Abstract
There are nearly 50 forkhead (FOX) transcription factors encoded in the human genome and, due to sharing a common DNA binding domain, they are all thought to bind to similar DNA sequences. It is therefore unclear how these transcription factors are targeted to specific chromatin regions to elicit specific biological effects. Here, we used chromatin immunoprecipitation followed by sequencing (ChIP-seq) to investigate the genome-wide chromatin binding mechanisms used by the forkhead transcription factor FOXM1. In keeping with its previous association with cell cycle control, we demonstrate that FOXM1 binds and regulates a group of genes which are mainly involved in controlling late cell cycle events in the G2 and M phases. However, rather than being recruited through canonical RYAAAYA forkhead binding motifs, FOXM1 binding is directed via CHR (cell cycle genes homology region) elements. FOXM1 binds these elements through protein-protein interactions with the MMB transcriptional activator complex. Thus, we have uncovered a novel and unexpected mode of chromatin binding of a FOX transcription factor that allows it to specifically control cell cycle-dependent gene expression.
INTRODUCTION
There are nearly 50 different forkhead transcription factors encoded in mammalian genomes, and these proteins all contain the conserved forkhead DNA binding domain (reviewed in references 1 and 2). Forkhead transcription factors are involved in controlling a wide range of biological processes and are aberrantly expressed or regulated in disease states, including cancer (reviewed in reference 2). However, due to sharing a common DNA binding domain, forkhead transcription factors are generally believed to bind to variations of the RYAAAYA motif. Hence, it is unclear how individual forkhead proteins are specifically recruited to the regulatory regions of different cohorts of target genes to control defined biological responses. One key process which is controlled by forkhead transcription factors is the cell cycle and, in particular, the G2-M transition. The initial links to G2-M control were made with the Saccharomyces cerevisiae forkhead protein Fkh2, which controls the temporal expression of a cluster of genes at this phase of the cell cycle (reviewed in reference 3). More recently, members of the FOXO and FOXM classes of forkhead transcription factors have been linked with controlling the same process in mammalian cells (4–6). In both cases, forkhead transcription factors coordinate the integration of signals from the cell cycle regulatory machinery to transcriptional outputs. This is exemplified by the links to the cell cycle regulated Polo-like kinase PLK1, which is recruited to cell cycle-regulated promoters through promoter elements bound by the forkhead transcription factors FOXM1 and Fkh2, albeit indirectly in the case of Fkh2 (7, 8).
In mammalian cells, the transcriptional control of a cluster of genes at the G2-M transition, is coordinated through promoter elements which typically contain CHR (cell cycle genes homology region) and CDE (cell cycle-dependent element) motifs. In addition, there are usually closely associated CCAAT boxes for the recruitment of the NF-Y transcription factor (reviewed in reference 9). The CHR is typically located at or close to the transcriptional start site. Recently, it was shown that the CHR element is bound by the DREAM and MMB transcriptional regulatory complexes, and these complexes play a role in controlling cell cycle-dependent transcription of genes expressed at the G2-M border (10). The DREAM and MMB complexes are functionally inter-related, and both contain the MuvB core complex, which includes LIN9, LIN37, LIN52, LIN54, and RBBP4 in addition to specific additional subunits in each case (11, 12). The DREAM complex is repressive in nature and contains additional subunits such as p130 and E2F4, whereas the MMB complex is thought to activate transcription and contains B-MYB (MYBL2). Although FOXM1 is known to play a role in controlling the expression of the same class of genes as bound by the DREAM and MMB complexes, it is unclear how these complexes interact. Indeed, there appear to be no conserved canonical forkhead binding motifs within proximal promoters of genes expressed during G2/M, that correspond to the classic RYAAAYA motif recognized by most members of this transcription factor family. Several studies have implicated upstream forkhead binding motifs in mediating the response of a subset of G2-M genes to FOXM1 (see, for example, reference 6), but this has not been systematically investigated across all of the genes activated during this part of the cell cycle.
Here, we have used chromatin immunoprecipitation followed by sequencing (ChIP-seq) to interrogate the chromatin binding profile of FOXM1 in a genome-wide manner. In keeping with its known role in cell cycle control, FOXM1 is generally bound to the regulatory regions of genes which encode proteins involved in mitotic regulation. However, surprisingly, FOXM1 binding is not associated with forkhead DNA binding motifs but instead coincides with CHR elements. Rather than binding to the CHR directly, FOXM1 interacts with the MMB complex and is recruited to chromatin via interactions with this complex. Thus, FOXM1 has an atypical mode of chromatin association which distinguishes it from other family members and allows it to have a specific role in cell cycle control.
MATERIALS AND METHODS
Plasmid constructs.
The human CCNB1 promoter reporter plasmids pAS3017 (pGL4.10-hCCNB1 WT) and pAS3019 (pGL4.10-hCCNB1 CHR mutant) were derived from hB1-Luci constructs that have been described earlier (13). pRL-CMV-vector (Promega), pAS188 (pCMV5 empty vector), and pAS1175 (pCMV5-FOXM1b) are used in luciferase assays. pAS1175 (encoding FOXM1b with Flag and hexahistidine C-terminal tags) was constructed by a two-step procedure, first by cloning a HindIII/XhoI-cut PCR product (primers ADS1177/ADS1178 on template IMAGE clone 3834244) into pAS728 to create pAS1171, followed by cloning a HindIII/XbaI fragment from pAS1171 into the same sites in pCMV5. The plasmids pAS3048 (p3×Flag-FOXM1b-WT) (a gift from Suyun Huang) and pAS3069 [p3×Flag-FOXM1b(Δ1-116)] were used in ChIP assays. The plasmids used in glutathione S-transferase (GST) pulldown experiments were pAS3059 [GST-FOXM1b(1-367)], pAS3060 [GST-FOXM1b(117-367)], pAS3061 [GST-FOXM1b(235-367], pAS3062 [GST-FOXM1b(235-490)], pAS3068 [GST-FOXM1b(451-748)], pAS3066 [GST-FOXM1b(1-130)], and pAS3067 [GST-FOXM1b(1-235)], which were generated by ligating EcoRI/XhoI-cut PCR products generated with the oligonucleotide pairs ADS2908/2911, ADS2909/2911, ADS2910/2911, ADS2910/2912, ADS2932/2913, ADS2908/2918, and ADS2908/2919, respectively, into pGEX-6p1 vector (GE Healthcare).
Tissue culture, transfection, reporter assay, and reverse transcription-PCR.
U2OS, HEK293/293T, NIH 3T3, and HCT116 cells were maintained in Dulbecco modified Eagle medium containing 10% fetal bovine serum. To block cells at G1/S, U2OS cells were cultured in medium containing 2 mM thymidine (Sigma; catalog no. T1895) for 16 h, followed by a release in fresh medium for 12 h and then in medium containing 2 mM thymidine for another 16 h.
