Abstract
Human immunodeficiency virus type 1 (HIV-1) transmission results from infection with one or a small number of variants from the donor quasispecies. Transmitted/founder (T/F) viruses have recently been identified from acutely infected patients, but the way in which they interact with primary targets of HIV-1 infection is poorly understood. We have conducted a biological characterization of a panel of subtype B T/F acute and chronic envelope (Env)-expressing chimeric virus in primary human target cells and mucosal tissues. Both acute and chronic Envs preferentially replicated in peripheral blood mononuclear cells (PBMC) and a CD4 T-cell line compared to monocyte-derived macrophages, or dendritic cells (DC). In a model of trans infection from monocyte-derived dendritic cells to T cells, chimeric virus from acute Envs achieved significantly lower titers compared to chronic Envs. Challenge of primary human mucosal tissues revealed significantly higher levels of replication in chronic Env-expressing virus in rectal tissue compared to cervical and penile tissues and enhanced replication in tonsillar tissue relative to acute Envs. In agreement with data from the DC to T-cell trans infection assay, chronic Env-chimeric virus pools were transmitted more efficiently by migratory cells from cervical and penile tissues to CD4+ T cells than individual acute Env chimeras. These data indicate that virus with HIV-1 Envs of transmitted acute infections preferentially replicate in T cells rather than macrophages or dendritic cells and are less efficiently transmitted from antigen-presenting cells to CD4 T cells than chronic Envs. Such properties together with chemokine (C-C motif) receptor 5 (CCR5) use may confer an advantage for transmission.
INTRODUCTION
The most common route of human immunodeficiency virus type 1 (HIV-1) transmission worldwide is by sexual intercourse (1). Identification and phenotypic characterization of transmitted and early founder virus may be pivotal in effectively targeting vaccine responses against the earliest event in mucosal HIV-1 infection. Previous studies have sought to identify and characterize the virus envelope in early phase HIV-1 infection, as it mediates entry into susceptible target cells, and as such is a common target for vaccine design. Characterization of env genes found early after infection has yielded conflicting results. Most groups have found evidence of limited diversity in envelope regions soon after infection, in patients prior to seroconversion (2), in recent seroconverters (3), and in mother-to-child transmission (4), suggesting there is a significant bottleneck leading to transmission of only one isolate or a small number of isolates. Despite contradictory evidence showing HIV-1 heterogeneity in envelope sequences from early heterosexual and homosexual transmission (5, 6), the extremely limited diversity of clones isolated close to transmission is in stark contrast to the high genetic diversity and swarm of HIV-1 clones found in later chronic disease. Longitudinal analysis of the V3 envelope region shows continual increases in HIV-1 genetic diversity during asymptomatic disease from just one or a few transmitted HIV-1 clones (7). Conflicting observations on the relative number of HIV-1 clones observed after the transmission bottleneck may be due to the mode of transmission, time of sample collection (up to 6 months postinfection), and the source of viral nucleic acids (either from plasma or peripheral blood mononuclear cells [PBMC]). The technique employed to characterize the sequences may also influence results; for example, PCR, cloning, and sequencing may result in Taq-induced PCR errors, and nonproportional representation of target sequences, whereas heteroduplex tracking assays do not generate sequence information for phylogenetic analysis (8).
Recent work utilizing single-genome amplification (SGA) to examine HIV from plasma viral RNA (vRNA) and direct sequencing of uncloned env amplicons has allowed transmitted virus to be enumerated in acutely infected patients. The stage of disease was classified in these patients using Fiebig stages, which classify phases of acute infection according to diagnostic tests for viral RNA and proteins (9). Data from 102 patients demonstrated that 76% of patients acutely infected with subtype B HIV-1 strains are infected with a single virus, while the remainder are infected with 2 to 5 isolates (8). Examination of env genes in subtype A and C transmission pairs (discordant couples) found that 18/20 (90%) were infected with a single isolate and that when multiple variants were transmitted, this was associated with genital inflammation in the newly infected partner (10). The SGA approach has also highlighted factors that influence transmission of multiple variants; 36% of a cohort of men who have sex with men (MSM) were infected by multiple variants, with the number of virus transmitted being between 2 and 10 (11). Despite these numerous studies describing limited genetic diversity of transmitted HIV-1, little is known about possible enhanced tropism for mucosal tissue, replication dynamics, or other phenotypic properties of the transmitted HIV-1 variants within the host or between hosts at the time of acute infection. Aside from preferential transmission of chemokine (C-C motif) receptor 5 (CCR5) over chemokine (C-X-C motif) receptor 4 (CXCR4) using HIV-1 clones (12), it is not clear whether a simple mechanical process involving limited barrier disruption results in a new infection by just a few HIV-1 clones or whether the infecting HIV-1 clones are selected due to specific phenotypic properties, i.e., not characteristic in the majority of HIV-1 clones within complex HIV-1 quasispecies of the donor. Recent studies examined possible differences in host cell entry efficiency of HIV-1 clones derived from acute infections versus chronic disease. Acute and chronic HIV-1 clones did not show a phenotypic difference in entry kinetics, CCR5 or CD4 affinity, and sensitivity to various entry inhibitors (13, 14).
Sexual transmission of HIV-1 requires the virus inoculum in semen or vaginal or rectal secretions to cross the mucosal epithelium and establish infection in susceptible target cells found in the epithelium and genital or rectal mucosal tissue. Multiple mechanisms have been proposed to explain how HIV-1 crosses the epithelial barrier, including direct infection of epithelial cells, transcytosis through epithelial cells or M-cells, transmigration of infected donor cells, uptake by intraepithelial Langerhans cells (LC), and physical breaches to the epithelium (1, 15, 16). Once virus has penetrated the epithelium, T cells, macrophages, and dendritic cells (DC) including Langerhans cells found in epithelial and subepithelia of genital and rectal mucosas are proposed as primary targets for HIV infection. However, the tropism of transmitted/founder (T/F) virus for such cells is poorly understood. Defining the mechanisms by which acute T/F virus interact with mucosal tissue models and primary target cells for HIV infection may provide important clues for vaccine design.
This study seeks to determine whether there is a phenotypic difference in infection fitness of acute T/F envelope chimeric virus relative to chronic virus in cellular and tissue models thought to reflect the potential range of primary cells at the mucosal portals of entry. To this end, we have conducted a biological characterization of chimeric virus harboring a panel of subtype B transmitted/founder envelope genes from multiple patients in acute infection (8, 17) and compared their properties to those of chronically infected individuals. We have characterized replication of acute and chronic Env-chimeric virus in the following: a CD4 T-cell line (PM-1); primary HIV-susceptible PBMC; macrophages; monocyte-derived dendritic cells (MDDC); a MDDC-to-T-cell transinfection model; and in primary human tissue explants of cervical, penile glans, rectal, and tonsillar origin. Viruses were titrated in each of the cellular and tissue models, and the titers were compared with chronic envelope chimeric viral pools and prototypic macrophage-tropic strains (Table 1). The presented data indicate that virus with HIV-1 Envs of transmitted acute infections preferentially replicate in T cells rather than macrophages or dendritic cells and are less efficiently transmitted from antigen-presenting cells to CD4 T cells than chronic Envs.
Table 1.
Acute and chronic Env-chimeric viruses
| Virus IDa | Patient designation | Infection group | Genderb | Transmissionc | Phenotype | HIV-1 subtype |
|---|---|---|---|---|---|---|
| B1 | 1054.TC4.1499 | Acute | M | SPD | R5 | B |
| B2 | 6244_13.B5.4576 | Acute | M | SPD | R5 | B |
| B3 | PRB926_04.A9.4237 | Acute | ? | SPD | R5 | B |
| B4 | WEAUd15.410.787 | Acute | M | MSM | X4/R5 | B |
| B7 | SC05.8C11.2344 | Acute | M | Heterosexual | R5 | B |
| B8 | 1059_09.A4.1460 | Acute | M | SPD | R5 | B |
| B9 | 6240_08.TA5.4622 | Acute | M | SPD | R5 | B |
| B14 | SC45.4B5.2631 | Acute | M | Heterosexual | R5 | B |
| B17 | REJO.D12.1972 | Acute | M | Heterosexual | R5 | B |
| B19 | 700010040.C9.4520 (CH040) | Acute | M | MSM | R5 | B |
| B20 | 63358.p3.4013 | Acute | ? | SPD | R5 | B |
| C1 | I10 | Chronic | M | MSM | R5 | B |
| C2 | 217 | Chronic | ? | ? | R5 | B |
| C3 | 801 | Chronic | ? | ? | R5 | B |
| C4 | K44 | Chronic | M | MSM | R5 | B |
| C5 | Q0 | Chronic | M | MSM | R5 | B |
| C6 | 9 | Chronic | ? | ? | R5 | B |
| C7 | 11 | Chronic | ? | ? | R5 | B |
| C8 | 17 | Chronic | ? | ? | R5 | B |
| C9 | 18 | Chronic | ? | ? | R5 | B |
| C10 | 24-2 | Chronic | ? | ? | R5 | B |
| C11 | 27 | Chronic | ? | ? | R5 | B |
| C12 | 29 | Chronic | ? | ? | R5 | B |
| C13 | 49 | Chronic | ? | ? | R5 | B |
| C14 | 78 | Chronic | ? | ? | R5 | B |
| YU2 | None (reference strain) | R5 | B |
ID, identification.
