Abstract
St. Louis encephalitis virus (SLEV) is a mosquito-borne flavivirus responsible for several human encephalitis outbreaks over the last 80 years. Mature flavivirus virions are coated with dimeric envelope (E) proteins that mediate attachment and fusion with host cells. E is a class II fusion protein, the hallmark of which is a distinct dimer-to-trimer rearrangement that occurs upon endosomal acidification and insertion of hydrophobic fusion peptides into the endosomal membrane. Herein, we report the crystal structure of SLEV E in the posfusion trimer conformation. The structure revealed specific features that differentiate SLEV E from trimers of related flavi- and alphaviruses. SLEV E fusion loops have distinct intermediate spacing such that they are positioned further apart than previously observed in flaviviruses but closer together than Semliki Forest virus, an alphavirus. Domains II and III (DII and DIII) of SLEV E also adopt different angles relative to DI, which suggests that the DI-DII joint may accommodate spheroidal motions. However, trimer interfaces are well conserved among flaviviruses, so it is likely the differences observed represent structural features specific to SLEV function. Analysis of surface potentials revealed a basic platform underneath flavivirus fusion loops that may interact with the anionic lipid head groups found in membranes. Taken together, these results highlight variations in E structure and assembly that may direct virus-specific interactions with host determinants to influence pathogenesis.
INTRODUCTION
St. Louis encephalitis virus (SLEV) was first discovered as the agent responsible for over 1,000 cases of encephalitis during a 1933 summer outbreak in St. Louis, MO (1, 2). Additional epidemics have occurred from 1964 to 2006 in the Americas, ranging from the United States (3–5) to Argentina and Brazil (6–8). However, reported infections have declined significantly in the United States since West Nile virus (WNV) emerged in 1999 (9), suggesting WNV has since outcompeted SLEV in North America. SLEV naturally cycles between Culex, Aedes, Anopheles, and Theobaldia genera (10, 11) mosquitoes and passerine birds, but humans may also become infected as “dead end” hosts through mosquito bites.
SLEV is a member of the Flavivirus genus of the Flaviviridae family. Other flaviviruses known to infect humans include dengue virus (DENV), Japanese encephalitis virus (JEV), and tick-borne encephalitis virus (TBEV). Each flavivirus is classified into a serocomplex with its own distinct tropism and pathogenesis, based on cross-neutralization tests (12, 13). More specifically, when polysera harvested from an animal infected with one virus neutralizes a different virus in vitro, the two are said to belong to the same serocomplex. SLEV, JEV, and WNV are members of the JEV serocomplex, while DENV and TBEV each represent prototypical members of other prominent serocomplexes. JEV serocomplex viruses are transmitted by mosquitoes and often cause asymptomatic infections, but severe cases can result in nausea, fever, and meningoencephalitis (14). TBEV, Langat virus, and Powassan virus make up the TBEV serocomplex and are transmitted by hard ticks. These viruses cause symptoms similar to those of the JEV serocomplex but spread to secondary tissue more slowly and are more frequently associated with severe symptoms (15). The DENV serocomplex contains 4 serotypes (DENV1 to -4) of mosquito-borne viruses. Infection with any DENV serotype can cause headaches, muscle pain, skin rash, and hemorrhagic fever.
The flavivirus virion is enveloped, and its surface is decorated with 180 transmembrane envelope (E) proteins (16, 17) that mediate cellular attachment and fusion with host membranes. Flavivirus E, along with the alphavirus E1 protein, represent class II viral fusion proteins based on their activation mechanisms and three-dimensional structures (18–20). Features that define class II proteins are a requisite dimer-to-trimer rearrangement prior to fusion and a distinct β-strand-rich, 3-domain architecture (18–20). The SLEV E ectodomain is 68% identical to serocomplex-related JEV E but only 46% and 40% identical to those of DENV2 and TBEV, viruses from different serocomplexes. Alphavirus E1, however, shares no appreciable sequence identity to SLEV E or any other flavivirus E protein. Despite differences in their amino acid sequences and arrangements on the viral particle, the structures of E and E1 are remarkably similar (16, 21–25). The conservation of three domains has been well documented in crystal structures of flavivirus E ectodomains from TBEV (24), DENV (26), JEV (27), and WNV (28, 29), as well as alphavirus E1 from Semliki Forest virus (SFV) (25) and Chikungunya virus (CHIKV) (30). Domain I (DI) is a β-barrel, DII is formed from two discontinuous loops that protrude from DI, and DIII is an Ig-like sandwich positioned on the opposite side of DI from DII. Antibodies that neutralize SLEV (31, 32), WNV (33–39), DENV (40–43), and JEV (44, 45) have been mapped to epitopes on all three domains. These antibodies can inhibit both attachment to cells and fusion in the endosome (46–48).
Cryo-electron microscopy (cryo-EM) reconstructions of mature WNV (17) and DENV (16) revealed antiparallel E homodimers arranged with icosahedral symmetry on the surface of the mature virion. This head-to-tail arrangement results in burial of the hydrophobic fusion loop at the dimer interface to prevent premature insertion into host membranes. Upon internalization, the acidic pH of the endosome disrupts the E dimers and transforms them into a fusogenic state (49, 50). E inserts its fusion loops into the endosomal membrane and collapses into a compact, trimeric conformation that forms when DIII swings in an arc to pack against DI beneath DII (21, 23, 49, 51–53). Alphavirus E1 forms heterodimers with chaperone protein E2 but dissociates and undergoes a similar conformational change when acidified in the late endosome (54). In either case, this transition is believed to provide the energy to fuse viral and host membranes, releasing the nucleocapsid into the cyotsol.
We previously established that prefusion E proteins of the JEV serocomplex have distinct biophysical characteristics, such as a small dimer interface and diminished dimerization propensity (27, 29). We therefore determined the crystal structure of acidified SLEV E to examine the structure and assembly of the trimeric, postfusion conformation. SLEV E served as a representative of the JEV serocomplex for comparative analysis with class II fusion proteins from DENV and TBEV serocomplexes and the distantly related alphavirus SFV. SLEV E did not crystallize in the prefusion conformation, so we utilized the structure of serocomplex-related JEV E as a model to evaluate probable differences between pre- and postfusion SLEV E. Our analyses revealed that postfusion envelopes possess a series of well-conserved trimer contacts yet exhibit substantial differences in domain orientation, fusion loop spacing, and electrostatic surface potential. These biophysical properties may influence pathogenesis by modulating interactions with environmental factors, such as the specific fusion pH or host membrane composition.
MATERIALS AND METHODS
Cloning, expression, and purification of SLEV E.