Lipofectamine RNAiMAX (Invitrogen) transfection reagent was used for small interfering RNA (siRNA) transfection; X-tremeGene HP DNA (Roche) and Polyfect (Qiagen) transfection reagent were used for plasmid transfections. At 24 to 48 h after transfection, the cells were harvested for further analyses. All siRNAs were ON-TARGETplus SMART pools from Dharmacon, and a final concentration of 20 nM was used, except for the siRNA against mouse FoxM1, which was described previously (6). Control nontargeting siRNAs (Dharmacon) were used throughout. For the luciferase assays, 12-well plates were used, and for each well the cells were transfected with 200 ng of reporters containing the appropriate promoters, 10 ng of Renilla luciferase plasmid pGL4.70, and 790 ng of empty or FOXM1 expression plasmids. At 24 h after transfection, NIH 3T3 cells were synchronized in G2/M by serum deprivation for 60 h, followed by restimulation as described previously (10), and harvested for a luciferase assay using a dual-luciferase reporter assay system (Promega) according to the manufacturer's instructions. Real-time reverse transcription-quantitative PCR (RT-qPCR) was carried out as described previously (14). The primer-pairs used for RT-qPCR experiments are available on request.
Western blotting, coimmunoprecipitation assays, immunofluorescence, and FACS analysis.
Western blotting was carried out with primary antibodies as follows: Flag M2 (F1804 or F3165; Sigma), FOXM1 (C-20; Santa Cruz), LIN9 (ab62329; Abcam), LIN37 (kindly provided by J. DeCaprio [11]), B-MYB (BMyb LX015.1; kindly provided by R. Watson [15]), p130 (C-20; Santa Cruz), E2F4 (C-20; Santa Cruz), and NFYA (G2; Santa Cruz). Coimmunoprecipitation was carried out using the Miltenyi green fluorescent protein (GFP) kit (Miltenyi Biotech) for GFP-tagged FOXM1 according to the manufacturer's instructions. For analyzing interactions with Flag-tagged FOXM1, the same protocol was used, but Flag-agarose beads (Sigma) were used instead. The proteins were detected either by chemiluminescence with SuperSignal West Dura substrate (Pierce) and visualized with a Fluor-S MultiImager (Bio-Rad) or for infrared dye-conjugated antibodies, the signal was collected with a Li-Cor Odyssey infrared imager. Immunofluorescence experiments and fluorescence-activated cell sorting (FACS) analyses of cell cycle were performed as previously described (16).
GST pulldown assays.
GST fusion proteins were purified from Escherichia coli, and pulldown assays were performed essentially as described previously (17), except that the source of the interacting proteins was total U2OS cell lysates. Ethidium bromide (200 μg/ml) was included in the binding and washing buffers where indicated.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays using control IgG (Millipore) or antibodies specific to FOXM1 (C-20; Santa Cruz) or Flag (M2; F3165; Sigma) were performed essentially as described previously (18) using 1 × 106 to 5 × 106 U2OS cells for a standard ChIP. ChIP assays after FOXM1 overexpression were performed as described earlier using HCT116 cells with integrated reporter constructs (10). Bound regions were detected by quantitative PCR using primers (sequences available upon request). Quantitative PCR was performed at least in duplicate, from at least two independent experiments, and analyzed as described previously (19).
ChIP-seq assays.
A total of 4.8 ×107 U2OS cells were subjected to a double thymidine block as described above. At 10 h after release from this double thymidine block, the cells were harvested for ChIP. Then, 4.5 μg of anti-FOXM1 (C-20) was used per experiment. The ChIPed DNA and input DNA were amplified as previously described (20); libraries were generated, and sequencing was performed on an Illumina GAIIx genome analyzer according to the manufacturer's protocols.
Bioinformatics analysis.
All software was run in their own default settings, unless otherwise indicated. Sequences from the ChIP-seq analysis (using 36-bp reads) were aligned against the National Center for Biotechnology Information build 36.3/hg18 of the human genome using MAQ (http://maq.sourceforge.net/), allowing up to two mismatches. Two biological replicas were performed yielding 36,900,198 and 11,465,752 reads in the two experiments. Only reads that were uniquely mapped to the genome were preserved, yielding 27,892,797 and 8,906,067 reads, respectively. After reads which were mapped to mitochondria genome were removed, the peaks were called by MACS (version 1.4.1) (21). The false discovery rate (FDR) cutoff was set as 0.1, and only peaks that were present in both replicas were preserved, giving a total of 277 peaks. The peaks containing >70% repeat sequences and having <3-fold enrichment over local background were then manually removed, yielding a total of 270 peaks.
Motif discovery was performed by HOMER (22) using the sequences within ±100 bp around the FOXM1 summits and the default background settings, i.e., sequences randomly selected from the genome with the same GC percentage content as the target sequences. Nearest genes were assigned to peaks by HOMER and Gene Ontology (GO) analyses were performed by GREAT. Genomic distributions were determined by using CEAS (23).
For comparison with LIN9 and B-MYB ChIP-seq, FOXM1 binding sites (hg18) and mapped reads (hg18) were converted to hg19 using UCSC LiftOver tool (http://hgdownload.cse.ucsc.edu/admin/exe/). The K-means linear clustering method was used, and heat maps were generated by seqMiner (24).
The recently published microarray data sets profiling gene expression of the HeLa cell cycle (25) were downloaded from Gene Expression Omnibus (accession number GSE27031). Among the genes in the microarray data set, a set of genes that are associated with FOXM1 binding regions were selected. Z scores of each of these selected genes throughout the expression profiles were computed to compare the expression patterns in the scale of statistical standard deviation. The scores were visualized in a heat map to present various expression patterns throughout the cell cycle.
GEO accession number.
The ChIP-seq data have been submitted to GEO under accession number GSE38170.
Statistical analysis.
Statistical analysis for qRT-PCR studies and ChIP assays were performed using the Student t test. The error bars in all graphs represent standard deviation.
RESULTS
FOXM1 binds at the promoter regions of genes involved in mitotic control.
