Abstract
Every known SWI/SNF chromatin-remodeling complex incorporates an ARID DNA binding domain-containing subunit. Despite being a ubiquitous component of the complex, physiological roles for this domain remain undefined. Here, we show that disruption of ARID1a-DNA binding in mice results in embryonic lethality, with mutant embryos manifesting prominent defects in the heart and extraembryonic vasculature. The DNA binding-defective mutant ARID1a subunit is stably expressed and capable of assembling into a SWI/SNF complex with core catalytic properties, but nucleosome substrate binding and promoter occupancy by ARID1a-containing SWI/SNF complexes (BAF-A) are impaired. Depletion of ARID domain-dependent, BAF-A associations at THROMBOSPONDIN 1 (THBS1) led to the concomitant upregulation of this SWI/SNF target gene. Using a THBS1 promoter-reporter gene, we further show that BAF-A directly regulates THBS1 promoter activity in an ARID domain-dependent manner. Our data not only demonstrate that ARID1a-DNA interactions are physiologically relevant in higher eukaryotes but also indicate that these interactions facilitate SWI/SNF binding to target sites in vivo. These findings support the model wherein cooperative interactions among intrinsic subunit-chromatin interaction domains and sequence-specific transcription factors drive SWI/SNF recruitment.
INTRODUCTION
The identification of DNase I-hypersensitive sites in active globin genes was the first evidence linking nucleosome conformation to transcriptional activity (1). Insight into how nucleosome conformation is controlled came from subsequent genetic studies that identified yeast mutants unable to switch mating types (SWI) or utilize sucrose as a carbon source (sucrose nonfermenter, or SNF) (2, 3). It was later found the majority of swi/snf-encoded proteins were components of a large, multiprotein complex (collectively referred to as the SWI/SNF chromatin-remodeling complex) that could utilize energy derived from ATP hydrolysis to alter nucleosome conformation and thus modulate DNA accessibility (4–6).
To remodel chromatin, the SWI/SNF complex must recognize and bind its nucleosomal target substrates. This complex binds DNA with high affinity, but the DNA binding domains responsible for mediating intrinsic subunit-DNA interactions bind with little to no sequence specificity in vitro—a property that makes them incompatible with a primary role in recruitment (7–14). Current models implicate subunit interactions with sequence-specific transcription factors in primary SWI/SNF recruitment (15). The precise function of non-sequence-specific DNA binding domains within SWI/SNF is currently unknown.
Of the DNA binding domains responsible for mediating non-sequence-specific subunit-DNA interactions, the ARID domain is noteworthy because all known SWI/SNF complexes incorporate a subunit belonging to the ARID family of proteins. In addition, the SWI/SNF complexes found in higher eukaryotes are known to incorporate two or more mutually exclusive ARID subunits, with each subunit representing a biochemically distinct complex subfamily. In mammals, ARID2 (BAF200) embodies the PBAF complex, while ARID1a (BAF250a) and the paralogous subunit ARID1b (BAF250b) embody BAF-A and BAF-B complexes, respectively (16–21). Additional evidence indicates that ARID subunit usage influences transcription factor associations with these complexes (22). These data are consistent with the notion that ARID subunits can tailor their respective complexes for specific roles (21, 22) although their exact contributions to SWI/SNF function have not been established.
Currently, of the three known mammalian ARID subunits, only ARID1a has been deemed essential. Homozygous Arid1a-null embryos manifest an early block in germ layer formation (23). ARID1a is also mutated in 50% of the ovarian clear cell carcinomas and 30% of the endometrioid carcinomas found in humans (24, 25). Since ARID1a-containing BAF-A complexes are essential, we surmised that genetic approaches aimed at disrupting the ARID domain of ARID1a would allow us to determine how ARID-DNA interactions contribute to SWI/SNF activity in vivo.
To address this question, we screened an N-ethyl-N-nitrosurea (ENU)-mutagenized mouse embryonic stem (ES) cell library for point mutations in the region encoding the ARID domain of ARID1a and isolated an ENU-induced mutation which leads to a valine-to-glycine substitution at amino acid position 1068 (herein referred to as V1068G). The mutant ARID1a protein is stably expressed, and it incorporates into a BAF-A complex with core chromatin remodeling function. However, its ability to bind DNA is severely impaired. Impaired ARID1a-DNA interactions result in a partial loss-of-function or hypomorphic allele because homozygous mutant embryos gastrulate and are recovered as late as embryonic day 13.5 (E13.5). Homozygous Arid1a embryos carrying the V1068G mutation (Arid1aV1068G/V1068G) manifest heart defects, fail to establish proper extraembryonic vasculature, and exhibit hemorrhaging. As a result of these phenotypes, Arid1aV1068G/V1068G embryos failed to establish proper circulation, culminating in ischemic arrest in utero. We show that disruption of ARID1a-DNA interactions reduces BAF-A affinity for nucleosomes. Consistent with this, mutant BAF-A occupancy at the known BAF-A target gene, THROMBOSPONDIN-1 (THBS1), was impaired, resulting in THBS1 upregulation. Finally, we show that BAF-A directly represses THBS1 promoter activity using luciferase reporter assays. Our data demonstrate that the ARID domain of ARID1a is essential for development and that ARID1a-DNA interactions are required for promoter occupancy by SWI/SNF.
MATERIALS AND METHODS
Library screening, generation of the V1068G mutant allele, and mouse husbandry.
E14 and CT129 ENU-mutagenized ES cell libraries were generated as previously described (26, 27). RNA isolated from 96-well ES cell libraries was reverse transcribed and used as the template for PCRs using 12 primer pairs spanning the mouse ARID1a cDNA (NCBI Reference Sequence NM_001080819.1). A series of denaturation and slow annealing cycles were performed to allow formation of heteroduplexes. Samples were then analyzed by denaturing high-performance liquid chromatography (dHPLC) using WAVE mutation detection software (Transgenomic). Screening temperatures were chosen based on analysis of the melting curve for each PCR fragment. Following detection of a mutation and confirmation by sequencing, cell lines were subcloned from a single cell suspension and verified by dHPLC to ensure clonality of the mutations. Karyotyping was performed to monitor the genomic integrity of cells prior to injection. Cells were injected into C57BL/6 blastocysts and implanted into pseudopregnant females. Germ line animals were detected by coat color and PCR/HPLC confirmation of tail DNA samples. They were then out-crossed onto a CD1 background for >10 generations. All mice were maintained at the University of North Carolina at Chapel Hill Animal Facility using standard techniques in accordance with protocols approved by the University of North Carolina at Chapel Hill Institutional Animal Care and Use Committee.
Genotyping.
Arid1a+/+; Arid1aV1068G/+; Arid1aV1068G/V1068G alleles were genotyped by a HincII restriction fragment length polymorphism induced by the T-to-G mutation (T is base 3203 in NCBI Reference Sequence NM_001080819.1) (Fig. 1A). Briefly, genomic DNAs harvested from mouse embryos or tail samples from live-born mice were PCR amplified using standard techniques and the following primer set: forward (F), 5′-GCCGTTTGAATTTGTCCTGT; reverse (R), 5′-TCTTGGGTTGGGACTTCTTG. PCR products were subsequently digested with HincII (NEB) and then resolved by 2% Tris-borate-EDTA agarose gel electrophoresis.
Fig 1.
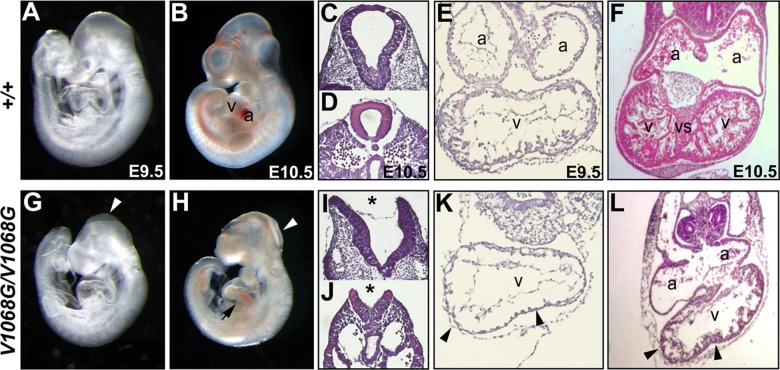
Arid1aV1068G/V1068G embryos manifest neural tube and heart defects. Whole-mount images of E9.5 and 10.5 Arid1a+/+ (A and B) or Arid1aV1068G/V1068G (G and H) littermate embryos are shown. E9.5 and E10.5 Arid1aV1068G/V1068G embryos display open head folds (G and H, arrowheads). E10.5 Arid1aV1068G/V1068G hearts are grossly abnormal and appeared developmentally delayed (H, arrows). Transverse histological sections through the hindbrain region (I, asterisk) or base of tail (J, asterisk) of E9.5 Arid1aV1068G/V1068G embryos indicate neural tube closure defects. Transverse paraffin sections of E9.5 (E and K) and E10.5 (F and L) Arid1a+/+ and Arid1aV1068G/V1068G hearts are shown. Trabeculation defects are prominent in E9.5 Arid1aV1068G/V1068G hearts (K and L, arrowheads), and these defects are present at E10.5. Mutant E10.5 hearts (L) have hypoplastic myocardial walls and severe ventricular septal defects. v, ventricles; a, atria; vs, ventricular septum.
Cell culture and siRNA transfections.
Arid1a+/+ and Arid1aV1068G/V1068G mouse embryonic fibroblasts (MEFs) were isolated from E9.5 embryos using standard procedures and cultured in Dulbecco's modified Eagle medium supplemented with 4.5 g/liter glucose, 2 mM sodium pyruvate, 0.5 g/liter l-glutamine, 15% fetal bovine serum, 200 units/liter penicillin G, and 200 g/liter streptomycin sulfate. Low-passage primary MEFs were transformed as previously described (28, 29). MEFs were transfected with 50 nM amounts of nontargeting (mock), BRG1, and BRM small interfering RNA (siRNA) smart pools (M-056591-00-005 and M-041135-01-0005; Dharmacon) using 2.5 μl/ml of RNAiMax transfection reagent (Invitrogen) according to the manufacturer's instructions. BRG1 and BRM depletion were verified by reverse transcription-PCR (RT-PCR) using gene-specific primer sets (data not shown). Mouse NIH 3T3 and human HEK 293T cells were cultured following ATCC standards.
Protein expression and DNA binding experiments.
The glutathione S-transferase (GST)–ARID protein fragments used in DNA binding experiments are the products of GST-DRI258–410 (DRI258–410 is the region of amino acids 258 to 410 encoded by the dead ringer gene, referred to as GST-DriARID) (30) and GST-p270600–1018 (GST fused to the region of amino acids 600 to 1018 of human p270, referred to as GST-hARID) (16). Valine-to-glycine amino acid substitutions were generated by PCR-based, site-directed mutagenesis of the parent GST-ARID plasmids, followed by blunt-ended circularization, using the following 5′ phosphorylated primers: GST-DRI258-410 F, 5′-(Phos)-GGCTTGGTGGATGGCATCAACAAGAA, and R, 5′-(Phos)-GCCGCGGGCTATCACCAGATTGTACAG; GST-p270600-1018 F, 5′-(Phos)-GATTGACTCAGGGCAACAAGAACAA, and R, 5′-(Phos)-CACCAATCTCCTTCACAGACACATAGAG. GST-DRI258-410 and GST-p270600-1018 proteins were expressed in Escherichia coli NEB Express (New England BioLabs) and Rosetta-2 (Merck) strains, respectively, and then purified essentially as previously described (31).