Residues 1 to 407 of the SLEV E ectodomain (strain MS1-7) were cloned into the pET21a(+) expression vector. The vector was transformed into Escherichia coli BL21-CodonPlus-DE3(RIL) cells. An individual colony was used to inoculate four 10-ml starter cultures that were transferred to 4 liters of methionine-deficient medium (Athena Enzyme Systems). Cultures were grown at 37°C until reaching an optical density (OD) of 0.6. Upon reaching this OD, cultures were supplemented with 0.5 g feedback inhibition stock (0.1 g l-lysine, 0.1 g l-threonine, 0.1 g l-phenylalanine, 0.05 g l-leucine,0.05 g l-isoleucine, 0.05 g l-valine, and 0.05 g l-selenomethionine) to prevent synthesis of methionine and initiate incorporation of selenomethionine (protocol adapted from that described by Stols et al. [55]). At 15 min after addition of this stock, 1 ml of 1 M isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to each 1-liter culture to induce protein expression. After 6 h, cultures were harvested and cells were pelleted. Cells were solubilized in solution buffer (50 mM Tris [pH 8.0], 25% sucrose, 10 mM dithiothreitol [DTT]), and an equal amount of lysis buffer (50 mM Tris [pH 8.0], 1% Triton X-100, 100 mM NaCl, 10 mM DTT) was added. Lysozyme was added to a final concentration of 0.8 mg/ml, and the mixture was sonicated to disrupt cellular membranes. This solution was then centrifuged at 10,000 rpm to pellet inclusion bodies. The inclusion bodies were washed 3 times with wash buffer (50 mM Tris [pH 8.0], 0.5% Triton X-100, 100 mM NaCl, 1 mM DTT) and sonicated after each wash. Finally, the mixtures were washed in buffer without Triton X-100 and then solubilized in 6 M guanidine-HCl, 10 mM Tris (pH 8.0), and 20 mM β-mercaptoethanol. Aliquots of this solution were added dropwise to a reservoir of refolding buffer containing 400 mM nondetergent sulfobetaine 201 (NDSB-201), 100 mM Tris (pH 8.0), 0.5 mM oxidized glutathione, and 5 mM reduced glutathione and allowed to fold overnight. Protein was then concentrated using an Amicon 400 concentrator with 30-kDa cutoff membrane and purified by size exclusion chromatography (SEC) and anion-exchange chromatography.
Crystallization.
Purified, soluble SLEV E was dialyzed into a buffer containing 0.05 M acetate (pH 5.5) and 10 mM sodium chloride and crystallized by hanging drop vapor diffusion at 20°C. Each drop was a mixture of 0.5 μl of protein solution at a 5-mg/ml concentration and 0.5 μl of mother liquor containing 0.1 M acetate (pH 5.5), 3% polyethylene glycol 8000 (PEG 8000), and 2% ethylene glycol. Diffraction quality crystals grew in 3 to 7 days. Crystals were rapidly dragged through a drop of 25% ethylene glycol for cryoprotection prior to cooling in liquid nitrogen. Data were collected at the APS (advanced photon source) 19-ID beamline and processed, integrated, and scaled using HKL2000 (56). SLEV E crystallized in space group I23, with unit cell dimensions of a = b = c = 177.5 and α = β = γ = 90°.
Structure determination.
A model of SLEV E in the postfusion conformation was utilized to solve the structure by molecular replacement (MR). This model was generated with the Phyre2 server (57) by threading the SLEV E sequence onto DI and DIII of DENV2 E in the postfusion trimer conformation (PDB ID 1OK8) and onto DII of JEV E (PDB ID 3P54). MR was performed with the PHENIX program (58). Refinement was carried out by using a variety of approaches. PHENIX refine was used to perform rigid body refinement of the three domains, followed by positional refinement incorporating secondary structure restraints, simulated annealing, and TLS (translation liberation screw) refinement (59) using three groups and B-factor refinement. The resultant partial model was used to carry out a combination of MR and selenomethionine single anomalous dispersion (MRSAD) phasing in the PHENIX Autosol program (58, 60). The improved maps generated by the incorporation of the experimental phase allowed for model building in Coot (61) with the assistance of B-factor sharpening using a value of 15Å2. Additional rounds of positional and B-factor refinement were performed using the jelly body function of refmac5 (62) or with PHENIX refine (58). The final structure had an Rwork of 22.2% and Rfree of 26.7% and contained ectodomain residues 2 to 145 and 163 to 404 of the 407 total encompassed by the construct.
Structural analysis.
Anomalous difference peaks were generated with PHENIX Autosol (58) and the CCP4 Fast Fourier transform (FFT) algorithm (63). First, SAD phasing was carried out in Autosol in the absence of a partial model to avoid model bias. Next, this solution and the SLEV E model were used as inputs for the CCP4 FFT to generate a map for use in visualization software. The resultant density was visualized in Pymol (64) at a contour level of 4.5σ. Interatomic distances between fusion loops were measured in Pymol. The PISA Web server (65) was utilized to determine the buried surface area and contacts between E protomers. ClustalW was used to align flavivirus E sequences (66). To determine angles between DI and DII or DIII, entire E proteins from DENV1, DENV2, TBEV, and SFV were superimposed onto SLEV DI by secondary structure matching in Coot (61). Next, artificial models of each E protein were generated by superimposing DI, DII, and DIII individually onto SLEV E to create a reference structure. The angles between DI and DII or DI and DIII of the native structure versus the reference structure were then calculated with Dyndom (63, 67). Electrostatic surface potentials were determined by using PDB2PQR (68, 69) to generate PQR files with protonation states assigned at pH 6.0 for use in the APBS (70) plugin for Pymol and visualized over a range of −3 kT/e to +3kT/e.
Protein structure accession number.
The coordinates for the protein structure have been deposited in the Protein Data Bank and assigned PDB ID number 4FG0.
RESULTS
SLEV E structure.
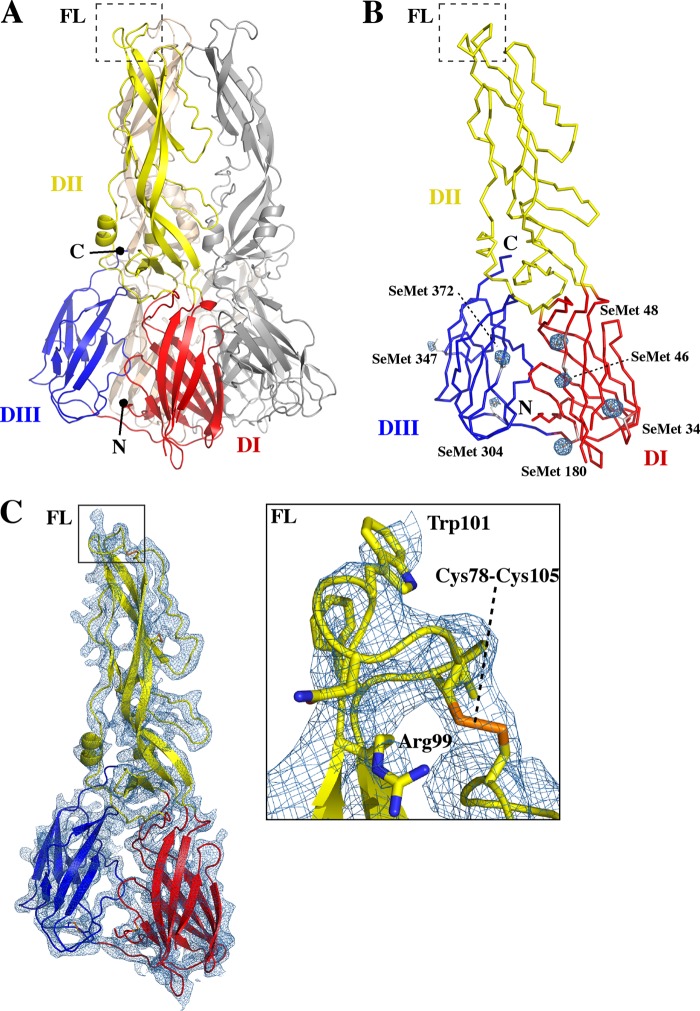
A selenomethionine (SeMet)-substituted SLEV E ectodomain from strain MS1-7 lacking the C-terminal stem region was overexpressed as bacterial inclusion bodies. These inclusion bodies were solubilized and oxidatively refolded to allow for proper disulfide formation. This strategy was employed because initial crystals of SLEV E diffracted poorly, and so the experimental phase information gained through this incorporation could be used to validate the structure through the identification of anomalous SeMet peaks in the electron density maps. Purified SLEV E crystallized in cubic space group I23, and phase was determined using a combination of MR and SAD data. There was one E monomer per asymmetric unit, but application of cubic symmetry generated the postfusion trimer (Fig. 1A). The structure was refined at 3.9-Å resolution with an Rwork of 22.2% and Rfree of 26.7% (Table 1), and it included ectodomain residues 2 to 145 and 163 to 404. Residues 146 to 162 of the loop between the Eo and Fo strands were disordered, as has been observed in other postfusion E structures (23, 51, 52).
