Abstract
The exon junction complex (EJC) is highly conserved in many organisms and is involved in various steps of mRNA metabolism. During the course of investigating the role of EJC in the germ line sex determination of the nematode Caenorhabditis elegans, we found that depletion of one of the three core subunits (Y14, MAG-1, and eukaryotic translation initiation factor 4III [eIF4AIII]) or one auxiliary subunit (UAP56) of EJC resulted in the cytoplasmic leakage of unspliced RNAs from almost all of the C. elegans protein-coding genes examined thus far. This leakage was also observed with the depletion of several splicing factors, including SF3b, IBP160, and PRP19, all of which genetically interacted with Y14. We also found that Y14 physically interacts with both pre-mRNA and spliceosomal U snRNAs, especially U2 snRNA, and that the interaction was abolished when both IBP160 and PRP19 were depleted. Our results strongly suggest that a specific set of EJC subunits is recruited onto introns and interacts with components of the spliceosome, including U2 snRNP, to provide a critical signal for the surveillance and nuclear retention of unspliced RNAs in C. elegans.
INTRODUCTION
The complete removal of introns from pre-mRNA before mRNA export to the cytoplasm is critical to ensure the proper expression of the protein product in higher eukaryotes. Premature export of intron-containing RNAs gives rise to abnormal protein products that are very harmful to various cell functions, although most of such abnormal RNAs are eliminated by mRNA quality control systems, including nonsense-mediated mRNA decay (NMD) (1–4). Thus, cells must have mechanisms to retain partially spliced pre-mRNA in the nucleus. Indeed, most intron-containing RNAs, except in the case of alternative splicing, are normally localized in the nucleus. One mechanism is spliceosome formation around introns, which is confined to the nucleus because most splicing factors are localized in the nucleus. In this mechanism, the spliceosome itself or one or more spliceosome-associated factors anchor intron-containing RNAs in the nucleus (5–7). Another mechanism for the nuclear retention of intron-containing RNAs is the splicing-dependent recruitment of mRNA export-related factors. The transcription and export (TREX) complex and exon junction complex (EJC) form on the 5′ region of the nascent transcript (8, 9) and 20 to 24 nucleotides upstream from splice junctions (10, 11), respectively. The TREX complex contains the proteins UAP56 and Aly/REF (12, 13), both of which are also subunits of EJC (14–16), suggesting the tight coupling of splicing with mRNA export and the involvement of TREX and EJC for preventing the premature export of unspliced RNAs. Both spliceosome-dependent and export-related-factor-coupled mechanisms for the nuclear retention of intron-containing RNAs may operate concurrently, but the molecular details remain to be elucidated.
EJC consists of four core subunits (Y14, Mago-nashi, eukaryotic translation initiation factor 4III [eIF4AIII], and Barentz/MLN51) and several auxiliary subunits (such as UAP56 and Aly/REF), and these components are highly conserved in higher eukaryotes (11, 17–20). Previous studies showed that EJC and its core subunits have multiple biological functions. In mammals, EJC is critical in defining the premature termination codon (PTC) in the NMD system (21), and the EJC core subunits are involved in translational regulation together with the partner of Y14 and Mago-nashi, PYM (22, 23). In Drosophila melanogaster, the EJC core subunits Y14/Tsunagi and Mago-nashi are required for properly localizing oskar mRNA (24–27), which is important for posterior formation, and for the efficient splicing of long introns in several genes, including the mitogen-activated protein (MAP) kinase gene mapk (28, 29). We previously reported that Y14 and MAG-1 (Mago-nashi homologue) are required for proper germ line sex determination in Caenorhabditis elegans hermaphrodites, based on the observation that depleting either Y14 or MAG-1 by RNA interference (RNAi) [referred to below as mag-1(RNAi) and Y14(RNAi)] prevents the switch from spermatogenesis to oogenesis, resulting in germ line masculinization in hermaphrodites (30, 31).
The switch in germ line sex determination from spermatogenesis to oogenesis in C. elegans hermaphrodites is regulated by a group of genes (see Fig. S1A in the supplemental material) in which functional tra-2 expression is critical for initiating oogenesis (32). It was shown previously that tra-2 expression is regulated via translational control by FOG-2 and GLD-1 subsequent to alternative nuclear export pathways of tra-2 mRNA (33–36). In this study, we analyzed how Y14 is involved in germ line sex determination in C. elegans. We first performed epistasis analysis and found that Y14 was required for proper tra-2 expression. Further analyses showed that depleting Y14 caused nonconventional cytoplasmic RNA splicing of prematurely exported tra-2 pre-mRNA. We also showed that the resultant abnormal TRA-2 protein translated from the aberrantly spliced tra-2 RNA inhibited the function of the normal TRA-2 protein. Similar premature export of intron-containing unspliced RNAs was observed for almost all C. elegans genes examined so far upon depletion of Y14, MAG-1, and eIF4AIII, three EJC core subunits, or UAP56, an auxiliary subunit, but not other EJC subunits. Interestingly, the premature export of intron-containing RNAs was dependent upon two export receptors, NXF-2 and XPO-1/CRM1 (37). The germ line masculinization phenotype resulting from the depletion of Y14 was enhanced by depleting several splicing factors, such as the U2 snRNP subunit SF3b, intron-binding protein IBP160, and ubiquitin ligase PRP19 (38). Moreover, we found that Y14 interacts with pre-mRNA and spliceosomal snRNAs, especially U2 snRNA, and that this interaction is dependent upon the presence of MAG-1, eIF4AIII, IBP160, and PRP19. Our results strongly suggest that a specific set of EJC subunits is recruited as pre-EJC onto introns that are committed to be spliced and interacts with some components of the spliceosome, including U2 snRNP, to provide a critical signal for the surveillance and nuclear retention of intron-containing unspliced RNAs in C. elegans.
MATERIALS AND METHODS
Strains and culture conditions.
C. elegans strains were cultured at 20°C using standard methods (39). Bristol N2 was used as the wild type. The mutant strains used in this study were the following: TR1331 smg-1(r861) I, TR1335 smg-5(r860) I, and NL2098 rrf-1(pk1417) I.
RNA interference.
Sense and antisense RNA corresponding to the coding or intron region of each gene were generated using T3 (Ambion) and T7 (Promega) RNA polymerases. As a control, we synthesized green fluorescent protein (GFP) gene double-stranded RNA (dsRNA) from pPD79.44 (Fire Lab C. elegans vector kit, 1995 plate). For postembryonic RNAi, first-larval-stage (L1) larvae were soaked in the dsRNA solution (1.6 μg/μl) for 24 h and then transferred to nematode growth medium (NGM) plates. The germ line phenotype was observed 72 h after recovery. The concentrations of dsRNA used differed for the following genes: xpo-1, 0.4 μg/μl (see Fig. 3 and 4); UAP56, 0.4 μg/μl (see Fig. 5B); SAP130, 0.2 μg/μl (see Fig. 6A), 0.4 μg/μl (see Fig. 6C), and 20 ng/μl (see Fig. 6D); Y14, 0.4 μg/μl; SAP130 and CDC5L, 0.08 μg/μl (see Fig. 6E); nxf-1, 20 ng/μl (see Fig. S4B in the supplemental material); U1-70k, 0.8 μg/μl (see Fig. S6C in the supplemental material); and PRP8, 0.8 μg/μl (see Fig. S6C). For immunoprecipitation experiments, we adopted the feeding RNAi method (40). Synchronized L1 larvae were fed with bacteria expressing dsRNA for each gene in liquid medium. Worms were harvested when they reached the young adult stage. As a control, worms were fed with bacteria containing the empty vector L4440.
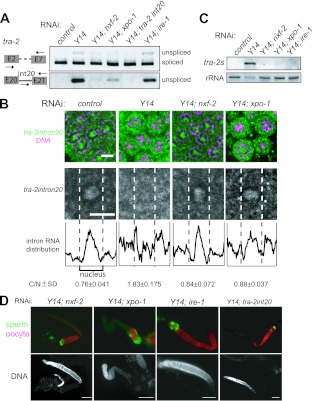
Fig 3.
NXF-2 and XPO-1 are responsible for the premature export of unspliced tra-2 RNA. (A) RT-PCR assays were performed to monitor tra-2 RNA expression in the adult hermaphrodites subjected to RNAi as indicated. The primers used for amplification are schematically shown on the left. The amplified products corresponding to the unspliced RNAs were excised and subjected to sequence analysis. Unspliced tra-2 RNA accumulated in Y14(RNAi) animals, which was suppressed by simultaneous RNAi of nxf-2, xpo-1, or tra-2 intron 20 but not ire-1. (B) In situ hybridization of mitotic cells within gonads dissected from adult hermaphrodites subjected to RNAi as indicated. Cells were probed with tra-2 intron 20 (green) followed by DNA staining (magenta), shown as merged views (top). Separate views of single cells probed with tra-2 intron 20 are also shown (middle). The fluorescence intensities in the middle panels were plotted to show the intracellular distribution of the intron (bottom). Dotted lines indicate the borders between the nucleus and cytoplasm. The ratios of the cytoplasmic to nuclear fluorescence intensities for tra-2 intron 20 for five cells were averaged to quantify the intracellular distribution of the intron, shown below (C/N [cytoplasm/nucleus]). Scale bars, 4 μm. (C) Northern blot analysis of tra-2s RNA expression in adult hermaphrodites subjected to RNAi as indicated. 40S rRNA was used as a loading control. (D) Gonad arms were dissected from adult hermaphrodites subjected to RNAi as indicated and stained with the anti-MSP antibody (green) and the anti-RME-2 antibody (red), shown as merged views (top). DAPI-stained views of the same gonad arms are also shown (bottom). Scale bars, 50 μm.
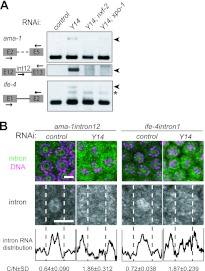
Fig 4.
