Abstract
We show that influenza A H1N1 virus infection leads to very low infectivity in mouse dendritic cells (DCs) in vitro compared with that in human DCs. This holds when H3 or H5 replaces H1 in recombinant viruses. Viruslike particles confirm the difference between mouse and human, suggesting that reduced virus entry contributes to lower mouse DC infectivity. Low infectivity of mouse DCs should be considered when they are used to study responses of DCs that are actually infected.
TEXT
Dendritic cells (DCs) bridge innate and adaptive immunity by recognizing pathogens, secreting cytokines, and migrating to lymph nodes to initiate adaptive immunity. Because tissue-resident DCs are not abundant, murine bone marrow-derived dendritic cells (BMDCs) or splenic DCs (sDCs) are commonly used for experiments (1).
BMDCs have been used to study the effects of influenza virus infection, in particular infection with the mouse-adapted H1N1 influenza virus strain A/Puerto Rico/8/34 (PR8) (2–4). Human DCs may be derived from monocytes extracted from blood buffy coats (1). Experimental results are often based on methods where the average expression of all cells in a sample is assayed, e.g., by enzyme-linked immunosorbent assay (ELISA), quantitative PCR (qPCR), or Western blotting (5, 6). If one were to compare the impact of different influenza A strains on BMDCs, or compare the antagonist response of the same virus in mouse and human DCs, the fact that any response measured comes from a combination of infected and paracrine-stimulated uninfected cells makes it desirable that all studies have similar levels of infection. The results of a comparison of viruses would be complicated if cellular rates of infection were significantly different, since the share of cytokine-induced gene expression levels contributed by uninfected cells would then differ significantly as well.
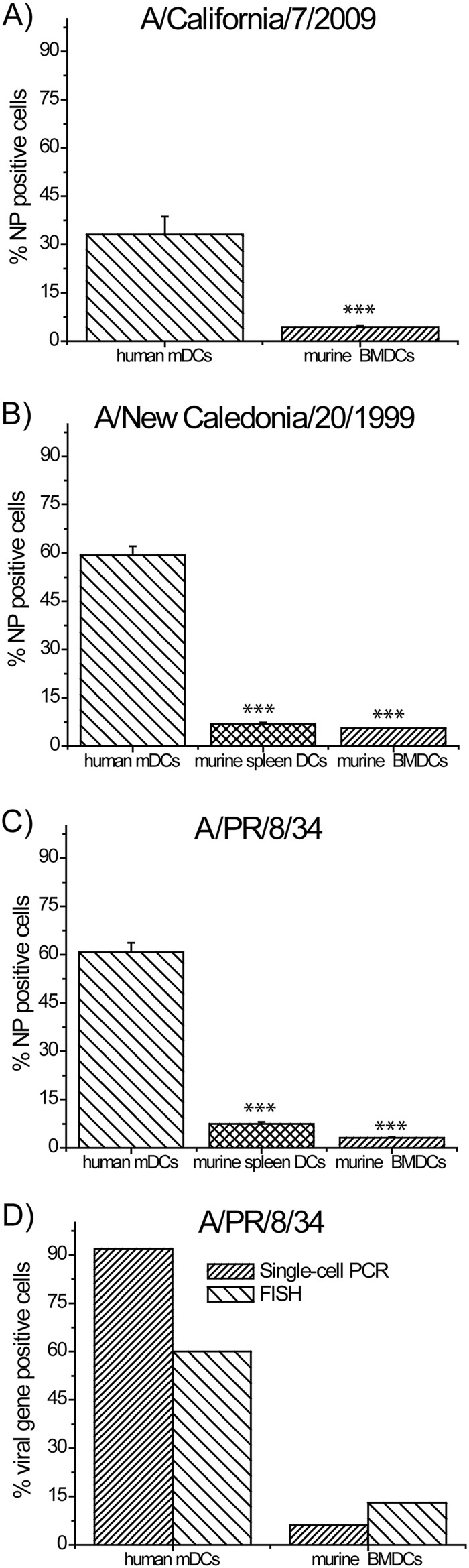
We accurately determined the fraction of DCs infected by virus, using a BSL-2 environment. We stained murine BMDCs, sDCs, and human monocyte-derived dendritic cells (mDCs) 8 h after infection at a multiplicity of infection (MOI) of 1 (by plaque assay in MDCK cells) with the pandemic A/California/7/2009 and seasonal A/New Caledonia/20/1999 influenza viruses using an Influenza virus nucleoprotein (NP)-specific antibody (Abcam). Infection rate quantified by flow cytometry revealed 33% NP-positive human mDCs versus 4% NP-positive murine BMDCs when infected with A/California/7/2009 (Fig. 1A) and 59% positive human mDCs versus 6% murine sDCs and 5% murine BMDCs when infected with A/New Caledonia/20/1999 (Fig. 1B). We obtained similar results with the mouse-adapted PR8 strain, namely, 63% human mDCs versus 5 to 8% murine BMDCs and sDCs (Fig. 1C). Similar results were obtained using mouse DCs derived from interferon I/III receptor-null mutant mice (data not shown). That mouse BMDCs show a striking lack of infectivity is consonant with recent results using PR8 reporting only 30 to 40% infectivity at an MOI as high as 30 (7). We confirmed these surprising results by two other sensitive techniques, multiprobe single-molecule fluorescence in situ hybridization (FISH) (8) and single-cell PCR assays (9) (Fig. 1D). It should be noted that in addition to the low percentage of infected cells, infection in DCs is abortive (7).