M, male; ?, not known.
Risk behavior where known. Subjects listed as “SPD” were source plasma donors who denied having sex for money, homosexual activity, and intravenous drug use (IVDU) or receiving a blood transfusion or a tattoo in the preceding year (18). MSM, men who have sex with men; ?, not known.
MATERIALS AND METHODS
Reagents.
All reagents were from Sigma-Aldrich UK, unless otherwise stated. Tissue culture flasks were from Greiner, and BD Falcon Microtest tissue culture plates were used.
Virus.
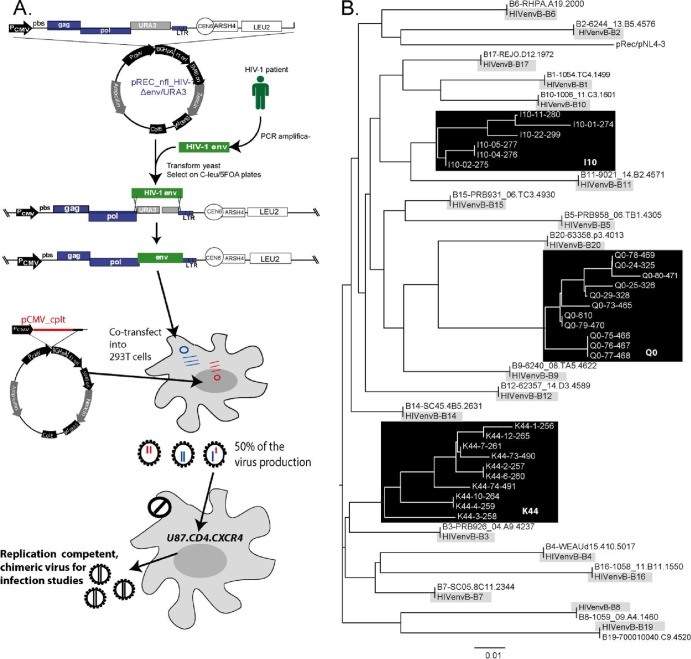
Eleven acute subtype B envelopes (Envs) were cloned from the Center for HIV/AIDS Vaccine Immunology (CHAVI) acute infection studies (8) (Table 1). In addition, 14 chronic Envs (isolated from patients infected for more than 1 year) were cloned from anonymous patient samples under institutional review board (IRB) approval in Eric Arts' laboratory at Case Western Reserve University (Table 1). All HIV-1 Envs were cloned using the protocol described in reference 17 and schematically depicted in Fig. 1A. Briefly, the approximately 3 μg of the 3-kbp env DNA (HXB2 numbering 5957 to 8904) from acute infection clones and 3 μg of the 3.1-kbp env DNA (HXB2 numbering 5701 to 8819) of chronic infections were transfected into Saccharomyces cerevisiae with linearized plasmids, pREC_nfl_NL4-3_ Δenv/URA3. Yeast colonies were selected on complete medium lacking leucine (C−Leu) plates supplemented with fluoroorotic acid (FOA). The resulting vectors, pREC_nfl_NL4-3_envpatientxclone, harbored in the FOA-resistant yeast colony were then transformed into bacteria to amplify the DNA plasmid for purification as described previously (17). It is important to note that for the chronic patient samples, the env PCR product harbored the amplified patient quasispecies, and as such, >100 yeast colonies were removed from Leu−/FOA plates for bulk plasmid purification and eventual reconstitution of sample quasispecies. In contrast, only a single env clone (provided by B. F. Keele and B. H. Hahn, University of Alabama) was subcloned into pREC_nfl_NL4-3_ Δenv/URA3 representing the limited genetic diversity in the acute sample. Each pREC_nfl_NL4-3_envpatientxclone of the yielded vectors and the supplementary vector, pCMV_cplt (CMV stands for cytomegalovirus) (17), were then cotransfected into 293T cells with Effectene transfection reagent (Qiagen) to produce virus, which were then propagated in U87.CD4.CCR5 cells as described previously (17) and schematically depicted in Fig. 1A. This procedure of highly efficient yeast-based recombination/cloning followed by 293T transfections and short propagations in U87 cells is thought to preserve the HIV-1 qausispecies population better than a similar approach using bacterial restriction enzyme cloning (17). Previous studies (including our own) on HIV-1 fitness/infectivity used prepared viruses, termed primary HIV-1 isolates, by PBMC cocultivation and subsequent virus expansion. This procedure results in high selection bias and genetic bottlenecks. For example, the presence of only 10% CXCR4-tropic virus in a quasispecies prior to PBMC propagation led to complete dominance of these X4 clones in the “primary isolate” within 2 to 3 weeks of PBMC propagation. However, we estimate a 100-fold reduction in the genetic bottleneck using our chimeric virus cloning/production procedure based on 454 pyrosequencing of the virus population in the patient sample prior to manipulations (R. M. Gibson, K. R. Henry, M. E. Quinones-Mateu, and E. J. Arts, unpublished data).
Fig 1.
Construction and sequence analyses of the HIV-1 env chimeric viruses derived from acute and chronic infection samples. (A) An HIV-1 env cassette was reverse transcription-PCR (RT-PCR) amplified as described in Materials and Methods and then transfected into yeast along with the linearized pREC_nfl_HIV-1Δenv/URA3. The URA3 gene is replaced by the env cassette via homologous recombination/gap repair, and this process is selected via yeast colony growth on plates lacking leucine but containing FOA. FOA is converted into a toxic anabolite by the uracil biosynthesis machinery encoded by URA3. The pREC_nfl_HIV-1 vectors with chronic or acute env genes are then purified and used to transfect 293T cells along with the complementing vector, pCMV_cplt by the method of Dudley et al. (17). Virus-containing supernatant from these transfections are propagated for a maximum of 2 weeks on U87.CD4.CCR5 cells to produce the virus employed in all subsequent assays. Abbreviations: PCMV, promoter for cytomegalovirus: pbs, primer-binding sequence; LTR, long terminal repeat; PCR amplifica-, PCR amplification. (B) The viruses amplified on the U87.CD4.CCR5 cells served as the templates for RT-PCR and DNA sequencing. The neighbor-joining phylogenetic tree was constructed based on the C2-V3 region of env within the propagated viruses. For each of the acute clones, the original HIV-1 env gene derived from SGA of the patient sample is provided to verify sequence identity to the cloned virus.
Cell-free supernatants of these HIV Env virus were expanded for 7 to 10 days in Jurkat-tat-R5 cells and quantified using the 50% tissue culture infective dose (TCID50) assay. These Env-chimeric virus were evaluated for differential tropism in cellular and explant tissue assays. The HIV-1 env gene was also reverse transcribed-PCR amplified, cloned into pCR-TOPOII vectors (Invitrogen), and sequenced in the C2-V3 region as previously described (19). Neighbor-joining phylogenetic trees (Fig. 1B) were constructed to compare env sequences of the single clones analyzed by SGA (8) to those cloned into these chimeric env viruses. The intrapatient diversity of the chronic env chimeric viruses is also shown for three representative chronic-Env virus: the Q0 virus derived from a patient sample following 67 months of infection; I10 from a patient sample following 48 months of infection, and K44 from a patient sample following 85 months of infection (19).
Cell isolation and culture.
PBMC were isolated from single donor buffy coats, obtained from the National Health Service (NHS) Blood and Transplant Service, using density gradient centrifugation with Histopaque-1077 Hybri-Max and washing 2 or 3 times with cold, sterile phosphate-buffered saline (PBS).
