Abstract
The replication of lepidopteran baculoviruses is characterized by the production of two progeny phenotypes: the occlusion-derived virus (ODV), which establishes infection in midgut cells, and the budded virus (BV), which disseminates infection to different tissues within a susceptible host. To understand the structural, and hence functional, differences between BV and ODV, we employed multiple proteomic methods to reveal the protein compositions and posttranslational modifications of the two phenotypes of Helicoverpa armigera nucleopolyhedrovirus. In addition, Western blotting and quantitative mass spectrometry were used to identify the localization of proteins in the envelope or nucleocapsid fractions. Comparative protein portfolios of BV and ODV showing the distribution of 54 proteins, encompassing the 21 proteins shared by BV and ODV, the 12 BV-specific proteins, and the 21 ODV-specific proteins, were obtained. Among the 11 ODV-specific envelope proteins, 8 either are essential for or contribute to oral infection. Twenty-three phosphorylated and 6 N-glycosylated viral proteins were also identified. While the proteins that are shared by the two phenotypes appear to be important for nucleocapsid assembly and trafficking, the structural and functional differences between the two phenotypes are evidently characterized by the envelope proteins and posttranslational modifications. This comparative proteomics study provides new insight into how BV and ODV are formed and why they function differently.
INTRODUCTION
Baculoviruses are insect-specific pathogens containing large circular double-stranded DNA genomes. Over millions of years of interdependence between viruses and their natural insect hosts, both have undergone a coevolution, such that lepidopteran baculoviruses have developed a unique biphasic replication cycle that generates two progeny phenotypes, the budded virus (BV) and the occlusion-derived virus (ODV). ODVs are embedded in occlusion bodies (OBs) that offer the virions a certain amount of protection against environmental degradation. Once ingested by a susceptible insect, ODVs are released from OBs within the larval midgut and initiate oral infection. After infecting midgut epithelial cells, BVs are synthesized and released to disseminate systemic infection of different tissues within the larval host. The two phenotypes have been used in a wide range of applications. Due to their expandable genome and the presence of very strong promoters, BVs have been established as successful vectors for the expression of thousands of proteins and have also been studied as potential vectors for gene therapy (1). The OBs of certain baculoviruses have been widely used in agriculture and forestry as viable alternatives to chemical insecticides in insect pest control (2).
The broad applications of baculoviruses provide a strong rationale for identifying the proteins associated with both phenotypes and for understanding their roles in baculovirus infection. While previous proteomic studies have elucidated the protein compositions of a few baculoviruses (3–8), a comprehensive investigation into the portfolio and landscape of viral proteins, as well as their associated protein modifications, has not been carried out. The family Baculoviridae contains four genera: Alphabaculovirus, Betabaculovirus, Gammabaculovirus, and Deltabaculovirus. The members of Alphabaculovirus are subdivided into two groups, I and II, based on phylogeny. The Helicoverpa armigera nucleopolyhedrovirus (HearNPV), which has a genome size of 131 kb and contains 135 open reading frames (ORFs), is a group II Alphabaculovirus and has been used successfully as a viral pesticide against cotton bollworms (9). In this study, we systematically investigated the HearNPV proteome using multiple proteomic techniques to reveal the protein compositions and posttranslational modifications of BV and ODV. Additionally, Western blotting and isobaric tags for relative and absolute quantitation (iTRAQ) were employed to analyze the locations of the structural proteins of BV and ODV. Our results show that the proteins that are shared by BV and ODV appear to be important for nucleocapsid formation and trafficking, but the two phenotypes differ markedly in their membrane protein compositions. The posttranslational protein modifications and the host proteins associated with BV and ODV are also significantly different. The detailed proteomics analyses exploring all the structural proteins of BV and ODV presented here shed light on the infection mechanisms used by the two distinct phenotypes.
MATERIALS AND METHODS
Cell cultures, virus infection, virion purification, and fractionation.
A Helicoverpa zea cell line, HzAM1 (10), was maintained at 27°C in Grace's insect medium supplemented with 10% fetal bovine serum (FBS). The HearNPV G4 strain (9) was used in this study. The BVs were collected from the supernatant of the infected HzAM1 cells and purified as described previously (4). Polyhedra were isolated and purified from infected H. armigera larvae (11), and the ODVs were released from polyhedra by alkaline treatment (pH 10.9) for 15 min and purified on continuous (25% to 65%) sucrose gradients. Purified BV and ODV were further separated into envelope (E) and nucleocapsid (NC) fractions (12). The NC and E fractions were precipitated with 3 volumes of 50% acetone-50% methanol-0.1% acetic acid and used for further SDS-PAGE and multiple proteomic analyses.
SDS-PAGE and in-gel digestion.
Purified BV, ODV, and their NC and E proteins were separated on a 12% SDS-PAGE gel. The protein bands or regions were excised from the BV lane, destained, reduced, alkylated, and subjected to in-gel trypsin digestion (11). The peptide mixtures obtained were further desalted by ZipTipC18 (Millipore) and dried by SpeedVac (11).
In-solution digestion and HILIC fractionation of peptides for shotgun proteomic analysis.
Shotgun proteomics is also known as multidimensional protein identification technology (MudPIT) (13). Precipitated E and NC proteins were resuspended in lysis buffer (8 M urea, 4 mM CaCl2, 0.2 M Tris-HCl [pH 8.0]), reduced, alkylated, and subjected to in-solution trypsin digestion as described previously (14). Peptides digested from 50 μg HearNPV virion (BV or ODV) proteins were further fractionated using hydrophilic interaction chromatography (HILIC) as described previously (14). Seven fractions for BV and 10 fractions for ODV were collected.
iTRAQ labeling and SCX fractionation of peptides for quantitative proteomic analyses.
Isobaric tags for relative and absolute quantitation (iTRAQ) labeling processing was conducted according to the iTRAQ protocol (Applied Biosystems). Peptides digested in solution from equivalent amounts (50 μg) of NC and E proteins were used in this study, and two replicate experiments were performed as follows. In one replicate, peptides digested from NC and E proteins were labeled with iTRAQ reagents 116 and 117, respectively. In the second replicate, peptides digested from NC and E proteins were labeled with iTRAQ reagents 117 and 116, respectively. The labeled peptides were mixed together and fractionated by strong cation exchange (SCX) (15), and seven fractions were collected.
Phosphopeptide enrichment using the IMAC-Fe3+ method.
In-solution-digested peptides from 1.5 mg of virion (BV or ODV) proteins were dissolved in 1% acetic acid and loaded onto immobilized metal affinity chromatography (IMAC)-Fe3+ resins to enrich phosphopeptides (14). The experiment was repeated three times.
Enrichment of N-linked glycopeptides by the solid-phase extraction method using hydrazide chemistry.
Peptides from 1 mg of in-solution-digested virion (BV or ODV) proteins were used for solid-phase extraction of N-linked glycopeptides (SPEG) as described previously (16) without isotope labeling. Two replicate experiments were performed.
Mass spectrometry and database search for protein identification and posttranslational modification assignments.
Peptides derived from in-gel digestion were subjected to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses using 4000 QTRAP. The mobile phase consisted of two components: (A) 2% acetonitrile with 0.1% formic acid and (B) 98% acetonitrile with 0.1% formic acid. Peptides were loaded onto the precolumn (0.5 mm by 2 mm; Michrom Bioresources, Inc.) at a flow rate of 5 μl/min and then were eluted into the analytic column at a flow rate of 300 nl/min with a gradient starting from 5% B held for 5 min, programmed to 60% B for 75 min, and held for another 5 min. The precursor ion's range was chosen from m/z 400 to m/z 1,600, and the product ion's range was chosen from m/z 50 to m/z 1,600. For the peptides derived from digestion in solution, LC-MS/MS analyses were conducted using QSTAR Elite as described previously (14).
Analyses of the raw MS spectra generated by LC-MS/MS analyses were performed with the ProteinPilot 4.0 software program (Applied Biosystems) using the Paragon algorithm (17) against the ORF database of HearNPV for viral protein identification and posttranslational modification assignments. A lepidopteran protein database (derived from GenBank) was utilized to identify host proteins. For the assignment of phosphorylation sites, the Scaffold PTM (posttranslational modification) (18) was used in combination with ProteinPilot software (Applied Biosystems). Only the phosphorylation sites identified by ProteinPilot and by Scaffold PTM (with a probability of ≥0.75) were considered significant.