Fig 1.
BMDC, sDC, and human DC infectivity rates. DCs infected with influenza A/California/7/2009 (A), influenza A/New Caledonia/20/1999 (B), and influenza A/PR/8/34 (C) assayed 8 h after infection for viral NP expression with flow cytometry (***, P < 0.005). NP expression reaches a plateau 6 to 8 h after infection (data not shown). (D) Human and murine DC viral gene expression rate assayed by FISH (influenza NA assayed) and single-cell qPCR (influenza NS1 assayed) measured 8 h and 6 h postinfection, respectively. Results shown in panels A to D were obtained using cells from four different donors.
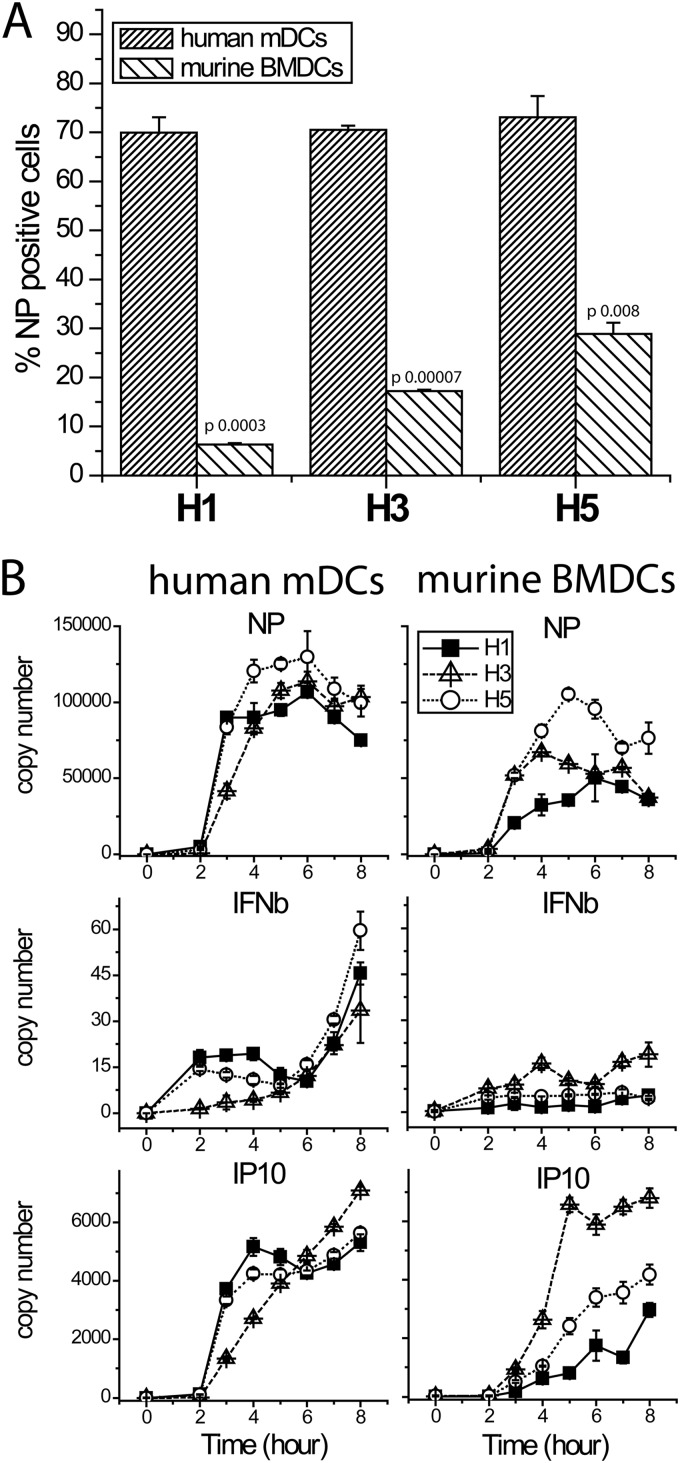
Hargadon et al. showed that respiratory dendritic cells isolated from mouse lungs have better infection rates with H2- and H3- than with H1-containing viruses (10). Therefore, we studied infection of human mDCs and murine BMDCs with PR8 and PR8 7:1 reassortant viruses bearing H3 from influenza A/Wyoming/3/2003 or H5 from influenza A/Vietnam/1203/04(HALO) (11). Again, although H5 showed somewhat better infectivity, for all three hemagglutinins (HAs), infectivity in murine BMDCs was significantly lower than in human mDCs (Fig. 2A). Beta interferon (IFN-β) and viral NP genes also showed attenuated expression levels in mouse DCs (Fig. 2B). The levels of IP10, an interferon-inducible gene, are influenced by levels of IFN and by the number of uninfected cells, which are the main source of IFN-induced genes, as they are free from viral immune antagonism. The low but detectable IFN-β expression seen in mouse DCs correlates with the low percentage of cells infected. Levels of IP10 expression in samples from mouse and human cells are more similar, presumably reflecting the offsetting effects in mouse of increased number of uninfected cells and lower interferon levels.
