Abstract
Tissue-specific gene expression requires modulation of nucleosomes, allowing transcription factors to occupy cis elements that are accessible only in selected tissues. Master transcription factors control cell-specific genes and define cellular identities, but it is unclear if they possess special abilities to regulate cell-specific chromatin and if such abilities might underlie lineage determination and maintenance. One prevailing view is that several transcription factors enable chromatin access in combination. The homeodomain protein CDX2 specifies the embryonic intestinal epithelium, through unknown mechanisms, and partners with transcription factors such as HNF4A in the adult intestine. We examined enhancer chromatin and gene expression following Cdx2 or Hnf4a excision in mouse intestines. HNF4A loss did not affect CDX2 binding or chromatin, whereas CDX2 depletion modified chromatin significantly at CDX2-bound enhancers, disrupted HNF4A occupancy, and abrogated expression of neighboring genes. Thus, CDX2 maintains transcription-permissive chromatin, illustrating a powerful and dominant effect on enhancer configuration in an adult tissue. Similar, hierarchical control of cell-specific chromatin states is probably a general property of master transcription factors.
INTRODUCTION
Cell identities and functions in multicellular organisms represent the outcome of interactions between transcription factors (TFs) and chromatin. Because nucleosomes restrict access of genomic DNA (1), TFs must overcome a steric barrier at the cis-regulatory elements of tissue-specific genes during development and preserve access in adult tissues. Sequence-specific DNA-binding proteins that initiate chromatin access at tissue-specific enhancers in embryos or differentiating cells are known as pioneer factors. Although pioneer factors recruit additional TFs, they do not necessarily affect long-term nucleosome organization, and they may be dispensable after tissues are specified (2–4). For example, FOXA proteins initiate liver development and facilitate hormone responses in other epithelia but are not required to maintain the adult liver (4). Indeed, the determinants of continuous, cell-type-specific DNA access within chromatin are unclear, and in one prevalent view, multiple TFs cooperate indirectly, without overt hierarchies, to maintain transcription-permissive chromatin (4–7).
Cell fate and function depend critically on a limited number of lineage-restricted TFs that control tissue-specific transcriptional programs. For example, MYOD1, MYOG, and MYF5 in muscle (8, 9) or GATA1, CEBPA, and SFPI1 in blood (10, 11) function at the apex of transcriptional hierarchies and regulate innumerable tissue-specific genes. TFs such as MYOD1, CEBPA, and SFPI1 also tailor cell-specific responses to extrinsic signals (12, 13), but the basis for the sum of these potent in vivo activities remains unclear. One hypothesis is that such TFs, which are often regarded as “master regulators,” control cell lineages by virtue of a particular role in inducing or maintaining open, active chromatin at tissue-specific cis elements (5).
The intestine-specific homeodomain protein CDX2 is one such TF that specifies embryonic intestinal epithelium (14), imparts intestinal character to stomach cells (15), and maintains adult intestinal function and identity (16–18). Consistent with the idea that these properties reflect coordinated control of many intestine-specific genes (19), we previously showed that CDX2 occupies more than 12,000 sites (P < 10−5; 2,885 high-confidence sites at P < 10−10) in the genome of adult mouse intestinal villus cells (18). Most of this binding occurs far from transcriptional start sites (TSSs) within a chromatin configuration indicative of active enhancers: two well-positioned nucleosomes enriched with histone H3 dimethylated on lysine 4 (H3K4me2) that flank an interval of apparent nucleosome depletion (20–22). Moreover, in human intestinal Caco-2 cells, CDX2 often binds DNA in close proximity to GATA6, HNF4A, and other tissue-restricted TFs (22). HNF4A, in particular, is highly expressed in mature intestinal villus cells and binds cis elements of genes associated with terminal cell differentiation (23, 24). The DNA sequence motif that HNF4A favors is also highly enriched at CDX2 binding regions in the mouse intestine (18). These observations in human and murine cells are consistent with the emerging view that small groups of lineage-restricted TFs assemble on distant cis-regulatory modules to regulate tissue-specific genes (25).
The activities of master TFs at tissue-specific chromatin elements in general and at cis-regulatory modules in particular remain unclear. In part, this is because reciprocal TF relationships with each other and with chromatin are challenging to investigate in vivo. Here, we use Cdx2, Hnf4a, and compound mutant mice to investigate individual TF requirements in facilitating binding of partner TFs and in maintaining chromatin access.
MATERIALS AND METHODS
ChIP.
Chromatin immunoprecipitation (ChIP) was performed as described previously (18). Epithelial cells were isolated from villi in the middle one-third of the mouse small intestine (jejunum) using EDTA to separate epithelium from underlying lamina propria, as reported previously (26). Villi were separated from crypts by retaining vortex fractions that failed to pass through a 70-μm-pore-size filter. For TF ChIP, villus fractions were first cross-linked using 1% formaldehyde in phosphate-buffered saline (PBS) for 15 min at 4°C and then for 35 min at room temperature, followed by sonication in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.1) using a Bioruptor until most DNA fragments were between 200 to 500 bp in length, as determined by agarose gel electrophoresis. Cell lysates were diluted 6- to 10-fold in binding buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris-HCl, pH 8.1), incubated with antibody (Ab)-coupled magnetic beads for 16 h at 4°C, and washed six times. DNA was recovered, and cross-links were reversed in 1% SDS and 0.1 M NaHCO3 at 65°C for 8 h. For H3K4me2 ChIP, mononucleosome fractions of isolated villus cells were prepared by treatment with MNase until most of the DNA appeared by agarose gel electrophoresis to have mononucleosome length. Samples were dialyzed in 1,000 volumes of radioimmunoprecipitation assay (RIPA) buffer and used in ChIP as described above, following published protocols (20, 22). Six micrograms of Ab was used for each ChIP: CDX2 (catalog item BL3194; Bethyl Laboratories), HNF4A (catalog item 6547; Santa Cruz Biotechnology), or H3K4me2 (catalog item 07-030; Millipore). DNA libraries were prepared for sequencing according to the manufacturer's recommendations (Illumina).
Motif analysis at TF binding sites detected by ChIP-Seq.
To analyze DNA motif content in TF-bound sites, we ran the motif-based interval screener using the PSSM (MISP) tool in the Cistrome project (http://cistrome.org/ap/) on CDX2-only, HNF4A-only, and cooccupied sites. This tool computes the log-fold difference between the probabilities of true and background motif enrichments at a given site, providing Motif-Scan scores along with their specific locations; the background frequency is obtained using a zero-order or first-order Markov chain. A Motif-Scan score of 100 was empirically selected as the lower limit; i.e., scores of <100 were regarded as insignificant for further consideration. Motif content was quantified as the fraction of binding sites with the presence of the motif at a given quantile-based score cutoff with respect to all binding sites (see Fig. S1 in the supplemental material). When multiple instances of a motif were detected in a TF binding site, only the one with the highest score was considered.
Determination of TF binding and cooccupancy.
After mapping onto the reference mouse genome (UCSC assembly mm9, NCBI build 37), ChIP with high-throughput sequencing (ChIP-Seq) fragments with two or fewer mismatches in each library were kept for identifying TF binding sites, using MACS (model-based analysis of ChIP-Seq), version 1.4.0 beta (27), and consideration of local chromatin bias. Resulting WIG files were visualized in the Integrative Genomics Viewer (28). TF cooccupancy was determined by considering the distance between nearest-neighbor binding summits (P < 10−5) at increasing distances (Fig. 1C).