Binding conditions for gel mobility shift assays were as previously described (19, 32). Binding isotherms were estimated by varying the protein concentrations by 2-fold dilutions (from 2 μM to 0.48 nM). 32P-end-labeled, double-stranded DNA probe at 50 pM was used per binding reaction. Forward sequences of the oligonucleotide probes used in the binding reactions are as follows: dri.16, 5′-CATCAATAAATTAGAATTAA; p270.18, ACCGGGGGTGCCACACCGTTGA (16). GST-DriARID-DNA complexes were resolved by 8% Tris-borate-EDTA acrylamide electrophoresis, and GST-hARID-DNA complexes were resolved by 8% Tris-acetate-EDTA (pH 7.8) acrylamide electrophoresis. Gels were dried and then imaged using a PhosphorImager (Molecular Dynamics, Inc.). Fractional occupancies were calculated from band densitometry measurements using the following formula: 1 − (DNAfree/DNAtotal).
Western blotting.
Western blotting was performed using standard procedures and the following antibodies: ARID1a (A301-040A or A301-041A; Bethyl Labs), ARID1b (sc-32762; Santa Cruz), ARID2 (A302-230A; Bethyl Labs), BRG1 (sc-17796; Santa Cruz), BRM (sc-6450; Santa Cruz), INI1/SNF5 (A301-087A; Bethyl Labs), BAF60a (611728; BD Biosciences), cullin-2 (ab1870; Abcam), THBS-1 (MS-421-P0; Thermo Scientific), hemagglutinin (HA) tag (A190-108A; Bethyl Labs), β-actin (ab8226; Abcam), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Sigma), and nucleolin (A300-711A; Bethyl Labs). Primary antibodies were detected using goat anti-rabbit, goat anti-mouse, or donkey anti-goat secondary antibodies conjugated to horseradish peroxidase (Santa Cruz). For fluorescent Western blot detection, Li-COR Bioscience Odyssey blocking buffer and fluorescent secondary antibodies were used according to the manufacturer's instructions.
Coimmunoprecipitations and immunoaffinity isolation of ARID1a-containing complexes.
Small-scale nuclear extracts used in immunoprecipitations were prepared from wild-type or Arid1aV1068G/V1068G MEFs as previously described (33, 34). Briefly, nuclear proteins were extracted from isolated nuclei with two sequential 30-min incubations in 0.6 volumes (per semidry cell pellet) of buffer C (20 mM HEPES-KOH [pH 7.9], 0.2 mM EDTA, 25% glycerol, 420 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, mammalian protease inhibitor cocktail) at 4°C. The final salt concentration of nuclear extracts was lowered to approximately 150 mM KCl by the addition of KCl-free buffer C. Nuclear extracts were clarified by centrifugation, snap frozen in liquid nitrogen, and then stored at −80°C. Antibodies specific for BRG1 (sc-10768; Santa Cruz) and BRM (ab15597; Abcam) or ARID1a (sc-32761; Santa Cruz) were preconjugated to bovine serum albumin (BSA)-blocked protein A/G-agarose (Santa Cruz). Immunoprecipitations (IPs) were performed in IP buffer (20 mM HEPES-KOH [pH 7.9], 0.2 mM EDTA, 10% glycerol, 0.1% Tween 20, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, mammalian protease inhibitor cocktail) supplemented with 150 mM KCl using 600 μg of precleared nuclear extracts. Immunoprecipitates were washed seven times with 20 volumes of IP buffer supplemented with 500 mM KCl. To lower the final salt concentration, the last wash was performed with IP buffer supplemented with 100 mM KCl. The immunoprecipitates were eluted with acidic glycine, and then the eluates were neutralized with 0.1 volumes of 1 M Tris-Cl (pH 8.0). The final eluates were resolved by SDS-PAGE and analyzed by Western blotting. For immunoaffinity isolation of ARID1a-containing complexes, the washed agarose bead–anti-ARID1a immune conjugates from above were equilibrated in IP buffer supplemented with 60 mM KCl following the last 100 mM KCl wash and used immediately or stored at 4°C. Similar high-salt immunopurification methods have been used to isolate SWI/SNF complexes from mammalian cells (35).
REA.
Restriction enzyme accessibility (REA) assays were performed as previously described (36). A mononucleosome-positioning DNA fragment from the pTPT plasmid (37) was radiolabeled with [α32P]dCTP by PCR using the following primers: F, ACGCGTCGGTGTTAGAGC, and R, GAATTCTCTAGACAGTGTCCCA. Mononucleosomes were assembled according to the “salt jump” method (38, 39) in the presence of core histones from HeLa cells and sonicated calf thymus carrier DNA. Crude mononucleosome assemblies were gel fractionated by 5% Tris-EDTA (pH 7.6) acrylamide electrophoresis and then gel purified by diffusion in 10 mM Tris-Cl (pH 7.6), 0.2 mM EDTA, 40 mM NaCl, 0.01% Igepal CA-380, and 0.5 mM phenylmethylsulfonyl fluoride at 4°C for 12 to 18 h. Mononucleosome assemblies were concentrated and then stored at 4°C in gel elution buffer supplemented with 15% glycerol, 0.3 mg/ml bovine serum albumin (BSA), 0.1% Igepal CA-380, and 1 mM dithiothreitol. Mononucleosomes were incubated at 37°C for 15 min prior to use. Gel-purified mononucleosomes (<1 nM) were incubated with either 2 mM ATP or ATPγS and saturating amounts of immunoaffinity-purified ARID1a-containing complexes on beads in a 20-μl reaction mixture containing 12 mM HEPES-KOH (pH 7.9), 2 mM Tris-Cl (pH 7.6), 60 mM KCl, 8 mM NaCl, 4 mM MgCl2, 0.2 mM dithiothreitol, 0.06 mg/ml bovine serum albumin (BSA), 0.02% Igepal CA-380, 3% glycerol, and 0.5 unit/μl PstI enzyme (NEB) for 10, 20, 30, or 40 min at 30°C. Reactions were quenched by the addition of 1.5 volumes (30 μl) of 20 mM Tris-Cl (pH 7.6), 50 mM EDTA, 2% SDS, 0.2 mg/ml proteinase K, and 0.75 mg/ml yeast tRNA (from Saccharomyces cerevisiae), followed by a 1-h incubation at 55°C. Samples were ethanol precipitated and resolved by 6% Tris-borate-EDTA acrylamide electrophoresis. Gels were dried and then exposed to film. Band densitometry measurements from triplicate samples containing independent preparations of BAF-A complexes were averaged, and the standard deviations of the triplicate values measured were plotted. Statistical differences were detected using a two-tailed Student t test.
Nucleosome pulldown assays.
Saturating amounts of immunoaffinity-purified ARID1a-containing, BAF-A complexes on beads were incubated with the internally labeled and gel-purified TPT mononucleosomes (<1 nM) from the procedure described above in a 20-μl reaction mixture containing 12 mM HEPES-KOH (pH 7.9), 2 mM Tris-Cl (pH 7.6), 60 mM KCl, 8 mM NaCl, 4 mM MgCl2, 0.2 mM dithiothreitol, 0.06 mg/ml bovine serum albumin (BSA), 0.02% Igepal CA-380, and 3% glycerol supplemented with 1.6 to 50 ng (from 2-fold dilutions) of sheared salmon sperm DNA for 30 min at 30°C. The competitor DNA titration range was determined in a previous experiment (data not shown). Beads containing nucleosome-bound BAF-A complexes were washed three times with 25 volumes of binding buffer to remove unbound nucleosomes, and the bound fractions were eluted by the addition of 1.5 volumes (30 μl) of 20 mM Tris-Cl (pH 7.6), 50 mM EDTA, 2% SDS, 0.2 mg/ml proteinase K, and 0.75 mg/ml yeast tRNA, followed by a 1-h incubation at 55°C. Samples were ethanol precipitated and resuspended in H2O. Each pulldown condition was performed in triplicate using independent preparations of BAF-A complexes. Independent samples were subjected to scintillation counting (as cpm) or resolved by 6% Tris-borate-EDTA acrylamide electrophoresis. Average cpm values and the standard deviations of the triplicate values measured were plotted. Statistical differences were detected using a two-tailed Student t test.
ChIP.
Wild-type or Arid1aV1068G/V1068G MEFs were fixed in growth medium containing formaldehyde for 10 min at room temperature, and the formaldehyde-cross-linked chromatin extracts were prepared as previously described (40). Cross-linked chromatin was sonicated on ice to an average DNA length between 300 and 700 bp in 10 mM Tris-Cl (pH 8.0), 1 mM EDTA, and 0.5 mM EGTA. Precleared chromatin (300 μg) was incubated with 5 μg of ARID1a (sc-32761; Santa Cruz), ARID1b (sc-32762; Santa Cruz), ARID2 (sc-98299; Santa Cruz), BRG1 (sc-10768; Santa Cruz), BRM (sc-6450; Santa Cruz), INI1/SNF5 (A301-087A; Bethyl Labs), or polymerase II (Pol II) (sc-899; Santa Cruz) antibodies in 1 ml of chromatin immunoprecipitation (ChIP) buffer (50 mM Tris-Cl [pH 7.6], 150 mM NaCl, 5 mM EDTA, 0.5% Igepal CA-380, 1% TX-100) (41) overnight at 4°C. Salmon sperm DNA-blocked protein A/G-agarose beads (Santa Cruz) were added for 3 h at 4°C, and then the agarose bead-anti-ARID1a immune conjugates were washed eight times with 20 volumes of ChIP buffer supplemented with 500 mM NaCl, followed by a final wash in low-salt TE buffer (10 mM Tris-Cl [pH 8.0], 1 mM EDTA, 50 mM NaCl). DNA was eluted, and cross-links were reversed by the addition of a 10% Chelex-100–H2O slurry (Bio-Rad) and a 15-min incubation at 95°C. Protein was digested with 20 μg/ml proteinase K at 55°C for 1 h. Real-time quantitative PCR using Ssofast PCR master mix (Bio-Rad) and a CFX96 thermocycler (Bio-Rad) was performed using the following gene-specific primers: for THBS1 −3.0 kb, TCTGAGCCCTCCAGAGGTAA and TCTGTTGCTCAGGACTGTGG; THBS1 −1.4 kb, CAAGAGGCACTGGCTGTGTA and CTTGTGTGGGAGGGAGTGTT; THBS1 −1.2 kb, GGATCCCTGGACTCAGAGC and GCTGCCTGATCCTTTGTCTC; THBS1 −0.4 kb, ACCCACCAGGATTGACTCAG and TTCCATGACAGAGACGCTTG; THBS1 transcription start site (TSS), TTAAAAGGTCCTGGGCTCCT and GAAGGACAAGGACGCTGAGT; THBS1 +1.0 kb, GCTCCTCACAGCCAGAACTT and TAGATCTTGGCCCTTCACCA; THBS1 down, TGTCTGTCTTCACGGGGTTA and TGGCTTCAGCCTATTCCTTG; ADAMTS1, CGTCACTCTGGGGGTGTAAG and CACTCTTTTCCCCAGAGCAG; CRABP1, CCAGGGGAGAGCAAGTTCC and CTTGAGTCGCTAGGGTAG; GAPDH (control), GGAAGCAGCATTCAGGTCTCT and CCTCCTCCCTCTCTTTGGAC; INSULIN (control), TGGATGCCCACCAGCTTTATAGTCC and AACTGGTTCATCAGGCCATCTGGTC. Data from three independent experiments were averaged and plotted as a percentage of the total input ± standard error of the mean (SEM; adjusted for a 50-fold input dilution). Statistical differences were detected using a two-tailed Student t test.
RT-PCR.
