Abstract
Chronic infection by hepatitis C virus (HCV) is a cause of the global burden of liver diseases. HCV entry into hepatocytes is a complicated and multistep process that represents a promising target for antiviral intervention. The recently reported amphipathic α-helical virucidal peptide (C5A) from the HCV NS5A protein suggests a new category of antiviral drug candidates. In this study, to identify C5A-like HCV inhibitors, synthetic peptides derived from the C5A-corresponding NS5 protein region of selected Flaviviridae viruses were evaluated for their anti-HCV activities. A peptide from GB virus A (GBV-A), but not other flaviviruses, demonstrated an inhibitory effect on HCV infection. Through a series of sequence optimizations and modifications of the peptide helicity and hydrophobicity, we obtained a peptide designated GBVA10-9 with highly potent anti-HCV activity. GBVA10-9 suppressed infection with both cell culture-derived and pseudotyped HCV in vitro, and the 50% cell culture inhibitory concentration ranged from 20 nM to 160 nM, depending on the genotypic origin of the envelope proteins. GBVA10-9 had no detectable effects on either HCV attachment to Huh7.5.1 cells or viral RNA replication. No virucidal activity was found with GBVA10-9, suggesting an action mechanism distinct from that of C5A. The inhibitory effect of GBVA10-9 appeared to occur at the postbinding step during viral entry. Taken together, the results with GBVA10-9 demonstrated a potent activity for blocking HCV entry that might be used in combination with other antivirals directly targeting virus-encoded enzymes. Furthermore, GBVA10-9 also provides a novel tool to dissect the detailed mechanisms of HCV entry.
INTRODUCTION
Hepatitis C virus (HCV) infects an estimated 170 million people worldwide. Failure to clear circulating HCV leads to a chronic infection that is frequently associated with the development of liver cirrhosis and hepatocellular carcinoma. The current standard-of-care therapy regimen for chronic HCV infection is based on a combination of PEGylated interferon (IFN) and ribavirin. However, the significant adverse effects and the limited efficacy in specific genotypes lead to poor tolerance and virus relapse (1). Recently, the FDA approved two direct-acting antiviral drugs (DAAs) that target HCV NS3/4A protease with an increased and sustained virological response (2–5). However, a monotarget antiviral strategy greatly increases the risk of rapidly emerging resistant mutations because of the high genetic heterogeneity of HCV. Successful treatment will probably require a combination therapy with multiple inhibitors directed at diverse viral targets. Therefore, development of more effective antiviral drugs is urgently needed.
HCV entry is the first step of the viral life cycle and is a promising target for inhibiting viral infection. The HCV genome encodes two envelope glycoproteins, E1 and E2, which associate as a noncovalent heterodimer and constitute the majority of the viral membrane-integrated proteins. HCV glycoproteins play pivotal roles during viral entry by interacting with cellular factors such as CD81, scavenger receptor class B type I, claudin-1, and occludin. HCV entry into hepatocytes is a complex and multistep process involving viral attachment, receptor binding, and endocytosis. The recently characterized peptide C5A, which has an amphipathic α-helical structure, showed significant inhibitory effects on HCV and HIV infection in vitro (6). C5A is derived from the membrane anchor domain of the HCV NS5A protein and shows virucidal activity, most likely related to the destabilization viral membranes. The discovery of C5A provides a unique therapeutic alternative for treatment of HCV infection.
Thus, because of the close structural and functional link between HCV and other Flaviviridae viruses, we hypothesized that peptides from the NS5A-homologous sequences in other viruses might also interfere with HCV infection. In this study, we identified a GB virus A (GBV-A)-derived synthetic peptide that inhibits HCV infection in the cell culture infection system. Importantly, in a series of structure-based de novo designs, the anti-HCV activity of the peptide was greatly enhanced compared to that of the parental peptide. Our data show that this peptide exerts its inhibitory effect by blocking the HCV entry process without virucidal activity.
MATERIALS AND METHODS
Cells and reagents.
The human hepatoma cell line Huh7.5.1 was provided by Francis V. Chisari (The Scripps Research Institute, La Jolla, CA) (7). The HCV genotype 1b replicon-containing cell line 2−3+ was provided by Stanley Lemon (University of Texas Medical Branch, Galveston, TX); G418 (500 μg/ml) was routinely added to the 2−3+ cells to maintain viral RNA replication. HeLa, HEK293T, and HepG2 cells were obtained from the ATCC. The above-listed cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with penicillin and streptomycin, 1% nonessential amino acids (NEAA), and 10% fetal bovine serum (FBS) (Gibco, Carlsbad, CA) at 37°C in a 5% CO2 atmosphere. PEGylated IFN-α2b (PEG-IFN-α2b) was obtained from Schering Plough (Kenilworth, NJ). Antibodies against HCV core and β-actin were obtained from Thermo Scientific (Waltham, MA) and Sigma-Aldrich (Buchs, Switzerland), respectively. Horseradish peroxidase (HRP)-conjugated secondary antibody was obtained from Jackson ImmunoResearch (West Grove, PA). Heparin was purchased from Sigma-Aldrich (St. Louis, MO).
Peptide synthesis and purification.
The peptides were synthesized by the solid-phase method using 9-fluorenylmethoxy carbonyl (Fmoc) chemistry. Fmoc-Lys(Boc)-Wang resin (193 mg, 0.1 mmol) was well swelled in dimethylformamide (DMF; 3 ml) for 3 h before the synthesis. The peptides were cleaved from the resin by treatment with trifluoroacetic acid (TFA)-1,2-ethanedithiol-(EDT)-triisopropylsilane (TIS)-H2O (5.7:0.7:0.3:0.3) for 2 h. The crude peptides were purified by preparative Shimadzu LC-6A reverse-phase high-performance liquid chromatography (RP-HPLC) using a Kromasil C18 column (250 mm by 10-mm inner diameter, 5-μm particle size, and 100-Å pore size; Akzo Nobel, Bohus, Sweden) with a linear AB gradient (0.1% acetonitrile/min) at a flow rate of 2 ml/min. Eluent A was 0.1% aqueous TFA in water, and eluent B was 0.1% TFA in acetonitrile. The peptides were further characterized by mass spectrometry and amino acid analysis. All peptides were initially dissolved in dimethyl sulfoxide (DMSO; Sigma-Aldrich) at a concentration of 2 mM for use as stocks for screening and treatment.
Analytical RP-HPLC of peptides.
The peptides were analyzed on a Shimadzu LC-20A high-performance liquid chromatograph. Runs were performed on a Zorbax 300 SB-C8 column (150 mm by 4.6-mm inner diameter, 5-μm particle size, and 300-Å pore size; from Agilent Technologies, Santa Clara, CA) using a linear AB gradient (1% acetonitrile/min) and a flow rate of 1 ml/min. Eluent A was 0.1% aqueous TFA, and eluent B was 0.1% TFA in acetonitrile.
