Abstract
Bats carry a variety of paramyxoviruses that impact human and domestic animal health when spillover occurs. Recent studies have shown a great diversity of paramyxoviruses in an urban-roosting population of straw-colored fruit bats in Ghana. Here, we investigate this further through virus isolation and describe two novel rubulaviruses: Achimota virus 1 (AchPV1) and Achimota virus 2 (AchPV2). The viruses form a phylogenetic cluster with each other and other bat-derived rubulaviruses, such as Tuhoko viruses, Menangle virus, and Tioman virus. We developed AchPV1- and AchPV2-specific serological assays and found evidence of infection with both viruses in Eidolon helvum across sub-Saharan Africa and on islands in the Gulf of Guinea. Longitudinal sampling of E. helvum indicates virus persistence within fruit bat populations and suggests spread of AchPVs via horizontal transmission. We also detected possible serological evidence of human infection with AchPV2 in Ghana and Tanzania. It is likely that clinically significant zoonotic spillover of chiropteran paramyxoviruses could be missed throughout much of Africa where health surveillance and diagnostics are poor and comorbidities, such as infection with HIV or Plasmodium sp., are common.
INTRODUCTION
Chiroptera (bats) is an ancient and diverse order of mammals that harbors a plethora of virus families, including paramyxoviruses (1, 2). Bat paramyxoviruses have various levels of impact on human and domestic animal health if and when they spill over into these populations, ranging from incidental serological findings in healthy humans to fatal zoonotic infections and epizootics notifiable to the World Organization for Animal Health (OIE). Recently, bats have been shown to harbor a great diversity of previously unknown paramyxoviruses, some of which are related to known zoonoses (2–5). These sequence data suggest that additional viruses with zoonotic potential exist in bat populations. Further investigation of this possibility through isolation, characterization, and epidemiological studies of these novel viruses is required to determine if any zoonotic spillover occurs.
The two earliest isolates of paramyxoviruses from bats appeared to have had relatively little impact on human and domestic animal health. The first, bat parainfluenza virus, was incidentally isolated from the Old World fruit bat Rousettus leschenaulti in India in 1966 (6, 7). Classified as a paramyxovirus on the basis of physical, biochemical, and immunological studies, bat parainfluenza virus has yet to be confirmed genetically. Although there was an indication that this virus had transmitted to humans (10% of human sera tested were positive in hemagglutination inhibition tests), the possibility of cross-reactivity was not ruled out (6, 7). The next paramyxovirus isolated from bats was Mapuera virus, isolated from the microbat frugivore Sturnira lilium in Brazil in 1979 (8). Although Mapuera virus has not been specifically linked to disease outbreaks, its close phylogenetic relationship with porcine rubulavirus (PorPV), which caused epizootic disease in Mexican pigs in the early 1980s (9), led to speculation about its potential for host switching (10).
Currently, the two bat paramyxoviruses with the highest levels of impact on human and domestic animal health are, undoubtedly, Hendra and Nipah viruses (HeV and NiV, respectively; within the genus Henipavirus). Henipaviruses were first detected in the 1990s and are harbored by Old World fruit bats of the genus Pteropus across Australia and southern and south-eastern Asia (11). HeV first emerged in Australia in 1994, killing more than a dozen horses and one of two infected humans (12, 13). Further spillover events have occurred almost annually and currently total 33, with 67 horse deaths and the deaths of four of seven people infected (14 and ProMED mail archive references 20111014.3075 and 20120106.1001359). NiV first emerged in Malaysia in 1998, when it caused an epidemic of respiratory and encephalitic disease in pigs and humans resulting in the deaths of over 150 people and the culling of over 1,000,000 pigs (15, 16). A variant of NiV continues to cause almost annual outbreaks of fatal encephalitis in humans in Bangladesh with worrying epidemiologic features, such as human-to-human transmission and, apparently, direct bat-to-human transmission (17).
In addition to these extremes of little or drastic consequences of spillover for human and domestic animal health, there are other bat paramyxoviruses from the genus Rubulavirus for which the impacts are intermediate or unknown. A rubulavirus derived from Australian Pteropus sp., Menangle virus (MenPV), was isolated in 1997 from an outbreak of reproductive disease in pigs (18). Two humans involved in the outbreak were found to have neutralizing antibodies against the virus, and both had a history of a severe flu-like illness (19). In 2001, Tioman virus (TioPV) was isolated from Malaysian Pteropus sp. and found to be closely related to MenPV and, more distantly, to other rubulaviruses (20). The possibility of zoonotic spillover of TioPV was investigated on Tioman island, and 3 of 169 asymptomatic humans had low-titer TioPV-neutralizing antibodies (21).
Finally, there have been genome sequences of bat paramyxoviruses characterized for which the impacts on animal and human health have not yet been investigated. Fecal samples collected in 2008 from Rousettus leschenaulti fruit bats in China were found to contain three main lineages of rubulavirus sequence, all most closely related to MenPV and TioPV. Full-length genome sequences were obtained from three representative samples and named the Tuhoko viruses (ThkPV1, ThkPV2, and ThkPV3) (5). In addition to ThkPVs' close phylogenetic relationship with MenPV and TioPV, other genomic features were found to be unique among these viruses within the genus Rubulavirus. Also, more recently, two genomes of mumps-like and henipavirus-like paramyxoviruses contained in samples collected from African fruit bats (Epomophorus sp. and Eidolon helvum, respectively) in 2009 were characterized (2). The clinical implications of these sequenced viruses are unknown.
The Old World fruit bat, Eidolon helvum, is common throughout sub-Saharan Africa and appears to be a reservoir for multiple paramyxoviruses. It has a high seroprevalence against henipaviruses (22, 23) and also harbors a wide diversity of cocirculating paramyxoviruses, some of which are phylogenetically related to MenPV, TioPV, and henipaviruses (2–4). There are multiple opportunities for direct and indirect contact between E. helvum and humans, as this bat species is widely hunted for bush meat in west Africa (24) and frequently feeds in fruit orchards. Furthermore, extremely large (∼1 million individuals) roosts occur in urban settings, such as in central Accra, Ghana (25), and in Muheza, Tanzania, where E. helvum animals roost in the grounds of hospitals.
The close phylogenetic relationship of E. helvum paramyxoviruses with viruses that are known to be zoonotic and evidence of prior infection of E. helvum with henipaviruses suggest the potential for spillover of paramyxoviruses to humans and domestic animals from these populations. However, further work is required to investigate if spillover occurs and establish the consequences of this for human and domestic animal health. Previously, urine has been shown to be a robust sample type for investigating paramyxoviruses in bat populations. Here, we describe the isolation and characterization of two novel paramyxoviruses from the urine of E. helvum in Accra, Ghana, and we report initial efforts to identify zoonotic spillover of the viruses.
MATERIALS AND METHODS
Ethics declaration.
All animal sampling events were preapproved by the Zoological Society of London's animal ethics committee. Collection and investigation of human samples in Ghana was approved by the Institutional Review Board of Noguchi Memorial Institute for Medical Research (reference number 002/10-11). Samples collected from Tanzania were approved by the National Institute for Medical Research (reference number NIMR/HQ/R.8a/Vol.IX/392) and the London School for Tropical Medicine and Hygiene.
Urine samples.
Urine samples were collected underneath an E. helvum roost in Accra, Ghana, in September-November 2010 as previously described (3). For each urine pool collected, 500 μl was used for virus isolation. Each sample was diluted 1:1 with virus transport medium (phosphate-buffered saline [PBSA] supplemented with 10% fetal calf serum and double-strength antibiotic/antimycotic [200 U/ml penicillin, 200 μg/ml streptomycin, and 0.5 μg/ml fungizone amphotericin B; Gibco]). Virus isolation was attempted on 38 of 72 collected samples (U34 to U72).
Sera from E. helvum and humans.
Sera collected from various species and locations were used in this study (see Table 4). All sera were heat treated at 56°C for 30 min prior to use in serological assays.
Table 4.