Fig 2.
Infection of murine BMDCs and human mDCs with PR8-based recombinant viruses expressing H1-, H3-, or H5-subtype hemagglutinin. (A) Infectivity rates measured as NP expression by flow cytometry 8 h postinfection. (B) Gene expression time courses measured by real-time reverse transcription (RT)-PCR.
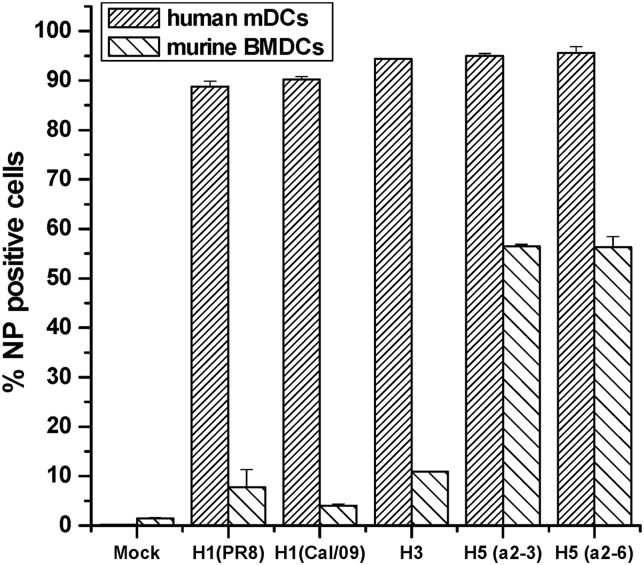
We have been unable to explain why H5 influenza viruses infect murine BMDCs somewhat better than H1 influenza viruses. H1 encoded by human influenza viruses binds preferentially to 2,6-linked sialic acids (SA2,6) (11). To test whether oligosaccharide linkage differences are responsible for higher H5 infectivity, we infected murine BMDCs with a recombinant A/Vietnam/1203/04 (HALO)H5N1 virus (a 6:2 virus with 6 PR8 segments) with two point mutations which altered the HA sialic acid binding preference from a 2,3 to a 2,6 linkage (11). We found no difference in infectivity between the H5 SA2,3 binding virus and the mutated SA2,6 binding virus (Fig. 3), excluding linkage differences as the cause of different levels of infectivity.
Fig 3.
Murine BMDC and human mDC infectivity rates 8 h postinfection with wild-type H1 (A/Puerto Rico/8/34), H1 (A/California/7/2009), H3 (A/Wyoming/3/2003), H5 (alpha 2,6-specific), or H5 (alpha 2,3-specific) recombinant A/Vietnam/1203/04 viruses (6:2 viruses with 6 PR8 segments). Infectivity was assayed by measuring viral NP expression by flow cytometry. All differences between human and murine infected DCs had a P value of ≤0.001.