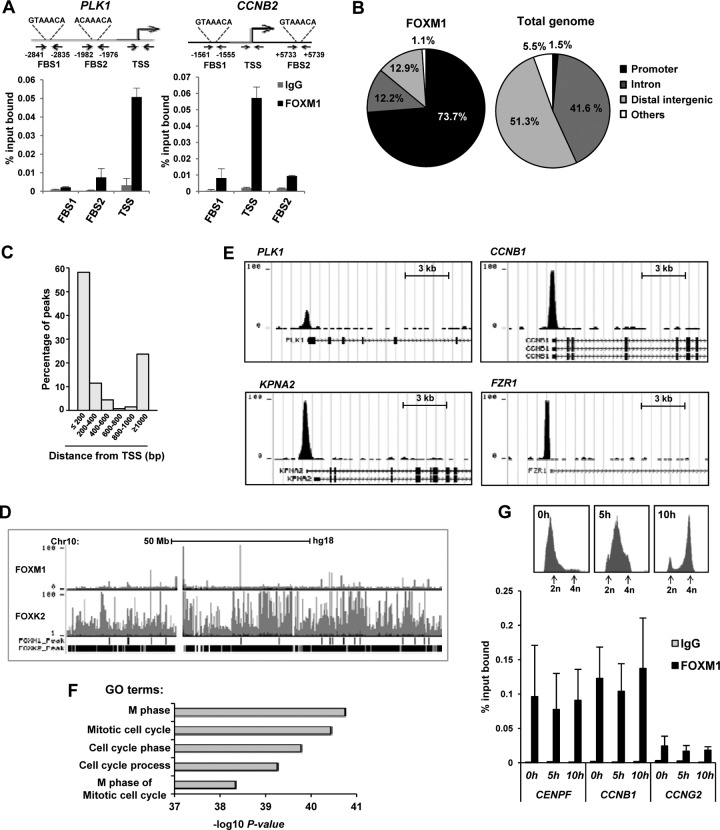
To gain further insight into how FOXM1 controls its target genes, we first used ChIP in U2OS cells to analyze the binding of endogenous FOXM1 to the promoter regions of two well-characterized targets, PLK1 and CCNB2. However, although we could detect weak binding to regions containing forkhead binding sites (FBS) in both cases, substantially more binding was observed in the vicinity of the transcriptional start sites (TSS) which lacked obvious FBS (Fig. 1A). Given this unexpected finding, we performed ChIP-seq analysis in U2OS cells to determine whether FOXM1 followed this type of binding pattern on a genome-wide scale. To maximize our chances of identifying target genes, we performed ChIP-seq on U2OS cells released from a double thymidine block for 10 h to allow the cells to accumulate in the late G2 and M phases. Using MACS (21), we identified 270 high-confidence binding regions for FOXM1 which were observed in two independent experiments (see Table S1 in the supplemental material). The majority of these regions are located in close proximity to the promoter, with 74% either in the 5′ untranslated region (UTR) or within 1 kb upstream from the TSS (Fig. 1B). More detailed analysis showed that 58% of the FOXM1 binding regions are located within 200 bp of the TSS (Fig. 1C) and in keeping with this analysis, 77% of FOXM1 binding regions are associated with CpG islands. The low number of binding regions was somewhat surprising, given the large numbers typically observed for transcription factors; however, when a different peak caller, HOMER (22), was used only 237 peaks were identified, and these showed a substantial overlap (74%) with those called by MACS. Moreover, comparison of the binding profiles for FOXM1 and a different forkhead transcription factor FOXK2 (26) across entire chromosomes demonstrated that the small number of FOXM1 binding regions is unlikely due to a failure of the peak calling algorithms (Fig. 1D). Binding regions were identified in the promoter regions of known target genes such as PLK1 and CCNB1 but also in the regulatory regions of novel targets such as FZR1 and KPNA2 (Fig. 1D). Interestingly, FZR1 (also known as CDH1) has a known role in late cell cycle control as part of the APC/C ubiquitin ligase complex (reviewed in reference 27), but although KPNA2 is characterized as a nuclear import receptor, little is known about function in this context. To establish whether more of the FOXM1 target genes might also encode proteins involved in controlling late cell cycle events, we used GREAT (28) to first associate the binding regions with potential target genes and then determine whether these genes are involved in common biological processes. All of the most significant gene ontology (GO) terms that we identified are associated with the cell cycle and in particular mitotic events (Fig. 1F). Given that FOXM1 binding appears to be specifically associated with genes involved in mitotic events, we also tested whether FOXM1 is also associated with regulatory regions at other phases of the cell cycle. FOXM1 binding was detected in the promoter regions of three different targets—CENPF, CCNB1, and CCNG2—in cells enriched in the G1 phase by treatment with thymidine and also in cells released from this block that had accumulated in the S or in the G2 and M phases of the cell cycle (Fig. 1G). Little difference in the magnitude of binding under each condition is observed. Thus, FOXM1 binds largely to proximal promoter regions in a manner that is largely independent of cell cycle phase and potentially plays a unique and widespread role in controlling late cell cycle events.
Fig 1.
FOXM1 binds to the promoter regions of genes involved in late cell cycle control. (A) ChIP analysis of FOXM1 binding to the PLK1 and CCNB2 promoters in asynchronously growing U2OS cells, using the indicated primer pairs. (B) Distribution of FOXM1 ChIP-seq regions (left) compared to the total genomic DNA distribution (right). The sector corresponding to the promoter includes sequences up to 1 kb upstream from the TSS or in the 5′ UTR. (C) Distribution of peaks summit distances from the TSS. (D) Screenshot from the UCSC browser showing the distribution of FOXM1 and FOXK2 binding peaks in U2OS cells across chromosome 10. (E) Example FOXM1 binding peak profiles for the indicated genes. (F) The top five overrepresented gene ontology (GO) terms in genes associated with FOXM1 binding regions. (G) ChIP analysis of FOXM1 binding to the PLK1 and CCNB2 promoters in U2OS cells released from a double thymidine block for the indicated times, using the indicated primer pairs. DNA content profiles of U2OS cells at the indicated time points following release from a double thymidine block are shown above the graph. DNA content is determined by propidium iodide (PI) staining and peaks corresponding to cells before (2n) and after (4n) DNA replication are shown.
FOXM1 activates a program of late cell cycle genes.
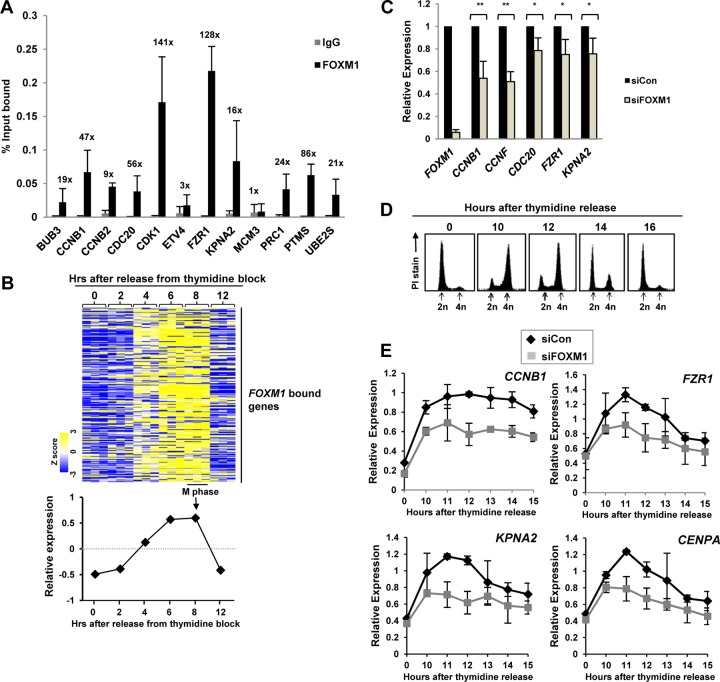
To validate the ChIP-seq data, we performed ChIP-qPCR on 13 different FOXM1 binding regions. Twelve of these regions showed enrichment for FOXM1 binding compared to nonspecific IgG (Fig. 2A). Next, we determined whether the FOXM1 target genes are cyclically expressed during the late G2 and M phases. We took advantage of a recent microarray study that analyzed the temporal gene expression patterns during the late cell cycle (25) and investigated the expression profiles of the 270 genes associated with the FOXM1 binding regions (Fig. 2B). These genes showed a strong cyclical expression pattern, with peak activity as the cells enter the late G2 and M phases, strongly suggesting that FOXM1 has a role in controlling their temporal cell cycle-dependent expression. To directly test this hypothesis, we first depleted FOXM1 and analyzed the expression of four of the newly identified target genes in asynchronously growing U2OS cells. Significant decreases in the expression of all of these genes were observed (Fig. 2C). Moreover, we also examined the effect of FOXM1 depletion on the temporal cell cycle-dependent activation kinetics of 10 of its target genes in U2OS cells released from a double thymidine block and allowed to progress through to the late G2 and M phases (Fig. 2D and E). As expected, FOXM1 depletion caused decreased expression of known targets such as CCNB1, but it also resulted in reduced cell cycle-dependent activation among all of the other targets tested (Fig. 2E and data not shown). Thus, FOXM1 both binds and regulates the target genes we identified by ChIP-seq.
