Abstract
Alternative splicing provides a critical and flexible layer of regulation intervening in many biological processes to regulate the diversity of proteins and impact cell phenotype. To identify alternative splicing differences that distinguish epithelial from mesenchymal tissues, we have investigated hundreds of cassette exons using a high-throughput reverse transcription-PCR (RT-PCR) platform. Extensive changes in splicing were noted between epithelial and mesenchymal tissues in both human colon and ovarian tissues, with many changes from mostly one splice variant to predominantly the other. Remarkably, many of the splicing differences that distinguish normal mesenchymal from normal epithelial tissues matched those that differentiate normal ovarian tissues from ovarian cancer. Furthermore, because splicing profiling could classify cancer cell lines according to their epithelial/mesenchymal characteristics, we used these cancer cell lines to identify regulators for these specific splicing signatures. By knocking down 78 potential splicing factors in five cell lines, we provide an extensive view of the complex regulatory landscape associated with the epithelial and mesenchymal states, thus revealing that RBFOX2 is an important driver of mesenchymal tissue-specific splicing.
INTRODUCTION
Transitions from epithelial to mesenchymal (EMT) and mesenchymal to epithelial (MET) states have important roles not only in normal tissue and organ development but in the pathogenesis of diseases including cancer (1). In normal tissues, epithelial cells display a cuboidal morphology and a polar organization maintained by tight cell-cell interconnections. Mesenchymal cells lack these features and display higher motility and invasiveness. During the process of carcinogenesis, EMT is thought to be crucial to elicit migration, resistance to apoptosis, and ultimately invasion and metastasis (1–5). Conversely, the reverse process, MET, is associated with the colonization of secondary sites by cells that have metastasized. Therefore, profiling the molecular differences between the epithelial and mesenchymal states may help us understand the underlying regulatory programs that establish these states and promote their interconversion.
The commonly accepted view is that EMT can be induced by growth factors, such as transcription growth factor β1 (TGF-β1), which trigger signaling pathways that ultimately activate a network of transcription regulators, including Snail, Slug, Twist, and others (6). This transcriptional reprogramming elicits the expression of mesenchymal markers (e.g., vimentin) and represses the expression of epithelial ones (e.g., E-cadherin) to impart distinctive properties such as motility and invasion (7–10). Alternative splicing control provides another layer of regulation that can contribute to EMT (11, 12). The tyrosine kinase Ron (MST1R) is alternatively spliced to produce an exon 11-lacking version that can promote invasion (13). Three RNA-binding proteins (RBPs), SRSF1, hnRNP H, and hnRNP A2/B1, affect the alternative splicing of Ron exon 11 (14–16). The extracellular signal-regulated kinase (ERK) signaling cascade impacts on this regulatory pathway since it can phosphorylate the RNA-binding protein SAM68, which in turn decreases SRSF1 expression, promoting exon 11 inclusion and inducing MET (17). Another alternative splicing event (ASE) affecting EMT occurs in the small GTPase Rac1, and this event is antagonistically regulated by SRSF1 and SRSF3 (18). Rac1 splice variants differentially affect the activity of matrix metalloproteinases that in turn enhance the expression of Snail1 and vimentin, thus promoting EMT (19, 20). Studies focusing on the alternative splicing of the FGFR2 gene have also yielded important insights into the contribution of alternative splicing in EMT. Distinct FGFR2 splice variants are produced in epithelial and mesenchymal tissues (21), and many splicing factors affect the production of these isoforms, including hnRNP A1, PTBP1, hnRNP F/H, and RBFOX2 (22–24). The RNA-binding proteins epithelial splicing regulatory protein 1 (ESRP1) and ESRP2 are also critical for controlling the cell type-specific splicing switch of FGFR2 variants. The expression of ESRPs is restricted to epithelial cells, and their downregulation abrogates epithelial tissue-specific alternative splicing, while their overexpression in mesenchymal cells induces MET (25). The ESRPs control the alternative splicing of many genes implicated in cell adhesion, polarity, and migration (26–28). One important target is the cell and matrix interactor CD44, which undergoes a splicing switch critical for EMT which is prevented by ESRP1 (29). ESRPs also control the alternative splicing of p120-catenin to favor the production of an isoform that promotes cell-cell adhesion, while the loss of ESRPs elicits the synthesis of a splice variant that encourages cell migration (30). Finally, ESRPs stimulate the production of the TCF7L2 alternatively spliced isoform, which reduces the expression of genes stimulated by β-catenin (25, 28). Additional changes in alternative splicing associated with EMT have been uncovered by using an in vitro model of EMT, and a few splicing differences were validated in breast cancer cell lines and tissues (31). A subset of these events were known to be regulated by RBFOX2 (32), and the depletion of RBFOX2 promoted phenotypic changes indicative of MET (31).
Since reversible transitions between epithelial and mesenchymal states appear critical for cancer invasion and metastasis and because alternative splicing is important for EMT/MET, we sought to identify some of the events, networks and regulatory programs that establish these states. Here we have identified alternative splicing events that differ significantly between epithelial and mesenchymal tissues in human colon and ovary. These changes overlapped significantly with the cancer-associated splicing differences previously uncovered by comparing ovarian cancers with their normal counterparts (33). To identify regulators that impose these splicing signatures, we followed a subset of these events in an extensive RNA interference (RNAi) approach targeting 78 RNA-binding proteins and potential splicing factors in 5 different cancer cell lines that display a spectrum of epithelial and mesenchymal characteristics. This strategy allowed us to document a key role for RBFOX2 in establishing the mesenchymal splicing signatures characteristic of normal tissues and cancer cell lines.
MATERIALS AND METHODS
Human fetal tissues.
Midgestation (17- to 20-week) human colon specimens were obtained from normal legal or therapeutic pregnancy termination with informed patient consent. No tissues were collected from cases associated with known fetal abnormalities or intrauterine fetal demise. Studies were approved by the Institutional Review Committee for the use of human material from the Centre Hospitalier Universitaire de Sherbrooke/Faculté de Médecine et des Sciences de la Santé. Specimens were processed for the separation of the epithelium from its underlying mesenchyme by using the nonenzymatic dissociation (see Fig. S1a in the supplemental material) Matrisperse-based method previously described for the isolation of pure preparations of epithelial and mesenchymal intestinal fractions (34). Matrisperse was from BD Biosciences.
Cell lines and siRNAs.
The cervical cancer cell line HeLa; the prostate cancer cell line PC-3, the breast cancer cell lines ZR-75-1, Hs578T, MCF-7, and MDA-MB-231; the ovarian cancer cell lines TOV-112D, OVC-116, OVCAR3, and SKOV3ip1; and the colon cancer cell line HCT116 have been described previously (35). PC-3 cells were grown, respectively, in Dulbecco modified Eagle medium (DMEM), Ham's F-12 medium, and α minimal essential medium (α-MEM) supplemented with 10% fetal bovine serum. The small interfering RNAs (siRNAs) used to knock down the expression of RNA-binding proteins were purchased from IDT (Coralville, IA), and their sequences are listed in Table S2c in the supplemental material. siRNAs were transfected into cells at a concentration of 100 nM using Lipofectamine 2000 (Invitrogen). RNA was extracted from mock-transfected and siRNA-transfected cells 96 h posttransfection.
qRT-PCR.