Monocyte-derived macrophages (MDM) were obtained from buffy coat-derived PBMC by adherence (1 × 106 PBMCs/well in 96-well plates) and matured for 5 to 7 days by culture in RPMI 1640 plus 10% human serum (HS) and 20 ng/ml of granulocyte-macrophage colony-stimulating factor (GM-CSF) (R&D Systems).
Culture of monocyte-derived dendritic cells.
Monocytes were isolated from PBMC by adherence to 175-cm tissue culture flasks. Cells were resuspended at 10 × 106/ml in RPMI 1640 plus 0.5% HS and incubated at 37°C for 2 h. Nonadherent cells were removed from culture by washing 4 times with sterile, room temperature PBS. Cells were cultured for a total of 6 days in 30 ml per flask of DC medium (RPMI 1640 plus 5% HS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine) supplemented with 40 ng/ml interleukin 4 (IL-4) (equivalent to 500 U/ml) and 20 ng/ml GM-CSF (equivalent to 1,000 U/ml) (R&D Systems). After 3 days, 20 ml of medium was removed from each flask and centrifuged at 300 × g for 5 min at room temperature, spent medium was discarded and replaced with 20 ml of fresh DC medium, cells were added back to their original flask and supplemented with cytokines as described above. Residual T cells were removed from DC cultures after 6 days by depletion using anti-CD3 Dynabeads according to the manufacturer's instructions. Cells were then phenotyped using fluorochrome-conjugated antibodies against human DC-SIGN, CD123, CD11c, HLA-DR, CD80, CD83, CD86 (BD Bioscience), and CD3 (Beckman Coulter) with isotype-matched control antibodies as negative controls. Fluorescence-activated cell sorting (FACS) data were acquired on an FC500 flow cytometer using CXP software. Phenotyping of immature monocyte-derived dendritic cells (MDDC) indicated high levels of expression of DC-SIGN, CD11c, and HLA-DR (means of 91.8%, 92.5%, and 98%, respectively) and low levels of CD123, CD80, and CD83 (6.5%, 9.5% and 6.0%, respectively). Cells were negative for CD3 (<1%; mean, 0.7%).
Cellular assays. (ii) Virus infectivity in cells measured by TCID50.
The relative replication of each virus across the different cell types was assessed by determining the TCID50/ml (tissue culture infective dose). This provided a direct assessment of the relative infectivity in the different models, removing the need to standardize viral input according to p24 content or infectivity in a specific model. In order to determine the TCID50/ml in PBMC cultures, cells from three different donors were activated with phytohemagglutinin (PHA) (5 μg/ml) for 3 days before being washed and plated out at 105 cells per well in a U-bottomed, 96-well plate. Virus was added at a 1 in 5 dilution, with serial 5-fold dilutions done down the plate, and left to incubate overnight at 37°C. Each virus dilution was tested in replicates of five. The virus was washed off the next day by centrifugation for 5 min at 350 × g, and cell culture medium (RPMI 1640 with 10% fetal calf serum [FCS], 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine [complete RPMI]) with IL-2 (100 U/ml) (Novartis UK) was added back at a total volume of 200 μl/well. The supernatant was collected on days 7 and 14 and assessed for viral replication by measuring HIV-1 p24 activity. For comparison, the TCID50/ml calculation was also performed for monocyte-derived macrophages (from three separate donors) and the PM-1 cell line in which cells and the virus were used at the same concentration as for PBMC. The TCID50/ml in TZM-bl cell lines was calculated using 5 × 104 cells/well and a starting dilution of 1 in 2 for each virus, with subsequent 4-fold serial dilutions. The cells were incubated overnight with the virus without the addition of dextran. HIV infectivity in the TZM-bl cells was then determined using a luciferase reporter gene assay, in which the cells were lysed and luciferase expression was assessed after the addition of substrate (luciferase assay system; Promega, United Kingdom) (14). HIV-1BaL and HIV-1YU-2 on a NL4-3 backbone (NL-BaL.ecto and NL-YU2.ecto, gifts of C. Ochsenbauer and J. C. Kappes) were used as a positive control.
(ii) Virus infectivity (TCID50) in MDDC and MDDC/PM-1 cocultures.
MDDC, from three different donors, were plated at 5 × 104/well in 100 μl DC medium in U-bottom 96-well plates, and 100 μl of virus was added at a 1/2 dilution with serial 2-fold dilutions down the plate, with 5 replicates per dilution. Cells were incubated with virus for 2 h at 37°C and then washed three times with sterile, room temperature PBS at 300 × g. Washed MDDC were then resuspended in 250 μl of DC medium. PM-1 cells were plated at 2 × 104 cells per well in U-bottom 96-well plates in 150 μl complete RPMI, and 50 μl of washed, infected MDDC was added to the PM-1 cells. Both MDDC and MDDC/PM-1 cultures were maintained for a total of 14 days at 37°C, virus replication was measured by HIV-1 p24 enzyme-linked immunosorbent assay (ELISA) at 7 and 14 days postinfection, and TCID50/ml was calculated as for other cell types. This culture period was selected to give any slow growing virus potential to lead to a positive p24 signal irrespective of replicative fitness in PM-1 cells.
Tissue assays.
Ex vivo genital, tonsillar, and rectal tissue samples were used for assessing the relative replication of each virus. Penile tissue was obtained following gender reassignment surgery at Charing Cross Hospital, London, United Kingdom. All participants had ceased hormonal therapy a minimum of 6 weeks prior to surgery, and tissues were collected with written consent according to local research committee (LRC) guidelines. Cervical, rectal, and tonsillar tissue samples were provided from the Duke Mucosal Tissue Repository, Duke Human Vaccine Institute, Duke University Medical Center, NC.
Cervical and penile tissue was cut into 2- to 3-mm3 explants comprising both epithelium and stroma and infected with either acute or chronic Env-chimeric virus (at a dilution of 1 in 2 for each virus) or HIV-1BaL for 2 h. The tissue was then washed to remove unbound virus and then cultured in complete RPMI medium. Migratory cells emigrating from tissue explants were harvested after overnight culture and cocultured with PM-1 CD4 T cells (4 × 104/well) for 21 days. Rectal and tonsillar tissue were dissected into 1- to 2-mm3 pieces infected with virus (Env-chimeric virus or HIV-1BaL) for 2 h and then washed in sterile PBS. The tissues were then cultured upon gel foam rafts (Pharmacia & Upjohn), 3 or 4 pieces per raft per well, in either RPMI 1640 supplemented with 15% FCS, 2 mM l-Glu, 200 units/ml of penicillin-streptomycin (Pen-Strep), 50 μg/ml gentamicin (Invitrogen, United Kingdom), and 1 μg/ml amphotericin B (Fungizone) (Invitrogen, United Kingdom) for tonsillar tissue or Dulbecco modified Eagle medium (DMEM) supplemented with 10% FCS, 2 mM l-Glu, 200 units/ml Pen-Strep, 80 μg/ml gentamicin, and 1 μg/ml amphotericin B for rectal tissue. For all tissues (cervical, penile, rectal, and tonsillar), the medium was changed every 3 or 4 days, and the tissue was cultured for a total of 21 days. For each tissue and virus, three different donors were used. Relative replication of each virus among the different tissues was assessed by measuring p24 release in supernatant over time. Cumulative HIV-1 production was calculated from the sum of p24 output determined at each time point.
HIV-p24 ELISA.
Cell and tissue culture supernatant was assessed for the production of p24 by using p24 ELISA kits (NCI-Frederick, MD) according to the manufacturer's instructions.
Statistical analysis.
Statistical analyses were performed on GraphPad Prism 5 or GraphPad InStat 3.10, utilizing the Mann-Whitney test to compare TCID50 or Spearman's rank correlation coefficient for comparison between macrophages and dendritic cells. P values of <0.05 were considered significant.
RESULTS
Cell tropism studies of acute and chronic subtype B Env-chimeric virus.