Putative signal peptide (SP) and transmembrane domains (TMs) were predicted, respectively, by the SignalP 4.0 (http://www.cbs.dtu.dk/services/SignalP/) and TopPred (http://mobyle.pasteur.fr/cgi-bin/MobylePortal/portal.py?form=toppred) software packages.
Statistical analyses of iTRAQ peptide data for protein localization.
The natural logarithm of the relative peptide ion ratios of the envelope versus the nucleocapsid, i.e., [log ratio = ln(E/NC)] obtained with ProteinPilot software, was used to infer protein localization. Any proteins with 3 or more identified peptides were evaluated by statistical analyses. A peptide ratio of either 0.000 or 9,999 indicates that only one peak was found, and these data were not considered further.
According to the ProteinPilot software manual, the average protein abundance (APA) ratio is estimated using the formula APA ratio = eweighted average of log ratio. The point estimate of the weighted average of log ratio, WLR, was calculated using the following equation:
where xi is log(peptide iTRAQ ratioi), n = the number of peptide ratios contributing to the estimate of a protein's average ratio, and wi is equal to 1/(% error)i, the weight for xi.
The 95% confidence intervals of the weighted average of the log ratio were evaluated using the nonparametric bootstrap bias-corrected and accelerated (BCa) method (19), implemented in the boot.ci function in the boot R package (20).
Western blot analyses.
The polyclonal antibodies against polyhedrin (POLH), HA9(49K), E18, HA66, GP41, P33, E25, VP39, 38K, P6.9, ODV-E66, C42, VP80, and the polyhedron envelope protein (PEP) of HearNPV were generated as described by Deng et al. (4). Previously generated polyclonal antibodies against HA44, HA100 and per os infectivity factor 5 (PIF5) (4), P74, PIF1, PIF2 and PIF3 (21), PIF4 (22), P78/83 (23), HA51 (24), FP25K (25), EC43 (26), the fibroblast growth factor (FGF) (27), and fusion protein (F protein) (28) were also used. Western blot analyses were conducted as described previously (4).
RESULTS
Proteins associated with BV and ODV.
In this study, we employed 5 proteomic methods to reveal the protein compositions and posttranslational modifications of HearNPV BV and ODV. These included (i) iTRAQ, which involves isobaric tags and identifies proteins with relative and absolute quantifications, (ii) shotgun proteomics, which involves microcapillary multidimensional chromatography and identifies peptides from protein mixtures, (iii) IMAC, which involves peptide enrichment by immobilized metal ion affinity chromatography and identifies phosphopeptides, (iv) SDS-PAGE and LC-MS/MS, which involve in-gel trypsin digestion and identify the proteins of SDS-PAGE bands, and (v) SPEG proteomics, which involves the solid-phase extraction of N-linked glycopeptides and identifies N-glycosylated proteins.
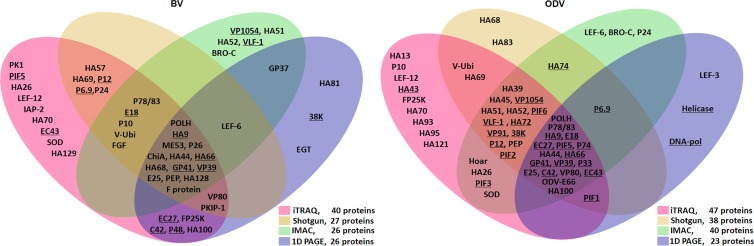
Purified BV, ODV, and their NC and E proteins were separated on a 12% SDS-PAGE gel as shown in Fig. S1 in the supplemental material. The details of the proteomic results are shown in Data Sets S1 and S2 in the supplemental material. A summary of the first four proteomic methods is presented in Fig. 1, while the SPEG results are shown in Table 1. In BV, 40 proteins were identified by iTRAQ, 27 proteins by shotgun proteomics, 26 proteins by IMAC, and 26 proteins by SDS-PAGE and LC-MS/MS (Fig. 1, left; see also Data Set S1 in the supplemental material). In addition, SPEG detected 6 proteins and further identified viral cathepsin (V-CATH) as a BV-associated protein (Table 1; see also Data Set S1). When the data from the various techniques were combined, a total of 51 proteins were identified from BV, of which 33 were identified by at least two proteomics methods.
Fig 1.
Summary of protein composition results for HearNPV BV (left) and ODV (right) compiled from multiple proteomic approaches. The proteins that were conserved in all baculoviruses are underlined. The proteins identified by iTRAQ-, shotgun-, IMAC-, and SDS-PAGE and LC-MS/MS (one-dimensional [1D] PAGE)-based proteomics are indicated in their respective regions. The 1D PAGE data of ODV were derived from Deng et al. (4).
Table 1.
Results of N-glycoproteomic analyses of HearNPV BV using the SPEG method
| Protein | HearNPV ORF | AcMNPV ORF | No. of AAsa | Molecular mass (kDa) | TMs and SPsb | Detected N-glycosylated peptide |
|||
|---|---|---|---|---|---|---|---|---|---|
| Peptide sequencec | Start (AA) | Stop (AA) | N-Glycosylated sites | ||||||
| E18 | 10 | 143 | 81 | 8.8 | TM, SP | GADTNAFAFQNPLN#ATMR | 51 | 68 | N64 |
| P26 | 22 | 136 | 267 | 30.5 | TM, SP | TVSINVIGHQSN#DSDTLDR | 46 | 64 | N57 |
| ChiA | 41 | 126 | 570 | 65.5 | TM, SP | FATFDYN#TSGR | 98 | 108 | N104 |
| AMLDQVQIQTN#R | 346 | 357 | N356 | ||||||
| V-CATH | 56 | 127 | 365 | 42 | TM | HFLQQYN#K | 57 | 64 | N63 |
| NNN#DSLSTSAQFGVNK | 99 | 114 | N101 | ||||||
| FGF | 113 | 32 | 301 | 34.4 | TM, SP | N#GTVWGITN#STDSHSVFYR | 49 | 64 | N49, N57 |
| NVLVN#SSGVHR | 138 | 148 | N142 | ||||||
| HSNVN#ATEGVN#QTNFY | 207 | 222 | N211, N217 | ||||||
| F protein | 133 | 23 | 677 | 78.2 | TM | NKN#LTSCENSETIFH | 102 | 114 | N104 |
| NNLLITEYVDMSSTFN#FSR | 511 | 529 | N526 | ||||||
| EINN#NTIFK | 568 | 576 | N571 | ||||||
AA, amino acid.
TMs denote putative transmembrane regions with a score of ≥0.95; SPs denote predicted signal peptides with a score of ≥0.8.
The # symbol indicates glycosylation (N).
Similarly, a total of 57 ODV-associated proteins were identified by different proteomics techniques, including 47 proteins by iTRAQ, 38 proteins by shotgun proteomics, 40 proteins by IMAC, and 23 proteins by SDS-PAGE and LC-MS/MS (Fig. 1, right; see also Data Set S2 in the supplemental material). Among these proteins, 40 were detected by at least two methods.
Localization of virion-associated proteins by iTRAQ.
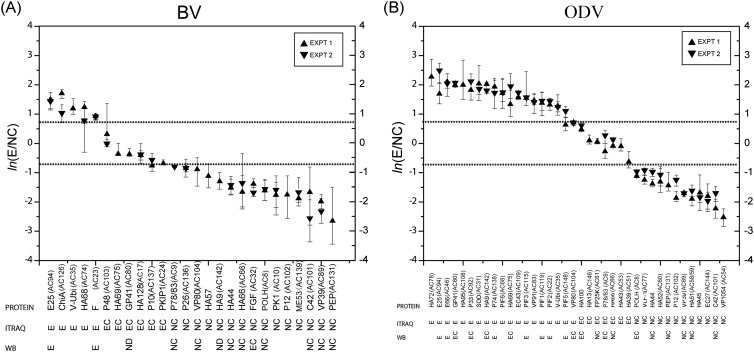
In order to identify protein locations, BV and ODV were separated into E and NC fractions and analyzed by quantitative mass spectrometry using iTRAQ labeling. Based on the relative protein abundances of E/NC in terms of the estimated protein abundance ratio for the envelope versus the nucleocapsid, BV and ODV proteins with 3 or more identified peptides were grouped into one of three categories: (i) envelope (E)-associated proteins (E/NC ≥ 2 [ln(E/NC) ≥ 0.69]), (ii) envelope-and-nucleocapsid (EC)-associated proteins (0.5 < E/NC < 2 [−0.69 < ln(E/NC) < 0.69]), and (iii) nucleocapsid (NC)-associated proteins (E/NC ≤ 0.5 [ln(E/NC) ≤ −0.69]) (see Table S1 in the supplemental material). In this way, a total of 26 BV proteins comprising 5 envelope-associated proteins, 6 envelope-and-nucleocapsid-associated proteins, and 15 nucleocapsid-associated proteins were identified (Fig. 2A; see also Table S1A in the supplemental material). For ODV, a total of 38 proteins consisting of 18 envelope-associated proteins, 8 envelope-and-nucleocapsid-associated proteins, and 12 nucleocapsid-associated proteins were identified (Fig. 2B; see also Table S1B in the supplemental material).