CD spectroscopy.
Circular dichroism (CD) spectra were measured with a 0.02-cm path length quartz cuvette on a J-810 spectropolarimeter (Jasco, Easton, MD) at 25°C. The data were collected from 250 to 190 nm at a sensitivity of 100 millidegrees, a response time of 1 s, a bandwidth of 1.0 nm, and a scan speed of 100 nm/min. The peptide concentration of 75 μM was measured in benign buffer (50 mM KH2PO4-K2HPO4, 100 mM KCl, pH 7) or benign buffer with 50% 2,2,2-trifluoroethanol (TFE). The mean residue molar ellipticities were calculated by the equation [θ] = θ/10lcMn, where θ is the ellipticity in millidegrees, l is the optical path length of the cuvette in centimeters, cM is the peptide concentration in M, and n is the number of residues in the peptide. The values of the mean residue molar ellipticities of the peptide analogs at 208 nm were used to determine the relative helicity of the peptides.
HCV grown in cell culture.
The production of JFH-1 (genotype 2a, provided by T. Wakita) HCV cell culture (HCVcc) virus was performed as previously described (8). The firefly luciferase reporter gene containing Jc1-luc chimeric HCVcc consists of core, E1, E2, p7, and the amino-terminal 33 amino acids of NS2 from the J6/CF isolate but the carboxy-terminal part of NS2 and the remaining proteins from JFH-1 (9). Infections were scored by determination of the viral genomic RNA copy number or by measuring the luciferase activity in the cell lysates using a firefly luciferase assay system from Promega (Madison, WI).
qRT-PCR.
Quantifications of HCV RNA were performed by quantitative real-time RT-PCR (qRT-PCR), as previously described (10). Briefly, the total intracellular RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. The primer pair 5′-GTCTAGCCATGGCGTTAGTA-3′ and 5′-CTCCCGGGGCACTCGCAAGC-3′ was used to quantify the highly conservative 5′ untranslated region (UTR) of the HCV genome (10). Real-time quantitative PCR was performed using the QuantiFast SYBR green RT-PCR kit (Qiagen, Germantown, MD) on an ABI Prism 7900 system.
HCV pseudotyped particles and the viral entry assay.
Packaging of HCV pseudotyped particles (HCVpp) or vesicular stomatitis virus G protein (VSV-G)-pseudotyped lentivirus (VSV-Gpp) was performed as previously described (11). Briefly, HEK293T cells were seeded 1 day before transfection at 2.5 × 106 cells in a 10-cm plate in 10 ml of DMEM containing 10% FBS. The next day, the cells were transfected using TransMax transfection reagent (Giantagen, Beijing, China). The transfecting DNA mixture (1 ml) was composed of 10 μg of pNL-4.3-Luc-E−R− and either 15 μg of the HCV E1E2 expression plasmid (genotypes 1a, 2a, 3a, 4c, and 6) (12) or 3 μg of pHEF-VSV-G (expressing VSV-G Env protein). Culture supernatants containing HCVpp or VSV-Gpp were collected at 48, 60, and 72 h posttransfection and filtered through a 0.22-μm syringe filter (11). To conduct the infection assay, Huh7.5.1 cells were seeded in a 48-well plate at a density of 5 × 104/well the day before infection. The next day, 200 μl of supernatant containing HCVpp or VSV-Gpp was added into each well in the presence of 8 μg/ml of Polybrene and 2 μl of 2 M HEPES (pH 7.55) and then spin infected for 1.5 h in a tabletop centrifuge (Beckman; 2,500 × g, 30°C), followed by another 1.5 h of incubation in a CO2 cell incubator. The cells were lysed at 48 h postinfection and assayed with the luciferase assay system (Promega) in a Modulus microplate luminometer (TurnerBioSystems, Sunnyvale, CA).
Cytotoxicity assay.
To investigate the cellular toxicity of peptide, Huh7.5.1, HepG2, or HeLa cells were seeded in a 96-well plate at a density of 1 × 105/well and kept at 37°C in an atmosphere of 5% CO2 for 24 h. The next day, all cell lines were treated with the peptide for 4 h or 24 h, and then 20 μl of 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) solution (5 mg/ml in phosphate-buffered saline [PBS]) was added to all wells. After incubation for 4 h at 37°C, the MTT was removed and 150 μl of DMSO was added. The mixture was shaken, and the formazan crystals were fully dissolved for approximately 10 min. The value of each well was measured at a test wavelength of 490 nm. The cell growth curve was plotted according to the A values.
HCVcc binding assay.
HCVcc was mixed with GBVA10-9 (2 μM), C5A (8 μM), scrambled peptide (2 μM), or heparin (200 μg/ml) for 1 h at 37°C and then added to precooled Huh7.5.1 cells that were seeded in triplicate in 12-well plates for 2 h at 4°C. The resulting cells were washed three times with cold PBS, followed by total RNA isolation with TRIzol reagent (Invitrogen). Quantification of the RNA was conducted using a QuantiFast SYBR green RT-PCR kit (Qiagen).
Velocity sedimentation ultracentrifugation.
HCVcc was concentrated by ultrafiltration using Amicon Ultra-15 centrifugal filter devices (Millipore) according to the manufacturer's protocol. The concentrated virus was divided into three tubes and treated with GBVA10-9 (2 μM), C5A (18 μM), or scrambled peptide (2 μM) for 2 h at 37°C. The resulting materials were loaded on a 10 to 60% sucrose gradient (12.5-ml total volume) and centrifuged at 200,000 × g for 1 h at 4°C in an SW41Ti rotor (Beckman). Finally, the sucrose gradient fractions (10 fractions in total) were carefully collected for HCV RNA quantification by RT-PCR, while the density of each fraction was weighed and recorded. At the same time, the virus-containing sucrose gradients of fractions 3 and 4 were transferred to naïve Huh7.5.1 cells and the infectivity titer of each fraction was evaluated 48 h thereafter.
Fusion assay.
The inhibition by peptides of HCV envelope protein-mediated cell fusion was determined as previously described (13).
Western blotting.
For cell lysate preparation, the monolayer cells were lysed with lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Nonidet P-40, 50 mM NaF, 1 mM Na3VO4, 5 mM β-glycerophosphate, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride) supplemented with a protease inhibitor cocktail (Sigma-Aldrich) on ice. The lysate was cleared by centrifugation at 14,000 × g for 20 min. The samples were boiled in 2× SDS loading buffer and loaded onto a 10% polyacrylamide gel. After electrophoresis, the separated proteins were transferred onto a nitrocellulose membrane (Bio-Rad, Hercules, CA). The resulting blots were blocked with 10% milk for 1 h and then incubated with a primary monoclonal antibody specific to HCV core or β-actin overnight at 4°C. The HRP-linked goat anti-mouse secondary antibody was applied to the immunoblot at a 1:2,000 dilution. An enhanced chemiluminscence (ECL) reagent (Amersham Biosciences, Piscataway, NJ) was used as the substrate for detection.
