Abstract
The p300, CBP, and pCAF lysine acetyltransferase (KAT) proteins have been reported to physically interact with bovine (BPV) and human (HPV) papillomavirus E2 proteins. While overexpression of these KAT proteins enhances E2-dependent transcription, the mechanism has not been determined. Using RNA interference (RNAi) to deplete these factors, we demonstrated that E2 transcriptional activity requires physiological levels of p300, CBP, and pCAF. Each protein appears to have a unique function in E2-dependent transcription, since overexpression of one KAT failed to compensate for RNAi knockdown of another KAT. Using an in vitro acetylation assay, we identified highly conserved lysines that are targeted by p300 for acetylation. The conservative changes of lysines at positions 111 and 112 to arginine were of particular interest. The K111R and the K111R/K112R mutants showed reduced transcriptional activity that was not responsive to p300 overexpression, while the K112R mutant retained activity. p300 and CBP were detected at the viral promoter; however, pCAF was not. We propose a model by which E2 transcriptional activity is controlled by p300-mediated acetylation of lysine 111. This model represents a novel mechanism regulating papillomavirus gene expression.
INTRODUCTION
The papillomavirus E2 protein controls transcription of viral early gene products by binding to specific DNA motifs (ACCN6GGT) critically placed within the viral genome (1). While viral gene expression is controlled by a variety of cellular transcription factors, including TFIID, Sp1, Oct1, and AP1 (2–6), expression of E2 results in increased transcriptional activation from the bovine papillomavirus (BPV) promoter and enhancer elements containing E2 binding sites (7, 8). DNA binding activity is conferred by the C-terminal DNA binding domain (DBD); however, the N-terminal transactivation domain (TAD) of E2 is also necessary for specific activity (9–12). The ability of the TAD to activate transcription in the absence of the DBD (13) indicates that its function is at least partly mediated through complex formation with cellular factors (10, 12).
Prior studies have identified E2-interacting proteins that facilitate E2-dependent transcription; however, the precise mechanisms through which these factors contribute to E2 activation of the viral early promoter remain unclear. Several reports have demonstrated that general transcription factors TFIIB and TFIID, including the TATA binding protein (TBP) and several TBP-associated factors, interact with E2 (14–18) and that TFIIB and TBP can enhance transcription by E2 (3, 16, 18). Cellular coactivators such as Brd4, hBrm, Gps2 (also known as AMF-1), and Tax1BP1 interact with E2 and enhance E2-dependent transcriptional activation (19–24). Brd4 and Tax1BP1 stabilize E2 protein, which partially explains how these factors increase E2 transcriptional activity (20, 24–26). Association with the Brm ATP-dependent chromatin remodeling complex enhances E2-dependent transcriptional activity specifically on episomally maintained templates (19). E2-dependent coactivation by either Tax1BP1 or Gps2 is enhanced further through complex formation with the cellular acetyltransferase p300 (22, 24).
Histone acetyltransferases (HATs), also known as lysine (K) acetyltransferases (KATs), catalyze the transfer of an acetyl group from acetyl coenzyme A (acetyl-CoA) molecules to the ε-amino group of lysine side chains. Each KAT is thought to recognize a distinct molecular surface around the target lysine and not necessarily a specific consensus peptide sequence (27, 28). Further specificity is conferred through the formation of higher-order complexes containing one or more KATs and a variety of subunits (28–30). Subunits commonly encode chromatin binding domains such as bromodomains or chromodomains that recognize acetylated (31–33) and methylated (34, 35) lysine residues, respectively. Through recognition of various combined modifications on histone tails, acetyltransferase complexes are recruited to specific promoter or enhancer regions where activity is required (36–40). Acetylation of nonhistone proteins, such as transcriptional cofactors, can affect substrate stability, subcellular localization, and interactions with other proteins or DNA (41).
Published data reveal that p300, CBP, and the p300/CBP-associated factor (pCAF) interact directly with E2 proteins of BPV type 1 (BPV-1) and human papillomavirus type 16 (HPV-16), HPV-18, HPV-11, and HPV-6b (22, 42–44). These KAT proteins have also been demonstrated to enhance E2-dependent transcription (22, 42–45). Further, p300 coexpressed with small amounts of HPV-16 E2 enhances transcriptional activity from the HPV-16 promoter, which is generally repressed at higher levels of E2 (45). The E2 coactivator Gps2, which also directly binds p300, was proposed to facilitate enhancement of BPV-1 E2 activity by p300 (22). The KIX domain of CBP, which is also the CREB binding region, interacts with the N-terminal transactivation domain of HPV-18 E2. Heterologous expression of the CBP KIX domain fused to the VP16 activation domain enhanced E2-dependent transcription (43). The N-terminal 390 amino acids of pCAF are necessary for interaction with HPV-6b, -11, -16, and -18 E2 (42). Enhancement of E2-dependent transcription by pCAF was further stimulated by coexpression of CBP (42), suggesting that these KAT proteins act through distinct pathways. These studies also showed that the acetyltransferase domains of CBP and pCAF were required for enhancement and that acetyltransferase-defective mutants were unable to enhance E2-dependent transcription (42, 43).
The functional interaction of KATs p300, CBP, and pCAF with E2 suggests a role for acetylation in the regulation of E2-dependent transcription. This could conceptually occur through histone alteration and subsequent chromatin remodeling, similar to the proposed role for hBrm, and/or through posttranslational modification of lysines within E2. We have examined the role of p300, CBP, and pCAF in E2-dependent transcription using selective RNA interference (RNAi) depletion and explored the possibility that E2 is a substrate for acetylation. The results shown here are consistent with distinct roles for each KAT protein in regulating E2 activity. We demonstrate that E2 is acetylated by p300 in vitro and that the mutation of conserved N-terminal lysines restricts E2 activity and uncouples E2-dependent transcription from the effects of p300 overexpression. These data provide the first evidence for the direct acetylation of E2 as a mechanism to regulate its transcriptional activity.
MATERIALS AND METHODS
Cells and transfections.
RPE-1 cells were cultured in a 1:1 mix of Dulbecco's modified Eagle's medium (DMEM) and Ham's F-12 medium (Invitrogen). C33a, RKO, C127, ID13, and C127-A3 cells were cultured in DMEM (Invitrogen). ID13 and C127-A3 cells are BPV-1-transformed cell lines. BPV genomes in C127-A3 cells contain three mutations within the E2 reading frame leading to increased E2 protein levels (46). All cell culture media were supplemented with penicillin-streptomycin solution (Invitrogen) as well as 10% fetal bovine serum (Atlas Biologicals and Sigma-Aldrich). Transfections of all cells except C33a were performed using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer's specifications. All DNA, small interfering RNA (siRNA), and transfection reagents for these experiments were diluted in Opti-MEM serum free medium (Invitrogen) in the absence of antibiotics. The calcium phosphate transfection method was used for experiments with C33a cells.
Plasmids and siRNAs.
pGL2-E24BS contains 4 high-affinity E2 binding sites upstream of a simian virus 40 (SV40) promoter which drives luciferase expression (47). Expression plasmids used include pCG-E2, pCMVβ-p300, pRSV-CBP, and pCI-pCAF. All E2 mutations from lysine to arginine (R) were generated with the Quikchange II site-directed mutagenesis system according to the manufacturer's specifications (Agilent Technologies) using pCG-E2 as a template. The ΔNcoI BPV-1 genome does not express E2 due to a deletion within the E2 reading frame (48). p300 (CAGAGCAGUCCUGGAUUAG), CBP (AAUCCACAGUACCGAGAAAUG), and pCAF (UCGCCGUGAAGAAAGCGCA) custom siRNAs and control predesigned siRNA (AM4635) were purchased from Ambion.
ChIP.
A total of 106 C127, ID13, or C127-A3 cells were plated onto 10-cm tissue culture dishes. Cells were lysed and sonified prior to chromatin immunoprecipitation (ChIP) as described previously (24). Cells to be probed for endogenous protein were harvested 24 h after plating; otherwise, cells were transfected the day after plating and harvested 24 h later. Immune complexes were captured using Dynabeads (Invitrogen), and DNA was then purified using a QIAquick PCR purification kit (Qiagen) according to the manufacturer's specifications. Purified samples were amplified by PCR using the following BPV-1 LCR primers: sense, AAAGTTTCCATTGCGTCTGG; antisense, GCTTTTTATAGTTAGCTGGCTATTTT.
Purified ΔNcoI BPV-1 DNA was quantified with an Eppendorf Mastercycler using the BPV-1 LCR primers listed above. A four-point, 10-fold dilution series was prepared from input DNA of each individual lysate for a reference curve. Threshold cycle (CT) values for each immunoprecipitated DNA sample were compared to the appropriate reference curves, and the amount of immunoprecipitated DNA was calculated as a percentage of input DNA. Two-way analysis of variance (ANOVA) was performed with Bonferroni's post hoc analysis using GraphPad Prism, version 5.01.
Immunofluorescence and immunoblotting.
