Abstract
Understanding how HIV-1 persists during effective antiretroviral therapy (ART) should inform strategies to cure HIV-1 infection. We hypothesize that proliferation of HIV-1-infected cells contributes to persistence of HIV-1 infection during suppressive ART. This predicts that identical or monotypic HIV-1 DNA sequences will increase over time during ART. We analyzed 1,656 env and pol sequences generated following single-genome amplification from the blood and sputum of six individuals during long-term suppressive ART. The median proportion of monotypic sequences increased from 25.0% prior to ART to 43.2% after a median of 9.8 years of suppressive ART. The proportion of monotypic sequences was estimated to increase 3.3% per year (95% confidence interval, 2.3 to 4.4%; P < 0.001). Drug resistance mutations were not more common in the monotypic sequences, arguing against viral replication during times with lower antiretroviral concentrations. Bioinformatic analysis found equivalent genetic distances of monotypic and nonmonotypic sequences from the predicted founder virus sequence, suggesting that the relative increase in monotypic variants over time is not due to selective survival of cells with viruses from the time of acute infection or from just prior to ART initiation. Furthermore, while the total HIV-1 DNA load decreased during ART, the calculated concentration of monotypic sequences was stable in children, despite growth over nearly a decade of observation, consistent with proliferation of infected CD4+ T cells and slower decay of monotypic sequences. Our findings suggest that proliferation of cells with proviruses is a likely mechanism of HIV-1 DNA persistence, which should be considered when designing strategies to eradicate HIV-1 infection.
INTRODUCTION
Models developed after the introduction of highly active antiretroviral (ARV) therapy (ART) theorized complete inhibition of HIV-1 replication and estimated that due to the short half-life of infected cells (1.6 days), infection would be cured after 3 years of therapy (1). Subsequent observations revised upwards the calculated half-life of HIV-1-infected cells to between 4.6 and 44 months (2, 3), making viral eradication with currently available ART improbable. A thorough understanding of the mechanisms that allow the virus to persist despite effective treatment may point to interventions that could cure HIV-1 infection.
HIV-1 persists primarily in resting memory CD4+ T cells (4–9), although other cell types (10), such as monocytic lineages, may also serve as reservoirs in blood (11) and tissues (12–14). Low-level plasma viremia, generally below the limit of detection of clinical assays, is common during ART (15–19). In a subset of individuals, increasing genetic distances indicate HIV-1 replication (19–25). However, the majority of individuals have no evidence of viral evolution (19, 26, 27), and often, the sequences generated from low-level viremias are monotypic (15, 19, 28–31). The latter observations led us to evaluate the hypothesis that proliferation of HIV-1-infected cells is a mechanism that contributes to HIV-1 persistence during suppressive ART.
Multiple observations support the hypothesis (4, 19, 27, 32) that HIV-1 persists due to proliferation of infected cells. First, populations of monotypic HIV-1 env sequences have been observed in plasma (15, 19, 29) and genital tract (33) specimens. Second, central memory CD4+ T cells have proliferative potential (34) and a relatively high rate of HIV-1 infection, and phylogenetic analyses of HIV-1 in these cells demonstrate genetic patterns consistent with cellular proliferation (4, 15, 19, 29).
Testable predictions of the hypothesis that cellular proliferation contributes to the persistence of HIV-1 infection are that during suppressive ART, monotypic HIV-1 DNA sequences will increase in proportion and that the concentration of this HIV-1 subpopulation will not decay over time. To test this prediction, HIV-1 DNA sequences were derived following single-genome amplification (SGA) from longitudinal specimens collected before and during a median of 10 years of suppressive ART and the concentrations of unique and monotypic sequences were estimated over time.
MATERIALS AND METHODS
Ethics statement.
This project was approved by the Human Subjects Committee of the Institutional Review Board at Seattle Children's Hospital. All participants and their guardians provided informed written consent and/or assent, as appropriate for their age.
Participants.
HIV-1-infected children and adolescents were enrolled into a prospective observational study, described in more detail previously (35). Peripheral blood and induced sputum specimens were collected annually to evaluate HIV-1 sequences in lymphocytes and alveolar macrophages. Participants who remained on effective ART (plasma HIV-1 RNA load, <30 to 50 copies/ml at >80% of clinic visits) and who had blood specimens available from before initiation of effective ART and blood and sputum available from ≥3 time points after initiation of effective ART were selected for this study.