AchPV serum neutralization testing results by species and location collected from 2006 to 2010
| Species | Country | Site | Date | Total | No. (%) positivea by SNT of: |
|
|---|---|---|---|---|---|---|
| AchPV1 | AchPV2 | |||||
| E. helvum | Ghana | Accra | Jan 2007 | 56 | 26 (46.4) | 8 (14.3) |
| Sep 2010 | 101 | 14 (13.9) | 7 (6.9) | |||
| Tanzania | Dar es Salaam | Aug 2010 | 25 | 3 (12.0) | 2 (8.0) | |
| Gulf of Guinea islandsb | Príncipe | Mar-May 2010 | 15 | 6 | 0 | |
| São Tomé | 26 | 13 | 1 | |||
| Annobón | 6 | 4 | 0 | |||
| H. sapiens | Ghana | Volta | Mar 2011 | 27 | 0 | 0 |
| Accra | Nov 2010 | 216 | 0 | 2 (0.9) | ||
| Tanzania | Muheza | Jun 2006 to May 2007 | 226 | 0 | 1 (0.4) | |
The number and percentage of neutralizing sera are shown, except for nonrandom samples (see Materials and Methods).
Serum samples were not random with respect to age and gender status.
Bat sera.
Sera collected from five different E. helvum populations were available for testing in this study. Serum samples from the Accra E. helvum population (from which the urine samples were collected) were taken at two different time points: January 2007 (n = 56; from reference 22; non-Accra E. helvum samples were removed from analyses) and September 2010 (n = 101). Sera collected in August 2010 (n = 25) from an east African roost of E. helvum in Dar es Salaam, Tanzania's principal city, and subsets of sera collected from each of three remote island populations of E. helvum in the Gulf of Guinea (sampled in March to May 2010), São Tomé (n = 26), Príncipe (n = 15), and Annobón (n = 6), were also analyzed. Samples from the Gulf of Guinea were subsets of samples collected for another study which had been previously determined to contain significant levels (a median fluorescence intensity [MFI] of >750 on Luminex serology) of anti-henipavirus antibody (described in references 26 and 23).
Bats were captured and bled as previously described (22, 23). Gender and age class were recorded. Three age classifications were used: fully grown bats with secondary sexual characteristics, i.e., nipple development or descended testes (adult), fully grown bats without secondary sexual characteristics (sexually immature), and bats not yet fully grown (juvenile).
Human sera.
Sera were collected from healthy and symptomatic humans in Ghana and Tanzania who worked at, lived adjacent to, or were patients in hospitals where large numbers of E. helvum roost. Also, in Ghana, samples were collected from bush meat hunters.
In Ghana, subsets of serum samples collected from healthy adult (>18 years) volunteers were used. In November 2010 (n = 216), samples were collected from individuals who worked or lived in the area of the Accra E. helvum roost. In March 2011, samples were collected from individuals involved in the hunting and/or butchering of bats (n = 27) within the Volta region of Ghana (Wli Falls, Biobio Island, and Kpando).
In Tanzania, serum samples (n = 226) were collected from febrile pediatric patients (aged 2 months to 13 years) who were admitted to Teule Hospital, Muheza, Tanga Region, Tanzania, between June 2006 and May 2007 as previously described (27).
Cell culture conditions.
All cell culture experiments described are either with Vero cells (ATCC CCL-81) or, where specified, Pteropus alecto primary kidney (PaKi) cells (28) grown at 37°C in 5% CO2. The growth medium used was Dulbecco's modified Eagle's medium supplemented with F12-Ham (Sigma), 10% fetal calf serum, double-strength antibiotic/antimycotic (200 U/ml penicillin, 200 μg/ml streptomycin, and 0.5 μg/ml fungizone amphotericin B; Gibco), and ciprofloxacin (10 μg/ml; MP Biomedicals).
Virus isolation.
Urine samples were thawed to room temperature and clarified by centrifugation at 10,000 × g for 5 min. Clarified sample supernatants were then diluted 1:6 with medium and clarified further by centrifugation at 3,000 × g for 5 min. The double-clarified sample (1 ml) was then added to confluent 25-cm2 Vero cell monolayers. Following incubation during gentle rocking for 1 h at 37°C, an additional 6 ml of medium was added to each flask, and monolayers were examined every 48 h for signs of cytopathic effect (CPE). If no CPE occurred, 1 ml of supernatant was passaged onto a new cell monolayer at day 7 postinfection (up to three passages). If CPE occurred, RNA was extracted from the supernatant (using an RNeasy kit; Qiagen) and was analyzed using a previously described consensus reverse transcription-PCR (RT-PCR) (29) to detect if paramyxovirus RNA was present.
Purification of viral parental stocks.
Virus stocks for use in subsequent studies were purified from the supernatant of CPE-positive flasks. Supernatant was purified by three rounds of limiting dilution. Briefly, each round consisted of supernatant undergoing serial 10-fold dilution, with 12 replicates of each dilution point being distributed across a 96-well cell culture plate to which a Vero cell suspension was added. Supernatant toward the endpoint of the dilution (i.e., from a well containing virus derived from a single infectious particle) was then used in the second and final rounds of purification. Parent stock of the purified viruses was then generated by infection of a 75-cm2 monolayer of Vero cells from which the supernatant was harvested at 72 h postinfection.
Growth analysis of novel viruses.
To analyze the growth of novel viruses in cell culture, Vero and PaKi cell monolayers were infected with virus at a multiplicity of infection (MOI) of 0.01 and incubated for 1 h at 37°C. Following three washes of the cell monolayer with PBS, growth medium (as described above) was added and samples for titration taken at 24-h intervals for 7 days postinfection. Growth interval samples were titrated on Vero cells.
Genome sequencing. (i) Preparation of viral genomic RNA for sequencing.
Confluent 75-cm2 Vero cell monolayers were infected with 50 μl of supernatant from CPE-positive isolation flasks. At 72 h postinfection, virus particles were harvested and semipurified through a sucrose cushion. Briefly, 15 ml of supernatant was ultracentrifuged at 35,000 × g for 1 h at 4°C through a 2-ml sucrose cushion (20% sucrose [wt/vol] in Tris [100 mM], 2.0 M NaCl, 10 mM EDTA, pH 7.4). Pellets were resuspended in 350 μl of buffer RLT, and nucleic acids were extracted using an RNeasy kit (Qiagen).
(ii) Random RT-PCR and pyrosequencing.
Randomly amplified DNA was prepared for pyrosequencing from viral genomic RNA (as described in reference 30). Briefly, 40 ng of genomic viral RNA was reverse transcribed (Superscript III; Invitrogen) using a random octamer-conjugated primer (5′ GTTTCCCAGTAGGTCTCNNNNNNNN 3′). Complementary-strand DNA was synthesized by a Klenow reaction. DNA polymerase buffer and 15 U of DNA polymerase I enzyme large fragment (Promega) were added, and incubation at 37°C for 30 min was followed by inactivation at 70°C for 15 min. A PCR using one or more Multiplex IDentifier (MID) tags was then performed (GoTaq Green Mastermix; Promega) using the following conditions: 95°C for 3 min; 40 cycles of 95°C for 1 min, 48°C for 1 min, and 72°C for 1 min; and 72°C for 7 min, hold 4°C. For AchPV1, MID1 (A*C*G*A*GTGCGTGTTTCCCAGTAGGTCTC) and MID5 (A*T*C*A*GACACGGTTTCCCAGTAGGTCTC) were used, and for AchPV2, MID2 (A*C*G*C*TCGACAGTTTCCCAGTAGGTCTC) was used (an asterisk denotes thiol modifications to facilitate barcoding). Products were visualized using a 1% agarose gel with SYBR safe stain (Invitrogen) and purified using a QIAquick PCR purification kit (Qiagen). Samples were prepared for 454 sequencing according to the Titanium series manual, Rapid library preparation, and the emPCR Lib-L SV manual (30).
(iii) Sequencing of genomic ends through end-to-end ligation of viral ends.