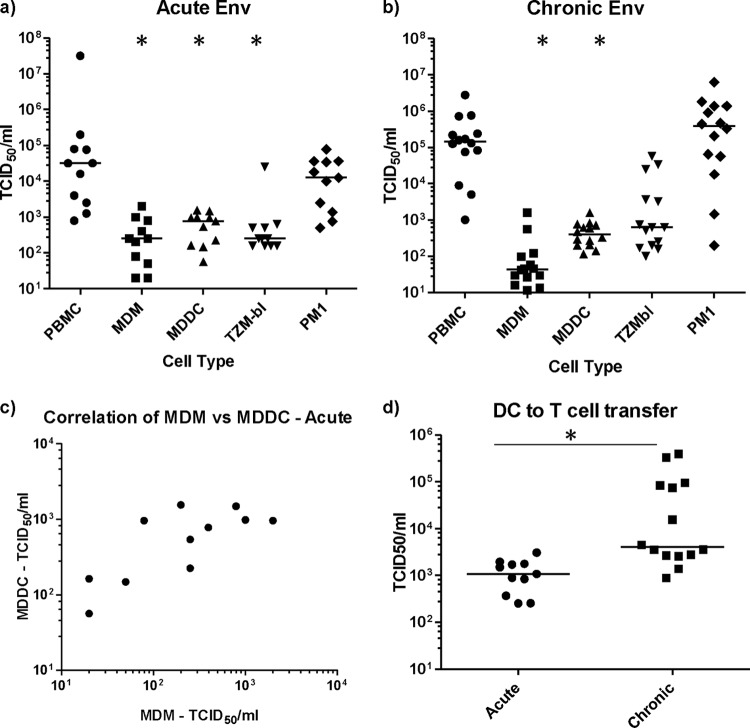
Potential target cells within the genital and colorectal compartments include antigen-presenting cells, dendritic cells (epithelial and subepithelial populations) and macrophages, and CD4+ T cells. Characterization of the cellular tropism of acute transmitted/founder virus Envs may provide important insights into the relevant contributions of these different cell types to the initial events determining transmission at the mucosal portals of entry. A panel of 11 acute Env-chimeric subtype B viruses (Table 1) were cloned from the CHAVI acute infection studies according to the protocol depicted in Fig. 1A. The fidelity of the chimeric virus to the original HIV-1 env gene derived from SGA is shown in Fig. 1B. Cell tropism of acute Env-chimeric virus was initially assessed by measuring replication in PBMC and in vitro monocyte-derived macrophages (MDM). All acute Env-chimeric virus demonstrated preferential infectivity of PBMC compared to macrophages (P = 0.0002) (Fig. 2). This was in sharp contrast to the two macrophage-tropic strains, HIV-1BaL and HIV-1NL4-3.YU2 that showed equal infectivity in both cell types. These data confirm previous studies using both MDM and CD4+ T cells (11, 18, 20, 21).
Fig 2.
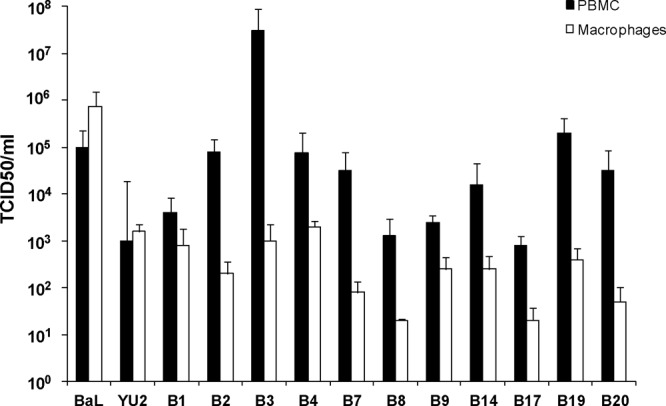
Acute subtype B Env-chimeric virus preferentially infects PBMC over macrophages. The relative TCID50/ml of subtype B acute Env-chimeric virus was determined in PBMC and monocyte-derived macrophages. Activated PBMC or MDM were cultured with acute virus (B1 to B20) or control virus for up to 14 days. Culture supernatant was collected and assessed for p24 release. HIV-1BaL and HIV-1YU-2 (macrophage-tropic strains) which were used as controls, showed equal replication in both cell types unlike the acute Env-chimeric virus, which showed preferential replication in PBMC.
Infectivity was then further assessed in a wider panel of cells, including PBMC, MDM, monocyte-derived dendritic cells (MDDC), and PM-1 CD4+ T cells, and in TZM-bl cells as a control (Fig. 3a). The T/F acute chimeric virus showed similar levels of infectious virus when assessed for reverse transcriptase assay providing an average virtual TCID50 of 105.3 (104.1 to 105.8). All acute Env-chimeric virus displayed preferential infectivity for PBMC and CD4+ PM-1 T cells. Infectivity of acute-Env virus for MDM, MDDC, and TZM-bl cells were significantly lower than for both PBMC and PM-1 cells (P < 0.0005). Interestingly, there was a positive correlation between infection of MDM and MDDC (P = 0.03 and r = 0.641) (Fig. 3c). In contrast, there was no correlation with any pairings between the other cell types tested. To determine whether the NL4-3 backbone may have reduced macrophage tropism, control experiments were performed using the env sequence of HIV-1BaL inserted into the same backbone. Here there was no observable difference between HIV-1BaL and BaL Env-chimeric virus with respect to infection of T cells and macrophages, suggesting that the NL4-3 backbone had little impact on macrophage tropism as previously shown (21).
Fig 3.
Replication of subtype B Env-chimeric virus across cell lines and primary cell models. (a) The relative TCID50/ml of the acute Env virus was determined in a variety of different cell types; the TCID50 values for PBMC were compared to those in MDM, MDDC, and TZM-bl cells, using a Mann-Whitney t test and 95% confidence interval (95% CI). Values that are significantly different (P < 0.0005) are indicated by an asterisk. (b) Subtype B chronic Env-chimeric virus (n = 14) were also evaluated for TCID50/ml and compared according to cell type. Values that are significantly different (P < 0.0001) by Mann-Whitney t test and 95% CI are indicated by an asterisk. The horizontal bars in panels a and b represent the median TCID50/ml for each cell type. (c) The TCID50s for the acute Env-chimeric virus in MDM and MDDC show a positive correlation (P = 0.037 and r = 0.641) by Spearman's rank correlation coefficient and 95% CI. (d) MDDC-to-CD4+-T-cell transmission of Env-chimeric virus. Acute and chronic Env-chimeric virus were titrated on MDDC, washed, and cocultured with PM-1 cells. Infectivity was measured in MDDC/PM-1 cocultures by HIV-1 Gag p24 ELISA, and TCID50/ml was calculated for each virus. MDDC were significantly better able to transmit chronic virus than acute virus to T cells. Values that are significantly different (P < 0.0003) by the Mann-Whitney t test and using 95% CI are indicated by an asterisk and long thin black line. The horizontal bars represent the median TCID50 for each virus type.
A panel of Env-chimeric viruses (Table 1) generated from chronically infected subtype B subjects was also evaluated (Fig. 3b). Unlike acute Env-chimeric virus, derived from a single SGA clone, chronic Env-chimeras represent the amplified and cloned patient quasispecies (Fig. 1B). The chronic Env-chimeric virus panel displayed a similar profile of cellular tropism with 2- to 3-log-unit-higher (significantly higher) infectivity for PBMC and PM-1 cells over MDM and MDDC (P < 0.0001). No significant differences were observed between acute and chronic Env-chimeric virus in any of the primary cell types examined (PBMC, MDM, and MDDC); however, chronic Env chimeric virus had significantly higher TCID50/ml than acute Env chimeric virus in the PM-1 CD4 T cell line (P < 0.006). Based on the results of previous studies (19, 22) and in relation to in vivo conditions, different clones in the chronic Env chimeric virus quasispecies may replicate more efficiently and dominate replication in the different primary cell types and tissues. However, it assumed that the acute Env chimeric virus may still be more proficient at infection in transmission models, since they have already emerged from this selection process.
Transmission of Env-chimeric virus from MDDC to CD4+ T cells.
Dendritic cells have long been proposed as playing an important role in transmission of HIV-1, as they are thought to be among the first cells that encounter the virus and thus facilitate the spread of the virus to CD4+ T cells. One of the ways they are thought to do this is through trans-infection, mediated by the C-type lectin, DC-SIGN, expressed on the surfaces of dendritic cells (23). Therefore, the efficiency of viral transmission of acute Env-chimeric virus, from MDDC to CD4+ T cells, was assessed. Here MDDC were pulsed with serial dilutions of virus for 2 h, washed to remove unbound virus, and then directly cocultured with CD4 indicator cells for 14 days, whereupon supernatant was collected and assessed for infectivity on the basis of p24 expression. The majority of acute Env-chimeric viruses were transmitted less efficiently compared to the chronic Env-chimeric virus as determined by TCID50 (P < 0.0003) (Fig. 3d). In order to further understand the potential mechanism behind this trend, we assessed the viral binding affinities of the Env-chimeric virus and lab-adapted virus (BaL, SF162, LAV, YU2, and RF) to the C-type lectin DC-SIGN (data not shown). Acute and chronic Env-virus showed similar affinity for DC-SIGN that was not significantly different (data not shown), suggesting that this mechanism was not responsible for the higher level of transmission of chronic Env-chimeric virus to T cells compared to acute Env virus observed in Fig. 3d. It is also important to note that other factors may play a role. Our previous reports suggest that acute HIV-1 Envs (derived from the same patients) do not show a mean difference in CD4 and/or CCR5 usage compared to the chronic HIV-1 Envs from typical progressors (13). However, the variations in receptor affinities, host cell entry kinetics, and sensitivity to entry inhibitors were much greater in acute HIV-1 Envs and more uniform for chronic Env-derived virus (13).