Quantitative real-time reverse transcription-PCR (qRT-PCR) assays for the validation of knockdowns were conducted with SyberGreen (Power SYBR green master mix 2X, ABI catalog no. 4367660). Aldolase A (RTPrimerDB ID: 915) was used as the housekeeping gene on the same samples. A total of 200 ng of RNA measured for integrity using the Agilent Lab-on-Chip station and quantified on a Thermo Scientific NanoDrop spectrophotometer was reverse transcribed with random hexamers (Roche catalog no. 11034731001) with Transcriptor reverse transcriptase (Roche catalog no. 03531317) in a final volume of 10 μl. Ten nanograms of cDNA was used for quantification in the presence of the specific primers at 0.2 μM in a 10-μl reaction in triplicate. The expression of the epithelial and stromal tissue-specific markers was normalized using the geometrical mean of three housekeeping genes (glyceraldehyde-3-phosphate dehydrogenase [GAPDH], MRPL19, and YWHAZ) (36). The sequences of oligonucleotides used for quantitative PCR (qPCR) are given in Table S2g in the supplemental material. Reactions were carried out in the ABI 7500 qPCR (Applied Biosystems, Foster City, CA) or Eppendorf Realplex system. A first cycle of 10 min at 95°C was followed by 40 cycles of 15 s at 94°C, 20 s at 55°C, and 20 s at 68°C.
RT-PCR assays.
Our collection of alternative splicing events was extracted from the RefSeq database. Sets of primers mapping in the exons flanking all the simple alternative splicing events were designed using Primer3 with default parameters. Total RNA was extracted using TRIzol and quantified using the Lab-on-Chip station (Agilent Inc., Santa Clara, CA). A total of 2 μg of RNA was reverse transcribed using a mix of random hexamers and oligo(dT) and Omniscript reverse transcriptase (Qiagen, Germantown, MD) in a final volume of 20 μl. Twenty nanograms of cDNA was amplified with 0.2 U/10 μl of HotStarTaq DNA polymerase (Qiagen) in the buffer provided by the manufacturer and in the presence of the specific primers (IDT) for each splicing unit (at concentrations ranging from 0.3 to 0.6 μM) and deoxynucleoside triphosphates (dNTPs). The list of ASEs and oligonucleotides and the expected sizes of RT-PCR products are shown in Table S2 in the supplemental material. Reactions were carried out in the GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA). A first cycle of 15 min at 95°C was followed by 35 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C. The reaction was ended with the extension step of 10 min at 72°C. Visualization and analysis of amplified products were done using the LabChip HT DNA assay on an automated microfluidic station (Caliper, Hopkinton, MA) (37).
Bioinformatic analysis.
The clustering analysis was done by R using the R Project for Statistical Computing (http://www.r-project.org/) using a Euclidean distance metric.
RESULTS
Alternative splicing differences between normal epithelial and mesenchymal tissues.
To identify alternative splicing events that distinguish epithelial from mesenchymal tissues, we used total RNA purified from human fetal colon samples, as it is possible to isolate relatively pure epithelial and mesenchymal samples. Epithelial and mesenchymal fractions were separated using a nonenzymatic method (see Fig. S1a in the supplemental material); the purity of the fractions was confirmed by monitoring the expression of the epithelial and stromal tissue-specific markers E-cadherin and vimentin, respectively (see Fig. S1b). We then assayed a set of 283 alternatively spliced cassette exons, between 18 and 309 nucleotides long, selected for the high quality of their PCRs. Primers were designed to cover alternatively spliced regions such that the sizes of the two PCR products derived from the two isoforms ranged between 100 and 500 bp. Following endpoint RT-PCR on our high-throughput platform (33, 37–39), the abundance and size of each amplicon were measured by capillary microfluidic fractionation using Caliper reading stations. For each reaction, the relative abundance of the products was converted to a “percent spliced in” (psi or ψ) value defined as the molarity of the long product divided by the combined molarities of the long and short products. Although alternative splicing changes on the order of 10 percentage points are often considered significant, we found that 26 of the 283 events shifted by more than 50 percentage points (i.e., from mostly one isoform to predominantly the other) (see Tables S1 and 2a in the supplemental material) (all raw data are shown at http://palace.lgfus.ca/data/related/1447).
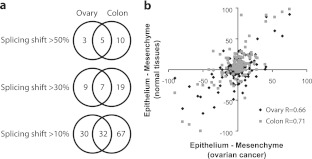
To determine if similar epithelial versus mesenchymal cell type-specific splicing differences could be observed in other tissues, we examined laser-captured and microdissected ovarian stromal tissue and compared it to fallopian tube epithelium. Similarly to the two colon samples, our ovarian and fallopian tube samples contained vimentin and cadherin levels consistent with pure mesenchyme and epithelium, respectively (see Fig. S1c in the supplemental material). The analysis was conducted with 4 samples for each tissue category for 178 of the alternative exons that had been examined in colon. This second mesenchyme/epithelium comparison identified 8 exons, 5 in common with the colon system, which display a “switch-like” behavior (a splicing shift of more than 50 percentage points) (Fig. 1a; see also Table S2a in the supplemental material). Overall, approximately one-third of all exons displayed greater than 10 percentage points of difference in each set of mesenchymal and epithelial tissues examined, and approximately half (32 exons) of the differences seen in ovarian samples also occurred in the colon (Fig. 1a).
Fig 1.
A majority of ovarian cancer-associated splicing changes correspond to changes between normal epithelial and mesenchymal tissues. (a) Common epithelial/mesenchymal tissue-specific alternative exons between the colon and the ovary systems. A total of 178 alternative exons were used for comparison between the colon and ovary systems. The Venn diagram illustrates the tissue distribution of the epithelial/mesenchymal alternative exons obtained for differences in percentage points that go in the same direction and for various cutoff values. (b) Scatter plot of differences in the alternative splicing of 178 alternative exons in normal epithelial versus mesenchymal fetal colon tissues as measured by endpoint RT-PCR (y axis, in gray). RT-PCR assays were also carried out for 178 alternative exons in dissected normal ovarian stroma and fallopian epithelial tissues (y axis, in black). The values are plotted as Δψ (ψepithelium − ψmesenchyme) relative to values obtained by comparing 21 normal ovaries versus 25 ovarian serous high-grade tumors (x axis; ovarian cancer) (33). The Pearson correlation between the splicing shift in normal ovarian and colon systems and the splicing shift in ovarian cancer is indicated on the bottom right of the graph.