The 3′ and 5′ ends of the genomes were sequenced using an end-to-end ligation protocol similar to that described in reference 31, where genomic viral RNA was ligated end to end, and inverted nested PCR was used to amplify across the ligated ends. Briefly, 5 ng of viral genomic RNA was incubated at 37°C for 1 h in RNA ligase buffer with 20 U of T4 RNA ligase (Promega). Primers for nested PCR amplification were designed ∼200 nucleotides (nt) (FWD1/REV1) and ∼150 nt (FWD2/REV2) from the 3′ (reverse complement) and 5′ (complement) ends, respectively (sequences are available on request). Ligated RNA was reverse transcribed using the REV1 primer (Superscript III; Invitrogen), and nested PCR was performed (Expand High Fidelity PCR system; Roche) under the following conditions: 94°C for 2 min; 40 cycles of 94°C for 15 s, 49°C for 30 s, and 72°C for 1 min; and then 72°C for 5 min, hold 4°C. PCR products were visualized using 1% agarose gel electrophoresis, extracted using a gel extraction kit (Qiagen), cloned (pGEM-T Easy; Promega), and sequenced.
(iv) Gap-filling RT-PCRs.
Specific RT-PCRs were conducted to fill in gaps (<300 bp) between contiguous sequences of the genomes. Primers were designed using the CLC genomics workbench (v 4.8; CLC Bio), and RT-PCR was performed on genomic viral RNA using the Superscript III Platinum Taq One-Step RT-PCR system (Invitrogen) according to the manufacturer's instructions.
Bioinformatic analysis. (i) Genome assembly.
Pyrosequencing reads were processed using Newbler software (v 2.5; 454 Life Sciences, Roche). Reads were binned according to their MID tags and then de novo assembled using default settings. Assembled contiguous sequences were then mapped with low stringency to Tuhoko virus 2 (GenBank accession number GU12808.1) using the CLC genomics workbench (v 4.8). Contiguous sequences were then aligned manually with PCR products from gap-filling and genome-end PCRs into a final consensus genome sequence using Se-Al (http://tree.bio.ed.ac.uk/software/seal/).
(ii) Gene annotations.
Genome features were annotated using the CLC genomics workbench and a combination of automated open reading frame (ORF) searches, comparison of nucleotide and encoded protein sequences to those of other Paramyxovirinae, and the conditions determined to be favorable for the initiation of translation (32).
(iii) Phylogenetic analysis.
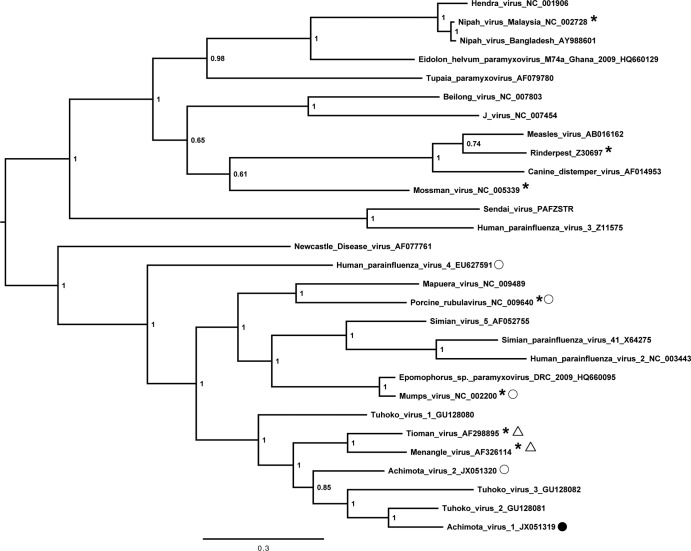
Reference genomes of previously sequenced paramyxoviruses were downloaded from GenBank (accession numbers are provided in figures), and protein sequences were extracted. Protein sequences were aligned using MUSCLE (33), and alignments were gap stripped prior to inference of Bayesian phylogenetic trees using Mr Bayes (34). Standard GTR+I+G nucleotide substitution was used for phylogenies based on nucleotide sequences, and default settings were used for the protein sequence-based phylogeny.
(iv) Protein sequence pairwise identities.
Pairwise amino acid identities were determined within the CLC genomics workbench following the global alignment of the protein sequences using Clustal W (35).
Serological methods. (i) Determination of serological cross-reactivity of Achimota viruses.
Control sera containing antibodies against other known paramyxoviruses were screened at dilutions of 1:10 to 1:1,000 against Achimota viruses to look for evidence of serum neutralization. These sera were horse serum raised against mumps virus, pig sera raised against Nipah virus and against Tioman virus, and rabbit sera raised against Mossman virus, Menangle virus, and Rinderpest virus and against a recombinant nucleoprotein from porcine rubulavirus (see Fig. 3). Eidolon helvum sera with various levels of neutralization activity against Achimota viruses (including those that neutralized both AchPVs and those with titers of 1:80 to at least one AchPV) were also tested at a dilution of 1:20 (in duplicate) for their ability to neutralize Menangle and Tioman viruses. In addition, polyclonal rabbit serum was raised against AchPV1, as described in reference 36, and was tested for neutralization against AchPV1, AchPV2, mumps virus, human parainfluenza virus 4 (hPIV4), and porcine rubulavirus (PorPV). In all cases, sera were challenged to neutralize 200 50% tissue culture infectious doses (TCID50) of virus.
Fig 3.
Paramyxovirinae nucleoprotein phylogenetic tree. The tree is Bayesian inferred from a gap-stripped 532-amino-acid alignment of nucleoproteins from viruses representative of the Paramyxovirinae. Node labels are posterior probability values, and the bar represents 0.3 amino acid substitutions per site. Accession numbers for reference viruses used in the alignment are as shown. The viruses whose antisera failed to neutralize AchPVs in serum neutralization testing (SNT) are marked with an asterisk. The viruses which AchPV1- and AchPV2-neutralizing E. helvum sera failed to neutralize are marked with a triangle. The viruses against which AchPV1-specific rabbit sera was tested for neutralization are marked with a circle. This sera was found to neutralize only AchPV1 (filled circle).
(ii) Serum neutralization testing (SNT) of sera from E. helvum and humans.
Sera were initially screened in duplicate at a dilution factor of 1:20. Positive sera were then retested and titrated in a 2-fold dilution series from 1:20 to 1:160 (except in the case of the positive E. helvum samples from the Gulf of Guinea, where limited serum volume precluded titration, and positive human sera, which were retested at 1:10 to 1:160). In addition to this, three AchPV2-neutralizing human sera (and three nonneutralizing human controls) were tested for neutralization against mumps virus and hPIV4. Serum dilutions were incubated with 200 TCID50 of virus (confirmed by parallel virus titration) for 30 min at 37°C prior to the addition of Vero cell suspension at an MOI equivalent to 0.01. Cell monolayers were assessed for evidence of virus neutralization 7 days postinfection.
Nucleotide sequence accession numbers.
The annotated genome sequences of Achimota viruses 1 and 2 have been deposited in GenBank under accession numbers JX051319 (AchPV1) and JX051320 (AchPV2).
RESULTS
Achimota virus isolation and behavior in cell culture.
Novel paramyxoviruses were isolated from Eidolon helvum and propagated in cell culture. Two urine samples produced CPE on Vero cell monolayers (syncytium formation and cell death): one on day 5 postinfection of the second pass (sample no. U46) and one on day 5 of the third pass (U69). The viruses isolated from samples U46 and U69 were called Achimota virus 1 (AchPV1) and Achimota virus 2 (AchPV2), respectively. Achimota is the name of a local area in the capital city of Ghana, Accra, where the samples were collected.
RNA extracted from each of the supernatants was found to contain paramyxovirus RNA using an RT-PCR. Phylogenetic analysis of supernatant-derived PCR products showed that AchPV1 had previously been detected in consensus PCR analysis of the sample from which it was derived (U46), whereas AchPV2 (from sample U69) had not been detected during the equivalent analysis (3) (see Fig. S1 in the supplemental material).