Fig 1.
Structure and assembly of SLEV E. SLEV E has the 3-domain architecture characteristic of class II viral fusion proteins. (A) The SLEV E postfusion trimer was generated through application of cubic symmetry and is displayed in a cartoon representation. DI, DII, and DIII are colored red, yellow, and blue in one E subunit, and the symmetry mates are colored gray and wheat, respectively. (B) Ribbon representation of one protomer of the SLEV E fusogenic trimer with DI, DII, and DIII, shown in red, yellow, and blue, respectively. Selenomethionine side chains are displayed in stick representation, and anomalous peaks generated from experimental phasing are shown in a mesh representation contoured at 4.5σ. C. The MR SAD electron density map is displayed as a blue mesh over an entire SLEV E monomer contoured at 1.5σ. The inset is a zoom box of the fusion loop with side chains displayed in stick representation. Well-resolved residues Arg99, Trp103, and the disulfide formed by Cys78 and Cys105 are labeled.
Table 1.
Data collection and refinement statistics
| Data parameter | Statistic(s) or value(s) |
|---|---|
| Space group | I23 |
| Cell dimensions | a = b = c = 177.5 +, α = β = γ = 90.00° |
| Resolution (Å) | 50.0–3.90 + (4.04–3.90 +)a |
| Completeness (%) | 98.9 (99.5)a |
| Redundancy | 22.1 (22.6) |
| I/σ | 30.4 (6.2) |
| R-merge (I) (%) | 11 (7.3) |
| Refinement | |
| Resolution (Å) | 34.8–3.90 + (4.14–3.90 +)a |
| No. of Rwork reflectionsb (F > 0) | 8,555 (1,228)a |
| No. of Rfree reflections | 847 (132) |
| Rwork | 0.222 (0.285) |
| Rfree | 0.267 (0.333) |
| SLEV E residues (no. of atoms) | 384 (2,944) |
| Mean isotropic B-value (Å2) | 152.6 |
| RMSD bond length (Å) | 0.002 |
| RMSD bond angle (°) | 0.52 |
| Estimated coordinate error | 1.01 |
| Ramachandran outliers, favored, allowed (%) | 0.00, 92.4, 7.6 |
| Rotamer outliers (%) | 0.00 |
Values in parentheses are for data in the highest-resolution shell.
Statistics are as defined in the Phenix program.
SLEV E adopted the three-domain architecture characteristic of class II viral fusion proteins (Fig. 1A). DI is a β-barrel composed of 8 strands, and DII is formed from two extended loops that protrude from DI. DII contains a long β-sheet made up of strands in both portions of this discontinuous domain that supports the fusion loop. At the opposite end of DI is a flexible linker that connects to DIII, a 7-stranded β-sandwich similar to the classical Ig fold. Since the structure was determined at low resolution, anomalous peaks generated through SAD phasing in the absence of a model were used for validation. Each of the 7 SLEV E selenomethionine residues localized to the density at these peaks (Fig. 1B), confirming the sequence register and accuracy of the model. The electron density map generated from the MR SAD phasing also corresponded well to the main chain and most side chains of the model, including those of the functionally important fusion loop region (Fig. 1C).
Differences in JEV serocomplex E structures.
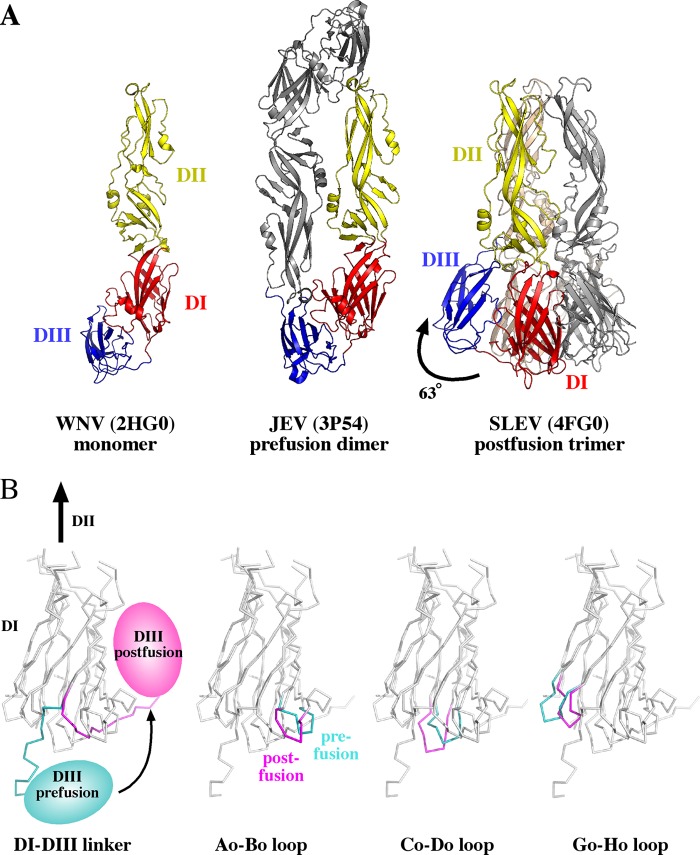
Crystal structures of JEV serocomplex E proteins from SLEV, WNV, and JEV provided a detailed structural perspective of the viral life cycle. While it would have been ideal to compare the pre- and postfusion conformations from SLEV E, extensive trials only yielded diffracting crystals of the postfusion trimer. In the absence of such a structure, we believe monomeric and prefusion E proteins from related serocomplex members WNV and JEV provided a suitable basis for comparison. WNV E crystallized in an unusual monomer conformation that may represent a structural transition that occurs during maturation or acidification. The WNV E proteins assembled in a perpendicular lattice with fusion loops buried in the same pocket they occupy in the mature dimer (29). JEV E formed the antiparallel prefusion dimer observed in cryo-EM structures of mature virions, with fusion loops buried in a cavity formed by DI and DIII of the opposing subunit (27). SLEV E, on the other hand, crystallized as a postfusion trimer (Fig. 2A). In this conformation, DIII swings 63° toward DI relative to its equivalent position in the JEV E dimer. This reorientation causes it to interact with its parent DI, instead of DII of a neighboring protomer.
Fig 2.
Structural differences between JEV serocomplex E proteins. (A) WNV, JEV, and SLEV E represent monomeric, dimeric, and trimeric assemblies of JEV serocomplex E proteins. DI, DII, and DIII are shown in red, yellow, and blue, respectively. The second protomer of the JEV dimer is shown in gray, and the second and third protomers of the SLEV E trimer are shown in gray and wheat. A curved arrow in the middle JEV panel highlights the shift in DIII position that leads to the trimeric structure in the SLEV panel on the right. (B) Structural differences in prefusion JEV E compared to postfusion SLEV E are highlighted in the 4 panels. Regions of DI with substantial structural differences are shown in teal in the prefusion structure and magenta in the postfusion structure. The specific structural region highlighted is indicated in the text below each panel.