Fig 1.

CDX2 and HNF4A bind intestinal epithelial cell DNA in close vicinity in areas of active chromatin. (A) CDX2 and HNF4A binding in intestinal villus cells from WT mouse jejunum, determined by ChIP-Seq peaks called at a P value of <10−5. Overlapping DNA occupancy was evident in 2,783 regions. (B) The HNF4A recognition motif was significantly enriched within HNF4A binding regions. (C) Cumulative plots showing the fraction of TF binding sites detected by ChIP-Seq (P < 10−5) within the indicated distance from the partner factor's binding summit. Beyond 300 bp (dotted line), the frequency with which partner factors occupy the same genomic region decreased to the same level as background binding across the genome. Accordingly, 300 bp was selected as a cutoff for considering an interval to be cooccupied by CDX2 and HNF4A. (D) Histogram depicting the frequency at which HNF4A ChIP-Seq peaks appear within a 50-bp window of the indicated distance from the summit of a CDX2 binding site. The two factors frequently bind within 300 bp of each other (gray bars), and such clustering is not evident for CDX2 and random genomic regions equal in number and length to HNF4A binding sites (black bars). (E) ChIP-Seq tags tallied over a representative 1.5-Mb region on mouse chromosome 1 illustrate binding of only HNF4A (blue), only CDX2 (red), or both TFs. (F) Genomic distribution of HNF4A occupancy without (left) and with (right) nearby CDX2 binding, showing that HNF4A largely binds DNA in adult mouse intestinal cells far from gene promoters. (G) Composite plots of the H3K4me2 ChIP-Seq signal surrounding TF binding summits. Average sequence counts were determined in relation to the centers of TF occupancy, indicated as 0, and plotted for each class of binding regions: CDX2 only, HNF4A only, or both TFs (cooccupied). H3K4me2 enrichment on two well-positioned nucleosomes that flank a region of diminished signal and TF occupancy delineate active chromatin in presumptive enhancers.
Calling positions of H3K4me2 mononucleosomes and identifying functional enhancers from different nucleosome dynamics in control and mutant mice.
To detect H3K4me2-marked nucleosomes, NPS (nucleosome positioning from sequencing) software (29) was applied to H3K4me2 ChIP-Seq data on mononucleosomes from wild-type (WT), Cdx2−/−, Hnf4a−/−, and Cdx2−/−; Hnf4a−/− intestinal epithelium. Distances of >2 kb from transcription start sites (TSSs) of RefSeq genes were considered to identify potential enhancers. Using the Tagtab application in the BINOCh software package (30), groups of two to three consecutively positioned nucleosomes with centers within a 450- to 600-bp span were identified from an H3K4me2 profile in WT intestines. Each nucleosome group was scored to quantify its change in mutant chromatin relative to the WT, i.e., as less or more accessible for TF binding. The nucleosome stabilization-destabilization (NSD) score, multiplied by 1,000, produces positive values for chromatin that is more accessible for TF binding under a condition and negative values when sites are less accessible.
Composite analysis of H3K4me2 nucleosomes in regions occupied by CDX2, HNF4A, or both TFs.
To assess the impact of TF deletion on chromatin represented by H3K4me2-marked nucleosomes, we plotted average ChIP-Seq signals for H3K4me2 in WT and mutant intestinal villus cells within the indicated distance from TF binding summits. ChIP-Seq wiggle traces were normalized with respect to the total of ∼2 × 107 uniquely mapped tags, and average traces were plotted using CEAS (31). This package was also applied to determine the binding distribution of HNF4A, CDX2, or both TFs with respect to TSSs (Fig. 1F). The SeqPos algorithm (32), available at http://cistrome.dfci.harvard.edu, was used to identify motifs enriched within HNF4A binding sites.
Immunodetection.
Immunoblotting and immunostaining were performed as described previously (18). CDX2 antibodies were used at dilutions of 1:1,000 (Bethyl Laboratories) or 1:20 (Biogenex Laboratories). HNF4A antibody (6547; Santa Cruz Biotechnology) was applied at a 1:1,000 dilution for immunoblotting and immunostaining.
Measuring significant changes in binding of one TF in the absence of another.
The average CDX2 and HNF4A ChIP-Seq signals at cooccupied sites were first obtained to determine the average width of binding sites. The average peak width, defined as the distance between the peak boundaries at 10% of the average peak height, was 370 bp for CDX2 and 350 bp for HNF4A. We therefore considered 360 bp as the average peak width for fair comparison between the two TFs. ChIP-Seq tags from wild-type and TF-depleted intestines were then counted 180 bp downstream and 180 bp upstream of the binding summit and log2 fold changes between wild-type and mutant ChIP-Seq tag counts were calculated to assess the impact of one TF's absence on the other factor's binding. In the following equations, FC is fold change, n and m represent ChIP-Seq tag counts for CDX2 and HNF4A, respectively, and KO (for knockout) refers to the mutant, TF-depleted intestine: log FCCDX2 = log2(nWT/nHNF4AKO) and log FCHNF4A = log2(mWT/mCDX2KO).
Defining background regions.
To estimate the direct impact of one TF's absence on the other TF's binding at sites they cooccupy in wild-type intestinal cells, it was necessary first to define an internal control set of regions occupied by only one TF; control regions that harbor low ChIP-Seq signals for the other TF enable precise capture of the primary impact of TF depletion and reduce spurious effects. To estimate the genomic background for residual TF binding, we measured ChIP-Seq signals for CDX2 and HNF4A on regions that showed high H3K4me2 enrichment but no evidence for binding of either TF (MACS P values of ≤10−5) and regarded the median ChIP-Seq values on those regions as the respective “noise thresholds.” Similarly, we defined “signal thresholds” for each TF by estimating the median ChIP-Seq signal in CDX2 and HNF4A binding sites (MACS P value of <10−5). Thus, binding sites unique to CDX2 show a HNF4A ChIP-Seq signal lower than the HNF4A noise threshold and a CDX2 ChIP-Seq signal higher than the CDX2 signal threshold; binding sites unique to HNF4A are defined similarly. Because we observed quantitative continua of TF binding, these thresholds allow more stringent categorization of binding events as solitary or cooccupied. Genomic regions with high H3K4me2 signal but no CDX2 or HNF4A binding were used to calculate the null distributions of scores for altered chromatin structure. These distributions, which represent the basal variation in measured chromatin under any condition, are not attributable to CDX2 or HNF4A binding.
Comparing differences in TF binding in regions cooccupied by CDX2 and HNF4A with those in sites occupied by only one of these TFs.
After computing the log fold changes in TF binding between cooccupied and singly bound regions, visual observation indicated that the distributions of the log fold changes were highly shifted toward the negative side. We therefore performed a nonparametric statistical test (Mann-Whitney) to assess the significance of reduced binding in cooccupied regions with respect to the reduced signal in singly bound sites.
Comparing changes in chromatin structure between WT and mutant intestines in sites cooccupied by CDX2 and HNF4A with those in sites bound by a single TF.