Replication of subtype B Env-chimeric virus in different tissue types.
We then assessed the ability of the panel of acute Env-chimeric virus to infect different types of mucosal tissue: cervical, penile glans, tonsillar, and rectal tissues. Tissue was dissected into 1- to 3-mm3 explants and infected with either acute or chronic Env-chimeric virus (average TCID50/ml of 4.4 and 4.3, respectively, in PBMC), or with HIV-1BaL used as a control. After 2 h, unbound virus was washed off, and the tissue was cultured for a period of 3 weeks during which supernatant was harvested and assessed for p24 levels. All of the acute Env-chimeric virus replicated with greater variation in all tissues, though there appeared to be a trend for more efficient replication in rectal tissue (Fig. 4a). This trend was significant with the chronic Env-chimeric viruses, which showed greater replication efficiency in rectal tissue than in penile and cervical tissue (P < 0.003) (Fig. 4b). There was no difference between the acute and chronic viruses for each tissue type, except for tonsillar tissue where higher replication was observed with chronic viruses (P < 0.007). Experiments were then performed to assess the potential dissemination of virus by migratory cells that emigrate out of tissue during the first 24 h of culture (24). Cervical and penile tissue explants were exposed to virus for 2 h and washed to remove unbound virus. Migratory cells from these tissues were collected 24 h later and cocultured with CD4+ PM-1 T cells for up to 21 days. Viral replication was assessed by measurement of p24 concentration in culture supernatant. Interestingly, migratory cells from cervical tissue inefficiently transmitted acute Env-chimeric virus to CD4+ T cells. This was also true for migratory cells from penile tissue, to the extent that only three acute viruses were transmitted to the PM-1 cells, with the other viruses not transmitted at all. In contrast, the chronic Env-chimeric viruses were very efficiently transmitted by migratory cells compared to acute Env-chimeric virus (P < 0.0001) (cervical and penile tissue) (Fig. 5). These data reflect a similar but more exaggerated pattern to the transfer of acute and chronic Env-chimeric virus by MDDC to T cells (Fig. 3d).
Fig 4.
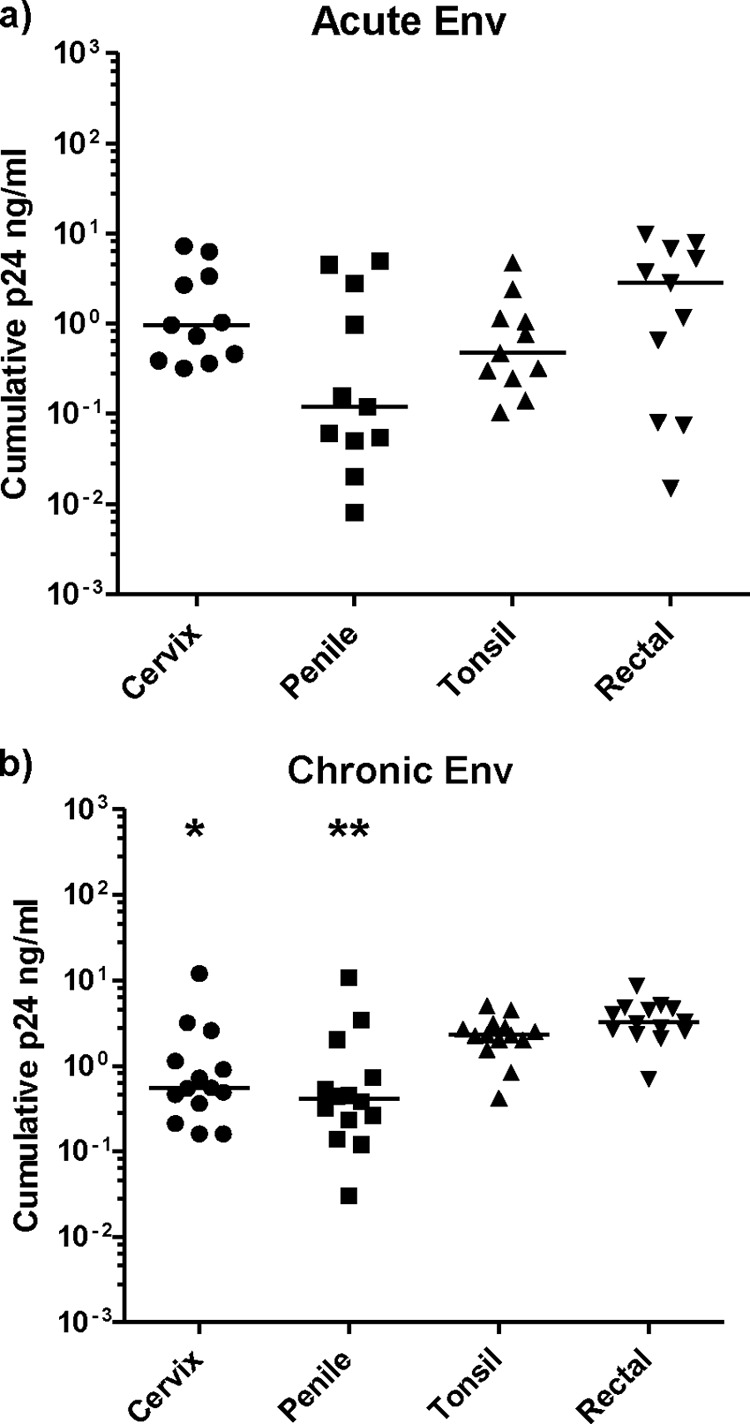
Replication of Env-chimeric virus in mucosal tissue explants. (a and b) Cervical, penile, tonsillar, and rectal tissue were dissected into 1- to 3-mm3 explants and after exposure to either acute (a) or chronic (b) Env-chimeric virus for 2 h, were cultured for a total of 21 days, and supernatant was collected every 3 or 4 days. HIV-1 p24 levels were then measured to assess relative viral replication. There was a significant difference between chronic p24 levels in rectal versus cervical or penile tissue infected with chronic Env virus. Values that were significantly different by the Mann-Whitney t test and using 95% CI are indicated as follows: *, P < 0.003; **, P = 0.0003. Horizontal bars represent the median p24 level for each tissue type.
Fig 5.
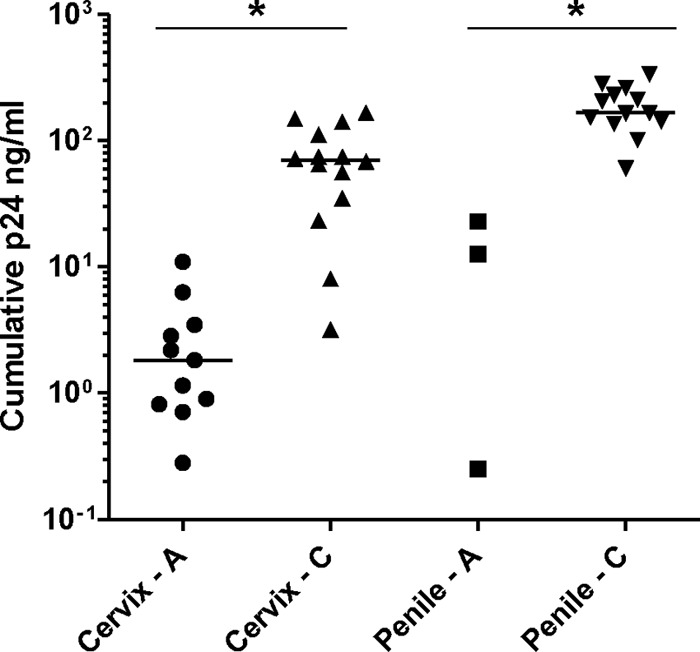
Relative transmission of Env-chimeric virus from migratory cells emigrating from genital explants to CD4+ T cells. Genital tissue explants were exposed to either acute or chronic Env-chimeric virus for 2 h and then washed to remove unbound virus. Migratory cells emigrating from tissue explants were harvested after overnight culture and cocultured with PM-1 CD4 T cells for 21 days. p24 levels were measured in collected culture supernatant by ELISA. Horizontal bars represent the median p24 level for each tissue type. Cervix - A and Penile - A are explants infected with acute Env-chimeric virus, and Cervix - C and Penile - C are explants infected with chronic Env-chimeric virus. Values that were significantly different (P < 0.0001) by the Mann-Whitney t test and using 95% CI are indicated by an asterisk and thin black line.