Y14 is required for the nuclear retention of unspliced RNAs. (A) RT-PCR assays were performed to monitor the expression of ama-1 and ife-4 in the adult hermaphrodites subjected to RNAi as indicated. The primers used for amplification are schematically shown on the left. Arrowheads indicate unspliced RNAs. The asterisk indicates a nonspecifically amplified product. (B) In situ hybridization of mitotic cells within gonad arms dissected from Y14(RNAi) hermaphrodites. Cells were probed with ama-1 intron 12 or ife-4 intron 1 (green), followed by DNA staining (magenta), shown as merged views (top). Separate views of single cells probed with the introns are also shown (middle). The intracellular distribution of the introns is shown as described in the legend to Fig. 3B (bottom). Scale bars, 4 μm.
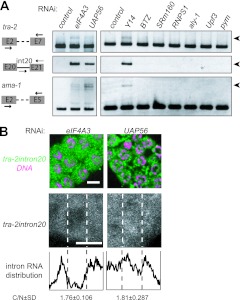
Fig 5.
The EJC subunits required for the nuclear retention of unspliced RNAs. (A) RT-PCR assays were performed to monitor the expression of tra-2 and ama-1 in the animals subjected to RNAi as indicated. The primers used for amplification are schematically shown on the left. Arrowheads indicate unspliced RNAs. (B) In situ hybridization of mitotic cells within gonad arms dissected from adult hermaphrodites subjected to RNAi as indicated. Cells were probed with tra-2 intron 20 (green), followed by DNA staining (magenta), shown as merged views (top). Separate views of single cells probed with tra-2 intron 20 are also shown (middle). The intracellular distribution of the intron is shown as described in the legend to Fig. 3B (bottom). Scale bars, 4 μm.
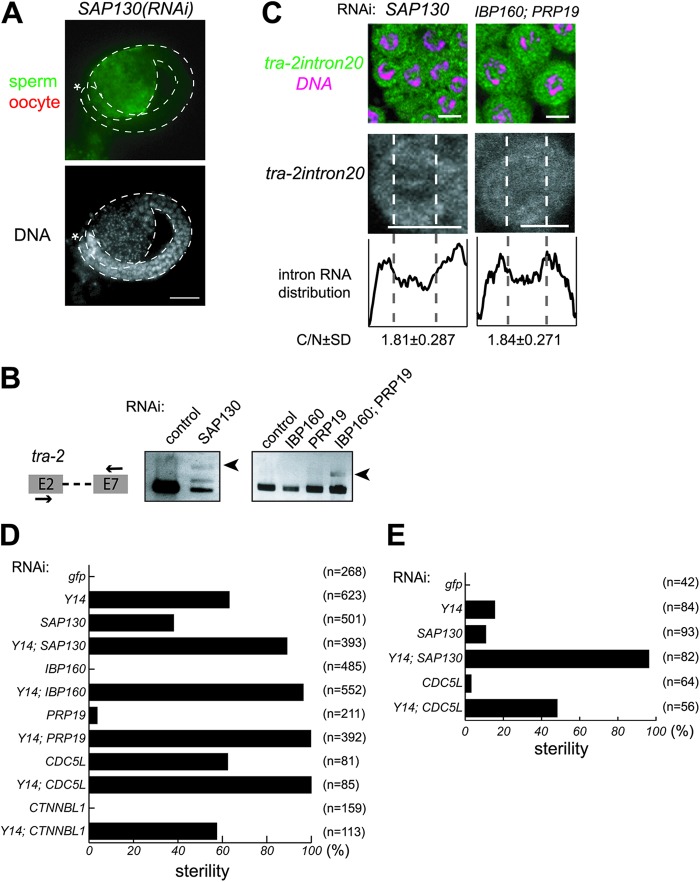
Fig 6.
The splicing factors required for the nuclear retention of unspliced RNAs and their interactions with Y14. (A) Gonad arms were dissected from SAP130(RNAi) hermaphrodites and stained with the anti-MSP antibody (green) and anti-RME-2 antibody (red) (top) and DAPI (bottom). Asterisks indicate the distal end of the gonad. Gonad is outlined by dashed lines. Scale bar, 20 μm. (B) RT-PCR assays were performed to monitor tra-2 expression in animals subjected to RNAi as indicated. The primers used for amplification are schematically shown on the left. Arrowheads indicate unspliced tra-2 RNA. (C) In situ hybridization of mitotic cells within gonad arms dissected from SAP130(RNAi) and IBP160(RNAi); PRP19(RNAi) hermaphrodites. Cells were probed with tra-2 intron 20 (green), followed by DNA staining (magenta), shown as merged views (top). Separate views of single cells probed with the intron are also shown (middle). The intracellular distribution of the intron is shown as described in the legend to Fig. 3B (bottom). Scale bars, 4 μm. (D) Percent sterility caused by RNAi as indicated. (E) Percent sterility caused by indicated RNAi treatment in the background of the rrf-1(pk1417) mutation.
Transgene construction.
The pie-1 promoter-containing vector pJH4.52 (41) was used to express the tra-2s transgene in the germ line. The sequence spanning BamHI to KpnI that contains GFP::His-11 and the pie-1 3′ untranslated region (3′UTR) was replaced with the sequence for XhoI, tra-2s, SmaI, hemagglutinin (HA), EcoRV, and the tbb-2 3′UTR. The tra-2s expression plasmid (0.5 ng/μl, NaeI cut), C. elegans genomic DNA (40 ng/μl, PvuII cut), and pJERI (0.5 ng/μl, myo-3 promoter::dsRed2, ScaI cut) were coinjected into N2 hermaphrodites. To construct the plasmid to express IBP160::HA, the eft-3 promoter and unc-54 3′UTR from the pDEST-eft-3p vector (42) were inserted into the KpnI-XhoI site and NotI-SacI site, respectively, of pBluescript II SK(+). The sequence spanning the SmaI to NotI sites of the resultant plasmid was replaced with the sequence for IBP160, SpeI, and HA. The IBP160::HA expression plasmid (50 ng/μl) and pJERI (50 ng/μl) were coinjected into N2 hermaphrodites. The strain expressing IBP160::HA was mated with the strain expressing either Y14-ZZ (IgG binding domain)-GFP or ZZ-GFP (30).
RT-PCR analysis.
Total RNA was extracted from adult worms that were subjected to postembryonic RNAi using Sepasol-RNA II super reagent (Nacalai Tesque). cDNA was synthesized using the Superscript III reverse transcriptase system with random hexamers (Invitrogen) from 0.5 μg of DNase-treated total RNA, and 1/20 of the cDNA product was used for each PCR reaction mixture with Ex Taq (TaKaRa). Detection of the lariat intron was performed according to previously described methods (43, 44). The primers used in this study are listed in Table S2 in the supplemental material.
qRT-PCR analysis.
The cDNA was synthesized using the PrimeScript reverse transcriptase reagent kit (TaKaRa) from 0.5 μg of DNase-treated total RNA. Quantitative reverse transcription (qRT)-PCR was performed with SYBR premix EX Taq II and the Thermal Cycler Dice real-time system II (TaKaRa) according to the standard protocol. Specific amplification of the PCR products was confirmed by analyzing the dissociation curve, running the products on an agarose gel, and sequencing. Each sample was measured as duplicates, and each experiment was repeated three times.
Subcellular fractionation.
Fractionation was performed as previously described (45) with some modifications. Fourth-larval-stage (L4) or young-adult-stage worms were washed in M9 buffer and collected by sedimentation. Worms were resuspended in ice-cold isotonic buffer (25 mM HEPES at pH 7.6, 10 mM KCl, 5% glycerol, 0.5 mM dithiothreitol [DTT], 1× Complete Mini EDTA-free protease inhibitor [Roche], 40 U/ml recombinant RNase inhibitor [TaKaRa]) and were cut with a 25-gauge needle. Worms were flash-frozen and lysed using a tissue grinder and pestle. Lysates were centrifuged at 40 × g for 30 s at 4°C. The supernatant was centrifuged at 2,000 × g for 5 min at 4°C. The pellet and supernatant were defined as the nuclear and cytoplasmic fractions, respectively. One-tenth of each fraction was used for Western blotting, and the rest was used for RT-PCR analysis.
Northern blot analysis.
Total RNA (0.5 μg per lane) was separated on a formaldehyde-containing 1% agarose gel, transferred to a nitrocellulose membrane, and hybridized with digoxigenin-labeled antisense RNA probes. The RNA probes were synthesized with digoxigenin-UTP using T3 RNA polymerase (Stratagene). The tra-2 probe (785 bp) corresponds to nucleotides 3643 to 4428 in its cDNA sequence. The Y14 and mag-1 probes correspond to their entire coding regions.
In situ hybridization.
Worms were cut with a 25-gauge needle in phosphate-buffered saline (PBS) on MAS-coated glass slides (Matsunami Glass). Freeze-cracked dissected gonads were fixed as described previously (46) with the exception that a modified fixative (100 mM K2HPO4 at pH 7.2, 0.25% glutaraldehyde, 3% formaldehyde) was used. Antisense RNA probes were synthesized with digoxigenin-UTP using T3 or T7 RNA polymerase. The tra-2 intron 20 probe (473 bp) corresponds to nucleotides 7985 to 8458 in C15F1.3. The ama-1 intron 12 probe (528 bp) corresponds to nucleotides 18318 to 18846 in F36A4. The ife-4 intron 1 probe (273 bp) corresponds to nucleotides 3259 to 3531 in C05D9. Hybridized digoxigenin-labeled probes were detected with antidigoxigenin-Cy3 antibody (Jackson ImmunoResearch Laboratory). Stained worms were mounted with FluorSave (Calbiochem). Fluorescence signals were analyzed by using NIH ImageJ software with the rectangular scan function. The y axis of the rectangle was positioned to intersect a nucleus. Vertically averaged pixel intensities were obtained using the Plot Profile feature and plotted with Adobe Illustrator. To quantify the ratios of the cytoplasmic to nuclear RNA, averages of the pixel intensities were calculated for the cytoplasm and nucleus of each of five cells in at least two independent staining experiments. The border between the nucleus and cytoplasm was determined by the fluorescence intensity of DRAQ5 (eBioscience).