Changes in chromatin structure were determined by the NSD scoring system (20), and scores were multiplied by 1,000. Briefly, NSD scores represent the difference between H3K4me2 signal on flanking nucleosomes relative to the adjacent center nucleosomes between WT and mutant conditions. Scores for the change in chromatin state were compared using a Mann-Whitney test for significance because the score distributions were highly asymmetric. NSD calculations were also applied to arrange genomic regions from most accessible (positive NSD scores) to least accessible (negative NSD scores) in Cdx2−/− intestine. Figure 6E plots the frequency of TF binding within each bin of 750 NSD-scored chromatin regions relative to the expected number of binding regions in each bin if no binding tendency is observed. Histograms in Fig. 7E enumerate the number of regions within each NSD-scored bin.
Detecting differential gene expression.
Epithelial cells were isolated from the middle one-third of WT and mutant small intestines, using the EDTA-based method described above, in the section on ChIP analysis; total RNA was isolated using TRIzol and processed and hybridized to Affymetrix Mouse Genome 430 2.0 arrays as described previously (22). RNA expression data from WT (n = 4), Cdx2−/− (n = 3), Hnf4a−/− (n = 2), and Cdx2−/−; Hnf4a−/− (n = 3) samples were preprocessed with the robust multiarray average (RMA) software within the Affymetrix package (version 1.24.2) of BioConductor (33) to determine the background and to perform quantile normalization for comparison across samples. The Limma package (34) was used to identify transcripts expressed at levels significantly different from controls at a 5% false discovery rate (FDR).
Association between differential gene expression and TF binding.
To infer the association of TF binding with changes in gene expression, we first counted the number of binding sites for CDX2, HNF4A, or both TFs within 20 kb of annotated TSSs for each RefSeq gene. Genes were then ranked according to the significance level for altered expression in mutant samples (P values of Limma analysis) from the most reduced to the most increased and grouped into bins of 100 genes each (see the red-green heat map in Fig. 3B). The average number of TF binding sites was calculated near the genes in each bin and normalized with respect to the average number of binding sites for genes that did not change in mutant samples, i.e., genes with FDR values larger than 50% (see the yellow heat map in Fig. 3B).
Microarray data accession number.
All data are deposited in the Gene Expression Omnibus (GEO) under accession number GSE34568.
RESULTS
In vivo cooccupancy of intestinal TFs on distant enhancer DNA.
We first examined HNF4A occupancy in cells freshly isolated from wild-type (WT) mouse intestinal villi, using chromatin immunoprecipitation and DNA sequencing (ChIP-Seq) to identify 22,327 binding sites (P < 10−5; 8,734 sites at P < 10−10) (Fig. 1A). The HNF4A consensus motif was highly enriched in these regions (Fig. 1B). DNA-binding motif analysis further revealed enrichment of CDX2 motifs in HNF4A binding regions detected by ChIP-Seq and vice versa, as well as enrichment of both motifs in regions that showed binding by both TFs (see Fig. S1 in the supplemental material). These findings imply that HNF4A and CDX2 come in direct contact with DNA but also allow for the possibility that some binding occurs through intermediary factors. To set a distance within which to regard regions as cooccupied by the two TFs, we determined the cumulative rate of nearby TF binding. We raised the distance from HNF4A or CDX2 binding summits incrementally and calculated the fraction of sites that bound the other TF within each interval (Fig. 1C). This analysis showed rapidly increasing cooccupancy at distances under 300 bp, whereas binding frequencies of each TF beyond this interval were similar to those in the genome at large. On this basis we selected 300 bp as an empirical cutoff distance for TF cooccupancy, noting several features. First, 300 bp approximates the length of DNA a displaced nucleosome might expose in accessible chromatin. Second, within the 300-bp interval, we observed no particularly favored distance between CDX2 and HNF4A binding summits (Fig. 1D). Third, more than 12% of HNF4A binding occurred near CDX2-occupied sites, and nearly a quarter of all CDX2 binding in vivo occurred within 300 bp of HNF4A and often closer (Fig. 1D). This degree of cooccupancy is similar to reports of TF convergence at cis elements in other contexts (35–37), and sample ChIP-Seq data illustrate both separate and joint occupancy of CDX2 and HNF4A in the intestinal cell genome (Fig. 1E). Most HNF4A occupancy, with or without nearby CDX2 binding, occurred more than 2 kb away from transcription start sites (TSSs), in intergenic regions and introns (Fig. 1F).
Cooccupancy in 300-bp windows far from gene promoters suggests that CDX2 and HNF4A bind functional enhancers (38). Additional features of active cis elements include heightened nuclease sensitivity, signifying nucleosome depletion, and histone modifications associated with transcriptional activation, such as histone H3 methylated at lysine 4 (H3K4me1), H3K4me2, and histone H3 acetylated at lysine 27 (H3K27Ac) (21, 39–42). To identify such enhancers, we performed ChIP-Seq for H3K4me2 on mononucleosome fractions of intestinal villus cell chromatin. The H3K4me2 mark appears on promoters as well as enhancers, and H3K4me2-marked nucleosome pairs are known to flank regulatory elements (20–22, 39). Among well-positioned H3K4me2-marked nucleosome pairs, nearly 150,000 pairs were located >2 kb from TSSs, that is, away from promoters and in candidate distant enhancers. At least 32% of CDX2 binding, 43% of HNF4A binding, and 59% of CDX2 and HNF4A cooccupancy in intestinal cells, all detected at a P value of <10−5, fell between such nucleosome pairs (Fig. 1G). The presence of thousands of H3K4me2-marked sites that CDX2 and HNF4A occupy in intestinal cells allowed us to adopt a genetic approach to investigate the requirement of each TF for the other factor's binding to DNA and in preserving chromatin marks.
Concerted activator functions of HNF4A and CDX2.