To analyze target gene transcript levels in wild-type or Arid1aV1068G/V1068G MEFs, total RNA was extracted using the TRIzol method (Invitrogen), followed by an RNA cleanup step and on-column DNA digestion using an RNeasy miniprep kit (Qiagen) according to the manufacturer's instructions. cDNA was prepared using Multiscribe reverse transcriptase (Applied Biosystems) and a random primer mix (NEB). Real-time quantitative PCR using Ssofast PCR master mix (Bio-Rad) and a CFX96 thermocycler (Bio-Rad) was performed using the following gene-specific primers: for THBS1, CATCTTTGAACTCATTGG and TAGGTCTTGGAACTTGTC; ADAMTS1, TACTCTGGCACGGTGAAC and ATGAAGAACTCCTCTCCTTGTAG; CRABP1, AATGAGAACAAGATTCACT and GCCAAATGTCAGGATTAG; BRG1, GCTTCTTTGTTTCCTGAG and TCTGGAGTAGACATCTTTAC; BRM, CTGATAACGAGTGACCAT and TAGATGACAATGAGGACG; β-actin gene (control), CGAGCGTGGCTACAGCTTCACC and CCGATCCACACAGAGTACTTGCG. Expression was calculated from three independent experiments using ΔCT (where CT is threshold cycle) or ΔΔCT methods. Expression levels were normalized to the β-actin gene. Statistical differences were detected using a two-tailed Student t test.
In situ hybridization and immunostaining.
In situ hybridizations and histology were performed exactly as previously described (42) using a probe specific for Peg1 (43). Whole-mount immunostaining for platelet endothelial cell adhesion molecule 1 (PECAM1; also CD-31) was performed as described using a monoclonal rat anti-mouse CD31 antibody (MEC13.3; BD Biosciences) (44). For direct THBS1 immunostaining of slides, embryos were fixed in 10% neutral buffered formalin (Sigma), washed in 1× phosphate-buffered saline (PBS), and then cryo-protected through a graded sucrose series. Specimens were then embedded on dry ice. Samples were sectioned at 12 μm and collected on Superfrost-Plus slides (Fisher). Frozen sections were briefly thawed on a 42°C slide warmer, equilibrated in 1× PBS, and then incubated for 45 min in 100 mM glycine and 1× PBS (pH 7.3). Block and antibody diluent consisted of 1× PBS–0.05% Tween 20 supplemented with 5% donkey serum (Jackson ImmunoResearch), 1% (wt/vol) BSA, and 0.1% (wt/vol) cold-water fish gelatin (Sigma). Slides were incubated in a 1:25 dilution of goat anti-THBS1 antibody (sc-12312; Santa Cruz Biotechnology), followed by detection with a species-specific fluorescent secondary antibody raised in donkey. Slides were mounted and coverslipped in 4′,6′-diamidino-2-phenylindole (DAPI)-containing Prolong-Gold medium (Invitrogen).
Construction of N-terminally HA-tagged ARID1a expression plasmids.
To construct the HA-tagged mouse ARID1a expression plasmids, we used a PCR-based strategy to clone ARID1a (isoform 2) from wild-type or Arid1aV1068G/V1068G MEFs. mRNA was reverse transcribed using Superscript III (Invitrogen), and the resulting cDNA was PCR amplified using Phusion ultra-high-fidelity polymerase (NEB) and the following primer set: F, ATGGATCAGATGGGAAAGATGAGACCTCAGC; R, TCATGACTGGCCAATCAAAAACAGTACATCACA (the start codon is underlined). The PCR products were gel purified and blunt cloned into the JET1.2 plasmid (Fermentas). Cloned cDNA inserts were extensively verified by sequencing. XhoI/SalI-digested cDNA fragments were ligated into the pcDNA3.1(−) expression plasmid (Invitrogen) so that a single hemagglutinin (HA) tag was fused to the N termini of wild-type and mutant ARID1a proteins using a GGS amino acid linker. Recombinant protein expression was verified by plasmid transfection into HEK 293T cells and by Western blotting using an anti-HA antibody (data not shown).
THBS1 promoter-luciferase assays.
The mouse THBS1 −2.8 kb promoter-luciferase reporter plasmid (plasmid 12409; Addgene) has been previously described (45). The THBS1 −0.48 kb THBS1 promoter-luciferase plasmid was constructed by BamHI digestion and recircularization of the THBS1 −2.8 kb promoter-luciferase plasmid. The parental pXP1 luciferase reporter plasmid was used as a negative control. NIH 3T3 cells at 50 to 60% confluence were transfected with 0.5 μg of reporter plasmid and 0 to 0.5 μg of wild-type or mutant HA-tagged mouse ARID1a (HA-mARID1a) expression plasmids using TransIT-2020 reagent (Mirus Bio) according to the manufacturer's instructions. The empty pcDNA3.1 expression plasmid was used as carrier DNA to bring the final DNA concentration to 0.5 μg or as a negative control. Transfections were normalized for efficiency using 2 ng of Renilla luciferase plasmid. Cells were harvested 48 h after transfection, and reporter activity was measured using a Dual Luciferase assay system (Promega) according to the manufacturer's instructions. Normalized luciferase measurements from triplicate transfections were averaged, and the standard deviations of the triplicate values measured were plotted. Statistical differences were detected using a two-tailed Student t test.
Microscopy, imaging, and bioinformatics.
Histological sections were analyzed and photographed on an Olympus BX51 microscope using bright-field optics and a DP series digital camera (Olympus). Whole-mount embryo and yolk sac specimens were imaged on a Leica MZ FLIII stereomicroscope using a DS Ri1 digital camera (Nikon). Fluorescence microscopy and image acquisition were performed using a Leica DM LB microscope and a Retiga-2000R digital camera (Q-Imaging). Virtual protein mutagenesis was performed using the MuSE virtual mutation module of PROTMAN software (46). Protein structures were rendered and visualized using PyMol software (Schrodinger, LLC). KaleidoGraph software (Synergy) was used for curve-fitting the electrophoretic mobility shift assay (EMSA) binding isotherms. Comparative genome analysis was performed using VISTA algorithms (47).
RESULTS
An ENU mutagenesis screen in mouse ES cells identifies a novel Arid1a hypomorphic allele.
To determine if ARID subunit-DNA interactions have essential roles in higher eukaryotes, we chose a phenotype-driven, ENU chemical mutagenesis strategy to isolate critical point mutations in the ARID domain of ARID1a. We screened our existing ENU-mutagenized mouse ES cell libraries for point mutations within this domain (26, 27, 48). Mutations were identified by heteroduplex detection using high-performance liquid chromatography (dHPLC). Two of the three missense mutations identified within the ARID coding region of ARID1a were transmitted through the germ line. The first mutation (T to A) resulted in a cysteine-to-serine substitution at amino acid 1100 (C1100S), while the second mutation (T to G) resulted in a nonconservative, valine-to-glycine substitution at amino acid 1068 (V1068G) in ARID1a (NP_001074288). Homozygous C1100S mice were phenotypically normal at weaning. Arid1aV1068G/+ mice on an outbred (CD1) background were also phenotypically normal at this stage. However, live-born Arid1aV1068G/V1068G offspring were not recovered from crosses between heterozygous mice, indicating that the V1068G mutation results in homozygous lethality.
To determine the timing of lethality in utero, embryos were collected at embryonic time points from crosses between heterozygous mice and genotyped. At E8.5, Arid1aV1068G/V1068G embryos were recovered with Arid1aV1068G/+ and Arid1a+/+ littermate offspring at normal Mendelian ratios (data not shown). At time points beyond E13.5, only Arid1aV1068G/+ and Arid1a+/+ embryos were recovered. Table 1 demonstrates that Arid1aV1068G/V1068G embryos were routinely recovered at E9.5, and of those recovered, fewer than half of them (16%) lacked beating hearts. Beyond E10.5, the majority of these mutant embryos were either dead or exhibited abated pulse rates based upon visual inspection, and the number of resorbed embryos observed in litters collected at these later time points tended to increase with age. Since Arid1aV1068G/V1068G embryos arrest much later in development than Arid1a-null embryos (23), this mutation manifests as a hypomorphic allele.
Table 1.
The V1068G mutation results in homozygous lethality
| Embryonic day | No. of V1068G/V1068G embryos (no. of dead embryos) | No. of resorbed embryos | Lethality (%) (lethality + resorption [%]) |
|---|---|---|---|
| 9.5 | 32 (5) | 4 | 16 (25) |
| 10.5 | 18 (7) | 4 | 39 (50) |
| 11.5 | 10 (8) | 4 | 80 (86) |
| 12.5 | 1 (1) | 9 | 100 |
| 13.5 | 2 (2) | 6 | 100 |
Arid1aV1068G/V1068G embryos are grossly abnormal and developmentally delayed at midgestation.
Arid1aV1068G/V1068G embryos died around midgestation. These mutant embryos are grossly indistinguishable from their wild-type littermates prior to E8.5, and their allantoic buds and early somites are visible (data not shown). At later time points, most mutant embryos are smaller and display gross abnormalities, but the overall presentation of their body axes is appropriate for their respective developmental stage as embryo turning and the associated inversion of germ layers appear to occur normally (Fig. 1G and H). Mutant embryos can be readily identified upon dissection by their open neural tubes starting around E9.0 (Fig. 1G and H, arrowheads). Failure of neural tube closure was consistently observed in the hindbrain region and, to a lesser extent, in a posterior region near the base of the tail (Fig. 1I and J, asterisks). Mutants also had small optic cups and forebrains compared to the wild type at E9.5 (Fig. 1G). In spite of their slightly reduced size and these overt defects, several aspects of embryonic development appear to proceed normally in Arid1aV1068G/V1068G embryos, such as the formation of otic placodes and branchial arches. However, by E10.5, most mutant embryos were clearly developmentally delayed, often resembling those collected at E9.5 in appearance and by limb bud staging criteria (Fig. 1H). This delay is strikingly evident in E10.5 mutant hearts, which appear grossly immature compared to those of the wild type (Fig. 1H, arrows). Beyond E10.5, pericardial effusion was noted in some mutants, further indicating defects in heart function at these later stages. Hemorrhaging was also observed in the pericardial sac and abdomens of a few E11.5 mutant embryos. These gross phenotypes are consistent with developmental arrest between E9.0 and 10.5.
Heart defects are associated with the V1068G mutation.
Our gross inspection of whole-mount specimens indicated that heart maturation was defective in mutant embryos. To examine these defects in greater detail, histological sections harvested from E9.5 and E10.5 mutant hearts were compared to similar sections from littermate controls. At E9.5, mutant hearts lacked ventricular trabeculation, and these defects persisted in E10.5 ventricles (Fig. 1K and L, arrowheads). By E10.5, ventricular septal defects were obvious in mutant hearts, indicating a block in cardiac chamber maturation (Fig. 1L). These heart defects bear a striking resemblance to those observed in heterozygous Arid1a-null embryos and in embryos completely lacking ARID1a in second heart field (SHF) derivatives (via Mef2c-AHF-Cre), which further supports a role for ARID1a-containing BAF-A complexes in cardiac progenitor differentiation (23, 49). In spite of these similarities, our data suggest that the midgestation lethality observed in Arid1aV1068G/V1068G embryos is due to the culmination of defects in multiple tissues because SHF-specific loss of ARID1a in the heart results in embryonic lethality at later time points (49).
ARID1a-containing BAF-A complexes are required for vascular branching and vessel integrity in the extraembryonic yolk sac and placenta.