Fig 2.
FOXM1 directly activates late cell cycle genes. (A) ChIP-qPCR validation of FOXM1 binding to regions associated with the indicated genes. Experiments were carried out with three biological repeats. Bars indicate the average percentages of input precipitated with the FOXM1 antibody or nonspecific IgG with the standard deviations. The numbers above each set of bars represent the fold increase in signal with FOXM1 over IgG. (B) Heat map from an expression microarray analysis performed in HeLa cells after release from a double thymidine block (25) of the 270 genes that are associated with FOXM1 binding regions. The graph below summarizes the average Z score of the expression profiles of this group of genes. (C and E) RT-qPCR analysis of the expression of the indicated genes in U2OS cells grown asynchronously (C) or subjected to a double thymidine block and released for the indicated times (E). Cells were pretreated with a nontargeting siRNA (siCon) or siRNA against FOXM1. The data are the averages of two (E) or three (C) experiments and are shown for each gene relative to its expression in the presence of the control siRNA (taken as 1 [C]) or the internal control gene HMBS (E). ** and * represent P values of <0.01 and <0.05, respectively. (D) DNA content profiles of U2OS cells at the indicated time points following release from a double thymidine block. DNA content is determined by propidium iodide (PI) staining, and peaks corresponding to cells before (2n) and after (4n) DNA replication are shown.
FOXM1 binds to regions containing CHR motifs.
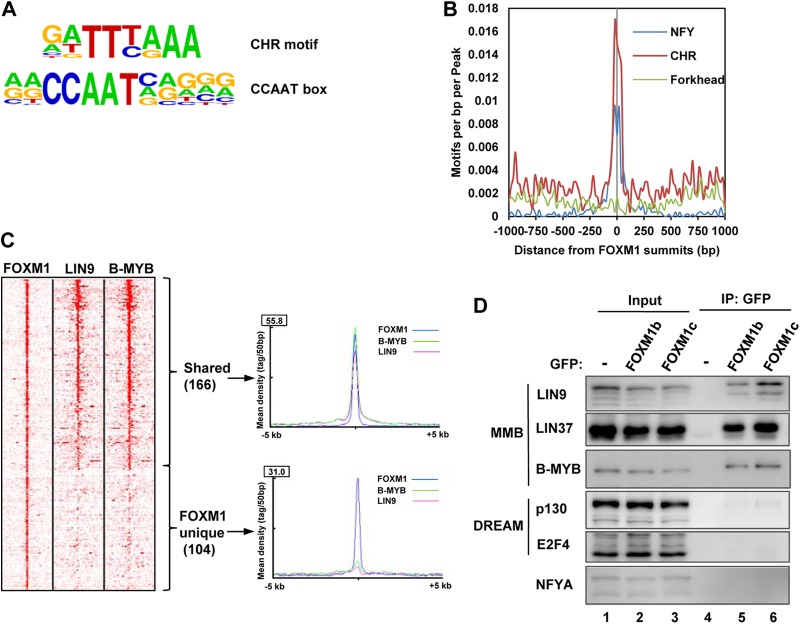
Next, we searched the FOXM1 binding regions for overrepresented DNA binding motifs using HOMER (22). We expected to find motifs related to the canonical FBS (RYAAAYA) bound by FOX proteins but instead two other motifs, the CCAAT-box and the CHR, were identified as significantly enriched (Fig. 3A) (P < 10−110). Matches to these motifs were found in >60% of all regions in both cases, and the motifs are located very close to the FOXM1 binding peak summits (Fig. 3B). Similar results were returned using two alternative motif finding tools, MEME and Weeder, and the same two elements were returned as the most enriched motifs in the FOXM1 binding regions. In contrast, none of these motif finding tools detected overrepresentation of motifs resembling the FBS, and the motifs present within the FOXM1 binding regions were dispersed throughout the region rather than being located close to the peak summit as would be expected (Fig. 3B). To further substantiate this finding, we searched for the occurrence of the FBS, RYAAAYA, within 200 bp of the summit of the FOXM1 binding regions and within 270 random sequences from the genome with a similar genomic distribution. Only 43 FOXM1 binding regions contained RYAAAYA, but 64 random sequences contained RYAAAYA, indicating a clear lack of enrichment of RYAAAYA sites within FOXM1 binding peaks.
Fig 3.
FOXM1 binds to CHR-containing regions and interacts with the MMB complex. (A) TFBS logos for the two top ranking motifs identified by HOMER when searching ±100 bp from the summits of the FOXM1-bound regions. (B) Frequency of CCAAT, CHR, and forkhead DNA binding motif occurrence in 20-bp bins relative to the summits of the FOXM1 binding regions. (C) Heat maps showing FOXM1, LIN9, and B-MYB ChIP-seq tag densities in the 10-kb region surrounding the summit of the FOXM1 binding regions. Regions are clustered using K-means linear clustering according to similar tag density profiles. The graphs on the right show the mean tag densities across all regions in either the “shared” or “unique” categories of binding regions for these proteins. (D) Coimmunoprecipitation analysis (IP) of GFP alone (−) or GFP-tagged FOXM1b or FOXM1c and the indicated endogenously expressed DREAM and MMB complex proteins from HEK293 cells.
Recently, the DREAM and MMB complexes were shown to bind to regions containing the CHR motif (10). We therefore compared the binding profile of FOXM1 to those of LIN9 (a core component of the DREAM and MMB complexes) and B-MYB (a MMB-specific component) from ChIP-seq experiments performed in proliferating HeLa cells (25). A large overlap between FOXM1 and both LIN9 and B-MYB binding was observed, with 62% of regions sharing the same binding profile (Fig. 3C), although a number of apparently unique FOXM1 binding regions were observed. The coincidental binding of FOXM1 with the DREAM and MMB complex subunits suggested that FOXM1 might interact with one or both of these complexes. We therefore performed coimmunoprecipitation analysis from cells transfected with enhanced GFP (EGFP) or EGFP fused to either the FOXM1b or FOXM1c isoforms (Fig. 3D). The two isoforms are both transcriptional activators and differ by the inclusion of an additional alternatively spliced exon (A1) in FOXM1c. We observed that both FOXM1 isoforms interacted with the MuvB core complex components LIN9 and LIN37, as well as with the MMB-specific component B-MYB. In contrast, little binding could be detected to the DREAM components p130 and E2F4. Similarly, little binding could be detected between FOXM1 and the CCAAT-box binding protein NF-YA (Fig. 3D), suggesting that the appearance of the CCAAT-box as an overrepresented motif is due to its common cooccurrence with the CHR in promoter regions (10).