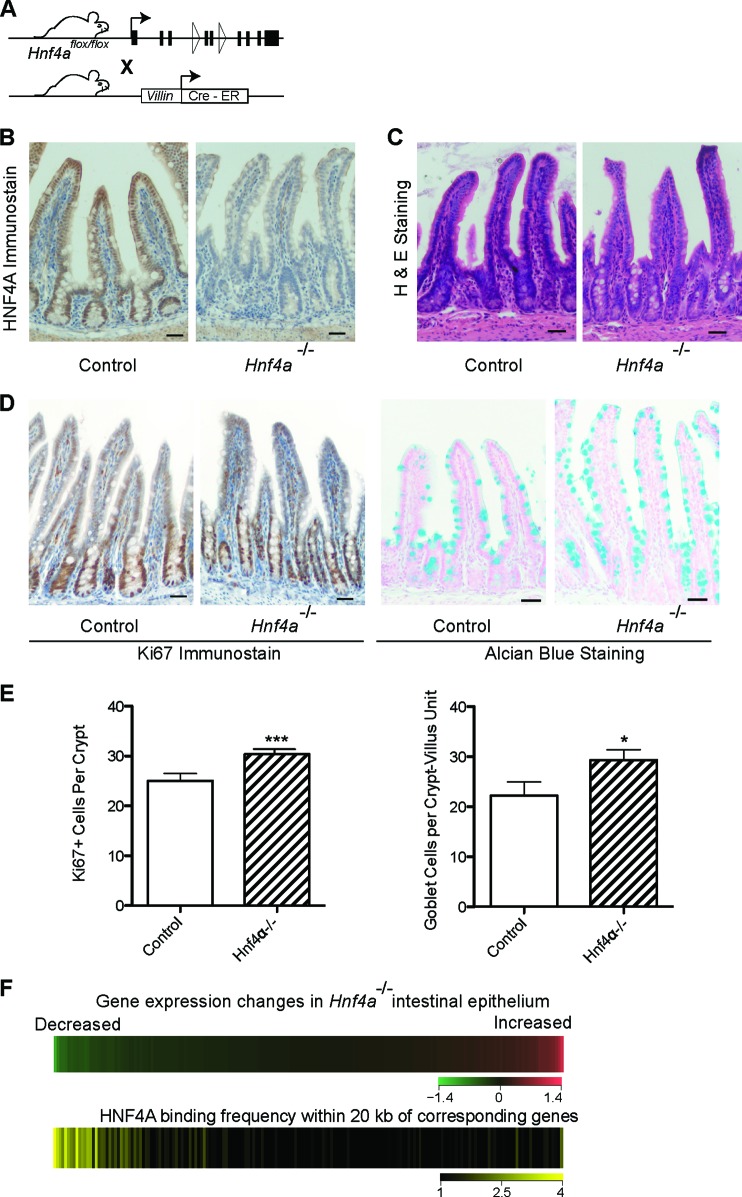
CDX2 primarily activates intestinal genes (18). To determine HNF4A activity, we crossed mice carrying a floxed Hnf4a allele (43) with Villin-CreER-T2 transgenic mice (44), which express tamoxifen-dependent Cre recombinase in the intestine (Fig. 2A). Tamoxifen efficiently depleted HNF4A from Hnf4aFl/Fl; Villin-CreER-T2 intestines (Fig. 2B), and, in agreement with published reports (45, 46), the mutant epithelium remained largely intact (Fig. 2C), with only small increases in goblet and replicating crypt cells (Fig. 2D and E). Next, we examined HNF4A occupancy in wild-type intestines with respect to gene expression changes in Hnf4a-null intestinal cells. HNF4A binding correlated with genes that were reduced in mutant mice but not with those that were increased or unaltered (Fig. 2F), implying that HNF4A also mainly activates genes.
Fig 2.
HNF4A functions as a transcriptional activator in intestinal epithelium. (A) Schema for inducible, intestine-specific depletion of HNF4A in vivo. (B) Immunostaining verifies efficient HNF4A loss in intestinal epithelium after tamoxifen-induced Cre recombination. (C) Hematoxylin and eosin staining shows that Hnf4a−/− gut epithelium is overtly intact. (D) The proportions of Ki67+ proliferating crypt cells and Alcian blue-avid goblet cells are slightly increased in Hnf4a−/− gut epithelium, as others have reported. Scale bar, 30 μm. (E) The small but significant increase in Ki67+ and goblet cells was quantified by counting labeled cells (data represent the mean cell counts ± standard deviations from >3 mice; *, P < 0.05; ***, P < 0.001). (F) HNF4A activates transcription in intestinal villi. We partitioned all transcripts, measured by microarray analysis (top heat map), in bins of 100, from those most reduced in Hnf4a−/− intestines (green) to those most increased (red) and calculated the HNF4A binding frequency near these groups of 100 genes in ChIP-Seq analysis of WT intestines (bottom heat map). HNF4A binding was highest near genes whose levels drop with HNF4A loss.
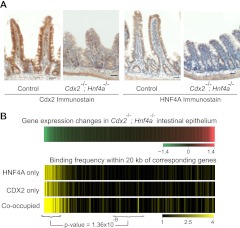
Extending this analysis to double mutant mice, we observed that loss of both HNF4A and CDX2 in Cdx2Fl/Fl; Hnf4aFl/Fl; Villin-CreER-T2 intestines affected villus height and structure (Fig. 3A) more profoundly than loss of either HNF4A (Fig. 2) or CDX2 (18) alone. The detailed phenotype of these compound mutant mice will be reported elsewhere (A. San Roman et al., unpublished data). More germane to the topic of the present study, the combined loss of CDX2 and HNF4A affected the levels of nearly 4,000 transcripts (Fig. 3B, top) (false discovery rate of 5%). Binding of both proteins in WT intestinal villi correlated only with transcripts that declined in the mutant gut (Fig. 3B, bottom), confirming their respective activator functions. Nearly 70% of genes with reduced expression in the double mutant intestines showed binding of CDX2, HNF4A, or both TFs within 20 kb (Fig. 3B), which implies that they stimulate transcription directly, often in concert; this likely contributes toward defective villus structure in double mutant intestines (Fig. 3A). Thus, CDX2 and HNF4A cooccupancy at hundreds of distant foci of active chromatin is associated with optimal expression of adjoining intestinal genes.
Fig 3.
(A) Immunostaining demonstrates that both CDX2 and HNF4A are efficiently depleted in Cdx2-Hnf4a double mutant mice after tamoxifen treatment. Scale bar, 30 μm. Note the reduced villus height in the mutant tissue relative to that of the control. (B) mRNA expression changes observed in Cdx2-Hnf4a double mutant intestines are depicted in a heat map, similar to that in Fig. 2F, with transcripts that decrease arrayed on the left (green) and those that increase represented to the right (red). Yellow heat maps show the corresponding frequency of normalized TF binding within 20 kb of these 100-gene binned groups. Association with TF binding was highly significant (Mann-Whitney test, P = 1.36 × 10−9 for cooccupied sites) for the top decile of transcripts that are altered in Cdx2−/−; Hnf4a−/− intestines, relative to those that are unchanged; this implies that both CDX2 and HNF4A activate genes.
Nonreciprocal requirements for TF binding in vivo.
We assessed DNA occupancy of each TF in mouse intestinal villi lacking the other protein. Absence of HNF4A did not materially alter CDX2 expression (Fig. 4A), and ChIP-Seq of WT and Hnf4a−/− intestines revealed little effect on global CDX2 occupancy, either at sites without nearby HNF4A binding or at sites where the two TFs normally cooccupy DNA within 300 bp (Fig. 4B). A site-by-site comparison of CDX2 binding in Hnf4a−/− intestines further revealed insignificant differences in occupancy between sites that ordinarily bind only CDX2 and those that engage both TFs; signals in both cases were similar in WT and mutant intestines (Fig. 4C and D).
Fig 4.
HNF4A binding is dispensable for CDX2 occupancy of intestinal epithelial cell DNA. Binding of CDX2 was assessed in the absence of HNF4A. (A) CDX2 immunostaining confirms that its expression is unaffected in Hnf4a−/− intestines. (B) Composite ChIP-Seq plots of all regions bound only by CDX2 (left) or by both CDX2 and HNF4A (right). DNA occupancy of CDX2 is similar in wild-type and Hnf4a−/− cells, with a very minor diminution in binding in mutant cells. (C) In a site-for-site comparison of normalized ChIP-Seq tag counts, the minor changes in CDX2 binding are similar among sites bound only by CDX2 or by both TFs (Mann-Whitney test, P = 0.13). Box plots show the log2 fold difference of the normalized CDX2 ChIP-Seq read counts between WT and Hnf4a-null intestines, at cooccupied and CDX2-only binding sites. The middle bar indicates the median of the distribution, and the lower and upper edges represent 25% and 75% quartiles, respectively. Whiskers represent the lower (−1.5 × interquantile range) and upper (+1.5 × interquantile range) edges. Circles beyond the whiskers are outliers. (D) A representative data trace from chromosome 16 position 22120000 to 22180000 illustrates preserved CDX2 binding in Hnf4a−/− cells.
