Abstract
Avian-origin influenza virus polymerase activity can be dramatically increased in human cells with the PB2 E627K mutation. Previously, others have proposed that this mutation increases the stability of the viral ribonucleoprotein complex (vRNP) measured by the interaction between PB2 and NP. However, we demonstrate here that a variety of PB2 adaptive mutations, including E627K, do not enhance the stability of the vRNP but rather increase the amount of replicated RNA that results in more PB2-NP coprecipitation.
TEXT
Avian influenza A viruses typically do not replicate efficiently or cause disease in humans (1) and are also restricted in mammalian cells. Host restriction involves multiple determinants; however, influenza virus polymerase is considered to play an important role. The heterotrimeric polymerase complex (PA, PB1, and PB2) associates with viral RNA (vRNA) and nucleoprotein (NP) to form a ribonucleoprotein (RNP) complex responsible for viral replication and transcription (reviewed in reference 2). Host-specific genetic signatures have been identified on all of the polymerase subunits and on NP, but the PB2 protein arguably carries the dominant determinants of host range (3–11). The introduction of mammalian host-adaptive PB2 mutations (e.g., E627K, D701N, and Q591K) into avian viruses has been shown to support enhanced viral replication and pathogenicity in mammalian model systems (6, 7, 12–14).
The molecular basis for polymerase host range adaptation is not fully elucidated, and it is possible that adaptation by different genetic mutations is achieved by distinct mechanisms. Certainly, other viral genes in addition to PB2 can adapt the avian virus polymerase for increased replication in mammalian cells. For example, mammalian-adaptive mutations in PA have been described, and recently, the second gene product of RNA segment 8, NEP, was shown to compensate for defective H5N1 RNA replication in cultured human cells (7, 15, 16). However, there is a body of evidence that mammalian host adaptation is dependent on the polymerase and/or NP adapting to interact with specific mammalian host factors to enable efficient viral transcription/replication (17–19).
The best-studied host-associated signature in PB2 is at amino acid position 627 (10, 13). Avian influenza viruses generally encode a glutamic acid at this site, whereas human isolates that circulated until 2009 typically encode a lysine. An idea popular in the literature is that the enhanced polymerase activity observed with PB2 E627K is due to an increased biochemical stability of the vRNP in human cells (20–23). We wished to investigate if this mechanism also explains how other naturally or experimentally occurring PB2 mutations adapt avian virus polymerase for mammalian replication. However, in the course of our study we established that the published approach measures the consequence of polymerase activity rather than the strength of the polymerase-NP interaction as previously suggested. Thus, we conclude that the instability of the polymerase-NP interaction in the vRNP is not in itself responsible for avian influenza virus polymerase restriction in human cells.
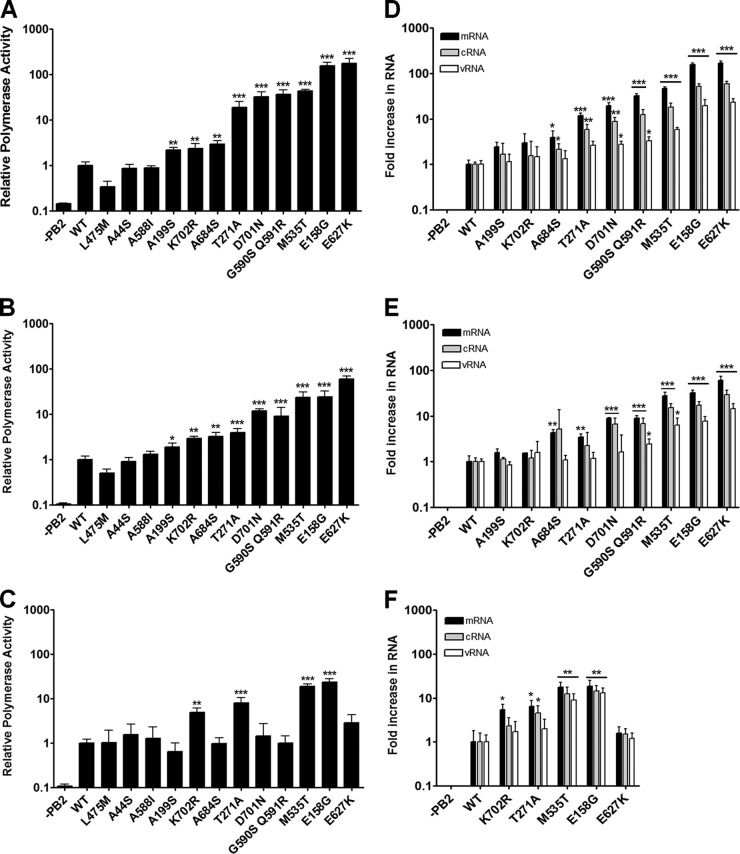