Statistical analysis.
The assays were performed in triplicate. Data from repeated experiments were expressed as means ± standard deviations (SD). The statistical analyses were performed using SigmaPlot 10 and Prism5, and P values of less than 0.05 (obtained by the Student t test) were considered statistically significant.
RESULTS
Screening of flavivirus-derived anti-HCV peptides.
The recently identified virucidal peptide C5A, derived from the N terminus of HCV NS5A protein, demonstrated a wide spectrum of activities against HCV and HIV (6, 14). In addition to HCV, the family Flaviviridae consists of West Nile virus (WNV), dengue virus (DEN), yellow fever virus (YFV), GBV, bovine viral diarrhea virus (BVDV), classical swine fever virus (CSFV), and several tick-borne encephalitis viruses. NS5 proteins of these viruses were inferred to have significant structural and functional similarities. To test the hypothesis that peptides from the C5A-homologous sequences in other flaviviruses might interfere with HCV infection, we synthesized eight peptides containing 18 amino acids of the corresponding C5A sequence (Table 1). The C5A peptide was derived from the N terminus of the HCV NS5A sequence and showed a perfect amphipathic characteristic. Based on these criteria, we analyzed the extreme N-terminal 18 to 22 amino acids of the selected Flaviviridae virus NS5 proteins. Some of the sequences adopt the predicted amphipathic α-helical-like structure (GBV-A, BVDV, and CSFV), and the most amphipathic portion was selected. However, some do not possess the similar structure (WNV, DEN, GBV-B, GBV-C, and YFV), and the first 18 amino acids were simply selected for those viruses.
Table 1.
Sequences and anti-HCV activities of C5A-corresponding peptides from different flaviviruses
| No. | Name | Sequence | Amphipathicity | IC50 (μM) | % inhibition at 20 μM |
|---|---|---|---|---|---|
| 1 | C5A | SWLRDIWDWICEVLSDFK | Yes | 2 | 98 |
| 2 | WNV | AKGSRAIWFMWLGARFLE | No | >20 | 0 |
| 3 | DEN | KREKKLGEFGKAKGSRAI | No | >20 | 0 |
| 4 | YFV | WVLNNPHMKDKTTVKEWR | No | >20 | 0 |
| 5 | GBV-A | LLDWCVRLGRYLLRRLKT | Yes | 10 | 86 |
| 6 | GBV-B | IWQYVCNFFVICFNVLKA | No | >20 | 3 |
| 7 | GBV-C | LWEWIMRQVRMVMSRLRA | No | >20 | 13 |
| 8 | BVDV | VLDLIYSLHKQINRGLKK | Yes | >20 | 7 |
| 9 | CSFV | ILELLYKFRDNIKSSVRE | Yes | >20 | 6 |
Using the Jc1-Luc HCVcc system, serially diluted peptides were mixed with the virus and inoculated into Huh7.5.1 cells. The 50% inhibitory concentrations (IC50) of all peptides were determined. As summarized in Table 1, a GBV-A-derived peptide (no. 5) showed antiviral activity with an IC50 of 10 μM, whereas the other peptides had activities against HCV infection with an IC50 higher than 20 μM.
Peptide design and antiviral activity.
To further enhance the anti-HCV activity of the GBV-A NS5A-derived peptide (no. 5), the tryptophan at position 4 and cysteine at position 5 were interchanged to reach a theoretically perfect amphipathic structure, and then a series of length and positional optimizations was performed (Table 2). The interchanged amino acids are underlined in the first sequence in Table 2. When tested for anti-HCV activity using the Jc1-Luc HCVcc system, peptide GBVA10 (no. 19) (obtained by removing from the parental peptide N-terminal 3 residues and extending the C terminus by 3 residues) achieved the most potent activity, by a factor of 5, with an IC50 of 2 μM (Table 2). Interestingly, GBVA10 (no. 19) still showed perfect amphipathic properties (Fig. 1A). In addition, peptides GBVA4, GBVA5, and GBVA8 were obtained, and they displayed slightly improved activity, but GBVA2, GBVA3, GBVA6, GBVA7, GBVA9, GBVA11, and GBVA 12 displayed less activity.
Table 2.
Optimization of the GBVA peptide sequence and antiviral activity
| No. | Name | Sequencea | Length (aab) | Description | IC50 (μM) | % inhibition at 20 μM |
|---|---|---|---|---|---|---|
| 10 | GBVA1 | LLDCWVRLGRYLLRRLKT | 18 | aa 1–18 | 10 | 88 |
| 11 | GBVA2 | LLDCWVRLGRYLLRRLKTPFTRL | 23 | aa 1–23 | 15 | 50 |
| 12 | GBVA3 | LLDCWVRLGRYLLRRLKTPFT | 21 | aa 1–21 | >20 | 0 |
| 13 | GBVA4 | LLDCWVRLGRYLLRRLKTP | 19 | aa 1–19 | 3 | 99 |
| 14 | GBVA5 | LLDCWVRLGRYLLRRLK | 17 | aa 1–17 | 5 | 99 |
| 15 | GBVA6 | LDCWVRLGRYLLRRLKTPFTRL | 22 | aa 2–23 | >20 | 0 |
| 16 | GBVA7 | CWVRLGRYLLRRLKTPFTRL | 20 | aa 4–23 | 12 | 89 |
| 17 | GBVA8 | LDCWVRLGRYLLRRLKTP | 18 | aa 2–19 | 4 | 99 |
| 18 | GBVA9 | DCWVRLGRYLLRRLKTPF | 18 | aa 3–20 | 12 | 90 |
| 19 | GBVA10 | CWVRLGRYLLRRLKTPFT | 18 | aa 4–21 | 2 | 100 |
| 20 | GBVA11 | WVRLGRYLLRRLKTPFTR | 18 | aa 5–22 | 12 | 58 |
| 21 | GBVA12 | VRLGRYLLRRLKTPFTRL | 18 | aa 6–23 | >20 | 0 |
The underlining in the first sequence indicates interchanged amino acids.
aa, amino acids.