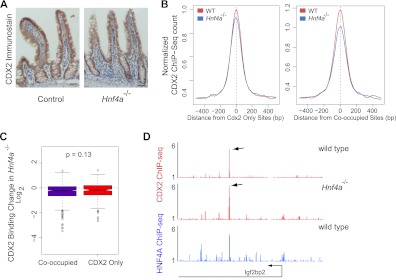
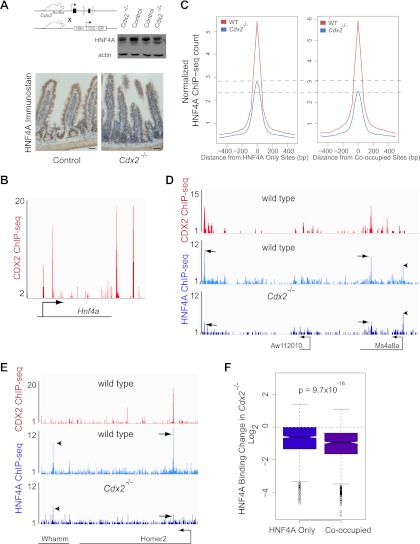
In contrast, CDX2 loss reduced HNF4A binding for two seemingly distinct reasons. First, extensive CDX2 binding at the Hnf4a locus in WT intestines (Fig. 5B) suggests that CDX2 regulates Hnf4a expression directly. In agreement with this idea, Hnf4a levels were slightly lower in the Cdx2−/− gut (62.8% of wild-type transcript levels by microarray analysis; P = 1.18 × 10−4 as computed by Limma), and both immunoblotting and immunostaining showed a commensurate decline in protein levels (Fig. 5A). Reflecting the lower protein level, HNF4A occupancy was modestly reduced at sites where it normally binds DNA without CDX2 nearby (Fig. 5C and F, left). Second, DNA binding was severely affected at sites where CDX2 and HNF4A ordinarily cooccupy DNA (Fig. 5C and F, right). For example, HNF4A binding was lost at the Homer2, Ms4a8a, and Aw112010 loci, where CDX2 and HNF4A cooccupy DNA, but barely reduced at the linked Whamm locus or Ms4a8a promoter, which do not bind CDX2 (Fig. 5D and E; see also Fig. S2A in the supplemental material). Indeed, site-by-site analysis indicated highly significant differences between binding of HNF4A at cooccupied regions and its binding at sites where it normally occupies DNA without nearby CDX2 (Fig. 5F). Taken together, these data show that HNF4A is dispensable for CDX2 binding but that CDX2 is necessary for HNF4A binding, especially at sites where the two proteins cooccupy DNA.
Fig 5.
CDX2 is necessary for HNF4A to bind intestinal epithelial cell DNA at cooccupied regions. (A) Immunoblotting and immunohistochemical evidence that HNF4A levels are reduced, approximately by half, in Cdx2−/− intestinal epithelium. (B) Four prominent CDX2 binding sites are observed in a 40-kb window at the murine Hnf4a locus, suggesting direct effects on Hnf4a expression (P < 10−5). (C) Composite plots of ChIP-Seq data from WT and Cdx2−/− intestinal cells, showing HNF4A occupancy at all regions where HNF4A binds DNA without (left box) or with (right box) nearby CDX2 binding. Aggregate HNF4A binding to DNA is reduced, probably owing to lower protein levels, and the compromise is substantially larger at sites cooccupied by CDX2 (right box). Dotted lines mark the difference in average HNF4A peak heights in WT and Cdx2−/− intestines. (D and E) Selective losses in HNF4A binding are shown at representative regions containing both HNF4A-only and cooccupied sites (D, chromosome 19 position 11085000 to 11160000; E, chromosome 7 position 88720000 to 88856000). In Cdx2−/− cells, HNF4A occupancy is affected less or not at all at sites without CDX2 binding (arrowheads) and severely compromised at sites where both TFs normally cooccupy DNA (arrows). (F) Statistical evidence (Mann-Whitney test) that changes in HNF4A binding in the absence of CDX2 are more severe at cooccupied than at singly bound sites.
Effect of CDX2 deficiency on chromatin at sites of HNF4A cooccupancy.
The spacing between CDX2 and HNF4A binding in vivo (Fig. 1D) or between CDX2 and HNF4A recognition motifs in cooccupied regions in silico varies considerably in a distribution that suggests no obvious mechanism for direct cooperativity in TF binding. Instead, TF occupancy commonly coincided with the space between flanking H3K4me2-marked nucleosomes (Fig. 1D and G), suggesting indirect cooperativity mediated through chromatin (7, 47). To examine this possibility, we used H3K4me2 ChIP-Seq to examine the chromatin configuration in mutant intestines, specifically assessing H3K4me2 signals near regions that bind both CDX2 and HNF4A in WT cells. Tamoxifen-induced HNF4A depletion had no effect on the active chromatin configuration at cooccupied regions (Fig. 6A); H3K4me2 marks on separated, well-positioned nucleosomes were preserved. In Cdx2−/− villi, however, this characteristic H3K4me2 profile around TF-binding sites was notably diminished at cooccupied regions (Fig. 6B). Furthermore, the nucleosomal H3K4me2 profiles in these regions were similar in Cdx2−/− and Cdx2−/−; Hnf4a−/− intestinal villus cells (Fig. 6C), indicating that HNF4A adds little to the dependence on CDX2. Thus, CDX2 but not HNF4A is necessary to maintain transcription-permissive chromatin at hundreds of distant intestinal cis elements in vivo. The finding that CDX2 loss reduces expression of most linked genes (Fig. 3B) suggests that this activity underlies its transcriptional regulatory function.
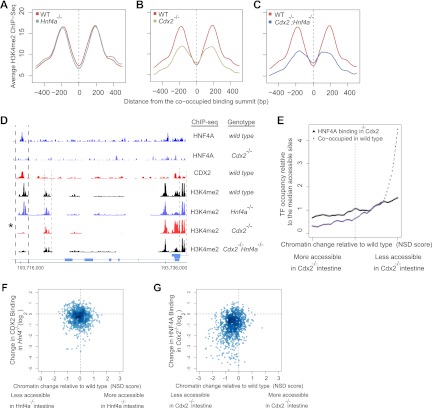
Fig 6.