To determine if alternative exons that differentiate normal epithelium and mesenchyme could be globally applied as EMT markers, we compared their splicing profiles to those reported in a recent study that used a human mammary epithelial cell line engineered to undergo EMT by activation of Twist (31). While only 6 of the 32 markers common to colon and ovary changed during EMT in that cell line (see Fig. S2 in the supplemental material), the majority of them (4/6) remain EMT markers when shifts greater than 30 percentage points are considered. Thus, while each epithelial/mesenchymal tissue pair displays a unique set of splicing events, a core set of events distinguishes epithelial from mesenchymal specimens irrespective of their source. Moreover, alternative exons with the largest splicing differences are more likely to behave as EMT markers.
The normal epithelial splicing profile is found in ovarian cancer.
We previously observed large-scale splicing differences between ovarian cancer specimens (an epithelial tissue-type cancer) and normal ovary (mostly comprising mesenchymal tissue) (33) and sought to establish if these differences reflect the cell type composition of those tissues. Figure 1b (diamonds) shows a comparison of the inclusion level differences between ovarian stroma and fallopian epithelium with the inclusion level differences between normal and cancer ovary for the 178 alternative exons. The comparison clearly indicates a high degree of similarity between splicing signatures (Pearson correlation R = 0.66), suggesting that the epithelial splicing signature is a common feature of both normal and malignant epithelial tissues.
We also compared the fetal colon splicing signature with the normal and cancer ovary splicing profiles for the same set of 178 alternative exons (Fig. 1b, squares). Remarkably, we found that the splicing differences between ovarian cancer and normal ovary were also very similar to the splicing differences between normal epithelial and mesenchymal colon tissues (Pearson correlation R = 0.71). Thus, the majority of the cancer-associated splicing events, identified previously by comparing ovarian cancer with normal ovary, reflect the epithelial/mesenchymal status of the cells. Despite this significant degree of overall similarity, a small subset of three alternative exons (MYO18A, CD46, and ABI1) differed (>30%) in their inclusion levels between epithelial normal and epithelial cancer tissues. These events may therefore be more closely related to the cancer status than to the epithelial status. Overall, these results show that the majority of the epithelial tissue-specific alternative exons of normal tissues are also regulated in ovarian cancer.
The splicing signatures of cancer cell lines match their epithelial/mesenchymal identities.
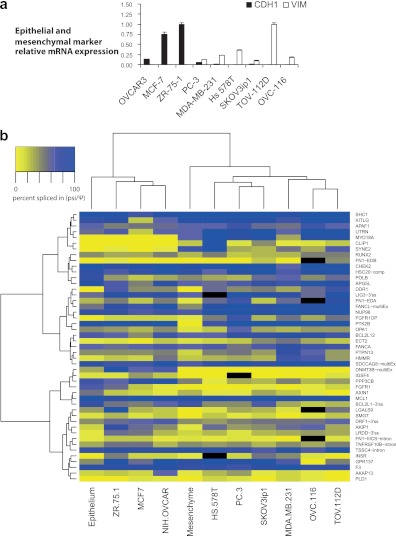
Cancer cell lines often serve as models to study regulatory features of EMT. Because of the distinctive epithelial and mesenchymal splicing signatures, we set out to use various cancer cell lines to explore the underlying regulatory landscape. It was recently shown that a set of 20 exons, regulated by the epithelial tissue-specific splicing factors (ESRPs), could discriminate between epithelial and mesenchymal cell lines (28). We used a panel of 9 cancer cell lines to determine if our set of alternative exons possess similar discriminatory power. The 9 cell lines display differences in their epithelial and mesenchymal characteristics based on the expression of vimentin and E-cadherin (Fig. 2a). The ovarian cancer cell line TOV-112D is the most mesenchymal based on its high level of vimentin and lack of E-cadherin, as detected by qRT-PCR. The other end of the spectrum is occupied by the estrogen-sensitive breast cancer cell line ZR-75-1 and the MCF-7 breast cancer cell line, which both display a highly epithelial phenotype based on E-cadherin expression and the lack of detectable vimentin. The prostate cancer cell line PC-3 shows an intermediate phenotype with some E-cadherin and vimentin expression. Remarkably, using our set of nearly 200 alternative exons, we found that the classification based on global splicing profiles matched the classification based on vimentin and E-cadherin expression (see Fig. S3a and Table S2a in the supplemental material; also see http://palace.lgfus.ca/data/related/1448).
Fig 2.
The epithelial-mesenchymal splicing signature of cancer cell lines matches their vimentin/E-cadherin signatures. (a) The relative expression levels of the epithelial marker CDH1 and the stromal mesenchymal marker VIM were monitored in nine cancer cell lines by quantitative RT-PCR. The geometrical mean of three housekeeping genes was used to calculate the normalization factor (see Materials and Methods). Bar graphs represent the means of three technical replicates, and error bars represent standard deviations. (b) Heat map showing the unsupervised hierarchical clustering of fetal colon epithelial and mesenchymal tissues along with 9 cancer cell lines using the 47 alternative splice events shown (see Table S2b in the supplemental material). Splicing (ψ) values are represented in shades of yellow (exon skipping) to blue (exon inclusion), as indicated in the color key histogram.
To create a resource to study the role of a large collection of putative splicing regulators in varied physiological states, we selected a set of 47 alternative splicing events (ASEs) from three previous studies based on PCR quality in a large number of samples (33, 37, 39) (http://palace.lgfus.ca/data/related/1452). These 47 ASEs displayed a wide range of splicing differences between 0 and >50 percentage points between normal epithelial and mesenchymal tissues (see Fig. S3b and Tables S1 and S2b in the supplemental material; see also http://palace.lgfus.ca/data/related/1444). Importantly, this smaller set of ASEs also distinguished cell lines according to their mesenchymal and epithelial status (Fig. 2b); the probability that the three cancer cell lines that cluster with the normal epithelium based on global splicing profiles are the same as those that cluster with it based on vimentin and E-cadherin expression is significant (Fisher's test, P = 0.01). Comparing the median values of the 3 epithelial with the 6 mesenchymal cell lines identified 27 responsive ASEs with shifts greater than 10 percentage points (7 with shifts greater than 30 percentage points; see column W in Table S2b). All 7 of the 30% ASEs shift in the same direction between colon epithelial and mesenchymal tissues (P = 0.01). Thus, this combination of ASEs and cell lines is appropriate to investigate the regulatory features of epithelial and mesenchymal splicing control.
Since our analysis in tissues showed that roughly one-third of ASEs displayed an epithelial/mesenchymal tissue-specific profile, it is likely that the clustering observed in Fig. 2b and in Fig. S3a in the supplemental material is driven by a subset of such ASEs. If this is the case, switch-like ASEs may represent particularly strong classifiers. We tested the discriminatory power of the 4 ASEs whose alternative splicing profile was changed by more than 30 percentage points in normal mesenchymal tissue versus epithelium for both the colon and the ovary and in the Twist-induced breast cell line (31) (see Fig. S2 in the supplemental material). These four ASEs are located in the ADD3, CSNK1G3, ENAH, and PBRM1 genes, and they provided a classifier that was as precise as the more extensive splicing signatures (see Fig. S3c).