Processing of sputum specimens.
Sputum was induced by ultrasonic nebulization (Ultra-Neb99; DeVilbiss, Somerset, PA) of hypertonic (3%) saline for five sequential 4-min episodes. Sputum was treated with 10% dithiothreitol (Calbiochem, San Diego, CA) for 1 to 2 h and resuspended in phosphate-buffered saline, and cells were quantified (Z1 Coulter Counter; Beckman, Brea, CA). Differential cell counts were performed after Wright-Giemsa staining. Peripheral blood mononuclear cells (PBMCs) were isolated from peripheral blood using Accuspin tubes (Sigma-Aldrich, St. Louis, MO).
Extraction, amplification, sequencing, and quantification of HIV-1 DNA.
DNA was extracted from PBMCs and sputum cells using a Gentra DNA purification system (Qiagen, Valencia, CA). The first round of PCR was multiplexed to amplify pol and env, followed by separate second-round reactions to amplify the reverse transcriptase (RT) and protease regions of pol and the C2-V5 region of env, as previously described (19, 35). Quantification of HIV-1 DNA was performed by nested PCR of serially diluted nucleic acid using the QUALITY program (36). The number of copies of HIV-1 DNA per ml of peripheral blood was estimated on the basis of the HIV-1 DNA load per million PBMCs divided by the CD4 cell percentage (based on the assumption that HIV-1 DNA was derived from CD4 lymphocytes) and then multiplied by the number of CD4 cells per ml of blood.
Sequencing and phylogenetics.
Bidirectional sequences were generated, edited, and screened for hypermutation as described previously (35). For each participant, any previously reported HIV-1 sequences were also included in the analysis (GenBank accession numbers FJ446721 to FJ446779, FJ446811 to FJ446835, FJ446888 to FJ446944, FJ446986 to FJ447082, FJ447104 to FJ447132, FJ447190 to FJ447267, FJ447310 to FJ447337) (35). Sequences were aligned with the ClustalW program (37), and maximum-likelihood phylogenetic trees were constructed using the DIVEIN web tool (http://indra.mullins.microbiol.washington.edu/DIVEIN/) (38) and the defaults standard as of November 2010. Sequences from the source of infection (the mother of each pediatric participant) were used to root the phylogenetic tree, when possible; otherwise, several reference sequences of the same subtype as the participant were selected from the Los Alamos National Laboratory HIV Sequence Database. The FigTree (version 1.3.1) program (http://tree.bio.ed.ac.uk/software/figtree/) was used to graphically represent the trees. Neighbor-joining (NJ) phylogenetic analysis of all sequences from all participants was performed using the Seaview program (39) to evaluate potential cross-contamination.
Screening for resampling and contamination.
Limiting dilution PCR precludes resampling of PCR-amplified genomes. The sequence data were generated over several years in at least three different physical locations, and all the labs had physical and protocol safeguards designed to prevent contamination. To investigate the possibility of contamination or sample mix-up, all of the sequences for each gene region (env and pol) were used to construct a single NJ tree. These trees were examined to confirm that all sequences from each participant formed separate distinct clusters.
Definition of monotypic sequences.
Monotypic sequences were defined, using the DIVEIN program output, as two or more sequences with a pairwise distance of zero. As defined by DIVEIN, sequences with an ambiguous base or shorter length could have a zero pairwise distance to two nonidentical sequences. To avoid this potential problem, 5 of 831 pol sequences that could not be placed into a single cluster of monotypic sequences were removed from the analysis. Clades of monotypic viruses were defined on the basis of a unique distance from the estimated most recent common ancestor (MRCA) of infection.
Identification of synonymous mutations, drug resistance mutations, coreceptor usage, unique amino acid patterns, glycosylation sites, and distance from MRCA of infection.
Synonymous sequences were defined as different nucleotide sequences that encode the same amino acid sequence. Major drug resistance mutations in RT were defined as per the International Antiviral Society-USA (http://www.iasusa.org). Coreceptor usage was predicted on the basis of the env sequence using an X4/R5 position-specific scoring matrix (PSSM) (40). Amino acids in the largest monotypic cluster that differed from the consensus of the other sequences were identified using the Viral Epidemiology Signature Pattern Analysis (VESPA) web tool at the Los Alamos National Laboratory HIV Sequence Database (41) and default settings. Potential N-glycosylation and mucin-type GalNAc O-glycosylation were identified using the NetNglyc (version 1.0) program (42) and the NetOGlyc (version 3.1) program (43) (both programs are hosted at http://www.cbs.dtu.dk/services/). Potential O-glycosylation sites were not identified in any of the env sequences. The pairwise distance from each sequence to the estimated MRCA was calculated by DIVEIN.