Western blot analysis.
RNAi-treated adult worms were washed with and sonicated in buffer A (50 mM Tris-HCl at pH 7.5, 25 mM KCl, 5 mM MgCl2). Worm extracts were centrifuged at 14,000 rpm for 10 min, and the concentration of the supernatants was determined by Bradford assay. The worm extracts (50 ng) were boiled for 2 to 3 min in SDS sample buffer, separated by electrophoresis on a 10% SDS-polyacrylamide gel, and transferred to an Immobilon P transfer membrane. Anti-TRA-2 ICD (0.2 μg/ml), antifibrillarin (1:500, 38F3; EnCor), anti-HA (1:1,000; Bethyl) and anti-histone H4 (0.02 μg/ml) primary antibodies were diluted in PBS, 1% skim milk and 3% goat serum. Peroxidase-conjugated secondary antibodies were used at a dilution of 1:5,000. Signals were developed using ImmunoStar LD (Wako) and detected using ImageQuant LAS4000 (GE Healthcare).
Immunostaining.
Immunostaining was performed as described previously (47). Briefly, dissected gonads were fixed with 3% formaldehyde in 100 mM K2HPO4 at pH 7.2 for 1 h, followed by cold methanol (−30°C) for 5 min. Fixed gonads were incubated overnight at room temperature with the following primary antibodies: anti-TRA-2 ICD (1 μg/ml), anti-RME-2 (1:50), anti-major sperm protein (anti-MSP) (1:40), and anti-HA tag (1:1,000) antibody, all diluted in PBS with 1 mg/ml bovine serum albumin. Alexa Fluor 488- or 546-conjugated secondary antibodies were used at a dilution of 1:800. Phalloidin CF488A (Biotium) was used at a dilution of 1:20. Stained worms were mounted in FluorSave (Calbiochem). The anti-TRA-2 antibody and the anti-RME-2 antibody were generously provided by Hiroyuki Kawahara (Tokyo Metropolitan University) and Barth Grant (Rutgers University), respectively.
Immunoprecipitation.
Immunoprecipitation was performed as described previously (30), with the following modifications. To detect the interaction between Y14 and RNAs, Complete Mini EDTA-free protease inhibitor (Roche), recombinant RNase inhibitor (TaKaRa), and Turbo DNase (Ambion) were added to IPP150 buffer (10 mM Tris-HCl at pH 8.0, 150 mM NaCl, 0.1% NP-40). To detect the interaction between Y14 and IBP160::HA, worms expressing Y14-ZZ-GFP (or ZZ-GFP) and IBP160::HA were lysed in the pulldown buffer (50 mM Tris-HCl at pH 8.0, 100 mM NaCl, 0.1% NP-40, Complete Mini EDTA-free protease inhibitor). The lysates were supplemented with 50 U/ml micrococcal nuclease and 1 mM CaCl2 and incubated at 4°C for 2 h.
Drug treatment.
RNAi-treated third-larval-stage (L3) animals were incubated with 100 ng/ml leptomycin B (LMB) and 1% ethanol in M9 buffer or with 1% ethanol in M9 buffer alone for 10 h at 20°C and then transferred to NGM plates and recovered overnight. L4 animals were incubated with 2.5 μg/ml spliceostatin A (SSA) and 2.5% methanol in M9 buffer or with 1% methanol in M9 buffer alone for 5 h at 20°C. LMB and SSA were generously provided by Minoru Yoshida (Riken Asi).
Microscopy.
An Olympus BX51 microscope or an Olympus FV1000 confocal microscope was used to acquire images. Adobe Photoshop CS4, Adobe Illustrator 10.0.3, and NIH ImageJ software were used to process the images.
RESULTS
Depletion of Y14 or MAG-1 inhibits the proper expression of the TRA-2 protein.
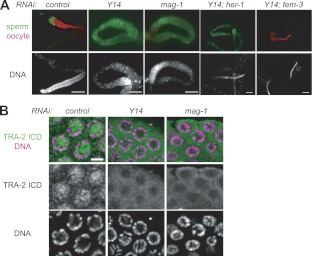
To examine how Y14 affects germ line sex determination in C. elegans, we performed epistasis RNAi analysis using Y14 and the known sex determination genes (see Fig. S1A in the supplemental material). Soaking RNAi treatment at the first larval stage (L1) was adopted in most RNAi experiments in this study to avoid embryonic lethality, because Y14 is essential for embryogenesis (30). Simultaneous RNAi of Y14 and her-1 [referred to here as Y14(RNAi); her-1(RNAi)] resulted in most animals producing only sperm, similar to Y14(RNAi) and mag-1(RNAi), whereas Y14(RNAi); fem-3(RNAi) resulted in most animals producing only oocytes, similar to fem-3(RNAi) (Fig. 1A; also see Fig. S1B and C in the supplemental material). These results indicate that Y14 acts between her-1 and fem-3 in germ line sex determination. Because tra-2 acts between her-1 and fem-3 to promote oogenesis (32), we tested whether Y14 is required for properly expressing the TRA-2 protein. TRA-2 is essentially a membrane-bound protein, and the TRA-2 intracellular domain (ICD) generated by cleavage with TRA-3 protease enters the nucleus and functions as an oogenesis-promoting transcriptional regulator (48–50). Immunofluorescence analysis of the gonads using an anti-TRA-2 ICD antibody clearly detected fluorescence in the nucleus in control GFP(RNAi) animals, reflecting the presence of a functional TRA-2 ICD in the nucleus (Fig. 1B). In contrast, fluorescence was not detected in the nucleus but was retained around the plasma membrane region in Y14(RNAi) and mag-1(RNAi) animals. The TRA-2 localization pattern in Y14(RNAi) and mag-1(RNAi) animals suggested the absence of the TRA-2 ICD in the germ cells of Y14(RNAi) and mag-1(RNAi) animals. Western blot analysis of the total proteins from the whole animals, however, showed that the TRA-2 ICD was present even in Y14(RNAi) and mag-1(RNAi) animals, although the amount was significantly lower than that in the control animals (see Fig. S1D in the supplemental material). These results indicate that the level of the TRA-2 ICD is not sufficient to promote oogenesis in the gonads of Y14(RNAi) and mag-1(RNAi) animals.
Fig 1.
Y14 and MAG-1 are required for proper tra-2 expression. (A) Adult gonad arms of adult hermaphrodites subjected to RNAi as indicated were dissected and stained with the anti-MSP antibody to visualize sperm (green) and the anti-RME-2 antibody to visualize oocytes (red), shown as merged views (top). DAPI (4′,6′-diamidino-2-phenylindole)-stained views of the same gonad arms are shown to visualize DNA (bottom). Scale bars, 50 μm. (B) Germ cells at the pachytene stage in adult gonad arms dissected from adult hermaphrodites that had been subjected to RNAi as indicated were stained with the anti-TRA-2 ICD antibody (green) and DAPI (magenta), shown as merged views (top). Separate views of the same cells are also shown (middle and bottom). Scale bar, 3 μm.
The unusual TRA-2S protein causes germ line masculinization.
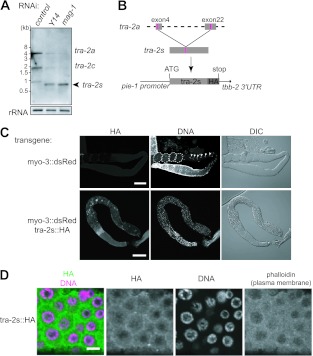
We examined tra-2 mRNA expression in Y14(RNAi) and mag-1(RNAi) animals by Northern blot analysis (Fig. 2A). Mature tra-2 mRNAs (full-length tra-2a and the oocyte-specific variant tra-2c) decreased significantly in Y14(RNAi) and mag-1(RNAi) animals compared to their amounts in the control animals, which is consistent with the reduced amount of the TRA-2 ICD in these animals (see Fig. S1D in the supplemental material). Instead, a very short tra-2 RNA of ∼800 nucleotides accumulated in Y14(RNAi) and mag-1(RNAi) animals. The short tra-2 RNA was detected with the probes containing exons 2 to 7 and exons 22 to 23 but not with the probe containing exons 16 to 19 (data not shown). We therefore cloned this short tra-2 RNA by RT-PCR using relevant primers followed by sequence analysis and found it to be an unusual RNA (referred to as tra-2s) that was spliced between the internal regions of exon 4 and exon 22 (see Fig. S2A in the supplemental material). There were no GU or AG sequences for conventional splicing in the donor and acceptor sites, but instead, a short bipartite sequence, UUAUXXXXXCACC, is directly repeated at the splice sites. Because tra-2s RNA potentially encodes a short aberrant TRA-2 protein (referred to as TRA-2S), we thought that the TRA-2S protein would be generated in Y14(RNAi) and mag-1(RNAi) animals, in which it would cause germ line masculinization. To test this possibility, a transgene expressing HA-tagged TRA-2S under the control of the germ line-specific pie-1 promoter (pie-1::tra-2s::HA) was introduced into hermaphrodites together with the transformation marker dsRed under the control of the muscle-specific myo-3 promoter (myo-3::dsRed), and their progeny were analyzed (Fig. 2B and C). When myo-3::dsRed alone was introduced as a control, all of the dsRed-positive progeny were fertile, which was clearly indicated by the presence of oocytes and embryos in the gonads. In contrast, when pie-1::tra-2s::HA was introduced with myo-3::dsRed, ∼40% of the dsRed-positive transformed progeny showed germ line masculinization and became sterile; in other words, they produced excess sperm but no oocytes or embryos, similar to Y14(RNAi) animals. Most of the rest of the transformed progeny were still fertile but showed less expression of the TRA-2S protein (see Fig. S2B in the supplemental material). Immunofluorescence analysis of the gonads using an anti-HA tag antibody revealed that TRA-2S was localized mainly around the plasma membrane region, as determined by phalloidin staining (Fig. 2D), similar to the pattern of endogenous TRA-2 in Y14(RNAi) and mag-1(RNAi) animals (Fig. 1B). In addition, immunofluorescence analysis using an anti-TRA-2 ICD antibody showed that endogenous TRA-2 was localized mostly around the plasma membrane region of the sterile transformed progeny expressing high levels of HA-tagged TRA-2S, whereas it was detected in the nucleus of the fertile transformed progeny expressing low levels of TRA-2S (see Fig. S2C). These results indicate that relatively high expression of TRA-2S inhibits the expression of the endogenous TRA-2 ICD and explain germ line masculinization in Y14(RNAi) and mag-1(RNAi) animals, which expressed a significant amount of tra-2s RNA.