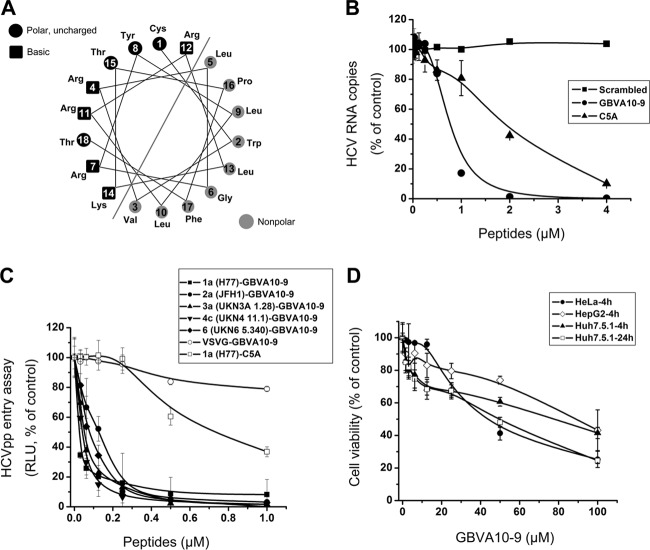
Fig 1.
Characterization of the anti-HCV activity of GBVA10-9. (A) The helical wheel analysis demonstrates that GBVA10 is amphipathic. (B) Serially diluted GBVA10-9 or C5A was mixed with HCVcc (multiplicity of infection = 0.1) and then immediately added to Huh7.5.1 cells and incubated for 12 h before the medium was changed. The HCV RNA was quantified at 48 h after infection. A scrambled peptide (TCFWLVTRKLLGRRRYLL) was included as a negative control. (C) VSV-Gpp and HCVpp packaged with envelope proteins from major genotypes were used to infect Huh7.5.1 cells in the presence of serially diluted GBVA10-9 for entry assay. A C5A-treated genotype 1a HCVpp was included as a control. GBVA10-9 inhibits multiple genotypes of HCVpp, with IC50 ranging from 0.02 to 0.15 μM. The IC50 of C5A is about 0.76 μM. The results are calculated relative to those for DMSO-treated cells. (D) Cytotoxicity of GBVA10-9 measured by MTT assay. The peptide was incubated either with HeLa, HepG2, or Huh7.5.1 cells for 4 h or with Huh7.5.1 cells for 24 h before measurements.
Next, we utilized peptide GBVA10 (no. 19) as a framework to systematically introduce amino acid substitutions to modulate its hydrophobicity, helicity, and electrical charge and evaluated their anti-HCV activity using genotype 1a HCVpp. For the hydrophobic group, two central locations at positions 9 and 6 were selected as the amino acid substitution sites because the central-location substitutions have a greater effect on the peptide secondary structure and hydrophobicity (15, 16). As shown in Table 3, several amino acids (Lys, Ser, Ala, Val, and Leu) representing a wide range of hydrophobicities were selected to replace the original amino acids to change the hydrophobicity and the number of i→i + 3 and i→i + 4 hydrophobic interactions. Here “i” means any position number (i) of an amino acid residue in the peptide sequence.
Table 3.
Design and sequences of α-helical anti-HCV peptides
| Group | No. | Peptide | No. of net charge | No. of hydrophobic interactions | Sequencea | IC50 (μM) for H77 HCVpp |
|---|---|---|---|---|---|---|
| Parental | 19 | GBVA10 | +5 | 5 | CWVRLGRYLLRRLKTPFT | 2 |
| Hydrophobic group | 22 | GBVA10-1(L9K) | +6 | 3 | CWVRLGRYKLRRLKTPFT | >20 |
| 23 | GBVA10-2(L9S) | +5 | 3 | CWVRLGRYSLRRLKTPFT | 18 | |
| 24 | GBVA10-3(L9A) | +5 | 3 | CWVRLGRYALRRLKTPFT | 2 | |
| 25 | GBVA10-4(L9V) | +5 | 5 | CWVRLGRYVLRRLKTPFT | 16 | |
| 26 | GBVA10-5(G6A) | +5 | 5 | CWVRLARYLLRRLKTPFT | 2.5 | |
| 27 | GBVA10-6(G6V) | +5 | 9 | CWVRLVRYLLRRLKTPFT | 1.25 | |
| 28 | GBVA10-7(G6L) | +5 | 9 | CWVRLLRYLLRRLKTPFT | 1.25 | |
| Helical group | 28 | GBVA10-7(G6L) | +5 | 9 | CWVRLLRYLLRRLKTPFT | 1.25 |
| 29 | GBVA10-8(G6L/P16L) | +5 | 10 | CWVRLLRYLLRRLKTLFT | 1.25 | |
| 30 | GBVA10-9(P16L) | +5 | 6 | CWVRLGRYLLRRLKTLFT | 0.16 | |
| 31 | GBVA10-10(L9I) | +5 | 5 | CWVRLGRYILRRLKTPFT | 10 | |
| 32 | GBVA10-11(L9I/L13I) | +5 | 5 | CWVRLGRYILRRIKTPFT | 0.63 | |
| 33 | GBVA10-12(L5I/L9I/L13I) | +5 | 5 | CWVRIGRYILRRIKTPFT | >20 | |
| Electric group | 34 | GBVA10-13(C1K/Y8K/T15K/T18K) | +9 | 5 | KWVRLGRKLLRRLKKPFK | >20 |
| 35 | GBVA10-14(C1K/Y8K/T15K) | +8 | 5 | KWVRLGRKLLRRLKKPFT | >20 | |
| 36 | GBVA10-15(Y8K/T15K) | +7 | 5 | CWVRLGRKLLRRLKKPFT | >20 | |
| 37 | GBVA10-16(Y8K) | +6 | 5 | CWVRLGRKLLRRLKTPFT | >20 | |
| 38 | GBVA10-17(C1E/Y8E/T15E) | +2 | 5 | EWVRLGRELLRRLKEPFT | >20 | |
| 39 | GBVA10-18(C1E/Y8E/T15E/T18E) | +1 | 5 | EWVRLGRELLRRLKEPFE | >20 | |
| 40 | GBVA10-19(Y8E/T15E) | +3 | 5 | CWVRLGRELLRRLKEPFT | >20 | |
| 41 | GBVA10-20(Y8E) | +4 | 5 | CWVRLGRELLRRLKTPFT | >20 |
Bold italic letters indicate the substituted amino acids of the parental peptide; all amino acids are l-amino acids.
The peptide hydrophobicity, helicity, and electric charge showed significant effects on anti-HCV activity (Table 3). The peptide hydrophobicity was determined by the retention times during RP-HPLC (Table 4). It is clear that decreasing the peptide hydrophobicity with the amino acid substitutions resulted in the loss of anti-HCV activity of GBVA10. Exhibiting the same anti-HCV activity, peptide GBVA10-3 (no. 24) is the only exception among the peptide analogs with a lower hydrophobicity than that of GBVA10, indicating that alanine has an effect similar to that of the original leucine on antiviral activity. In contrast, peptide anti-HCV activity was generally improved by increasing the peptide hydrophobicity.
Table 4.