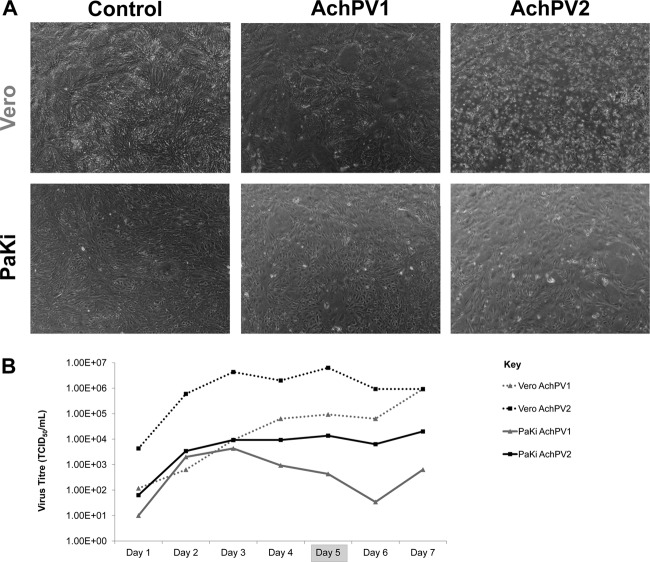
In addition to being genetically distinct, these two viruses behaved differently in cell culture. Although purified stocks of each virus were able to infect both Vero and PaKi cells, the CPE of each virus differed (Fig. 1A), and AchPV2 consistently grew to a higher titer than AchPV1 (Fig. 1B).
Fig 1.
Growth of AchPVs in cell culture. (A) Cytopathic effect of AchPVs on Vero and PaKi cell monolayers (4× magnification) at day 5 postinfection and (B) daily viral titers of AchPV1 (gray markers) and AchPV 2 (black markers) recovered from both Vero (dashed lines) and PaKi (solid lines) cell monolayers over a seven-day infection period.
Genomic characterization of AchPV1 and AchPV2.
The full-length genomes of both AchPV1 and AchPV2 were sequenced. AchPV1 was 15,624 and AchPV2 was 15,504 nt in length, with both viruses appearing to follow the structural constraint that restricts the genomes of paramyxoviruses to lengths divisible by six (37). Both viruses had 3′ leader sequences of 55 nt in length. The first 14 nt of the leader sequences followed the motif conserved among known rubulaviruses, with the exception of a GA couplet (instead of AG) at positions 5 and 6. These 14 nt were also the reverse complement of the final 14 nt of the trailer sequence, excepting the AG/GA couplet in positions 5 and 6 (Fig. 2).
Fig 2.
Fifty-five-nucleotide 3′ leaders and 5′ trailers (in reverse complement) of Achimota viruses (AchPV), Tuhoko viruses (ThkPV), Menangle virus (MenPV), Tioman virus (TioPV), porcine rubulavirus (PorPV), and mumps virus (MuV). Residues identical to the 3′ leader of AchPV have been marked as dots. The gray boxes highlight those viruses in which the reverse complementary nature of the 5′ trailer is compromised by an AG couplet at positions 5 and 6.
The AchPV genomes contained 6 genes, N/P/M/F/HN/L, with enhanced coding capacity through mRNA editing of the P gene (Table 1). As for other rubulaviruses, the AchPV P genes encoded three proteins (V/P/W) via mRNA editing at a conserved editing site (TTTAAGAGGGG in both viruses) (Table 1). Unedited mRNA transcripts resulted in ORFs encoding V proteins, and insertion of a single and two guanine residues at the editing site resulted in ORFs encoding putative W and P proteins, respectively.
Table 1.
Nucleotide positions and length of gene features of AchPV1 and AchPV2
| Gene and feature | AchPV1 |
AchPV2 |
||||
|---|---|---|---|---|---|---|
| Start | End | Length (aa) | Start | End | Length (aa) | |
| N | ||||||
| 3′ leader | 1 | 55 | 1 | 55 | ||
| mRNA | 56 | 1846 | 56 | 1818 | ||
| ORF | 165 | 1736 | 524 | 165 | 1706 | 514 |
| P | ||||||
| mRNA | 1856 | 3310 | 1826 | 3288 | ||
| V ORF | 1962 | 2687 | 242 | 1932 | 2639 | 236 |
| W ORF | 1962 | 2461 | 167 | 1932 | 2431 | 167 |
| P ORF | 1962 | 3165 | 402 | 1932 | 3123 | 398 |
| mRNA editing site | 2439 | 2449 | 2403 | 2413 | ||
| M | ||||||
| mRNA | 3319 | 4756 | 3313 | 4747 | ||
| ORF | 3354 | 4484 | 377 | 3347 | 4477 | 377 |
| F | ||||||
| mRNA | 4826 | 6649 | 4772 | 6647 | ||
| ORF | 4907 | 6508 | 534 | 4869 | 6464 | 532 |
| HN | ||||||
| mRNA | 6692 | 8645 | 6655 | 8586 | ||
| ORF | 6734 | 8521 | 596 | 6746 | 8497 | 584 |
| L | ||||||
| mRNA | 8736 | 15600 | 8610 | 15481 | ||
| ORF | 8744 | 15559 | 2,272 | 8618 | 15439 | 2,274 |
| 5′ Trailer | 15560 | 15624 | 15440 | 15504 | ||
Genes were bound by conserved transcriptional start and stop signals and separated by intergenic regions (IGR) that varied in length and sequence composition (Table 2). Similar to MenPV, TioPV, and ThkPV2, AchPV predicted transcriptional start signals most commonly contained a guanine residue in the +1 position (instead of the typical adenine in other rubulaviruses and other PMVs), although this was not confirmed experimentally. Notably, two start codons were identified in the +1 translation frame of the AchPV1 HN mRNA at positions 34 and 43. The former translation start codon had T, A, and T residues in the −3, −1, and +4 positions, respectively, compared to A, C, and G in the latter. The latter site thus was annotated as the translational start site based on its enhanced suitability according to the Kozak consensus sequence (32). Although it was not experimentally confirmed whether the AchPV attachment proteins cause hemagglutination or possess neuraminidase activity, they were annotated as HN by convention with closely related viruses.
Table 2.
Gene boundary information for AchPV1 and AchPV2, including sequence boundaries for the start and finish of transcription for each gene and length and sequence boundaries of IGRsa
| Virus sequence | Start | Gene | Stop | IGR sequence boundaries | Size (nt) |
|---|---|---|---|---|---|
| AchPV1 | gGGCCcGaac | Consensus | ttttTTTAAGAAAAAa | ||
| AGGCCCGAAAGT | N | TTTTAAGAAAAAA | TTGAAATTT | 9 | |
| GGGCCCGAAG | P | TTTAAGAAAAAA | CCAAAAGT | 8 | |
| GGGCCCGGAC | M | TTTTAAGAAAAAA | CTTGAGGATATATA.........GAAGAAAAGAAGAAT | 69 | |
| GGGCCCGGAC | F | TTTAAGAAAAAA | CTGATAAGTTGAGG.........AAGATAATCAAACAT | 42 | |
| GGGCCCGACC | HN | TTTAAGAAAAAA | GTTGAGTAGAAGTG.........CAGGACAATAATAAT | 90 | |
| GGGCCAGAAT | L | TTTTTTTAAGAAAAA | CGACTTATTGATTTT ... 5′ trailer | ||
| AchPV2 | gGGCCcGAa | Consensus | tTTTAAGAAAAAa | ||
| AGGCCCGAATGT | N | TTTTAAAGAAAAA | CAAAGAT | 7 | |
| GGGCCCGAAT | P | TTTAAGAAAAAA | CTTATAAACTGATACACTAAAAGT | 24 | |
| GGGCCAGAAC | M | TTTAAGAAAAAA | TGTAACCATCTTTAAGTGCAAGTT | 24 | |
| GGGCCCGACC | F | TTTAAGAAAAAA | CTATGCT | 7 | |
| GGGCGCGAAC | HN | TTTAAGAAAAAA | TCTAATAATGATCATATAACCCT | 23 | |
| GGGCCAGAAT | L | ATTAAGAAAAAA | CTTATTCATTTTCCC ... 5′ trailer |
Uppercase letters denote the base where conserved across all genes; lowercase letters show the majority base where site is variable across the genome.