Statistical analysis of the proportion and concentration of monotypic sequences over time.
The overall prevalences of monotypic sequences were compared using data from time points from which all specimen types were available. A linear longitudinal mixed model with random intercepts was used to estimate the rate of change in the percentage of monotypic sequences over time and to calculate the confidence interval for the estimated slope to determine whether the slope was significantly different from zero. Models were assessed for interactions between genes and the slope of the percentage of monotypic sequences over time. For each model examined, the interaction P values were >0.1, so a weighted average of env and pol sequences was used for statistical analysis. Two similar analyses restricted to sequences from either PBMCs or sputa alone were performed. In those analyses, the sequences had to be identical to sequences derived from the same cell type in order to be defined as monotypic. The numbers of monotypic and unique sequences per ml of blood were calculated and used to estimate changes in total HIV DNA, using linear longitudinal mixed models with random intercepts to assess the slope over time.
Analysis of alternative hypotheses.
Descriptive information is provided for alternative hypotheses. Analyses of phenotypic properties was restricted to the largest cluster of identical sequences, as these clusters were less likely to have occurred by chance and more likely to have unique phenotypic properties if viral replication was responsible for the increase in identical sequences. Statistical analysis was performed to compare the distance of monotypic versus nonmonotypic sequences from the estimated MRCA using a Wilcoxon rank-sum (Mann-Whitney) analysis within each subject.
All statistical analyses were performed using the Stata SE (version 12) program (Stata Corp., College Station, TX). Two-sided P values of <0.05 were considered significant.
Nucleotide sequence accession numbers.
The pol and env sequences obtained in this study were submitted to GenBank and can be found under accession numbers KC314011 to KC315287.
RESULTS
Study population.
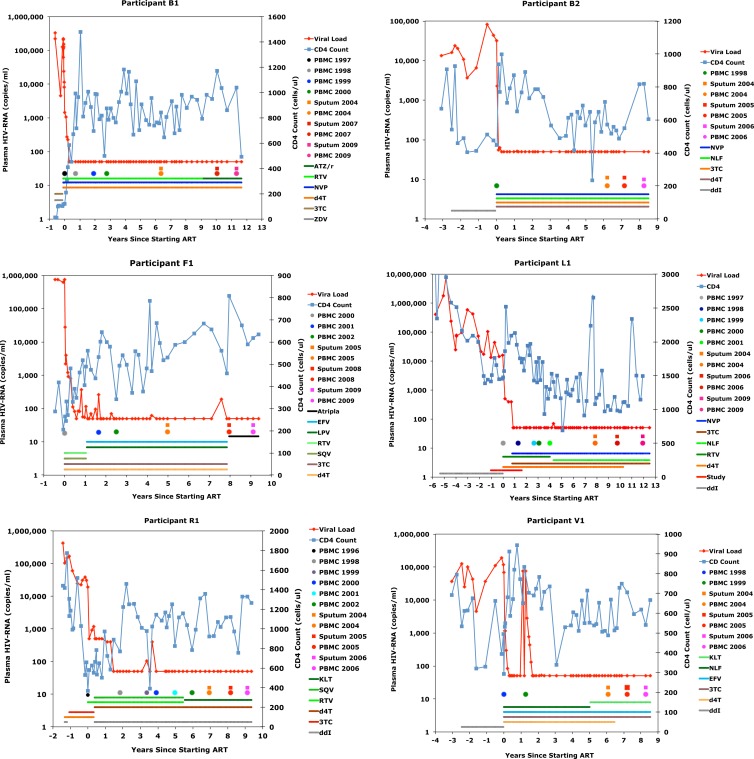
A total of 15 participants enrolled in the study. Analysis was limited to six participants with at least three blood and sputum specimens collected during ART suppression of plasma viremia. Figure 1 summarizes the clinical data for each participant. Plasma HIV-1 RNA was analyzed a median of 5.2 times per year and was undetectable (<30 to 50 copies/ml) at a median of 96% (range, 85 to 100%) of clinic visits over a median of 9.8 years (range, 8.3 to 11.4 years). Detectable HIV-1 RNA occurred at isolated clinic visits, with a median of 1.8 episodes per decade (range, 0 to 7 episodes per decade). The median detectable value was 130 copies/ml (range, 51 to 400 copies/ml), excluding a brief (<1-month) treatment interruption by participant V1 during the 2nd year of ART. The CD4 lymphocyte cell populations of all participants increased over the study; the median increase was 392 CD4 cells/μl (range, 24 to 922 CD4 cells/μl).