A total of 3.5 × 105 RKO cells or 2.5 × 105 RPE-1 cells were plated on collagen-coated coverslips (BD Biosciences) and transfected the following day with expression plasmids. Twenty-four hours later, cells were fixed for 30 min with 3.7% paraformaldehyde (Electron Microscopy Sciences), permeabilized for 10 min with 0.2% Triton X-100, and then incubated in blocking buffer (5% normal goat serum, 1% bovine serum albumin [BSA], and 0.05% Triton X-100 in phosphate-buffered saline [PBS]) overnight. Primary and secondary antibodies were diluted in blocking buffer and incubated on the coverslips for 1 h at approximately 20°C. Coverslips were washed with 0.05% Triton X-100 in PBS and then mounted on glass slides using ProLong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen). Cells were visualized and images collected using a Leica TSC SP2 AOBS confocal microscope with Leica imaging software. For quantification, 50 E2-positive cells were counted per coverslip and a minimum of 4 coverslips were counted for wild-type E2 and each mutant. Each coverslip represents an individual experiment. Counted cells were sorted into one of four categories, and each category was presented as a percentage of total cells counted.
RKO and RPE-1 cells for immunoblot analysis were plated onto six-well dishes at the above-stated densities and transfected the following day with expression plasmid alone or cotransfected with siRNA. Twenty-four hours posttransfection or 48 h after siRNA transfection, cells were lysed using 2% SDS in 50 mM Tris-HCl (pH 8.0) and 1 mM dithiothreitol (DTT). Following protein estimation using bicinchoninic acid (BCA) protein assay reagent (Thermo Scientific), equal amounts of protein were loaded and separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). Gels were transferred to polyvinylidene fluoride (PVDF) membranes (Millipore), blocked, and then probed with specific antibodies. Signal was detected using Super Signal enhanced chemiluminescence substrates (Thermo Scientific).
Antibodies used for both immunofluorescence and immunoblot experiments included BPV-1 E2 B201; Santa Cruz p300 N-15 (sc-583), CBP A-22 (sc-369), and pCAF E-8 (sc-13124); and Sigma-Aldrich actin (A-2668). Secondary antibodies included Alexafluor 488 and 555 (Invitrogen) for immunofluorescence and horseradish peroxidase conjugates (Jackson Laboratory) for immunoblotting.
In vitro acetylation assay.
Two hundred nanograms of protein substrate was mixed with 50 ng of acetyltransferase in a 10-μl reaction mixture containing 40 mM Tris-HCl (pH 8.0), 75 mM potassium chloride (KCl), and 10 μM acetyl-CoA (Sigma-Aldrich). Each reaction was carried out at 30°C for 1 h and terminated by adding SDS-PAGE sample buffer and heating the samples to 95°C for 5 min. Proteins were resolved by SDS-PAGE and transferred to a PVDF membrane, which was probed for acetylated substrates using an anti-acetyl lysine antibody (Cell Signaling Technology; catalog number 9441). Duplicate samples were separated by SDS-PAGE, and protein input was visualized using Coomassie brilliant blue R250. Substrates included BPV-1 E2 (49), p53, and histone protein (50), which were incubated with p300, pCAF, and Gcn5 (50) acetyltransferases. All proteins were purified from bacteria or baculovirus-infected Sf9 insect cells except for histone protein, which was purified from HeLa cells.
Luciferase reporter assay.
A total of 3.5 × 105 RKO cells or 2.5 × 105 RPE-1 cells were plated onto six-well dishes and transfected in triplicate the following day with pGL2-E24BS, expression plasmids, and 10 nM siRNA as described previously (47). Cells were lysed 24 h posttransfection or 48 h after siRNA transfection using reporter lysis buffer (Promega), and luciferase activity was detected on an EnVision multilabel plate reader (PerkinElmer) after addition of luciferase assay reagent or Steady-Glo luciferase assay reagent (Promega) according to the manufacturer's specifications. Means were averaged from at least four independent experiments, and error bars in figures represent standard errors of the means (SEM). Wild-type E2 transcriptional activation in the presence or absence of control siRNA was set to 100%, and each experimental value was calculated as a percentage of the wild type or control. A two-tailed t test or one-way ANOVA with Dunnet's or Bonferroni's post hoc analysis was performed with GraphPad Prism, version 5.01.
Mass spectrometry and data analysis.
Following acetylation with p300, BPV-1 E2 protein was excised from a polyacrylamide gel stained with Coomassie brilliant blue R250 (Fisher). The gel band was destained and digested with trypsin, chymotrypsin, or GluC overnight at 30°C. Peptides from each sample were injected onto a Symmetry C18 trapping cartridge (Waters, Inc.) using a NanoAquity autosampler (Waters, Inc.) and separated by in-line gradient elution onto a 75-μm (internal diameter) by 10-cm column packed with a BEH 130 stationary phase (Waters). Samples were separated using a linear gradient from 3% to 90% of solvent B where solvent A was 2% acetonitrile and solvent B was 98% acetonitrile, both solvents containing 0.1% formic acid and 0.01% trifluoroacetic acid. During the gradient elution, data-dependent scans were performed with 8 scan events per cycle consisting of one full mass spectra (MS) from m/z 400 to 2,000 followed by product ion scans (collision-induced dissociation = 35%) on the 10 most intense ions in the full scan. Precursor ions used for product ion scans were dynamically excluded for 30 s. Proteins were identified from the product ion spectra using SEQUEST (Thermo Scientific) and X!Tandem search engines across the entire Swiss-Prot database (51); the results from each search were combined using Scaffold, version 3.00 (Proteome Software, Portland, OR).
RESULTS
E2 transcriptional activity is dependent on physiological levels of p300, CBP, and pCAF.
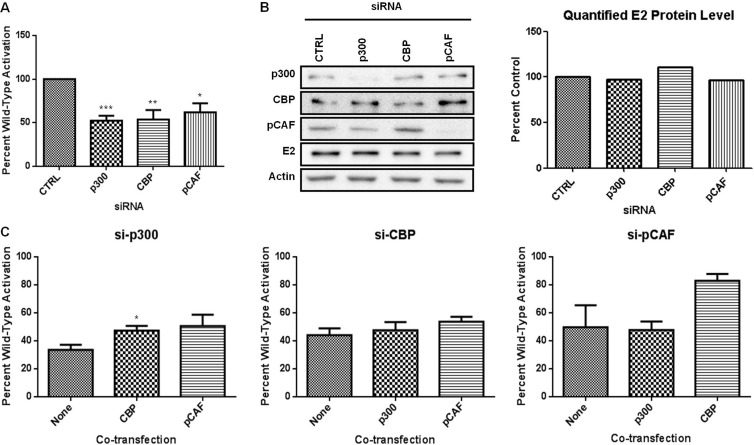
Transfection of expression vectors for p300, CBP, and pCAF has been reported to increase E2-dependent transcription (22, 42–44). How this occurs is not known. To first investigate dependence on each KAT, we used RNAi to examine the consequences of selective KAT depletion on activation of an E2-responsive luciferase reporter. BPV-1 E2 was cotransfected with a reporter plasmid and either control siRNA or siRNA targeting p300, CBP, or pCAF. E2 stimulated reporter expression nearly 80-fold, which is consistent with previous reports (47). Individual siRNA knockdown of p300, CBP, or pCAF resulted in approximately 40 to 50% reduction of wild-type E2 activity compared to that in samples transfected with the scrambled siRNA (Fig. 1A). A minimum of 50% reduction in KAT protein levels was confirmed by immunoblot analysis (Fig. 1B). Depletion of p300, CBP, and pCAF had no significant effect on E2 protein levels (Fig. 1B).
Fig 1.
E2 transcriptional activity is dependent on physiological levels of p300, CBP, and pCAF. (A) RPE-1 cells were transfected with a luciferase reporter plasmid containing four E2 binding sites, E2, and a control siRNA or siRNA specific for p300, CBP, or pCAF. Luciferase activity was measured 48 h posttransfection. Results are presented as a percentage of wild-type activity with control siRNA. Student's t test was performed comparing knockdown to control. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B) Lysates from transfected cells were prepared for SDS-PAGE and immunoblotted for E2 protein. The graph represents quantified E2 protein in the presence of the indicated siRNA. (C) Luciferase assay was performed as for panel A except that p300, CBP, and pCAF were overexpressed in the presence of siRNA specific for p300, CBP, or pCAF. Each graph represents one transfected siRNA, and results are presented as a percentage of wild-type activity with control siRNA. Student's t test was performed comparing HAT transfection to siRNA alone. *, P < 0.05.