RBFOX2 drives mesenchymal tissue-specific splicing.
To identify RNA-binding proteins (RBPs) and associated splicing factors that confer epithelial and mesenchymal tissue-specific splicing, we knocked down 97 potential splicing regulators in five cell lines (MCF-7, NIH-OVCAR3, MDA-MB-231, SKOV3ip1, and PC-3) displaying a spectrum of epithelial and mesenchymal markers based on Fig. 2a. We monitored the splicing of the 47 ASEs tested in Fig. 2b because they provide a balanced mixture of events that either do not change, change slightly, or change by up to 80 percentage points when splicing between normal epithelial and mesenchymal tissues is compared (see Fig. S3b in the supplemental material). Including in the analysis a range of epithelial/mesenchymal tissue-responsive as well as nonresponsive ASEs is important to ensure the specificity of responses to the various knockdowns. Each potential splicing factor was knocked down with two distinct siRNAs (see Table S2c, column C, in the supplemental material). We considered only samples with knockdown efficiencies that were greater than 50% for both siRNAs, as assessed by quantitative PCR (qPCR) (see Table S2c, column D). For 78 of the 97 proteins, we achieved a knockdown efficiency of at least 50% with both siRNAs in at least one cell line (see Table S2d and e). The effects of the knockdowns on the splicing of the 47 alternative events are shown in Fig. S4 in the supplemental material and at http://palace.lgfus.ca/data/related/1451. One of the most notable features, unrelated to epithelial and mesenchymal states, was the impact of depleting the constitutive U2 snRNP-associated splicing factors SF3A1, SF3A2, SF3A3, and SF3B4, which promoted exclusion of a large group of exons (box a in Fig. S4), as well as the inclusion of introns and the use of upstream 3′ splice sites (box b in Fig. S4). Thus, depleting the levels of these constitutive splicing factors can be limiting for the inclusion of alternative exons but also for the exclusion of some introns, as would be expected. The nearly identical impact of knocking down physically associated core spliceosomal components (i.e., the four different SF3 subunit proteins) confirms the accuracy and sensitivity of our high-throughput RT-PCR approach. A clear signature is also visible in this overview for 8 exons enhanced by the alternative splicing factor RBFOX2 (box c in Fig. S4).
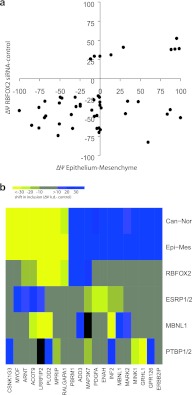
To establish which of these knockdowns best matched the splicing differences that distinguish normal mesenchymal from epithelial cells, we looked at the 14 of the total of 47 alternative splice events that shifted more than 20% between epithelium and mesenchyme. Of the knockdowns that shifted splicing of any of these events to this extent (>20%), the highest correlation (R = 0.75) was seen for knockdowns of the splicing factor RBFOX2 in MDA-MB-231 and SKOV3ip cells (P < 0.001) (see Fig. S5 in the supplemental material).
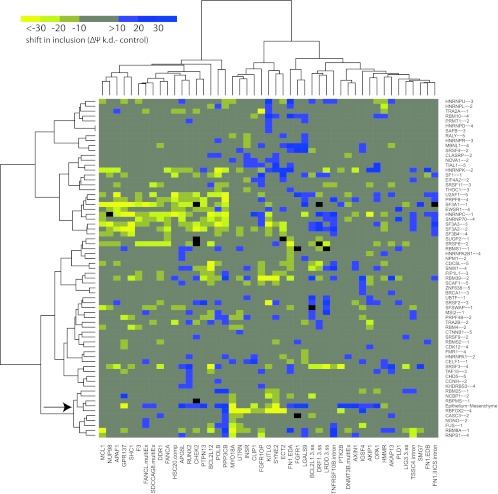
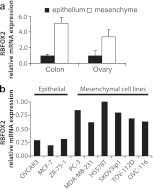
To condense the data and facilitate visual inspection, we then plotted the median percent spliced in values of all cell lines for each RBP (Fig. 3; see also Table S2e in the supplemental material). Here again, the RBFOX2 knockdowns clustered with the epithelial-to-mesenchymal splicing differences. These results indicate that many epithelial or mesenchymal tissue-specific ASEs are regulated by RBFOX2 and that additional constitutive and/or regulatory RBPs are likely contributing to their regulation. Consistent with a role for RBFOX2 in contributing to mesenchymal tissue-specific splicing, we found that the expression level of Rbfox2, as determined by quantitative RT-PCR, was 5 times higher in normal mesenchymal tissue than in epithelial colon tissue and more than 3 times higher in normal ovary than in fallopian epithelium (Fig. 4a). Likewise, the levels of Rbfox2 correlated well with the mesenchymal and epithelial identity of cancer cell lines (Fig. 4b).
Fig 3.
The impact of knocking down RBFOX2 most closely matches the differences between epithelium and mesenchyme. Unsupervised hierarchical clustering of the median change in splicing (Δψ knockdown [k.d.] − control) for 47 alternative splicing events in each of the 68 splicing factor knockdowns for which the median splicing value changed by at least 10 percentage point for at least one alternative splice event. The number next to the protein indicates the number of cell lines in which the RBP was successfully knocked down (the criterion being >50% downregulation for both siRNAs as assessed by qPCR). The results are clustered along with the splicing differences between epithelium and mesenchyme colon tissues. Splicing shifts are in bins of 10 percentage points from less than −30 percentage points (bright yellow) to greater than +30 percentage points (darkest blue). The RBFOX2 (siRNA control) (below) and the epithelial-mesenchymal (above) splicing differences are indicated by the arrow.
Fig 4.
Rbfox2 expression data in normal epithelium and mesenchyme. (a) Rbfox2 expression in the epithelium and mesenchyme samples isolated from human fetal colon and ovary/fallopian tissues. The expression of Rbfox2 was monitored by quantitative RT-PCR in colon (technical triplicate) and ovarian (biological quadruplicate) tissues. Values were normalized to the geometrical means of three housekeeping genes. The results were plotted in the form of bar graphs. (b) Quantitative RT-PCR measurement of Rbfox2 in cancer cell lines. Cell lines are organized according to their epithelial/mesenchymal characteristics.