Fig 2.
Summary of the protein localization results from iTRAQ and Western blotting (WB) for BV (A) and ODV (B). The proteins are ordered along the x axis according to their estimated ln(E/NC) ratios, which are found along the y axis. The protein names are shown on the first row below the plot, protein classifications based on iTRAQ data are shown on the second row, and Western blot results are shown on the third row below the plot. Ninety-five percent confidence intervals are shown as vertical lines on the plot. The ORF numbers of the AcMNPV homologues for all the HearNPV ORFs are indicated in brackets.
Identification and localization of proteins by Western blotting.
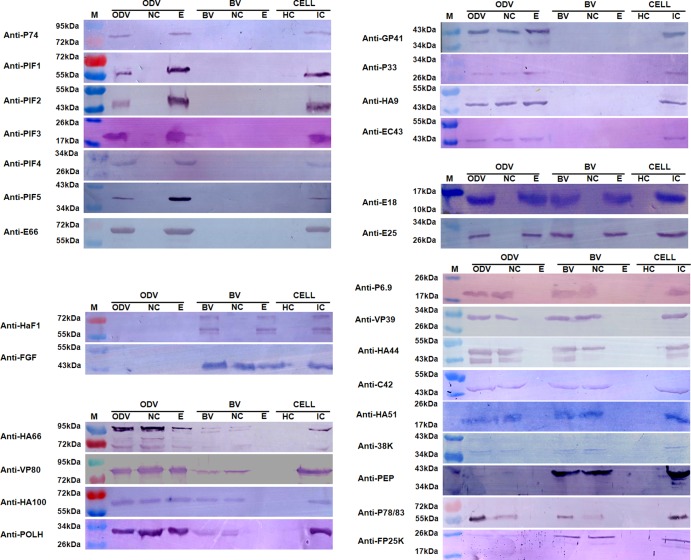
To further confirm the locations of the structural proteins, Western blotting was also performed. Polyclonal antibodies against 28 HearNPV proteins were used in Western blot analyses to identify and confirm the iTRAQ-based protein localization data (Fig. 3). The protein localization results are outlined in Table S1 in the supplemental material, and they show the proteins that were identified by at least two methods to total 35 for BV and 41 for ODV. The Western blot protein localization results are summarized at the bottom of the iTRAQ plots in Fig. 2.
Fig 3.
Western blot analyses of protein localization in the nucleocapsid and envelope fractions of HearNPV BV and ODV. Purified BV and ODV, as well as their nucleocapsid (NC) and envelope (E) fractions, were loaded for Western blot analyses. Healthy cells (HC) and virus-infected cells (IC) were used as negative and positive controls, respectively. The antibodies used for the Western blots are indicated to the left of each blot diagram. M indicates molecular mass markers.
The Western blot analyses detected 17 BV-associated proteins (Fig. 3), which corroborated the iTRAQ results for the localization of 10 proteins, including 2 envelope-associated proteins (E25 and membrane fusion protein F) and 8 nucleocapsid-associated proteins (P78/83, VP80, HA44, HA66, POLH, C42, VP39, and PEP) (Fig. 2A). The Western blots designated FGF as an envelope-and-nucleocapsid-associated protein, while iTRAQ showed it to be predominantly in the nucleocapsid, since the signal from the nucleocapsid fraction was significantly higher than that from the envelope fraction. We therefore conclude that FGF is present in both envelope and nucleocapsid fractions, but it is proportionately richer in the nucleocapsid. In addition, Western blot analyses detected E18 in the envelope fraction of BV and P6.9, HA51, 38K, HA100, and FP25K in the nucleocapsid fraction of BV (Fig. 3). However, either no peptides or fewer than three peptides were identified by iTRAQ for these proteins and, therefore, could not be evaluated by this method. Although iTRAQ detected the presence of GP41 in both the envelope and nucleocapsid fractions and HA9 in the nucleocapsid fraction, neither was observed by Western blotting; this is probably because the amounts in BV were too small to be detected under our Western blot conditions.
Twenty-six proteins were detected to be associated with ODV by Western blotting (Fig. 3). Seven proteins (E25, E66, P74, PIF3, PIF1, PIF2, and PIF5) were located in the envelope, 3 proteins (VP80, HA100, and HA66) were located in the envelope and nucleocapsid fractions, and 5 proteins (HA44, PEP, VP39, HA51, and C42) were located in the nucleocapsid fraction. These data are in agreement with the iTRAQ results (Fig. 2B). Of the remaining proteins, 4 (GP41, P33, HA9, and EC43) were detected in both the envelope and nucleocapsid fractions by Western blotting but were identified as envelope-associated proteins by iTRAQ. Following our interpretation of the BV results, we conclude that these 4 proteins are envelope-and-nucleocapsid-associated proteins, but proportionately higher quantities are present in the envelope relative to the nucleocapsid. For FP25K and P78/83, Western blotting detected them in the nucleocapsid fraction, while iTRAQ located them in both the envelope and nucleocapsid fractions. We therefore conclude that these proteins are located in both the nucleocapsid and the envelope, but the amounts in the latter component were so small that they could not be detected by Western blotting. Finally, our Western blot analyses identified E18 as an envelope protein, P6.9 and 38K as nucleocapsid proteins, and PIF4 as an envelope protein in ODVs (Fig. 3). No peptides or fewer than three peptides were identified by iTRAQ in these proteins, so they could not be evaluated.
Comparative model of protein compositions and localizations of BV and ODV.
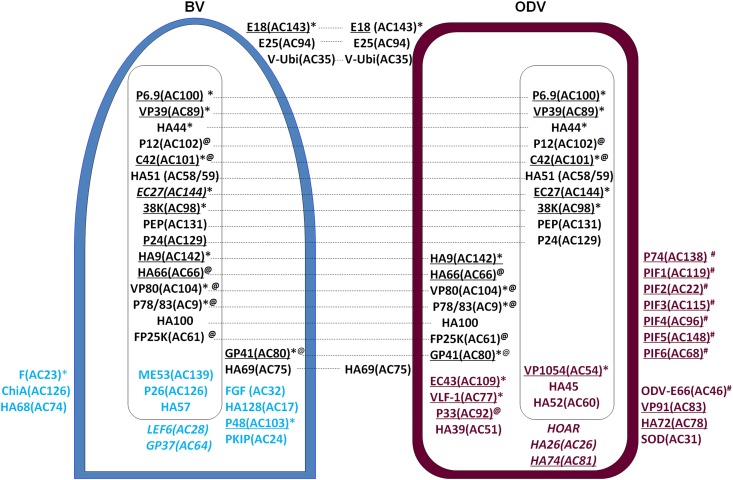
By combining all the proteomics (iTRAQ, shotgun MS/MS, IMAC, SDS-PAGE and LC-MS/MS, SPEG, and Western blotting) and protein localization (Western blotting and iTRAQ) data, the comparative protein portfolios of BV and ODV are presented in Fig. 4. As Autographa californica multiple nucleopolyhedrovirus (AcMNPV) is the type species of the baculovirus family, the AcMNPV homologues of the HearNPV ORFs are also indicated in Fig. 4 for general information. Only the proteins identified by at least two independent methods are included in Fig. 4, except for PIF4, which was detected only by Western blotting. We also removed POLH and P10 from Fig. 4, because these are the two most heavily expressed proteins at the late stages of baculovirus infection, but they are not related to BV or ODV formation.
Fig 4.