Fig 1.
Summary of clinical data from the six participants. Keys define the symbols and colors used for antiretrovirals, plasma HIV-1 RNA, and blood CD4+ cell concentrations. The multicolored circles and squares in the middle of the graphs indicate the dates on which specimens were collected. ATZ/r, atazanavir-ritonavir; RTV, ritonavir; NVP, nevirapine; d4T, stavudine; 3TC, lamivudine; ZDV, zidovudine; NLF, nelfinavir; ddI, dideoxyinosine; EFV, efavirenz; LPV, lopinavir; SQV, saquinavir; TNF, tenofovir.
Viral sequences analyzed.
A total of 1,656 bidirectional single-genome sequences were analyzed from the six participants, including 830 env (C2-V5 region) and 826 pol (encoding RT) sequences (Table 1). A total of 339 env and pol sequences (20%) were from specimens collected before ART; the remainder was from specimens collected during suppressive ART. Sputum specimens, all collected after the initiation of ART, contributed a total of 548 (33%) sequences. The majority of env and pol sequences (921 [56%]) were concordant, defined as being generated from the same multiplexed first-round PCR, and presumably came from the same viral template. Phylogenetic trees constructed with all sequences did not reveal contamination.
Table 1.
Summary of env and pol sequences generated from each participanta
| Participant | Total no. of sequences |
env sequence |
pol (RT) sequence |
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. | % |
No. | % |
||||||
| Pre-ART | Sputum | Concordant | Pre-ART | Sputum | Concordant | ||||
| B1 | 324 | 182 | 9 | 33 | 41 | 142 | 8 | 35 | 61 |
| B2 | 179 | 100 | 11 | 41 | 49 | 79 | 22 | 38 | 97 |
| F1 | 269 | 120 | 10 | 29 | 64 | 149 | 15 | 35 | 60 |
| L1 | 295 | 104 | 7 | 26 | 44 | 191 | 13 | 21 | 51 |
| R1 | 400 | 210 | 10 | 34 | 37 | 190 | 12 | 33 | 56 |
| V1 | 189 | 114 | 11 | 43 | 61 | 75 | 17 | 43 | 93 |
| Median | 282 | 117 | 10 | 32 | 47 | 145.5 | 14 | 34 | 61 |
| Total | 1,656 | 830 | 826 | ||||||
Concordant sequences are defined by amplification of both env and pol in the same first-round PCR. Because the region of env sequenced is longer and more variable than the RT sequence, more env sequencing reactions failed, resulting in a greater percentage of concordant RT sequences.
Characterization of monotypic sequences.