Many different mammalian host range determinants have been localized to the PB2 protein. Some of these have arisen naturally and have also been identified in bioinformatic studies, and others have appeared through the serial passage of avian viruses in mice. We first investigated whether a panel of mutations exclusively influence polymerase activity in a host-dependent manner, as we and others have previously shown for the mutation at position 627. A series of mammalian host-specific PB2 signatures (A44S, E158G, A199S, T271A, M535T, A588I, G590S Q591R, E627K, A684S, D701N, and K702R) (4, 7, 9, 10, 24–26) located throughout the protein were singly introduced into the A/Turkey/England/50-92/91 (50-92) (H5N1) PB2 gene by site-directed mutagenesis on the pCAGGS PB2 expression plasmid. 50-92 is a typical poultry-adapted avian influenza virus which has no evidence of mammalian polymerase adaptation and displays host range restriction in mammalian cells (19, 27, 28). Human 293T, swine NPTr, or avian DF-1 cells were transfected with plasmids that use species-specific polymerase I promoters to direct synthesis of minigenomes containing the firefly luciferase open reading frame flanked by segment 8 terminal noncoding sequences together with 50-92 NP, PA, PB1, and wild-type (WT) or mutant PB2 protein expression plasmids (29). Firefly luciferase activities were obtained 12 h after transfection and normalized to an internal control (Renilla luciferase expressed from a cotransfected polymerase II expression plasmid) (Fig. 1A to C).
Fig 1.
Polymerase activity supported by PB2 mutants. (A to C) 293T (A), NPTr (B), and DF-1 (C) cells were transfected with pCAGGS 5092 PB1, PA, NP, and WT or mutated PB2 as well as pCAGGS Renilla and a virus-like firefly luciferase minigenome-expressing plasmid. Luciferase production was measured 12 h posttransfection. Values were normalized to Renilla expression and to the activity of the WT polymerase. (D to F) The levels of viral mRNA, cRNA, and vRNA synthesized by different RNA polymerase PB2 mutants normalized to that of the WT polymerase in 293T (D), NPTr (E), and DF-1 (F) cells. Expression of 50-92 NP mRNA was used as an internal control. Results are from one experiment, undertaken in triplicate, representative of three independent experiments. The statistical significance of differences in polymerase activity or RNA synthesis from that of WT was assessed by a two-tailed, unpaired Student t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Almost all of the introduced PB2 mutations significantly enhanced 50-92 polymerase activity in human and swine cells, whereas only a subset of the PB2 mutations significantly enhanced polymerase activity in avian cells. For example, introducing E158G dramatically enhanced polymerase activity in human, swine, and avian cells, whereas the introduction of E627K resulted in enhancement in activity in only human and swine cells. The observation that some PB2 mutations enhance polymerase activity in a mammalian host-specific manner whereas others affect activity in human, swine, and avian cells suggests that there are different mechanisms of enhancing activity. This concept is supported by studies in which certain cumulative PB2 mutations enhance polymerase activity in an additive manner (24, 27, 30).
To assess the effects of the PB2 host range mutations on viral genome replication and transcription, the levels of viral mRNA, cRNA, and vRNA were separately measured by reverse transcription and real-time quantitative PCR (qRT-PCR) as previously described (31). Briefly, at 12 h posttransfection total RNA was extracted and vRNA was reverse transcribed with a primer complementary to the luciferase coding region. cRNA was amplified with a primer complementary to the 3′ portion of the segment 8 cRNA, and oligo(dT)20 was used to reverse transcribe mRNA. Real-time quantitative PCR was performed with primers specific to the luciferase coding region, and NP mRNA transcribed from the expression plasmid was used as an internal control. Regardless of whether the mutation was host specific or not, an increase in polymerase activity correlated with proportionally enhanced synthesis of all 3 viral RNA species in human, avian, and swine cells (Fig. 1D to F).