Comparative schematics showing protein compositions and localizations of HearNPV BV and ODV based on experimental results. The envelope proteins are shown in the figure on the surfaces of BV and ODV, the nucleocapsid proteins are shown inside each rectangle, and the EC proteins are located in the space between the envelope and the nucleocapsid. The proteins shared by BV and ODV are in black, and the proteins located in both BV and ODV are linked by dashed lines; BV-specific proteins are shown in blue, and the ODV-specific proteins are shown in maroon. The underlined proteins are conserved baculovirus core genes, an asterisk denotes proteins essential for BV production, @ denotes proteins involved in nucleocapsid and/or protein traffic, and # denotes proteins involved in oral infection. The BV and ODV proteins with unknown locations are shown in italics. The ORF numbers of the AcMNPV homologues for all the HearNPV ORFs are indicated in brackets.
Of the 54 proteins presented in Fig. 4, 21 proteins are shared by BV and ODV (shown in black), 12 proteins are specific to BV- (shown in blue), and 21 proteins are specific to ODV (shown in maroon).
Among the proteins shared by BV and ODV, 3 proteins were located in the envelope of both BV and ODV (E18, E25, and V-ubiquitin [V-Ubi]), and 8 proteins were located in the nucleocapsid of both BV and ODV (P6.9, VP39, HA44, P12, C42, HA51 [AC59], 38K, and PEP). EC27 was located in the nucleocapsid of ODV, but its location in HearNPV BV was not identified in this study. Six proteins were located in the nucleocapsid fraction of BV (HA9 [AC142], HA66 [AC66], VP80, P78/83, HA100, and FP25K) but appeared to be EC proteins in ODV. One protein was located in both the nucleocapsid and envelope fractions of BV and ODV (GP41). One protein was an EC protein in BV but an envelope component of ODV (HA69 [AC75]).
Of the 13 BV-specific proteins, 3 proteins were in the envelope (F protein, chitinase [ChiA], and HA68 [AC74]), 3 proteins were in the nucleocapsid (ME53, P26, and HA57), 4 proteins were EC proteins (FGF, HA128 [AC17], P48, and protein kinase-interacting protein [PKIP-1]), and 3 proteins had no localization data from iTRAQ or Western blots (P24, late expression factor 6 [LEF6], and GP37).
Of the 21 ODV-specific proteins, 11 were in the envelope, including 8 proteins (P74, PIF1, PIF2, PIF3, PIF4, PIF5, PIF6, and ODV-E66) involved in oral infection (denoted by the # symbol in Fig. 4) (21, 22, 29–33). Recently, the truncated ODV-E66 protein was identified as a chondroitin lyase that can efficiently digest chondroitin and chondroitin 6-sulfate (34). The remaining envelope proteins were VP91, HA72 (AC78), and superoxide dismutase (SOD). In addition, 3 proteins were located in the nucleocapsid (VP1054, HA45, and HA52 [AC60]), 4 proteins were located in both the envelope and the nucleocapsid fractions (EC43, very late expression factor 1 [VLF-1], P33, and HA39 [AC51]), and 3 proteins were of unknown location (HOAR, HA26 [AC26], and HA74 [AC81]).
Posttranslational modifications of virion proteins.
Posttranslation modifications play important roles in protein function, and in this study, N-glycosylation, phosphorylation, and N-terminal acetylation of BV and ODV proteins were analyzed.
N-glycosylation of virion proteins was detected by SPEG proteomics. Although there are many predicted N-glycosylation sites, a total of 14 were detected in 12 different glycopeptides from 6 BV-associated proteins (E18, P26, ChiA, V-CATH, FGF, and F protein) (Table 1). No ODV proteins were found to be N-glycosylated.
A total of 43 phosphorylation sites in 36 phosphopeptides from 23 viral proteins were identified (Table 2), yielding a Ser/Thr/Tyr phosphorylation ratio of 31:10:2 (72.1%/22.3%/4.6%). Among all the phosphorylation sites, 38 sites were in ODV-associated proteins, while 4 sites were in BV-associated proteins; only 1 site was identified in both BV and ODV (Table 2), indicating that BV and ODV have different phosphorylation profiles. Twenty-nine phosphorylation sites and their surrounding motifs were matched with consensus substrate sequences (motifs) for specific eukaryotic protein kinases by PHOSIDA (Phosphorylation Site Database) analysis (35) (Table 2).
Table 2.
Summary of protein phosphorylation in HearNPV BV and ODV identified by IMAC
| Protein | Phosphopeptide sequencea | Phosphorylated site | Site determination |
Motif (eukaryotic kinase)b | |
|---|---|---|---|---|---|
| BV | ODV | ||||
| POLH | MpYTRYpSYpSPpTLGK | Y2 | No | Yes | |
| S6 | No | Yes | PKA (R.p[ST]) | ||
| S8 | Yes | No | |||
| T10 | No | Yes | |||
| NLDpSLDKYLVAEDPFLGPGK | S50 | No | Yes | ||
| NQKLpTLFK | T71 | No | Yes | Aurora ([RK].p[ST][ILV]) | |
| IKEFAPDAPLYTGPApY | Y246 | No | Yes | ||
| P78/83 | NQLNQRLpSNAQTQQISAK | S311 | No | Yes | PKA (R.p[ST]) |
| E18 | NNPFVNpTPQRpTMM | T75 | No | Yes | CDK2 (p[ST]P.[KR]), ERK ([VX]p[ST]P), CDK1 (p[ST]P.[KR]) |
| T79 | No | Yes | |||
| EC27 | TVpTEIINADEK | T15 | No | Yes | CAMK2 (R . . p[ST]) |
| PIF5 | RQpSVKpTNFPSTNTR | S118 | No | Yes | PKA (R.p[ST], R[RK].p[ST]), CAMK2 (R . . p[ST], R . . p[ST]V), Aurora ([RK].p[ST][ILV]), Aurora-A ([RKN]R.[ST][MLVI]) |
| T121 | No | Yes | CK1 (S . . p[ST]) | ||
| LEF6 | STDRIpSEIGDWCR | S140 | No | Yes | PKA (R.p[ST]), CK1 ([ST] … pS) |
| HA44 | SLGLMpSR | S23 | No | Yes | |
| RPPpSPMNVSESTTIVPQSQR | S138 | Yes | Yes | CAMK2 (R . . p[ST]), ERK (P.p[ST]P) | |
| VLF1 | HLpSVDDLNTLIK | S221 | No | Yes | CAMK2 (R . . p[ST], R . .p[ST]V) |
| LRpSTIGLK | S236 | No | Yes | ||
| GP41 | QLQESSpSDAWTNK | S25 | No | Yes | |
| FDpSDENLIK | S82 | No | Yes | PKA (KR . . p[ST]), CAMK2 (R . . p[ST]), PKD ([LVI].[RK] . . p[ST]), CHK1 ([MILV].[RK] . . p[ST]) | |
| RPpSLFQGATFLNAPSSNGSNVEQNR | S269 | No | Yes | PKA (R.p[ST]), Aurora ([RK].p[ST][ILV]), Aurora-A ([RKN]R.p[ST][MLVI]) | |
| VP39 | ITpSEGLLASVR | S225 | No | Yes | CAMK2 (R . . p[ST]) |
| E25 | PLpTYpSEIIDEGNR | T80 | No | Yes | CK2 (p[ST] . . E), CAMK2 (R . . p[ST]) |
| S82 | No | Yes | NEK6 (L . . p[ST]) | ||
| TVGANCVFMGTISEPSQTSTLNQQQQQQQSAGSpSLPTTANR | S124 | No | Yes | CK1 ([ST] … pS) | |
| VpTANFDIK | T133 | No | Yes | PKA (R.p[ST]) | |
| C42 | QPpSRGNLLNLFR | S98 | No | Yes | CAMK2 (R . . p[ST]) |
| VP80 | NEpSETLPASTSIR | S121 | No | Yes | CAMK2 (R . . p[ST]) |
| TRpSPTTSNEK | S345 | No | Yes | GSK3 (pS . . S), CAMK2 (R . . p[ST]); AKT (R[RST].p[ST].[ST]) | |
| BRO-C | RHpSNAGDVYSAQQVVHAMK | S435 | No | Yes | PKA (R.p[ST], R[RK].p[ST]), CAMK2 (R . . p[ST]) |
| PEP | CWpSDFK | S55 | Yes | No | PKA (KR . . p[ST]), CAMK2 (R . . p[ST]) |
| HOAR | QEPVDMSDVECLNLPLpSPAR | S698 | No | Yes | CDK2 (p[ST]P.[KR]), ERK (P.p[ST]P), CDK1 (p[ST]P.[KR]), NEK6 (L . . p[ST]) |
| HA39 | ISSLSLRApSV | S193 | No | Yes | PKA (R.p[ST]) |
| HA45 | SDpSTSGLSNDSDIFDVIR | S4 | No | Yes | |
| HA72 | LAIpSDFNVDAADLSNNTDDSSTK | S26 | No | Yes | NEK6 (L . . p[ST]) |
| pSILGHNDLTSYR | S46 | No | Yes | CK1 (S . . p[ST], [ST] … pS) | |
| HA74 | HYpSVTASAASK | S228 | No | Yes | GSK3 (pS … S), CAMK2 (R . . p[ST], R . . p[ST]V) |
| VP91 | FVEQYGpTNIHK | T605 | No | Yes | |
| PIF2 | YFAGPQNMpSQVAGR | S171 | No | Yes | DNA damage response kinase (p[ST]Q) |
| NTFRpSHWDELLPDGTR | S210 | No | Yes | ||
| VTHIVPGDKTpSMCAAVVDR | T290 | No | Yes | ||
| F protein | pSKDpSFDQYDEHKL | S665 | Yes | No | |
| S668 | Yes | No | CK1 (S . . p[ST]) | ||
ND, not detectable.