These data imply that each KAT protein has a unique function that cannot be compensated for by the remaining KAT proteins. This was unexpected, particularly for p300 and CBP, which are highly homologous and similar in function (52, 53). The residual E2-dependent transcriptional activity following depletion of each KAT might be due to the reduced but persistent levels of the targeted KAT. While we considered using p300 or CBP null mouse embryonic fibroblasts (MEFs), unrecognized compensatory changes might make results difficult to interpret. We decided to explore potential redundancy among p300, CBP, and pCAF by measuring the ability of each KAT to restore E2-dependent transcriptional activity following depletion of another KAT protein. In these experiments, we observed that overexpression of pCAF was unable to restore E2 transcriptional activity following p300 depletion (Fig. 1C); however, overexpression of CBP resulted in a small but significant increase in activity. E2-dependent transcriptional activity following siRNA silencing of CBP or pCAF was not functionally replaced through overexpression of another KAT protein (Fig. 1C). There was an increase in E2 activity following expression of CBP in pCAF-depleted cells; however, this activity was not significantly different from that in pCAF-depleted cells without overexpression and was 20% lower than E2 activity in the presence of control siRNA.
p300 and CBP interact with the BPV-1 genome.
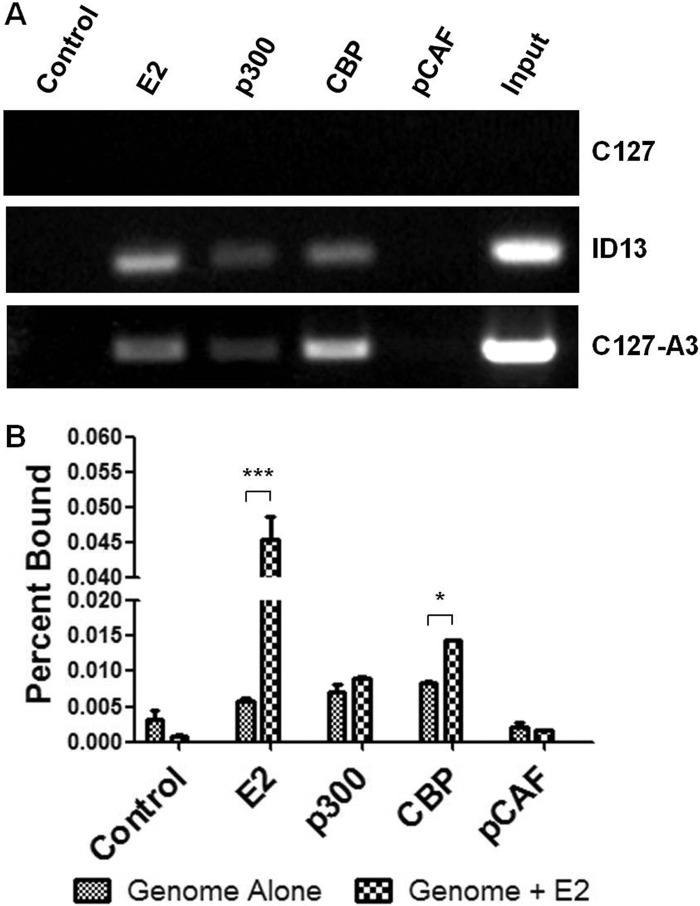
A previous report demonstrated that the presence of E2 increased pCAF-mediated acetylation of histone H3 using a reporter plasmid system (42). While this report indicates that E2 may recruit KATs, specifically pCAF, to DNA containing E2 binding sites, it did not directly examine the presence of the KAT protein at the promoter. We investigated the ability of endogenous KAT proteins to interact with the BPV-1 LCR through ChIP assay. As expected, we readily detected E2 in complex with the BPV-1 genomes present in ID13 cells and C127-A3 cells but not in parental C127 cells that do not contain BPV DNA (Fig. 2A). ChIP assay using p300 and CBP antibodies revealed their interaction with the viral genomes, specifically with the LCR, in both of the BPV-transformed cell lines (Fig. 2A). Interestingly, pCAF antibody failed to coimmunoprecipitate BPV-1 DNA in cross-linked lysates of ID13 or C127-A3 cells (Fig. 2A).
Fig 2.
p300 and CBP but not pCAF interact with the BPV-1 genome. (A) Untransfected C127, ID13, and C127-A3 cell lysates were prepared for ChIP assay and immunoprecipitated using either a control antibody or antibodies recognizing BPV-1 E2, p300, CBP, or pCAF. Precipitated DNA was amplified by PCR using BPV-1 LCR primers. (B) C127 cells were transfected with ΔNcoI genomes in the presence or absence of BPV-1 E2. Lysates were prepared for ChIP assay and immunoprecipitated using control antibody or antibodies directed at BPV-1 E2, p300, CBP, or pCAF. Precipitated DNA was quantified using real-time PCR. Two-way ANOVA with Bonferroni's post hoc analysis was performed comparing genome alone and genome plus E2 for each immunoprecipitation. *, P < 0.05; ***, P < 0.001.
While it is difficult to evaluate quantitative differences between samples using traditional PCR methods, ChIP of the viral genome with CBP antibodies appeared to be greater with the C127-A3 cells, in which E2 protein levels are higher (Fig. 2A). To address this observation, the ability of E2 to recruit KATs to the LCR was investigated using a modified ChIP assay. C127 cells were transfected with mutant BPV-1 genomes that do not express E2, and the viral DNA in complex with the KAT was quantified by real-time PCR. Following transfection of E2, the amount of viral DNA coprecipitated by E2 antibodies increased 8-fold over background (Fig. 2A). There was a 2-fold increase in the amount of genome coprecipitated with CBP antibodies in E2-transfected samples (Fig. 2B). However, no such increases were observed following immunoprecipitation with p300 or pCAF antibodies. The amount of pCAF associated with the genome was comparable to background levels detected following control immunoprecipitation (Fig. 2B).
E2 is acetylated by p300.
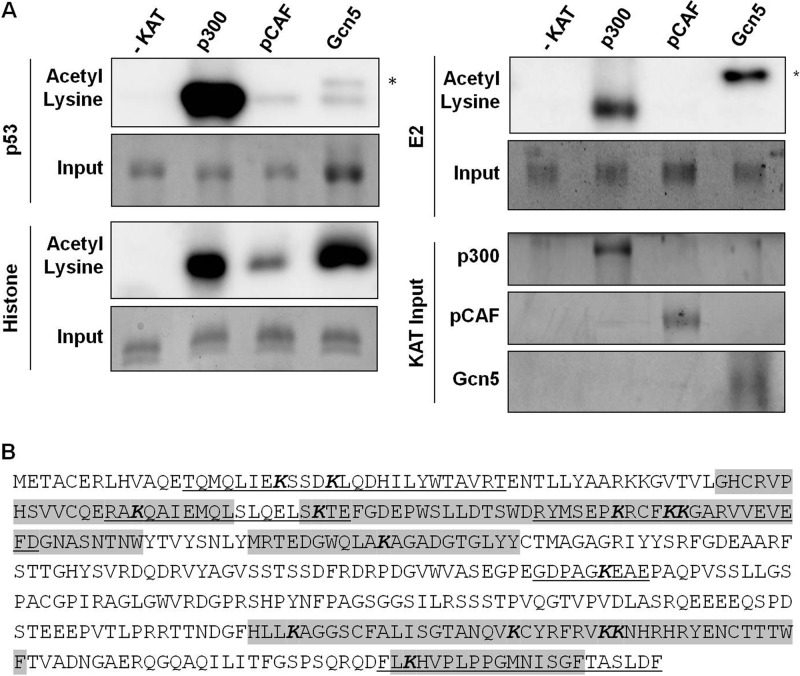
Previous reports investigating the functional interactions between p300, CBP, and pCAF with BPV and HPV E2 proteins have focused on the effects of KAT overexpression on E2-dependent transcriptional activation (22, 42–45). While these reports provide evidence that these proteins affect E2-dependent transcription, little is known about their mechanism of action. Despite evidence that acetyltransferase activity is necessary for enhancement by CBP and pCAF (42, 43), acetylation of E2 has not been reported. Previous attempts to identify acetylated BPV-1 E2 in vitro and in cultured cells were inconsistent or unsuccessful (data not shown) (22). These results were likely hampered by low levels of E2 proteins being immunoblotted with insensitive anti-acetyl lysine antibodies. We performed in vitro acetylation reactions using p300, pCAF, and Gcn5 (a pCAF paralog) proteins that were bacterially expressed or purified from baculovirus-infected Sf9 cells. Purified KAT and E2 proteins were combined in the presence of acetyl-CoA, and resulting acetylated protein was detected by immunoblot analysis with an acetyl lysine antibody. Purified histones and p53 were included as positive controls for acetylation. Robust acetylation was detected on histone substrates by each KAT (Fig. 3A). p300 mediated strong acetylation of p53, while pCAF and Gcn5 were much less active (Fig. 3A). This may reflect the fact that p300 acetylates p53 on several more lysines than does pCAF or Gcn5 (54, 55) or an epitope preference for the acetyl lysine antibody. After incubation of BPV-1 E2 with p300, a strong band corresponding to acetylated E2 was observed. Acetylation of E2 was not detected in reaction mixtures containing either pCAF or Gcn5 (Fig. 3A). Autoacetylation of the short isoform of Gcn5 used in this experiment is detectable in these blots at a migration slightly slower than those of E2 and p53 (50).
Fig 3.