Previously, coimmunoprecipitation experiments demonstrated that the interaction between PB2 627E and NP when expressed either alone or in the context of a reconstituted RNP complex in human cells was weak. The introduction of a lysine at position 627 in PB2 was shown to strengthen this interaction and thus presented as a mechanism to facilitate mammalian host range adaptation (20). In addition, Ng et al. recently reported differences in the interactions of the C-terminal sequence of PB2 with either 627E or 627K and NP by using purified components and biophysical techniques (22). In contrast, Mehle and Doudna showed that PB2 harboring the avian signature glutamic acid at position 627 bound NP as well as did PB2 627K when just these two proteins were expressed but that the interaction between PB2 627E and NP in the context of a reconstituted RNP complex was diminished in human cells (21). The latter findings suggested that the species-restricted activity of PB2 627E was the result of a defect(s) in vRNP assembly or conformation that precludes binding to NP within the RNP complex.
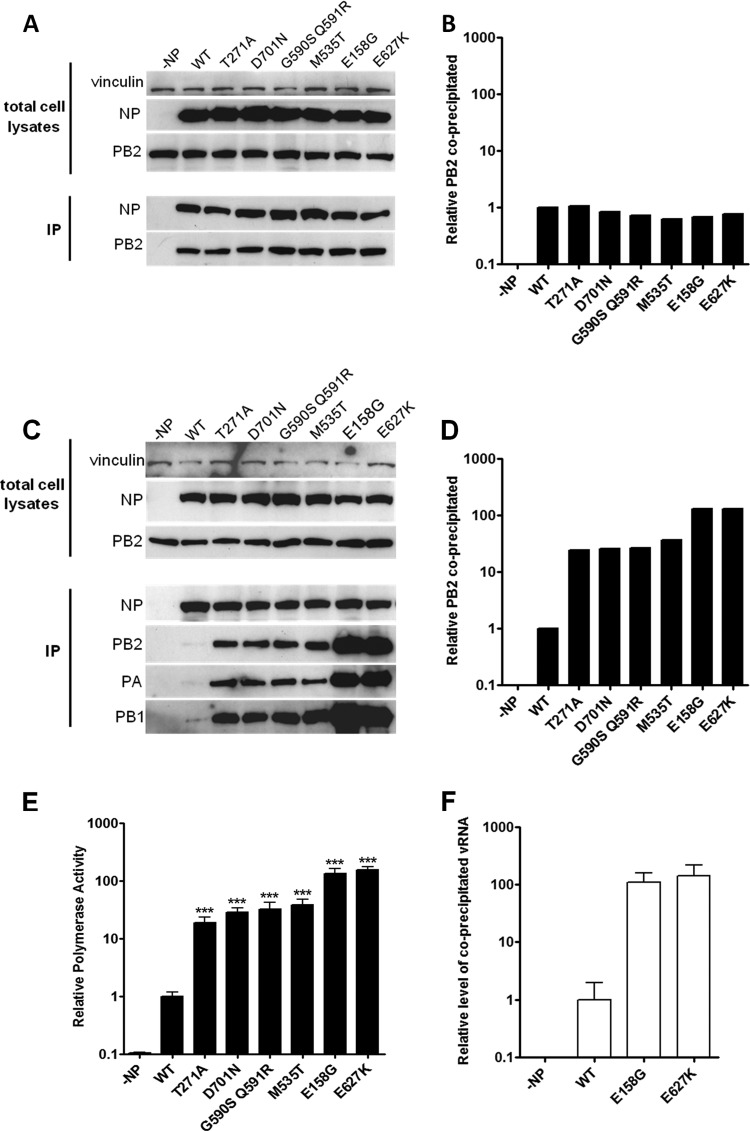
We aimed to investigate if other PB2 host range determinants stabilized the PB2-NP interaction in human cells in the manner reported for E627K. The PB2 mutations which enhanced polymerase activity most dramatically in human cells (E158G, T271A, M535T, G590S Q591R, E627K, and D701N) were introduced into FLAG-tagged PB2 in the pCAGGS expression vector. The C-terminal FLAG tag ensured that all PB2 mutants were readily and equally detected regardless of the introduction of host range mutations (see Fig. 3A and C), and the presence of the tag reduced polymerase activity only 2-fold (Fig. 2A and B).