DISCUSSION
Sexual transmission of HIV-1 across mucosal surfaces is the most common route of transmission worldwide (1). During mucosal transmission, virus crosses the epithelial barrier and encounters potential target cells: CD4 T cells, DC subsets, and macrophages, all of which can be productively infected with HIV-1. The founder population is thought to undergo local amplification and then disseminates to the draining lymph nodes, establishing a systemic infection (15, 25, 26). Although CD4 T cells are the main source of HIV replication and dissemination, it has been suggested that DCs mediate the spread of HIV to CD4 T cells (25, 27, 28). Understanding the interactions between the transmitted virus and the initial target cells during this short window after virus challenge could be critical in identifying requirements of vaccine-mediated protection.
We have characterized a panel of subtype B acute and chronic HIV-1 envelope chimeric viruses in susceptible target cells and primary human tissue explant models. Our data demonstrate that both acute and chronic Env-chimeric virus have significantly higher titers in PBMC and PM-1 CD4+ T cells than in either MDM or MDDC. In agreement with our data, lower levels of replication in macrophages compared to CD4 T cells have already been documented with both Env-expressing and full-length subtype C transmitted virus (18, 20) and full-length subtype B clones (11, 21). However, we found no difference in tropism between chronic and acute Envs when primary cells (PBMC, MDM, and MDDC) were directly infected, which has also been shown in subtype C-infected transmission pairs (20).
Historically, transmitted virus has been characterized as “macrophage-tropic”; however, this may more accurately reflect CCR5 coreceptor use, rather than preferential replication in macrophages (29). In agreement with data on transmission of R5-dependent HIV variants (12), transmitted Envs have been shown to be predominantly R5-tropic (8, 13, 18), although a small proportion of R5/X4 isolates have been observed. However, none of the acute virus tested here demonstrated the preferential macrophage tropism seen with prototypic macrophage-tropic strains HIV-1BaL and HIVYU2.
The preferential tropism of acute Env-chimeric virus for CD4 T cells rather than macrophages or dendritic cells is in agreement with data from ex vivo tissue models and nonhuman primate models, indicating that CD4 T cells are the primary target of acute HIV infection. Intravaginal simian immunodeficiency virus (SIV) challenge in rhesus macaques have shown that almost all infected cells during primary infection are CD4 T cells (30, 31), while data from ex vivo female genital tissue models also found that the earliest detected infected cells are CD4+ T cells (32, 33) and that these cells have the effector memory phenotype (32, 34).
Interestingly, infection of the primary human tissue models (cervical, penile, tonsillar, and rectal tissues) did not reveal any significant differences between the tissue types when infected with acute Env-chimeric virus, although there was a trend for higher levels of replication in rectal tissue compared to other tissue types. Infection with chronic Env-chimeric virus resulted in significantly higher replication in rectal tissue compared to either cervical or penile tissue. These data are in keeping with the documented higher rates of infection for male-to-male transmission (1) and the higher frequencies of susceptible CD4+ CCR5+ T cells in rectal tissue (35–37). The route of transmission is not known for all of the Env-chimeric viruses (Table 1); however, for those where it was recorded, there was no apparent difference in cellular or tissue tropism between virus acquired by heterosexual (viruses B7, B14, and B17) or MSM (viruses B4 and B19) transmissions. Interestingly, chronic Env-chimeric virus infected PM-1 CD4 T cells more efficiently than acute virus (P < 0.006) and displayed higher replication in tonsillar tissue (P < 0.007).
When MDDC were pulsed with serial dilutions of virus, washed, and directly cocultured with PM-1 CD4 T cells, a significant difference in titer was observed. Acute Env-chimeric virus achieved significantly lower titers compared to chronic Env-chimeric viral pools, suggesting lower levels of MDDC-to-T-cell transmission. As infectivity (TCID50) was determined on the basis of detectable p24 production in MDDC/PM-1 cocultures, this would be minimally influenced by relative differences in replicative fitness for PM-1 cells per se. Furthermore, there was no correlation between infectivity for PM-1 cells and MDDC/PM-1 cocultures. Given that MDDC were directly cocultured with PM-1 CD4 T cells following viral exposure, this model would be expected to detect both trans- and cis-mediated infection of CD4 T cells. However, similar fitness for direct infection of MDDC (Fig. 3a and b) indicate that differences in cis infection of MDDCs are unlikely to account for the observed differences in MDDC-to-T-cell transfer. The relative inefficient transmission of acute Env-chimeric virus to PM-1 CD4 T cells was also observed in the migratory cells from cervical and penile tissue. We cannot exclude the possibility that more-fit clones from chronically derived virus could be preferentially transferred from migratory cells in this model; nevertheless, no correlation was seen between dissemination of virus in this model (acute and chronic) and replicative infectivity of PM-1 cells alone. Previous data from our laboratory have characterized migratory cells from cervical tissues to be either CD3+ CD4+ or CD3− HLA-DR+ (of which the majority also expressed DC-SIGN). Both the CD3+ population and the HLA-DR+ population were shown to be capable of transmitting infection to CD4+ T cells; however, the level of replication observed was higher in HLA-DR+ cell cocultures and accounted for the majority of the dissemination (24). We have assessed the population of migratory cells that emigrate from penile tissue by flow cytometry and these display a similar population of CD3+ CD4+ T cells and CD3− CD4+ DC-SIGN+ CD11c+ dendritic cells (data not shown).
One important distinction between the HIV-1 derived from acute Envs compared to chronic Envs utilized in this study is the diversity in the two viral populations. We first contemplated performing these studies with selected clones from the diverse HIV-1 population found in chronic infections and to compare them with single acute clones. In the case of the acute HIV-1 Envs, a single HIV-1 clone was generally identified (8) in these patient samples, and as consequence, only a single HIV-1 Env-chimeric virus was constructed (Fig. 1). In the case of the chronic derived virus, the quasispecies are quite diverse (in relation to a consensus virus for each patient), and as such, the reconstructed virus harbored a minimum of 100 distinct Env clones. Our previous studies have compared the replicative fitness of HIV-1 Env chimeric virus clones within an intrapatient population (19, 22, 38). Approximately 30 to 50% of all Env clones from a chronic patient sample yields dead virus, whereas the remaining 50 to 70% Env genes display significant variations in entry efficiency (22, 38). Thus, the random selection of individual HIV-1 clones from a chronic virus population may introduce a large experimental bias. Although controversial, a virus quasispecies or swarm is thought to represent one virus entity recapitulating the actual fitness/phenotype within that patient sample at that time (39). Previous studies have shown that a diverse virus population is substantially more fit for replication in tissue culture models than single virus clones (19, 40–43). Nonetheless, all tissues and primary cells used in this study likely introduce a genetic bottleneck on the chronic Env-chimeric viruses such that some clones will replicate better than others. We could attempt to identify and characterize these chronic clones, but it is important to note that this study focused on identifying unique phenotypic features of the acute Env-chimeric viruses. There is an assumption that the infecting HIV-1 clones during acute infection may reflect a selected phenotype favoring mucosal infection or virus transfer within the targeted tissue (e.g., genital and rectal mucosa). We found no evidence for selection of acute Env sequences promoting enhanced DC-to-T-cell transfer compared to chronic Env-chimeric virus. The inefficient tropism of both acute and chronic Env-chimeric virus for DC infection does not support a dominant role for this pathway in mucosal transmission.