E2 is acetylated by p300. (A) In vitro acetylation of p53 (top left), purified histone (bottom left), and BPV-1 E2 (top right) proteins, using purified acetyltransferase p300, pCAF, or Gcn5. Immunoblots used an anti-acetyl lysine antibody. The higher-molecular-weight band indicated by the asterisk, visible in p53 and BPV-1 E2 acetylation reactions, is due to autoacetylation of truncated Gcn5. Input for each substrate as well as each acetyltransferase (bottom right) is presented as an identical reaction processed in parallel with the gel stained for total protein using Coomassie brilliant blue. (B) Acetylated peptide sequence coverage of BPV-1 E2 obtained from proteomic analysis of digestion with chymotrypsin (underlined) and GluC (gray shading). Detected acetylated lysine residues are in bold italic type.
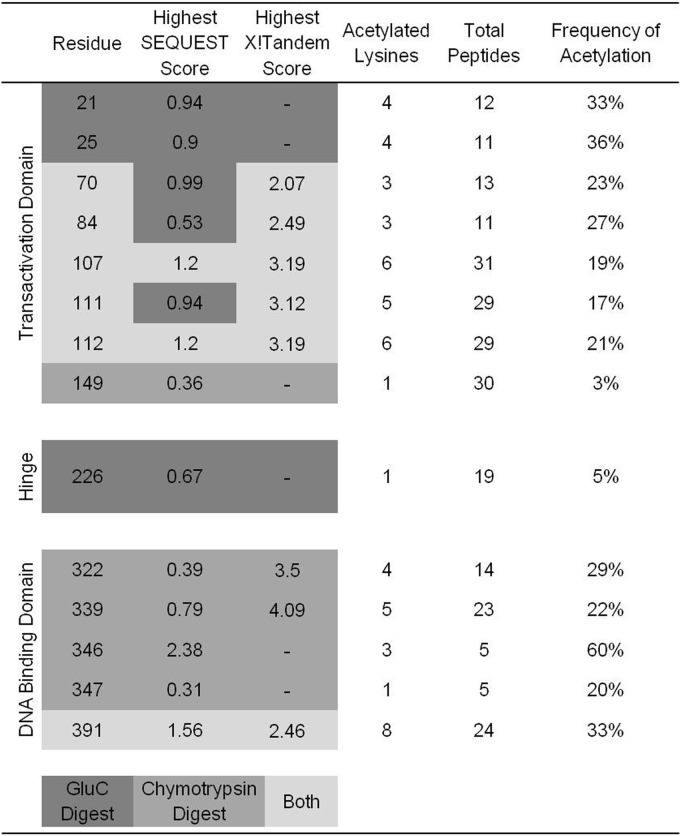
Lysines in BPV-1 E2 that were targeted by p300 were identified using a proteomic approach. BPV-1 E2 was acetylated in vitro and digested, and the resulting peptides were analyzed by mass spectrometry. Each peptide was compared to the entire Swiss-Prot database and assembled using both the SEQUEST and X!Tandem search engines. These two utilities directly compare uninterpreted tandem mass spectra to protein databases using different algorithms, resulting in protein identification. Acetylated peptides of BPV-1 E2 were not identified following trypsin digestion. This is likely due to the inability of trypsin to cleave after acetylated lysines. Analysis of BPV-1 E2 digested with chymotrypsin yielded 97% total sequence coverage and identified 11 acetylated lysine residues (Fig. 3B). Analysis of GluC digestion yielded 89% sequence coverage and nine acetylated lysines, of which three were unique to GluC digestion (Fig. 3B). Nine acetylated lysines were found in the N-terminal TAD, five in the C-terminal DBD, and one in the central hinge region. Acetylated lysines were discovered in an average of 22% of each unique E2 peptide identified. The frequency of lysine acetylation in each unique peptide ranged from 3% to 60%. The low average frequency of acetylation could explain the difficulty detecting acetylated E2 in vivo. A summary of data compiled from chymotrypsin and GluC digests, including SEQUEST and X!Tandem correlation scores, is presented in Table 1.
Table 1.
Proteomic analysis of acetylated E2
E2 lysine mutations display transcriptional defects.
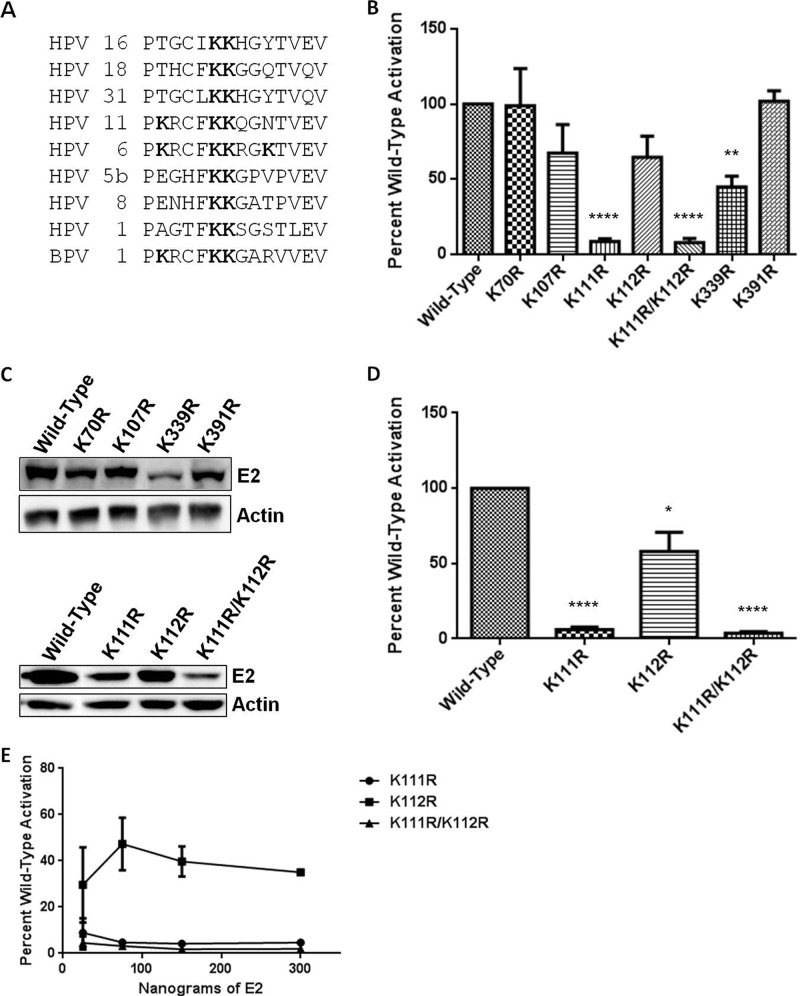
We next sought to characterize the functional significance of E2 acetylation by p300 in vivo. Six lysines identified in the proteomic analysis were selected for mutation on the bases of sequence conservation among BPV-1 and several HPV types, frequency with which the modified lysine was identified in proteomic analysis, and SEQUEST and X!Tandem correlation scores. Lysines 107, 111, and 112 are part of a putative BPV-1 E2 N-terminal nuclear localization signal (NLS) (56). Lysines 111 and 112 are conserved throughout eight HPV types, including high-risk 16, 18, and 31 (Fig. 4A), and lysine 339 is a critical residue that mediates interaction with E2 DNA binding sites (57). These lysines were conservatively mutated to arginine to avoid effects related to variations of residue charge. Each mutant was screened for its ability to activate transcription from an E2-responsive reporter and for protein expression levels.
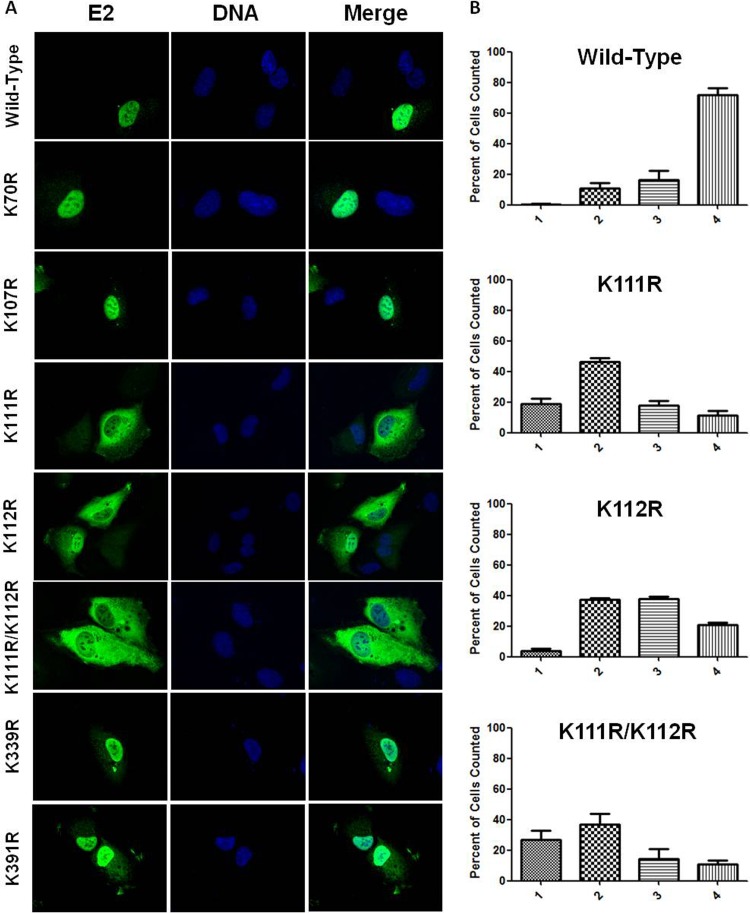
Fig 4.