PKA, protein kinase A; CDK2, cyclin-dependent kinase 2; ERK, extracellular signal-related kinase; CDK1, cyclin-dependent kinase 1; CAMK2, Ca2+/calmodulin-dependent protein kinase; CK1, casein kinase 1; PKD, protein kinase; CHK1, checkpoint kinase protein 1; NEK6, NIMA-related kinase 6; GSK3, glycogen synthase kinase 3; AKT, protein kinase B.
The MS/MS data from multiple proteomic analyses supplied additional information on N-terminal acetylation of HearNPV proteins. We discovered that 12 proteins contained acetylated N termini (Table 3). Seven proteins (ME53, P10, GP41, C42, HA128, HA45, and VP39) were found to be acetylated at the second amino acid, indicating that their N-terminal methionine was removed after translation. Eukaryotes contain at least six different N-terminal acetyltransferases (Nats), NatA to NatF, each with a different substrate specificity (36, 37). The Nat predictions are listed in Table 3, and the results indicate that NatA, NatB, NatD, and NatF participate in baculovirus N-terminal acetylation.
Table 3.
Summary of N-terminal acetylation detected in HearNPV BV and ODV by multiple proteomic methods
| BV or ODV | Protein | N-terminal acetylated peptide |
Detected byb: |
||||
|---|---|---|---|---|---|---|---|
| Peptide sequence | Start to end | Predicted host Nata | Shotgun | iTRAQ | IMAC | ||
| BV | POLH | MYTRYSYSPTLGK | 1–13 | NatF/? | − | − | + |
| ME53 | ATTSTAASLVNQHR | 2–15 | NatB | + | + | + | |
| P10 | SQNILLVIR | 2–10 | NatA | − | − | + | |
| GP41 | SQPHAVTTALQHQQHQK | 2–18 | NatA | + | + | − | |
| C42 | SGVMLFLEIENMK | 2–18 | NatD | − | − | + | |
| HA128 | SSNETTTVLSPR | 2–13 | NatA | − | − | + | |
| ODV | POLH | MYTRYSYSPTLGKTYVYDNK | 1–20 | NatF/? | − | − | + |
| HA9 | MNLDENKVALER | 1–12 | NatB | + | − | + | |
| E18 | MDDLRGTTTTGAGR | 1–14 | NatB | − | − | − | |
| HA45 | (M)SDSTSGLSNDSDIFDVIR | 2–19 | NatA | + | − | + | |
| HA72 | MNLDIPYDR | 1–9 | NatB | − | − | + | |
| GP41 | (M)SQPHAVTTALQHQQHQK | 2–18 | NatA | + | + | + | |
| VP39 | (M)ALVTVPTATTR | 2–12 | NatA | + | − | + | |
| C42 | (M)SGVMLFLEIENMK | 2–14 | NatD | − | − | + | |
| HA90 | MDLIETIDSVEAR | 1–14 | NatB | + | − | + | |
The N-terminal acetyltransferase targeting the peptide sequence was predicted according to the references 36 and 37. NatF/? means the N-terminal methionine may be acetylated by NatF or by unknown N-terminal acetyltransferases (Nats).
+ or − indicates that the N-terminal acetylated peptide was detected or not detected in the proteomic analyses, respectively.
Virion-associated host proteins.
Some viruses contain specific host proteins in their virions to facilitate infection. We identified 101 BV-associated and 21 ODV-associated host proteins that were classified based on their cellular functions, including cytoskeleton assembly, vesicle trafficking, signaling, metabolism, and posttranslational modification (see Table S2 in the supplemental material). Of all the HearNPV-associated host proteins, 52 have been detected previously in several other envelope viruses (see Table S2 in the supplemental material), implying that enveloped viruses may use the same or similar cellular pathways during their infection processes. The roles of these virion-associated host proteins in baculovirus replication are still unknown.
DISCUSSION
We have applied multiple proteomic approaches to investigate protein composition and localization in BV and ODV, and we provide detailed comparative schematics of the two phenotypes (Fig. 4). Our results should help clarify functional differences between these distinct phenotypes and shed light on how the virus switches from BV to ODV production during its life cycle.
BV and ODV share common proteins important for nucleocapsid assembly and trafficking.
Our study identified 21 proteins shared by BV and ODV, 6 of which are essential for nucleocapsid formation: P6.9, VP39, C42, AC142, EC27, and 38K. During baculovirus infection, nucleocapsids assemble in the virogenic stroma (VS) in the nucleus, where DNA replication takes place. The detailed mechanisms of nucleocapsid assembly are largely unknown, but the generally accepted model is that the viral genome is prepackaged with the basic DNA binding protein P6.9 (core protein), and the nucleoprotein complex is then inserted into preformed tube-like capsid sheaths composed of VP39 (38). This process is somehow interrupted when P6.9 (39), AC142, C42, EC27 (40), or 38K (41) is deleted.
In AcMNPV, very late factor 1 (VLF-1), VP1054, and AC109 (EC43) were also found to be components of BV and ODV and are essential for nucleocapsid assembly (42–44). In our study, VLF-1, VP1054, and EC43 were identified as ODV components, but their presence was also detected in BV by one of the proteomics methods (see Data Set 1 in the supplemental material). The demarcation from the AcMNPV data could be due to (i) the proteins being shared by both BV and ODV, while our requirement of being detected by at least two methods might have been too stringent, or (ii) these proteins being required during nucleocapsid assembly while their integration into the nucleocapsid may not be necessary.
Cellular actin cytoskeleton is used during different stages of baculovirus infection, and direct interactions of actin with nucleocapsid proteins are involved in (i) the transportation of the nucleocapsid from the cytoplasm to the nucleus at the early stages of infection (45), (ii) nucleocapsid assembly at VS (46), (iii) the transportation of a newly formed nucleocapsid from VS to the ring zone region (RZ) periphery of the inner nuclear membrane (47), and (iv) egress of the nucleocapsid from the nucleus to the cytoplasm and budding through the cytoplasmic membrane (45). Therefore, it is not surprising that we found that many proteins involved in actin interaction are shared by BV and ODV. As examples, P12 is involved in the nuclear localization of globular actin (G-actin) (48), P78/83 and C42 are involved in actin polymerization and nucleocapsid trafficking (45, 49, 50), and VP80 interacts with filamentous actin (F-actin) and is involved in the transportation of a nucleocapsid from VS to RZ (47). AC66, which contains an actin-binding domain, is involved in nucleocapsid egress from the nucleus to the cytoplasm and in ODV occlusion (51). The deletion of AC66 resulted in the accumulation of nucleocapsid in VS (51). Therefore, it is likely that AC66 is also involved in nucleocapsid transportation from the VS to the RZ.
After being transported to the RZ, either the nucleocapsids egress from the nucleus to form BV in the early stage of infection or they become enveloped to form ODV in the late stage of infection. During ODV formation, abundantly induced intranuclear microvesicles are derived from the inner nuclear membrane, which act as either direct precursors or assembly foci for the ODV envelope. How nucleocapsids egress from the nucleus and what regulates the nucleocapsid to switch from producing BV to forming ODV are not clear, but several BV-ODV shared proteins (FP25K, GP41, E18, and E25) may be involved in these processes. FP25K assists in trafficking ODV membrane proteins to the inner nuclear membrane and microvesicles (52). When FP25K is deleted, BV production is increased and ODV synthesis is decreased (53). GP41 is a tegument protein of AcMNPV ODV that is modified with O-linked N-acetylglucosamine (O-GlcNAc) (54). GP41 is essential for BV production, as nucleocapsids failed to egress from the nucleus when gp41 was mutated (55). E18 and E25 are BV-ODV envelope proteins in AcMNPV (56, 57); both are associated with virus-induced intranuclear microvesicles (56, 57). E18 is essential for BV production (58), and the deletion of E25 resulted in a lack of ODV production and a significant reduction in BV synthesis (59).