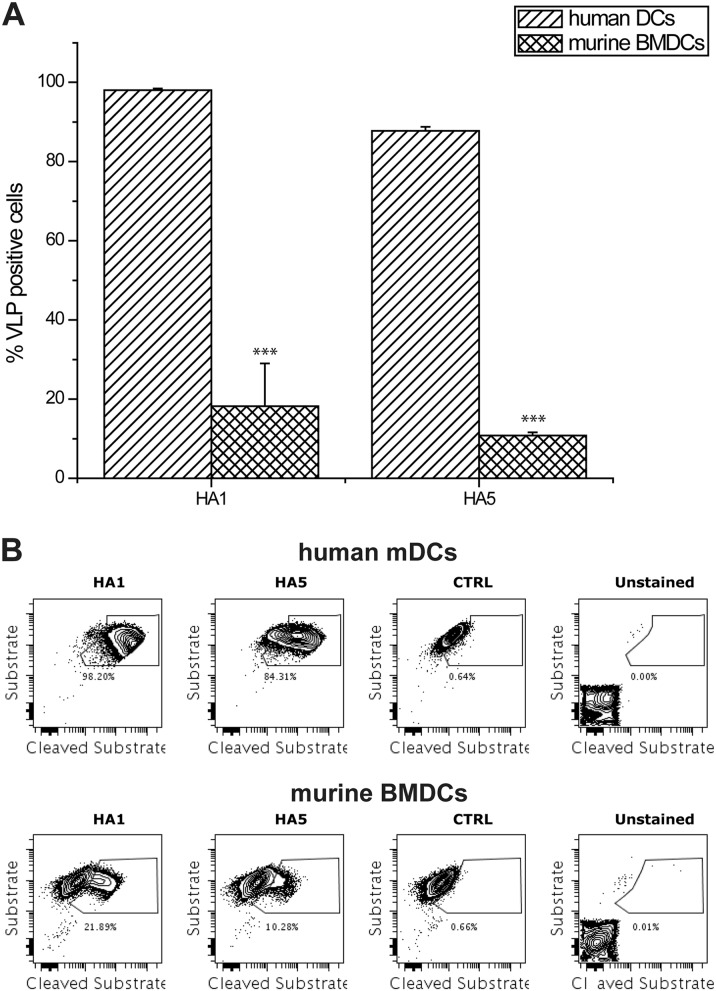
To examine whether HA contributes to reduced viral entry in mouse DCs compared to that in human DCs, we constructed beta lactamase containing influenza viruslike particles (VLPs) with different HAs on the surface (12). VLP constructs consist of HA from A/Puerto Rico/8/34 (H1) or A/Vietnam/1203/2004 (H5), with, in both cases, an NA from A/WSN/33 (H1) and beta lactamase (BLA) enzyme-bearing VP40 from Ebola virus. We found that viral entry of both VLPs was reduced for mouse BMDCs (Fig. 4), suggesting that reduced viral entry may be one factor contributing to the differences in mouse and human infectivity. Although for mouse BMDCs H5 reassortant 7:1 and 6:2 virus (with a PR8 backbone) infectivity appears higher than for wild-type PR8 independently of sialic acid receptor binding preference (for the 6:2 recombinants), this higher infectivity does not appear to be due to a larger rate of cell penetration. The reasons for better infectivity of human DCs than of mouse DCs are not fully explained and will require further study.
Fig 4.
Entry assay with murine BMDCs and human mDCs with H1- and H5-bearing VLPs in a 1:2 dilution. VLPs consist of HA from either A/Puerto Rico/8/34 (H1) or A/Vietnam/1203/2004 (H5), NA from A/WSN/33 (H1), and Beta-lactamase enzyme-bearing VP40 from Ebola virus. Cells were infected with the VLPs and then incubated with a dual fluorochrome-bearing substrate 4 h after infection. When VLPs enter the cell, the BLA enzyme cleaves the substrate, reducing the FRET signal. VLP concentrations of H1 and H5 are, respectively, 2 and 3 log2 units of HA. (A) Percentage of cells with cleaved substrate in human mDCs and BMDCs. Concentrations were adjusted to give approximately the same levels in human DCs. (B) Sample plots of the VLP assays of unstained, mock-infected, and H1- and H5-infected human mDCs and murine BMDCs. (***, P ≤ 0.005 between H5 and H1 in corresponding solution).