Relationships of AchPVs within the Paramyxovirinae.
Phylogenetic analysis of the full-length nucleoprotein sequence showed that the AchPVs were rubulaviruses and formed a phylogenetic cluster with each other and with Menangle virus (MenPV), Tioman virus (TioPV), and the recently described Tuhoko viruses (ThkPVs) (Fig. 3). This relationship was also confirmed using alignments of the P, V, M, F, HN, and L proteins, as well as a concatenation of these sequences (data not shown). The viruses were also related to, but distinct from, numerous rubulavirus fragments detected in a wide range of bat species throughout Africa (2) (see Fig. S2 in the supplemental material).
These phylogenetic relationships were reflected in the relatedness of AchPV1 and AchPV2 proteins to those from other Paramyxovirinae. AchPV proteins had their highest amino acid identity with orthologous proteins from other fruit bat-derived rubulaviruses, intermediate amino acid identities to those from the remaining rubulaviruses, and their lowest amino acid identities to proteins from non-rubulavirus Paramyxovirinae (Table 3).
Table 3.
Pairwise amino acid identities of Achimota viruses with orthologous proteins from other members of the Paramyxovirinaeb
| Genus and species | % Identity to protein of: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AchPV1 |
AchPV2 |
|||||||||||||
| N | P | V | M | F | HN | L | N | P | V | M | F | HN | L | |
| Unclassified (rubulavirus-like) | ||||||||||||||
| AchPV1 | 64 | 41 | 31 | 57 | 44 | 31 | 62 | |||||||
| AchPV2 | 64 | 41 | 31 | 57 | 44 | 31 | 62 | |||||||
| ThkPV1 | 58 | 40 | 40 | 55 | 43 | 25 | 58 | 60 | 33 | 26 | 54 | 40 | 26 | 60 |
| ThkPV2 | 77 | 54 | 50 | 75 | 57 | 48 | 70 | 65 | 39 | 33 | 57 | 47 | 33 | 63 |
| ThkPV3 | 61 | 47 | 42 | 55 | 43 | 37 | 61 | 60 | 36 | 27 | 51 | 38 | 33 | 59 |
| MenPV | 63 | 44 | 35 | 57 | 44 | 24 | 56 | 68 | 40 | 33 | 58 | 43 | 25 | 57 |
| TioPV | 64 | 41 | 35 | 55 | 45 | 27 | 56 | 68 | 37 | 31 | 56 | 43 | 26 | 57 |
| Rubulavirus | ||||||||||||||
| MuV | 45 | 24 | 24 | 41 | 35 | 22 | 51 | 46 | 22 | 19 | 44 | 37 | 24 | 53 |
| MapPV | 43 | 24 | 21 | 46 | 34 | 21 | 51 | 44 | 25 | 20 | 44 | 37 | 24 | 52 |
| SimV5 | 41 | 24 | 25 | 37 | 33 | 20 | 51 | 41 | 24 | 20 | 38 | 36 | 24 | 51 |
| hPIV2 | 41 | 26 | 25 | 38 | 31 | 20 | 50 | 41 | 24 | 20 | 38 | 36 | 23 | 51 |
| SimPV5 | 44 | 26 | 25 | 36 | 34 | 20 | 50 | 44 | 24 | 22 | 40 | 38 | 25 | 50 |
| PorPV | 44 | 22 | 18 | 42 | 35 | 22 | 51 | 46 | 25 | 20 | 45 | 37 | 23 | 51 |
| hPIV4 | 41 | 20 | 17 | 42 | 34 | 21 | 48 | 41 | 22 | 16 | 43 | 37 | 23 | 48 |
| Morbillivirus | ||||||||||||||
| RPV | 21 | 12 | NDa | 19 | 22 | 8 | 27 | 21 | 8 | ND | 16 | 30 | 9 | 34 |
| MeV | 21 | 11 | ND | 18 | 23 | 8 | 27 | 21 | 10 | ND | 16 | 22 | 9 | 27 |
| CDV | 22 | 9 | ND | 16 | 18 | 8 | 27 | 20 | 10 | ND | 16 | 22 | 9 | 27 |
| Henipavirus | ||||||||||||||
| HeV | 24 | 9 | 12 | 19 | 23 | 13 | 26 | 24 | 8 | 9 | 17 | 19 | 13 | 27 |
| NiV (M) | 24 | 9 | 11 | 19 | 24 | 14 | 27 | 24 | 8 | 10 | 17 | 25 | 13 | 26 |
| NiV (B) | 24 | 9 | 11 | 19 | 24 | 14 | 27 | 24 | 8 | 9 | 17 | 25 | 13 | 27 |
| Avulavirus | ||||||||||||||
| NDV | 30 | 17 | ND | 25 | 26 | 17 | 34 | 30 | 17 | ND | 24 | 25 | 20 | 27 |
| Respirovirus | ||||||||||||||
| SeV | 17 | 7 | ND | 16 | 21 | 17 | 27 | 18 | 7 | ND | 14 | 25 | 18 | 26 |
| Unclassified | ||||||||||||||
| BeiV | 22 | 11 | 11 | 17 | 23 | 13 | 28 | 23 | 8 | 12 | 17 | 22 | 13 | 27 |
| JPV | 20 | 11 | 12 | 18 | 24 | 14 | 27 | 20 | 7 | 12 | 17 | 23 | 15 | 27 |
| MosV | 24 | 10 | 17 | 16 | 22 | 11 | 27 | 23 | 9 | 17 | 15 | 25 | 9 | 27 |
| TPMV | 21 | 9 | 14 | 16 | 24 | 13 | 26 | 21 | 9 | 13 | 17 | 22 | 13 | 27 |
ND, not determined.
MuV, mumps virus; MapPV, Mapuera virus; SimPV41, simian parainfluenza virus 41; hPIV2, human parainfluenza virus 2; SimV5, simian virus 5; hPIV4, human parainfluenza virus 4; RPV, rinderpest virus; MeV, measles virus; CDV, canine distemper virus; NDV, Newcastle disease virus; SeV, Sendai virus; BeiV, Beilong virus; JPV, J virus; MosV, Mossman virus; TPMV, Tupaia paramyxovirus.
No serological cross-reactivity of Achimota viruses with other paramyxoviruses was observed. Sera raised against a variety of closely related and more divergent paramyxoviruses did not neutralize AchPVs (Fig. 3, stars). Additionally, E. helvum sera that neutralized AchPVs (see below) did not neutralize MenPV or TioPV (ThkPVs have not yet been isolated and were not tested) (Fig. 3, triangles). Polyclonal rabbit sera raised against AchPV1 were found not to neutralize mumps virus, PorPV, hPIV4, or AchPV2 but had a neutralizing titer of 1:40 against the homologous virus (Fig. 3, circles). In addition to this, there was no correlation between antibody titers against AchPV1 and AchPV2 in neutralizing E. helvum sera (see below; also see Fig. 5).
Fig 5.
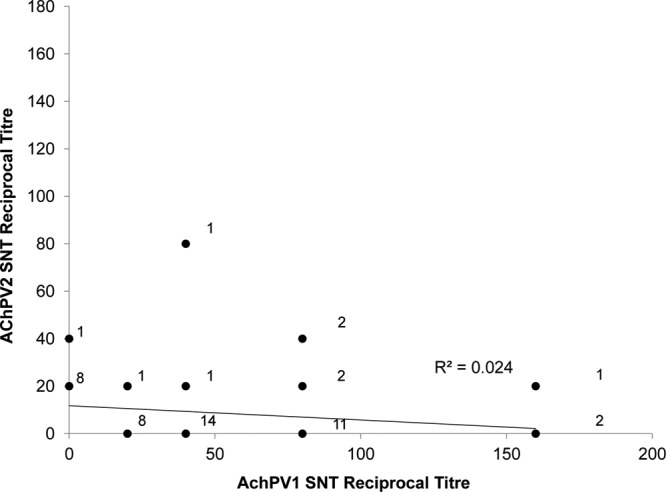
Relationship between reciprocal titers of AchPV1 and AchPV2 neutralization for 52 E. helvum serum samples from Ghana and Tanzania that neutralized either virus. The number of samples is adjacent to each marker. Linear regression (line) with residual sum of squares (R2) is shown.