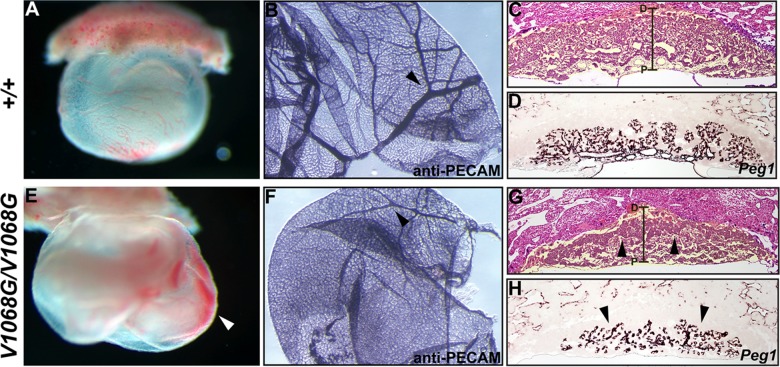
Embryo viability beyond E10.5 is known to be critically dependent on proper extraembryonic circulation (50). This dependence, coupled with our recent findings demonstrating yolk sac angiogenic remodeling defects in Brg1fl/fl; Tie2-Cre+/0 embryos, led us to examine this tissue in greater detail (44, 51). Indeed, the yolk sacs of Arid1aV1068G/V1068G embryos were always abnormal in comparison to those of littermate controls. Evidence of hemorrhaging in the exocoelomic space was frequently observed in these individuals starting as early as E9.5 (Fig. 2E, arrowhead). The extent of blood loss observed varied from a few patchy spots to large pools of blood. By E10.5, wild-type yolk sacs displayed a hierarchical, branch-like pattern of vessels, whereas mutant yolk sac vasculature was poorly developed, often resembling an immature, primary plexus consisting of small capillaries and very few large, vitelline vessels (Fig. 2B and F). A scarcity of large vessels coupled with blood loss also contributed to a frequently observed pallor of mutant yolk sacs upon dissection. In spite of these prominent yolk sac vascular defects, primitive hematopoiesis and flow patterns must have been established in mutants because nucleated, fetal blood can be visualized throughout the embryo proper and yolk sac or pooled in the extraembryonic voids. Furthermore, these primitive fetal blood cells most likely originated from the yolk sac because definitive erythropoiesis in embryonic livers does not initiate until around E11.5. Overall, the presentation of these phenotypes as early as E9.5 indicates that yolk sac vascular dysfunction greatly contributes to mutant embryo mortality.
Fig 2.
Prominent extraembryonic vascular abnormalities are present in Arid1aV1068G/V1068G concepti. Whole-mount images of E9.5 Arid1a+/+ (A) and Arid1aV1068G/V1068G (E) yolk sacs are shown. Blood pools in the exocoelomic space of Arid1aV1068G/V1068G (E, arrowhead) are indicative of hemorrhaging. Whole-mount PECAM staining of E10.5 Arid1a+/+ (B) and Arid1aV1068G/V1068G (F) yolk sacs was performed. Hierarchical vascular branching patterns are evident in wild-type yolk sacs (B, arrowhead), whereas highly branched large vessels are scarce in mutant yolk sacs (F, arrowhead). Hematoxylin- and eosin-stained paraffin sections of E10.5 Arid1a+/+ (C) and Arid1aV1068G/V1068G (G) placentas are shown. False-colored, yellow shading identifies the placenta. Proximal (P) to distal (D) placental axes are demarcated by brackets. Mutant placentas display thick spongiotrophoblast layers (G, arrowheads). Near-adjacent paraffin sections of E10.5 Arid1a+/+ (D) and Arid1aV1068G/V1068G (H) placentas hybridized with the pan-mesodermal marker Peg1 are shown. Vascular branching is reduced in mutant placentas (H, arrowheads).
Improper placental development also leads to death around midgestation in mice (50). Development of the placenta ensues shortly after the attachment of allantois to the chorion around E8.5. Chorioallantoic attachment and subsequent formation of the umbilical cord take place at these early stages in mutants (data not shown). At later stages, the trophoblast layers of mutant placentas were discernible by histology, often having thick spongiotrophoblast layers (Fig. 2G, arrowheads). In general, mutant placentas also appeared smaller than their wild-type counterparts, despite having large maternal lacunae. The extent of vascularization in both mutant and wild-type placentas was determined by the expression of the extraembryonic mesoderm marker, Peg1 (Fig. 2D and H) (43). Vascular density was reduced, and minimal branching was observed in E10.5 mutant placentas (Fig. 2H, arrowheads). Specifically, the parental vessels appear to elongate and bifurcate, but smaller, capillary-like vessels were scarce. This reduction in vascular branching indicates that fetal-maternal exchange is defective in V1068G mutants. Therefore, proper extraembryonic vascular development requires normal BAF-A complex function.
Mutant protein is stably expressed, and it incorporates into a SWI/SNF complex with core catalytic function.
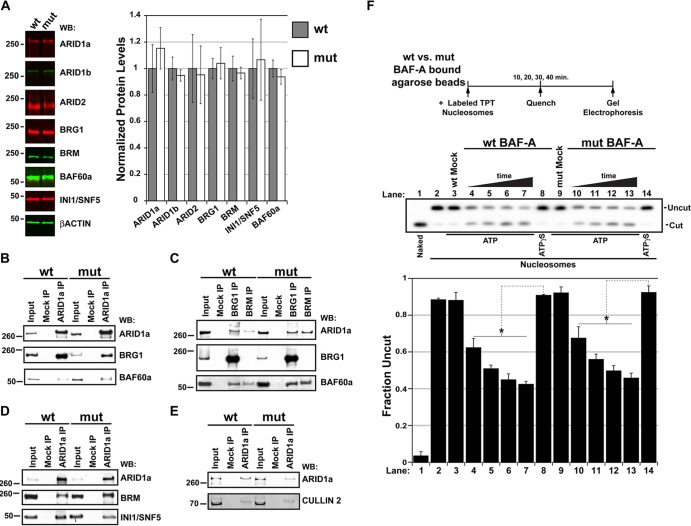
ARID domains do not appear to be required for complex integrity, nor do they appear to be required for the core catalytic properties of the complex (52, 53). However, point mutations like V1068G could influence protein stability and/or conformation, leading to loss of multiple interaction surfaces. The presence of normal ARID1a protein levels in mutant E9.5 whole-embryo lysates indicates that this point mutation does not influence mutant protein expression and/or stability (Fig. 3A). We extended our analysis of potential subunit expression differences in wild-type versus Arid1aV1068G/V1068G E9.5 embryos by quantitative Western blotting using antibodies that recognize the alternative ARID subunits ARID1b and ARID2, the catalytic subunits BRG1 and BRM, and the accessory subunits INI1/SNF5 and BAF60a. Significant differences in subunit expression were not detected when age-matched, whole-embryo protein lysates from Arid1aV1068G/V1068G embryos were compared to those of the wild type (Fig. 3A).
Fig 3.
The V1068G mutation does not affect the stability or the catalytic activity of BAF-A complexes. (A) Quantitative fluorescent Western blots (WB) of Arid1a+/+ and Arid1aV1068G/V1068G E9.5 whole-embryo lysates probed with SWI/SNF subunit-specific antibodies. Graph represents the average ± SEM of the normalized band intensity from three age- or littermate-matched whole-embryo lysates. Band intensity measurements are plotted as a ratio to β-actin signals, and wild-type (wt) measurements were set at 1. mut, mutant. (B to E) Coimmunoprecipitation of SWI/SNF complex subunits or cullin-2 from wild-type or Arid1aV1068G/V1068G MEFs. Shown are Western blot panels containing input protein, mock-precipitated (protein A/G-agarose bead only) protein, or protein precipitated with antibodies specific for ARID1a, BRG1, or BRM. Western blots were probed with antibodies specific for ARID1a, BRG1, BRM, INI1/SNF5, BAF60a, and cullin-2. (F) Mononucleosome disruption by wild-type and mutant ARID1a-containing complexes increases PstI restriction site exposure. Fixed amounts of anti-ARID1a immunopurified fractions from either wild-type or Arid1aV1068G/V1068G MEFs were incubated with limiting amounts of gel-purified, radiolabeled mononucleosomes over a 40-min time course in the presence of ATP. Reaction mixtures containing wild-type or mutant anti-ARID1a fractions were quenched at 10-min intervals. Reaction mixtures containing ATPγS or mock-purified (protein A/G-agarose bead only) fractions and incubated over the entire time course served as controls. Naked DNA (lane 1) or mononucleosomes (lane 2) incubated without immunopurified material are shown. Uncut fractions were plotted as a ratio of the total DNA. Error bars in panel F represent the standard deviations, and significant differences were calculated using a two-tailed Student t test (*, P < 0.05).
In order to determine if the endogenous mutant protein still incorporates into the SWI/SNF complex, immunoprecipitations were performed using antibodies that precipitate either ARID1a or the catalytic subunits, BRG1 and BRM (Fig. 3B to D). These experiments were performed using native nuclear extracts from either wild-type or Arid1aV1068G/V1068G mouse embryonic fibroblasts (MEFs). Since an ARID1a-specific antibody is capable of precipitating BRG1 and BRM, as well as the accessory subunits BAF60a and INI1/SNF5, complex integrity does not appear to be affected by the V1068G substitution in ARID1a (Fig. 3B and D). In reciprocal immunoprecipitations using antibodies that precipitate either BRG1 or BRM, we further corroborated the interactions with ARID1a and BAF60a (Fig. 3C). We extended our analysis to include a noncanonical ARID1a-interaction partner, cullin-2, which can also form stable interactions with mutant ARID1a (Fig. 3E) (54). Together, these findings suggest that the V1068G mutation does not influence the ability of ARID1a to stably associate with canonical SWI/SNF subunits or noncanonical interaction partners.
To determine if mutant ARID1a-containing, BAF-A complexes still maintain core catalytic function, we subjected reconstituted mononucleosomes to a PstI restriction enzyme accessibility assay in the presence of wild-type or mutant immunopurified BAF-A complexes with either ATP or the nonhydrolyzable analog, ATPγS (36). Reactions were performed using saturating amounts of immobilized BAF-A complexes (on beads), and the rates of PstI site exposure were examined over a 40-min time course, with samples quenched every 10 min. Both wild-type and mutant ARID1a-containing complexes increased the rate of PstI site exposure to similar extents with approximately half of the nucleosome particles undergoing remodeler-dependent cleavage after 20 min, suggesting that these complexes are catalytically active in the presence of ATP (Fig. 3F, lanes 5 and 11). Importantly, BAF-A-dependent PstI site-accessible fractions were not observed in reaction mixtures containing mock-purified samples (Fig. 3F, lanes 3 and 9) or ATPγS (lanes 8 to 14). These findings are reminiscent of the intact core catalytic activities of yeast SWI/SNF complexes lacking an ARID domain (53). Altogether, these data strongly suggest that BAF-A complex integrity and core catalytic properties are unaffected by the V1068G mutation within the ARID domain of ARID1a.
The V1068G mutation abrogates ARID-DNA binding.
The lethal phenotypes observed in Arid1aV1068G/V1068G embryos demonstrate that the V1068 residue is critical for normal ARID1a function. In general, critical protein residues are likely to be conserved throughout evolution. Based on primary sequence homology, the minimal ARID domain of ARID1a is phylogenetically similar to the Drosophila melanogaster Retained/Dri (Retn/Dri) and Mus musculus ARID3a/BRIGHT minimal ARIDs (55). The mutated valine (V1068) residue (Fig. 4A, red) is conserved among all ARID proteins belonging to the Dri/BRIGHT clade, including the D. melanogaster and Saccharomyces cerevisiae putative ARID1a homologs, Osa and SWI1, respectively. This residue is also invariant among all ARID proteins found within the mammalian SWI/SNF complexes, including ARID1b and ARID2 (10). Notably, all known ARID proteins have a nonpolar residue (valine or isoleucine) corresponding to this position (55).
Fig 4.