Our results imply that for mouse dendritic cells studied in vitro, because of a poor rate of infection, particularly for H1N1 viruses, the levels of interferon-induced genes are dominated by uninfected cells. In this case, the effects of viral antagonism of the interferon-induced signaling pathway and the responses occurring in infected cells may be difficult to infer from experiments where only a small percentage of cells are infected with virus.
ACKNOWLEDGMENTS
We thank Irene Ramos and Ana Fernandez-Sesma for their gift of mutated H5 viruses and for helpful discussions.
This work was supported by NIAID contract HHSN272201000054C. This work was also partly supported by the Center for Research on Influenza Pathogenesis (CRIP) and the NIAID-funded Center of Excellence for Influenza Research and Surveillance, contract HHSN266200700010C, and by grant U01AI095611 from the NIAID Mucosal Immunity Study Team (MIST) Program to A.G.-S.
Footnotes
Published ahead of print 28 November 2012
REFERENCES
- 1. Inaba K, Swiggard WJ, Steinman RM, Romani N, Schuler G, Brinster C. 2009. Isolation of dendritic cells. 86:3.7.1–3.7.19 In Coligan JE, Bierer BE, Margulies DH, Shevach EM, Strober W. (ed), Current protocols in immunology. John Wiley and Sons, Hoboken, NJ: [DOI] [PubMed] [Google Scholar]
- 2. Diebold SS, Montoya M, Unger H, Alexopoulou L, Roy P, Haswell LE, Al-Shamkhani A, Flavell R, Borrow P, Reis e Sousa C. 2003. Viral infection switches non-plasmacytoid dendritic cells into high interferon producers. Nature 424:324–328 [DOI] [PubMed] [Google Scholar]
- 3. Ichinohe T, Pang IK, Iwasaki A. 2010. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat. Immunol. 11:404–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lopez CB, Garcia-Sastre A, Williams BR, Moran TM. 2003. Type I interferon induction pathway, but not released interferon, participates in the maturation of dendritic cells induced by negative-strand RNA viruses. J. Infect. Dis. 187:1126–1136 [DOI] [PubMed] [Google Scholar]
- 5. Noone CM, Lewis EA, Frawely AB, Newman RW, Mahon BP, Mills KH, Johnson PA. 2005. Novel mechanism of immunosuppression by influenza virus haemagglutinin: selective suppression of interleukin 12 p35 transcription in murine bone marrow-derived dendritic cells. J. Gen. Virol. 86:1885–1890 [DOI] [PubMed] [Google Scholar]
- 6. Pejawar SS, Parks GD, Alexander-Miller MA. 2005. Abortive versus productive viral infection of dendritic cells with a paramyxovirus results in differential upregulation of select costimulatory molecules. J. Virol. 79:7544–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ioannidis LJ, Verity EE, Crawford S, Rockman SP, Brown LE. 2012. Abortive replication of influenza virus in mouse dendritic cells. J. Virol. 86:5922–5925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Raj A, Tyagi S. 2010. Detection of individual endogenous RNA transcripts in situ using multiple singly labeled probes. Methods Enzymol. 472:365–386 [DOI] [PubMed] [Google Scholar]
- 9. Hu J, Sealfon SC, Hayot F, Jayaprakash C, Kumar M, Pendleton AC, Ganee A, Fernandez-Sesma A, Moran TM, Wetmur JG. 2007. Chromosome-specific and noisy IFNB1 transcription in individual virus-infected human primary dendritic cells. Nucleic Acids Res. 35:5232–5241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hargadon KM, Zhou H, Albrecht RA, Dodd HA, García-Sastre A, Braciale TJ. 2011. Major histocompatibility complex class II expression and hemagglutinin subtype influence the infectivity of type A influenza virus for respiratory dendritic cells. J. Virol. 85:11955–11963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ramos I, Bernal-Rubio D, Durham N, Belicha-Villanueva A, Lowen AC, Steel J, Fernandez-Sesma A. 2011. Effects of receptor binding specificity of avian influenza virus on the human innate immune response. J. Virol. 85:4421–4431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tscherne DM, Garcia-Sastre A. 2011. An enzymatic assay for detection of viral entry. 51:26.12.1–26.12.10 In Bonifacino JS, Dasso M, Harford JB, Lippincott-Schwartz J, Yamada KM. (ed) , Current protocols in cell biology. John Wiley and Sons, Hoboken, NJ: [DOI] [PubMed] [Google Scholar]