CDX2 loss adversely affects the active chromatin configuration at sites of HNF4A cooccupancy. (A to C) Nucleosomal H3K4me2 profiles were determined by ChIP-Seq of MNase-digested chromatin from WT and TF-depleted intestines. Aggregate plots of the ChIP-Seq signal are centered on the 1,657 sites that CDX2 and HNF4A cooccupy in WT mice and also carry the H3K4me2 mark of active enhancers. Loss of HNF4A did not affect the nucleosomal H3K4me2 profile (A), whereas CDX2 loss prominently reduced H3K4me2 signals (B). Additional loss of HNF4A did not further affect the H3K4me2 profile of Cdx2−/− tissue (C). (D) Representative ChIP-Seq data from a region on chromosome 1. CDX2 loss compromised both chromatin structure and HNF4A binding, specifically at a cooccupied site (bold dashed lines, asterisk), sparing the H3K4me2 mark where it appears without CDX2 binding (light dashed lines), both within and far from a promoter. See also Fig. S2 in the supplemental material. (E) Frequency of HNF4A-CDX2 cooccupied sites in WT (purple) and of HNF4A binding sites in Cdx2−/− (black) intestinal cells plotted in relation to the nucleosome stabilization-destabilization (NSD) score, a measure of altered H3K4me2-marked chromatin in Cdx2−/− intestines. Regions with reduced chromatin access in the absence of CDX2, represented to the right, correspond to those that CDX2 and HNF4A cooccupy in WT intestines. Regions that are unchanged (center) or have become more accessible in Cdx2−/− gut, represented to the left, show no correlation with CDX2 and HNF4A binding. (F and G) Changes in nucleosomal H3K4me2 configuration at the 1,657 cooccupied regions were plotted against the corresponding change in binding of CDX2 or HNF4A in mouse intestines lacking the other TF. Hnf4a-null intestine (F) showed little change in CDX2 occupancy (y axis) or chromatin configuration (x axis), whereas active chromatin and HNF4A binding were proportionately compromised in Cdx2−/− intestine (G). Thus, CDX2 protects active chromatin at hundreds of regions, and HNF4A binding is preferentially diminished in the regions of highest chromatin effect in Cdx2−/− intestines.
ChIP-Seq traces from Cdx2−/− intestinal villi (Fig. 6D; see also Fig. S2B in the supplemental material) illustrate concomitant disruption of HNF4A binding and H3K4me2 signals at sites of CDX2 cooccupancy. Importantly, in these and other examples, HNF4A binding and H3K4me2 are not affected at sites where CDX2 does not bind DNA. To determine if the association between disrupted HNF4A binding and disrupted H3K4me2 is general, we assessed the concordance between the two variables. In the graph in Fig. 6E, all paired nucleosomes are arrayed serially in bins of 750 pairs according to the nucleosome stabilization-destabilization score (NSD), which ranks each nucleosome pair according to the magnitude of difference between WT and Cdx2−/− intestinal epithelium (20); pairs that showed loss of the H3K4me2 configuration in Cdx2−/− intestine are represented to the right, and those showing increased H3K4me2 signal in Cdx2−/− intestine are shown to the left. In WT intestines, HNF4A and CDX2 most often cooccupy DNA in regions that require CDX2 for an active H3K4me2 configuration (Fig. 6E, purple curve increasing to the right). In Cdx2−/− intestinal cells, HNF4A occupancy was dramatically reduced in regions with altered chromatin but was largely preserved in areas where the H3K4me2 configuration was unaffected (Fig. 6E, black line). These data suggest an intimate association between HNF4A occupancy and CDX2-dependent chromatin states.
To examine this relationship site by site, we generated scatter plots of TF binding and chromatin H3K4me2 at every site of CDX2 and HNF4A cooccupancy. Each dot in Fig. 6F and G represents the effect at a single site of loss of one TF on the nucleosome H3K4me2 configuration (x axis) and on binding of the partner TF (y axis). Thus, the minimal effect of HNF4A loss on the H3K4me2 signal or CDX2 binding results in even distribution of dots around the (0, 0) coordinate, reflecting intrinsic signal variance with few outliers (Fig. 6F). In contrast, Cdx2−/− dots are highly skewed toward the lower left quadrant (Fig. 6G). The strong association between reduced HNF4A binding and loss of active chromatin in Cdx2−/− intestines implies that HNF4A relies indirectly on a CDX2-dependent chromatin state for access to DNA. Of note, loss of HNF4A binding in Cdx2−/− intestines had no relationship to the distance between CDX2 and HNF4A binding sites.
General effect of CDX2 deficiency on intestinal cell chromatin in vivo.
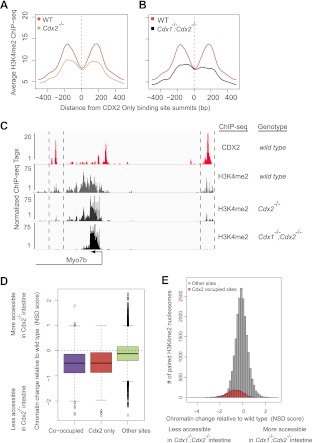
The dependence of chromatin states on CDX2 has important implications for its function as a master regulator of intestinal fate and function (14–18), especially if this dependence represents a general mechanism for chromatin access at intestinal genes. To address this possibility, we considered the more than 9,000 sites that CDX2 occupies without nearby HNF4A binding in intestinal villus cells (Fig. 1A). We noted first that, although CDX2 provides the bulk of Caudal-family protein activity in the gut (19, 48), phenotypic defects are more severe in Cdx1−/−; Cdx2−/− than in Cdx2−/− intestine (18), indicating some functional redundancy between CDX2 and CDX1. To determine whether CDX1 contributes to maintaining enhancer chromatin structure at CDX2-occupied sites, we measured nucleosomal H3K4me2 in mouse intestines deficient in both CDX1 and CDX2. H3K4me2 enrichment flanking CDX2 binding sites, already reduced in Cdx2−/− intestines, was further diminished in Cdx1−/−; Cdx2−/− intestinal cells (Fig. 7A to C). Thus, CDX factors jointly maintain permissive chromatin at thousands of intestinal enhancers in vivo, including those that HNF4A does not occupy.
Fig 7.
CDX1 and CDX2 jointly maintain the chromatin state at thousands of intestinal enhancers, beyond those occupied by HNF4A. (A and B) Aggregate plots of enhancer H3K4me2 profiles in mutant intestines at CDX2-bound regions that lack HNF4A binding. The H3K4me2 mark on positioned nucleosomes flanking CDX2 occupancy is considerably depleted in Cdx2−/− (A) and further compromised in Cdx1−/−; Cdx2−/− (B) intestinal cells, revealing a profound requirement for Caudal family proteins in maintaining active chromatin in the adult gut. (C) A representative genomic region (chromosome 18 position 32187000 to 32215000) containing Myo7b illustrates progressive reduction in the H3K4me2 mark upon loss of CDX proteins. This is evident at regions that flank CDX2 binding sites both upstream and downstream (regions bracketed by dotted lines) of the TSS, whereas H3K4me2 levels at the Myo7b promoter (arrow) are unaffected. (D) Distributions of the magnitude of changes in H3K4me2-marked chromatin following CDX2 loss. Sites that bind CDX2 without HNF4A are affected as much by CDX2 loss as those that CDX2 and HNF4 cooccupy; both classes of binding regions suffer compared to regions where neither TF binds DNA. (E) CDX2 effects on chromatin are local rather than global. Regions that normally contain positioned H3K4me2-marked nucleosome pairs but do not bind CDX2 or HNF4A show a normal distribution of changes after CDX2 loss (gray histogram). Although most areas of the genome were unchanged and chromatin was marked more strongly at many sites (NSD of >0), regions that bind CDX2 in WT intestines (red shading) had the most severely affected chromatin in Cdx1−/−; Cdx2−/− intestinal cells. The difference in distributions of the two histograms has a P value of 2.67 × 10−156 by the two-sample t test.