Epidemiological investigations. (i) Distribution of Achimota viruses in Eidolon helvum.
Our serological studies indicate that Achimota viruses were distributed throughout the geographical range of E. helvum in 2010. Eidolon helvum sampled in September 2010 in Ghana (n = 101) had seroprevalences of 14% (95% confidence interval [CI], 8 to 22%) and 7% (95% CI, 3 to 14%) against AchPV1 and AchPV2, respectively (Table 4 and Fig. 4A). Similar seroprevalences were found in E. helvum sampled in Tanzania in August 2010 (n = 25), with seroprevalences being 12% (95% CI, 4 to 30%) and 8% (95% CI, 2 to 25%) against AchPV1 and AchPV2, respectively (Table 4). In addition to these large, urban populations, sera from smaller, more isolated island populations in the Gulf of Guinea sampled in March to May 2010 were tested against AchPVs. Sera neutralizing AchPV1 (13/26) and AchPV2 (1/26) were detected in E. helvum sampled on the island São Tomé. Sera neutralizing AchPV1 were also detected on the islands of Príncipe (6/15) and the smallest, most remote island, Annobón (4/6) (Table 4). Seroprevalence calculations for island samples are not shown, as the samples were nonrandom (all were selected for antibodies against henipaviruses; see Materials and Methods), thus invalidating comparison of seroprevalences between these populations.
Fig 4.
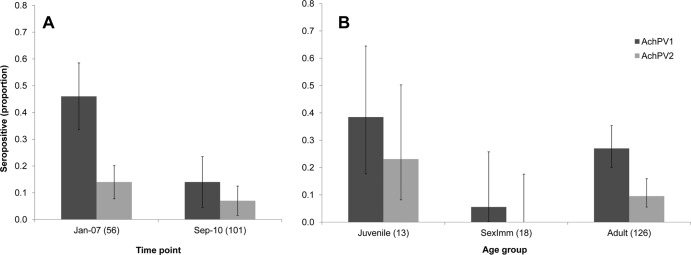
AchPV1 and AchPV2 seroprevalence for 157 E. helvum samples from Accra, Ghana, by time of collection (A) and bat age group (B). The error bars represent 95% confidence intervals. Numbers in parentheses represent the sample size of each group.
(ii) Historical presence and epidemiology of AchPVs in the E. helvum roost in Accra, Ghana.
Sera collected from a single roost at two time points allowed the detection of AchPVs over a long period of time and initial insights into their dynamics in the natural host. Sera collected in Accra in January 2007 (n = 56) had a higher seroprevalence against both AchPV1 (46%; 95% CI, 34 to 59%) and AchPV2 (14%; 95% CI, 7 to 26%) than those collected in 2010 (Fig. 4A). The combined seroprevalence results from these two time points enabled the examination of epidemiological patterns among the 157 E. helvum samples from Accra (Table 5 and Fig. 4B). No significant difference in seroprevalence attributable to gender for either virus was observed. AchPV1 seroprevalence in adult males (28%; 95% CI, 19 to 38%) and adult females (26%; 95% CI, 15 to 40%) were similar, and the same was found for AchPV2 (adult male seroprevalence was 11% [95% CI, 6 to 20%], and adult female seroprevalence was 6% [95% CI, 2 to 17%]). An association with age for seropositivity against AchPV1 was detected where seropositivity in the sexually immature (intermediate) age group (6%; 95% CI, 0 to 26%) was significantly different from that of juveniles (38%; 95% CI, 18 to 64%; P < 0.05) and adults (27%; 95% CI, 20 to 35%; P < 0.05). Due to lower seroprevalence, the same significance could not be established for AchPV2 with these sample numbers (reflected by the greater overlap of confidence intervals for AchPV2 seroprevalences). However, the pattern of age-related seroprevalence for both viruses appeared to be similar (Table 5 and Fig. 4B).
Table 5.
AchPV serum neutralization testing results by gender and age of 157 E. helvum samples collected in Accra, Ghana, in January 2007 and September 2010
| Gender and age | Total | No. positivea by SNT of: |
|
|---|---|---|---|
| AchPV1 | AchPV2 | ||
| Female | |||
| Adult | 47 | 12 | 3 |
| Sexually immature | 10 | 0 | 0 |
| Juvenile | 7 | 4 | 3 |
| Male | |||
| Adult | 79 | 22 | 9 |
| Sexually immature | 8 | 1 | 0 |
| Juvenile | 6 | 1 | 0 |
| Total | 157 | 40 | 15 |
The numbers of neutralizing sera are shown.
(iii) Differences in Eidolon helvum immunity to AchPVs.
Different levels of immunological reactivity were observed against AchPV1 and AchPV2 in bat populations and in individuals. In the populations of E. helvum examined, seroprevalences against AchPV2 were consistently lower than those seen with AchPV1 (Table 4 and Fig. 4). The endpoint dilutions of antibody titers against the two viruses were also markedly different. The 52 samples from Ghana or Tanzania that neutralized either or both viruses for which titer information was available show that titers were higher against AchPV1 than AchPV2 (Fig. 5). Eight sera neutralized both AchPV1 and AchPV2, amounting to a weak statistical association (P = 0.08 by Fisher's exact test), but no correlation of neutralization titer with that of the heterologous virus was observed (Fig. 5).
(iv) Evidence for the possible zoonotic spillover of Achimota viruses.
Sera collected from both healthy and febrile humans neutralized AchPV2. Sera from 2 of 216 healthy adult volunteers sampled in Ghana neutralized AchPV2 at a dilution of 1:20 (in duplicate) on initial and repeat testing, and 1 of 226 Tanzanian samples neutralized AchPV2 at a dilution of 1:20 on initial testing and 1:10 on repeat testing. AchPV2 was not neutralized by any sera collected from 27 bush meat hunters in Ghana, and no human sera were found to neutralize AchPV1. Of the three AchPV2-neutralizing human sera, only one was found to neutralize mumps virus to a titer of 1:20, while the others (and three non-AchPV2-neutralizing human sera) were negative for neutralization against this virus. These six human sera also failed to neutralize hPIV4.
DISCUSSION
Bats harbor a number of paramyxoviruses of varied importance to human and domestic animal health. Here, we have reported the isolation, characterization, and initial investigations of the spillover potential of two novel rubulaviruses from the straw-colored fruit bat, Eidolon helvum: Achimota viruses 1 (AchPV1) and 2 (AchPV2). Despite being isolated from urine samples previously investigated for the presence of paramyxoviruses (3) and other interrogations of this host species for paramyxoviruses (2, 4), AchPV2 had not been detected prior to this study. Each Achimota virus had distinct behaviors in cell culture, with AchPV2 growing to higher titers and having slightly different CPEs, in both Vero and PaKi cell lines, compared to those of AchPV1. In addition to being distinct in cell culture, the two viruses appeared to be serologically distinct from each other, as well as from other paramyxoviruses, with respect to neutralization. No cross-neutralization of these viruses by sera raised against a variety of paramyxoviruses was observed, and AchPV-neutralizing sera (including a specifically raised AchPV1 polyclonal serum) was not observed to cross-neutralize other paramyxoviruses. There was also no evidence for cross-neutralization between the two novel viruses, with AchPV1-specific sera being nonneutralizing against AchPV2, and there was no correlation of neutralization titers in 52 AchPV-neutralizing E. helvum sera.
The viruses were characterized by full-length genome sequencing, providing insight into their place within, and relationships with, known members of the Paramyxoviridae. Achimota viruses were rubulaviruses that phylogenetically clustered (and shared their highest protein sequence identities) with other rubulaviruses from Old World fruit bats: MenPV, TioPV, and ThkPVs. Members of this cluster also shared genomic features that distinguished them from other rubulaviruses (although inconsistently throughout the group), such as alternative and noncomplementary leader sequences, the use of guanine instead of adenine in the +1 position of transcriptional start sites, and variations in the sialic acid-binding motif. These commonalities in genomic organization further support a more recent common ancestor for these viruses relative to other groups.