These results suggest that FOXM1 is recruited to regulatory regions through the CHR motif, and its interactions with the MMB complex suggest that this regulatory complex might be important in mediating this effect.
FOXM1 is recruited to chromatin via the CHR element.
To establish the importance of the CHR in recruiting FOXM1 to regulatory regions, we first examined the role of this element in FOXM1-mediated transcriptional activation. Overexpression of FOXM1 resulted in the expected activation of a CCNB1 promoter-driven luciferase reporter gene (Fig. 4A). However, this activation was severely curtailed upon mutation of the CHR element. Conversely, depletion of FoxM1 caused a decrease in the activity of a Ccnb1 promoter-driven reporter construct, but this decrease was lost when a mutated reporter lacking an intact CHR element was tested (Fig. 4B). These results pointed to an important role for the CHR element, and this conclusion was further substantiated by the observation that mutation of the CHR element lead to a decrease in the binding of FOXM1 to an integrated Ccnb2 promoter-driven transgene in vivo (Fig. 4C). This observation gives strong evidence that it is in fact the CHR and not another site serving as the element through which a FOXM1-containing complex contacts the DNA.
Fig 4.
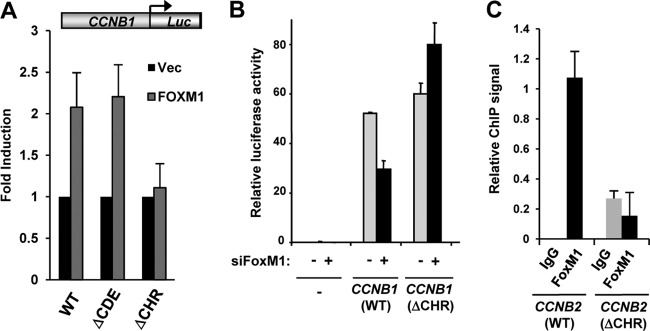
FOXM1 is recruited via the CHR element. (A) Transient reporter gene assay on wild-type (WT) and mutant (ΔCDE and ΔCHR) CCNB1-driven luciferase reporters in U2OS cells in the presence or absence (Vec) of FOXM1. The data are averages of three independent experiments. (B) Transient reporter gene assays in NIH 3T3 cells with vector control (−) or wild-type (WT) or mutant (ΔCHR) Ccnb1-driven luciferase genes. NIH 3T3 cells were treated with control siRNAs (siFoxM1: −) or siRNA against FoxM1 (+) and blocked in G0 by serum deprivation and released for 24 h to accumulate in G2-M. The data are averages of duplicate experiments. (C) ChIP assays of transiently transfected Flag-tagged FOXM1 in HCT116 cells containing wild-type (WT) or mutant (ΔCHR) stably integrated mouse Ccnb2 transgenes. ChIP was performed with nonspecific IgG or anti-Flag antibodies; the data are the averages of two experiments and are presented as binding to each transgene relative to binding to the endogenous locus.
The MMB binding domain in FOXM1 is required for its recruitment to chromatin.
Since the CHR is also important for recruitment of the MMB complex, we examined the relationship between FOXM1 and MMB binding to chromatin. Depletion of LIN9 in U2OS cells caused reductions of FOXM1 binding to a panel of different FOXM1 target genes (Fig. 5A). However, a slight decrease on FOXM1 expression was also observed (Fig. 5B). We therefore expressed exogenous FOXM1 in HEK293 cells due to their higher transfection efficiencies and examined whether LIN9 and/or B-MYB depletion affected FOXM1 binding in this context (Fig. 5C). Again, significant reductions of FOXM1 binding to many of its target genes were observed, suggesting a role for components in the MMB complex in FOXM1 recruitment. Since FOXM1 is thought to act as a transcriptional activator, it would be predicted that depletion of LIN9 and the loss of FOXM1 binding would be accompanied by reductions in gene activation. Indeed, this appears to be the case, since siRNA-mediated LIN9 depletion caused reductions in the expression of all six of the FOXM1 targets that we tested (Fig. 5D).
Fig 5.
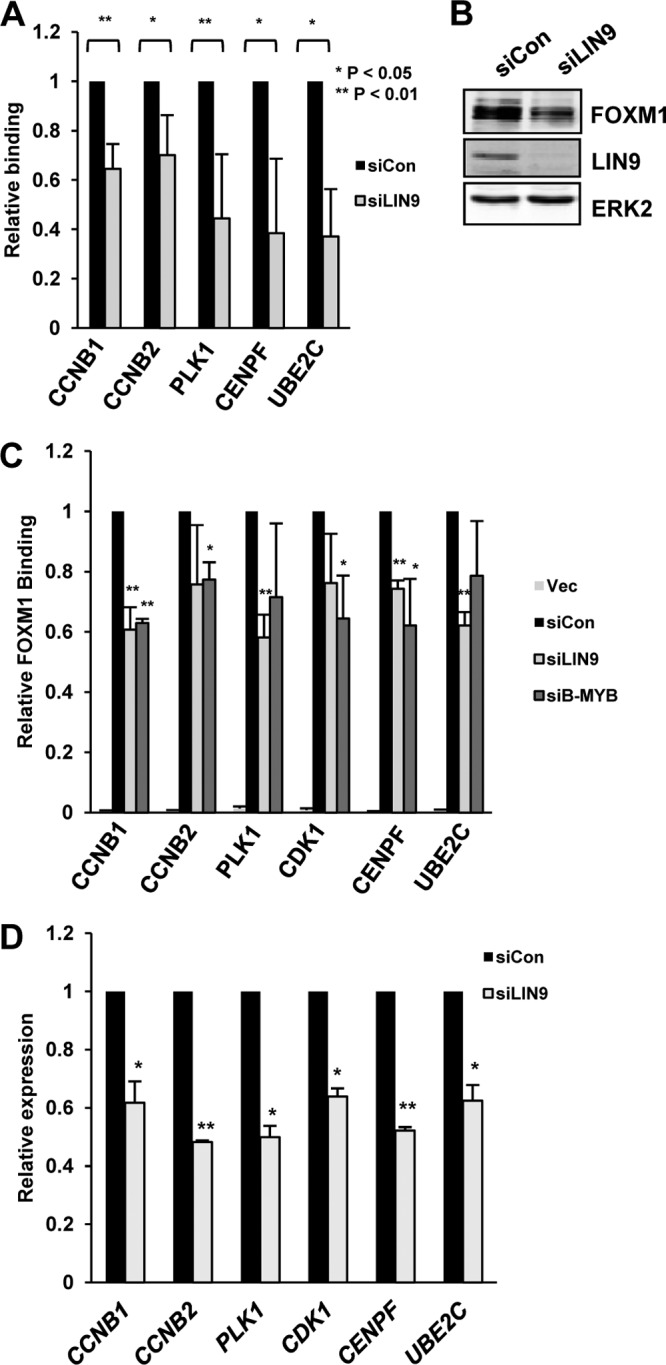
FOXM1 recruitment depends on the MMB complex. (A) ChIP assay of endogenous FOXM1 was performed in U2OS cells on the promoters of the indicated genes after knockdown of LIN9 or in the presence of a nontargeting control siRNA (siCon). (B) Western blot of FOXM1 and LIN9 levels after knockdown of LIN9 or in the presence of a nontargeting control siRNA (siCon). ERK2 represents a loading control. (C) ChIP assays of transiently transfected Flag-tagged FOXM1 in HEK293T cells. FOXM1 ChIP was performed on promoters of the indicated genes after knockdown of LIN9 or B-MYB or in the presence of a nontargeting control siRNA (siCon). “Vec” represents cells transfected with empty vector. The data for both ChIP experiments are the average of two independent experiments. ** and * represent P values of <0.01 and <0.05, respectively. (D) RT-qPCR analysis of the expression of the indicated genes in asynchronously growing U2OS cells. Cells were treated with a nontargeting siRNA (siCon) or a siRNA against LIN9. The data are the average of two experiments and are shown for each gene relative to its expression in the presence of the control siRNA (taken as 1). ** and * represent P values of <0.001 and <0.01, respectively.