Fig 2.
Aberrant tra-2s RNA causes germ line masculinization. (A) Northern blot analysis of tra-2 RNA expression in the adult hermaphrodites subjected to RNAi as indicated. Two major mRNAs, tra-2a (∼4.7 kb) and tra-2c (∼1.8 kb), which encode the functional TRA-2 isoforms, were observed in the control worms. tra-2s RNA (∼800 nucleotides) appeared specifically in Y14(RNAi) and mag-1(RNAi) animals. 40S rRNA was used as a loading control. (B) Schematic representation of the tra-2s transgene. The coding sequence of tra-2s cDNA was fused with the HA tag sequence and cloned into an expression vector containing the pie-1 promoter and the tbb-2 3′ untranslated region (UTR). (C) Gonad arms were dissected from transgenic animals expressing dsRed alone (top) or both dsRed and TRA-2S::HA (bottom) and stained with anti-HA tag antibody (left) and DAPI (middle). Nomarski views of the same gonad arms are also shown (right). Embryos are surrounded by dotted lines, and oocyte nuclei are indicated by arrowheads. Scale bars, 60 μm. Note that excess sperm production is observed as small bright dots in the DAPI-stained gonad arm from transgenic animals expressing TRA-2S::HA. (D) Germ cells at the pachytene stage in gonad arms dissected from transgenic animals expressing TRA-2S::HA were stained with the anti-HA tag antibody (green) and DAPI (magenta), shown as a merged view. Separate views of the same cells are also shown, as well as a phalloidin-stained view to show the plasma membrane. Scale bar, 4 μm.
NXF-2 and XPO-1 are responsible for the premature export of unspliced tra-2 RNA to the cytoplasm.
To examine how tra-2s RNA is produced, we analyzed tra-2 RNAs by RT-PCR using primers for exons 2 and 7. We found that in addition to mature tra-2 mRNA, completely unspliced tra-2 RNA that contained all of the introns in the amplified region accumulated in Y14(RNAi) animals (Fig. 3A), although detailed sequence analysis of the amplified products showed that partially spliced RNAs that contained some of the introns were also detected. Unspliced tra-2 RNA with intron 20 was also detected in Y14(RNAi) animals. Because we adopted excess amplification conditions of RT-PCR in these assays in order to see unspliced RNAs clearly, the amount of spliced tra-2 mRNAs in Y14(RNAi) animals appeared to be equal to that in the control animals. However, we confirmed significant decrease of the spliced tra-2 mRNAs in Y14(RNAi) and mag-1(RNAi) animals under moderate amplification conditions of RT-PCR and qRT-PCR, as observed in the Northern blot analysis (see Fig. S3A and B in the supplemental material). To examine the intracellular localization of the unspliced RNA, we performed in situ hybridization analysis using tra-2 intron 20 as a probe. Enhanced visualization revealed that the signal for this intron was restricted within the nucleus, as expected. In contrast, an intense signal for this intron was detected in the cytoplasm of both germ line and somatic cells in Y14(RNAi) animals (Fig. 3B; also see Fig. S3C in the supplemental material). Consistently, unspliced tra-2 RNA disappeared with simultaneous RNAi of tra-2 intron 20 (Fig. 3A). When tra-2 intron 1 was used as a probe, a similar intense signal for intron 1 was detected in the cytoplasm in Y14(RNAi) animals (see Fig. S3D). As expected by the same germ line phenotype and appearance of tra-2s RNA, as in Y14(RNAi) animals, unspliced tra-2 RNA accumulation and an intense intron signal in the cytoplasm were observed in mag-1(RNAi) animals also (see Fig. S3E and F). To confirm that the cytoplasmic intron signals are derived from tra-2 unspliced RNA, we prepared the nuclear and cytoplasmic fractions from Y14(RNAi) and mag-1(RNAi) animals using two nuclear proteins, histone H4 (see Fig. S4A in the supplemental material) and fibrillarin (data not shown) as fractionation controls and examined tra-2 RNA in these fractions (see Fig. S4B). As expected, a significant amount of unspliced tra-2 RNA was detected in the cytoplasmic fractions prepared from Y14(RNAi) and mag-1(RNAi) animals. These results indicate that unspliced tra-2 RNA is leaked out to the cytoplasm when either Y14 or MAG-1 is depleted.
To gain insight into the mechanism by which unspliced tra-2 RNA leaks out to the cytoplasm, we examined the effect of RNAi on five genes encoding C. elegans export receptors: NXF-1 (homologue of mammalian TAP) (51), NXF-2 (NXF-1-like protein) (37), XPO-1 (homologue of exportin-1/CRM1), XPO-2 (homologue of exportin-2/CAS), and XPO-3 (homologue of exportin-t) (52). Interestingly, the accumulation of unspliced tra-2 RNA decreased significantly in Y14(RNAi); nxf-2(RNAi) and Y14(RNAi);xpo-1(RNAi) animals (Fig. 3A). In contrast, Y14(RNAi); xpo-2(RNAi) and Y14(RNAi); xpo-3(RNAi) animals did not show this decrease in unspliced tra-2 RNA (see Fig. S5A in the supplemental material). In the case of Y14(RNAi); nxf-1(RNAi) animals, we could not analyze the level of unspliced tra-2 RNA using RT-PCR because the animals showed severe growth retardation and lethality. To confirm the involvement of NXF-2 and XPO-1 in the leakage of unspliced tra-2 RNA to the cytoplasm, we performed in situ hybridization using tra-2 intron 20 as a probe and found that the cytoplasmic intron 20 signal decreased significantly, to the control level, in Y14(RNAi); nxf-2(RNAi) and Y14(RNAi); xpo-1(RNAi) animals but not in Y14(RNAi); nxf-1(RNAi), Y14(RNAi); xpo-2(RNAi), and Y14(RNAi); xpo-3(RNAi) animals (Fig. 3B; also see Fig. S5B in the supplemental material). In addition, treatment of Y14(RNAi) animals with leptomycin B (LMB), a specific inhibitor of XPO-1 (53), suppressed the cytoplasmic accumulation of intron 20 (see Fig. S5C). The involvement of XPO-1 in premature export of unspliced tra-2 RNA was also supported by the fact that depletion of the nuclear cap-binding protein CBP80, which is known to be required for U snRNA export by the XPO-1 orthologue CRM1 (54, 55), suppresses the premature export of unspliced RNAs caused by Y14 depletion (see Fig. S5D). These results suggest that unspliced tra-2 RNA in Y14(RNAi) animals is mistakenly exported to the cytoplasm by NXF-2 and XPO-1.
tra-2s RNA is produced by aberrant cytoplasmic splicing.
We hypothesized that tra-2s RNA is produced from unspliced tra-2 RNA that is prematurely exported to the cytoplasm. In support of this idea, tra-2s RNA disappeared in Y14(RNAi); nxf-2(RNAi) and Y14(RNAi); xpo-1(RNAi) animals (Fig. 3C), indicating that unusual splicing of tra-2s occurs in the cytoplasm. It is known that cytoplasmic splicing by interferon gene regulatory element 1 (IRE-1) during the unfolded protein response (UPR) is well conserved in many organisms, including C. elegans (56). We therefore thought that IRE-1 may be involved in the production of tra-2s RNA and examined the effect of IRE-1 depletion. As expected, tra-2s RNA disappeared in Y14(RNAi); ire-1(RNAi) animals (Fig. 3C). Moreover, germ line masculinization was suppressed in these double-RNAi-treated animals, similar to Y14(RNAi); tra-2 intron 20(RNAi) animals (Fig. 3D), and fertility was restored, except in Y14(RNAi); xpo-1(RNAi) animals, which showed underdeveloped gonads and fewer germ cells. It should be noted that nxf-2(RNAi), xpo-1(RNAi), and ire-1(RNAi) animals did not show germ line sex determination abnormalities (see Fig. S5E in the supplemental material). It is well known that xbp-1 mRNA is a normal target of IRE-1 and that the cytoplasmically spliced xbp-1 mRNA encodes a key transcriptional regulator for the UPR. Thus, suppression of germ line masculinization by ire-1(RNAi) could be a secondary effect of the UPR (57). However, this is probably not the case, because Y14(RNAi); xbp-1(RNAi) animals still showed sterility, as in Y14(RNAi) animals, whereas the Y14(RNAi) sterility phenotype was suppressed by ire-1(RNAi) (see Fig. S5F). It should also be noted that unspliced tra-2 RNA was still observed in Y14(RNAi); ire-1(RNAi) animals (Fig. 3A), indicating that IRE-1 is not involved in the premature export of unspliced tra-2 RNA per se. These results indicate that in the absence of Y14, unspliced tra-2 RNA is misexported to the cytoplasm and then spliced to tra-2s RNA, possibly by IRE-1.