The interrelationship of these bat-derived rubulaviruses highlights the increasingly apparent and complex reservoir role that Chiroptera appear to play for mammalian Paramyxoviridae (2, 3). Within the phylogenetic cluster are viruses detected in three genera of Old World fruit bats sampled in China, Malaysia, Australia, and Ghana with no apparent clustering of viruses by host species or geography. This phylogenetic admixing of viruses from genetically distinct and geographically separated host species was similarly seen in a recent study of paramyxoviral sequences in bat species worldwide (2). This study and others indicate that there are far more extant viruses in this bat-derived rubulavirus lineage yet to be characterized (2, 3, 5). For this reason, it is crucial that the potential consequences and risk of zoonotic spillover of such viruses be investigated further.
Sera collected from E. helvum and humans in Ghana and Tanzania, as well as those from populations of E. helvum on islands in the Gulf of Guinea, neutralized AchPVs. As no evidence for neutralizing cross-reactivity with other paramyxoviruses and between AchPVs had been observed, we inferred prior AchPV infection in E. helvum and other species through detection of AchPV-neutralizing antibodies. Infection with AchPV1 and AchPV2 was widely distributed throughout the geographical range of E. helvum in 2010, with antibodies against both viruses being detected in all regions under study.
Human sera were also screened for evidence of previous infection with AchPVs. Out of a total of 442 sera tested, we detected three samples that neutralized AchPV2: two from healthy adults and one from a febrile pediatric patient. This likely indicates the presence of neutralizing antibodies, especially as (with the exception of one sample, which also neutralized mumps virus) these AchPV2-neutralizaing sera were not found to neutralize the human rubulaviruses mumps virus or hPIV4. None of the 442 human sera neutralized AchPV1, which would be expected to occur at a similar rate if the AchPV2 neutralization had resulted from nonspecific factors, further supporting the presence of neutralizing antibodies in these samples. These antibodies are unlikely to have arisen from a serologically cross-reactive virus, as no known human paramyxoviruses exist within this unique lineage of Old World fruit bat-derived rubulaviruses. It is possible that these antibodies resulted from infection with another zoonotic virus from this unique lineage that is serologically cross-reactive; however, these have yet to be characterized. The clinical implications of infection with such viruses are currently unknown. However, the neutralization titers of human sera against AchPV2 in this study (1:10 to 1:20) were closer to the titers observed in asymptomatic adults against TioPV (1:5 to 1:10 [21]) than those observed against MenPV in humans with a history of clinical illness (1:128 and 1:512 [19]).
Critical to understanding the dynamics of potential zoonotic spillover of AchPVs are longitudinal studies of naturally infected bat populations, and the sera analyzed from the bat population in Accra provided some initial insight into AchPV epidemiology. We detected seropositivity to both AchPVs in the population in samples collected in 2007 and 2010, but there was a marked decline in seroprevalence over this time period. It is not known if this represents a natural cycle of endemic infection or if it is indicative of a waning epidemic. It is notable, however, that both AchPVs were isolated from the Accra bat colony from samples collected in 2010, confirming virus circulation at that time. Also, the age-specific seroprevalence data for both AchPVs is suggestive (though limited by sample numbers) of horizontal transmission, with waning of maternal antibodies in juveniles and a subsequent increase in seroprevalence with age. Horizontal transmission of AchPVs seems plausible given the isolation of the viruses from urine and that this mode of transmission is common for paramyxoviruses (38), including henipaviruses in other Old World fruit bat species (39). The studies at other sites also provide insight into these dynamics with infection on small island populations, supporting the hypothesis that large populations capable of reaching high critical community sizes are not required to support paramyxovirus infection in bats (23).
Here, we have described the isolation of novel rubulaviruses from the urine of African straw-colored fruit bats. The isolated viruses were phylogenetically closely related to each other and other Old World fruit bat-derived rubulaviruses despite the urine samples containing phylogenetically diverse members of the Paramyxovirinae (3). This possibly indicates that isolation of paramyxoviruses in other genera requires more-specialized approaches than those used here. The viruses are widely distributed across E. helvum populations in Africa and may be capable of zoonotic host switching. The consequences of Achimota virus infection in all species is currently unknown and requires further investigation, in parallel with ongoing studies of infection in the natural host to determine spillover dynamics.
Supplementary Material
ACKNOWLEDGMENTS
We thank Andres Fernandes-Loras for assistance with sampling and Andy Kwabena Alhassan and the Veterinary Services Directorate, Ghana. We thank Anthony Fooks at the Animal Health and Veterinary Laboratories Agency. We also thank Jackie Pallister and Reuben Klein for the provision of anti-PorPV-nucleoprotein sera and Volker Haring for assistance with 454 sequencing assembly. We also thank two anonymous reviewers for their helpful comments on the manuscript and Christian Drosten and Felix Drexler for their helpful discussions on the content.
K.B. and P.R.M. are supported by the Wellcome Trust. J.L.N.W. is supported by the Alborada Trust and the Research and Policy for Infectious Disease Dynamics (RAPIDD) program of the Science and Technology Directorate, Department of Homeland Security. D.T.S.H. is supported by RAPIDD and a David H. Smith Conservation Research Fellowship. A.A.C. is supported by a Royal Society Wolfson Merit award.
Footnotes
Published ahead of print 14 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01202-12.
REFERENCES
- 1. Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. 2006. Bats: important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 19:531–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Drexler JF, Corman VM, Muller MA, Maganga GD, Vallo P, Binger T, Gloza-Rausch F, Rasche A, Yordanov S, Seebens A, Oppong S, Sarkodie YA, Pongombo C, Lukashev AN, Schmidt-Chanasit J, Stocker A, Carneiro AJ, Erbar S, Maisner A, Fronhoffs F, Buettner R, Kalko EK, Kruppa T, Franke CR, Kallies R, Yandoko ER, Herrler G, Reusken C, Hassanin A, Kruger DH, Matthee S, Ulrich RG, Leroy EM, Drosten C. 2012. Bats host major mammalian paramyxoviruses. Nat. Commun. 3:796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baker KS, Todd S, Marsh G, Fernandez-Loras A, Suu-Ire R, Wood JL, Wang LF, Murcia PR, Cunningham AA. 2012. Co-circulation of diverse paramyxoviruses in an urban African fruit bat population. J. Gen. Virol. 93:850–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Drexler JF, Corman VM, Gloza-Rausch F, Seebens A, Annan A, Ipsen A, Kruppa T, Muller MA, Kalko EK, Adu-Sarkodie Y, Oppong S, Drosten C. 2009. Henipavirus RNA in African bats. PLoS One 4:e6367 doi:10.1371/journal.pone.0006367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lau SK, Woo PC, Wong BH, Wong AY, Tsoi HW, Wang M, Lee P, Xu H, Poon RW, Guo R, Li KS, Chan KH, Zheng BJ, Yuen KY. 2010. Identification and complete genome analysis of three novel paramyxoviruses, Tuhoko virus 1, 2 and 3, in fruit bats from China. Virology 404:106–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hollinger FB, Pavri KM. 1971. Bat parainfluenza virus: immunological, chemical, and physical properties. Am. J. Trop. Med. Hyg. 20:131–138 [PubMed] [Google Scholar]