Structural implications of the conserved valine 1068 residue. (A) Clustal alignment of minimal ARID domains from M. musculus (Mm) ARID1a/BAF250a aligned with similar regions from D. melanogaster (Dm) Osa and S. cerevisiae (Sc) SWI1 ARID subunits, as well as D. melanogaster Retn/Dri/Bright and M. musculus ARID3a/BRIGHT proteins. The mutated valine 1068 residue (red) is highlighted in yellow. (B, left) PyMol rendition of all eight ARID domain NMR conformers (PDB code 1RYU) for the ARID domain structure of H. sapiens ARID1a. α-Helices are depicted in red. V1068 is located in α-helix 3. (B, right) Close-up view of the V1068 side chain contact neighborhood. Favorable, nearest-neighbor V1068, V1059, Y1097, L1077, and K1094 (blue boldface in panel A) side chain interactions form a hydrophobic pocket in the center of a multi-α-helical bundle (H2 to H5).
Nonpolar amino acids typically stabilize proteins by sequestering their side chains in hydrophobic environments. To gather molecular information about the critical V1068 residue, we simulated the effect of the valine-to-glycine substitution in V1068G using a computational algorithm called simplicial neighborhood analysis of protein packing (SNAPP), which is useful for estimating how mutational changes in hydrophobic residues affect protein stability (46, 56–58). In general, a mutation that lowers the total SNAPP score negatively contributes to protein stability (56). We calculated the SNAPP scores for all eight 120-amino-acid conformers derived from the previously reported nuclear magnetic resonance (NMR) solution structure of the ARID domain (hARID) of Homo sapiens ARID1a (Protein Data Bank [PDB] code 1RYU) (59). Our analysis yielded an average SNAPP score of 268.7 for all wild-type hARID conformers. By substituting valine for a glycine residue, this average score is reduced by 2.8 log-likelihood units, suggesting a reduction in thermodynamic stability. Scoring of individual residue contacts within the V1068 side chain contact neighborhood indicated that this negative difference is attributable to the effect of the glycine on the collapse of a hydrophobic pocket created by side chains emanating from a multi-α-helical bundle consisting of α-helices 2 to 5 (H2 to H5) (Fig. 4B). Of the 14 amino acids conserved throughout all Dri/BRIGHT minimal ARIDs, V1068 and two other nonpolar residues (V1059 and Y1097) constitute this hydrophobic residue cluster, whereas the other two residues found in this region (L1077 and K1094) show conservative changes across most Dri/BRIGHT ARIDs (Fig. 4A, blue boldface, and B, inset). Consistent with nonpolar side chain sequestering tendencies, the single polar side chain identified in this motif, K1094, is oriented toward the edge of this cluster, while the aliphatic valine and leucine side chains (V1059, V1068, and L1077) and the bulky aromatic ring of tyrosine 1097 are sequestered toward its center (Fig. 4B). We predict that the V1068G mutation disrupts favorable van der Waals contacts within this pocket region, leading to ARID domain instability and diminished DNA binding potentials.
To test this hypothesis, single-component saturating electrophoretic mobility shift assays (ssEMSAs) were performed using bacterially expressed GST fusion protein fragments containing either a wild-type or mutant (V → G) ARID domain from human ARID1a (GST-hARID) (Fig. 2). In ssEMSAs, the apparent Kd (dissociation constant), or Kapp, equals the protein concentration for which 50% of the DNA is bound when the DNA probe concentration is limiting. Binding isotherms were estimated from reaction mixtures containing titrated amounts of protein and limiting amounts (50 pM) of the radiolabeled dri.16 probe (16). As shown in Fig. 5A and C, saturating amounts of wild-type GST-hARID protein are capable of binding subsaturating amounts of DNA. Curve fitting calculated a Kapp of 92.2 nM for the wild-type GST-hARID protein fragment (Fig. 5C). However, even high concentrations (2 μM) of mutant protein fragments carrying the valine-to-glycine substitution are not capable of binding DNA (Fig. 5B and C). Similar results were observed in reaction mixtures containing saturating amounts (1 μM) of wild-type or mutant GST-hARID protein fragments and an alternate DNA probe or reaction buffer (data not shown). Thus, the V1068G substitution causes a severe reduction in ARID1a-DNA interactions.
Fig 5.
The V1068G mutation leads to loss of DNA binding. (A and B) Single-component saturating EMSAs (ssEMSAs) using titrated amounts of wild-type (wt) or mutant (mut; V → G) recombinant protein fragments containing the non-sequence-specific ARID domain of H. sapiens (Hs) ARID1a. (D and E) Similar ssEMSAs using titrated amounts of wild-type or mutant (V → G) recombinant protein fragments containing the sequence-specific ARID domain of D. melanogaster Retn/Dri/Bright. Subsaturating amounts of the dri.16 probe (50 pM) were used in each DNA binding reaction. (C and F) Fractional occupancies were calculated from band densitometry measurements and plotted versus log protein concentrations. Blue dotted lines in panels C and F represent curves fitted to EMSA results using Kaleidagraph software.
Since the residues comprising the ARID hydrophobic pocket show conservation among Dri/BRIGHT ARID proteins, we next wanted to determine if this substitution would also abrogate the DNA binding potentials of a functionally unrelated, sequence-specific ARID domain within this clade. Similar ssEMSAs were performed using a GST fusion protein fragment encompassing the Drosophila Retn/Dri ARID domain (GST-DriARID) and a radiolabeled probe (dri.16) containing its cognate binding site (16, 30). Consistent with previously reported binding affinity measurements, wild-type GST-DriARID fragments are capable of binding DNA with a Kapp of 50.2 nM (Fig. 5D and F) (32). However, mutant GST-DriARID fragments carrying the valine-to-glycine substitution are not capable of saturation binding at protein concentrations greater than the Kapp of the wild-type fragment. Likewise, since less than 20% of the free DNA is bound in reaction mixtures containing micromolar amounts of mutant GST-DriARID, the Kapp for this fragment could not be estimated (Fig. 5E and F). Taken together, these data strongly suggest that a central hydrophobic pocket formed by a multi-α-helical bundle within the ARID domain is required for both non-sequence- and sequence-specific DNA interactions.
Loss of ARID1a-DNA interactions reduces BAF-A affinity for nucleosomes.
In addition to primary recruitment by transcription factors, it has been proposed that the full binding capacity of SWI/SNF for its nucleosomal substrate requires the summation of multiple sequence-nonspecific subunit-chromatin interaction surfaces (60). To test whether impaired ARID1a-DNA interactions reduce BAF-A binding affinity for nucleosomes, we performed competitive nucleosome pulldown assays using natively purified wild-type or mutant BAF-A complexes. Saturating amounts of immobilized (on beads) complexes were incubated with limiting amounts of radiolabeled nucleosomes in reaction mixtures containing titrated amounts of cold competitor DNA (Fig. 6A). Under competitive binding conditions, differences in the amount of nucleosome bound correlates with differences in the relative binding affinities of the complex. In reaction mixtures containing no competitor DNA, nucleosome binding by mutant BAF-A complexes was comparable to that of the wild type (Fig. 6A and B). However, a significant reduction in mutant BAF-A binding affinity was observed in reaction mixtures containing increasing amounts of competitor DNA, with approximately 1.5-fold less nucleosomal substrate bound to mutant complexes at competitor DNA concentrations as low as 3.1 ng (Fig. 6A and B). No binding was observed in reaction mixtures containing mock-purified samples (Fig. 6A and B). Altogether, these data strongly suggest that a severe reduction in ARID1a-DNA interactions leads to a moderate decrease in BAF-A affinity for nucleosomes, which would be consistent with a role for the ARID domain in contributing to some, but not all, SWI/SNF subunit-chromatin interactions.
Fig 6.

Loss of sequence-nonspecific ARID1a-DNA interactions reduces BAF-A affinity for nucleosomes. (A) Saturating amounts of immobilized wild-type or mutant BAF-A complexes were incubated with limiting amounts of radiolabeled mononucleosomes in reaction mixtures containing increasing amounts of cold competitor DNA, and then the beads were subjected to a pulldown and washing. Mock-purified fractions from wild-type or mutant cells were used as negative controls. (B) Graph depicts the average scintillation counts measured from independent binding reactions. The numbers above the graphical columns denote the fold difference in average scintillation counts of the wild type compared to those of the mutant. Significant differences were calculated using a two-tailed Student t test (*, P < 0.05).
The ARID domain of ARID1a is required for repressive BAF-A associations at THBS1.
To determine if the V1068G lesion augments ARID1a occupancy at gene promoters, we performed chromatin immunoprecipitation (ChIP) assays on formaldehyde-cross-linked chromatin from exponentially growing wild-type and mutant Arid1aV1068G/V1068G MEFs. We decided to use MEFs in these experiments because they can be efficiently cultured at a scale amenable to biochemical fractionation and because they can easily be isolated from embryos prior to E10.0. The following set of putative ARID1a target genes was chosen because they are widely expressed throughout the embryo and because they are targets of BRG1 or BRM: THBS1, ADAMTS1, and CRABP1 (61–63). Two unlinked promoter control elements from a constitutive (GAPDH) or silent (INSULIN-1 or INS-1) gene were used as negative genomic controls (Fig. 7A). ARID1a was enriched over these control regions at all three putative targets in wild-type MEFs (Fig. 7A). In Arid1aV1068G/V1068G MEFs, mutant ARID1a occupancy was reduced to nearly background levels at all target sites examined, indicating that this mutant protein does not efficiently bind chromatin (Fig. 7A). This reduction also correlates with concurrent changes in target gene expression (Fig. 7B).
Fig 7.

Loss of DNA binding correlates with reduced promoter occupancy and concurrent changes in SWI/SNF target gene expression. (A) Formaldehyde-cross-linked chromatin from wild-type and Arid1aV1068G/V1068G MEFs was immunoprecipitated using an ARID1a-specific antibody. DNA was amplified by quantitative PCR to determine if mutant ARID1a occupancy was reduced at known SWI/SNF target gene promoters (THBS1, ADAMTS1, and CRABP1). Promoter control elements for a constitutive (GAPDH) or silent (INS-1) gene were used as negative genomic controls. Data were plotted as the percentage of total input or chromatin bound. (B) cDNA synthesized from RNA isolated from wild-type or Arid1aV1068G/V1068G MEFs was used in a quantitative PCR to examine target gene expression differences. Error bars in both panels represent the SEMs, and significant differences between the wild type and mutant were calculated using a two-tailed Student t test (*, P < 0.05).