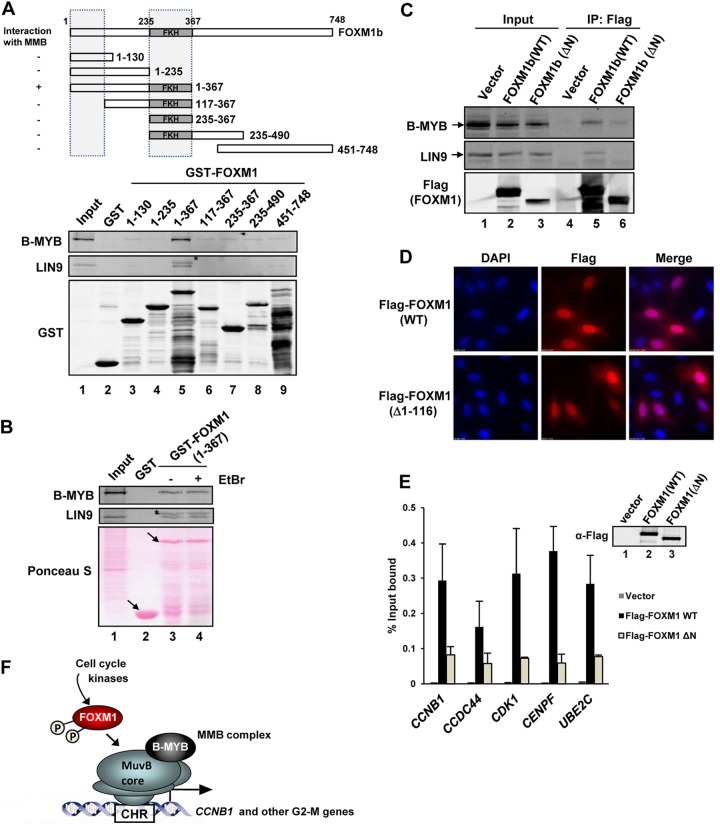
Together, these results therefore can be combined in a model wherein the MMB complex binds and recruits FOXM1 to chromatin through the CHR element. To test this, we first mapped the MMB interacting region(s) on FOXM1 using GST pulldown assays. Both the forkhead domain and a region located in the N-terminal 116 amino acids of FOXM1 are required for MMB complex binding and the minimum region of FOXM1 which is sufficient for interaction encompasses amino acids 1 to 367 (Fig. 6A). Interaction with the MMB complex via nonspecific DNA binding through the Forkhead domain was ruled out by repeating the assays in the presence of ethidium bromide (Fig. 6B). To establish whether the same region of FOXM1 is important in binding to the MMB complex in vivo, we performed coimmunoprecipitation analysis in U2OS cells transfected with wild-type FOXM1 or with an N-terminally truncated FOXM1 mutant that lacks the MMB binding region [FOXM1(Δ1-116)]. Consistent with the effects seen in vitro, deletion of the N-terminal region of FOXM1 led to reduced binding to both B-MYB and LIN9 (Fig. 6C, lanes 5 and 6). We then examined the ability of the N-terminally truncated FOXM1 mutant to be recruited to chromatin in vivo. This mutant protein is expressed equivalently to the wild-type protein and localizes to the nucleus (Fig. 6D and E) but, importantly, the binding of FOXM1(Δ1-116) to a range of FOXM1 target genes was drastically reduced (Fig. 6E). Thus, a major mechanism for recruitment of FOXM1 to its regulatory regions in chromatin is through protein-protein interactions with the MMB complex rather than the canonical mechanism used by FOX proteins in binding to RYAAAYA DNA elements.
Fig 6.
FOXM1 is recruited indirectly via the MMB complex to CHR elements. (A and B) GST pulldown assay using the indicated GST-FOXM1 fusion proteins and total cell extracts from U2OS cells. Ethidium bromide (EtBr) was added to the GST pulldown reactions where indicated. Interacting LIN9 and B-MYB proteins were revealed by Western blotting (upper panels) and Coomassie or Ponceau S stained input GST fusion proteins are shown below. A total of 3% cell lysate input is shown (lane 1). Arrows represent bands corresponding to full-length GST fusion proteins. Asterisks indicate a cross-reacting GST fusion protein band. (C) Coimmunoprecipitation (IP) analysis from U2OS cells expressing the indicated Flag-tagged FOXM1 fusion proteins. Precipitated and coprecipitated proteins were detected by immunoblotting with the indicated antibodies (left). A total of 3% cell lysate input is shown (lanes 1 to 3). (D) FOXM1(Δ1-116) is localized to the nucleus. U2OS cells were transfected with plasmids encoding Flag-tagged version of full-length wild-type (WT) and a Δ1-116 version of FOXM1, and expression was detected by immunofluorescence with anti-Flag antibody. Nuclei are revealed by using DAPI (4′,6′-diamidino-2-phenylindole) staining. A merge of the two stains is shown on the right. (E) ChIP assays of transiently transfected Flag-tagged FOXM1 in HEK293T cells. Cells were transfected with empty vector or wild-type (WT) or mutant (Δ1-116/ΔN) versions of FOXM1 prior to ChIP for Flag-tagged FOXM1 on the indicated genes. Expression levels were revealed by Western analysis with anti-flag antibody (inset). The data are the averages of two independent experiments. (F) Model showing recruitment of FOXM1 to the CHR element through MMB complex binding. FOXM1 is linked to the cell cycle through activation by cell cycle-regulated kinases.