Several loops that protrude from the DI β-barrel toward DIII have specific conformations that accommodate the pre- or postfusion DI-DIII linker. The JEV prefusion linker lies between the Co-Do and Go-Ho strands (labeled as in Fig. 2B) and emerges from DI as an essentially linear peptide. In the postfusion structure, this peptide kinks at residues D291/K292 and threads back toward DI between the Ao-Bo and Co-Do loops (Fig. 2B). The position of the prefusion linker results in a shift of the Go-Ho loop outward from the DI core relative to the postfusion structure. However, the postfusion DI Ao-Bo and Co-Do loops are splayed apart to permit this alternate trajectory.
Trimer packing and assembly.
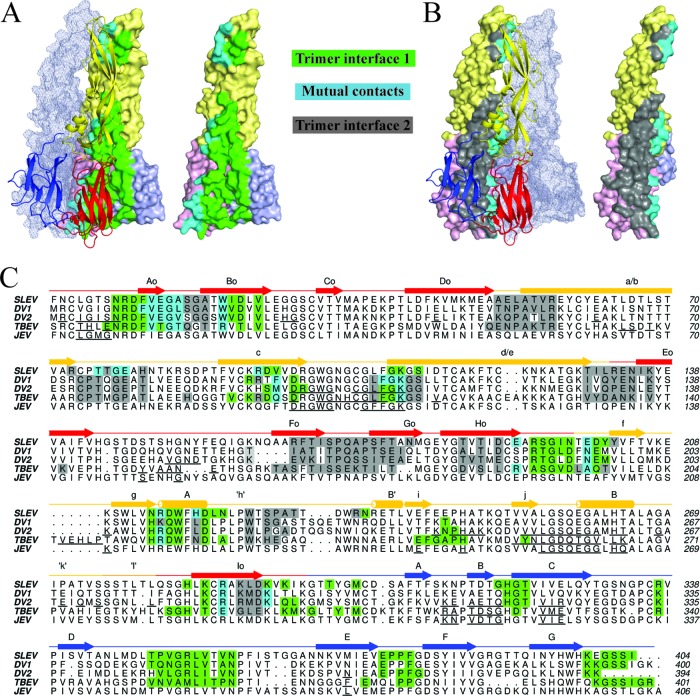
Comparison of trimer and dimer contacts from a variety of flavivirus E structures revealed two unique interfaces, with the exception of a few overlapping residues near the fusion loop (Fig. 3B). Thus, the chemical changes induced by the acidic pH of the endosome enable the formation of entirely new interfaces. SLEV E trimers are stabilized by a series of interactions involving all three domains. The lateral edge of DI from one protomer packs into a groove formed by DI and DIII of its neighbor to secure the base of the trimer (Fig. 3A). Additionally, the edge of DII contacts residues of an adjacent DII near the DI-DII joint and beneath the fusion loop. SLEV trimers were generated by application of 3-fold crystallographic symmetry, so that the two interfaces were identical on each protomer.
Fig 3.
Conservation of flavivirus E trimer interfaces. (A) The left panel displays trimer interface 1, formed by two E protomers. Residues contacted by the cartoon representation are highlighted on the surface-rendered protein. Green residues represent contacts exclusive to the surface-rendered protein, while cyan residues are contacts in both molecules at the interface. The third E protomer that does not make contacts at the highlighted interface is shown as a transparent mesh envelope. (B) The right panel displays trimer interface 2. Contacts in both molecules in this panel are also cyan, while residues exclusive to the surface-rendered protein are shown in gray. The protomer that does not make contacts at this interface is also shown in mesh representation. (C) A ClustalW sequence alignment of SLEV, DENV1, DENV2, and TBEV E. SLEV E domains I, II, and III are shown in red, yellow, and blue, respectively. The SLEV E secondary structure is displayed above the sequence with a straight line for coils/loops, arrows for strands, and cylinders for helices. Contact residues are highlighted in the same color scheme described for panels A and B. For JEV, DENV2, and TBEV E proteins with dimeric structures available, the dimer contacts are underlined in black.
We previously reported that JEV E possesses substantially less buried surface area than other dimeric E structures and does not favor dimerization at prefusion (neutral) pH, as evaluated by multiangle light scattering (27). SLEV E and WNV E also did not dimerize under these conditions, suggesting that the icosahedral assembly of JEV serocomplex dimers is stabilized by lateral contacts present in mature virions. We therefore compared contact residues and the buried surface area of SLEV E relative to DENV1/2 and TBEV to investigate serocomplex-specific features of postfusion trimer structure and stability. While JEV E dimers buried ∼50% less surface area than DENV or TBEV E, the extensive SLEV E trimer interface buried 2,100 Å2, ∼10% more than DENV1/2 E and the same amount as TBEV E or SFV E1 (Table 2).
Table 2.
Comparisons of trimer structures and assemblies
| Virus (trimer) | Distance between fusion loops (Å) | DI-DII angle (°) relative to SLEV E | DI-DIII angle (°) relative to SLEV E | Buried surface area (Å2)a |
|---|---|---|---|---|
| SLEV (4FG0) | 35 | 2,103 | ||
| DENV1 (3G7T) | 27 | 10.7 | 2.5 | 1,893 |
| DENV2 (1OK8) | 26 | 10.8 | 6.8 | 1,968 |
| TBEV (1URZ) | 26 | 8.7 | 16.1 | 2,105* |
| SFV (1RER) | 47 | 9.9 | 65.2 | 2,139* |
*, the buried surface area was averaged over each trimer subunit.
Class II trimer relative domain orientations.
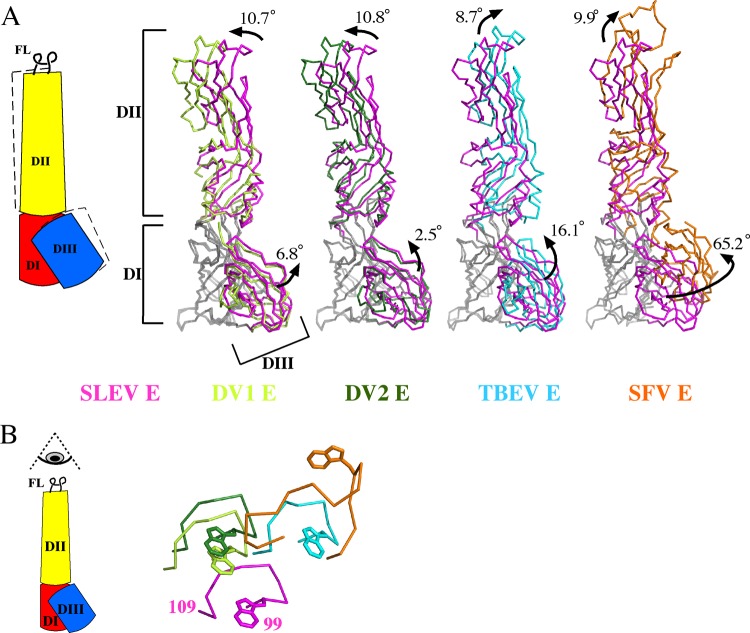
Class II envelope proteins are comprised of three rigid domains linked by two joints (24, 30, 71, 72). These flexible regions connect the central DI to DII on one side and DIII on the other. Conformational changes borne out of twisting or bending of domains about these residues are vital to many stages of the flavivirus life cycle. SLEV DI was used as a reference for structural alignments to compare relative domain orientations in postfusion envelope structures from TBEV, DENV1, DENV2, and SFV (Fig. 4A).