Fig 3.
Effect of PB2 mutations on polymerase-NP interaction in 293T cells. (A) 293T cells were transfected with pCAGGS 50-92 NP and WT or mutated PB2-FLAG. At 12 h posttransfection, cell lysates were prepared and subjected to NP immunoprecipitation prior to Western blot analysis. Results are from one experiment representative of three independent experiments. (B) The levels of coimmunoprecipitated PB2 in panel A were normalized to precipitated NP using ImageJ. (C) 293T cells were transfected with pCAGGS 50-92 NP, PB1, PA, and WT or mutated PB2-FLAG and a virus-like firefly luciferase reporter plasmid. At 12 h posttransfection, cell lysates were prepared and subjected to NP immunoprecipitation prior to Western blot analysis. Results are from one experiment representative of three independent experiments. (D) The levels of coimmunoprecipitated PB2 in panel C were normalized to precipitated NP using ImageJ. (E) Polymerase activity supported by mutated PB2-FLAG compared to WT PB2-FLAG. Values were normalized to Renilla expression and to the activity of the WT polymerase. Results shown are the means ± standard deviations from three independent experiments. The statistical significance of differences in polymerase activity compared to that of WT was assessed by a two-tailed, unpaired Student t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (F) Level of vRNA isolated from cell immunoprecipitates analyzed in panel C for PB2 E158G-FLAG and PB2 E627K-FLAG normalized to PB2 WT.
Fig 2.
Activity of 50-92 with untagged or FLAG-tagged PB2. 293T cells were transfected with pCAGGS 50-92 NP, PB1, PA, and WT (A) or E627K (B) PB2 (untagged or FLAG tagged), as well as a virus-like firefly luciferase reporter plasmid and pCAGGS Renilla. Firefly luciferase production was measured 12 h posttransfection. Values were normalized to Renilla expression and to the activity of the untagged polymerase. Results shown are the means ± standard deviations from three independent experiments.
293T cells were transfected as in the minireplicon assay to reconstitute active vRNPs. At 12 h posttransfection, cells were resuspended in 250 μl of cell lysis buffer (50 mM Tris-HCl [pH 8.0], 200 mM NaCl, 1 mM dithiothreitol [DTT], 2 mM EDTA, 0.75% Igepal CA-630 [Sigma], 25% glycerol, one Complete Mini EDTA-free protease inhibitor cocktail tablet [Roche]/10 ml, 5 μl/ml RNasin [Promega]) and incubated for 1 h at 4°C. Immunoprecipitations were performed with protein A-agarose (Santa Cruz) and 3 μl of a rabbit polyclonal antibody specific for NP, kindly provided by P. Digard. Immunoprecipitated proteins were washed, dissolved in SDS sample buffer, and analyzed by Western blotting using rabbit polyclonal antibodies directed against NP and PB1 and a mouse monoclonal antibody specific for PA. Antibodies for PB1 and PA were kindly provided by J. Ortín. PB2 was detected using an anti-FLAG antibody (Sigma).