To further assess the role of RBFOX2 in setting the mesenchymal splicing pattern, we investigated the behavior of the 48 most RBFOX2-responsive exons, identified from our previous study (33), in the epithelial and mesenchymal samples from normal fetal colon. We find that the majority of these alternative exons were affected, with 18 switching >50 percentage points between epithelial and mesenchymal cells (Fig. 5a; see also Table S2f in the supplemental material and http://palace.lgfus.ca/data/related/683). In total, therefore, we have identified 43 switch-like splicing events that shift from predominantly one isoform to nearly exclusively the other between epithelial and mesenchymal cells: 29/283 from the original screen in normal tissues, 3 more from the 47 cancer and apoptosis ASEs, and a further 11 RBFOX2 targets not included in the above screens (summarized in Table S1 in the supplemental material with splicing details in Table S2a, b, and f). Notably, all the alternative exons for which inclusion was more prevalent in the normal mesenchyme than in the epithelium were previously reported as positively enhanced targets of RBFOX2, whereas exons included more frequently in the normal epithelium than in the mesenchyme can be either repressed or stimulated by RBFOX2 (33). We conclude that RBFOX2, either alone or in combination with other factors, plays an important role in setting the mesenchymal tissue-specific splicing profile.
Fig 5.
RBFOX2 contributes to complex regulation of splicing in mesenchymal cells. (a) Scatter plot comparing the changes of percent-spliced-in values (Δψ) for 48 RBFOX2 targets (33) with their Δψ from the epithelial and mesenchymal colon comparison. Eighteen switch-like alternative exons (Δψ, >50 percentage points) can be identified from the epithelium and mesenchymal colon comparison. (b) Twenty-six switch-like exons from Table S2a in the supplemental material were subjected to further analysis in cells knocked down for four RNA-binding proteins implicated in EMT (see the text). The heat map shows data for 20 exons that shifted between cancer and normal tissue and gave good data for ESRP1 and RBFOX knockdowns. Splicing shifts are shown in bins of 10 percentage points, in increasingly strong shades of blue (inclusion) or yellow (skipping) for changes greater than 10, 20, or 30 percentage points, respectively.
RBFOX2, MBNL1, and ESRPs are major contributors to the mesenchymal splicing signature.
While it is clear that RBFOX2 is important to set the mesenchymal tissue-specific splicing profile, we wished to compare its role relative to those of three other RNA-binding protein families that have been implicated in the epithelial and mesenchymal states. We therefore analyzed the top epithelial/mesenchymal tissue-discriminatory alternative exons from Fig. 1b in cell lines where expression of each RBP was high. MBNL1 was knocked down in MDA-MB-231, as we noted some overlap between its knockdown and epithelial splicing (Fig. 3). PTBP1 regulates many EMT exons, and its expression drops during EMT (31). We therefore performed a double PTBP1/PTBP2 knockdown in SKOV3ip1. Finally, we used the epithelial ZR-75-1 cell line to perform ESRP1/ESRP2 double knockdown because these proteins are expressed in epithelial cells and control epithelial tissue-specific splicing decisions (25, 31). The results can be viewed at http://palace.lgfus.ca/data/related/1456 and in Table S2a in the supplemental material. The 20 ASEs that shifted in our ovarian and fetal colon systems give good data in the various cell lines used (in the ESRP and RBFOX2 samples) and are shown in Fig. 5b. Of the four splicing factors, RBFOX2 knockdowns correlated with epithelial status (R = 0.68), as did MBNL1 knockdown (R = 0.41), whereas ESRP and PTBP knockdowns were anticorrelated (R = −0.63 and R = −0.29, respectively). Overall, 95% of the splicing events that distinguish between mesenchyme and (cancerous or normal) epithelium are affected by one of these four classes of splicing factors, and changes in RBFOX and ESRP proteins together affect 70% of the exons. MBNL1 appears to cooperate with RBFOX2 in some cases of mesenchymal splicing (Fig. 5b). Remarkably, the alternative splicing of PLOD2, which was also previously shown to form part of a signature that distinguishes luminal from basal cell-like breast cancer cell lines (31), is controlled by RBFOX2, MBNL1, and PTBP1/2 knockdown. MBNL1 and RBFOX2 also both enhanced exon inclusion in LRRFIP2, and both promoted exon skipping in ADD3. The direction of the shift imposed by MBNL1 on PLOD2 and INF2 is antagonized by the PTBPs. Note that the three RBFOX2 target exons in LRRFIP2, MAP3K7, and ADD3 were previously assayed for RBFOX2 and ESRP responsiveness (26), and our results are entirely consistent with this report (see Table S2a). Other regulated splicing events are displayed in Fig. S6 in the supplemental material.
DISCUSSION
In the last 10 years, global studies of alternative splicing have revealed many switch-like changes (i.e., from mainly one isoform to predominantly the other) between normal mammalian tissues (40) and also between normal and cancer tissues (33). While similar tissues, such as skeletal muscle/heart, kidney/liver, and different brain regions, form clusters with similar splicing signatures likely reflecting similar mixtures of cell types in these tissues, the splicing signatures of specific cell types within a tissue have received less attention. To remedy this, we have studied splicing on a large scale in microdissected epithelial and mesenchymal cell types from fetal colon and also from ovarian stroma and fallopian tube, which are almost completely composed of mesenchymal and epithelial cells, respectively.
Using our high-throughput RT-PCR platform to carry out the largest RNA interference (RNAi) screen of mammalian RNA-binding proteins ever undertaken, we have identified 43 alternative exons that are strongly associated with the epithelium and the mesenchymal identities of normal cells from human fetal colon and ovary (see Table S1 in the supplemental material). While several of these switch-like changes (>50 percentage points) in alternative splicing occur in genes involved in processes related to EMT (see below), other events are in genes not known to be transcriptionally affected during EMT. Thus, the layers of transcription and alternative splicing regulation can be, but are not always, interconnected to achieve definition of the epithelial and mesenchymal states. These observations are consistent with a recent study conducted in vitro using a model breast cell line engineered to undergo EMT by activation of the transcription factor Twist (31). The limited overlap between our studies may be due to the complementary techniques; RNA-Seq at limited sequencing depth will focus on highly expressed genes. Importantly, most of the splicing changes that we uncovered also occurred in cancer tissues and cell lines, suggesting that these events are relevant to the epithelial/mesenchymal identity of cancer tissues and that they may be used to follow the transit between these states during tumor invasion and dissemination. Since a large expanse of the characteristic epithelial/mesenchymal splicing program that we observed in ovary was shared by fetal colon, its implementation is likely to be a hallmark of the epithelial and mesenchymal identity in a large variety of normal and cancer tissues.
The splicing signature that distinguishes normal epithelial from mesenchymal tissues and cancers of epithelial origin from normal stromal tissues was also capable of categorizing cancer cell lines according to their epithelial-mesenchymal status. In fact, the splicing differences between the most epithelial and the most mesenchymal cell lines of our set were qualitatively and quantitatively similar to differences between normal epithelial and normal mesenchymal tissues, even though the cell lines and the tissues were of different origin (see Fig. S3c in the supplemental material). This result confirms cancer cell lines as suitable models to investigate the underlying regulatory splicing programs that control the epithelial and mesenchymal identities in normal and cancer-driven situations.