DISCUSSION
Members of the forkhead transcription factor family are all thought to bind to variants of the core RYAAAYA motif. This is exemplified by both in vitro studies for several members (see, for example, reference 29) and also in vivo ChIP-seq studies for members of the FOXA, FOXP, and FOXK subclasses (26, 30 –32). This presents a conundrum for producing regulatory specificity among different family members. FOXM1 appears to have solved this problem by diverging from the other family members. It weakly binds to the canonical core forkhead DNA binding motif (33) but, rather than binding such motifs, is instead recruited to a different promoter element, the CHR motif, through an indirect mechanism that involves binding to the MMB complex (Fig. 6F). This enables it to have a specific role in controlling the expression of genes during the G2-M phase of the cell cycle. The uniqueness of this binding mode is emphasized by the observation that only 59 of the binding regions occupied by FOXP3 overlap with those occupied by FOXM1, despite the fact that FOXP3 shows a strong tendancy to bind at promoters and binds to over 40,000 sites in the genome (30).
Our results are broadly consistent with two recent studies that demonstrate functional interactions between FOXM1 and the MMB complex in G2/M transcriptional control (25, 34). In the first of these studies, DeCaprio and coworkers start from ChIP-seq studies for the MMB complex components LIN9 and B-MYB which uncover a tight link to regulating gene expression in the G2 and M phases of the cell cycle. This in turn leads them to implicate FOXM1 in the same process (25). Here, our data have converged on the same finding of collaboration between the MMB complex and FOXM1, but starting from FOXM1 ChIP-seq data. Importantly, in agreement with our findings, that previous study also found that the MMB complex is required for FOXM1 recruitment to chromatin and provided important re-ChIP data demonstrating cooccupancy of FOXM1 and components of the MMB complex. Moreover, the authors also showed that FOXM1 does not control binding of the MMB complex. However, there are important differences since, in contrast to our findings and those of others (35), more substantive fluctuations in FOXM1 binding through the cell cycle were observed. It is not clear why this is the case, but it could be due to the differing cell types used by the different groups or the epitopes recognized by the different antibodies used in ChIP experiments. It is important to note in this context that in quiescent cells in the G0 phase of the cell cycle, FOXM1 binding to chromatin is likely reduced due its reduced expression under these conditions, and we find that this is indeed the case in T98G cells (data not shown). In a second study by Watson and coworkers (34), the role of the B-Myb subunit of the MMB complex was probed, and it was shown that this was absolutely required for FOXM1 recruitment to chromatin, again consistent with a role for the MMB complex in this recruitment process. However, that study concluded that B-Myb was working to recruit FOXM1 through B-Myb binding sites, at least on the Birc5 promoter, rather than to the CHR elements identified here and by the DeCaprio groups (25). Despite the general similarities in our findings, there are several important differences between our study and these two other studies; one study concludes that FOXM1 is recruited at least partially via canonical RYAAAYA motifs (25), while the other proposes that B-MYB acts as a pioneering factor to permit FOXM1 recruitment without a direct interaction of FOXM1 with MMB (34). These interpretations stand in clear contrast to the observation that in a genome-wide analysis, FOXM1 binding is observed in promoter regions mostly lacking canonical forkhead binding sites. Here, we resolve the resulting contradiction by suggesting a new mechanism for recruitment of a forkhead factor to chromatin. We demonstrate that protein-protein interactions between FOXM1 and MMB complex components are central to the FOXM1 recruitment mechanism to CHR elements. This binding occurs independently from canonical RYAAAYA DNA motifs. Although a previous study identified RYAAAYA motifs as overrepresented in MMB and DREAM complex binding sites and suggested specific DNA-based interactions with FOXM1 (25), these motifs are not overrepresented in the FOXM1 cistrome, and those that are presented are not localized at the summit of the binding peaks (Fig. 3B). This might point to the existence of alternative forkhead transcription factors which functionally interact with these complexes at a different subset of their target genes. Moreover, it is possible that FOXM1 recruitment to subsets of target genes is mediated by alternative mechanisms such as the one proposed for the Birc5 promoter (34).
It is currently unclear how the MMB complex and FOXM1 work together to mediate cell cycle control. However, the mechanism is likely to be complex, since the repressive DREAM complex can also bind to CHR elements, likely via MuvB components that are shared with the MMB complex (10, 36). FOXM1 is linked to intrinsic cell cycle regulatory networks by acting as a target for the Cdk-cyclin and PLK1 protein kinases (8, 35, 37). These kinases promote the transcriptional activation properties of FOXM1 at the late G2 and M phases of the cell cycle. Since FOXM1 binding to chromatin appears not to vary greatly during the cell cycle from late G1 to the late G2 and M phases (Fig. 1G) (35), this link to cell cycle-dependent kinases provides a neat mechanism for the activation of a preassembled transcriptional regulatory complex. Conversely, the DREAM complex acts to repress transcription and functions antagonistically to FOXM1 at CHR elements to keep its target genes repressed during the early phases of the cell cycle (10, 25). In this case, dissociation of the DREAM complex-specific subunits, leaving the MMB complex associated with the promoter, accompanies the activation process. Thus, the MuvB complex plays a critical role in directing the temporal expression of G2/M-expressed genes through the CHR element. Interestingly, the N-terminal region of FOXM1 which interacts with the MMB complex also acts as an autoinhibitory region. It is therefore tempting to speculate that interaction with the MMB complex might also contribute to FOXM1 activation. Future studies are required to uncover the regulatory activities that govern the temporal exchange of transcriptional regulators to the CHR element during the cell cycle and lead to the precise cyclical control of transcription of its associated genes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Karren Palmer for excellent technical assistance. We thank Peter March in the Bioimaging facility and Andy Hayes, Claire Haslam, and Ian Donaldson at the Genomic Technologies and Bioinformatics facilities. We thank Catherine Millar, Shen-Hsi Yang, and members of our laboratories for helpful comments on the manuscript and stimulating discussions. We thank Roger Watson for B-Myb monoclonal antibody, James DeCaprio for LIN37 antibody, and Suyun Huang for reagents.
M.Q. is the recipient for a graduate fellowship from the Freistaat Sachsen. This study was supported by grants from the Wellcome Trust and a Royal Society-Wolfson award to A.D.S. and from the IZKF/BMBF to K.E.
All of the authors contributed to writing the manuscript. X.C., G.A.M., M.Q., M.F., and B.S. performed, designed, and interpreted the experiments. N.H. contributed to the bioinformatics analysis. K.E. and A.D.S. devised the project and helped with experimental design and interpretation.
Published ahead of print 29 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00881-12.