Three core subunits and one auxiliary subunit of EJC are required for the nuclear retention of unspliced RNAs.
To determine whether the leakage of unspliced RNA to the cytoplasm under Y14(RNAi) or mag-1(RNAi) conditions is specific for tra-2, we examined the splicing of other housekeeping genes. To our surprise, similar unspliced RNA accumulation was observed in Y14(RNAi) animals for the ama-1 gene encoding the RNA polymerase II large subunit and the ife-4 gene encoding translation initiation factor eIF4E (Fig. 4A; also see Fig. S4B in the supplemental material). Moreover, the unspliced RNA accumulation of these genes was suppressed by simultaneous RNAi of nxf-2 or xpo-1. We performed in situ hybridization using intron probes of these genes and confirmed that, similar to the case of tra-2, the unspliced RNAs of these genes leaked out to the cytoplasm in Y14(RNAi) animals (Fig. 4B). Subcellular fractionation revealed that unspliced RNAs were clearly detected in the cytoplasmic fractions prepared from Y14(RNAi) and mag-1(RNAi) animals but not in the cytoplasmic fraction prepared from the control animals. In addition, the excised intron 12 lariat of ama-1 was confined even in the nuclear fractions prepared from Y14(RNAi) and mag-1(RNAi) animals (see Fig. S4B). These results confirm that the in situ cytoplasmic intron signals were derived from unspliced RNAs. Unspliced RNA accumulation was also observed with almost all genes examined in Y14(RNAi) animals, irrespective of single-intron- or multi-intron-containing genes (see Fig. S6 in the supplemental material).
It is unlikely that these unspliced RNAs in Y14(RNAi) animals accumulate by impairment of the NMD pathway, because it was reported that EJC is not required for NMD in C. elegans (58). We also confirmed the dispensability of Y14 for NMD in C. elegans; Y14 is not required for the NMD-regulated rsp-6 expression (see Fig. S7A in the supplemental material) (59). On the contrary, cytoplasmically leaked unspliced RNAs seem to escape NMD, because the amount of unspliced tra-2 RNA did not change significantly in the smg mutants, which are deficient for NMD (60) (see Fig. S7B). In addition, the cytoplasmic leakage of unspliced RNAs is not due to simple splicing defects, because unspliced tra-2 RNA accumulated but was retained in the nucleus when U1-70k (a U1 snRNP component) or PRP8 (a U5 snRNP component) was depleted (see Fig. S7C and D).
To uncover the contribution of other EJC subunits to the nuclear retention of unspliced RNAs, we identified 10 EJC subunit homologues in addition to Y14 and MAG-1 in C. elegans and examined the effect of RNAi on these genes (Fig. 5A; also see Table S1 in the supplemental material). The accumulation of unspliced RNAs was observed in eIF4AIII(RNAi) and UAP56(RNAi) animals but not in other RNAi-treated animals. In situ hybridization using tra-2 intron 20 as a probe confirmed that the unspliced RNAs were leaked to the cytoplasm in eIF4AIII(RNAi) and UAP56(RNAi) animals (Fig. 5B). Because eIF4AIII(RNAi) and UAP56(RNAi) animals showed severe lethality, we performed RNAi for these genes under the rrf-1 mutant background to minimize the RNAi effect in somatic cells and examined the germ line phenotype (61, 62). Under the rrf-1 mutant background, the germ line masculinization phenotype was observed in eIF4AIII(RNAi) animals (see Fig. S8A in the supplemental material), but the germ cells did not enter meiosis in UAP56(RNAi) animals, suggesting an important role of UAP56 in the mitosis-meiosis transition. Taken together, these results indicate that at least three core subunits (Y14, MAG-1, and eIF4AIII) and one auxiliary subunit (UAP56) of EJC are required for the nuclear retention of unspliced RNAs in C. elegans.
It was recently shown that a splicing factor, CWC22, promotes the assembly of eIF4AIII to EJC in mammalian cells and Drosophila (63, 64). We therefore examined the effect of depletion of C. elegans CWC22 and found that CWC22(RNAi) animals showed germ line masculinization and cytoplasmic leakage of unspliced RNA under the rrf-1 background (see Fig. S8B, C, and D in the supplemental material), like Y14(RNAi), mag-1(RNAi), and eIF4AIII(RNAi) animals, supporting an important role of EJC core subunits in the nuclear retention of unspliced RNAs.
Interactions of Y14 with several splicing factors and pre-mRNA.
It was reported that inhibition of SF3b, which is an essential component of U2 snRNP, resulted in unspliced RNA leakage to the cytoplasm in yeast and mammalian cells (65–67). To determine whether this occurs in C. elegans, we depleted the SAP130 homologue, a component of SF3b. Although SAP130(RNAi) resulted in severe growth retardation and morphological abnormalities, we observed germ line masculinization and leakage of unspliced tra-2 RNA into the cytoplasm in SAP130(RNAi) animals (Fig. 6A, B, and C), similar to Y14(RNAi) animals. Consistently, treatment of wild-type animals with spliceostatin A, a specific inhibitor of SF3b (65), showed essentially the same results as the depletion of SAP130 (see Fig. S9A and B in the supplemental material). It was also reported previously that a weak loss-of-function mutation of SAP130 caused germ line masculinization (68). These results indicate that SF3b is required for the nuclear retention of unspliced RNAs in C. elegans as well. We therefore examined the interaction between Y14 and SAP130. To do so, we took advantage of the sterility phenotype upon RNAi (Fig. 6D). At a very low concentration of SAP130 dsRNA, SAP130(RNAi) animals grew to adulthood and ∼40% showed sterility. At the same dsRNA concentration, SAP130(RNAi) enhanced the sterility phenotype of Y14(RNAi) animals.
It was also reported that IBP160, an intron-associated protein, is required for EJC loading on mRNA in mammalian cells (38). We thus examined the effect of RNAi of the IBP160 homologue on the sterility phenotype of Y14(RNAi) animals (Fig. 6D). IBP160(RNAi) animals grew to adulthood and showed no sterility, but IBP160(RNAi) strongly enhanced the sterility phenotype conferred by Y14(RNAi). We also found that depletion of a ubiquitin ligase PRP19 homologue showed a similar enhancing effect on the sterility phenotype conferred by Y14(RNAi) (Fig. 6D). PRP19 is known to be required for remodeling snRNA interactions during splicing and to associate with other proteins, including a stable interaction with CDC5L and a weak interaction with CTNNBL1 (69–71). We therefore examined the effects of the two PRP19-interacting protein homologues and found that depleting the CDC5L homologue showed sterility by itself and enhanced the sterility phenotype resulting from Y14(RNAi), similar to the depletion of SAP130, while depleting the CTNNBL1 homologue did not show any effect on sterility, suggesting an interaction of the stable PRP19 complex with Y14 (Fig. 6D). Moreover, accumulation and leakage of unspliced tra-2 RNA to the cytoplasm were observed in IBP160(RNAi); PRP19(RNAi) animals (Fig. 6B and C), although the double-RNAi-treated animals showed severe growth retardation and arrest in larval stages, indicating involvement of IBP160 and PRP19 in the nuclear retention of unspliced RNA.
To confirm the functional interaction of Y14 with SAP130 and CDC5L, we further examined the sterility phenotype under lower concentrations of double-stranded RNAs using the rrf-1 mutant strain, in which the effect of RNAi can be reduced in somatic cells (Fig. 6E). Under these conditions, Y14(RNAi), SAP130(RNAi), and CDC5L(RNAi) animals showed 16%, 11%, and 3% sterility, respectively. In contrast, Y14(RNAi); SAP130(RNAi) and Y14(RNAi);CDC5L(RNAi) animals showed 96% and 48% sterility, respectively. The much higher extent of sterility in double-RNAi-treated than in single-RNAi-treated animals indicates that interactions of Y14 with SAP130 and CDC5L are synergistic. Taken together, these results strongly suggest that Y14 functionally interacts with SF3b, IBP160, PRP19, and CDC5L to achieve nuclear retention of unspliced RNAs.
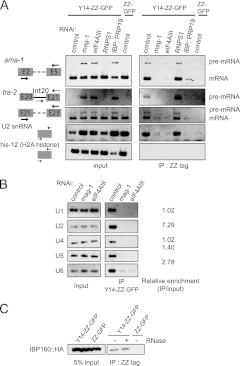
We next examined the physical interactions of Y14 with spliceosome components using a strain stably expressing the Y14-ZZ-GFP fusion protein, which can be functionally substituted for endogenous Y14 (30). Consistent with the requirement for nuclear retention of unspliced RNAs, cell lysates from mag-1(RNAi), eIF4AIII(RNAi), and IBP160(RNAi); PRP19(RNAi) animals accumulated unspliced RNAs for both ama-1 and tra-2 (Fig. 7A, left). Pulldown of Y14-ZZ-GFP using the affinity of its ZZ tag to IgG coprecipitated spliced mRNA for both ama-1 and tra-2 but not histone H2A mRNA, whose gene has no intron (Fig. 7A, right). The interaction of Y14 with mRNAs of genes that have introns reflects the well-known nature of EJC. Interestingly, Y14-ZZ-GFP also coprecipitated both pre-mRNA and spliceosomal snRNAs; U2 snRNA was highly enriched in the precipitates, indicating that of the spliceosomal snRNPs, U2 snRNP preferentially interacts with Y14 (Fig. 7A and B). This is consistent with the finding that three EJC core subunits, including Y14, are already present in the B and C splicing complexes (72). Splice leader RNAs for trans-splicing in C. elegans were hardly detectable in the Y14 precipitates (data not shown). These results suggest that Y14 seems to be located near the branch point and the 3′ splice site of pre-mRNA. Importantly, coprecipitation of pre-mRNAs and snRNAs with Y14-ZZ-GFP did not occur upon depletion of either MAG-1 or eIF4AIII or depletion of both IBP160 and PRP19; however, depletion of RNPS1, one of the auxiliary subunits of EJC, did not affect the coprecipitation of pre-mRNA and snRNAs, indicating that the Y14 interaction with both pre-mRNAs and snRNAs is relevant and closely correlated with the nuclear retention of unspliced RNAs by specific EJC subunits (Fig. 5A).