Lastly, we examined the degree to which maintaining active chromatin is a general activity of CDX2, beyond regions where it binds DNA near HNF4A. Comparative analysis of WT and Cdx2−/− intestinal cells revealed as profound a deficiency of H3K4me2-marked chromatin in regions of CDX2 binding without HNF4A as in those with HNF4A cooccupancy (Fig. 7D, left and center columns). This marked deficit of active chromatin in Cdx2−/− intestines does not represent a global disturbance because thousands of H3K4me2-enriched nucleosomes were preserved in mutant Cdx2−/− intestines (Fig. 7D, right column). Moreover, active chromatin appeared at hundreds of new sites (Fig. 7E, note the H3K4me2 chromatin distribution with NSD scores of >0), and the distribution of CDX2-occupied regions in WT cells (Fig. 7E, red bars) coincided significantly with that of negative NSD scores, signifying less marked chromatin, in Cdx mutants. Furthermore, DNA binding and chromatin modulation are probably sequence-specific actions because CDX2-dependent chromatin regions were significantly enriched for CDX2-binding motifs. In contrast, regions with increased H3K4me2-marked nucleosomes in Cdx2−/− intestines lacked enrichment of CDX2-binding motifs and probably represent the actions of other transcriptional regulators upon CDX2 loss.
DISCUSSION
Some TFs are expressed briefly during development and impart competence for tissue-restricted transcriptional programs. Such TFs operationally resemble pioneer factors, which specify cell lineages but seem to have a limited role in maintaining stable cellular or chromatin states beyond development (4). Instead, transcription-permissive chromatin at cell-specific cis-regulatory modules in adult tissues is thought to reflect the cooperative activity of multiple TFs, with no single factor dominating materially over others that occupy the same enhancer (4–7). In an important sense, however, the well-studied examples that support this view are not easily reconciled with the seemingly dominant biological activities of lineage-specifying TFs. Such tissue-restricted factors are classically exemplified by myogenic and hematopoietic TFs (9, 11) but are also recognized in most other tissues; they often help specify a cell lineage during development and maintain the same cell type in adults. CDX2 represents this class of master regulator TFs in the intestine because it induces an intestine-specific transcriptional program in heterologous stomach cells in vivo (15, 49), specifies the developing intestine (14), and maintains adult intestinal integrity and function (16–18). Recent work highlights the additional activity of master regulator TFs in tailoring tissue-specific transcriptional programs in response to ubiquitous intercellular signals such as Wnt and transforming growth factor β (TGF-β) (12, 13, 50).
Together, the powerful biological activities of lineage specification, cell maintenance, and cell-specific response suggest the possibility of a dominant role for such proteins within TF hierarchies. However, it is unclear how that role is enacted at the level of regulatory cis elements, nor has a role been demonstrated unambiguously with respect to chromatin and other TFs in mammalian cells in vivo. Our results reveal a cardinal requirement for CDX2 in maintaining enhancer chromatin in an active form at thousands of sites in the genome of adult mouse intestinal cells. CDX2 protects or provides the active H3K4me2-marked chromatin configuration near most of its TSS-distal binding sites that show enhancer properties. Furthermore, nearby HNF4A binding, present at about one in four such sites, is markedly compromised when the H3K4me2 mark is attenuated in the absence of CDX2. Whereas the Saccharomyces cerevisiae PHO5 promoter offered a precedent for a similar TF function in enabling chromatin access (51, 52), our study reveals the genome-wide activity of a lineage-specifying TF in a mammalian tissue in vivo.
Loss of CDX2's companion factor HNF4A did not materially affect nucleosomal H3K4me2 marks at thousands of sites where it occupies DNA, regardless of nearby CDX2 binding. Rather, our data reveal a hierarchy in which CDX2 governs access of other TFs by controlling enhancer chromatin structure. Through this powerful effect on chromatin, which we observed at the majority of CDX2-bound sites in intestinal villus epithelial cells, CDX2 might also enable TFs other than HNF4A to access DNA. However, our experiments do not address if CDX2 only maintains permissive chromatin or is also responsible for initiating chromatin access in nascent epithelial cells or in the embryonic gut, i.e., whether it behaves as a bona fide pioneer factor. This is an important topic for future investigation, together with the question of whether CDX2 controls chromatin in a manner similar to the yeast PHO5 promoter (51, 52) or through mechanisms that may be unique to complex mammalian enhancers. Most importantly, CDX2's profound role in preserving enhancer activity in an adult organ in vivo suggests that master regulators of other cell lineages may control tissue-specific genes by similarly maintaining chromatin in an active configuration at distant, lineage-restricted enhancers.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sylvie Robine and Peter Gruss for providing Villin-Cre and Cdx1−/− mice, respectively, and Cliff Meyer and Myles Brown for helpful discussions and comments on the manuscript.
This work was supported by National Institutes of Health grants R01DK082889 (R.A.S.), R01HG4069 (X.S.L.), K01DK088868 (M.P.V.), and P50CA127003 and fellowship number 1987 from the Crohn's and Colitis Foundation of America (M.P.V.).
We declare that we have no conflicts of interest.
M.P.V. was responsible for the study design, experiments, data analysis, and writing the manuscript; A.K.S.R. was responsible for experiments; H.S. and X.S.L. were responsible for data analysis and editing the manuscript; R.A.S. was responsible for study design and supervision, data analysis, and writing the manuscript.
Footnotes
Published ahead of print 5 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01185-12.
REFERENCES
- 1. Struhl K. 1999. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell 98: 1–4 [DOI] [PubMed] [Google Scholar]
- 2. Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. 2002. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 9: 279–289 [DOI] [PubMed] [Google Scholar]
- 3. Smale S. 2010. Pioneer factors in embryonic stem cells and differentiation. Curr. Opin. Genet. Dev. 20: 519–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zaret KS, Carroll JS. 2011. Pioneer transcription factors: establishing competence for gene expression. Genes Dev. 25: 2227–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Biggin MD. 2011. Animal transcription networks as highly connected, quantitative continua. Dev. Cell 21: 611–626 [DOI] [PubMed] [Google Scholar]
- 6. Boyes J, Felsenfeld G. 1996. Tissue-specific factors additively increase the probability of the all-or-none formation of a hypersensitive site. EMBO J. 15: 2496–2507 [PMC free article] [PubMed] [Google Scholar]
- 7. Polach KJ, Widom J. 1996. A model for the cooperative binding of eukaryotic regulatory proteins to nucleosomal target sites. J. Mol. Biol. 258: 800–812 [DOI] [PubMed] [Google Scholar]
- 8. Davis RL, Weintraub H, Lassar AB. 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51: 987–1000 [DOI] [PubMed] [Google Scholar]
- 9. Olson EN. 1990. MyoD family: a paradigm for development? Genes Dev. 4: 1454–1461 [DOI] [PubMed] [Google Scholar]
- 10. Kulessa H, Frampton J, Graf T. 1995. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 9: 1250–1262 [DOI] [PubMed] [Google Scholar]
- 11. Graf T, Enver T. 2009. Forcing cells to change lineages. Nature 462: 587–594 [DOI] [PubMed] [Google Scholar]
