- 11. Lopez PH, Schnaar RL. 2009. Gangliosides in cell recognition and membrane protein regulation. Curr. Opin. Struct. Biol. 19:549–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Regina Todeschini A, Hakomori SI. 2008. Functional role of glycosphingolipids and gangliosides in control of cell adhesion, motility, and growth, through glycosynaptic microdomains. Biochim. Biophys. Acta 1780:421–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zeng G, Yu RK. 2008. Cloning and transcriptional regulation of genes responsible for synthesis of gangliosides. Curr. Drug Targets 9:317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Neu U, Stehle T, Atwood WJ. 2009. The Polyomaviridae: contributions of virus structure to our understanding of virus receptors and infectious entry. Virology 384:389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Taube S, Jiang M, Wobus CE. 2010. Glycosphingolipids as receptors for non-enveloped viruses. Viruses 2:1011–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Villar E, Barroso IM. 2006. Role of sialic acid-containing molecules in paramyxovirus entry into the host cell: a minireview. Glycoconj. J. 23:5–17 [DOI] [PubMed] [Google Scholar]
- 17. Schmidt M, Chiorini JA. 2006. Gangliosides are essential for bovine adeno-associated virus entry. J. Virol. 80:5516–5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bergelson LD, Bukrinskaya AG, Prokazova NV, Shaposhnikova GI, Kocharov SL, Shevchenko VP, Kornilaeva GV, Fomina-Ageeva EV. 1982. Role of gangliosides in reception of influenza virus. Eur. J. Biochem. 128:467–474 [DOI] [PubMed] [Google Scholar]
- 19. Taube S, Perry JW, Yetming K, Patel SP, Auble H, Shu L, Nawar HF, Lee CH, Connell TD, Shayman JA, Wobus CE. 2009. Ganglioside-linked terminal sialic acid moieties on murine macrophages function as attachment receptors for murine noroviruses. J. Virol. 83:4092–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rolsma MD, Kuhlenschmidt TB, Gelberg HB, Kuhlenschmidt MS. 1998. Structure and function of a ganglioside receptor for porcine rotavirus. J. Virol. 72:9079–9091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Proia RL. 2003. Glycosphingolipid functions: insights from engineered mouse models. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358:879–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tettamanti G. 2004. Ganglioside/glycosphingolipid turnover: new concepts. Glycoconj. J. 20:301–317 [DOI] [PubMed] [Google Scholar]
- 23. Delorme C, Brussow H, Sidoti J, Roche N, Karlsson KA, Neeser JR, Teneberg S. 2001. Glycosphingolipid binding specificities of rotavirus: identification of a sialic acid-binding epitope. J. Virol. 75:2276–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu L, Crawford SE, Czako R, Cortes-Penfield NW, Smith DF, Le Pendu J, Estes MK, Prasad BV. 2012. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature 485:256–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang P, Xia M, Tan M, Zhong W, Wei C, Wang L, Morrow A, Jiang X. 2012. Spike protein VP8* of human rotavirus recognizes histo-blood group antigens in a type-specific manner. J. Virol. 86:4833–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu Y, Huang P, Tan M, Biesiada J, Meller J, Castello AA, Jiang B, Jiang X. 2012. Rotavirus VP8*: phylogeny, host range, and interaction with histo-blood group antigens. J. Virol. 86:9899–9910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zarate S, Espinosa R, Romero P, Guerrero CA, Arias CF, Lopez S. 2000. Integrin alpha2beta1 mediates the cell attachment of the rotavirus neuraminidase-resistant variant nar3. Virology 278:50–54 [DOI] [PubMed] [Google Scholar]
- 28. Gutierrez M, Isa P, Sanchez-San Martin C, Perez-Vargas J, Espinosa R, Arias CF, Lopez S. 2010. Different rotavirus strains enter MA104 cells through different endocytic pathways: the role of clathrin-mediated endocytosis. J. Virol. 84:9161–9169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Isa P, Realpe M, Romero P, Lopez S, Arias CF. 2004. Rotavirus RRV associates with lipid membrane microdomains during cell entry. Virology 322:370–381 [DOI] [PubMed] [Google Scholar]
- 30. Guerrero CA, Zarate S, Corkidi G, Lopez S, Arias CF. 2000. Biochemical characterization of rotavirus receptors in MA104 cells. J. Virol. 74:9362–9371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Winer J, Jung CK, Shackel I, Williams PM. 1999. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal. Biochem. 270:41–49 [DOI] [PubMed] [Google Scholar]
- 32. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 33. Carreno-Torres JJ, Gutierrez M, Arias CF, Lopez S, Isa P. 2010. Characterization of viroplasm formation during the early stages of rotavirus infection. Virol. J. 7:350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sanchez-San Martin C, Lopez T, Arias CF, Lopez S. 2004. Characterization of rotavirus cell entry. J. Virol. 78:2310–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Diaz-Font A, Chabas A, Grinberg D, Vilageliu L. 2006. RNAi-mediated inhibition of the glucosylceramide synthase (GCS) gene: a preliminary study towards a therapeutic strategy for Gaucher disease and other glycosphingolipid storage diseases. Blood Cells Mol. Dis. 37:197–203 [DOI] [PubMed] [Google Scholar]
- 36. Reference deleted.
- 37. Maruri-Avidal L, Lopez S, Arias CF. 2008. Endoplasmic reticulum chaperones are involved in the morphogenesis of rotavirus infectious particles. J. Virol. 82:5368–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fishman PH. 1982. Role of membrane gangliosides in the binding and action of bacterial toxins. J. Membr. Biol. 69:85–97 [DOI] [PubMed] [Google Scholar]
- 39. Kimura M, Hidari KI, Suzuki T, Miyamoto D, Suzuki Y. 2001. Engagement of endogenous ganglioside GM1a induces tyrosine phosphorylation involved in neuron-like differentiation of PC12 cells. Glycobiology 11:335–343 [DOI] [PubMed] [Google Scholar]
- 40. Dector MA, Romero P, Lopez S, Arias CF. 2002. Rotavirus gene silencing by small interfering RNAs. EMBO Rep. 3:1175–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dahiya R, Brown MD, Brasitus TA. 1986. Distribution of glycosphingolipids of monkey small and large intestinal mucosa. Lipids 21:107–111 [DOI] [PubMed] [Google Scholar]
- 42. Superti F, Donelli G. 1991. Gangliosides as binding sites in SA-11 rotavirus infection of LLC-MK2 cells. J. Gen. Virol. 72(Part 10):2467–2474 [DOI] [PubMed] [Google Scholar]
- 43. Rolsma MD, Gelberg HB, Kuhlenschmidt MS. 1994. Assay for evaluation of rotavirus-cell interactions: identification of an enterocyte ganglioside fraction that mediates group A porcine rotavirus recognition. J. Virol. 68:258–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Keranen A. 1975. Gangliosides of the human gastrointestinal mucosa. Biochim. Biophys. Acta 409:320–328 [DOI] [PubMed] [Google Scholar]
- 45. Qian M, Cai D, Verhey KJ, Tsai B. 2009. A lipid receptor sorts polyomavirus from the endolysosome to the endoplasmic reticulum to cause infection. PLoS Pathog. 5:e1000465 doi:10.1371/journal.ppat.1000465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chester MA. 1999. IUPAC-IUB joint commission on biochemical nomenclature (JCBN) nomenclature of glycolipids: recommendations 1997. J. Mol. Biol. 286:963–970 [DOI] [PubMed] [Google Scholar]