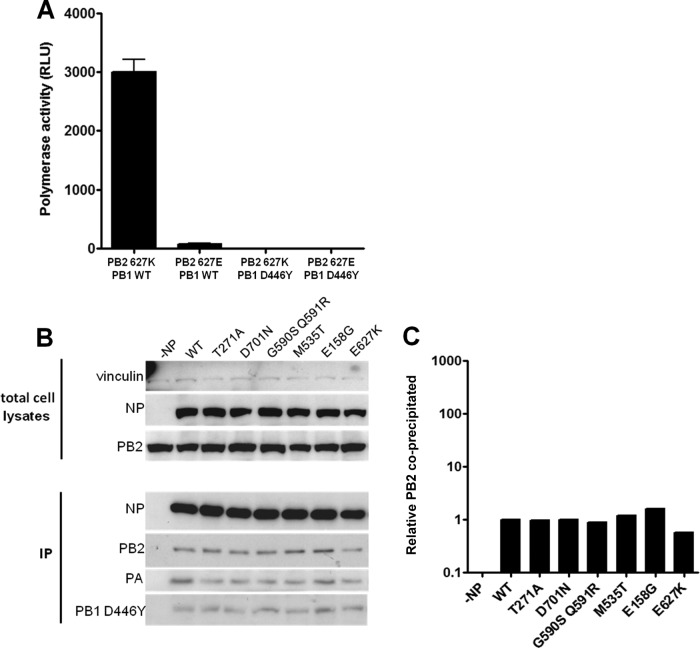
Initially, we demonstrated that the introduced PB2 host range determinants did not affect the amount of PB2 precipitated by NP when these two proteins were expressed alone in 293T cells (Fig. 3A and B), in accordance with the work of Mehle and Doudna (21). However, in the reconstituted RNP complex, those PB2 mutants that resulted in increased polymerase activity also increased the amount of PB2 precipitated by NP. The two results were proportionally correlated for all the introduced PB2 mutations (Fig. 3C to E). Moreover, a proportionate increase in precipitated PB1 and PA was also observed. It was confirmed by qRT-PCR that the precipitated RNP complex also contained a proportionate increase in negative-sense viral RNA-like RNA (Fig. 3F). Thus, it appeared that, as reported by others, mutations in PB2 that adapted avian virus polymerase for increased activity in mammalian cells did so by enhancing the interaction between NP and polymerase in the vRNP complex. However, it occurred to us that the increased signal of precipitated polymerase proteins might also reflect an increased amount of vRNP produced by more active polymerases as more vRNA molecules are made. To test this, we engineered a mutation into the conserved SDD motif of the catalytic site of the 50-92 polymerase subunit PB1 D446Y (32) that would prevent transcription and replication of the template viral RNA-like RNAs. In this case, vRNP complexes formed in transfected cells could be formed only from input RNA and expressed proteins rather than from amplified RNA products. The activity of a 50-92 polymerase harboring PB1 D446Y was abolished in a minireplicon assay in 293T cells (Fig. 4A). When the coimmunoprecipitation assay was repeated with PB1 D446Y, the PB2 host range mutations did not affect the amount of PB2 precipitated by NP compared with WT avian virus PB2 (Fig. 4B and C). This demonstrated that the PB2 host range mutations did not affect the stability of the PB2-NP interaction. Without amplification of viral RNA, there was no increase in RNP complex formation, and precipitation of the heterotrimeric polymerase complex by NP was not enhanced over that seen with vRNP complexes formed with wild-type avian virus PB2.
Fig 4.
In the absence of transcription/replication, PB2 mutations do not affect the amount of PB2 precipitated by NP. (A) PB1 mutation D446Y inhibits 50-92 polymerase activity. 293T cells were transfected with pCAGGS 50-92 NP, FLAG-tagged PB2 WT or 627K, PA, and PB1 WT or PB1 D446Y as well as a virus-like firefly luciferase reporter plasmid and pCAGGS Renilla. Firefly luciferase production was measured 12 h posttransfection. Values were normalized to Renilla expression. Results shown are the means ± standard deviations from three independent experiments. RLU, relative light units. (B) 293T cells were transfected with pCAGGS 50-92 NP, PB1 D446Y, PA, and WT or mutated PB2-FLAG and a virus-like firefly luciferase reporter plasmid. At 12 h posttransfection, cell lysates were prepared and subjected to NP immunoprecipitation prior to Western blot analysis. Results are from one experiment representative of three independent experiments. (C) The levels of coimmunoprecipitated PB2 in panel A were normalized to precipitated NP using ImageJ.