Fig 4.
Relative domain orientations of class II trimers. (A) DI of E or E1 proteins in the postfusion conformation was aligned to SLEV DI, and the angles between DI-DII and DI-DIII were measured. A cartoon describing shifts in the position of DII and DIII is displayed (left). Alignments are displayed as ribbons, with DI of each envelope shown in gray and SLEV E DII and DIII shown in magenta. DENV1 (lime), DENV2 (green), and TBEV (cyan) E or SFV E1 (orange) are superimposed onto SLEV E. (B) A top-down view of the fusion loops emphasizes the different relative positions of each DII (colored as for panel A).
The angles relating DI-DII and DI-DIII varied considerably among the trimeric structures. DII from DENV1 and DENV2 adopted similar conformations, shifted ∼11° relative to SLEV DII (Table 2). TBEV and SFV exhibited ∼9 to 10° differences in the DI-DII angle relative to SLEV but are tilted in a different direction than those of DENV1/2 (Fig. 4A; Table 2). When viewing the fusion loops from a top-down perspective, DII does not appear to have crystallized in positions along a single trajectory but instead pivots about DI (Fig. 4B). These variable DII angles suggest that the DI-DII joint operates like a ball and socket rather than a hinge.
The relative positions of DIII vary over an even larger range than those of DII. DENV1 and DENV2 DIII are tilted 6.5° and 2.5° in comparison to that of SLEV, while TBEV DIII is rotated 16.5° in this same direction about DI (Fig. 4A). SFV DIII was rotated 65° relative to SLEV DIII, representing a dramatic difference in the conformation of postfusion alphavirus versus flavivirus envelopes. This twisting of DIII alters the overall trimer geometry, interaction surface with DI, and potentially the direction the stem-loop projects from its C terminus.
Fusion loop separation.
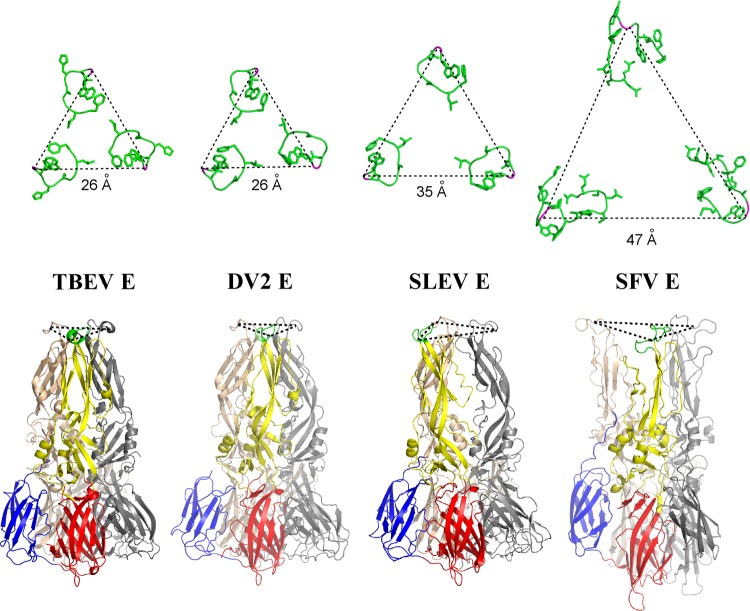
The different DI-DII angles and trimer assemblies observed among flavi- and alphavirus envelopes alter the distance between fusion loops. SFV E1 fusion peptides are positioned 47 Å apart and adopt an uncoiled conformation structurally distinct from those of the flaviviruses (Fig. 5). A length of 26 Å separates the loops of DENV1, DENV2, and TBEV E, while SLEV E fusion loops are situated at an intermediate distance of 35 Å (Fig. 5; Table 2). Trimer interfaces are well conserved in these E proteins, so this variable fusion loop spacing can be attributed to changes in DII tertiary structure and pivot positions about DI. It should be noted that the recombinant SLEV E is selenomethionine substituted, but any perturbations due to this incorporation will be local and unlikely to result in changes in domain orientation.
Fig 5.
Fusion loop separation. Fusion loops are shown in green with side chain sticks, and the conserved glycine used as a reference point for distance measurement across loops is shown in magenta (top row). Postfusion trimers from TBEV, DENV2, SLEV, and SFV are displayed in cartoon representation; DENV1 loops were equivalently spaced to DENV2 but are not shown. DI, DII, DIII, and the fusion loop of one E subunit are shown in red, yellow, blue, and green, respectively (bottom row). The second and third protomers are shown in gray and wheat.
Trimer electrostatics.
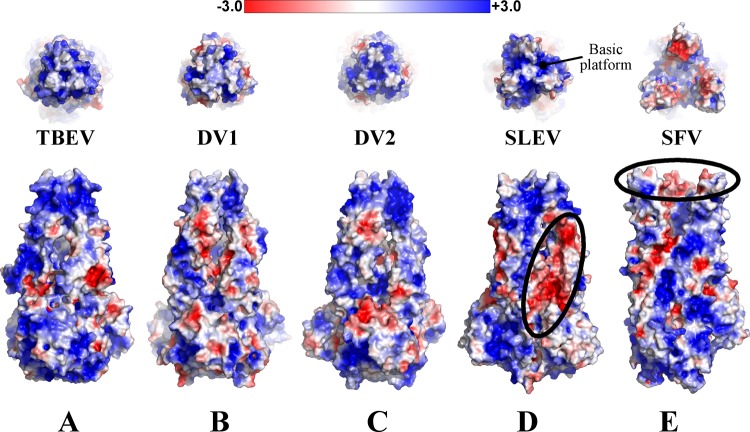
To investigate electrostatic features of E trimers that may contribute to structural changes or interactions with host determinants, we visualized the surfaces on postfusion E or E1 from DENV1, DENV2, TBEV, SLEV, and SFV. Protonation states were assigned at pH 6.0 to estimate the acidic environment of the endosome. In the flavivirus trimers, uncharged crowns of fusion loops lie atop a basically charged platform (Fig. 6A to D). Given that the fusion loops project into the hydrophobic portion of the bilayer, it seems reasonable to propose that the basic platform stabilizes the membrane-trimer complex by interacting with negatively charged lipid head groups. An acidic ridge of DII was also visible between SLEV E protomers. Repulsive forces exerted at this interface could serve to distort these domains (Fig. 6D) and position the fusion loops further apart than those of DENV1/2 or TBEV. An acidic patch is also present on the interior face of SFV fusion loops facing the central axis (Fig. 6E). These regions may repulse one another to splay fusion loops apart in the flower-like arrangement observed in the SFV trimer (21).
Fig 6.
Trimer electrostatics. TBEV, DENV1, DENV2, SLEV (E) and SFV (E1) envelope surface representations are colored based on surface potential, with red as negative potentia and blue as positive. The top panel is a top-down view of the fusion loops, and the bottom panel is a side view. SLEV and SFV have regions of negative charge that may be relevant to trimer structure or assembly (circled in black). The basic platform beneath flavivirus fusion loops is also labeled in panel D.