REFERENCES
- 1. Carlsson P, Mahlapuu M. 2002. Forkhead transcription factors: key players in development and metabolism. Dev. Biol. 250: 1–23 [DOI] [PubMed] [Google Scholar]
- 2. Hannenhalli S, Kaestner KH. 2009. The evolution of Fox genes and their role in development and disease. Nat. Rev. Genet. 10: 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Breeden LL. 2000. Cyclin transcription: timing is everything. Curr. Biol. 10: R586–R588 [DOI] [PubMed] [Google Scholar]
- 4. Alvarez B, Martinez AC, Burgering BM, Carrera AC. 2001. Forkhead transcription factors contribute to execution of the mitotic programme in mammals. Nature 413: 744–747 [DOI] [PubMed] [Google Scholar]
- 5. Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, Clevers H, Medema RH. 2005. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 7: 126–136 [DOI] [PubMed] [Google Scholar]
- 6. Wang IC, Chen YJ, Hughes D, Petrovic V, Major ML, Park HJ, Tan Y, Ackerson T, Costa RH. 2005. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol. Cell. Biol. 25: 10875–10894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Darieva Z, Bulmer R, Pic-Taylor A, Doris KS, Geymonat M, Sedgwick SG, Morgan BA, Sharrocks AD. 2006. Polo kinase controls cell-cycle-dependent transcription by targeting a coactivator protein. Nature 444: 494–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fu Z, Malureanu L, Huang J, Wang W, Li H, van Deursen JM, Tindal DJ, Chen J. 2008. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat. Cell Biol. 10: 1076–1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Muller GA, Engeland K. 2010. The central role of CDE/CHR promoter elements in the regulation of cell cycle-dependent gene transcription. FEBS J. 277: 877–893 [DOI] [PubMed] [Google Scholar]
- 10. Muller GA, Quaas M, Schumann M, Krause E, Padi M, Fischer M, Litovchick L, DeCaprio JA, Engeland K. 2012. The CHR promoter element controls cell cycle-dependent gene transcription and binds the DREAM and MMB complexes. Nucleic Acids Res. 40: 1561–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA. 2007. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol. Cell 26: 539–551 [DOI] [PubMed] [Google Scholar]
- 12. Schmit F, Cremer S, Gaubatz S. 2009. LIN54 is an essential core subunit of the DREAM/LINC complex that binds to the cdc2 promoter in a sequence-specific manner. FEBS J. 276: 5703–5716 [DOI] [PubMed] [Google Scholar]
- 13. Wasner M, Tschop K, Spiesbach K, Haugwitz U, Johne C, Mossner J, Mantovani R, Engeland K. 2003. Cyclin B1 transcription is enhanced by the p300 coactivator and regulated during the cell cycle by a CHR-dependent repression mechanism. FEBS Lett. 536: 66–70 [DOI] [PubMed] [Google Scholar]
- 14. O'Donnell A, Yang SH, Sharrocks AD. 2008. MAP kinase-mediated c-fos regulation relies on a histone acetylation relay switch. Mol. Cell 29: 780–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tavner F, Frampton J, Watson RJ. 2007. Targeting an E2F site in the mouse genome prevents promoter silencing in quiescent and post-mitotic cells. Oncogene 26: 2727–2735 [DOI] [PubMed] [Google Scholar]
- 16. Marais A, Ji Z, Child ES, Krause E, Mann DJ, Sharrocks AD. 2010. Cell cycle-dependent regulation of the forkhead transcription factor FOXK2 by CDK-cyclin complexes. J. Biol. Chem. 285: 35728–35739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shore P, Sharrocks AD. 1994. The transcription factors Elk-1 and serum response factor interact by direct protein-protein contacts mediated by a short region of Elk-1. Mol. Cell. Biol. 14: 3283–3291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee TI, Johnstone SE, Young RA. 2006. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 1: 729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Boros J, Donaldson IJ, O'Donnell A, Odrowaz ZA, Zeef L, Lupien M, Meyer CA, Liu XS, Brown M, Sharrocks AD. 2009. Elucidation of the ELK1 target gene network reveals a role in the coordinate regulation of core components of the gene regulation machinery. Genome Res. 19: 1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT. 2009. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods 48: 240–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, Liu XS. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shin H, Liu T, Manrai AK, Liu XS. 2009. CEAS: cis-regulatory element annotation system. Bioinformatics 25: 2605–2606 [DOI] [PubMed] [Google Scholar]
- 24. Ye T, Krebs AR, Choukrallah MA, Keime C, Plewniak F, Davidson I, Tora L. 2011. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res. 39: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sadasivam S, Duan S, DeCaprio JA. 2012. The MuvB complex sequentially recruits B-Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 26: 474–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ji Z, Donaldson IJ, Liu J, Hayes A, Zeef LA, Sharrocks AD. 2012. The forkhead transcription factor FOXK2 promotes AP-1-mediated transcriptional regulation. Mol. Cell. Biol. 32: 385–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Skaar JR, Pagano M. 2009. Control of cell growth by the SCF and APC/C ubiquitin ligases. Curr. Opin. Cell Biol. 21: 816–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. 2010. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28: 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pierrou S, Hellqvist M, Samuelsson L, Enerback S, Carlsson P. 1994. Cloning and characterization of seven human forkhead proteins: binding site specificity and DNA bending. EMBO J. 13: 5002–5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Katoh H, Qin ZS, Liu R, Wang L, Li W, Li X, Wu L, Du Z, Lyons R, Liu CG, Liu X, Dou Y, Zheng P, Liu Y. 2011. FOXP3 orchestrates H4K16 acetylation and H3K4 trimethylation for activation of multiple genes by recruiting MOF and causing displacement of PLU-1. Mol. Cell 44: 770–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Motallebipour M, Ameur A, Reddy Bysani MS, Patra K, Wallerman O, Mangion J, Barker MA, McKernan KJ, Komorowski J, Wadelius C. 2009. Differential binding and co-binding pattern of FOXA1 and FOXA3 and their relation to H3K4me3 in HepG2 cells revealed by ChIP-seq. Genome Biol. 10: R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wallerman O, Motallebipour M, Enroth S, Patra K, Bysani MS, Komorowski J, Wadelius C. 2009. Molecular interactions between HNF4a, FOXA2 and GABP identified at regulatory DNA elements through ChIP-sequencing. Nucleic Acids Res. 37: 7498–7508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Littler DR, Alvarez-Fernandez M, Stein A, Hibbert RG, Heidebrecht T, Aloy P, Medema RH, Perrakis A. 2010. Structure of the FoxM1 DNA-recognition domain bound to a promoter sequence. Nucleic Acids Res. 38: 4527–4538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Down CF, Millour J, Lam EW, Watson RJ. 2012. Binding of FoxM1 to G2/M gene promoters is dependent upon B-Myb. Biochim. Biophys. Acta 1819: 855–862 [DOI] [PubMed] [Google Scholar]
- 35. Laoukili J, Alvarez M, Meijer LA, Stahl M, Mohammed S, Kleij L, Heck AJ, Medema RH. 2008. Activation of FoxM1 during G2 requires cyclin A/Cdk-dependent relief of autorepression by the FoxM1 N-terminal domain. Mol. Cell. Biol. 28: 3076–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schmit F, Korenjak M, Mannefeld M, Schmitt K, Franke C, von Eyss B, Gagrica S, Hanel F, Brehm A, Gaubatz S. 2007. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle 6: 1903–1913 [DOI] [PubMed] [Google Scholar]
- 37. Major ML, Lepe R, Costa RH. 2004. Forkhead box M1B transcriptional activity requires binding of Cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/CBP coactivators. Mol. Cell. Biol. 24: 2649–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.