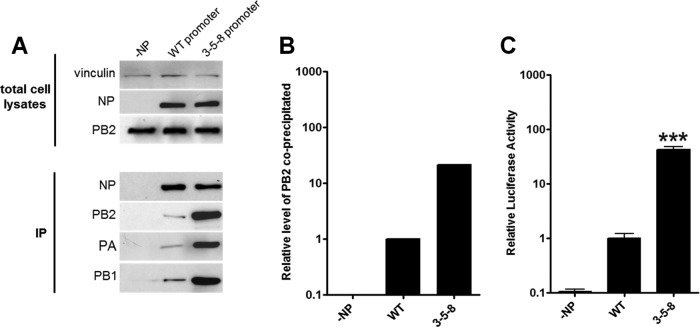
The conclusion that this coimmunoprecipitation assay was simply a measure of the number of vRNP complexes formed was reiterated using a different approach. The G3A, U5C, and C8U mutations (3-5-8 mutations) were introduced into the 3′ arm of the viral promoter of the luciferase reporter. These mutations have been shown to enhance the amplification and expression of the minigenome driven by an avian virus polymerase in human cells (33, 34). This was confirmed in a minireplicon assay with 50-92 polymerase in 293T cells: a 43-fold enhancement in luciferase reported signal was observed with the 3-5-8 reporter compared with the WT reporter (Fig. 5C). The 3-5-8 mutations result in perfect base pairing between the terminal nucleotides of the 3′ and 5′ ends of vRNA, which is likely to alter the structure of both vRNA and cRNA promoters by stabilizing the panhandle conformation and/or melting stem-loop structures in the corkscrew conformation (34). The coimmunoprecipitation assay was undertaken in the presence of the WT and mutated 3-5-8 luciferase reporters. The amount of PB2, PA, and PB1 precipitated by NP was much greater when vRNP complexes were formed with the 3-5-8 promoter mutant viral RNA-like RNA compared with the WT minigenome (Fig. 5A and B).
Fig 5.
Mutated 3-5-8 virus-like firefly luciferase reporter enhances luciferase activity and the amount of PB2 precipitated by NP. (A) 293T cells were transfected with pCAGGS 50-92 NP, PB1, PA, and PB2 (WT) as well as a WT or mutated 3-5-8 firefly luciferase reporter plasmid. At 12 h posttransfection, cell lysates were prepared and subjected to NP immunoprecipitation prior to Western blot analysis. Results are from one experiment representative of three independent experiments. (B) The levels of coimmunoprecipitated PB2 in panel A were normalized to precipitated NP using ImageJ. (C) Luciferase activity supported by 50-92 polymerase with 3-5-8 luciferase reporter. Values were normalized to Renilla expression and to the activity of the WT polymerase. Results shown are the means ± standard deviations from three independent experiments. The statistical significance of differences in polymerase activity compared to that of WT was assessed by a two-tailed, unpaired Student t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Taken together, our results can be explained by the idea that in human cells where avian virus polymerase is only poorly active, few vRNA molecules are produced and few nascent vRNP complexes form and, therefore, only a small amount of the heterotrimeric polymerase is precipitated by NP. However, by introducing mutations into the PB2 protein which increase polymerase activity or by mutating the viral promoter sequence, more RNP complexes are assembled around the RNA promoter and more PB2, PA, and PB1 will be precipitated with NP.
Our findings suggest that previous interpretations of this assay by other groups working in this field were preliminary because they did not take into account the possibility of an increase in replication resulting in an increase in the numbers of polymerase complexes (20–23). Unlike Labadie et al. (20), we did not find a primary defect in the interaction of PB2 with 627E and NP when expressed alone, out of context of the rest of the vRNP components. Similarly, Mehle and Doudna (21) also did not find a difference in the direct PB2-NP interaction with PB2 627E or 627K. Different strains of virus were used as sources of polymerase in these three studies, as well as different protein tags and different orders and conditions for precipitation and detection of interacting partners, and these variables may explain the discrepant results. Although Ng et al. (22), like Labadie and coworkers (20), found a difference in PB2-NP interaction dependent on residue 627, their work utilized fragments of PB2 and was not performed in the context of a host cell and so it may not be directly comparable with these other studies. In conclusion, we suggest that the restricted activity of an avian virus polymerase in human cells is not explained by the instability of the PB2-NP complex but rather by the lack of compatibility with a host cell factor required by influenza virus polymerase to replicate new vRNA molecules from cRNA templates, as recently suggested by Manz et al. (16).
ACKNOWLEDGMENTS
This work was supported by the Wellcome Trust grant 089870/Z/09/Z and by the European Union FP7 FLUPIG award.
We thank M. Mura for construction of pCAGGS PB2 M535T. We thank J. Ortín (Centro Nacional de Biotecnologia, Spain) for rabbit polyclonal anti-PB1 antibody and mouse monoclonal anti-PA antibody and P. Digard (Roslin Institute, Scotland) for rabbit polyclonal antibody directed against NP.