Taking these data together, it seems clear that the majority of the shared BV and ODV proteins are involved in nucleocapsid assembly and trafficking.
The membrane proteins distinguish BV and ODV and highlight their different functions.
The results of this study emphasize the differences between BV and ODV in their envelope components. There are 3 BV-specific envelope proteins and 11 ODV-specific envelope proteins. The membrane fusion protein F is the most abundant BV membrane protein (see Table S1 in the supplemental material), and it is crucial for BV entry into host cells. It binds to an unknown cellular receptor and mediates low-pH-induced fusion after endocytosis (60). The homologue of HA68, another BV-specific membrane protein, was found to play a role in BV production (61). Among 11 ODV-specific envelope proteins, 7 proteins are essential for oral infectivity (P74, PIF1, PIF2, PIF3, PIF4, PIF5, and PIF6), and 1 protein plays an important role in infection (ODV-E66) (21, 22, 30–34). In addition to the proteins discussed above, VP91 was also found to be a component of the PIF complex (62). Therefore, the majority of the identified ODV-specific envelope proteins are involved in and responsible for oral infectivity, whereas the major BV membrane protein is essential for BV infection.
Effects of gene knockout on the BV- and ODV-associated proteins.
Gene knockouts assist with the interpretation of whether a protein is a structural protein. A summary of the reported gene knockout effects of the homologues of HearNPV BV- and ODV-associated proteins is given in Table S3 in the supplemental material. The gene knockout effects are denoted by the letters a (lethal with malformed or absent nucleocapsids), b (lethal), c (inactivation of ODV), d (some effects), and e (no effects). Based on the gene knockout effects, the BV- and ODV-associated proteins are further interpreted (i) to be structural proteins, (ii) to very likely be structural proteins, (iii) to maybe be either structural or trapped proteins, or (iv) to maybe be trapped proteins (see Table S3 in the supplemental material). Since many of the gene knockout experiments have been done in vitro, the functions of the BV- and ODV-associated proteins in vivo still need to be examined.
N-Glycosylation is found only in BV-associated proteins.
Strikingly, SPEG analysis showed that all the detected N-glycosylated proteins belonged to BV, and no N-linked glycoproteins were identified in HearNPV ODVs. Since BVs are formed relatively early in the infection process in comparison to ODVs, it is possible that the host N-glycosylation system may not be functional at the late stages of infection. However, as all the identified N-glycosylated proteins contained signal peptides (Table 1), it is probable that only proteins with signal peptides could enter the N-glycosylation pathway. This should be important when the baculovirus system is used to express N-glycosylated proteins. As NPVs assemble in the nucleus while N-glycosylation occurs in the endoplasmic reticulum (ER), it is not surprising that nucleocapsid proteins are not N-glycosylated. However, there is evidence that many ODV envelope proteins containing the inner nuclear membrane sorting motif (INM-SM) are transported from the ER to the INM (52). We propose that those proteins may be transported from the cytoplasmic face of the ER membrane to the nucleus without entering the ER lumen for N-glycosylation. This would explain the lack of N-glycosylated proteins in ODVs.
Phosphorylation of viral proteins appears to be a common phenomenon during baculovirus infection.
Phosphorylation is a common posttranslational protein modification that plays a regulatory role in almost every aspect of the life of a cell. Phosphorylation or dephosphorylation is often associated with protein activation or inactivation, respectively. Baculovirus proteins undergo phosphorylation in infected cells (63, 64), and multiple phosphoproteins were detected in BV and ODV (12, 63, 64). A total of 23 baculoviral proteins were identified as phosphoproteins in our study (Table 3). With the exception of P78/83 (65), PEP (66), and POLH (67), the remaining proteins have not been reported to be phosphoproteins. The number of identified proteins indicates that phosphorylation plays important roles in the infection process. Exploring the regulation of the essential phosphoproteins is of interest to us for future studies.
The host proteins associated with baculovirus virions imply that common pathways are shared by many enveloped viruses.
In this study, 21 and 101 host proteins were identified to be associated with ODV and BV, respectively (see Table S2 in the supplemental material). The lower number of ODV-associated host proteins may be due to the stringent selection of proteins for ODV assembly and the specific incorporation of ODVs into OBs (68). In contrast, BV apparently was able to trap nonvirion proteins randomly (69), and efficient incorporation of foreign proteins has been shown by baculovirus surface display systems (70). Although we cannot rule out the possibility of contamination by host proteins during the virion purification process, the identified host proteins may have roles in baculovirus infection. Among the identified host proteins, 52 proteins have also been identified in other enveloped viruses (see Table S2 in the supplemental material), and some have been shown to be incorporated specifically into the virion. For example, cyclophilin A has been shown to be recruited into the virion for the infectivity of HIV-1 (71). It would be interesting to use techniques, such as RNA interference (RNAi), to investigate whether the identified host proteins are involved in baculovirus infection, as has been shown for other viruses (72).
The methodologies for identifying envelope and nucleocapsid proteins are applicable to other envelope viruses.
In our study, protein localization was initially identified by iTRAQ and subsequently validated by Western blotting. The former is a convenient technology for determining the localization of multiple proteins at one time. However, one of the challenges with iTRAQ is how to determine an appropriate cutoff ratio for the localization of a protein. In this study, we set a ratio of 2 as the cutoff point for assigning a protein to either the envelope or the nucleocapsid component. A protein was assigned to the envelope when the E/NC ratio was ≥2 [or ln(E/NC) was ≥0.69] and to the nucleocapsid when the E/NC ratio was ≤0.5 [or ln(E/NC) was ≤−0.69]. Although the classification appears to be arbitrary, it was based on the location of the apparent turning points of the curves in Fig. 2 and was corroborated by Western blots. We propose that the iTRAQ method and the E/NC cutoff value could find widespread use when investigating the protein localizations of other envelope viruses. It is worth mentioning that when a protein exists in both envelope and nucleocapsid fractions but predominantly in one fraction, the calculated E/NC ratio of iTRAQ sometimes places the protein in that fraction while Western blots may show that it actually existed in both fractions. Thus, Western blotting is recommended for amending the results of iTRAQ.
In summary, the data reported here represent the most comprehensive study of the protein compositions and modifications of BV and ODV to date, and they expand our understanding of how baculovirus structure relates to the distinct functions of both phenotypes. The data will also impact the application of baculoviruses. For example, as we know now that many ODV-specific genes are not essential for BV synthesis, with the help of synthetic biology, it will be possible to manufacture a mini-BV genome which is more efficient in protein expression and will likely provide a better baculovirus system for vaccine production. On the other hand, the ODV-specific proteins responsible for oral infection can be replaced with homologue proteins to produce biocontrol agents that are deleterious to a variety of target insect pests in agriculture and forestry.
Supplementary Material
ACKNOWLEDGMENTS
We thank Manli Wang for analyzing host proteins and Cunye Qiao and Yan Bai for their valuable assistance with the analysis of the MS data. We also thank Basil Arif and James Adams for the scientific editing of the manuscript.
This work was supported in part by the Ministry of Science and Technology of China (grant 2009CB118903 to Z.H.), the National Science Foundation of China (grants 30060002 and 31130058 to Z.H. and 30921001 to L.G.), the 111 Project of China (grant B06018 to Wuhan University), and a PSA project from the Ministry of Science and Technology of China and the Royal Academy of Sciences of the Netherlands (grant 2008DFB30220 to Z.H.).
Footnotes
Published ahead of print 31 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02329-12.