Alternative splicing changes associated with the epithelial and mesenchymal states.
While few switch-like changes had previously been identified and validated between epithelial and mesenchymal cells, the list of alternative transcripts whose alternative splicing has been associated with EMT has been growing steadily in recent years. Starting from the original member FGFR2 (41), the list now encompasses CD44, p120-catenin, ENAH, and nearly 40 other validated targets (with shifts greater than 5 percentage points) for ESRP1 and ESRP2, which contribute to the epithelial splicing program (25, 27–29, 42, 43). Also, an RNA-Seq analysis on a Twist-induced EMT model system has recently identified approximately 1,500 genes undergoing changes in alternative splicing during EMT, 64 of which display switch-like behavior (31). Our analysis, performed in normal tissues, has identified 43 alternative exons associated with epithelial-mesenchymal identity that display a switch-like behavior between these tissue types (gene functions are summarized in Table S1 and splicing details are summarized in Table S2, both in the supplemental material). Many of these have functions in specific pathways relevant to EMT. For example, EXOC1, MYOF, PLOD2, and SNX14 are associated with vesicle-mediated transport, a process intimately connected with the maintenance of cell polarity and adhesion (44). Other events occurred in genes associated with signaling pathways, including Wnt (LRRFIP2 and CSNK1G3), mitogen-activated protein (MAP) kinase (MAP3K7, PTK2B, and APBB2), Ras (ERBB2IP), and p38 (MINK1), while others have been previously associated with cancer (PBRM1, ARNT, ST7, PTPRD, and CA12).
The link with Wnt is particularly intriguing since Wnt is a regulator of beta-catenin, a component of the cadherin complex that controls cell-cell adhesion and influences cell migration (45), thus raising the possibility that alternative splicing changes in components of the Wnt pathway also mediate a subset of the phenotypic differences between the epithelial and mesenchymal cells. Components of the Wnt receptor signaling pathway were also enriched when the splicing profiles of human mammary epithelial cells were compared before and after Twist induction (31). In addition to changes that may have a functional impact and that have been identified here and previously in the study by Shapiro et al. (31) (e.g., MBNL1 and ENAH epithelial exon inclusion), most of the above events had never been linked to EMT. In most cases, the functional contribution of the state-specific splice variants to EMT/MET remains unclear and will require approaches that can deplete specific isoforms and/or reprogram splicing decisions, for example, using antisense strategies (46).
RBFOX2 and other regulators specify the mesenchymal tissue-specific splicing profile.
Studies on the FGFR2, Rac1, and Ron kinases have pioneered the quest for regulatory factors controlling the alternative splicing of EMT-relevant genes (14, 16, 18, 22–24, 47). To gain insights into the network of regulators that control the alternative splicing of genes associated with epithelial and mesenchymal identity, we used RNA interference to knock down 78 RNA-binding proteins (RBPs) and potential splicing factors in 5 different cell lines. This unbiased analysis yielded a rich collection of associations hinting at the combinatorial and complex regulatory programs underpinning the epithelial and mesenchymal splicing programs. Expanding from the Rac1 study (18), our analysis confirmed a role for SRSF2 and SRSF3 in regulating the splicing of many EMT-relevant genes (Fig. 3; see also Fig. S6 in the supplemental material).
Among the set of RBPs that we tested, the one whose knockdown most closely matched the differences between epithelial and mesenchymal tissues was RBFOX2. A role for RBFOX2 in cancer had previously been proposed, since we had observed that the levels of RBFOX2 were significantly lower in epithelial ovarian cancer than in normal tissues (33). Decreasing the level of RBFOX2 in cancer cell lines shifted the alternative splicing of many events in the same direction as that in breast and ovarian cancer tissues (33). A subsequent report demonstrated that RBFOX2 regulated subtype-specific splicing in a panel of breast cancer cell lines (15). Our observations that RBFOX2 regulates events that differ between the epithelial and mesenchymal cancer cell lines suggest that these subtypes reflect their epithelial/mesenchymal status. Overall, our data imply that RBFOX2 is important to specify the mesenchymal tissue-specific splicing profiles both in normal and in cancer tissues. Consistent with this proposition, Rbfox2 transcripts are 3- to 5-fold more abundant in normal mesenchymal tissue than in epithelial ovary and colon tissues (Fig. 4a). Moreover, cell lines classified as epithelial based on the E-cadherin/vimentin ratio expressed substantially lower levels of Rbfox2 than did cells classified as mesenchymal (Fig. 4b). Using a mammary cell line, inducible for Twist expression, Shapiro and collaborators recently identified RBFOX2 as a potential driver of the mesenchymal splicing signature (31). Although the impact of a depletion of RBFOX2 on splicing profiles was not investigated per se, the loss of RBFOX2 in mesenchymal cells provoked a partial reversion of the epithelial phenotype, with reduced levels of vimentin, changes in morphology, and restricted migration (31).
Continued efforts aimed at deciphering the complex splicing regulatory programs associated with the transitions between epithelial and mesenchymal status will likely become useful to develop strategies to control the metastatic invasion and implantation of tumor cells at secondary sites. Because EMT also offers resistance to apoptosis (7), specific changes in alternative splicing profiles may help predict the outcome of treatments whose goal is to promote the demise of cancer cells by apoptosis. In addition, intervening productively to reprogram a specific splicing event, or possibly several simultaneously, by altering the function of a common splicing regulator is a strategy worth considering. This approach could render tumor cells more sensitive to current therapeutic regimens. A precedent for RBFOX-target gene therapy was recently demonstrated through the use of antisense oligonucleotides that block RBFOX-binding sites and hence splicing of zebrafish RBFOX-target transcripts (48).
Mesenchyme-upregulated exons were usually RBFOX2 enhanced. However, RBFOX2-inhibited exons were either up- or downregulated in the mesenchyme (Fig. 5a), suggesting that RBFOX2 may be sufficient to promote exon inclusion but that exon skipping in mesenchyme may require an interplay between RBFOX2 and other factors, as suggested recently (49, 50) and consistent with recent reports indicating that many RBFOX protein-regulated exons are also targets of other splicing factors, including Nova, PTBP, and ESRP proteins (26, 51–53).
In conclusion, we have identified a large collection of splicing changes characteristic of the epithelial and mesenchymal phenotypes. While transitions between these states may be crucial for the migration and invasion of cancer cells, for example, by modifying cell signaling and cellular organization during EMT, we have shown that a majority of alternative events associated with epithelial/mesenchymal status are not cancer specific since they are qualitatively and quantitatively matched by differences between normal epithelial and mesenchymal tissues. Nevertheless, transient changes in the expression of RBFOX2 in cancer cells may be important to reconfigure transcript architecture and implement EMT/MET transitions that are adapted to the environment of the tumor.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sonia Couture, Ulrike Froehlich, Anne Bramard, and Julien Gervais-Bird for technical support.