Footnotes
Published ahead of print 31 October 2012
REFERENCES
- 1. Beare AS, Webster RG. 1991. Replication of avian influenza viruses in humans. Arch. Virol. 119:37–42 [DOI] [PubMed] [Google Scholar]
- 2. Naffakh N, Tomoiu A, Rameix-Welti MA, van der Werf S. 2008. Host restriction of avian influenza viruses at the level of the ribonucleoproteins. Annu. Rev. Microbiol. 62:403–424 [DOI] [PubMed] [Google Scholar]
- 3. Almond JW. 1977. A single gene determines the host range of influenza virus. Nature 270:617–618 [DOI] [PubMed] [Google Scholar]
- 4. Chen G. 2006. Genomic signatures of human versus avian influenza A viruses. Emerg. Infect. Dis. 12:1353–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Finkelstein DB, Mukatira S, Mehta PK, Obenauer JC, Su X, Webster RG, Naeve CW. 2007. Persistent host markers in pandemic and H5N1 influenza viruses. J. Virol. 81:10292–10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gabriel G, Dauber B, Wolff T, Planz O, Klenk H-D, Stech J. 2005. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc. Natl. Acad. Sci. U. S. A. 102:18590–18595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mehle A, Doudna JA. 2009. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc. Natl. Acad. Sci. U. S. A. 106:21312–21316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Miotto O, Heiny A, Tan T, August JT, Brusic V. 2008. Identification of human-to-human transmissibility factors in PB2 proteins of influenza A by large-scale mutual information analysis. BMC Bioinformatics 9:S18 doi:10.1186/1471-2105-9-S9-S18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Naffakh N, Massin P, Escriou N, Crescenzo-Chaigne B, van der Werf S. 2000. Genetic analysis of the compatibility between polymerase proteins from human and avian strains of influenza A viruses. J. Gen. Virol. 81:1283–1291 [DOI] [PubMed] [Google Scholar]
- 10. Subbarao E, London W, Murphy B. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 67:1761–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tamuri AU, Dos Reis M, Hay AJ, Goldstein RA. 2009. Identifying changes in selective constraints: host shifts in influenza. PLoS Comput. Biol. 5(11):e1000564 doi:10.1371/journal.pcbi.1000564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gabriel G, Abram M, Keiner B, Wagner R, Klenk HD, Stech J. 2007. Differential polymerase activity in avian and mammalian cells determines host range of influenza virus. J. Virol. 81:9601–9604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hatta M, Gao P, Halfmann P, Kawaoka Y. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840–1842 [DOI] [PubMed] [Google Scholar]
- 14. Yamada S, Hatta M, Staker BL, Watanabe S, Imai M, Shinya K, Sakai-Tagawa Y, Ito M, Ozawa M, Watanabe T, Sakabe S, Li C, Kim JH, Myler PJ, Phan I, Raymond A, Smith E, Stacy R, Nidom CA, Lank SM, Wiseman RW, Bimber BN, O'Connor DH, Neumann G, Stewart LJ, Kawaoka Y. 2010. Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS Pathog. 6(8):e1001034 doi:10.1371/journal.ppat.1001034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bussey KA, Desmet EA, Mattiacio JL, Hamilton A, Bradel-Tretheway B, Bussey HE, Kim B, Dewhurst S, Takimoto T. 2011. PA residues in the 2009 H1N1 pandemic influenza virus enhance avian influenza virus polymerase activity in mammalian cells. J. Virol. 85:7020–7028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Manz B, Brunotte L, Reuther P, Schwemmle M. 2012. Adaptive mutations in NEP compensate for defective H5N1 RNA replication in cultured human cells. Nat. Commun. 3:802 doi:10.1038/ncomms1804 [DOI] [PubMed] [Google Scholar]