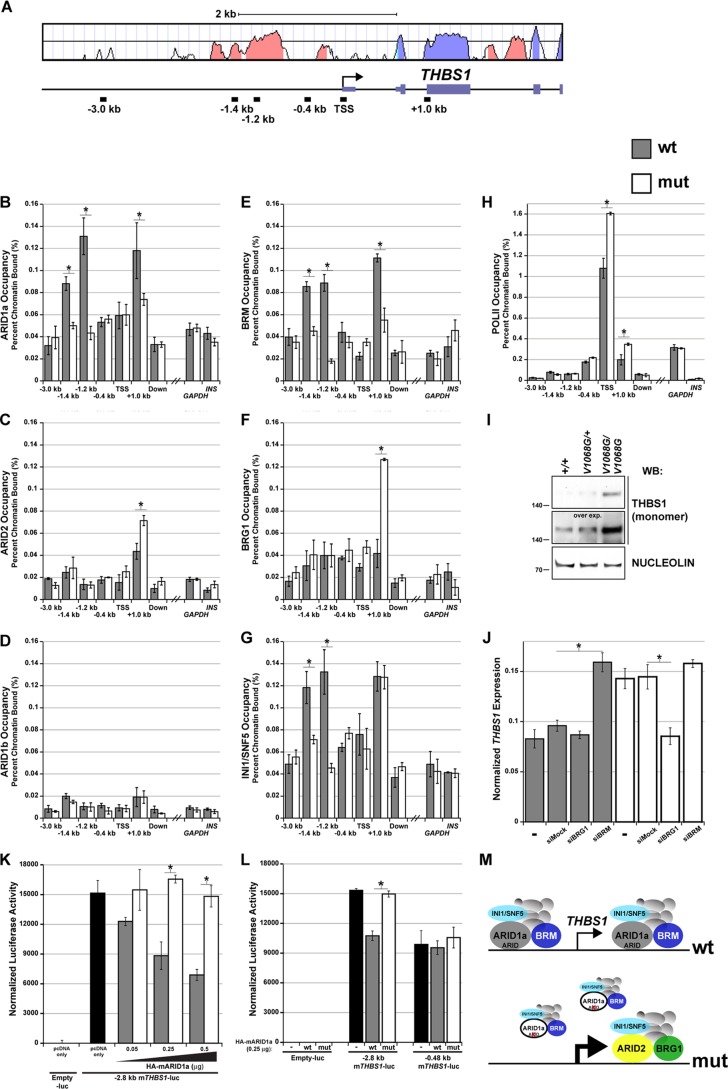
Since the V1068G mutation in ARID1a lowers BAF-A affinity for nucleosomes and since ARID1a occupancy is reduced in mutant cells, we reasoned that mutant BAF-A occupancy would also be affected by this mutation. To test this idea, we performed additional subunit-specific ChIP experiments to determine if promoter occupancy by the BAF-A complex is dependent on the ARID domain. Among the ARID1a target genes examined, THBS1 emerged as a good candidate for these experiments because transcriptional control elements within 2.8 kb upstream of its promoter have been extensively characterized in mouse embryonic fibroblast cell lines (45, 64, 65). Thus, THBS1 served as a model BAF-A target gene to further probe ARID domain function. We chose a ChIP-scanning approach to narrow regions of SWI/SNF enrichment across a 4-kb interval spanning this promoter fragment, largely focusing on genomic “landmarks” such as conserved regions or transcription start sites (Fig. 8A). BAF-A complexes are known to incorporate either BRG1 or BRM, and BRG1 incorporates into PBAF complexes containing ARID2. Therefore, to adequately survey all potential SWI/SNF complex configurations within this interval, we performed ChIP using antibodies that recognize all of the mammalian ARID subunits, ARID1a, ARID1b, and ARID2, as well as the catalytic subunits, BRG1 and BRM. We also included the core subunit INI1/SNF5, which is common to all mammalian SWI/SNF complexes. A cis-linked and nonconserved intergenic region located approximately 20 kb downstream of the THBS1 transcription start site (TSS) and the promoter control elements, GAPDH and INS-1, were used as negative genomic controls.
Fig 8.
The ARID domain of ARID1a is required for BAF-A occupancy at THBS1. (A) Mouse/human VISTA alignment of the THBS1 promoter and 5′ transcribed region. Salmon-colored peaks denote evolutionarily conserved regions, whereas lavender peaks denote exons. The locations of ChIP amplicons within this interval are plotted. (B to H) Formaldehyde-cross-linked chromatin from wild-type and Arid1aV1068G/V1068G MEFs was immunoprecipitated with ARID1a, ARID1b, ARID2, BRG1, BRM, INI1/SNF5, or Pol II antibodies. DNA was amplified by quantitative PCR to determine if loss of ARID-DNA binding leads to changes in BAF-A occupancy across THBS1. An intergenic, nonconserved downstream region (located at approximately kb +20) and two unlinked promoter control elements (GAPDH and INS-1) were used as negative genomic controls. Data were plotted as the percentage of total input or chromatin bound. (I) Whole-embryo protein lysates from triplicate pooled samples were used to examine THBS1 protein (reduced, monomeric form) expression differences by Western blotting. An overexposed image of the Western blot was also included to further emphasize these expression differences. The constitutive nuclear matrix protein, nucleolin, was used as a loading control. (J) cDNA synthesized from RNA isolated from wild-type or Arid1aV1068G/V1068G MEFs was used in a quantitative PCR to examine THBS1 expression differences following transfection with mock (nontargeting control), BRG1, or BRM siRNA. (K) Normalized luciferase activity of the THBS1 −2.8 kb promoter-luciferase fragment cotransfected with 0.05 to 0.5 μg of wild-type or mutant HA-mARID1a expression plasmids into NIH 3T3 cells. Cells cotransfected with the empty luciferase vector (−luc) or THBS1 −2.8 kb promoter-luciferase fragment and with empty pcDNA expression plasmids served as negative and positive controls, respectively. (L) Normalized luciferase activity of THBS1 −2.8 kb and −0.48 kb promoter-luciferase reporter plasmids cotransfected with 0.25 μg of pcDNA only (−) or wild-type or mutant HA-mARID1a expression plasmids. Empty luciferase reporter plasmid was used as a negative control. (M) Summary model of ChIP and expression data. Error bars in panels B to H and in panel J represent the SEMs. Error bars in panels K and L represent the standard deviations. Significant differences were calculated using a two-tailed Student t test (*, P < 0.05).
In wild-type MEFs, ARID1a enrichment was detected at a conserved upstream promoter element and at a downstream genic region located at kb +1.0 (Fig. 8B). ARID1a enrichment is coincident with BRM and INI1/SNF5 enrichment at these sites in wild-type cells (Fig. 8E and G). In Arid1aV1068G/V1068G MEFs, BRM-containing BAF-A complexes were depleted from these sites, as indicated by the reduction in ARID1a and BRM occupancy (Fig. 8B and E). INI1/SNF5 was also depleted from the upstream site, but its enrichment remained unaltered at the downstream kb +1.0 site in these cells (Fig. 8G). We detected BRG1 and the PBAF-specific ARID subunit, ARID2, at this downstream site only in mutant cells, which accounts for INI1/SNF5 occupancy at this site (Fig. 8C and F). We did not detect ARID1b enrichment at these sites in either wild-type or Arid1aV1068G/V1068G cells (Fig. 8D). We conclude that loss of ARID-DNA interactions in V1068G mutants perturbs proper BAF-A associations at THBS1, and these losses can influence chromatin associations by additional SWI/SNF complexes.
To determine if these changes in promoter occupancy correlated with concurrent changes in THBS1 expression, we profiled THBS1 expression in MEFs or embryos. In Arid1aV1068G/V1068G MEFs, THBS1 was upregulated by quantitative RT-PCR (qRT-PCR), compared to wild-type levels (Fig. 8J). THBS1 protein levels from E9.5 whole-embryo lysates (triplicate pooled) for all genotypic classes were examined by Western blotting. THBS1 protein levels were increased in E9.5 Arid1aV1068G/V1068G versus wild-type embryos, which is consistent with our qRT-PCR results (Fig. 8I). We also detected a slight increase in THBS1 protein in Arid1aV1068G/+ embryos (Fig. 8I). We further verified THBS1 upregulation by Western blotting and direct immunostaining of tissue sections from E10.5 Arid1aV1068G/V1068G versus wild-type embryos (data not shown). RNA polymerase II (Pol II) occupancy was also increased over the TSS and transcribed, genic region (kb +1) of THBS1 in mutant MEFs, suggesting that ARID-dependent regulation of THBS1 by BAF-A is likely occurring at the level of transcriptional repression (Fig. 8H).
To further explore this idea, we transfected wild-type and Arid1aV1068G/V1068G MEFs with small interfering RNAs (siRNAs) that target BRG1 or BRM. In wild-type MEFs, siRNA depletion of BRM, but not BRG1, led to THBS1 upregulation, which is consistent with repression by BRM-containing, BAF-A complexes (Fig. 8J). In contrast, BRG1 depletion in mutant cells resulted in THBS1 downregulation, suggesting that PBAF occupancy at the downstream site likely contributes to the transcriptional upregulation we observed in mutant cells (Fig. 8J). These results further suggest that BRM-containing BAF-A complexes directly repress THBS1 transcription in an ARID-dependent manner.
To test this model, we cotransfected increasing amounts of wild-type or mutant (V1068G) HA-tagged mouse ARID1a cDNA expression plasmids with a mouse THBS1 −2.8 kb promoter-luciferase reporter plasmid into NIH 3T3 cells. Overexpression of wild-type ARID1a repressed the THBS1 −2.8 kb promoter at plasmid concentrations at or above 0.25 μg (Fig. 8K). However, this level of repression was not observed in cells transfected with similar amounts of mutant ARID1a cDNA (Fig. 8K). To determine if ARID1a directly regulates this promoter in a manner that is consistent with BAF-A binding to the upstream conserved element, we performed a separate set of cotransfections using a minimal THBS1 −0.48 kb promoter-luciferase fragment and 0.25 μg of wild-type or mutant ARID1a expression plasmids. As expected, ARID1a overexpression repressed the THBS1 −2.8 kb promoter fragment in an ARID domain-dependent manner, whereas neither wild-type nor mutant ARID1a had an effect on the activity of the THBS1 −0.48 kb fragment in parallel assays (Fig. 8L). These data strongly suggest that the upstream element is a site of THBS1 promoter repression by BAF-A. ChIP and expression data for THBS1 are summarized in Fig. 8M. Taken together, these data demonstrate that the ARID domain of ARID1a is both necessary and sufficient for repressive BRM-containing BAF-A associations at THBS1.
DISCUSSION
In this study, we report the molecular and phenotypic characterization of an ENU-induced point mutation within the ARID DNA binding domain of ARID1a. This single amino acid substitution abrogates the DNA binding capacities of ARID1a but does not influence ARID1a protein levels, nor does it compromise the integrity or the core catalytic properties of the BAF-A complex. Loss of ARID1a-DNA binding led to a convergence of heart and vascular defects in mutant embryos, and the sum of defects culminated in ischemic arrest around midgestation. Our data indicate that the function of the SWI/SNF ARID domain is to facilitate complex binding to target gene promoters. Genetic evidence supporting such a role has remained nonexistent until now.
The importance of valine 1068 in both sequence-dependent and -independent ARID-DNA interactions has been unexplored because this residue does not contact DNA (32, 59). Our data indicate that this highly conserved, nonpolar amino acid specifically influences ARID domain stability and/or conformation through hydrophobic side chain packing although we cannot rule out the possibility that a yet untested protein-protein interaction is also perturbed. The functional significance of these hydrophobic interactions can be applied to most ARID proteins found in higher eukaryotes because they all have nonpolar residues corresponding to ARID1a positions 1059, 1068, 1077, and 1097 within this packing motif (32, 55, 59). In addition, at least one human cancer cell line (MDA-MB-361) has a valine-to-glycine substitution in amino acid 1059 (1058 in human) in ARID1a (66). We predict that this substitution also perturbs the ARID domain hydrophobic pocket, leading to loss of DNA binding. This highlights how protein structure-function-based approaches in the mouse can be used to predict the functional impact of missense mutations in humans.
SWI/SNF ARIDs are predicted to bind within the major groove, but they have lost the ability to bind specific bases within this region because the major groove-interacting, helix-turn-helix-like (H4-L2-H5) subdomain has been modified, and amino acids found at base-specific contact positions within this subdomain have diverged (32, 59). Less is known about how the flexible, minor groove- and phosphate backbone-interacting L1 loop and C terminus or the divergent extended ARID regions influence binding affinity and/or sequence preference. However, these interaction surfaces are also involved in DNA recognition because mutations in key major groove contact residues do not completely abolish DNA binding (10). The V1068G mutation reduced the DNA binding capacities of ARID fragments from Dri/BRIGHT and ARID1a proteins to nonphysiological levels. Therefore, this mutation likely perturbs multiple DNA interaction surfaces within the ARID domain, which is consistent with a severe reduction in ARID domain function.