Biophysical data of the peptide analogs
| Peptidea | Benignb |
50% TFEc |
tR (min)e | ||
|---|---|---|---|---|---|
| [θ]222 | % helixd | [θ]222 | % helixd | ||
| GBVA10 | −1,700 | 5.59 | −15,400 | 50.57 | 37.47 |
| GBVA10-1(L9K) | −1,700 | 5.59 | −8,600 | 28.33 | 31.47 |
| GBVA10-2(L9S) | −1,250 | 4.12 | −6,650 | 21.88 | 32.59 |
| GBVA10-3(L9A) | −950 | 3.12 | −8,750 | 28.80 | 33.57 |
| GBVA10-4(L9V) | −2,250 | 7.45 | −14,550 | 47.79 | 35.72 |
| GBVA10-5(G6A) | −3,200 | 10.60 | −20,000 | 65.73 | 40.85 |
| GBVA10-6(G6V) | −3,550 | 11.61 | −25,300 | 83.10 | 43.88 |
| GBVA10-7(G6L) | −4,050 | 13.39 | −26,050 | 85.84 | 44.94 |
| GBVA10-7(G6L) | −4,050 | 13.39 | −26,050 | 85.84 | 44.94 |
| GBVA10-8(G6L/P16L) | −6,400 | 21.14 | −30,400 | 100.00 | 51.59 |
| GBVA10-9(P16L) | −2,050 | 6.79 | −24,900 | 81.91 | 43.72 |
| GBVA10-10(L9I) | −2,900 | 9.56 | −15,550 | 51.06 | 37.46 |
| GBVA10-11(L9I/L13I) | −2,550 | 8.32 | −13,200 | 43.37 | 36.46 |
| GBVA10-12(L5I/L9I/L13I) | −4,400 | 14.48 | −11,350 | 37.31 | 36.09 |
| GBVA10-13(C1K/Y8K/T15K/T18K) | −750 | 2.50 | −9,900 | 32.47 | 28.97 |
| GBVA10-14(C1K/Y8K/T15K) | −450 | 1.52 | −12,000 | 39.41 | 30.89 |
| GBVA10-15(Y8K/T15K) | −1,450 | 4.74 | −20,300 | 66.68 | 33.67 |
| GBVA10-16(Y8K) | −2,900 | 9.52 | −15,250 | 50.20 | 34.53 |
| GBVA10-17(C1E/Y8E/T15E) | −2,350 | 7.71 | −17,950 | 58.96 | 36.28 |
| GBVA10-18(C1E/Y8E/T15E/T18E) | −4,300 | 14.16 | −13,000 | 42.80 | 36.44 |
| GBVA10-19(Y8E/T15E) | −4,600 | 15.16 | −18,600 | 61.23 | 37.16 |
| GBVA10-20(Y8E) | −2,200 | 7.24 | −18,250 | 59.94 | 37.33 |
Peptides are ordered by relative hydrophobicity during RP-HPLC at 25°C.
The mean residue molar ellipticities, [θ]222 (degree · cm2 · dmol−1) at a wavelength of 222 nm, were measured at 25°C in KP buffer (100 mM KCl, 50 mM PO4; pH 7.0).
The mean residue molar ellipticities, [θ]222 (degree · cm2 · dmol−1) at a wavelength of 222 nm, were measured at 25°C in KP buffer diluted 1:1 (vol/vol) with TFE.
The helical content (percentage) of a peptide relative to the molar ellipticity value of peptide G6L/P16L in 50% TFE.
tR, retention time at 5°C and 80°C during the RP-HPLC temperature profiling.
In this study, the helicity of GBVA10 was optimized through amino acid substitutions. As shown in Table 4, the peptide helicity was determined by the values of the molar ellipticity at 222 nm. In general, increasing the peptide helicity resulted in much better anti-HCV activity than decreasing the helicity, indicating the crucial role of peptide helicity for the inhibition of HCV (Tables 3 and 4). By substituting proline for the helix-favoring amino acid leucine, peptide GBVA10-9 (no. 30) was obtained, which displayed extremely high anti-HCV activity (IC50 = 0.16 μM). It is interesting that GBVA10-11 (no. 32) showed good anti-HCV activity (IC50 = 0.63 μM), but with a lower helicity than that of the parental peptide. This phenomenon may be attributed to the synergistic effect of the residues at positions 9 and 13 in the amino acid sequence.
The peptide charge also played an important role during the inhibition of HCV. In this study, it is clear that only peptide analogs with a net charge of 5 showed anti-HCV activity; increasing or decreasing the peptide electric charge resulted in the loss of anti-HCV activity (Tables 3 and 4).
In summary, by de novo design, we obtained a novel peptide GBVA10-9 (no. 30) that displayed improved anti-HCV activity at low (nanomolar) concentrations. In contrast, a scrambled peptide as a negative control failed to inhibit HCV.
GBVA10-9 blocks HCV entry.
GBVA10-9 inhibited HCVcc infection at IC50 of 0.75 μM, while the IC50 of C5A is about 2 μM in our system (Fig. 1B). To investigate in detail the steps influenced by GBVA10-9 during the HCV life cycle, the IC50 of GBVA10-9 was determined using HCVpp carrying envelope proteins derived from major HCV genotypes. The IC50 of GBVA10-9 ranged from 0.02 to 0.15 μM, depending on the genotypic origin of the envelope proteins (Fig. 1C). However, GBVA10-9 had little effect on the entry of VSV-G-pseudotyped lentivirus, suggesting a specific inhibition mechanism (Fig. 1C).
To rule out any confounding effects due to cytotoxicity, three distinct cell lines (HepG2, HeLa, and Huh7.5.1) were used to determine the 50% lethal concentration (LC50) of GBVA10-9. The LC50s of GBVA10-9 for 4 h of treatment were determined to be 40 μM for HeLa and 90 μM for both Huh7.5.1 and HepG2 cells. For a 24-h peptide treatment, the LC50 was 50 μM for Huh7.5.1 cells (Fig. 1D). The therapeutic index (LC50/IC50) of GBVA10-9 ranged from 250 to 560, depending on the cell types used. These results suggest that GBVA10-9 was able to suppress HCV at noncytotoxic concentrations.