DISCUSSION
The crystal structure of SLEV E revealed distinct biochemical features that may influence the interaction of JEV serocomplex viruses with host determinants, and therefore pathogenesis. In our analyses, SLEV E served as a basis for a structural comparison between all postfusion class II structures. Domains II and III of these envelope proteins are able to accommodate a range of orientations while remaining part of the trimeric assembly. Additionally, orientation of these domains contributed to variations of distances between the triangular crowns of fusion loops. Fusion loops of SLEV E were separated by an intermediate length compared to the tight spacing of DENV1, DENV2, and TBEV E or the wide set loops of SFV. Models of flavivirus fusion propose that the stem region truncated in recombinant E lies between trimers and splays them apart so that they reach an alphavirus-like conformation (23, 51, 73, 74). Recombinant SLEV E does not contain this stem, but it crystallized with wider spacing than other E trimers. This fusion loop separation could thus represent a characteristic feature of SLEV E or the JEV serocomplex.
Despite variations in relative domain positions, trimeric SLEV, DENV1, DENV2, and TBEV E proteins interact through at a series of structurally conserved positions (Fig. 3B). Prefusion dimers also assemble by forming contacts at conserved positions, but the dimer interface is generally smaller and appears to vary over a larger range. Also, SLEV E was previously found to dimerize less readily than DENV2 E (27), an observation that suggests the forces governing dimer and trimer assembly are not necessarily correlated. Trimer interfaces have similar buried surface area values, from 1,850 to 2,100 Å2, while dimers buried at 840 to 1,930 Å2. This wide range of calculated dimer surface area values may be explained by the presence of additional lateral interactions present outside the dimer interface in mature virions. On the other hand, the more conserved trimer interfaces may be necessary to rigidify E to allow for efficient fusion.
Ultimately, the functional reason fusion loops are exposed atop the E trimer is to drive interactions with the host lipid bilayer and merge it with that of the virion. This role led to the hypothesis that lipids or detergents would be required for trimer formation, and so the initial crystallization of postfusion E proteins from DENV2 (23) and TBEV E (51, 75) was carried out by purifying trimeric E complexes in detergents to simulate their membrane environment. However, our SLEV E (pH 5.5) structure and a previous structure of DENV1 E (pH 6.5) (52) have now been crystallized in the postfusion state without detergent, indicating that high protein concentration and an acidic environment are sufficient for trimerization. Interestingly, crystals grown in the absence of detergent (SLEV E,, 3.9 Å; DENV1 E, 3.5Å) have so far diffracted to lower resolutions than those crystallized in the presence of detergent (DENV2 E, 2.0 Å; TBEV E, 2.7 Å). Since weak diffraction is often a result of molecular motion or vibration, it is possible that detergents enhance trimer stability but are not an absolute requirement.
Biochemical features of class II envelope proteins may be finely tuned for interaction with environmental conditions, such as specific lipid compositions of membranes or pH values. Such interactions can direct viruses to certain host cells or cellular compartments. For example, alphavirus SFV requires cholesterol and sphingolipids for efficient fusion (76, 77), while fusion of flaviviruses DENV2 and DENV4 is dependent upon anionic lipids bis(monoacylglycero)phosphate and phosphatidylserine, which are found only in the late endosome (78). Analysis of trimer surface potentials revealed that DENV2 has a basic platform beneath its fusion loops, providing a possible means for its interaction with anionic lipids. Sphingolipids are also negatively charged, but it is less clear how they could interact with basic regions on SFV E1. Alpha- and flaviviruses also vary in pH requirements for efficient fusion, ranging from pH of ∼6.2 (TBEV and SFV) (79, 80) to ∼6.8 (DENV2) (81). It has been reported that flaviviruses with increased thermostability more efficiently infect certain hosts (82), and so it is conceivable that changes in optimal fusion pH might similarly affect transmission or tropism.
Comparative analysis of envelope structures from several viral serocomplexes and families provides valuable insights toward the understanding of class II fusion. The structure of SLEV E highlights several emerging serocomplex-specific structural features of E proteins. An understanding of the unique properties of these proteins should pave the way for structure-based experiments to illuminate the biochemical basis for flavivirus interactions with host determinants.
ACKNOWLEDGMENTS
This work was supported in part by NIH/NIAID U54 AI057160 (MRCE) and the Center for Structural Genomics of Infectious Diseases (CSGID) under contract numbers HHSN272200700058C and HHSN272201200026C.
We thank Melissa Barrow, Kelly Smith, Jad Abimansour, and Bill McCoy for helpful discourse and experimental suggestions and David Gohara for insightful discussions.
Footnotes
Published ahead of print 31 October 2012
REFERENCES
- 1. Bredeck JF. 1933. The story of the epidemic of encephalitis in St. Louis. Am. J. Public Health Nations Health 23:1135–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leake JP. 1933. Epidemiology of encephalitis: with special reference to the 1933 epidemic. Am. J. Public Health Nations Health 23:1140–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jones SC, Morris J, Hill G, Alderman M, Ratard RC. 2002. St. Louis encephalitis outbreak in Louisiana in 2001. J. Louisiana State Med. Soc. 154:303–306 [PubMed] [Google Scholar]
- 4. McCaig LF, Janowski HT, Gunn RA, Tsai TF. 1994. Epidemiologic aspects of a St. Louis encephalitis outbreak in Fort Walton Beach, Florida in 1980. Am. J. Trop. Med. Hyg. 50:387–391 [DOI] [PubMed] [Google Scholar]
- 5. Meehan PJ, Wells DL, Paul W, Buff E, Lewis A, Muth D, Hopkins R, Karabatsos N, Tsai TF. 2000. Epidemiological features of and public health response to a St. Louis encephalitis epidemic in Florida, 1990–1. Epidemiol. Infect. 125:181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Diaz LA, Nemeth NM, Bowen RA, Almiron WR, Contigiani MS. 2011. Comparison of argentinean Saint Louis encephalitis virus non-epidemic and epidemic strain infections in an avian model. PLoS Negl. Trop. Dis. 5:e1177 doi:10.1371/journal.pntd.0001177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mondini A, Cardeal ILS, Lázaro E, Nunes SH, Moreira CC, Rahal P, Maia IL, Franco C, Góngora DVN, Góngora-Rubio F, Cabrera EMS, Figueiredo LTM, Guimarães da Fonseca F, Bronzoni RVM, Chiaravalloti-Neto F, Nogueira ML. 2007. Saint Louis encephalitis virus, Brazil. Emerg. Infect. Dis. 13:176–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spinsanti LI, Díaz LA, Glatstein N, Arselán S, Morales MA, Farías AA, Fabbri C, Aguilar JJ, RÉ V, Frías M, Almirón WR, Hunsperger E, Siirin M, Da Rosa AT, Tesh RB, Enría D, Contigiani M. 2008. Human outbreak of St. Louis encephalitis detected in Argentina, 2005. J. Clin. Virol. 42:27–33 [DOI] [PubMed] [Google Scholar]
- 9. Lanciotti RS, Roehrig JT, Deubel V, Smith J, Parker M, Steele K, Crise B, Volpe KE, Crabtree MB, Scherret JH, Hall RA, MacKenzie JS, Cropp CB, Panigrahy B, Ostlund E, Schmitt B, Malkinson M, Banet C, Weissman J, Komar N, Savage HM, Stone W, McNamara T, Gubler DJ. 1999. Origin of the West Nile virus responsible for an outbreak of encephalitis in the northeastern United States. Science 286:2333–2337 [DOI] [PubMed] [Google Scholar]