E2 lysine mutations display transcriptional defects. (A) Sequence alignment of the region between amino acids 106 and 119 of BPV-1 E2. Bold designates lysine (K) in sequence alignment. Alignment was prepared using the ClustalW multiple alignment tool (73). (B) RPE-1 cells were cotransfected with an E2-responsive luciferase reporter and wild-type E2 or one of a series of lysine-to-arginine mutants. Luciferase activity was measured 24 h posttransfection, and results are presented as a percentage of wild-type E2 activation. t tests were performed comparing each mutant to wild-type E2. **, P < 0.01; ****, P < 0.0001. (C) Lysates of wild-type and mutant E2-transfected RPE-1 cells were prepared for SDS-PAGE, and immunoblot analysis was performed to determine the steady-state protein level for each of the E2 K/R mutants (top panel). RPE-1 cells were transfected with 3 μg of K111R, K112R, and K111R/K112R proteins (bottom panel). (D) C33a cells were cotransfected with an E2-responsive luciferase reporter and wild-type E2, E2 K111R, K112R, or K111R/K112R protein. Luciferase activity at 24 h posttransfection is presented as a percentage of wild-type activation. t tests were performed comparing each mutant to wild-type E2. *, P < 0.05; ****, P < 0.0001. (E) C33a cells were cotransfected with an E2-responsive luciferase reporter and four concentrations of each E2 mutant, including 25 ng, 75 ng, 150 ng, and 300 ng. Luciferase assay was performed as for panel D, and a dose-response curve was generated. Each point represents a percentage of wild-type E2 activity at that concentration.
Mutation of BPV-1 E2 lysines (K) 70 and 391 to arginine (R) resulted in transcriptional activity equivalent to that observed with wild-type E2 in RPE-1 cells (Fig. 4B) despite protein levels approximately 30% lower than that of the wild type (Fig. 4C). The K107R E2 mutant protein was present at ∼85% of the wild type, with a proportional decrease in its transcriptional activity (Fig. 4B and C). K112R E2 was present at levels similar to those of K107R E2 and displayed comparable transcriptional activity. This activity is not significantly different from that of wild-type E2 in RPE-1 cells; however, when K112R is transfected into C33a cells, the mean transcriptional activity is slightly lower (64.5% for RPE-1 cells and 58.1% in C33a cells), resulting in a significant difference (Fig. 4B and D). The K339R E2 mutant was impaired in its ability to activate transcription in RPE-1 cells (Fig. 4B). This may be partially attributable to protein levels at 45% of that of wild-type E2; however, transfection of increasing amounts of the K339R mutant resulted in a peak of transcriptional activity at less than 50% of that observed for wild-type E2 (data not shown). K111R and double-mutant K111R/K112R proteins were expressed at lower levels (Fig. 4c). While all other E2 mutants tested precipitated the BPV genome at levels comparable to those of wild-type E2, precipitation of the K111R and K111R/K112R mutants with the genome was reduced (data not shown). The K111R and K111R/K112R mutants were both severely impaired in their ability to activate transcription in RPE-1 and C33a cells (Fig. 4B and D). The transcriptional defects of the K111R and K111R/K112R mutants were not due to reduced protein levels; transfection of increasing amounts of either construct had no effect on transcriptional activity (Fig. 4E).
E2 lysines 111 and 112 are important for nuclear retention.
A region that is rich in basic amino acids in the N-terminal transactivation domain (TAD) of BPV-1 E2 has been reported to function as a NLS in the absence of the highly conserved C-terminal NLS (56). Four of the six lysines mutated in this study are within these two NLS sequences and prompted examination of mutant E2 localization. The subcellular distributions of four mutants, the K70R, K107R, K339R, and K391R mutants, were nearly indistinguishable from that of wild-type E2; nuclear localization was observed in nearly all cases (Fig. 5A). The three remaining K111R, K112R, and K111R/K112R mutant proteins deviated from wild-type localization and exhibit a spectrum of subcellular distributions ranging from exclusively cytosolic to exclusively nuclear (Fig. 5A). The severity of mislocalization, which was characterized as the extent of observed nuclear exclusion, was different for each mutant protein. The K112R mutant was observed to be more nuclear than the K111R/K112R double mutant, which was predominantly cytosolic. The K111R mutant displayed an intermediate phenotype, although its increased cytosolic localization more closely resembled that observed for the K111R/K112R mutant than for the K112R mutant (Fig. 5A).
Fig 5.
E2 lysine-to arginine-mutants mislocalize. (A) RPE-1 cells plated on coverslips were transfected with wild-type E2 or lysine-to-arginine mutants. Twenty-four hours posttransfection, cells were fixed and stained using an antibody to E2 (green), and DNA was visualized using DAPI (blue). (B) The degree of mislocalization was scored into four categories as follows: 1, exclusively cytosolic; 2, even distribution throughout the cytosol and nucleus; 3, diffuse staining with greater intensity in the nucleus; 4, exclusively nuclear staining. Results are presented as a percentage of total cells counted.
Quantifying the extent of mislocalization was necessary to fully characterize these mutants due to the spectrum of localization patterns observed for the K111R, K112R, and K111R/K112R mutants. The localization spectrum was divided into four discrete categories. E2 mutant-expressing cells were visually inspected, and each cell was assigned to a category according to predetermined selection criteria. All three mutant proteins were considerably mislocalized to the cytosol (Fig. 5B). The K111R and K111R/K112R mutants were most commonly found to be evenly distributed throughout both the nuclear and cytosolic compartments (Fig. 5B). While the localization profile of the K111R/K112R mutant was similar to that of the K111R mutant, it was found to be completely excluded from the nucleus more often than either of the other mutants (Fig. 5B; compare second and fourth graphs). While expression of the K112R mutant resulted in cytosolic accumulation, E2 staining was more prevalent in the nuclear compartment (Fig. 5B). The increase in cytosolic localization for each mutant is not due to Crm-1-mediated nuclear export; treatment of mutant-transfected cells with leptomycin B (LMB) had no effect on the distribution of E2 mutant proteins (data not shown).
p300 cannot enhance the transcriptional activity of E2 K111 mutants.
The mechanism by which p300 enhances E2-dependent transcription is unclear (22, 44, 45). The K111R and K111R/K112R E2 mutants retain their overall charge but cannot be acetylated and were severely defective transcriptionally. The relationship between K111 and p300 was further addressed in RKO cells that do not express endogenous p300 (58). Coexpression of p300 with wild-type E2 enhanced activation of E2-dependent transcription in RKO cells an average of 2-fold (Fig. 6). The K111R and K111R/K112R E2 mutants showed 10 to 20% of the transcriptional activity of wild-type E2, and this activity was not enhanced by coexpression of p300 (Fig. 6).
Fig 6.
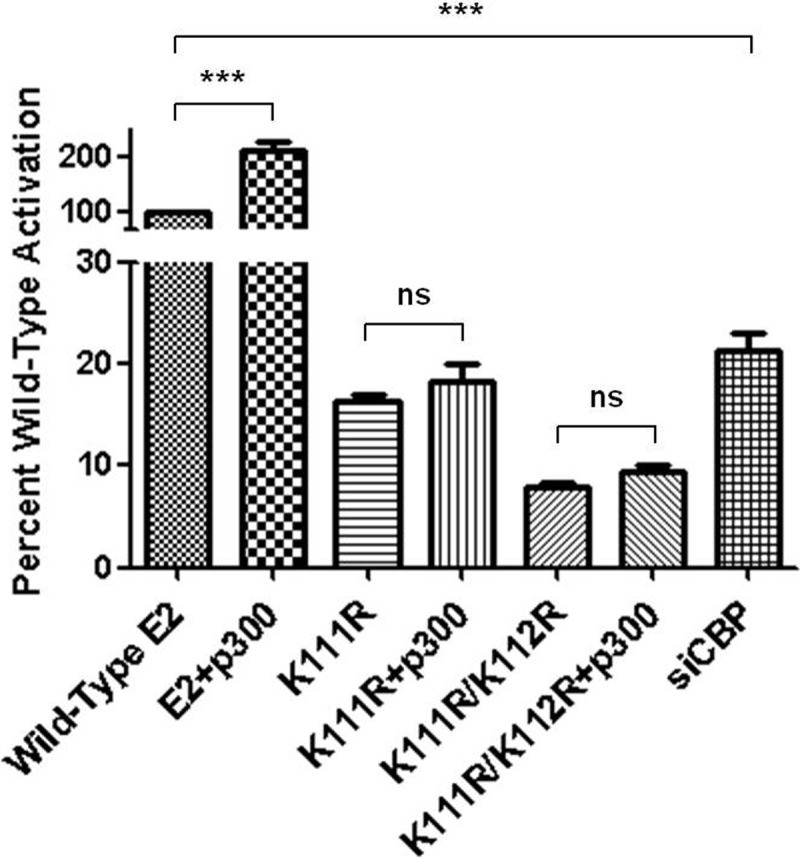
p300 does not enhance the transcriptional activity of E2 K111R mutants. RKO cells were transfected with an E2-responsive luciferase reporter, wild-type or mutant E2, and a p300 expression construct or empty vector. The sample transfected with wild-type E2 was cotransfected with control siRNA for comparison to the siCBP-transfected sample. Results are presented as a percentage of wild-type E2 activity. One-way ANOVA was performed with Bonferroni's post hoc analysis comparing all means. ***, P < 0.001. ns, not significant.