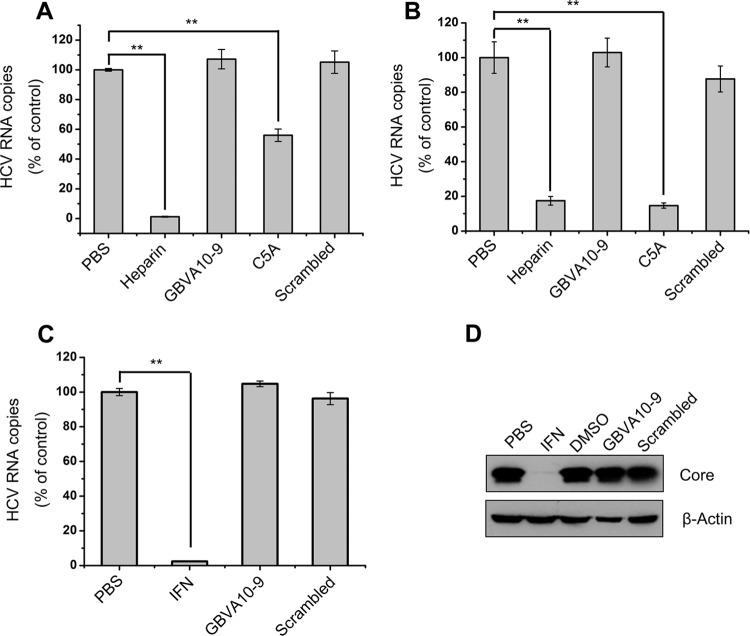
HCV entry into hepatocytes includes virion apolipoprotein-mediated attachment and receptor-mediated cell entry. Accordingly, we assessed the effect of GBVA10-9 on HCV attachment and found that HCVcc binding at low temperatures (4°C) to Huh7.5.1 cells was not affected by GBVA10-9 (Fig. 2A). After extensive washing off of the peptides, the Huh7.5.1 cells with attached HCV at 4°C were moved back to 37°C for 2 additional days of culture, and virus infection was evaluated. As shown in Fig. 2B, GBVA10-9 had no inhibitory effect on HCV infection that was significantly distinct from those of C5A and the positive control heparin. These results suggest that the virus attachment step is not the action target of GBVA10-9.
Fig 2.
GBVA10-9 does not affect HCV attachment or viral RNA replication. (A) HCVcc was mixed with GBVA10-9 (2 μM), C5A (8 μM), scrambled peptide (2 μM), or the HCV attachment inhibitor heparin (200 μg/ml) and then added to precooled Huh7.5.1 cells and incubated for 2 h at 4°C. The cells were then thoroughly washed in PBS and subjected to RNA isolation. Cell surface-bound HCV was quantified by qRT-PCR. **, P < 0.005. (B) After virus attachments and washes at 4°C as described above, a set of parallel cells were moved back to 37°C and incubated for 2 additional days. The intracellular HCV RNA titer was measured. (C) GBVA10-9 (2 μM), scrambled peptide (2 μM), or IFN-α2b (100 U/ml) was added to 2−3+ cells for 12 h before the medium was changed. At 72 h postinfection, intracellular HCV RNA was quantified by qRT-PCR. (D) Huh7.5.1 cells were infected with HCVcc (multiplicity of infection = 0.01) for 12 h and then treated with GBVA10-9 (2 μM), scrambled peptide (2 μM), DMSO (0.5%), PBS, or IFN-α2b (100 U/ml) for an additional 3 days, during which fresh peptides or IFN-α2b was added every 24 h. At 72 h postinfection, immunoblotting was performed with an anti-HCV core antibody.
GBVA10-9 does not inhibit HCV RNA replication.
To explore the possible involvement of GBVA10-9 in HCV genome replication, we used an HCV 1b subgenotype replicon (2−3+) to evaluate the effect of GBVA10-9. As shown in Fig. 2C, treatment of the replicon cells with 2 μM GBVA10-9 did not result in a reduction of the intracellular viral RNA abundance, whereas 100 U/ml of IFN-α2b strongly inhibited HCV replication. Correspondingly, the intracellular HCV core protein was inhibited by IFN-α2b but not GBVA10-9 in replicon cells (data not shown).
Next, we assessed the effect of GBVA10-9 on persistent HCV infection. Huh7.5.1 cells that were already infected with HCVcc were supplemented with GBVA10-9. In this study, IFN-α2b and a scrambled peptide were used as positive and negative controls, respectively. All analytes were retained in the medium for 72 h; the HCV core protein content was then detected. As shown in Fig. 2D, the HCV core protein was not altered by GBVA10-9, suggesting that GBVA10-9 does not suppress established HCV infection.
Inhibitory mechanisms of GBVA10-9.
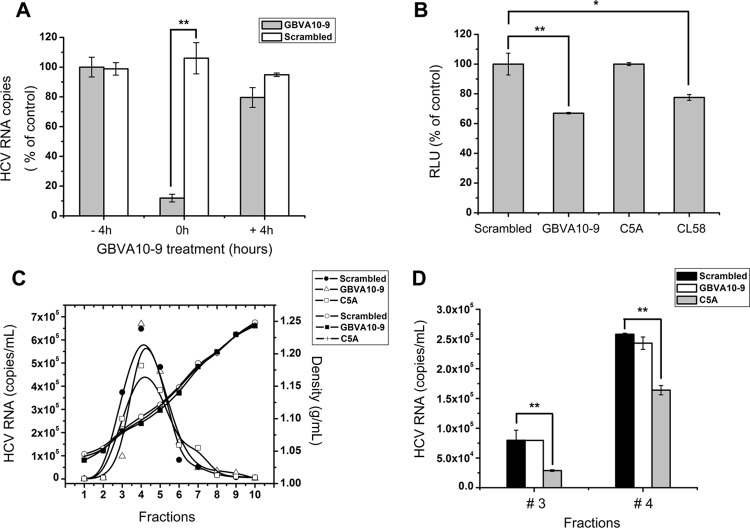
To gain more insight into the mechanism of GBVA10-9-dependent inhibition, we first assessed the possible targets upon which GBVA10-9 acts through a time-of-addition experiment. As shown in Fig. 3A, it was observed that GBVA10-9 inhibited HCV infection only when it was added to the cells together with the virus (0h) and not when it was added 4 h prior to or after infection, thus confirming the inhibitory effect of GBVA10-9 on viral entry. Pretreating the cells with GBVA10-9 for 4 h and then washing out the peptide and infecting the cells with the virus (− 4h) did not result in any inhibition of HCV, suggesting that GBVA10-9 did not exert its virus suppression effect via modifying cellular events (Fig. 3A). Low-pH-dependent HCV membrane fusion, which is a critical step during virus entry, requires both viral envelope proteins and cellular factors. To determine the effect of GBVA10-9 on HCV envelope protein-mediated membrane fusion, we set up a sensitive cell-cell fusion assay as previously described (13). The addition of GBVA10-9 significantly reduced the cell-cell fusion (Fig. 3B), which is similar to results obtained with our previously identified fusion-inhibitory peptide CL58 (13). In contrast, the control peptide, C5A, failed to exert any effect (Fig. 3B). These results support the idea that GBVA10-9 inhibits HCV entry at late steps.
Fig 3.