- 10. Hammon WM, Reeves WC. 1943. Laboratory Transmission of St. Louis encephalitis virus by three genera of mosquitoes. J. Exp. Med. 78:241–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nayar JK, Rosen L, Knight JW. 1986. Experimental vertical transmission of Saint Louis encephalitis virus by Florida mosquitoes. Am. J. Trop. Med. Hyg. 35:1296–1301 [DOI] [PubMed] [Google Scholar]
- 12. Calisher CH, Karabatsos N, Dalrymple JM, Shope RE, Porterfield JS, Westaway EG, Brandt WE. 1989. Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. J. Gen. Virol. 70:37–43 [DOI] [PubMed] [Google Scholar]
- 13. De Madrid AT, Porterfield JS. 1974. The flaviviruses (group B arboviruses): a cross-neutralization study. J. Gen. Virol. 23:91–96 [DOI] [PubMed] [Google Scholar]
- 14. Muckenfuss RS. 1934. Clinical observations and laboratory investigations on the 1933 epidemic of encephalitis in St. Louis. Bull. N. Y. Acad. Med. 10:444–453 [PMC free article] [PubMed] [Google Scholar]
- 15. Reiss CS. 2008. Neurotropic viral infections. Cambridge University Press, Cambridge, England [Google Scholar]
- 16. Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, Baker TS, Strauss JH. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108:717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mukhopadhyay S, Kim Chipman B-SPR, Rossmann MG, Kuhn RJ. 2003. Structure of West Nile virus. Science 302:248. [DOI] [PubMed] [Google Scholar]
- 18. Harrison SC. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kielian M. 2006. Class II virus membrane fusion proteins. Virology 344:38–47 [DOI] [PubMed] [Google Scholar]
- 20. Schibli DJ, Weissenhorn W. 2004. Class I and class II viral fusion protein structures reveal similar principles in membrane fusion. Mol. Membr. Biol. 21:361–371 [DOI] [PubMed] [Google Scholar]
- 21. Gibbons DL, Vaney Roussel M-CA, Vigouroux A, Reilly B, Lepault J, Kielian M, Rey FA. 2004. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 427:320–325 [DOI] [PubMed] [Google Scholar]
- 22. Mancini EJ, Clarke M, Gowen BE, Rutten T, Fuller SD. 2000. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol. Cell 5:255–266 [DOI] [PubMed] [Google Scholar]
- 23. Modis Y, Ogata S, Clements D, Harrison SC. 2004. Structure of the dengue virus envelope protein after membrane fusion. Nature 427:313–319 [DOI] [PubMed] [Google Scholar]
- 24. Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375:291–298 [DOI] [PubMed] [Google Scholar]
- 25. Roussel A, Lescar J, Vaney Wengler M-CG, Wengler G, Rey FA. 2006. Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus. Structure 14:75–86 [DOI] [PubMed] [Google Scholar]
- 26. Modis Y, Ogata S, Clements D, Harrison SC. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 100:6986–6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Luca VC, AbiMansour J, Nelson CA, Fremont DH. 2012. Crystal structure of the Japanese encephalitis virus envelope protein. J. Virol. 86:2337–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kanai R, Kar K, Anthony K, Gould LH, Ledizet M, Fikrig E, Marasco WA, Koski RA, Modis Y. 2006. Crystal structure of West Nile virus envelope glycoprotein reveals viral surface epitopes. J. Virol. 80:11000–11008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nybakken GE, Nelson CA, Chen BR, Diamond MS, Fremont DH. 2006. Crystal structure of the West Nile virus envelope glycoprotein. J. Virol. 80:11467–11474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Voss JE, Vaney Duquerroy M-CS, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, Rey FA. 2010. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468:709–712 [DOI] [PubMed] [Google Scholar]
- 31. Mathews JH, Roehrig JT. 1984. Elucidation of the topography and determination of the protective epitopes on the E glycoprotein of Saint Louis encephalitis virus by passive transfer with monoclonal antibodies. J. Immunol. 132:1533–1537 [PubMed] [Google Scholar]
- 32. Roehrig JT, Mathews JH, Trent DW. 1983. Identification of epitopes on the E glycoprotein of Saint Louis encephalitis virus using monoclonal antibodies. Virology 128:118–126 [DOI] [PubMed] [Google Scholar]
- 33. Diamond MS, Pierson TC, Fremont DH. 2008. The structural immunology of antibody protection against West Nile virus. Immunol. Rev. 225:212–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gould LH, Sui J, Foellmer H, Oliphant T, Wang T, Ledizet M, Murakami A, Noonan K, Lambeth C, Kar K, Anderson JF, de Silva AM, Diamond MS, Koski RA, Marasco WA, Fikrig E. 2005. Protective and therapeutic capacity of human single-chain Fv-Fc fusion proteins against West Nile virus. J. Virol. 79:14606–14613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oliphant T, Engle M, Nybakken GE, Doane C, Johnson S, Huang L, Gorlatov S, Mehlhop E, Marri A, Chung KM, Ebel GD, Kramer LD, Fremont DH, Diamond MS. 2005. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat. Med. 11:522–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oliphant T, Nybakken GE, Engle M, Xu Q, Nelson CA, Sukupolvi-Petty S, Marri A, Lachmi Olshevsky B-EU, Fremont DH, Pierson TC, Diamond MS. 2006. Antibody recognition and neutralization determinants on domains I and II of West Nile virus envelope protein. J. Virol. 80:12149–12159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pierson TC, Fremont DH, Kuhn RJ, Diamond MS. 2008. Structural insights into the mechanisms of antibody-mediated neutralization of flavivirus infection: implications for vaccine development. Cell Host Microbe 4:229–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Throsby M, Geuijen C, Goudsmit J, Bakker AQ, Korimbocus J, Kramer RA, Clijsters-van der Horst M, de Jong M, Jongeneelen M, Thijsse S, Smit R, Visser TJ, Bijl N, Marissen WE, Loeb M, Kelvin DJ, Preiser W, ter Meulen J, de Kruif J. 2006. Isolation and characterization of human monoclonal antibodies from individuals infected with West Nile virus. J. Virol. 80:6982–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Vogt MR, Moesker B, Goudsmit J, Jongeneelen M, Austin SK, Oliphant T, Nelson S, Pierson TC, Wilschut J, Throsby M, Diamond MS. 2009. Human monoclonal antibodies against West Nile virus induced by natural infection neutralize at a postattachment step. J. Virol. 83:6494–6507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brien JD, Austin SK, Sukupolvi-Petty S, O'Brien KM, Johnson S, Fremont DH, Diamond MS. 2010. Genotype-specific neutralization and protection by antibodies against dengue virus type 3. J. Virol. 84:10630–10643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Crill WD, Chang G-JJ. 2004. Localization and characterization of flavivirus envelope glycoprotein cross-reactive epitopes. J. Virol. 78:13975–13986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lai C-J, Goncalvez AP, Men R, Wernly C, Donau O, Engle RE, Purcell RH. 2007. Epitope determinants of a chimpanzee dengue virus type 4 (DENV-4)-neutralizing antibody and protection against DENV-4 challenge in mice and rhesus monkeys by passively transferred humanized antibody. J. Virol. 81:12766–12774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sukupolvi-Petty S, Austin SK, Engle M, Brien JD, Dowd KA, Williams KL, Johnson S, Rico-Hesse R, Harris E, Pierson TC, Fremont DH, Diamond MS. 2010. Structure and function analysis of therapeutic monoclonal antibodies against dengue virus type 2. J. Virol. 84:9227–9239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Goncalvez AP, Chien Tubthong C-HK, Gorshkova I, Roll C, Donau O, Schuck P, Yoksan S, Wang Purcell S-DRH, Lai C-J. 2008. Humanized monoclonal antibodies derived from chimpanzee Fabs protect against Japanese encephalitis virus in vitro and in vivo. J. Virol. 82:7009–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wu K-P, Wu C-W, Tsao Y-P, Kuo T-W, Lou Y-C, Lin C-W, Wu S-C, Cheng J-W. 2003. Structural basis of a flavivirus recognized by its neutralizing antibody: solution structure of the domain III of the Japanese encephalitis virus envelope protein. J. Biol. Chem. 278:46007–46013 [DOI] [PubMed] [Google Scholar]