- 12. Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, Dekoter RP, Young RA. 2011. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell 147: 565–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, Dibiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, LI P, Durand EM, Mosimann C, Heffner GC, Daley GQ, Paulson RF, Young RA, Zon LI. 2011. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell 147: 577–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gao N, White P, Kaestner KH. 2009. Establishment of intestinal identity and epithelial-mesenchymal signaling by Cdx2. Dev. Cell 16: 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Silberg DG, Sullivan J, Kang E, Swain GP, Moffett J, Sund NJ, Sackett SD, Kaestner KH. 2002. Cdx2 ectopic expression induces gastric intestinal metaplasia in transgenic mice. Gastroenterology 122: 689–696 [DOI] [PubMed] [Google Scholar]
- 16. Hryniuk A, Grainger S, Savory JG, Lohnes D. 2012. Cdx function is required for maintenance of intestinal identity in the adult. Dev. Biol. 363: 426–437 [DOI] [PubMed] [Google Scholar]
- 17. Stringer EJ, Duluc I, Saandi T, Davidson I, Bialecka M, Sato T, Barker N, Clevers H, Pritchard CA, Winton DJ, Wright NA, Freund JN, Deschamps J, Beck F. 2012. Cdx2 determines the fate of postnatal intestinal endoderm. Development 139: 465–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Verzi MP, Shin H, Ho LL, Liu XS, Shivdasani RA. 2011. Essential and redundant functions of caudal family proteins in activating adult intestinal genes. Mol. Cell. Biol. 31: 2026–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Beck F. 2004. The role of Cdx genes in the mammalian gut. Gut 53: 1394–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. He HH, Meyer CA, Shin H, Bailey S, Wei G, Wang Q, Zhany Y, Xu K, Ni M, Lupien M, Mieczkowski P, Lieb JD, Zhao K, Brown MA, Liu XS. 2010. Positioned nucleosomes flanking a labile nucleosome characterize transcriptional enhancers. Nat. Genet. 42: 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39: 311–318 [DOI] [PubMed] [Google Scholar]
- 22. Verzi MP, Shin H, He HH, Sulahian R, Meyer CA, Montgomery RK, Fleet JC, Brown M, Liu XS, Shivdasani RA. 2010. Differentiation-specific histone modifications reveal dynamic chromatin interactions and partners for the intestinal transcription factor CDX2. Dev. Cell 19: 713–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Olsen L, Bressendorff S, Troelsen JT, Olsen J. 2005. Differentiation-dependent activation of the human intestinal alkaline phosphatase promoter by HNF-4 in intestinal cells. Am. J. Physiol. Gastrointest Liver Physiol. 289: G220–226 [DOI] [PubMed] [Google Scholar]
- 24. Stegmann A, Hansen M, Wang Y, Larsen JB, Lund LR, Ritie L, Nicholson JK, Quistorff B, Simon-Assmann P, Troelsen JT, Olsen J. 2006. Metabolome, transcriptome, and bioinformatic cis-element analyses point to HNF-4 as a central regulator of gene expression during enterocyte differentiation. Physiol. Genomics 27: 141–155 [DOI] [PubMed] [Google Scholar]
- 25. Davidson EH, Levine MS. 2008. Properties of developmental gene regulatory networks. Proc. Natl. Acad. Sci. U. S. A. 105: 20063–20066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weiser MM. 1973. Intestinal epithelial cell surface membrane glycoprotein synthesis. I. An indicator of cellular differentiation. J. Biol. Chem. 248: 2536–2541 [PubMed] [Google Scholar]
- 27. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, Liu XS. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137 doi:10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat. Biotechnol. 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Y, Shin H, Song JS, Lei Y, Liu XS. 2008. Identifying positioned nucleosomes with epigenetic marks in human from ChIP-Seq. BMC Genomics 9: 537 doi:10.1186/1471-2164-9-537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meyer CA, He HH, Brown M, Liu XS. 2011. BINOCh: binding inference from nucleosome occupancy changes. Bioinformatics 27: 1867–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shin H, Liu T, Manrai AK, Liu XS. 2009. CEAS: cis-regulatory element annotation system. Bioinformatics 25: 2605–2606 [DOI] [PubMed] [Google Scholar]
- 32. Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, Shin H, Wong SS, Ma J, Lei Y, Pape UJ, Poidinger M, Chen Y, Yeung K, Brown M, Turpaz Y, Liu XS. 2011. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 12: R83 doi:10.1186/gb-2011-12-8-r83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5: R80 doi:10.1186/gb-2004-5-10-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smyth GK. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: Article3. [DOI] [PubMed] [Google Scholar]
- 35. Bulger M, Groudine M. 2010. Enhancers: the abundance and function of regulatory sequences beyond promoters. Dev. Biol. 339: 250–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. MacArthur S, Li XY, Li J, Brown JB, Chu HC, Zeng L, Grondona BP, Hechmer A, Simirenko L, Keranen SV, Knowles DW, Stapleton M, Bickel P, Biggin MD, Eisen MB. 2009. Developmental roles of 21 Drosophila transcription factors are determined by quantitative differences in binding to an overlapping set of thousands of genomic regions. Genome Biol. 10: R80 doi:10.1186/gb-2009-10-7-r80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Panne D. 2008. The enhanceosome. Curr. Opin. Struct. Biol. 18: 236–242 [DOI] [PubMed] [Google Scholar]
- 38. Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- 40. Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U. S. A. 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. 2001. Hepatocyte nuclear factor 4α (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell. Biol. 21: 1393–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, Robine S. 2004. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39: 186–193 [DOI] [PubMed] [Google Scholar]
- 45. Ahn SH, Shah YM, Inoue J, Morimura K, Kim I, Yim S, Lambert G, Kurotani R, Nagashima K, Gonzalez FJ, Inoue Y. 2008. Hepatocyte nuclear factor 4α in the intestinal epithelial cells protects against inflammatory bowel disease. Inflamm. Bowel Dis. 14: 908–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Babeu JP, Darsigny M, Lussier CR, Boudreau F. 2009. Hepatocyte nuclear factor 4α contributes to an intestinal epithelial phenotype in vitro and plays a partial role in mouse intestinal epithelium differentiation. Am. J. Physiol. Gastrointest. Liver Physiol. 297: G124–134 [DOI] [PubMed] [Google Scholar]
- 47. Taylor IC, Workman JL, Schuetz TJ, Kingston RE. 1991. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 5: 1285–1298 [DOI] [PubMed] [Google Scholar]
- 48. Subramanian V, Meyer BI, Gruss P. 1995. Disruption of the murine homeobox gene Cdx1 affects axial skeletal identities by altering the mesodermal expression domains of Hox genes. Cell 83: 641–653 [DOI] [PubMed] [Google Scholar]
- 49. Mutoh H, Hakamata Y, Sato K, Eda A, Yanaka I, Honda S, Osawa H, Kaneko Y, Sugano K. 2002. Conversion of gastric mucosa to intestinal metaplasia in Cdx2-expressing transgenic mice. Biochem. Biophys. Res. Commun. 294: 470–479 [DOI] [PubMed] [Google Scholar]
- 50. Verzi MP, Hatzis P, Sulahian R, Philips J, Schuijers J, Shin H, Freed E, Lynch JP, Dang DT, Brown M, Clevers H, Liu XS, Shivdasani RA. 2010. TCF4 and CDX2, major transcription factors for intestinal function, converge on the same cis-regulatory regions. Proc. Natl. Acad. Sci. U. S. A. 107: 15157–15162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Svaren J, Schmitz J, Horz W. 1994. The transactivation domain of Pho4 is required for nucleosome disruption at the PHO5 promoter. EMBO J. 13: 4856–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Venter U, Svaren J, Schmitz J, Schmid A, Horz W. 1994. A nucleosome precludes binding of the transcription factor Pho4 in vivo to a critical target site in the PHO5 promoter. EMBO J. 13: 4848–4855 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.