- 17. Bortz E, Westera L, Maamary J, Steel J, Albrecht RA, Manicassamy B, Chase G, Martinez-Sobrido L, Schwemmle M, Garcia-Sastre A. 2011. Host- and strain-specific regulation of influenza virus polymerase activity by interacting cellular proteins. mBio 2(4):00151–00111. doi:10.1128/mBio.00151-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gabriel G, Herwig A, Klenk HD. 2008. Interaction of polymerase subunit PB2 and NP with importin alpha1 is a determinant of host range of influenza A virus. PLoS Pathog. 4(2):e11 doi:10.1371/journal.ppat.0040011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moncorge O, Mura M, Barclay WS. 2010. Evidence for avian and human host cell factors that affect the activity of influenza virus polymerase. J. Virol. 84:9978–9986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Labadie K, Dos Santos Afonso E, Rameix-Welti M-A, van der Werf S, Naffakh N. 2007. Host-range determinants on the PB2 protein of influenza A viruses control the interaction between the viral polymerase and nucleoprotein in human cells. Virology 362:271–282 [DOI] [PubMed] [Google Scholar]
- 21. Mehle A, Doudna JA. 2008. An inhibitory activity in human cells restricts the function of an avian-like influenza virus polymerase. Cell Host Microbe 4:111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ng AK, Chan WH, Choi ST, Lam MK, Lau KF, Chan PK, Au SW, Fodor E, Shaw PC. 2012. Influenza polymerase activity correlates with the strength of interaction between nucleoprotein and PB2 through the host-specific residue K/E627. PLoS One 7(5):e36415 doi:10.1371/journal.pone.0036415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rameix-Welti M-A, Tomoiu A, Dos Santos Afonso E, van der Werf S, Naffakh N. 2009. Avian influenza A virus polymerase association with nucleoprotein, but not polymerase assembly, is impaired in human cells during the course of infection. J. Virol. 83:1320–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bussey KA, Bousse TL, Desmet EA, Kim B, Takimoto T. 2010. PB2 residue 271 plays a key role in enhanced polymerase activity of influenza A viruses in mammalian host cells. J. Virol. 84:4395–4406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ilyushina NA, Khalenkov AM, Seiler JP, Forrest HL, Bovin NV, Marjuki H, Barman S, Webster RG, Webby RJ. 2010. Adaptation of pandemic H1N1 influenza viruses in mice. J. Virol. 84:8607–8616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou B, Li Y, Halpin R, Hine E, Spiro DJ, Wentworth DE. 2011. PB2 residue 158 is a pathogenic determinant of pandemic H1N1 and H5 influenza A viruses in mice. J. Virol. 85:357–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Foeglein A, Loucaides EM, Mura M, Wise HM, Barclay W, Digard P. 2011. Influence of PB2 host range determinants on the intranuclear mobility of the influenza A virus polymerase. J. Gen. Virol. 92:1650–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Howard W, Hayman A, Lackenby A, Whiteley A, Londt B, Banks J, McCauley J, Barclay W. 2007. Development of a reverse genetics system enabling the rescue of recombinant avian influenza virus A/Turkey/England/50-92/91 (H5N1). Avian Dis. 51:393–395 [DOI] [PubMed] [Google Scholar]
- 29. Moncorge O, Long JS, Cauldwell AV, Zhou H, Lycett SJ, Barclay WS. 17 October 2012. Investigation of influenza polymerase activity in pig cells. J. Virol. doi:10.1128/JVI.01633-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li J, Ishaq M, Prudence M, Xi X, Hu T, Liu Q, Guo D. 2009. Single mutation at the amino acid position 627 of PB2 that leads to increased virulence of an H5N1 avian influenza virus during adaptation in mice can be compensated by multiple mutations at other sites of PB2. Virus Res. 144:123–129 [DOI] [PubMed] [Google Scholar]
- 31. Obayashi E, Yoshida H, Kawai F, Shibayama N, Kawaguchi A, Nagata K, Tame JR, Park SY. 2008. The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature 454:1127–1131 [DOI] [PubMed] [Google Scholar]
- 32. Biswas SK, Nayak DP. 1994. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J. Virol. 68:1819–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Crescenzo-Chaigne B, van der Werf S, Naffakh N. 2002. Differential effect of nucleotide substitutions in the 3′ arm of the influenza A virus vRNA promoter on transcription/replication by avian and human polymerase complexes is related to the nature of PB2 amino acid 627. Virology 303:240–252 [DOI] [PubMed] [Google Scholar]
- 34. Neumann G, Hobom G. 1995. Mutational analysis of influenza virus promoter elements in vivo. J. Gen. Virol. 76:1709–1717 [DOI] [PubMed] [Google Scholar]