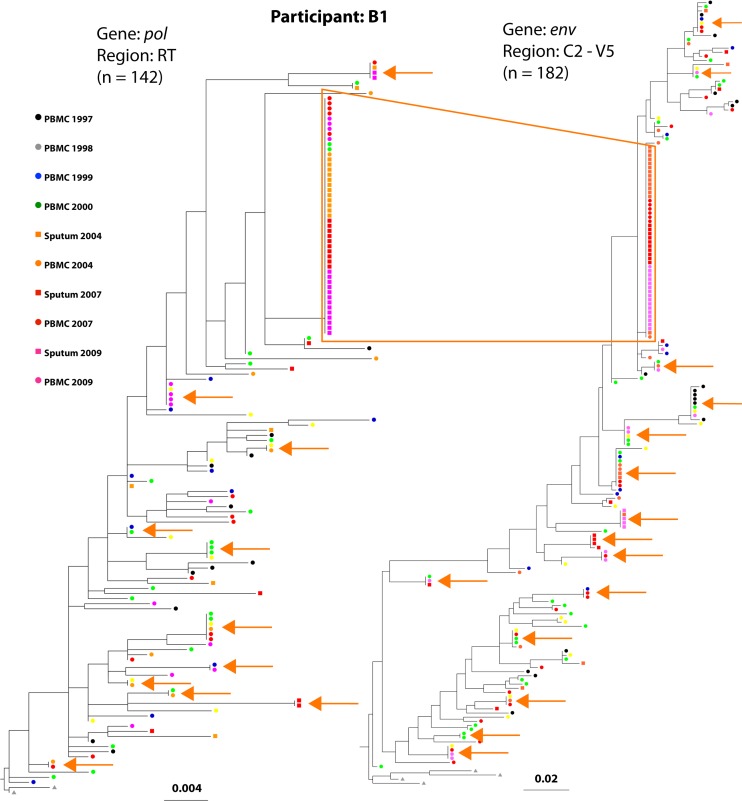
Sequences identical to at least one other sequence were defined as monotypic sequences and comprised 44% of all sequences. Analysis of all sequences across the six participants showed 182 clusters of monotypic sequences, 75% of which spanned multiple time points. In addition, all six individuals had at least one large clade of monotypic env and pol sequences that included more than 5% of their sequences. Each of these large clades included sequences from multiple time points and/or cell types (PBMCs or sputum cells). From time points with both PBMC and sputum sequences, similar proportions of sputum and PBMC sequences were monotypic (49.1% versus 45.7%; P = 0.41), as were the proportions of pol and env sequences (54.8 versus 48.5%; P = 0.28). Although the number and size of the monotypic clades varied across participants, phylogenetic analyses revealed monotypic sequences distributed throughout each participant's tree, from near the MRCA to the tips of the branches (results for participant B1 are shown in Fig. 2, and those for the remaining five participants are shown in Fig. S1A to J in the supplemental material).
Fig 2.
Phylogenetic trees of participant B1's HIV sequences demonstrate a large cluster of monotypic sequences. The phylogenetic trees were modeled on pol and env sequences from participant B1, who had the largest cluster of monotypic sequences (box outlined in orange). The tree constructed with pol sequences encoding reverse transcriptase is shown on the left, and the env tree is shown on the right. The tips of the trees are color coded to correspond to the dates and tissue source (see key). Concordant env and pol sequences, generated from the same multiplexed first-round reaction of the specimen diluted beyond a single template per reaction (single genome amplification), comprised 57% of the sequences in the largest monotypic cluster. Smaller clusters of monotypic sequences are indicated with orange arrows.
Increasing proportion of monotypic sequences through time.
The prevalence of monotypic pol and env sequences increased steadily in each individual during suppressive ART. Curves for individual participants and the linear-fit curve are shown in Fig. 3. The proportion of monotypic env and pol sequences increased at rates of 3.2% per year (see Fig. S2A in the supplemental material) and 3.5% per year (see Fig. S2B in the supplemental material) (interaction P = 0.1), and the weighted average increased 3.3% per year (95% confidence interval [CI], 2.3 to 4.4%; P < 0.001) (Fig. 3). At the final study time point, the proportion of monotypic sequences ranged from 21 to 90%. In separate analyses of the weighted average in PBMC sequences alone or sputum sequences alone, similar patterns were observed. The proportion of monotypic sequences in PBMCs increased at a rate of 2.5% per year (95% CI, 1.4 to 3.6%; P < 0.001) (see Fig. S2C in the supplemental material). Sputum was not collected until after the start of ART, limiting the analysis of sputa to a narrower time period, but the proportion of monotypic sputum sequences increased at a rate of 7.2% per year (95% CI, 2.3 to 11.9%; P = 0.003) (see Fig. S2D in the supplemental material).
Fig 3.
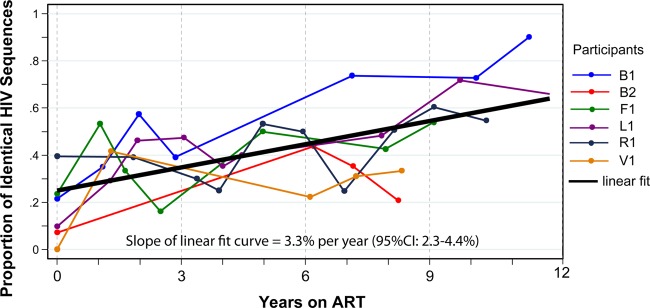
The proportions of monotypic HIV-1 sequences increased over a decade of suppressive ART. The proportions of the monotypic sequences (weighted average of env and pol) for individual participants are shown in different colors, and the linear-fit curve is shown in black. The slope and confidence interval for the linear-fit curve are given at the bottom of the graph.