An important caveat in describing a lack of preferential tropism by these acute Env-chimeric virus is the focus entirely on the HIV-1 env gene as opposed to the entire primary HIV-1 isolates. It is quite possible that other genomic regions of the acute virus may provide the key for a distinct tropism. For example, sequences outside env may dictate the level of envelope incorporation into budding virus (N. Parrish, J. Easlick, M. O'Brien, J. Decker, J. Salazar-Gonzalez, H. Li, T. Dokland, N. N. Bhardwaj, G. Shaw, and B. H. Hahn, presented at the 18th Conference on Retroviruses and Opportunistic Infections, 2011). In this study, we can only indicate that the envelope glycoproteins from acute T/F virus inserted into a common HIV backbone do not confer an enhanced tropism. In previous studies, we have shown that replicative fitness of primary HIV-1 isolates in PBMC, T cells, or macrophages is controlled by the efficiency of host cell entry and maps to the env gene (44, 45). Nonetheless, other virus factors in addition to or aside from the HIV-1 envelope glycoproteins may play an important role for enhanced tropism of acute-derived HIV-1 isolates for DC or macrophage cells embedded in mucosal tissues. Finally, it should be noted that the acute-derived HIV-1 compared to chronic-derived HIV-1 contained a slightly shortened env cassette (171 nucleotides [nt] shorter). For the acute virus constructs, the env cassette started just downstream of the first rev exon and ended in the gp41 transmembrane domain. The chronic virus constructs, in addition to the acute env cassette, included the first exon of tat. The acute env cassette was limited in size due to primer used in the previous SGAs.
Taken together, our data and that of others argue against a major role for macrophages and dendritic cells as primary target cells for initiating mucosal infection and strongly support the growing evidence that subtype B acute Env sequences preferentially target mucosal CD4+ T cells. More-efficient replication of chronic Env virus in PM-1 CD4 T cells and tonsillar tissue may reflect selection of more-fit clones from the chronic quasispecies that are not representative of the acute Env clones. The relative inefficient direct infection of MDDC by acute Env-chimeric virus supports previous studies suggesting infectious passage through subepithelial DC and macrophages is unlikely to be critical for establishment of infection (32, 33, 46–48). The observation that acute Env chimeric virus was less efficiently transmitted than chronic Envs by cellular emigrants from cervical and penile tissue suggests this property is not actively selected within the inoculating viral population. It is unclear whether this represents increased uptake of chronic virus by tissue emigrants or differential retention of acute Env virus within the tissue itself. Further study will be required to determine whether the observed less-efficient transfer of acute virus from MDDC to CD4 T cells confers a direct selective advantage for transmission or whether this represents a surrogate marker for an envelope phenotype that provides a selective advantage through an alternative mechanisms.
This study has specifically focused on the contribution of T/F Env, as expressed in chimeric virus with a common HIV-1 backbone, on infection of mucosal tissue and associated cells. Additional studies using full-length T/F infectious molecular clones are needed to determine whether sequences outside Env (independently or conjointly) have additional influence on these early events.
ACKNOWLEDGMENTS
This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), Division of AIDS (DAIDS), U.S. Department of Health and Human Services (HHS) and Center for HIV/AIDS Vaccine Immunology (CHAVI) U19 AI067854-05. We gratefully acknowledge an equipment grant from Dormeur Investment Service Ltd. that provided funding to purchase the plate reader and washer used in these studies.
We also acknowledge the laboratories of B. H. Hahn and B. F. Keele for their work in identifying and characterizing the transmitted/founder (T/F) virus. We thank Abbey Evans and Naomi Armanasco for technical assistance in the culture of penile glans tissue. We are most grateful to James Bellringer, Consultant Urologist and Gender Surgeon of Charing Cross Hospital, London, United Kingdom, for provision of penile tissue samples.
Footnotes
Published ahead of print 7 November 2012
REFERENCES
- 1. Shattock RJ, Moore JP. 2003. Inhibiting sexual transmission of HIV-1 infection. Nat. Rev. Microbiol. 1:25–34 [DOI] [PubMed] [Google Scholar]
- 2. Zhang LQ, MacKenzie P, Cleland A, Holmes EC, Brown AJ, Simmonds P. 1993. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J. Virol. 67:3345–3356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhu T, Mo H, Wang N, Nam DS, Cao Y, Koup RA, Ho DD. 1993. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 261:1179–1181 [DOI] [PubMed] [Google Scholar]
- 4. Wolinsky SM, Wike CM, Korber BT, Hutto C, Parks WP, Rosenblum LL, Kunstman KJ, Furtado MR, Muñoz JL. 1992. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science 255:1134–1137 [DOI] [PubMed] [Google Scholar]
- 5. Poss M, Martin HL, Kreiss JK, Granville L, Chohan B, Nyange P, Mandaliya K, Overbaugh J. 1995. Diversity in virus populations from genital secretions and peripheral blood from women recently infected with human immunodeficiency virus type 1. J. Virol. 69:8118–8122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ritola K, Pilcher CD, Fiscus SA, Hoffman NG, Nelson JAE, Kitrinos KM, Hicks CB, Eron JJ, Jr, Swanstrom R. 2004. Multiple V1/V2 env variants are frequently present during primary infection with human immunodeficiency virus type 1. J. Virol. 78:11208–11218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McNearney T, Hornickova Z, Markham R, Birdwell A, Arens M, Saah A, Ratner L. 1992. Relationship of human immunodeficiency virus type 1 sequence heterogeneity to stage of disease. Proc. Natl. Acad. Sci. U. S. A. 89:10247–10251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping L-H, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105:7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fiebig EW, Wright DJ, Rawal BD, Garrett PE, Schumacher RT, Peddada L, Heldebrant C, Smith R, Conrad A, Kleinman SH, Busch MP. 2003. Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS 17:1871–1879 [DOI] [PubMed] [Google Scholar]
- 10. Haaland RE, Hawkins PA, Salazar-Gonzalez J, Johnson A, Tichacek A, Karita E, Manigart O, Mulenga J, Keele BF, Shaw GM, Hahn BH, Allen SA, Derdeyn CA, Hunter E. 2009. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 5:e1000274 doi:10.1371/journal.ppat.1000274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li H, Bar KJ, Wang S, Decker JM, Chen Y, Sun C, Salazar-Gonzalez JF, Salazar MG, Learn GH, Morgan CJ, Schumacher JE, Hraber P, Giorgi EE, Bhattacharya T, Korber BT, Perelson AS, Eron JJ, Cohen MS, Hicks CB, Haynes BF, Markowitz M, Keele BF, Hahn BH, Shaw GM. 2010. High multiplicity infection by HIV-1 in men who have sex with men. PLoS Pathog. 6:e1000890 doi:10.1371/journal.ppat.1000890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Margolis L, Shattock R. 2006. Selective transmission of CCR5-utilizing HIV-1: the ‘gatekeeper’ problem resolved? Nat. Rev. Microbiol. 4:312–317 [DOI] [PubMed] [Google Scholar]
- 13. Hu Q, Huang X, Shattock RJ. 2010. C-C chemokine receptor type 5 (CCR5) utilization of transmitted and early founder HIV-1 envelopes and sensitivity to small-molecule CCR5 inhibitors. J. Gen. Virol. 91(Pt 12):2965–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haase AT. 2010. Targeting early infection to prevent HIV-1 mucosal transmission. Nature 464:217–223 [DOI] [PubMed] [Google Scholar]
- 16. Wu L. 2008. Biology of HIV mucosal transmission. Curr. Opin. HIV AIDS 3:534–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dudley DM, Gao Y, Nelson KN, Henry KR, Nankya I, Gibson RM, Arts EJ. 2009. A novel yeast-based recombination method to clone and propagate diverse HIV-1 isolates. Biotechniques 46:458–467 [DOI] [PubMed] [Google Scholar]