REFERENCES
- 1. Hu YC. 2006. Baculovirus vectors for gene therapy. Adv. Virus Res. 68:287–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bonning BC, Nusawardani T. 2007. Introduction to the use of baculoviruses as biological insecticides. Methods Mol. Biol. 388:359–366 [DOI] [PubMed] [Google Scholar]
- 3. Braunagel SC, Russell WK, Rosas-Acosta G, Russell DH, Summers MD. 2003. Determination of the protein composition of the occlusion-derived virus of Autographa californica nucleopolyhedrovirus. Proc. Natl. Acad. Sci. U. S. A. 100:9797–9802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Deng F, Wang R, Fang M, Jiang Y, Xu X, Wang H, Chen X, Arif BM, Guo L, Hu Z. 2007. Proteomics analysis of Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus identified two new occlusion-derived virus-associated proteins, HA44 and HA100. J. Virol. 81:9377–9385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Perera O, Green TB, Stevens SM, Jr, White S, Becnel JJ. 2007. Proteins associated with Culex nigripalpus nucleopolyhedrovirus occluded virions. J. Virol. 81:4585–4590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang R, Deng F, Hou D, Zhao Y, Guo L, Wang H, Hu Z. 2010. Proteomics of the Autographa californica nucleopolyhedrovirus budded virions. J. Virol. 84:7233–7242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang XF, Zhang BQ, Xu HJ, Cui YJ, Xu YP, Zhang MJ, Han YS, Lee YS, Bao YY, Zhang CX. 2011. ODV-associated proteins of the Pieris rapae granulovirus. J. Proteome Res. 10:2817–2827 [DOI] [PubMed] [Google Scholar]
- 8. Xu F, Ince IA, Boeren S, Vlak JM, van Oers MM. 2011. Protein composition of the occlusion derived virus of Chrysodeixis chalcites nucleopolyhedrovirus. Virus Res. 158:1–7 [DOI] [PubMed] [Google Scholar]
- 9. Chen X, Ijekl WF, Tarchini R, Sun X, Sandbrink H, Wang H, Peters S, Zuidema D, Lankhorst RK, Vlak JM, Hu Z. 2001. The sequence of the Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus genome. J. Gen. Virol. 82:241–257 [DOI] [PubMed] [Google Scholar]
- 10. Mcintosh AH, Ignoffo CM. 1983. Characterization of five cell lines established from species of Heliothis. Appl. Entomol. Zool. 18:262–269 [Google Scholar]
- 11. Yu JL, Guo L. 2011. Quantitative proteomic analysis of Salmonella enterica serovar Typhimurium under PhoP/PhoQ activation conditions. J. Proteome Res. 10:2992–3002 [DOI] [PubMed] [Google Scholar]
- 12. Braunagel SC, Summers MD. 1994. Autographa californica nuclear polyhedrosis virus, PDV, and ECV viral envelopes and nucleocapsids: structural proteins, antigens, lipid and fatty acid profiles. Virology 202:315–328 [DOI] [PubMed] [Google Scholar]
- 13. Yates JR, Ruse CI, Nakorchevsky A. 2009. Proteomics by mass spectrometry: approaches, advances, and applications. Annu. Rev. Biomed. Eng. 11:49–79 [DOI] [PubMed] [Google Scholar]
- 14. Yao Q, Li H, Liu BQ, Huang XY, Guo L. 2011. SUMOylation-regulated protein phosphorylation, evidence from quantitative phosphoproteomics analyses. J. Biol. Chem. 286:27342–27349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen X, Wu D, Zhao Y, Wong BH, Guo L. 2011. Increasing phosphoproteome coverage and identification of phosphorylation motifs through combination of different HPLC fractionation methods. J. Chromatogr. B Analyt. Technol. Biomed. Life. Sci. 879:25–34 [DOI] [PubMed] [Google Scholar]
- 16. Tian Y, Zhou Y, Elliott S, Aebersold R, Zhang H. 2007. Solid-phase extraction of N-linked glycopeptides. Nat. Protoc. 2:334–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. 2007. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6:1638–1655 [DOI] [PubMed] [Google Scholar]
- 18. Vincent-Maloney N, Searle B, Turner M. 2011. Probabilistically assigning sites of protein modification with scaffold PTM. J. Biomol. Tech. 22:S36 [Google Scholar]
- 19. Canty A, Ripley B. 2008. boot: Bootstrap R (S-Plus) functions. R package version 1.2-34. http://www.r-project.org
- 20. Davison AC, Hinkley DV. 1997. Bootstrap methods and their applications. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 21. Song J, Wang R, Deng F, Wang H, Hu Z. 2008. Functional studies of per os infectivity factors of Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus. J. Gen. Virol. 89:2331–2338 [DOI] [PubMed] [Google Scholar]
- 22. Huang H, Wang M, Deng F, Wang H, Hu Z. 2012. ORF85 of HearNPV encodes the per os infectivity factor 4 (PIF4) and is essential for the formation of the PIF complex. Virology 427:217–223 [DOI] [PubMed] [Google Scholar]
- 23. Nie Y, Wang Q, Liang C, Fang M, Yu Z, Chen X. 2006. Characterization of ORF2 and its encoded protein of the Helicoverpa armigera nucleopolyhedrovirus. Virus Res. 116:129–135 [DOI] [PubMed] [Google Scholar]
- 24. Zheng F, Huang Y, Long G, Sun X, Wang H. 2011. Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus ORF51 is a ChaB homologous gene involved in budded virus production and DNA replication. Virus Res. 155:203–212 [DOI] [PubMed] [Google Scholar]
- 25. Wu D, Deng F, Hu ZH, Yuan L, Sun XL. 2004. Molecular expression of the fp25k gene of HearNPV in E. coli and preparation of its antiserum. Virol. Sin. 19:380–384 [Google Scholar]
- 26. Fang M, Wang Ha, Wang Hu, Yuan L, Chen X, Vlak JM, Hu Z. 2003. Open reading frame 94 of Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus encodes a novel conserved occlusion-derived virion protein, ODV-EC43. J. Gen. Virol. 84:3021–3027 [DOI] [PubMed] [Google Scholar]
- 27. Li X, Liang C, Song J, Chen X. 2008. The ORF 113 of Helicoverpa armigera single nucleopolyhedrovirus encodes a functional fibroblast growth factor. Virol. Sin. 23:321–329 [Google Scholar]
- 28. Long G, Westenberg M, Wang H, Vlak JM, Hu Z. 2006. Function, oligomerization and N-linked glycosylation of the Helicoverpa armigera single nucleopolyhedrovirus envelope fusion protein. J. Gen. Virol. 87:839–846 [DOI] [PubMed] [Google Scholar]
- 29. Haas-Stapleton EJ, Washburn JO, Volkman LE. 2004. P74 mediates specific binding of Autographa californica M nucleopolyhedrovirus occlusion-derived virus to primary cellular targets in the midgut epithelia of Heliothis virescens larvae. J. Virol. 78:6786–6791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nie Y, Fang M, Erlandson MA, Theilmann DA. 2012. Analysis of the Autographa californica multiple nucleopolyhedrovirus overlapping gene pair lef3 and ac68 reveals that AC68 is a per os infectivity factor and that LEF3 is critical, but not essential, for virus replication. J. Virol. 86:3985–3994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ohkawa T, Washburn JO, Sitapara R, Sid E, Volkman LE. 2005. Specific binding of Autographa californica M nucleopolyhedrovirus occlusion-derived virus to midgut cells of Heliothis virescens larvae is mediated by products of pif genes Ac119 and Ac022 but not by Ac115. J. Virol. 79:15258–15264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sparks WO, Harrison RL, Bonning BC. 2011. Autographa californica multiple nucleopolyhedrovirus ODV-E56 is a per os infectivity factor, but is not essential for binding and fusion of occlusion-derived virus to the host midgut. Virology 409:69–76 [DOI] [PubMed] [Google Scholar]
- 33. Xiang X, Chen L, Hu X, Yu S, Yang R, Wu X. 2011. Autographa californica multiple nucleopolyhedrovirus odv-e66 is an essential gene required for oral infectivity. Virus Res. 158:72–78 [DOI] [PubMed] [Google Scholar]
- 34. Sugiura N, Setoyama Y, Chiba M, Kimata K, Watanabe H. 2011. Baculovirus envelope protein ODV-E66 is a novel chondroitinase with distinct substrate specificity. J. Biol. Chem. 286:29026–29034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gnad F, Gunawardena J, Mann M. 2011. PHOSIDA 2011: the posttranslational modification database. Nucleic. Acids Res. 39:D253–D260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Arnesen T. 2011. Towards a functional understanding of protein N-terminal acetylation. PLoS Biol. 9:e1001074 doi:10.1371/journal.pbio.1001074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Polevoda B, Arnesen T, Sherman F. 2009. A synopsis of eukaryotic Nalpha-terminal acetyltransferases: nomenclature, subunits and substrates. BMC Proc. 3(Suppl 6):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Williams GV, Faulkner P. 1997. Cytological changes and viral morphogenesis during baculovirus infection, p 61–107 In Miller LK. (ed), The baculoviruses. Plenum, New York, NY [Google Scholar]