This work was funded by grants from the Canadian Cancer Society Research Institute and the Canadian Health Research Institute. S. Abou Elela is a Chercheur National of the Fonds de Recherche du Québec-Santé. B. Chabot is the Canada Research Chair in Functional Genomics.
Footnotes
Published ahead of print 12 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01174-12.
REFERENCES
- 1. Thiery JP, Acloque H, Huang RY, Nieto MA. 2009. Epithelial-mesenchymal transitions in development and disease. Cell 139:871–890 [DOI] [PubMed] [Google Scholar]
- 2. Barrallo-Gimeno A, Nieto MA. 2005. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development 132:3151–3161 [DOI] [PubMed] [Google Scholar]
- 3. Klymkowsky MW, Savagner P. 2009. Epithelial-mesenchymal transition: a cancer researcher's conceptual friend and foe. Am. J. Pathol. 174:1588–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Polyak K, Weinberg RA. 2009. Transitions between epithelial and mesenchymal states: acquisition of malignant and stem cell traits. Nat. Rev. Cancer 9:265–273 [DOI] [PubMed] [Google Scholar]
- 5. Yilmaz M, Christofori G. 2009. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 28:15–33 [DOI] [PubMed] [Google Scholar]
- 6. Mani SA, Yang J, Brooks M, Schwaninger G, Zhou A, Miura N, Kutok JL, Hartwell K, Richardson AL, Weinberg RA. 2007. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc. Natl. Acad. Sci. U. S. A. 104:10069–10074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang J, Weinberg RA. 2008. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell 14:818–829 [DOI] [PubMed] [Google Scholar]
- 8. Taube JH, Herschkowitz JI, Komurov K, Zhou AY, Gupta S, Yang J, Hartwell K, Onder TT, Gupta PB, Evans KW, Hollier BG, Ram PT, Lander ES, Rosen JM, Weinberg RA, Mani SA. 2010. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc. Natl. Acad. Sci. U. S. A. 107:15449–15454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schmalhofer O, Brabletz S, Brabletz T. 2009. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev. 28:151–166 [DOI] [PubMed] [Google Scholar]
- 10. Micalizzi DS, Farabaugh SM, Ford HL. 2010. Epithelial-mesenchymal transition in cancer: parallels between normal development and tumor progression. J. Mammary Gland Biol. Neoplasia 15:117–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Warzecha CC, Carstens RP. 2012. Complex changes in alternative pre-mRNA splicing play a central role in the epithelial-to-mesenchymal transition (EMT). Semin. Cancer Biol. 22:417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Biamonti G, Bonomi S, Gallo S, Ghigna C. 2012. Making alternative splicing decisions during epithelial-to-mesenchymal transition (EMT). Cell. Mol. Life Sci. 65:2515–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lu Y, Yao HP, Wang MH. 2007. Multiple variants of the RON receptor tyrosine kinase: biochemical properties, tumorigenic activities, and potential drug targets. Cancer Lett. 257:157–164 [DOI] [PubMed] [Google Scholar]
- 14. Tauler J, Zudaire E, Liu H, Shih J, Mulshine JL. 2010. hnRNP A2/B1 modulates epithelial-mesenchymal transition in lung cancer cell lines. Cancer Res. 70:7137–7147 [DOI] [PubMed] [Google Scholar]
- 15. Lapuk A, Marr H, Jakkula L, Pedro H, Bhattacharya S, Purdom E, Hu Z, Simpson K, Pachter L, Durinck S, Wang N, Parvin B, Fontenay G, Speed T, Garbe J, Stampfer M, Bayandorian H, Dorton S, Clark TA, Schweitzer A, Wyrobek A, Feiler H, Spellman P, Conboy J, Gray JW. 2010. Exon-level microarray analyses identify alternative splicing programs in breast cancer. Mol. Cancer Res. 8:961–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G. 2005. Cell motility is controlled by SF2/ASF through alternative splicing of the Ron protooncogene. Mol. Cell 20:881–890 [DOI] [PubMed] [Google Scholar]
- 17. Valacca C, Bonomi S, Buratti E, Pedrotti S, Baralle FE, Sette C, Ghigna C, Biamonti G. 2010. Sam68 regulates EMT through alternative splicing-activated nonsense-mediated mRNA decay of the SF2/ASF proto-oncogene. J. Cell Biol. 191:87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goncalves V, Matos P, Jordan P. 2009. Antagonistic SR proteins regulate alternative splicing of tumor-related Rac1b downstream of the PI3-kinase and Wnt pathways. Hum. Mol. Genet. 18:3696–3707 [DOI] [PubMed] [Google Scholar]
- 19. Radisky DC, Levy DD, Littlepage LE, Liu H, Nelson CM, Fata JE, Leake D, Godden EL, Albertson DG, Nieto MA, Werb Z, Bissell MJ. 2005. Rac1b and reactive oxygen species mediate MMP-3-induced EMT and genomic instability. Nature 436:123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Egeblad M, Werb Z. 2002. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2:161–174 [DOI] [PubMed] [Google Scholar]
- 21. Oltean S, Sorg BS, Albrecht T, Bonano VI, Brazas RM, Dewhirst MW, Garcia-Blanco MA. 2006. Alternative inclusion of fibroblast growth factor receptor 2 exon IIIc in Dunning prostate tumors reveals unexpected epithelial mesenchymal plasticity. Proc. Natl. Acad. Sci. U. S. A. 103:14116–14121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mauger DM, Lin C, Garcia-Blanco MA. 2008. hnRNP H and hnRNP F complex with Fox2 to silence fibroblast growth factor receptor 2 exon IIIc. Mol. Cell. Biol. 28:5403–5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Del Gatto F, Plet A, Gesnel MC, Fort C, Breathnach R. 1997. Multiple interdependent sequence elements control splicing of a fibroblast growth factor receptor 2 alternative exon. Mol. Cell. Biol. 17:5106–5116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Carstens RP, Wagner EJ, Garcia-Blanco MA. 2000. An intronic splicing silencer causes skipping of the IIIb exon of fibroblast growth factor receptor 2 through involvement of polypyrimidine tract binding protein. Mol. Cell. Biol. 20:7388–7400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. 2009. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol. Cell 33:591–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dittmar KA, Jiang P, Park JW, Amirikian K, Wan J, Shen S, Xing Y, Carstens RP. 2012. Genome-wide determination of a broad ESRP-regulated posttranscriptional network by high-throughput sequencing. Mol. Cell. Biol. 32:1468–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Warzecha CC, Shen S, Xing Y, Carstens RP. 2009. The epithelial splicing factors ESRP1 and ESRP2 positively and negatively regulate diverse types of alternative splicing events. RNA Biol. 6:546–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Warzecha CC, Jiang P, Amirikian K, Dittmar KA, Lu H, Shen S, Guo W, Xing Y, Carstens RP. 2010. An ESRP-regulated splicing programme is abrogated during the epithelial-mesenchymal transition. EMBO J. 29:3286–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, Cheng C. 2011. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J. Clin. Invest. 121:1064–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yanagisawa M, Huveldt D, Kreinest P, Lohse CM, Cheville JC, Parker AS, Copland JA, Anastasiadis PZ. 2008. A p120 catenin isoform switch affects Rho activity, induces tumor cell invasion, and predicts metastatic disease. J. Biol. Chem. 283:18344–18354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, Burge CB, Gertler FB. 2011. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 7:e1002218 doi:10.1371/journal.pgen.1002218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. 2009. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 16:130–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Venables JP, Klinck R, Koh C, Gervais-Bird J, Bramard A, Inkel L, Durand M, Couture S, Froehlich U, Lapointe E, Lucier JF, Thibault P, Rancourt C, Tremblay K, Prinos P, Chabot B, Elela SA. 2009. Cancer-associated regulation of alternative splicing. Nat. Struct. Mol. Biol. 16:670–676 [DOI] [PubMed] [Google Scholar]
- 34. Perreault N, Beaulieu JF. 1998. Primary cultures of fully differentiated and pure human intestinal epithelial cells. Exp. Cell Res. 245:34–42 [DOI] [PubMed] [Google Scholar]
- 35. Patry C, Bouchard L, Labrecque P, Gendron D, Lemieux B, Toutant J, Lapointe E, Wellinger R, Chabot B. 2003. Small interfering RNA-mediated reduction in heterogeneous nuclear ribonucleoparticule A1/A2 proteins induces apoptosis in human cancer cells but not in normal mortal cell lines. Cancer Res. 63:7679–7688 [PubMed] [Google Scholar]
- 36. Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. 2007. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8:R19 doi:10.1186/gb-2007-8-2-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Klinck R, Bramard A, Inkel L, Dufresne-Martin G, Gervais-Bird J, Madden R, Paquet ER, Koh C, Venables JP, Prinos P, Jilaveanu-Pelmus M, Wellinger R, Rancourt C, Chabot B, Abou Elela S. 2008. Multiple alternative splicing markers for ovarian cancer. Cancer Res. 68:657–663 [DOI] [PubMed] [Google Scholar]
- 38. Venables JP, Klinck R, Bramard A, Inkel L, Dufresne-Martin G, Koh C, Gervais-Bird J, Lapointe E, Froehlich U, Durand M, Gendron D, Brosseau JP, Thibault P, Lucier JF, Tremblay K, Prinos P, Wellinger RJ, Chabot B, Rancourt C, Elela SA. 2008. Identification of alternative splicing markers for breast cancer. Cancer Res. 68:9525–9531 [DOI] [PubMed] [Google Scholar]
- 39. Venables JP, Koh CS, Froehlich U, Lapointe E, Couture S, Inkel L, Bramard A, Paquet ER, Watier V, Durand M, Lucier JF, Gervais-Bird J, Tremblay K, Prinos P, Klinck R, Elela SA, Chabot B. 2008. Multiple and specific mRNA processing targets for the major human hnRNP proteins. Mol. Cell. Biol. 28:6033–6043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pan Q, Shai O, Misquitta C, Zhang W, Saltzman AL, Mohammad N, Babak T, Siu H, Hughes TR, Morris QD, Frey BJ, Blencowe BJ. 2004. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell 16:929–941 [DOI] [PubMed] [Google Scholar]
- 41. Savagner P, Valles AM, Jouanneau J, Yamada KM, Thiery JP. 1994. Alternative splicing in fibroblast growth factor receptor 2 is associated with induced epithelial-mesenchymal transition in rat bladder carcinoma cells. Mol. Biol. Cell 5:851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pino MS, Balsamo M, Di Modugno F, Mottolese M, Alessio M, Melucci E, Milella M, McConkey DJ, Philippar U, Gertler FB, Natali PG, Nistico P. 2008. Human Mena+11a isoform serves as a marker of epithelial phenotype and sensitivity to epidermal growth factor receptor inhibition in human pancreatic cancer cell lines. Clin. Cancer Res. 14:4943–4950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Keirsebilck A, Bonne S, Staes K, van Hengel J, Nollet F, Reynolds A, van Roy F. 1998. Molecular cloning of the human p120ctn catenin gene (CTNND1): expression of multiple alternatively spliced isoforms. Genomics 50:129–146 [DOI] [PubMed] [Google Scholar]
- 44. Mellman I, Nelson WJ. 2008. Coordinated protein sorting, targeting and distribution in polarized cells. Nat. Rev. Mol. Cell Biol. 9:833–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nelson WJ, Nusse R. 2004. Convergence of Wnt, beta-catenin, and cadherin pathways. Science 303:1483–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Prinos P, Garneau D, Lucier JF, Gendron D, Couture S, Boivin M, Brosseau JP, Lapointe E, Thibault P, Durand M, Tremblay K, Gervais-Bird J, Nwilati H, Klinck R, Chabot B, Perreault JP, Wellinger RJ, Elela SA. 2011. Alternative splicing of SYK regulates mitosis and cell survival. Nat. Struct. Mol. Biol. 18:673–679 [DOI] [PubMed] [Google Scholar]
- 47. Lefave CV, Squatrito M, Vorlova S, Rocco GL, Brennan CW, Holland EC, Pan YX, Cartegni L. 2011. Splicing factor hnRNPH drives an oncogenic splicing switch in gliomas. EMBO J. 30:4084–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gallagher TL, Arribere JA, Geurts PA, Exner CR, McDonald KL, Dill KK, Marr HL, Adkar SS, Garnett AT, Amacher SL, Conboy JG. 2011. Rbfox-regulated alternative splicing is critical for zebrafish cardiac and skeletal muscle functions. Dev. Biol. 359:251–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Huang SC, Ou AC, Park J, Yu F, Yu B, Lee A, Yang G, Zhou A, Benz EJ., Jr 2012. RBFOX2 promotes protein 4.1R exon 16 selection via U1 snRNP recruitment. Mol. Cell. Biol. 32:513–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sun S, Zhang Z, Fregoso O, Krainer AR. 2012. Mechanisms of activation and repression by the alternative splicing factors RBFOX1/2. RNA 18:274–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang C, Frias MA, Mele A, Ruggiu M, Eom T, Marney CB, Wang H, Licatalosi DD, Fak JJ, Darnell RB. 2010. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science 329:439–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Takeuchi A, Hosokawa M, Nojima T, Hagiwara M. 2010. Splicing reporter mice revealed the evolutionally conserved switching mechanism of tissue-specific alternative exon selection. PLoS One 5:e10946 doi:10.1371/journal.pone.0010946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Llorian M, Schwartz S, Clark TA, Hollander D, Tan LY, Spellman R, Gordon A, Schweitzer AC, de la Grange P, Ast G, Smith CW. 2010. Position-dependent alternative splicing activity revealed by global profiling of alternative splicing events regulated by PTB. Nat. Struct. Mol. Biol. 17:1114–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.