- 18. Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, Li H, Decker JM, Wang S, Baalwa J, Kraus MH, Parrish NF, Shaw KS, Guffey MB, Bar KJ, Davis KL, Ochsenbauer-Jambor C, Kappes JC, Saag MS, Cohen MS, Mulenga J, Derdeyn CA, Allen S, Hunter E, Markowitz M, Hraber P, Perelson AS, Bhattacharya T, Haynes BF, Korber BT, Hahn BH, Shaw GM. 2009. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 206:1273–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Troyer RM, Collins KR, Abraha A, Fraundorf E, Moore DM, Krizan RW, Toossi Z, Colebunders RL, Jensen MA, Mullins JI, Vanham G, Arts EJ. 2005. Changes in human immunodeficiency virus type 1 fitness and genetic diversity during disease progression. J. Virol. 79:9006–9018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Isaacman-Beck J, Hermann EA, Yi Y, Ratcliffe SJ, Mulenga J, Allen S, Hunter E, Derdeyn CA, Collman RG. 2009. Heterosexual transmission of human immunodeficiency virus type 1 subtype C: macrophage tropism, alternative coreceptor use, and the molecular anatomy of CCR5 utilization. J. Virol. 83:8208–8220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ochsenbauer C, Edmonds TG, Ding H, Keele BF, Decker J, Salazar MG, Salazar-Gonzalez JF, Shattock R, Haynes BF, Shaw GM, Hahn BH, Kappes JC. 2012. Generation of transmitted/founder HIV-1 infectious molecular clones and characterization of their replication capacity in CD4 T lymphocytes and monocyte-derived macrophages. J. Virol. 86:2715–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Troyer RM, McNevin J, Liu Y, Zhang SC, Krizan RW, Abraha A, Tebit DM, Zhao H, Avila S, Lobritz MA, McElrath MJ, Le Gall S, Mullins JI, Arts EJ. 2009. Variable fitness impact of HIV-1 escape mutations to cytotoxic T lymphocyte (CTL) response. PLoS Pathog. 5:e1000365 doi:10.1371/journal.ppat.1000365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Geijtenbeek TBH, Kwon DS, Torensma R, van Vliet SJ, van Duijnhoven GCF, Middel J, Cornelissen ILMHA, Nottet HSLM, KewalRamani VN, Littman DR, Figdor CG, van Kooyk Y. 2000. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell 100:587–597 [DOI] [PubMed] [Google Scholar]
- 24. Hu Q, Frank I, Williams V, Santos JJ, Watts P, Griffin GE, Moore JP, Pope M, Shattock RJ. 2004. Blockade of attachment and fusion receptors inhibits HIV-1 infection of human cervical tissue. J. Exp. Med. 199:1065–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Miller CJ, Shattock RJ. 2003. Target cells in vaginal HIV transmission. Microbes Infect. 5:59–67 [DOI] [PubMed] [Google Scholar]
- 26. Pope M, Haase AT. 2003. Transmission, acute HIV-1 infection and the quest for strategies to prevent infection. Nat. Med. 9:847–852 [DOI] [PubMed] [Google Scholar]
- 27. Rinaldo CR, Piazza P. 2004. Virus infection of dendritic cells: portal for host invasion and host defense. Trends Microbiol. 12:337–345 [DOI] [PubMed] [Google Scholar]
- 28. Wu L, KewalRamani VN. 2006. Dendritic-cell interactions with HIV: infection and viral dissemination. Nat. Rev. Immunol. 6:859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Grivel JC, Shattock RJ, Margolis LB. 2011. Selective transmission of R5 HIV-1 variants: where is the gatekeeper? J. Transl. Med. 9(Suppl 1):S6 doi:10.1186/1479-5876-9-S1-S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li Q, Estes JD, Schlievert PM, Duan L, Brosnahan AJ, Southern PJ, Reilly CS, Peterson ML, Schultz-Darken N, Brunner KG, Nephew KR, Pambuccian S, Lifson JD, Carlis JV, Haase AT. 2009. Glycerol monolaurate prevents mucosal SIV transmission. Nature 458:1034–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang ZQ, Schuler T, Zupancic M, Wietgrefe S, Staskus KA, Reimann KA, Reinhart TA, Rogan M, Cavert W, Miller CJ, Veazey RS, Notermans D, Little S, Danner SA, Richman DD, Havlir D, Wong J, Jordan HL, Schacker TW, Racz P, Tenner-Racz K, Letvin NL, Wolinsky S, Haase AT. 1999. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science 286:1353–1357 [DOI] [PubMed] [Google Scholar]
- 32. Gupta P, Collins KB, Ratner D, Watkins S, Naus GJ, Landers DV, Patterson BK. 2002. Memory CD4+ T cells are the earliest detectable human immunodeficiency virus type 1 (HIV-1)-infected cells in the female genital mucosal tissue during HIV-1 transmission in an organ culture system. J. Virol. 76:9868–9876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hladik F, Sakchalathorn P, Ballweber L, Lentz G, Fialkow M, Eschenbach D, McElrath MJ. 2007. Initial events in establishing vaginal entry and infection by human immunodeficiency virus type-1. Immunity 26:257–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saba E, Grivel JC, Vanpouille C, Brichacek B, Fitzgerald W, Margolis L, Lisco A. 2010. HIV-1 sexual transmission: early events of HIV-1 infection of human cervico-vaginal tissue in an optimized ex vivo model. Mucosal Immunol. 3:280–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Anton PA, Elliott J, Poles MA, McGowan IM, Matud J, Hultin LE, Grovit-Ferbas K, Mackay CR, Chen ISY, Giorgi JV. 2000. Enhanced levels of functional HIV-1 receptors on human mucosal T cells demonstrated using intestinal biopsy tissue. AIDS 14:1761–1765 [DOI] [PubMed] [Google Scholar]
- 36. Lapenta C, Boirivant M, Marini M, Santini SM, Logozzi M, Viora M, Belardelli F, Fais S. 1999. Human intestinal lamina propria lymphocytes are naturally permissive to HIV-1 infection. Eur. J. Immunol. 29:1202–1208 [DOI] [PubMed] [Google Scholar]
- 37. Poles MA, Elliott J, Taing P, Anton PA, Chen ISY. 2001. A preponderance of CCR5+ CXCR4+ mononuclear cells enhances gastrointestinal mucosal susceptibility to human immunodeficiency virus type 1 infection. J. Virol. 75:8390–8399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lassen KG, Lobritz MA, Bailey JR, Johnston S, Nguyen S, Lee B, Chou T, Siliciano RF, Markowitz M, Arts EJ. 2009. Elite suppressor-derived HIV-1 envelope glycoproteins exhibit reduced entry efficiency and kinetics. PLoS Pathog. 5:e1000377 doi:10.1371/journal.ppat.1000377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Domingo E, Sheldon J, Perales C. 2012. Viral quasispecies evolution. Microbiol. Mol. Biol. Rev. 76:159–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lázaro E, Escarmís C, Pérez-Mercader J, Manrubia SC, Domingo E. 2003. Resistance of virus to extinction on bottleneck passages: study of a decaying and fluctuating pattern of fitness loss. Proc. Natl. Acad. Sci. U. S. A. 100:10830–10835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Novella IS, Quer J, Domingo E, Holland JJ. 1999. Exponential fitness gains of RNA virus populations are limited by bottleneck effects. J. Virol. 73:1668–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sierra S, Davila M, Lowenstein PR, Domingo E. 2000. Response of foot-and-mouth disease virus to increased mutagenesis: influence of viral load and fitness in loss of infectivity. J. Virol. 74:8316–8323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yuste E, Sanchez-Palomino S, Casado C, Domingo E, Lopez-Galindez C. 1999. Drastic fitness loss in human immunodeficiency virus type 1 upon serial bottleneck events. J. Virol. 73:2745–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ball SC, Abraha A, Collins KR, Marozsan AJ, Baird H, Quinones-Mateu ME, Penn-Nicholson A, Murray M, Richard N, Lobritz M, Zimmerman PA, Kawamura T, Blauvelt A, Arts EJ. 2003. Comparing the ex vivo fitness of CCR5-tropic human immunodeficiency virus type 1 isolates of subtypes B and C. J. Virol. 77:1021–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marozsan AJ, Moore DM, Lobritz MA, Fraundorf E, Abraha A, Reeves JD, Arts EJ. 2005. Differences in the fitness of two diverse wild-type human immunodeficiency virus type 1 isolates are related to the efficiency of cell binding and entry. J. Virol. 79:7121–7134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. de Witte L, Nabatov A, Geijtenbeek TBH. 2008. Distinct roles for DC-SIGN+-dendritic cells and Langerhans cells in HIV-1 transmission. Trends Mol. Med. 14:12–19 [DOI] [PubMed] [Google Scholar]
- 47. de Witte L, Nabatov A, Pion M, Fluitsma D, de Jong MAWP, de Gruijl T, Piguet V, van Kooyk Y, Geijtenbeek TBH. 2007. Langerin is a natural barrier to HIV-1 transmission by Langerhans cells. Nat. Med. 13:367–371 [DOI] [PubMed] [Google Scholar]
- 48. Tsegaye TS, Pöhlmann S. 2010. The multiple facets of HIV attachment to dendritic cell lectins. Cell. Microbiol. 12:1553–1561 [DOI] [PMC free article] [PubMed] [Google Scholar]