Fig 7.
Y14 interacts with pre-mRNA, spliceosomal snRNAs, and IBP160. (A) Pulldown assays for the identification of RNAs associated with Y14. Y14-ZZ-GFP was pulled down from cell lysates of animals subjected to RNAi as indicated, using the affinity of the ZZ tag to IgG, and the precipitates were analyzed by RT-PCR. The primer pairs used for amplification are schematically shown on the left. (B) Precipitates from the experiment whose results are shown in panel A were analyzed using the primers for five spliceosomal snRNAs. The relative enrichment of each snRNA in the precipitate from the control animals is shown on the right. (C) Pulldown assays to detect the physical interaction of Y14 with IBP160. Y14-ZZ-GFP was pulled down from cell lysates of animals expressing HA-tagged IBP160 in the presence or absence of RNase treatment, followed by Western blot analysis using the anti-HA tag antibody. IP, immunoprecipitate.
To examine whether Y14 physically interacts with IBP160, we established a transgenic strain expressing HA-tagged IBP160. Immunoprecipitation of HA-tagged IBP160 coprecipitated introns, including a lariat intron as well as U2 snRNA but not intronless histone H2A mRNA (see Fig. S10 in the supplemental material), indicating that IBP160 associates with introns during pre-mRNA splicing in C. elegans, the same as in mammalian cells (38). We then established a transgenic strain expressing both HA-tagged IBP160 and Y14-ZZ-GFP and examined the interaction between these proteins. Pulldown of Y14-ZZ-GFP coprecipitated HA-tagged IBP160 irrespective of the presence of RNase (Fig. 7C).
Taken together, these results indicate that three core subunits (Y14, MAG-1, and eIF4AIII) and possibly an auxiliary subunit (UAP56) of EJC form a functional complex and interact with the spliceosome, possibly through the association with U2 snRNP, depending on the presence of IBP160 and PRP19. Therefore, we suggest that functional interactions of the EJC subunits and specific splicing factors are required for the nuclear retention of unspliced RNAs in C. elegans.
DISCUSSION
Aberrant cytoplasmic splicing of tra-2 RNA.
In this study, we clarified the reason why the two EJC core subunits Y14 and MAG-1 are required for proper germ line sex determination in C. elegans. In the absence of either Y14 or MAG-1, intron-containing tra-2 RNA is leaked out to the cytoplasm and then aberrantly spliced to tra-2s RNA, possibly by IRE-1 (Fig. 2 and 3). It seems that tra-2s RNA is translated into a dominant-negative form of TRA-2 (TRA-2S), which inhibits the proper expression of the TRA-2 ICD in germ cells (Fig. 2; also see Fig. S2 in the supplemental material), leading to germ line masculinization as a result of blocking the switch from spermatogenesis to oogenesis. Although we do not know the detailed mechanism by which TRA-2S inhibits the expression of the TRA-2 ICD, TRA-2S might inhibit the protease activity of TRA-3 as a pseudosubstrate to prevent the production of the TRA-2 ICD. Alternatively, TRA-2S might anchor the TRA-2 ICD around the plasma membrane region to block its transcriptional regulation activity in the nucleus. Despite the dominant-negative function of TRA-2S, the depletion of Y14 affected the sexual differentiation of only germ line cells and not somatic cells. This result may be explained by the fact that germ line cells begin to proliferate during postembryonic stages and should be susceptible to the effects of RNAi at L1, while the sexual differentiation of somatic cells is mostly determined during embryonic stages (73).
Aberrant cytoplasmic splicing of unspliced tra-2 RNA occurs between two short bipartite direct repeat sequences, UUAUXXXXXCACC, so that the upstream repeat sequence is retained in the spliced product, as determined by sequence comparison between tra-2s RNA and its corresponding genomic region (see Fig. S2A in the supplemental material). This aberrant cytoplasmic splicing seems to be catalyzed by IRE-1, because production of tra-2s RNA was suppressed by depletion of IRE-1 (Fig. 3C). However, we did not find any distinct similarity in the primary sequences or secondary structures between the tra-2 gene and the xbp-1 gene, an authentic target gene of IRE-1, although short repeat sequences exist around the splice sites of xbp-1 RNA in humans, mice, Drosophila, and C. elegans (74, 75).
Surveillance and nuclear retention of intron-containing RNAs by pre-EJC.
Our results also reveal a critical and important function of three EJC core subunits for the nuclear retention of intron-containing RNAs in C. elegans. Depletion of Y14, MAG-1, or eIF4AIII does not seem to affect splicing per se, because fully spliced product was observed with all genes examined in this study. Nevertheless, completely unspliced or partially spliced RNAs were prematurely exported to the cytoplasm in the absence of the core subunits, except for Barentz/MLN51, or an auxiliary subunit of EJC, UAP56 (Fig. 3 to 5; also see Fig. S3 in the supplemental material), which suggests that these EJC subunits form a complex and participate in a surveillance system for splicing completion in the nucleus. In this respect, UAP56 was shown to function in coupling pre-mRNA splicing with subsequent mRNA export through its interaction with another EJC subunit, Aly/REF (76–79). In C. elegans, UAP56 was shown to be required for mRNA export as well (80, 81). It was shown that Barentz participates in EJC after splicing, whereas three other EJC core subunits are recruited to the spliceosome before splicing (16, 18). These findings suggest that Barentz has a slightly different function than other EJC core subunits.
It is very likely that interactions between the four EJC subunits and several splicing factors are responsible for the surveillance system, because IBP160, PRP19, and SF3b are involved in the nuclear retention of unspliced RNAs and interact functionally with Y14 (Fig. 6). Furthermore, Y14 associates with pre-mRNA and spliceosomal snRNAs, especially U2 snRNA, and this Y14 association depends on IBP160 and PRP19, as well as two EJC core subunits, MAG-1 and eIF4AIII (Fig. 7A and B). In this respect, it was recently shown that a splicing factor, CWC22, promotes the assembly of eIF4AIII to EJC (63, 64) and that the recruitment of CWC22 to the spliceosome is dependent upon the PRP19 complex (82). We also show that C. elegans CWC22 is involved in the nuclear retention of unspliced RNAs (see Fig. S8 in the supplemental material). These findings consequently support our results that PRP19 is involved in the nuclear RNA surveillance system. During the methylation of Sm proteins of U snRNPs, association of Y14 with spliceosomal snRNAs was also reported in a mammalian cell line, but in this case, preferential association of U2 snRNA was not observed (83). The EJC subunits may interact with the spliceosome through U2 snRNP at a very early stage in pre-mRNA splicing, because UAP56 was shown to be involved in U2 snRNP recruitment to pre-mRNA (76). We also showed that Y14 physically interacts with IBP160 independent of the presence of RNA (Fig. 7C). The results strongly suggest that Y14 associates with introns through the intron-binding activity of IBP160 during pre-mRNA splicing (see Fig. S10 in the supplemental material). We therefore propose a model in which three EJC core subunits and UAP56 form a complex (referred to as pre-EJC) that is recruited with the help of IBP160 and PRP19 onto introns that are committed to be spliced and interacts with U2 snRNP in C. elegans (Fig. 8). In this model, the interaction of pre-EJC with the spliceosome through U2 snRNP is an important signal for the nuclear retention of unspliced RNAs, although the mechanism possibly needs additional factors that recognize and anchor the complex of pre-EJC and the spliceosome within the nucleus. Once the introns are removed, spliceosomes containing a lariat intron dissociate from the spliced exons, and pre-EJCs translocate onto the exon junction to form EJCs with other auxiliary factors. Finally, conversion of all pre-EJC to EJC, which means that splicing is completed for all introns, permits TAP/NXF-1-dependent mRNA export to the cytoplasm. It should be noted that in this model, alternatively regulated introns are free from nuclear retention, because the spliceosome formation and pre-EJC recruitment would not occur on such introns.
Fig 8.
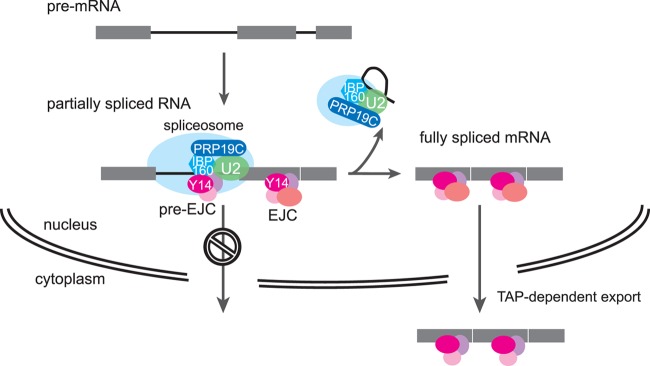
Model for the nuclear retention of unspliced RNAs through the interaction between pre-EJC and the spliceosome. Pre-EJC (Y14, MAG-1, eIF4AIII, and UAP56) is recruited onto introns that are committed to be spliced with the help of the IBP160 and PRP19 complex. Pre-EJC interacts with the spliceosome through U2 snRNP, and this interaction is recognized by one or more factors that anchor the complex in the nucleus. After removing all introns to be spliced, the spliceosomes release, and at the same time, the pre-EJCs translocate onto exon junctions as EJCs. Here, nuclear retention through pre-EJC is cancelled, and the fully spliced mRNA is exported by the nuclear export receptor TAP/NXF-1.
Premature export of unspliced RNAs by NXF-2 and XPO-1.