- 46. He RT, Innis BL, Nisalak A, Usawattanakul W, Wang S, Kalayanarooj S, Anderson R. 1995. Antibodies that block virus attachment to Vero cells are a major component of the human neutralizing antibody response against dengue virus type 2. J. Med. Virol. 45:451–461 [DOI] [PubMed] [Google Scholar]
- 47. Nybakken GE, Oliphant T, Johnson S, Burke S, Diamond MS, Fremont DH. 2005. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature 437:764–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thompson BS, Moesker B, Smit JM, Wilschut J, Diamond MS, Fremont DH. 2009. A therapeutic antibody against west nile virus neutralizes infection by blocking fusion within endosomes. PLoS Pathog. 5:e1000453 doi:10.1371/journal.ppat.1000453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Allison SL, Schalich J, Stiasny K, Mandl CW, Kunz C, Heinz FX. 1995. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 69:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cherrier MV, Kaufmann B, Nybakken GE, Lok Warren S-MJT, Chen BR, Nelson CA, Kostyuchenko VA, Holdaway HA, Chipman PR, Kuhn RJ, Diamond MS, Rossmann MG, Fremont DH. 2009. Structural basis for the preferential recognition of immature flaviviruses by a fusion-loop antibody. EMBO J. 28:3269–3276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bressanelli S, Stiasny K, Allison SL, Stura EA, Duquerroy S, Lescar J, Heinz FX, Rey FA. 2004. Structure of a flavivirus envelope glycoprotein in its low-pH-induced membrane fusion conformation. EMBO J. 23:728–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nayak V, Dessau M, Kucera K, Anthony K, Ledizet M, Modis Y. 2009. Crystal structure of dengue virus type 1 envelope protein in the postfusion conformation and its implications for membrane fusion. J. Virol. 83:4338–4344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Trainor NB, Crill WD, Roberson JA, Chang G-JJ. 2007. Mutation analysis of the fusion domain region of St. Louis encephalitis virus envelope protein. Virology 360:398–406 [DOI] [PubMed] [Google Scholar]
- 54. Wahlberg JM, Garoff H. 1992. Membrane fusion process of Semliki Forest virus. I: low pH-induced rearrangement in spike protein quaternary structure precedes virus penetration into cells. J. Cell Biol. 116:339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stols L, Millard CS, Dementieva I, Donnelly MI. 2004. Production of selenomethionine-labeled proteins in two-liter plastic bottles for structure determination. J. Struct. Funct. Genomics 5:95–102 [DOI] [PubMed] [Google Scholar]
- 56. Minor W, Otwinowski Z. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326 [DOI] [PubMed] [Google Scholar]
- 57. Kelley LA, Sternberg MJE. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 58. Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58:1948–1954 [DOI] [PubMed] [Google Scholar]
- 59. Merritt EA, Painter J. TLSMD web server for the generation of multi-group TLS models. J. Appl. Crystallogr. 39:109–111 [Google Scholar]
- 60. Terwilliger TC, Adams PD, Read RJ, McCoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, Hung LW. 2009. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D Biol. Crystallogr. 65:582–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132 [DOI] [PubMed] [Google Scholar]
- 62. Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. 2004. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 60:2184–2195 [DOI] [PubMed] [Google Scholar]
- 63. Collaborative Computational Project, Number 4 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763 [DOI] [PubMed] [Google Scholar]
- 64. PyMOL 2000. The PyMOL molecular graphics system. Schrödinger, LLC, Mannheim, Germany: http://www.pymol.org [Google Scholar]
- 65. Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797 [DOI] [PubMed] [Google Scholar]
- 66. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 67. Lee RA, Razaz M, Hayward S. 2003. The DynDom Database of protein domain motions. Bioinformatics 19:1290–1291 [DOI] [PubMed] [Google Scholar]
- 68. Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. 2007. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35:W522–W525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. 2004. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 32:W665–W667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 98:10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lescar J, Roussel A, Wien MW, Navaza J, Fuller SD, Wengler G, Wengler G, Rey FA. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137–148 [DOI] [PubMed] [Google Scholar]
- 72. Zhang Y, Zhang W, Ogata S, Clements D, Strauss JH, Baker TS, Kuhn RJ, Rossmann MG. 2004. Conformational changes of the flavivirus E glycoprotein. Structure 12:1607–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Allison SL, Schalich J, Stiasny K, Mandl CW, Heinz FX. 2001. Mutational evidence for an internal fusion peptide in flavivirus envelope protein E. J. Virol. 75:4268–4275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pangerl K, Heinz FX, Stiasny K. 2011. Mutational analysis of the zippering reaction during flavivirus membrane fusion. J. Virol. 85:8495–8501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Stiasny K, Allison SL, Schalich J, Heinz FX. 2002. Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH. J. Virol. 76:3784–3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kielian MC, Helenius A. 1984. Role of cholesterol in fusion of Semliki Forest virus with membranes. J. Virol. 52:281–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nieva JL, Bron R, Corver J, Wilschut J. 1994. Membrane fusion of Semliki Forest virus requires sphingolipids in the target membrane. EMBO J. 13:2797–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Zaitseva E, Yang Melikov S-TK, Pourmal S, Chernomordik LV. 2010. Dengue virus ensures its fusion in late endosomes using compartment-specific lipids. PLoS Pathog. 6:e1001131 doi:10.1371/journal.ppat.1001131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Guirakhoo F, Heinz FX, Mandl CW, Holzmann H, Kunz C. 1991. Fusion activity of flaviviruses: comparison of mature and immature (prM-containing) tick-borne encephalitis virions. J. Gen. Virol. 72:1323–1329 [DOI] [PubMed] [Google Scholar]
- 80. Kielian MC, Keränen S, KÄäriäinen L, Helenius A. 1984. Membrane fusion mutants of Semliki Forest virus. J. Cell Biol. 98:139–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Goncalvez AP, Purcell RH, Lai C-J. 2004. Epitope determinants of a chimpanzee Fab antibody that efficiently cross-neutralizes dengue type 1 and type 2 viruses map to inside and in close proximity to fusion loop of the dengue type 2 virus envelope glycoprotein. J. Virol. 78:12919–12928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kilpatrick AM, Meola MA, Moudy RM, Kramer LD. 2008. Temperature, viral genetics, and the transmission of West Nile virus by Culex pipiens mosquitoes. PLoS Pathog. 4:e1000092 doi:10.1371/journal.ppat.10000992 [DOI] [PMC free article] [PubMed] [Google Scholar]