Antiviral mechanism of GBVA10-9. (A) GBVA10-9 (2 μM) and scrambled peptide (2 μM) were added to Huh7.5.1 cells 4 h prior to (− 4h) or after (+ 4h) inoculation of HCVcc (multiplicity of infection = 0.1) or together with the virus (0h). For the − 4h cells, peptides were washed away before virus inoculation, whereas the 0h and + 4h cells were incubated with peptides along with the virus for a total of 12 h before the medium was changed. At 72 h postinfection, the intracellular HCV RNA was quantified by qRT-PCR. (B) Fusion inhibition assay. 293T cells expressing Stop-Luc and HCV receptor CLDN1 (recipient cells) were mixed at a 1:1 ratio with donor cells expressing Cre and HCV E1E2 (H77, genotype 1a) to initiate cell-cell fusion. The indicated peptides were added, and luciferase activity was measured 24 h thereafter. (C) Concentrated HCVcc was equally divided into PBS containing GBVA10-9 (2 μM), C5A (18 μM), or scrambled peptide (2 μM), incubated at 37°C for 2 h, and then analyzed by velocity sedimentation ultracentrifugation. After weighing, the sucrose gradient fractions were collected for RNA extraction. The amount of HCV RNA was quantified by qRT-PCR and plotted on the left y axis, while the density of each fraction was plotted on the right y axis. (D) The virus-containing sucrose gradients of fraction 3 and 4 as described for panel C were transferred to naïve Huh7.5.1 cells, and the infectivity titer of each fraction was evaluated 48 h thereafter.
To investigate whether GBVA10-9 was directly virucidal to HCV, the concentrated HCVcc was pretreated with either GBVA10-9, C5A, or scrambled peptide for 2 h at 37°C and then loaded onto a 10 to 60% sucrose gradient to measure the sedimentation velocity by rate zonal ultracentrifugation, as previously described (13). Each sucrose gradient fraction was weighed and then analyzed for the HCV load by quantitative reverse transcription real-time PCR (qRT-PCR). As shown in Fig. 3C, GBVA10-9 and the scrambled peptide displayed similar profiles of peaks of viral RNA, but C5A demonstrated a distinct pattern, with significantly lower virion abundance in some fractions. The densities of each fraction among treatments were nearly identical (Fig. 3C). In addition, the HCV infectivity titer of each fraction in the density gradient was evaluated by infecting naïve Huh7.5.1 cells. The infectivities in fractions 3 and 4 are shown in Fig. 3D. The infectivity in C5A-treated fractions was significantly lower than in those treated with GBVA10-9 or scrambled peptide (Fig. 3D). These results suggest that GBVA10-9 did not disrupt the structural integrity of HCV. Collectively, the results show that the antiviral mechanism of GBVA10-9 is different from that of C5A.
DISCUSSION
Approximately 2.2 to 3% of the world's population is chronically infected with HCV. Depending on the genotype of the infecting virus, 30 to 50% of chronically infected patients cannot clear the virus upon treatment with PEG-IFN combined with ribavirin. HCV entry is a complicated process that represents a promising target for novel anti-HCV drug development. In the present study, we designed and identified a new peptide inhibitor of HCV entry that might represent a leading candidate for HCV therapeutic drug design. We demonstrated that peptide GBVA10-9 inhibits HCVcc as well as HCVpp entry with a pan-genotypic method. Importantly, GBVA10-9 inhibits HCV infection at a low (nanomolar), noncytotoxic level. We also demonstrated that GBVA10-9 does not suppress HCV attachment to hepatocytes and may act at the postbinding step of entry.
Synthetic peptides represent promising candidates for antiviral investigations. T20 and congeneric peptides have been successfully applied clinically for the treatment of HIV-infected patients (17–20). Recently, Cheng and colleagues reported a novel virucidal peptide named C5A with a typical amphipathic α-helix structure that can inhibit both the initiation of HCV infection and ongoing HCV infection (6). The unique mechanism of C5A action suggests that it might not select resistant mutations. Evidence that C5A was derived from the membrane anchor domain of the HCV NS5A protein served as the impetus for the search for C5A-like HCV-inhibitory peptides from other flaviviruses. Through extensive screening and optimization, we identified and designed a peptide from the GBV-A NS5A protein with improved anti-HCV properties.
Deinhardt and colleagues reported in 1967 a new infectious agent that caused human viral hepatitis and could be passaged in marmosets (21). The transmissible agent was named the “GB agent” after the initials of the original patient; this agent was studied extensively as a putative cause of non-A, non-B hepatitis. The currently known GB viruses are GBV-A, GBV-B, GBV-C (hepatitis G virus), and GBV-D, and they are taxonomically closely related to HCV. Like HCV infection, GBV-A infection frequently leads to persistent viremia, resulting in lifelong infection (22–24). Although primary amino acid sequences differed considerably, a putative amphipathic α-helical structure feature was predicted in the N-terminal domains of the NS5A proteins from GBV-A and HCV (25). To our surprise, after the initial suggestion of a GBV-A NS5A-derived peptide (no. 5) with anti-HCV activity, a highly potent HCV entry-inhibitory peptide, GBVA10-9 (no. 30), was obtained by de novo design by improving amphipathicity and modifying helicity. GBVA10-9 blocks virus entry in both HCVcc and HCVpp models at a low (nanomolar) concentration. Importantly, the effect of GBVA10-9 on HCV entry is genotype independent. Further modifications of the peptide to improve its bioavailability and antiviral activity and to decrease its possible cytotoxicity may be necessary to develop a novel category of potent anti-HCV drugs in human therapy.
In this study, we attempted to investigate the inhibitory mechanisms of GBVA10-9 on HCV. Unlike C5A, GBVA10-9 does not disrupt the structural integrity of HCV. Our data demonstrate that GBVA10-9 can inhibit virus entry at a postattachment step, whereas GBVA10-9 affects neither HCV binding to Huh7.5.1 cells nor viral RNA replication and does not suppress established HCV infection. We speculate that GBVA10-9 may work at the moment that the virus binds to the host cell, or it may influence virus entry by affecting the integrity of host cell and virus membranes. Detailed mechanistic studies are urgently needed to understand its inhibitory nature; these investigations will be helpful to further improve the peptide for use as a therapeutic agent.
In conclusion, we identified a novel and potent GBV-A NS5A-derived peptide that interferes with entry of multiple genotypes of HCV. This new category of virus entry inhibitors might be potentially used to prevent de novo HCV infection during liver transplantation and unsafe blood practices.
ACKNOWLEDGMENTS
We thank T. Wakita, C. Rice, F. Chisari, G. Luo, and F. Cosset for providing cell lines and reagents.
This work was supported by grants from the Chinese Science and Technology Key Project (2012ZX10002007-003 and 2008ZX10002-014), grants from the National Basic Research Program of China (2011CB504800) and the National Natural Science Foundation of China (81271831 and 30970156) to W.Y., a grant from the Natural Science Foundation of Jilin Province (201015103) to Y.C., and grants from the Youth Foundation of Jilin Province (20100126) and the China Postdoctoral Science Foundation (20110491293), as well as a Basic Scientific Research Grant of Jilin University, to Y.H.