Although SWI/SNF ARIDs lack the major groove consensus binding convention used by most sequence-specific binding proteins, they may show increased binding affinity for a yet undetermined DNA sequence or higher-order DNA structure, or their recognition of DNA may be facilitated through the exploitation of sequence-dependent DNA deformability. Interactions like these may be driven, in part, by minor groove interactions, which are often accompanied by DNA bending. At least one example of ARID-induced DNA bending has been reported (67). Additional, high-resolution binding studies using full-length ARID subunits or approaches based on ChIP with high-throughput sequencing (ChIP-seq) may provide additional information.
A previous report has identified an upstream element between kb −1.3 and −1.1 of the mouse THBS1 promoter that is strongly repressive in NIH 3T3 cells (65). Consistent with these findings, our data suggest that ARID domain-dependent BAF-A enrichment within this element is necessary for THBS1 repression. Additional sequences within 2.8 kb upstream of the THBS1 promoter likely contain activating elements because the basal activity of this fragment is greater than that of the minimal THBS1 −0.48 kb fragment tested, despite having the upstream repressive element. Moreover, THBS1 transcripts and protein were detected in wild-type cells and embryos, respectively. We speculate that BAF-A activity at the upstream element may buffer the transcription output of THBS1, yet “prime” this locus for rapid induction in response to external stimuli, which would be consistent with its role as an immediate-early response gene (68, 69). The identification of factor binding sites that are necessary for BAF-A-mediated repression of THBS1 promoter activity or stimuli required to relieve this repression may allow us to further explore these possibilities.
Unexpectedly, we observed an ARID-dependent switch from a BAF-A to a PBAF configuration at a separate, and previously uncharacterized, downstream site located within the transcribed region (intron 2) of THBS1. Indeed, our data suggest that BAF-A binding to this downstream site may also regulate THBS1 activity. Recently, a similar switch from BAF-A to PBAF was observed on a single nucleosome following phorbol myristate acetate (PMA) stimulation of the HIV long terminal repeat (LTR) (70). At the THBS1 downstream site, we speculate that, similar to what has been proposed for the HIV LTR, repressive nucleosome positioning by BAF-A may act as a barrier to PBAF recruitment, perhaps limiting THBS1 elongation rates. Although it is still unclear whether PBAF can be recruited to this site in wild-type cells following THBS1 stimulation or if the switch we observed occurs on a single nucleosome, our data would also suggest that PBAF activity at this site contributes to the THBS1 upregulation we observed. Such antagonism may be an important regulatory step if the remodeling activity of one complex modulates the activity or recruitment of another, especially considering that two or more distinct complexes are often present in the same cell. This premise may integrate multiple chromatin remodelers into the regulatory decisions at a given locus. More data are needed to determine if complex switching or antagonism is a frequent event or if it is implicated in a particular transcriptional regulatory process.
The hypomorphic phenotypes observed in Arid1aV1068G/V1068G embryos indicate that some BAF-A activities are maintained following a severe reduction in ARID domain function. These partial loss-of-function phenotypes would be expected if mutant ARID1a had some residual binding activity in vivo or if there were partial redundancy between the ARID domain and other recruitment factors at select sites. Alternatively, BAF-A interactions with RNA polymerase II or other macromolecular complexes may circumvent the need for this targeting domain in additional processes that require SWI/SNF chromatin remodeling activities (71, 72). The phenotypic severity of the V1068G mutation parallels mutations in yeast SWI/SNF that remove ARID or bromodomain function as these domains are not required for full complex activity in vivo (53, 73). These rather limited targeting domain requirements suggest that multiple factors influence SWI/SNF recruitment in vivo.
The ARID domain is not the first target site-indiscriminate chromatin interaction domain implicated in SWI/SNF recruitment processes. For example, acetylated histone tails can increase SWI/SNF binding affinity for nucleosomes in a bromodomain-dependent manner, and this bromodomain-acetyl-histone H3 tail interaction is required for promoter occupancy by SWI/SNF (73, 74). These findings suggest that subunit interactions with sequence-specific transcription factors alone are not sufficient for SWI/SNF recruitment and that proper SWI/SNF associations with chromatin require intrinsic ARID-DNA and subunit-histone interactions. Additional site-indiscriminate subunit-DNA or subunit-histone interaction modules within SWI/SNF may perform similar roles, and their use, in combination, may further enhance recruitment or guide target site selection. Indeed, the moderate differences we observed in the relative nucleosome binding affinities for wild-type versus ARID-defective BAF-A complexes are similar to the differences observed for bromodomain-dependent SWI/SNF binding to acetylated versus unmodified histone H3 tails, suggesting that the combined effect of multiple subunit-chromatin interaction surfaces would titrate the binding affinities well above what would be expected from a single point of interaction (74). These interactions may also influence the residence time or off rate of the complex, which would stabilize site-specific associations (73). It is possible that ARID-DNA interactions with chromatin also induce conformational or allosteric changes in the complex, perhaps in a manner similar to the separable role proposed for bromodomain-acetyl-histone H3 tail interactions in H2A/H2B dimer displacement (74). Nonetheless, our data support the idea that cooperative interactions among intrinsic subunit-chromatin interaction domains and sequence-specific transcription factors drive SWI/SNF recruitment in vivo. Such cooperativity may allow the complex, or complex-associated transcription factor, interrogate low-affinity, nucleosome-bound sites while at the same time confer combinatorial control of target genes through differential subunit and transcription factor usage (60). These concepts are crucial for understanding how remodelers or other chromatin-modifying enzymes recognize and bind their naturally repressive substrates.
The high prevalence of inactivating ARID1a mutations in ovarian clear cell and endometrioid tumors strongly suggests that loss of ARID1a contributes to the etiology of ovarian cancer (24, 25). Several of these mutations occur only in the heterozygous state, supporting the known gene dose-dependent requirements of ARID1a in the mouse (23). Whether Arid1aV1068G/+ mice are more susceptible to ovarian tumors will require additional experiments. Nonetheless, we are optimistic that this mutant allele will advance our understanding of this disease.
ACKNOWLEDGMENTS
We thank members of the Magnuson lab and Scott Bultman for helpful discussions. We thank Randall Bowen and Della Yee for technical assistance, Anna Spagnoli for generously sharing equipment, Elizabeth Moran for sharing the GST-Dri and GST-p270 protein expression plasmids, and Myriam Hemberger for sharing the Peg1 probe template plasmid. Denis Fourches and Alex Tropsha performed the protein structure simulations and virtual mutagenesis.
This work was supported in part by an American Cancer Society Postdoctoral Fellowship to R.L.C. (PF-09-116-01-CCG) and NIH grants to C.P. (R01HL61656) and T.M. (R01HD036655).
Footnotes
Published ahead of print 5 November 2012
REFERENCES
- 1. Weintraub H, Groudine M. 1976. Chromosomal subunits in active genes have an altered conformation. Science 193: 848–856 [DOI] [PubMed] [Google Scholar]
- 2. Neigeborn L, Carlson M. 1984. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 108: 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stern M, Jensen R, Herskowitz I. 1984. Five SWI genes are required for expression of the HO gene in yeast. J. Mol. Biol. 178: 853–868 [DOI] [PubMed] [Google Scholar]
- 4. Cairns BR, Kim YJ, Sayre MH, Laurent BC, Kornberg RD. 1994. A multisubunit complex containing the SWI1/ADR6, SWI2/SNF2, SWI3, SNF5, and SNF6 gene products isolated from yeast. Proc. Natl. Acad. Sci. U. S. A. 91: 1950–1954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cote J, Quinn J, Workman JL, Peterson CL. 1994. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science 265: 53–60 [DOI] [PubMed] [Google Scholar]
- 6. Peterson CL, Dingwall A, Scott MP. 1994. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc. Natl. Acad. Sci. U. S. A. 91: 2905–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bourachot B, Yaniv M, Muchardt C. 1999. The activity of mammalian brm/SNF2α is dependent on a high-mobility-group protein I/Y-like DNA binding domain. Mol. Cell. Biol. 19: 3931–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Collins RT, Furukawa T, Tanese N, Treisman JE. 1999. Osa associates with the Brahma chromatin remodeling complex and promotes the activation of some target genes. EMBO J. 18: 7029–7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cote J, Peterson CL, Workman JL. 1998. Perturbation of nucleosome core structure by the SWI/SNF complex persists after its detachment, enhancing subsequent transcription factor binding. Proc. Natl. Acad. Sci. U. S. A. 95: 4947–4952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Patsialou A, Wilsker D, Moran E. 2005. DNA-binding properties of ARID family proteins. Nucleic Acids Res. 33: 66–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Quinn J, Fyrberg AM, Ganster RW, Schmidt MC, Peterson CL. 1996. DNA-binding properties of the yeast SWI/SNF complex. Nature 379: 844–847 [DOI] [PubMed] [Google Scholar]
- 12. Singh M, D'Silva L, Holak TA. 2006. DNA-binding properties of the recombinant high-mobility-group-like AT-hook-containing region from human BRG1 protein. Biol. Chem. 387: 1469–1478 [DOI] [PubMed] [Google Scholar]
- 13. Wang W, Chi T, Xue Y, Zhou S, Kuo A, Crabtree GR. 1998. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc. Natl. Acad. Sci. U. S. A. 95: 492–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wilsker D, Patsialou A, Zumbrun SD, Kim S, Chen Y, Dallas PB, Moran E. 2004. The DNA-binding properties of the ARID-containing subunits of yeast and mammalian SWI/SNF complexes. Nucleic Acids Res. 32: 1345–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sudarsanam P, Winston F. 2000. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 16: 345–351 [DOI] [PubMed] [Google Scholar]
- 16. Dallas PB, Pacchione S, Wilsker D, Bowrin V, Kobayashi R, Moran E. 2000. The human SWI-SNF complex protein p270 is an ARID family member with non-sequence-specific DNA binding activity. Mol. Cell. Biol. 20: 3137–3146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Inoue H, Furukawa T, Giannakopoulos S, Zhou S, King DS, Tanese N. 2002. Largest subunits of the human SWI/SNF chromatin-remodeling complex promote transcriptional activation by steroid hormone receptors. J. Biol. Chem. 277: 41674–41685 [DOI] [PubMed] [Google Scholar]
- 18. Kato H, Tjernberg A, Zhang W, Krutchinsky AN, AN W, Takeuchi T, Ohtsuki Y, Sugano S, de Bruijn DR, Chait BT, Roeder RG. 2002. SYT associates with human SNF/SWI complexes and the C-terminal region of its fusion partner SSX1 targets histones. J. Biol. Chem. 277: 5498–5505 [DOI] [PubMed] [Google Scholar]