The ability of wild-type E2 to activate transcription in RKO cells suggests that p300-mediated acetylation of lysine 111 is not necessary for this function. We also considered that in these cells, the p300 paralog CBP may be compensating for the absence of p300. Interestingly, RNAi depletion of CBP in RKO cells dramatically reduced the activity of wild-type E2, comparable to levels observed with the K111R and K111R/K112R E2 mutants (Fig. 6).
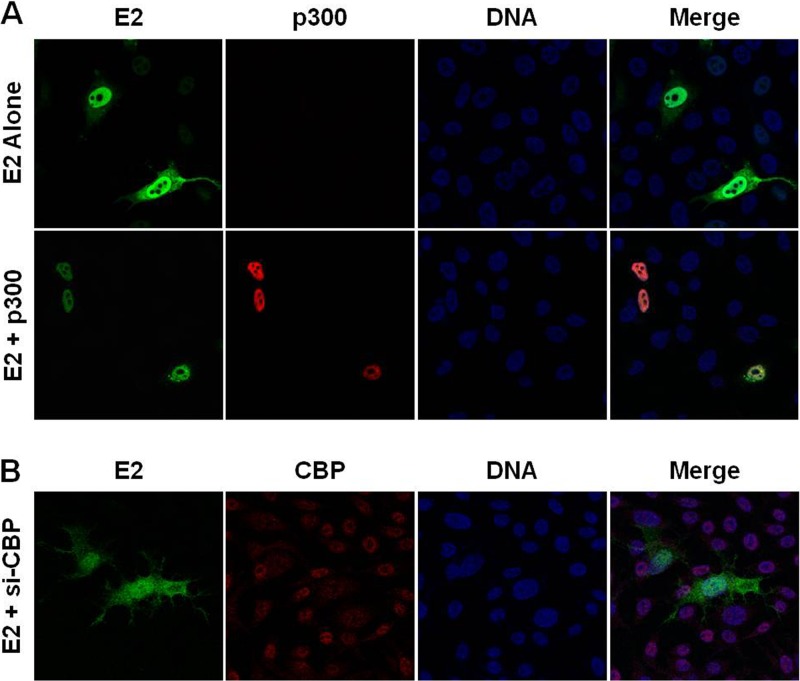
p300 and CBP promote E2 nuclear localization.
While we demonstrate that E2 is acetylated by p300 on K111 and K112, the role of acetylation in the mislocalization of the E2 mutants is unclear. The contribution of p300 to E2 localization was investigated in RKO cells. Expression of wild-type E2 in these p300-deficient cells resulted in accumulation of E2 protein in the cytosol (Fig. 7A), resembling the pattern observed for the K112R mutant in RPE-1 cells (Fig. 5A). Transfection of p300 restored nuclear localization of E2 (Fig. 7A). Introduction of p300 had no effect on the cytosolic mislocalization of the K111R, K112R, and K111R/K112R E2 mutants in RKO cells (data not shown). The reduction of E2 transcriptional activity in RKO cells following depletion of CBP prompted investigation into a compensatory role for this protein in E2 nuclear localization. Following depletion of CBP, wild-type E2 was present diffusely throughout the cytosol and nucleus (Fig. 7B). This phenotype was similar to that of the K111R mutant in RPE-1 cells (Fig. 5A).
Fig 7.
p300 and CBP facilitate E2 nuclear retention. (A) RKO cells plated on coverslips were transfected with wild-type E2 and a p300 expression construct or empty vector. Twenty-four hours posttransfection, the cells were fixed and stained using antibodies to E2 (green) and p300 (red), and DNA was visualized using DAPI (blue). (B) RKO cells were plated on coverslips and transfected with E2 and either control siRNA or siRNA to CBP. Forty-eight hours posttransfection, cells were fixed and stained using antibodies to E2 (green) and CBP (red), and DNA was visualized using DAPI (blue).
DISCUSSION
The lysine acetyltransferases p300, CBP, and pCAF have been reported to interact with the E2 proteins from several papillomaviruses (22, 42, 43). In each instance, overexpression of the KAT protein was found to increase E2-dependent transcription. In contrast, we used RNAi to specifically deplete p300, CBP, and pCAF under conditions in which the other KAT proteins remained present at endogenous levels. Interestingly, knockdown of each KAT reduced E2-dependent transcriptional activation. This demonstrates that the functions of the silenced KATs were not being replaced by the other nontargeted KAT proteins and implies that each KAT mediates nonoverlapping activities. The requisite KAT activities were then investigated. We demonstrated that p300 and CBP are present at the BPV-1 LCR by chromatin immunoprecipitation assay and that CBP was enriched at the LCR in the presence of E2 about 2-fold. We suggest that E2 recruitment of CBP to the viral promoter may serve to acetylate histones and lead to increased chromatin access for the RNA polymerase complex.
Lysine acetylation of a transcription factor leading to its activation is well documented (41); however, this has not been reported for any papillomavirus E2 protein. Because we were unable to isolate a sufficient amount of BPV E2 protein for microsequencing from mammalian cells with replicating BPV-1 genomes, we performed in vitro reactions using purified E2 and KAT proteins. Several E2 lysine residues were found to be acetylated. To test their biological relevance, each lysine was replaced with arginine to maintain charge at each position. Mutations of K111 alone and in combination with K112 resulted in substantial decreases in transcriptional activation.
Several reports have described mutation of E2 at lysines 111 and 112, which are highly conserved (59–62). Replacement of these lysines with alanines in BPV-1 E2 results in proteins that are transcriptionally defective in several cell types (59, 61). Additionally, the same mutations resulted in E2 mislocalization to the perinuclear region (59), indicating a possible deficiency in nuclear import. The conservative substitution of BPV-1 E2 lysine 111 to arginine reportedly results in severely impaired transcriptional activity, and K112R mutation was shown to activate transcription to near wild-type levels (60). These results are consistent with our data for the K111R mutant, and while we demonstrate that K112R mutant activity is lower than was previously shown, it is not significantly different from that of wild-type E2 in RPE-1 cells. The activity for the K112R mutant in C33a cells, however, is slightly lower, resulting in a significant difference that may also be attributable to variations in activity between cell types, which we suspect may relate to different KAT protein availability.
K111R and K111R/K112R mutant proteins were expressed at reduced levels. This likely reflects their lack of transcriptional activity; we routinely observe that E2 stimulates its own transcription in transient assays (63, 64). However, titration of increasing amounts of the K111R mutant did not result in increased activation, indicating that the deficiency is not due to a dosage effect (Fig. 4E). The reduction of transcriptional activity with K111 mutation to arginine suggests that acetylation of this residue is necessary for transcriptional activation.
The subcellular distribution of E2 may also be modulated through acetylation of lysines 111 and 112. This is supported by our finding that conservative mutation of these two residues to arginine resulted in E2 accumulation in the cytosol. Similar cytosolic accumulation was observed with wild-type E2 in cells in which p300 and CBP were absent. Our results for the K111R and K112R mutants are consistent with previous studies examining the localization of E2 proteins with deletions that disrupted the N-terminal NLS including lysines 111 and 112 (56) and for mutants with changes of lysine to alanine at both of these positions (59). Incomplete inclusion or exclusion of the mutants suggests that either inefficient nuclear import due to a weakened NLS or passive diffusion may be responsible. A defect in Crm-1-dependent nuclear export may be excluded given that LMB had no effect on E2 mutant localization. Another possibility is that K111 acetylation favors E2 retention within the nucleus. Notably, it was recently proposed that pCAF-mediated acetylation of lysines within the NLS of the retinoblastoma proteins promotes its nuclear retention (65). This scenario can be envisioned by the entry of E2 into a macromolecular KAT complex that prevents its loss to the cytosol.