Footnotes
Published ahead of print 21 November 2012
REFERENCES
- 1. Di Bisceglie AM, McHutchison J, Rice CM. 2002. New therapeutic strategies for hepatitis C. Hepatology 35:224–231 [DOI] [PubMed] [Google Scholar]
- 2. Jacobson IM, McHutchison JG, Dusheiko G, Di Bisceglie AM, Reddy KR, Bzowej NH, Marcellin P, Muir AJ, Ferenci P, Flisiak R, George J, Rizzetto M, Shouval D, Sola R, Terg RA, Yoshida EM, Adda N, Bengtsson L, Sankoh AJ, Kieffer TL, George S, Kauffman RS, Zeuzem S. 2011. Telaprevir for previously untreated chronic hepatitis C virus infection. N. Engl. J. Med. 364:2405–2416 [DOI] [PubMed] [Google Scholar]
- 3. Zeuzem S, Andreone P, Pol S, Lawitz E, Diago M, Roberts S, Focaccia R, Younossi Z, Foster GR, Horban A, Ferenci P, Nevens F, Mullhaupt B, Pockros P, Terg R, Shouval D, van Hoek B, Weiland O, Van Heeswijk R, De Meyer S, Luo D, Boogaerts G, Polo R, Picchio G, Beumont M. 2011. Telaprevir for retreatment of HCV infection. N. Engl. J. Med. 364:2417–2428 [DOI] [PubMed] [Google Scholar]
- 4. Poordad F, McCone J, Jr, Bacon BR, Bruno S, Manns MP, Sulkowski MS, Jacobson IM, Reddy KR, Goodman ZD, Boparai N, DiNubile MJ, Sniukiene V, Brass CA, Albrecht JK, Bronowicki JP. 2011. Boceprevir for untreated chronic HCV genotype 1 infection. N. Engl. J. Med. 364:1195–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bacon BR, Gordon SC, Lawitz E, Marcellin P, Vierling JM, Zeuzem S, Poordad F, Goodman ZD, Sings HL, Boparai N, Burroughs M, Brass CA, Albrecht JK, Esteban R. 2011. Boceprevir for previously treated chronic HCV genotype 1 infection. N. Engl. J. Med. 364:1207–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cheng G, Montero A, Gastaminza P, Whitten-Bauer C, Wieland SF, Isogawa M, Fredericksen B, Selvarajah S, Gallay PA, Ghadiri MR, Chisari FV. 2008. A virocidal amphipathic α-helical peptide that inhibits hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 105:3088–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 102:9294–9299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Koutsoudakis G, Kaul A, Steinmann E, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. 2006. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 80:5308–5320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Komurian-Pradel F, Paranhos-Baccala G, Sodoyer M, Chevallier P, Mandrand B, Lotteau V, Andre P. 2001. Quantitation of HCV RNA using real-time PCR and fluorimetry. J. Virol. Methods 95:111–119 [DOI] [PubMed] [Google Scholar]
- 11. Yang W, Qiu C, Biswas N, Jin J, Watkins SC, Montelaro RC, Coyne CB, Wang T. 2008. Correlation of the tight junction-like distribution of Claudin-1 to the cellular tropism of hepatitis C virus. J. Biol. Chem. 283:8643–8653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bartosch B, Dubuisson J, Cosset FL. 2003. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Si Y, Liu S, Liu X, Jacobs JL, Cheng M, Niu Y, Jin Q, Wang T, Yang W. 2012. A human claudin-1-derived peptide inhibits hepatitis C virus entry. Hepatology 56:507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bobardt MD, Cheng G, de Witte L, Selvarajah S, Chatterji U, Sanders-Beer BE, Geijtenbeek TB, Chisari FV, Gallay PA. 2008. Hepatitis C virus NS5A anchor peptide disrupts human immunodeficiency virus. Proc. Natl. Acad. Sci. U. S. A. 105:5525–5530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen Y, Guarnieri MT, Vasil AI, Vasil ML, Mant CT, Hodges RS. 2007. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob. Agents Chemother. 51:1398–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen Y, Mant CT, Farmer SW, Hancock RE, Vasil ML, Hodges RS. 2005. Rational design of alpha-helical antimicrobial peptides with enhanced activities and specificity/therapeutic index. J. Biol. Chem. 280:12316–12329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wild C, Oas T, McDanal C, Bolognesi D, Matthews T. 1992. A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. U. S. A. 89:10537–10541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kilby JM, Hopkins S, Venetta TM, DiMassimo B, Cloud GA, Lee JY, Alldredge L, Hunter E, Lambert D, Bolognesi D, Matthews T, Johnson MR, Nowak MA, Shaw GM, Saag MS. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302–1307 [DOI] [PubMed] [Google Scholar]
- 19. Chen CH, Matthews TJ, McDanal CB, Bolognesi DP, Greenberg ML. 1995. A molecular clasp in the human immunodeficiency virus (HIV) type 1 TM protein determines the anti-HIV activity of gp41 derivatives: implication for viral fusion. J. Virol. 69:3771–3777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. He Y, Xiao Y, Song H, Liang Q, Ju D, Chen X, Lu H, Jing W, Jiang S, Zhang L. 2008. Design and evaluation of sifuvirtide, a novel HIV-1 fusion inhibitor. J. Biol. Chem. 283:11126–11134 [DOI] [PubMed] [Google Scholar]
- 21. Deinhardt F, Holmes AW, Capps RB, Popper H. 1967. Studies on the transmission of human viral hepatitis to marmoset monkeys. I. Transmission of disease, serial passages, and description of liver lesions. J. Exp. Med. 125:673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hoofnagle JH, Lombardero M, Wei Y, Everhart J, Wiesner R, Zetterman R, Yun AJ, Yang L, Kim JP. 1997. Hepatitis G virus infection before and after liver transplantation. Liver Transplantation Database. Liver Transpl. Surg. 3:578–585 [DOI] [PubMed] [Google Scholar]
- 23. Lauer GM, Walker BD. 2001. Hepatitis C virus infection. N. Engl. J. Med. 345:41–52 [DOI] [PubMed] [Google Scholar]
- 24. Simons JN, Pilot-Matias TJ, Leary TP, Dawson GJ, Desai SM, Schlauder GG, Muerhoff AS, Erker JC, Buijk SL, Chalmers ML, et al. 1995. Identification of two flavivirus-like genomes in the GB hepatitis agent. Proc. Natl. Acad. Sci. U. S. A. 92:3401–3405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Brass V, Pal Z, Sapay N, Deleage G, Blum HE, Penin F, Moradpour D. 2007. Conserved determinants for membrane association of nonstructural protein 5A from hepatitis C virus and related viruses. J. Virol. 81:2745–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]