- 19. Nie Z, Xue Y, Yang D, Zhou S, Deroo BJ, Archer TK, Wang W. 2000. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol. Cell. Biol. 20: 8879–8888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nie Z, Yan Z, Chen EH, Sechi S, Ling C, Zhou S, Xue Y, Yang D, Murray D, Kanakubo E, Cleary ML, Wang W. 2003. Novel SWI/SNF chromatin-remodeling complexes contain a mixed-lineage leukemia chromosomal translocation partner. Mol. Cell. Biol. 23: 2942–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yan Z, Cui K, Murray DM, Ling C, Xue Y, Gerstein A, Parsons R, Zhao K, Wang W. 2005. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 19: 1662–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nagl NG, Jr, Wang X, Patsialou A, Van Scoy M, Moran E. 2007. Distinct mammalian SWI/SNF chromatin remodeling complexes with opposing roles in cell-cycle control. EMBO J. 26: 752–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. 2008. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc. Natl. Acad. Sci. U. S. A. 105: 6656–6661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, Roden R, Glas R, Slamon D, Diaz LA, Jr, Vogelstein B, Kinzler KW, Velculescu VE, Papadopoulos N. 2010. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330: 228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, Zeng T, Senz J, McConechy MK, Anglesio MS, Kalloger SE, Yang W, Heravi-Moussavi A, Giuliany R, Chow C, Fee J, Zayed A, Prentice L, Melnyk N, Turashvili G, Delaney AD, Madore J, Yip S, McPherson AW, Ha G, Bell L, Fereday S, Tam A, Galletta L, Tonin PN, Provencher D, Miller D, Jones SJ, Moore RA, Morin GB, Oloumi A, Boyd N, Aparicio SA, Shih Ie M, Mes-Masson AM, Bowtell DD, Hirst M, Gilks B, Marra MA, Huntsman DG. 2010. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 363: 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen Y, Vivian JL, Magnuson T. 2003. Gene-based chemical mutagenesis in mouse embryonic stem cells. Methods Enzymol. 365: 406–415 [DOI] [PubMed] [Google Scholar]
- 27. Chen Y, Yee D, Dains K, Chatterjee A, Cavalcoli J, Schneider E, Om J, Woychik RP, Magnuson T. 2000. Genotype-based screen for ENU-induced mutations in mouse embryonic stem cells. Nat. Genet. 24: 314–317 [DOI] [PubMed] [Google Scholar]
- 28. Aaronson SA, Todaro GJ. 1968. Development of 3T3-like lines from Balb-c mouse embryo cultures: transformation susceptibility to SV40. J. Cell. Physiol. 72: 141–148 [DOI] [PubMed] [Google Scholar]
- 29. Aaronson SA, Todaro GJ. 1968. SV40 T antigen induction and transformation in human fibroblast cell strains. Virology 36: 254–261 [DOI] [PubMed] [Google Scholar]
- 30. Gregory SL, Kortschak RD, Kalionis B, Saint R. 1996. Characterization of the dead ringer gene identifies a novel, highly conserved family of sequence-specific DNA-binding proteins. Mol. Cell. Biol. 16: 792–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smith DB, Johnson KS. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67: 31–40 [DOI] [PubMed] [Google Scholar]
- 32. Iwahara J, Iwahara M, Daughdrill GW, Ford J, Clubb RT. 2002. The structure of the Dead ringer-DNA complex reveals how AT-rich interaction domains (ARIDs) recognize DNA. EMBO J. 21: 1197–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dignam JD, Lebovitz RM, Roeder RG. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li YC, Ross J, Scheppler JA, Franza BR., Jr 1991. An in vitro transcription analysis of early responses of the human immunodeficiency virus type 1 long terminal repeat to different transcriptional activators. Mol. Cell. Biol. 11: 1883–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chi T, Yan Z, Xue Y, Wang W. 2004. Purification and functional analysis of the mammalian SWI/SNF-family of chromatin-remodeling complexes. Methods Enzymol. 377: 299–316 [DOI] [PubMed] [Google Scholar]
- 36. Fan HY, He X, Kingston RE, Narlikar GJ. 2003. Distinct strategies to make nucleosomal DNA accessible. Mol. Cell 11: 1311–1322 [DOI] [PubMed] [Google Scholar]
- 37. Sif S, Saurin AJ, Imbalzano AN, Kingston RE. 2001. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 15: 603–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Duband-Goulet I, Ouararhni K, Hamiche A. 2004. Methods for chromatin assembly and remodeling. Methods 33: 12–17 [DOI] [PubMed] [Google Scholar]
- 39. Stein A, Whitlock JP, Jr, Bina M. 1979. Acidic polypeptides can assemble both histones and chromatin in vitro at physiological ionic strength. Proc. Natl. Acad. Sci. U. S. A. 76: 5000–5004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Solomon MJ, Larsen PL, Varshavsky A. 1988. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell 53: 937–947 [DOI] [PubMed] [Google Scholar]
- 41. Nelson JD, Denisenko O, Bomsztyk K. 2006. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1: 179–185 [DOI] [PubMed] [Google Scholar]
- 42. Chandler RL, Chandler KJ, McFarland KA, Mortlock DP. 2007. Bmp2 transcription in osteoblast progenitors is regulated by a distant 3′ enhancer located 156.3 kilobases from the promoter. Mol. Cell. Biol. 27: 2934–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Screen M, Dean W, Cross JC, Hemberger M. 2008. Cathepsin proteases have distinct roles in trophoblast function and vascular remodelling. Development 135: 3311–3320 [DOI] [PubMed] [Google Scholar]
- 44. Griffin CT, Brennan J, Magnuson T. 2008. The chromatin-remodeling enzyme BRG1 plays an essential role in primitive erythropoiesis and vascular development. Development 135: 493–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shingu T, Bornstein P. 1994. Overlapping Egr-1 and Sp1 sites function in the regulation of transcription of the mouse thrombospondin 1 gene. J. Biol. Chem. 269: 32551–32557 [PubMed] [Google Scholar]
- 46. Tropsha A, Carter CW, Jr, Cammer S, Vaisman II. 2003. Simplicial neighborhood analysis of protein packing (SNAPP): a computational geometry approach to studying proteins. Methods Enzymol. 374: 509–544 [DOI] [PubMed] [Google Scholar]
- 47. Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32: W273–W279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chen Y, Yee D, Magnuson T. 2006. A novel mouse Smad4 mutation reduces protein stability and wild-type protein levels. Mamm. Genome. 17: 211–219 [DOI] [PubMed] [Google Scholar]
- 49. Lei I, Gao X, Sham MH, Wang Z. 2012. SWI/SNF Protein component BAF250a regulates cardiac progenitor cell differentiation by modulating chromatin accessibility during second heart field development. J. Biol. Chem. 287: 24255–24262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Copp AJ. 1995. Death before birth: clues from gene knockouts and mutations. Trends Genet. 11: 87–93 [DOI] [PubMed] [Google Scholar]
- 51. Griffin CT, Curtis CD, Davis RB, Muthukumar V, Magnuson T. 2011. The chromatin-remodeling enzyme BRG1 modulates vascular Wnt signaling at two levels. Proc. Natl. Acad. Sci. U. S. A. 108: 2282–2287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Inoue H, Giannakopoulos S, Parkhurst CN, Matsumura T, Kono EA, Furukawa T, Tanese N. 2011. Target genes of the largest human SWI/SNF complex subunit control cell growth. Biochem. J. 434: 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Prochasson P, Neely KE, Hassan AH, Li B, Workman JL. 2003. Targeting activity is required for SWI/SNF function in vivo and is accomplished through two partially redundant activator-interaction domains. Mol. Cell 12: 983–990 [DOI] [PubMed] [Google Scholar]
- 54. Li XS, Trojer P, Matsumura T, Treisman JE, Tanese N. 2010. Mammalian SWI/SNF—a subunit BAF250/ARID1 is an E3 ubiquitin ligase that targets histone H2B. Mol. Cell. Biol. 30: 1673–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kortschak RD, Tucker PW, Saint R. 2000. ARID proteins come in from the desert. Trends Biochem. Sci. 25: 294–299 [DOI] [PubMed] [Google Scholar]
- 56. Carter CW, Jr, LeFebvre BC, Cammer SA, Tropsha A, Edgell MH. 2001. Four-body potentials reveal protein-specific correlations to stability changes caused by hydrophobic core mutations. J. Mol. Biol. 311: 625–638 [DOI] [PubMed] [Google Scholar]
- 57. Krishnamoorthy B, Tropsha A. 2003. Development of a four-body statistical pseudo-potential to discriminate native from non-native protein conformations. Bioinformatics 19: 1540–1548 [DOI] [PubMed] [Google Scholar]
- 58. Sherman DB, Zhang S, Pitner JB, Tropsha A. 2004. Evaluation of the relative stability of liganded versus ligand-free protein conformations using simplicial neighborhood analysis of protein packing (SNAPP) method. Proteins 56: 828–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kim S, Zhang Z, Upchurch S, Isern N, Chen Y. 2004. Structure and DNA-binding sites of the SWI1 AT-rich interaction domain (ARID) suggest determinants for sequence-specific DNA recognition. J. Biol. Chem. 279: 16670–16676 [DOI] [PubMed] [Google Scholar]
- 60. Wu JI, Lessard J, Crabtree GR. 2009. Understanding the words of chromatin regulation. Cell 136: 200–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Harikrishnan KN, Chow MZ, Baker EK, Pal S, Bassal S, Brasacchio D, Wang L, Craig JM, Jones PL, Sif S, El-Osta A. 2005. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat. Genet. 37: 254–264 [DOI] [PubMed] [Google Scholar]
- 62. Park SW, Li G, Lin YP, Barrero MJ, Ge K, Roeder RG, Wei LN. 2005. Thyroid hormone-induced juxtaposition of regulatory elements/factors and chromatin remodeling of Crabp1 dependent on MED1/TRAP220. Mol. Cell 19: 643–653 [DOI] [PubMed] [Google Scholar]
- 63. Stankunas K, Hang CT, Tsun ZY, Chen H, Lee NV, Wu JI, Shang C, Bayle JH, Shou W, Iruela-Arispe ML, Chang CP. 2008. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev. Cell 14: 298–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fouladkou F, Lu C, Jiang C, Zhou L, She Y, Walls JR, Kawabe H, Brose N, Henkelman RM, Huang A, Bruneau BG, Rotin D. The ubiquitin ligase Nedd4-1 is required for heart development and is a suppressor of thrombospondin-1. J. Biol. Chem. 285:6770–6780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Volpert OV, Pili R, Sikder HA, Nelius T, Zaichuk T, Morris C, Shiflett CB, Devlin MK, Conant K, Alani RM. 2002. Id1 regulates angiogenesis through transcriptional repression of thrombospondin-1. Cancer Cell 2: 473–483 [DOI] [PubMed] [Google Scholar]
- 66. Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P, Jr, de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA. 2012. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483: 603–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kaplan MH, Zong RT, Herrscher RF, Scheuermann RH, Tucker PW. 2001. Transcriptional activation by a matrix associating region-binding protein. contextual requirements for the function of bright. J. Biol. Chem. 276: 21325–21330 [DOI] [PubMed] [Google Scholar]
- 68. Kobayashi S, Eden-McCutchan F, Framson P, Bornstein P. 1986. Partial amino acid sequence of human thrombospondin as determined by analysis of cDNA clones: homology to malarial circumsporozoite proteins. Biochemistry 25: 8418–8425 [DOI] [PubMed] [Google Scholar]
- 69. Majack RA, Mildbrandt J, Dixit VM. 1987. Induction of thrombospondin messenger RNA levels occurs as an immediate primary response to platelet-derived growth factor. J. Biol. Chem. 262: 8821–8825 [PubMed] [Google Scholar]
- 70. Rafati H, Parra M, Hakre S, Moshkin Y, Verdin E, Mahmoudi T. 2011. Repressive LTR nucleosome positioning by the BAF complex is required for HIV latency. PLoS Biol. 9: e1001206 doi:10.1371/journal.pbio.1001206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Brown SA, Imbalzano AN, Kingston RE. 1996. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 10:1479–1490 [DOI] [PubMed] [Google Scholar]
- 72. Wilson CJ, Chao DM, Imbalzano AN, Schnitzler GR, Kingston RE, Young RA. 1996. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell 84: 235–244 [DOI] [PubMed] [Google Scholar]
- 73. Hassan AH, Prochasson P, Neely KE, Galasinski SC, Chandy M, Carrozza MJ, Workman JL. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111: 369–379 [DOI] [PubMed] [Google Scholar]
- 74. Chatterjee N, Sinha D, Lemma-Dechassa M, Tan S, Shogren-Knaak MA, Bartholomew B. 2011. Histone H3 tail acetylation modulates ATP-dependent remodeling through multiple mechanisms. Nucleic Acids Res. 39: 8378–8391 [DOI] [PMC free article] [PubMed] [Google Scholar]