Our results also indicate that in the absence of pre-EJC, unspliced RNAs are prematurely exported to the cytoplasm, an event that depends on the activity of XPO-1 and NXF-2 (Fig. 3 and 4). XPO-1 is the C. elegans homologue of CRM1, which is known to be the export receptor for nuclear export signal-containing proteins, U snRNAs, and the large and small subunits of the ribosome in many organisms (52, 84). It is possible that XPO-1 mistakenly recognizes and exports unspliced RNAs as authentic cargo U snRNAs, which is consistent with the requirement of the nuclear cap binding protein CBP80 for the premature export of unspliced RNAs (Fig. 4D). However, different from the U snRNA export, the leakage of unspliced RNAs by pre-EJC dysfunction does not require PHAX, an adaptor protein for cap-dependent U snRNA export (data not shown). In this respect, it was reported that XPO-1 and CBP80 but not PHAX are involved in the biogenesis of microRNAs in C. elegans (52), raising an interesting possibility of misrecognition of unspliced RNAs by the pre-microRNA export machinery. Considering that the premature export of unspliced RNAs was suppressed by depletion of either XPO-1 or NXF-2, the two export receptors may act together to recognize and prematurely export unspliced RNAs if the RNA surveillance system by the pre-EJC formation does not work. NXF-2 was reported as an export receptor for tra-2 mRNA, whose depletion leads to the derepression of tra-2 mRNA translation, thereby leading to germ line feminization in C. elegans (35). In our hands, however, depletion of NXF-2 at the postembryonic stage did not show any visible germ line phenotypes (see Fig. S5E in the supplemental material). Interestingly, prematurely exported unspliced RNAs did not seem to undergo NMD (see Fig. S7B in the supplemental material), implicating TAP/NXF-1-dependent mRNA export in the efficiency of subsequent NMD.
Supplementary Material
ACKNOWLEDGMENTS
We thank Hiroyuki Kawahara for the anti-TRA-2 ICD antibody, Barth D. Grant for the anti-RME-2 antibody, Hiroshi Kimura for the anti-histone H4 antibody, Minoru Yoshida for leptomycin B and spliceostatin A, Asako Sugimoto for the pJH4.52 vector, Hidehito Kuroyanagi for the pDEST-eft-3p vector, and the Caenorhabditis Genetics Center for the C. elegans strains. We are grateful to Tatsuaki Goh for his technical help and guidance with the confocal microscopy and Naoyuki Kataoka for his helpful comments on the manuscript. We also thank Taizo Kawano, Teruaki Takasaki, Yuichiro Mishima, Kazuhiro Fukumura, Toshinobu Fujiwara, Yumi Sasano, Misato Gohara, and our laboratory members for their assistance and helpful discussions.
This work was supported by research grants to H.S. from JSPS, to K.I. from MEXT, JSPS, and Daiichi-Sankyo Foundation of Life Science, and to M.S. from JSPS (research fellow).
Footnotes
Published ahead of print 12 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01298-12.
REFERENCES
- 1. Behm-Ansmant I, Kashima I, Rehwinkel J, Sauliere J, Wittkopp N, Izaurralde E. 2007. mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 581: 2845–2853 [DOI] [PubMed] [Google Scholar]
- 2. Bhuvanagiri M, Schlitter AM, Hentze MW, Kulozik AE. 2010. NMD: RNA biology meets human genetic medicine. Biochem. J. 430: 365–377 [DOI] [PubMed] [Google Scholar]
- 3. Hwang J, Maquat LE. 2011. Nonsense-mediated mRNA decay (NMD) in animal embryogenesis: to die or not to die, that is the question. Curr. Opin. Genet. Dev. 21: 422–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rebbapragada I, Lykke-Andersen J. 2009. Execution of nonsense-mediated mRNA decay: what defines a substrate. Curr. Opin. Cell Biol. 21: 394–402 [DOI] [PubMed] [Google Scholar]
- 5. Dziembowski A, Ventura AP, Rutz B, Caspary F, Faux C, Halgand F, Laprevote O, Seraphin B. 2004. Proteomic analysis identifies a new complex required for nuclear pre-mRNA retention and splicing. EMBO J. 23: 4847–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rutz B, Seraphin B. 2000. A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J. 19: 1873–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Takemura R, Takeiwa T, Taniguchi I, McCloskey A, Ohno M. 2011. Multiple factors in the early splicing complex are involved in the nuclear retention of pre-mRNAs in mammalian cells. Genes Cells 16: 1035–1049 [DOI] [PubMed] [Google Scholar]
- 8. Katahira J, Yoneda Y. 2009. Roles of the TREX complex in nuclear export of mRNA. RNA Biol. 6: 149–152 [DOI] [PubMed] [Google Scholar]
- 9. Reed R, Cheng H. 2005. TREX, SR proteins and export of mRNA. Curr. Opin. Cell Biol. 17: 269–273 [DOI] [PubMed] [Google Scholar]
- 10. LE Hir H, Izaurralde E, Maquat LE, Moore MJ. 2000. The spliceosome deposits multiple proteins 20-24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19: 6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tange TO, Nott A, Moore MJ. 2004. The ever-increasing complexities of the exon junction complex. Curr. Opin. Cell Biol. 16: 279–284 [DOI] [PubMed] [Google Scholar]
- 12. Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. 2006. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 127: 1389–1400 [DOI] [PubMed] [Google Scholar]
- 13. Nojima T, Hirose T, Kimura H, Hagiwara M. 2007. The interaction between cap-binding complex and RNA export factor is required for intronless mRNA export. J. Biol. Chem. 282: 15645–15651 [DOI] [PubMed] [Google Scholar]
- 14. Gatfield D, Le Hir H, Schmitt C, Braun IC, Kocher T, Wilm M, Izaurralde E. 2001. The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr. Biol. 11: 1716–1721 [DOI] [PubMed] [Google Scholar]
- 15. Le Hir H, Gatfield D, Izaurralde E, Moore MJ. 2001. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 20: 4987–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tange TO, Shibuya T, Jurica MS, Moore MJ. 2005. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA 11: 1869–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bono F, Gehring NH. 2011. Assembly, disassembly and recycling: the dynamics of exon junction complexes. RNA Biol. 8: 24–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gehring NH, Lamprinaki S, Hentze MW, Kulozik AE. 2009. The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol. 7: e1000120 doi:10.1371/journal.pbio.1000120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kataoka N, Diem MD, Yoshida M, Hatai C, Dobashi I, Dreyfuss G, Hagiwara M, Ohno M. 2011. Specific Y14 domains mediate its nucleo-cytoplasmic shuttling and association with spliced mRNA. Sci. Rep. 1:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Le Hir H, Andersen GR. 2008. Structural insights into the exon junction complex. Curr. Opin. Struct. Biol. 18:112–119 [DOI] [PubMed] [Google Scholar]
- 21. Chang YF, Imam JS, Wilkinson MF. 2007. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 76: 51–74 [DOI] [PubMed] [Google Scholar]
- 22. Diem MD, Chan CC, Younis I, Dreyfuss G. 2007. PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat. Struct. Mol. Biol. 14: 1173–1179 [DOI] [PubMed] [Google Scholar]
- 23. Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW. 2009. Disassembly of exon junction complexes by PYM. Cell 137: 536–548 [DOI] [PubMed] [Google Scholar]
- 24. Ghosh S, Marchand V, Gaspar I, Ephrussi A. 2012. Control of RNP motility and localization by a splicing-dependent structure in oskar mRNA. Nat. Struct. Mol. Biol. 19: 441–449 [DOI] [PubMed] [Google Scholar]
- 25. Hachet O, Ephrussi A. 2001. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr. Biol. 11: 1666–1674 [DOI] [PubMed] [Google Scholar]
- 26. Hachet O, Ephrussi A. 2004. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature 428: 959–963 [DOI] [PubMed] [Google Scholar]
- 27. Mohr SE, Dillon ST, Boswell RE. 2001. The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes Dev. 15: 2886–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ashton-Beaucage D, Udell CM, Lavoie H, Baril C, Lefrancois M, Chagnon P, Gendron P, Caron-Lizotte O, Bonneil E, Thibault P, Therrien M. 2010. The exon junction complex controls the splicing of MAPK and other long intron-containing transcripts in Drosophila. Cell 143: 251–262 [DOI] [PubMed] [Google Scholar]
- 29. Roignant JY, Treisman JE. 2010. Exon junction complex subunits are required to splice Drosophila MAP kinase, a large heterochromatic gene. Cell 143: 238–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kawano T, Kataoka N, Dreyfuss G, Sakamoto H. 2004. Ce-Y14 and MAG-1, components of the exon-exon junction complex, are required for embryogenesis and germline sexual switching in Caenorhabditis elegans. Mech. Dev. 121: 27–35 [DOI] [PubMed] [Google Scholar]
- 31. Li W, Boswell R, Wood WB. 2000. mag-1, a homolog of Drosophila mago nashi, regulates hermaphrodite germ-line sex determination in Caenorhabditis elegans. Dev. Biol. 218: 172–182 [DOI] [PubMed] [Google Scholar]
- 32. Ellis R, Schedl T. 2007. Sex determination in the germ line. WormBook 5 March 2007: 1–13 doi/10.1895/wormbook.1.82.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Burdach J, O'Connell MR, Mackay JP, Crossley M. 2012. Two-timing zinc finger transcription factors liaising with RNA. Trends Biochem. Sci. 37: 199–205 [DOI] [PubMed] [Google Scholar]
- 34. Clifford R, Lee MH, Nayak S, Ohmachi M, Giorgini F, Schedl T. 2000. FOG-2, a novel F-box containing protein, associates with the GLD-1 RNA binding protein and directs male sex determination in the C. elegans hermaphrodite germline. Development 127: 5265–5276 [DOI] [PubMed] [Google Scholar]