How E2 acetylation on K111 stimulates its transcriptional activity requires further study. One possibility is that this creates an additional site for docking to the bromodomain of Brd4, which facilitates E2-dependent transcriptional activation (20, 23, 66, 67). While bromodomains have affinity for most acetylated lysines, they do exhibit binding preference (68, 69). The bromodomain interaction preference on histone H3 and H4 peptides is for diacetylated lysines (70, 71). This would indicate that acetylation of a combination of K111, K112, and possibly K107 would result in a higher-affinity interaction. A cocrystal of the N-terminal region of E2 with a short C-terminal peptide of Brd4 revealed the site of their interaction to span the three alpha helices and not the K111-containing peptide, which is at the N terminus of the beta sheet region (66). However, HPV-11 E2 has been demonstrated to interact with a region containing the second bromodomain of Brd4 in addition to the CTD (12). This supports the possibility that a second region of E2 may also participate in this interaction. Although the structures of several functional domains on Brd4, including both bromodomains and the C-terminal domain that interacts with E2, have been solved, the full-length protein structure has not been reported.
We previously reported that the transcriptional coactivator Gps2 is necessary for E2 transcription and directly binds to both p300 and E2 (22). E2 association with p300 using bacterially expressed proteins was weak and barely above background (22). Gps2 binding was mapped to residues 134 to 216 on the BPV-1 E2 TAD (47). We hypothesize that the interaction between E2 and Gps2 facilitates complex formation with p300, and this leads to acetylation of lysine 111. This, in turn, may stabilize the association of Brd4 in the transcription complex.
Here, we have outlined potential roles for the cellular acetyltransferases p300 and CBP in E2-dependent transcription. A specific role for pCAF in E2-dependent transcriptional control was not identified in this study. Acetylation of E2 by pCAF or Gcn5 may occur in vivo despite our inability to detect it using the acetyl lysine antisera in in vitro reactions. It is possible that modification of E2 by these KATs occurs only under specific conditions. While data indicate that p300, CBP, and pCAF may function independently, there does appear to be some biochemical redundancy. The reduction in transcriptional activation in RKO cells following CBP depletion suggests that CBP may be actively compensating for the loss of p300 in these cells. This is also consistent with a small but significant increase in activity following overexpression of CBP in RPE-1 cells that are depleted of p300.
It has been reported that p300 levels are low in basal keratinocytes and may increase with differentiation (44). As these epithelial cells transit into the suprabasal layers, increased expression of p300 favors acetylation of p53 and induction of p21Waf1/CIP1-mediated cell cycle arrest, further promoting commitment to keratinocyte differentiation (72). Our results suggest that elevated p300 expression in differentiating cells could also induce K111 acetylation, thereby stimulating E2-dependent transcription. The resulting increase in early gene protein levels, specifically E1, could induce viral DNA amplification, linking it to the differentiated status of the infected cell. The role of K111 acetylation by p300 in regulation of the viral replicative program is currently under active investigation.
ACKNOWLEDGMENTS
This work was supported by NIH grants R01CA58376 to Elliot J. Androphy and R01CA103867 and R01CA124760 to Cheng-Ming Chiang. Sara P. Culleton was supported by the NIH (T32 AI 060519).
Footnotes
Published ahead of print 14 November 2012
REFERENCES
- 1. Androphy EJ, Lowy DR, Schiller JT. 1987. Bovine papillomavirus E2 trans-activating gene product binds to specific sites in papillomavirus DNA. Nature 325:70–73 [DOI] [PubMed] [Google Scholar]
- 2. Chong T, Apt D, Gloss B, Isa M, Bernard HU. 1991. The enhancer of human papillomavirus type 16: binding sites for the ubiquitous transcription factors oct-1, NFA, TEF-2, NF1, and AP-1 participate in epithelial cell-specific transcription. J. Virol. 65:5933–5943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ham J, Steger G, Yaniv M. 1994. Cooperativity in vivo between the E2 transactivator and the TATA box binding protein depends on core promoter structure. EMBO J. 13:147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sandler AB, Baker CC, Spalholz BA. 1996. Sp1 is critical for basal and E2-transactivated transcription from the bovine papillomavirus type 1 P89 promoter. J. Gen. Virol. 77(Part 2):189–198 [DOI] [PubMed] [Google Scholar]
- 5. Thierry F, Spyrou G, Yaniv M, Howley P. 1992. Two AP1 sites binding JunB are essential for human papillomavirus type 18 transcription in keratinocytes. J. Virol. 66:3740–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang WM, Wu SY, Lee AY, Chiang CM. 2011. Binding site specificity and factor redundancy in activator protein-1-driven human papillomavirus chromatin-dependent transcription. J. Biol. Chem. 286:40974–40986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Spalholz BA, Lambert PF, Yee CL, Howley PM. 1987. Bovine papillomavirus transcriptional regulation: localization of the E2-responsive elements of the long control region. J. Virol. 61:2128–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Spalholz BA, Yang YC, Howley PM. 1985. Transactivation of a bovine papilloma virus transcriptional regulatory element by the E2 gene product. Cell 42:183–191 [DOI] [PubMed] [Google Scholar]
- 9. Giri I, Yaniv M. 1988. Structural and mutational analysis of E2 trans-activating proteins of papillomaviruses reveals three distinct functional domains. EMBO J. 7:2823–2829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haugen TH, Turek LP, Mercurio FM, Cripe TP, Olson BJ, Anderson RD, Seidl D, Karin M, Schiller J. 1988. Sequence-specific and general transcriptional activation by the bovine papillomavirus-1 E2 trans-activator require an N-terminal amphipathic helix-containing E2 domain. EMBO J. 7:4245–4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lambert PF, Dostatni N, McBride AA, Yaniv M, Howley PM, Arcangioli B. 1989. Functional analysis of the papilloma virus E2 trans-activator in Saccharomyces cerevisiae. Genes Dev. 3:38–48 [DOI] [PubMed] [Google Scholar]
- 12. Wu SY, Lee AY, Hou SY, Kemper JK, Erdjument-Bromage H, Tempst P, Chiang CM. 2006. Brd4 links chromatin targeting to HPV transcriptional silencing. Genes Dev. 20:2383–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haugen TH, Cripe TP, Ginder GD, Karin M, Turek LP. 1987. Trans-activation of an upstream early gene promoter of bovine papilloma virus-1 by a product of the viral E2 gene. EMBO J. 6:145–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Benson JD, Lawande R, Howley PM. 1997. Conserved interaction of the papillomavirus E2 transcriptional activator proteins with human and yeast TFIIB proteins. J. Virol. 71:8041–8047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carrillo E, Garrido E, Gariglio P. 2004. Specific in vitro interaction between papillomavirus E2 proteins and TBP-associated factors. Intervirology 47:342–349 [DOI] [PubMed] [Google Scholar]
- 16. Rank NM, Lambert PF. 1995. Bovine papillomavirus type 1 E2 transcriptional regulators directly bind two cellular transcription factors, TFIID and TFIIB. J. Virol. 69:6323–6334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu SY, Chiang CM. 2001. TATA-binding protein-associated factors enhance the recruitment of RNA polymerase II by transcriptional activators. J. Biol. Chem. 276:34235–34243 [DOI] [PubMed] [Google Scholar]
- 18. Yao JM, Breiding DE, Androphy EJ. 1998. Functional interaction of the bovine papillomavirus E2 transactivation domain with TFIIB. J. Virol. 72:1013–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kumar RA, Naidu SR, Wang X, Imbalzano AN, Androphy EJ. 2007. Interaction of papillomavirus E2 protein with the Brm chromatin remodeling complex leads to enhanced transcriptional activation. J. Virol. 81:2213–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee AY, Chiang CM. 2009. Chromatin adaptor Brd4 modulates E2 transcription activity and protein stability. J. Biol. Chem. 284:2778–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McPhillips MG, Oliveira JG, Spindler JE, Mitra R, McBride AA. 2006. Brd4 is required for E2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J. Virol. 80:9530–9543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peng YC, Breiding DE, Sverdrup F, Richard J, Androphy EJ. 2000. AMF-1/Gps2 binds p300 and enhances its interaction with papillomavirus E2 proteins. J. Virol. 74:5872–5879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schweiger MR, You J, Howley PM. 2006. Bromodomain protein 4 mediates the papillomavirus E2 transcriptional activation function. J. Virol. 80:4276–4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang X, Naidu SR, Sverdrup F, Androphy EJ. 2009. Tax1BP1 interacts with papillomavirus E2 and regulates E2-dependent transcription and stability. J. Virol. 83:2274–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gagnon D, Joubert S, Senechal H, Fradet-Turcotte A, Torre S, Archambault J. 2009. Proteasomal degradation of the papillomavirus E2 protein is inhibited by overexpression of bromodomain-containing protein 4. J. Virol. 83:4127–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zheng G, Schweiger MR, Martinez-Noel G, Zheng L, Smith JA, Harper JW, Howley PM. 2009. Brd4 regulation of papillomavirus protein E2 stability. J. Virol. 83:8683–8692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kimura A, Horikoshi M. 1998. How do histone acetyltransferases select lysine residues in core histones? FEBS Lett. 431:131–133 [DOI] [PubMed] [Google Scholar]