- 39. Wang M, Tuladhar E, Shen S, Wang H, van Oers MM, Vlak JM, Westenberg M. 2010. Specificity of baculovirus P6.9 basic DNA-binding proteins and critical role of the C terminus in virion formation. J. Virol. 84:8821–8828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vanarsdall AL, Pearson MN, Rohrmann GF. 2007. Characterization of baculovirus constructs lacking either the Ac 101, Ac 142, or the Ac 144 open reading frame. Virology 367:187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wu W, Lin T, Pan L, Yu M, Li Z, Pang Y, Yang K. 2006. Autographa californica multiple nucleopolyhedrovirus nucleocapsid assembly is interrupted upon deletion of the 38K gene. J. Virol. 80:11475–11485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vanarsdall AL, Okano K, Rohrmann GF. 2006. Characterization of the role of very late expression factor 1 in baculovirus capsid structure and DNA processing. J. Virol. 80:1724–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Olszewski J, Miller LK. 1997. Identification and characterization of a baculovirus structural protein, VP1054, required for nucleocapsid formation. J. Virol. 71:5040–5050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lin L, Wang J, Deng R, Ke J, Wu H, Wang X. 2009. ac109 is required for the nucleocapsid assembly of Autographa californica multiple nucleopolyhedrovirus. Virus Res. 144:130–135 [DOI] [PubMed] [Google Scholar]
- 45. Ohkawa T, Volkman LE, Welch MD. 2010. Actin-based motility drives baculovirus transit to the nucleus and cell surface. J. Cell Biol. 190:187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Volkman LE. 1988. Autographa californica MNPV nucleocapsid assembly: inhibition by cytochalasin D. Virology 163:547–553 [DOI] [PubMed] [Google Scholar]
- 47. Marek M, Merten OW, Galibert L, Vlak JM, van Oers MM. 2011. Baculovirus VP80 protein and the F-actin cytoskeleton interact and connect the viral replication factory with the nuclear periphery. J. Virol. 85:5350–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gandhi KM, Ohkawa T, Welch MD, Volkman LE. 2012. Nuclear localization of actin requires AC102 in Autographa californica multiple nucleopolyhedrovirus-infected cells. J. Gen. Virol. 93:1795–1803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Goley ED, Ohkawa T, Mancuso J, Woodruff JB, D'Alessio JA, Cande WZ, Volkman LE, Welch MD. 2006. Dynamic nuclear actin assembly by Arp2/3 complex and a baculovirus WASP-like protein. Science 314:464–467 [DOI] [PubMed] [Google Scholar]
- 50. Li K, Wang Y, Bai H, Wang Q, Song J, Zhou Y, Wu C, Chen X. 2010. The putative pocket protein binding site of Autographa californica nucleopolyhedrovirus BV/ODV-C42 is required for virus-induced nuclear actin polymerization. J. Virol. 84:7857–7868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ke J, Wang J, Deng R, Wang X. 2008. Autographa californica multiple nucleopolyhedrovirus ac66 is required for the efficient egress of nucleocapsids from the nucleus, general synthesis of preoccluded virions and occlusion body formation. Virology 374:421–431 [DOI] [PubMed] [Google Scholar]
- 52. Braunagel SC, Cox V, Summers MD. 2009. Baculovirus data suggest a common but multifaceted pathway for sorting proteins to the inner nuclear membrane. J. Virol. 83:1280–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wu D, Deng F, Sun X, Wang H, Yuan L, Vlak JM, Hu Z. 2005. Functional analysis of FP25K of Helicoverpa armigera single nucleocapsid nucleopolyhedrovirus. J. Gen. Virol. 86:2439–2444 [DOI] [PubMed] [Google Scholar]
- 54. Whitford M, Faulkner P. 1992. A structural polypeptide of the baculovirus Autographa californica nuclear polyhedrosis virus contains O-linked N-acetylglucosamine. J. Virol. 66:3324–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Olszewski J, Miller LK. 1997. A role for baculovirus GP41 in budded virus production. Virology 233:292–301 [DOI] [PubMed] [Google Scholar]
- 56. Braunagel SC, He H, Ramamurthy P, Summers MD. 1996. Transcription, translation, and cellular localization of three Autographa californica nuclear polyhedrosis virus structural proteins: ODV-E18, ODV-E35, and ODV-EC27. Virology 222:100–114 [DOI] [PubMed] [Google Scholar]
- 57. Russell RL, Rohrmann GF. 1993. A 25-kDa protein is associated with the envelopes of occluded baculovirus virions. Virology 195:532–540 [DOI] [PubMed] [Google Scholar]
- 58. McCarthy CB, Theilmann DA. 2008. AcMNPV ac143 (odv-e18) is essential for mediating budded virus production and is the 30th baculovirus core gene. Virology 375:277–291 [DOI] [PubMed] [Google Scholar]
- 59. Chen L, Hu X, Xiang X, Yu S, Yang R, Wu X. 2012. Autographa californica multiple nucleopolyhedrovirus odv-e25 (Ac94) is required for budded virus infectivity and occlusion-derived virus formation. Arch. Virol. 157:617–625 [DOI] [PubMed] [Google Scholar]
- 60. Pearson MN, Russell RL, Rohrmann GF. 2001. Characterization of a baculovirus-encoded protein that is associated with infected-cell membranes and budded virions. Virology 291:22–31 [DOI] [PubMed] [Google Scholar]
- 61. Guo ZJ, Qiu LH, An SH, Yao Q, Park EY, Chen KP, Zhang CX. 2010. Open reading frame 60 of the Bombyx mori nucleopolyhedrovirus plays a role in budded virus production. Virus Res. 151:185–191 [DOI] [PubMed] [Google Scholar]
- 62. Peng K, van Lent JW, Boeren S, Fang M, Theilmann DA, Erlandson MA, Vlak JM, van Oers MM. 2012. Characterization of novel components of the baculovirus per os infectivity factor complex. J. Virol. 86:4981–4988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kelly DC, Lescott T. 1984. Baculovirus replication: phosphorylation of polypeptides synthesized in Trichoplusia ni nuclear polyhedrosis virus-infected cells. J. Gen. Virol. 65:1183–1191 [DOI] [PubMed] [Google Scholar]
- 64. Maruniak JE, Summers MD. 1981. Autographa californica nuclear polyhedrosis virus phosphoproteins and synthesis of intracellular proteins after virus infection. Virology 109:25–34 [DOI] [PubMed] [Google Scholar]
- 65. Vialard JE, Richardson CD. 1993. The 1,629-nucleotide open reading frame located downstream of the Autographa californica nuclear polyhedrosis virus polyhedrin gene encodes a nucleocapsid-associated phosphoprotein. J. Virol. 67:5859–5866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Whitt MA, Manning JS. 1988. A phosphorylated 34-kDa protein and a subpopulation of polyhedrin are thiol linked to the carbohydrate layer surrounding a baculovirus occlusion body. Virology 163:33–42 [DOI] [PubMed] [Google Scholar]
- 67. Summers MD, Smith GE. 1975. Trichoplusia ni granulosis virus granulin: a phenol-soluble, phosphorylated protein. J. Virol. 16:1108–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ji X, Sutton G, Evans G, Axford D, Owen R, Stuart DI. 2010. How baculovirus polyhedra fit square pegs into round holes to robustly package viruses. EMBO J. 29:505–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Carbonell LF, Miller LK. 1987. Baculovirus interaction with nontarget organisms: a virus-borne reporter gene is not expressed in two mammalian cell lines. Appl. Environ. Microbiol. 53:1412–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Makela AR, Oker-Blom C. 2006. Baculovirus display: a multifunctional technology for gene delivery and eukaryotic library development. Adv. Virus Res. 68:91–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Franke EK, Yuan HE, Luban J. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372:359–362 [DOI] [PubMed] [Google Scholar]
- 72. Spurgers KB, Alefantis T, Peyser BD, Ruthel GT, Bergeron AA, Costantino JA, Enterlein S, Kota KP, Boltz RC, Aman MJ, Delvecchio VG, Bavari S. 2010. Identification of essential filovirion-associated host factors by serial proteomic analysis and RNAi screen. Mol. Cell. Proteomics 9:2690–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.