- 35. Kuersten S, Segal SP, Verheyden J, LaMartina SM, Goodwin EB. 2004. NXF-2, REF-1, and REF-2 affect the choice of nuclear export pathway for tra-2 mRNA in C. elegans. Mol. Cell 14: 599–610 [DOI] [PubMed] [Google Scholar]
- 36. Lee MH, Schedl T. 2001. Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes Dev. 15: 2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kerkow DE, Carmel AB, Menichelli E, Ambrus G, Hills RD, Jr, Gerace L, Williamson JR. 2012. The structure of the NXF2/NXT1 heterodimeric complex reveals the combined specificity and versatility of the NTF2-like fold. J. Mol. Biol. 415: 649–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ideue T, Sasaki YT, Hagiwara M, Hirose T. 2007. Introns play an essential role in splicing-dependent formation of the exon junction complex. Genes Dev. 21: 1993–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Timmons L, Fire A. 1998. Specific interference by ingested dsRNA. Nature 395: 854. [DOI] [PubMed] [Google Scholar]
- 41. Reese KJ, Dunn MA, Waddle JA, Seydoux G. 2000. Asymmetric segregation of PIE-1 in C. elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol. Cell 6:445–455 [DOI] [PubMed] [Google Scholar]
- 42. Kuroyanagi H, Ohno G, Sakane H, Maruoka H, Hagiwara M. 2010. Visualization and genetic analysis of alternative splicing regulation in vivo using fluorescence reporters in transgenic Caenorhabditis elegans. Nat. Protoc. 5: 1495–1517 [DOI] [PubMed] [Google Scholar]
- 43. Lorsch JR, Bartel DP, Szostak JW. 1995. Reverse transcriptase reads through a 2′-5′linkage and a 2′-thiophosphate in a template. Nucleic Acids Res. 23: 2811–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A. 2006. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 34: e63 doi:10.1093/nar/gkl151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zisoulis DG, Kai ZS, Chang RK, Pasquinelli AE. 2012. Autoregulation of microRNA biogenesis by let-7 and Argonaute. Nature 486:541–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sasano Y, Hokii Y, Inoue K, Sakamoto H, Ushida C, Fujiwara T. 2008. Distribution of U3 small nucleolar RNA and fibrillarin during early embryogenesis in Caenorhabditis elegans. Biochimie 90: 898–907 [DOI] [PubMed] [Google Scholar]
- 47. Lee MH, Schedl T. 2004. Translation repression by GLD-1 protects its mRNA targets from nonsense-mediated mRNA decay in C. elegans. Genes Dev. 18: 1047–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lum DH, Kuwabara PE, Zarkower D, Spence AM. 2000. Direct protein-protein interaction between the intracellular domain of TRA-2 and the transcription factor TRA-1A modulates feminizing activity in C. elegans. Genes Dev. 14: 3153–3165 [PMC free article] [PubMed] [Google Scholar]
- 49. Sokol SB, Kuwabara PE. 2000. Proteolysis in Caenorhabditis elegans sex determination: cleavage of TRA-2A by TRA-3. Genes Dev. 14: 901–906 [PMC free article] [PubMed] [Google Scholar]
- 50. Wang S, Kimble J. 2001. The TRA-1 transcription factor binds TRA-2 to regulate sexual fates in Caenorhabditis elegans. EMBO J. 20: 1363–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tan W, Zolotukhin AS, Bear J, Patenaude DJ, Felber BK. 2000. The mRNA export in Caenorhabditis elegans is mediated by Ce-NXF-1, an ortholog of human TAP/NXF and Saccharomyces cerevisiae Mex67p. RNA 6: 1762–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bussing I, Yang JS, Lai EC, Grosshans H. 2010. The nuclear export receptor XPO-1 supports primary miRNA processing in C. elegans and Drosophila. EMBO J. 29: 1830–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kudo N, Wolff B, Sekimoto T, Schreiner EP, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242: 540–547 [DOI] [PubMed] [Google Scholar]
- 54. Izaurralde E, Lewis J, Gamberi C, Jarmolowski A, McGuigan C, Mattaj IW. 1995. A cap-binding protein complex mediating U snRNA export. Nature 376: 709–712 [DOI] [PubMed] [Google Scholar]
- 55. Kataoka N, Ohno M, Kangawa K, Tokoro Y, Shimura Y. 1994. Cloning of a complementary DNA encoding an 80 kilodalton nuclear cap binding protein. Nucleic Acids Res. 22: 3861–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, Kaufman RJ. 2001. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107: 893–903 [DOI] [PubMed] [Google Scholar]
- 57. Martinon F, Glimcher LH. 2011. Regulation of innate immunity by signaling pathways emerging from the endoplasmic reticulum. Curr. Opin. Immunol. 23: 35–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Longman D, Plasterk RH, Johnstone IL, Caceres JF. 2007. Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev. 21: 1075–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Morrison M, Harris KS, Roth MB. 1997. smg mutants affect the expression of alternatively spliced SR protein mRNAs in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U. S. A. 94: 9782–9785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pulak R, Anderson P. 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 7: 1885–1897 [DOI] [PubMed] [Google Scholar]
- 61. Kumsta C, Hansen M. 2012. C. elegans rrf-1 mutations maintain RNAi efficiency in the soma in addition to the germline. PLoS One 7: e35428 doi:10.1371/journal.pone.0035428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A. 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107:465–476 [DOI] [PubMed] [Google Scholar]
- 63. Barbosa I, Haque N, Fiorini F, Barrandon C, Tomasetto C, Blanchette M, Le Hir H. 2012. Human CWC22 escorts the helicase eIF4AIII to spliceosomes and promotes exon junction complex assembly. Nat. Struct. Mol. Biol. 19: 983–990 [DOI] [PubMed] [Google Scholar]
- 64. Steckelberg AL, Boehm V, Gromadzka AM, Gehring NH. 2012. CWC22 connects Pre-mRNA splicing and exon junction complex assembly. Cell Rep. 2: 454–461 [DOI] [PubMed] [Google Scholar]
- 65. Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, Tani T, Horinouchi S, Yoshida M. 2007. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 3: 576–583 [DOI] [PubMed] [Google Scholar]
- 66. Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, Mizui Y. 2007. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol. 3: 570–575 [DOI] [PubMed] [Google Scholar]
- 67. Lo CW, Kaida D, Nishimura S, Matsuyama A, Yashiroda Y, Taoka H, Ishigami K, Watanabe H, Nakajima H, Tani T, Horinouchi S, Yoshida M. 2007. Inhibition of splicing and nuclear retention of pre-mRNA by spliceostatin A in fission yeast. Biochem. Biophys. Res. Commun. 364: 573–577 [DOI] [PubMed] [Google Scholar]
- 68. Mantina P, MacDonald L, Kulaga A, Zhao L, Hansen D. 2009. A mutation in teg-4, which encodes a protein homologous to the SAP130 pre-mRNA splicing factor, disrupts the balance between proliferation and differentiation in the C. elegans germ line. Mech. Dev. 126: 417–429 [DOI] [PubMed] [Google Scholar]
- 69. Chan SP, Kao DI, Tsai WY, Cheng SC. 2003. The Prp19p-associated complex in spliceosome activation. Science 302: 279–282 [DOI] [PubMed] [Google Scholar]
- 70. Grote M, Wolf E, Will CL, Lemm I, Agafonov DE, Schomburg A, Fischle W, Urlaub H, Luhrmann R. 2010. Molecular architecture of the human Prp19/CDC5L complex. Mol. Cell. Biol. 30: 2105–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wahl MC, Will CL, Luhrmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718 [DOI] [PubMed] [Google Scholar]
- 72. Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R. 2008. Isolation of an active step I spliceosome and composition of its RNP core. Nature 452: 846–850 [DOI] [PubMed] [Google Scholar]
- 73. Zarkower D. 2006. Somatic sex determination. WormBook 10 February 2006: 1–12 doi:10.1895/wormbook.1.84.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. 2002. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415: 92–96 [DOI] [PubMed] [Google Scholar]
- 75. Plongthongkum N, Kullawong N, Panyim S, Tirasophon W. 2007. Ire1 regulated XBP1 mRNA splicing is essential for the unfolded protein response (UPR) in Drosophila melanogaster. Biochem. Biophys. Res. Commun. 354: 789–794 [DOI] [PubMed] [Google Scholar]
- 76. Fleckner J, Zhang M, Valcarcel J, Green MR. 1997. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 11: 1864–1872 [DOI] [PubMed] [Google Scholar]
- 77. Jensen TH, Boulay J, Rosbash M, Libri D. 2001. The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr. Biol. 11: 1711–1715 [DOI] [PubMed] [Google Scholar]
- 78. Luo ML, Zhou Z, Magni K, Christoforides C, Rappsilber J, Mann M, Reed R. 2001. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413: 644–647 [DOI] [PubMed] [Google Scholar]
- 79. Taniguchi I, Ohno M. 2008. ATP-dependent recruitment of export factor Aly/REF onto intronless mRNAs by RNA helicase UAP56. Mol. Cell. Biol. 28: 601–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. MacMorris M, Brocker C, Blumenthal T. 2003. UAP56 levels affect viability and mRNA export in Caenorhabditis elegans. RNA 9: 847–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Shen H. 2009. UAP56—a key player with surprisingly diverse roles in pre-mRNA splicing and nuclear export. BMB Rep. 42: 185–188 [DOI] [PubMed] [Google Scholar]
- 82. Yeh TC, Liu HL, Chung CS, Wu NY, Liu YC, Cheng SC. 2011. Splicing factor Cwc22 is required for the function of Prp2 and for the spliceosome to escape from a futile pathway. Mol. Cell. Biol. 31: 43–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chuang TW, Peng PJ, Tarn WY. 2011. The exon junction complex component Y14 modulates the activity of the methylosome in biogenesis of spliceosomal small nuclear ribonucleoproteins. J. Biol. Chem. 286: 8722–8728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kohler A, Hurt E. 2007. Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8: 761–773 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.