- 28. Kimura A, Matsubara K, Horikoshi M. 2005. A decade of histone acetylation: marking eukaryotic chromosomes with specific codes. J. Biochem. 138:647–662 [DOI] [PubMed] [Google Scholar]
- 29. Lee KK, Workman JL. 2007. Histone acetyltransferase complexes: one size doesn't fit all. Nat. Rev. Mol. Cell Biol. 8:284–295 [DOI] [PubMed] [Google Scholar]
- 30. Utley RT, Cote J. 2003. The MYST family of histone acetyltransferases. Curr. Top. Microbiol. Immunol. 274:203–236 [DOI] [PubMed] [Google Scholar]
- 31. Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491–496 [DOI] [PubMed] [Google Scholar]
- 32. Jacobson RH, Ladurner AG, King DS, Tjian R. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288:1422–1425 [DOI] [PubMed] [Google Scholar]
- 33. Kasten M, Szerlong H, Erdjument-Bromage H, Tempst P, Werner M, Cairns BR. 2004. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 23:1348–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120–124 [DOI] [PubMed] [Google Scholar]
- 35. Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116–120 [DOI] [PubMed] [Google Scholar]
- 36. Lee JS, Smith E, Shilatifard A. 2010. The language of histone crosstalk. Cell 142:682–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Martin DG, Baetz K, Shi X, Walter KL, MacDonald VE, Wlodarski MJ, Gozani O, Hieter P, Howe L. 2006. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol. Cell. Biol. 26:7871–7879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Taverna SD, Ilin S, Rogers RS, Tanny JC, Lavender H, Li H, Baker L, Boyle J, Blair LP, Chait BT, Patel DJ, Aitchison JD, Tackett AJ, Allis CD. 2006. Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell 24:785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. 2009. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 138:1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zippo A, Serafini R, Rocchigiani M, Pennacchini S, Krepelova A, Oliviero S. 2009. Histone crosstalk between H3S10ph and H4K16ac generates a histone code that mediates transcription elongation. Cell 138:1122–1136 [DOI] [PubMed] [Google Scholar]
- 41. Glozak MA, Sengupta N, Zhang X, Seto E. 2005. Acetylation and deacetylation of non-histone proteins. Gene 363:15–23 [DOI] [PubMed] [Google Scholar]
- 42. Lee D, Hwang SG, Kim J, Choe J. 2002. Functional interaction between p/CAF and human papillomavirus E2 protein. J. Biol. Chem. 277:6483–6489 [DOI] [PubMed] [Google Scholar]
- 43. Lee D, Lee B, Kim J, Kim DW, Choe J. 2000. cAMP response element-binding protein-binding protein binds to human papillomavirus E2 protein and activates E2-dependent transcription. J. Biol. Chem. 275:7045–7051 [DOI] [PubMed] [Google Scholar]
- 44. Müller A, Ritzkowsky A, Steger G. 2002. Cooperative activation of human papillomavirus type 8 gene expression by the E2 protein and the cellular coactivator p300. J. Virol. 76:11042–11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Krüppel U, Muller-Schiffmann A, Baldus SE, Smola-Hess S, Steger G. 2008. E2 and the co-activator p300 can cooperate in activation of the human papillomavirus type 16 early promoter. Virology 377:151–159 [DOI] [PubMed] [Google Scholar]
- 46. Lehman CW, King DS, Botchan MR. 1997. A papillomavirus E2 phosphorylation mutant exhibits normal transient replication and transcription but is defective in transformation and plasmid retention. J. Virol. 71:3652–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Breiding DE, Sverdrup F, Grossel MJ, Moscufo N, Boonchai W, Androphy EJ. 1997. Functional interaction of a novel cellular protein with the papillomavirus E2 transactivation domain. Mol. Cell. Biol. 17:7208–7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Parish JL, Bean AM, Park RB, Androphy EJ. 2006. ChlR1 is required for loading papillomavirus E2 onto mitotic chromosomes and viral genome maintenance. Mol. Cell 24:867–876 [DOI] [PubMed] [Google Scholar]
- 49. Hou SY, Wu SY, Chiang CM. 2002. Transcriptional activity among high and low risk human papillomavirus E2 proteins correlates with E2 DNA binding. J. Biol. Chem. 277:45619–45629 [DOI] [PubMed] [Google Scholar]
- 50. Thomas MC, Chiang CM. 2005. E6 oncoprotein represses p53-dependent gene activation via inhibition of protein acetylation independently of inducing p53 degradation. Mol. Cell 17:251–264 [DOI] [PubMed] [Google Scholar]
- 51. Apweiler R, Bairoch A, Wu CH. 2004. Protein sequence databases. Curr. Opin. Chem. Biol. 8:76–80 [DOI] [PubMed] [Google Scholar]
- 52. McManus KJ, Hendzel MJ. 2003. Quantitative analysis of CBP- and P300-induced histone acetylations in vivo using native chromatin. Mol. Cell. Biol. 23:7611–7627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schiltz RL, Mizzen CA, Vassilev A, Cook RG, Allis CD, Nakatani Y. 1999. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 274:1189–1192 [DOI] [PubMed] [Google Scholar]
- 54. Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8:1243–1254 [DOI] [PubMed] [Google Scholar]
- 55. Prives C, Manley JL. 2001. Why is p53 acetylated? Cell 107:815–818 [DOI] [PubMed] [Google Scholar]
- 56. Skiadopoulos MH, McBride AA. 1996. The bovine papillomavirus type 1 E2 transactivator and repressor proteins use different nuclear localization signals. J. Virol. 70:1117–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Prakash SS, Grossman SR, Pepinsky RB, Laimins LA, Androphy EJ. 1992. Amino acids necessary for DNA contact and dimerization imply novel motifs in the papillomavirus E2 trans-activator. Genes Dev. 6:105–116 [DOI] [PubMed] [Google Scholar]
- 58. Ionov Y, Nowak N, Perucho M, Markowitz S, Cowell JK. 2004. Manipulation of nonsense mediated decay identifies gene mutations in colon cancer cells with microsatellite instability. Oncogene 23:639–645 [DOI] [PubMed] [Google Scholar]
- 59. Abroi A, Kurg R, Ustav M. 1996. Transcriptional and replicational activation functions in the bovine papillomavirus type 1 E2 protein are encoded by different structural determinants. J. Virol. 70:6169–6179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Brokaw JL, Blanco M, McBride AA. 1996. Amino acids critical for the functions of the bovine papillomavirus type 1 E2 transactivator. J. Virol. 70:23–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ferguson MK, Botchan MR. 1996. Genetic analysis of the activation domain of bovine papillomavirus protein E2: its role in transcription and replication. J. Virol. 70:4193–4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sakai H, Yasugi T, Benson JD, Dowhanick JJ, Howley PM. 1996. Targeted mutagenesis of the human papillomavirus type 16 E2 transactivation domain reveals separable transcriptional activation and DNA replication functions. J. Virol. 70:1602–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Breiding DE, Grossel MJ, Androphy EJ. 1996. Genetic analysis of the bovine papillomavirus E2 transcriptional activation domain. Virology 221:34–43 [DOI] [PubMed] [Google Scholar]
- 64. Grossel MJ, Sverdrup F, Breiding DE, Androphy EJ. 1996. Transcriptional activation function is not required for stimulation of DNA replication by bovine papillomavirus type 1 E2. J. Virol. 70:7264–7269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pickard A, Wong PP, McCance DJ. 2010. Acetylation of Rb by PCAF is required for nuclear localization and keratinocyte differentiation. J. Cell Sci. 123:3718–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Abbate EA, Voitenleitner C, Botchan MR. 2006. Structure of the papillomavirus DNA-tethering complex E2:Brd4 and a peptide that ablates HPV chromosomal association. Mol. Cell 24:877–889 [DOI] [PubMed] [Google Scholar]
- 67. Sénéchal H, Poirier GG, Coulombe B, Laimins LA, Archambault J. 2007. Amino acid substitutions that specifically impair the transcriptional activity of papillomavirus E2 affect binding to the long isoform of Brd4. Virology 358:10–17 [DOI] [PubMed] [Google Scholar]
- 68. Li T, Du Y, Wang L, Huang L, Li W, Lu M, Zhang X, Zhu WG. 2012. Characterization and prediction of lysine (K)-acetyl-transferase (KAT) specific acetylation sites. Mol. Cell. Proteomics 11(1):M111.011080. doi:10.1074/mcp.M111.011080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mujtaba S, Zeng L, Zhou MM. 2007. Structure and acetyl-lysine recognition of the bromodomain. Oncogene 26:5521–5527 [DOI] [PubMed] [Google Scholar]
- 70. Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc. Natl. Acad. Sci. U. S. A. 100:8758–8763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Morinière J, Rousseaux S, Steuerwald U, Soler-Lopez M, Curtet S, Vitte AL, Govin J, Gaucher J, Sadoul K, Hart DJ, Krijgsveld J, Khochbin S, Muller CW, Petosa C. 2009. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature 461:664–668 [DOI] [PubMed] [Google Scholar]
- 72. Wong PP, Pickard A, McCance DJ. 2010. p300 alters keratinocyte cell growth and differentiation through regulation of p21(Waf1/CIP1). PLoS One 5:e8369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]