- 7. Pavri KM, Singh KR, Hollinger FB. 1971. Isolation of a new parainfluenza virus from a frugivorous bat, Rousettus leschenaulti, collected at Poona, India. Am. J. Trop. Med. Hyg. 20:125–130 [DOI] [PubMed] [Google Scholar]
- 8. Zeller HG, Karabatsos N, Calisher CH, Digoutte JP, Murphy FA, Shope RE. 1989. Electron microscopy and antigenic studies of uncharacterized viruses. I. Evidence suggesting the placement of viruses in families Arenaviridae, Paramyxoviridae, or Poxviridae. Arch. Virol. 108:191–209 [DOI] [PubMed] [Google Scholar]
- 9. Stephan HA, Gay GM, Ramirez TC. 1988. Encephalomyelitis, reproductive failure and corneal opacity (blue eye) in pigs, associated with a paramyxovirus infection. Vet. Rec. 122:6–10 [DOI] [PubMed] [Google Scholar]
- 10. Wang LF, Hansson E, Yu M, Chua KB, Mathe N, Crameri G, Rima BK, Moreno-Lopez J, Eaton BT. 2007. Full-length genome sequence and genetic relationship of two paramyxoviruses isolated from bat and pigs in the Americas. Arch. Virol. 152:1259–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chua KB, Hooi LKCPS, Wee KF, Khong JH, Chua BH, Chan YP, Lim ME, Lam Sai Kit K. 2002. Isolation of Nipah virus from Malaysian island flying-foxes. Microbes Infect. 4:145–151 [DOI] [PubMed] [Google Scholar]
- 12. Murray PK, Selleck PW, Hooper PT, Hyatt AD, Gould AR, Gleeson LJ, Westbury HA, Hiley L, Selvey LA, Rodwell BJ, Ketterer PJ. 1995. A morbillivirus that caused fatal disease in horses and humans. Science 268:94–97 [DOI] [PubMed] [Google Scholar]
- 13. Selvey LA, Wells RM, McCormack JG, Ansford AJ, Murray PK, Rogers RJ, Lavercombe PS, Selleck PW, Sheridan JW. 1995. Infection of humans and horses by a newly described morbillivirus. Med. J. Australia 162:642–645 [DOI] [PubMed] [Google Scholar]
- 14. Smith I, Broos A, de Jong C, Zeddeman A, Smith C, Smith G, Moore F, Barr J, Crameri G, Marsh G, Tachedjian M, Yu M, Kung YH, Wang LF, Field H. 2011. Identifying hendra virus diversity in pteropid bats. PLoS One 6:e25275 doi:10.1371/journal.pone.0025275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chua KB, Goh KJ, Wong KT, Kamarulzaman A, Tan PSK, Ksiazek TG, Zaki SR, Paul G, Lam Sai Kit K, Tan CT. 1999. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet 354:1257–1259 [DOI] [PubMed] [Google Scholar]
- 16. Mohd Nor MN, Gan CH, Ong BL. 2000. Nipah virus infection of pigs in peninsular Malaysia. Rev. Sci. Tech. 19:160–165 [DOI] [PubMed] [Google Scholar]
- 17. Luby SP, Hossain MJ, Gurley ES, Ahmed BN, Banu S, Khan SU, Homaira N, Rota PA, Rollin PE, Comer JA, Kenah E, Ksiazek TG, Rahman M. 2009. Recurrent zoonotic transmission of Nipah virus into humans, Bangladesh, 2001–2007. Emerg. Infect. Dis. 15:1229–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Philbey AW, Kirkland PD, Ross AD, Davis RJ, Gleeson AB, Love RJ, Daniels PW, Gould AR, Hyatt AD. 1998. An apparently new virus (family paramyxoviridae) infectious for pigs, humans and fruit bats. Emerg. Infect. Dis. 4:269–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chant K, Chan R, Smith M, Dwyer DE, Kirkland PD, Group NE. 1998. Probable human infection with a newly described virus in the family paramyxoviridae. Emerg. Infect. Dis. 4:273–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chua KB, Wang LF, Lam Sai Kit K, Crameri GS, Yu M, Wise T, Boyle DB, Hyatt AD, Eaton BT. 2001. Tioman virus, a novel paramyxovirus isolated from fruit bats in Malaysia. Virology 283:215–229 [DOI] [PubMed] [Google Scholar]
- 21. Yaiw KC, Crameri G, Wang L, Chong HT, Chua KB, Tan CT, Goh KJ, Shamala D, Wong KT. 2007. Serological evidence of possible human infection with Tioman virus, a newly described paramyxovirus of bat origin. J. Infect. Dis. 196:884–886 [DOI] [PubMed] [Google Scholar]
- 22. Hayman DT, Suu-Ire R, Breed AC, McEachern JA, Wang L, Wood JL, Cunningham AA. 2008. Evidence of henipavirus infection in West African fruit bats. PLoS One 3:e2739 doi:10.1371/journal.pone.0002739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peel AJ, Baker KS, Crameri G, Barr JA, Hayman DT, Wright E, Broder CC, Fernandez-Loras A, Fooks AR, Wang LF, Cunningham AA, Wood JL. 2012. Henipavirus neutralising antibodies in an isolated island population of African fruit bats. PLoS One 7:e30346 doi:10.1371/journal.pone.0030346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kamins AO, Restif O, Ntiamoa-Baidu Y, Suu-Ire R, Hayman DTS, Cunningham AA, Wood JL, Rowcliffe JM. 2011. Uncovering the fruit bat bushmeat commodity chain and the true extent of fruit bat hunting in Ghana, West Africa. Biol. Cons. 144:3000–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hayman DTS, McCrea R, Restif O, Suu-Ire R, Fooks AR, Wood JLN, Cunningham AA, Rowcliffe MJ. 2012. Demography of straw-colored fruit bats in Ghana. J. Mammalogy 93:1393–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peel AJ. 2012. The epidemiology of Lagos bat virus and henipaviruses in straw coloured fruit bats (Eidolon helvum), using population genetics to infer population connectivity. University of Cambridge, Cambridge, United Kingdom [Google Scholar]
- 27. Nadjm B, Amos B, Mtove G, Ostermann J, Chonya S, Wangai H, Kimera J, Msuya W, Mtei F, Dekker D, Malahiyo R, Olomi R, Crump JA, Whitty CJ, Reyburn H. 2010. WHO guidelines for antimicrobial treatment in children admitted to hospital in an area of intense Plasmodium falciparum transmission: prospective study. BMJ 340:c1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Crameri G, Todd S, Grimley S, McEachern JA, Marsh GA, Smith C, Tachedjian M, De Jong C, Virtue ER, Yu M, Bulach D, Liu JP, Michalski WP, Middleton D, Field HE, Wang LF. 2009. Establishment, immortalisation and characterisation of pteropid bat cell lines. PLoS One 4:e8266 doi:10.1371/journal.pone.0008266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tong S, Chern SW, Li Y, Pallansch MA, Anderson LJ. 2008. Sensitive and broadly reactive reverse transcription-PCR assays to detect novel paramyxoviruses. J. Clin. Microbiol. 46:2652–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marsh GA, de Jong C, Barr JA, Tachedjian M, Smith C, Middleton D, Yu M, Todd S, Foord AJ, Haring V, Payne J, Robinson R, Broz I, Crameri G, Field HE, Wang LF. 2012. Cedar virus: a novel henipavirus isolated from Australian bats. PLoS Pathog. 8:e1002836 doi:10.1371/journal.ppat.1002836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chua KB, Wang LF, Lam Sai Kit K, Eaton BT. 2002. Full length genome sequence of Tioman virus, a novel paramyxovirus in the genus Rubulavirus isolated from fruit bats in Malaysia. Arch. Virol. 147:1323–1348 [DOI] [PubMed] [Google Scholar]
- 32. Kozak M. 1991. An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol. 115:887–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 35. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thalmann CM, Cummins DM, Yu M, Lunt R, Pritchard LI, Hansson E, Crameri S, Hyatt A, Wang LF. 2010. Broome virus, a new fusogenic orthoreovirus species isolated from an Australian fruit bat. Virology 402:26–40 [DOI] [PubMed] [Google Scholar]
- 37. Kolakofsky D, Pelet T, Garcin D, Hausmann S, Curran J, Roux L. 1998. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J. Virol. 72:891–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pringle CR. 1991. The order Mononegavirales. Arch. Virol. 117:137–140 [PubMed] [Google Scholar]
- 39. Plowright RK, Field HE, Smith C, Divljan A, Palmer C, Tabor G, Daszak P, Foley JE. 2008. Reproduction and nutritional stress are risk factors for Hendra virus infection in little red flying foxes (Pteropus scapulatus). Proc. Biol. Sci. 275:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.