Investigation of alternative hypotheses.
Phenomena other than cellular proliferation that could increase the proportion of monotypic HIV-1 DNA sequences during suppressive ART include (i) ongoing replication of a virus selected for a unique phenotype, (ii) greater relative persistence of homogeneous founder viruses, (iii) greater relative persistence of more homogeneous viruses that predominated immediately prior to ART initiation, and (iv) faster clearance of cells containing replication-competent proviruses with unique HIV-1 genotypes.
The possibility that ongoing replication of viruses produced monotypic HIV-1 subpopulations due to selection of variants with unique phenotypes was investigated by analyzing the largest monotypic cluster from each participant. First, nonmonotypic sequences were examined for accumulation of synonymous nucleotide sequences that encode the same amino acid sequence as the monotypic cluster, which would suggest ongoing viral replication with purifying selection. Among all of the nonmonotypic sequences across the participants, only 2 of 483 env (0.4%) and 5 of 441 pol (1.1%) sequences encoded the same amino acid sequence as the largest monotypic nucleotide sequence cluster from the respective participants.
Second, the pol sequences encoding RT were examined for selection of ARV resistance mutations that might explain the increase in monotypic sequence over time. Three participants administered antiretroviral monotherapy and/or dual therapy before initiating ART (participants B1, L1, and R1) had drug resistance detected in specimens collected both before and after starting ART (described previously [35]), but no new mutations in pol emerged during suppressive ART in any of the participants. Furthermore, the proportion of pol sequences with drug resistance mutations did not increase during ART, and there were no drug resistance mutations that were unique to the largest monotypic clusters.
Third, env sequences were examined for patterns in predicted coreceptor usage and glycosylation that might explain the observed increases in monotypic sequences over time. The HIV-1 env sequences of three participants (L1, R1, V1) were predicted to exclusively use the CCR5 coreceptor. The env sequences from the other three participants (B1, B2, F1) were predicted to use a mixture of CXCR4 and CCR5 coreceptors. Among these three participants, the proportion of X4 variants did not change during suppressive ART, and the predicted coreceptor usage of the largest monotypic clusters differed (X4 in participant B1 and F1 and R5 in participant B2). The amino acids encoded by the env sequences of each participant's largest monotypic cluster were compared to the consensus of his/her nonmonotypic sequences, which identified a median of 8 (range, 5 to 25) amino acid differences per participant. These amino acids were not contiguous and differed across the participants. Seven of these amino acid differences encoded N-glycosylation sites, all in the nonmonotypic sequences.
The possibility that monotypic HIV-1 variants persisted from the time of primary infection or from the time immediately prior to the initiation of ART was investigated using pairwise genetic distances. The genetic distances of monotypic and nonmonotypic sequences to the calculated MRCA of each participant were similar in 9 of 12 comparisons (Table 2). The monotypic sequences diverged slightly less than the nonmonotypic sequences in L1 pol, R1 env, and V1 pol. However, the multiple monotypic clades evident in the phylogenetic trees of these three participants do not concentrate at either the roots or the tips of the trees (see Fig. 1A, E, and I in the supplemental material), as would be expected if the monotypic viruses were the founders or those replicating at the time that ART was initiated.
Table 2.
Genetic distance of monotypic versus nonmonotypic sequences from calculated MRCA of each participant's infection
| Participant |
env |
pol (RT) |
||||
|---|---|---|---|---|---|---|
| Median (IQRa) genetic distance |
P value | Median (IQR) genetic distance |
P value | |||
| Monotypic clusters | Unique sequences | Monotypic clusters | Unique sequences | |||
| B1 | 0.059 (0.033–0.063) | 0.059 (0.041–0.065) | 0.4 | 0.010 (0.010–0.019) | 0.014 (0.010–0.017) | 1.0 |
| B2 | 0.065 (0.061–0.069) | 0.061 (0.044–0.069) | 0.3 | 0.010 (0.006–0.013) | 0.011 (0.009–0.015) | 0.2 |
| F1 | 0.035 (0.025–0.047) | 0.048 (0.036–0.051) | 0.06 | 0.007 (0.003–0.009) | 0.007 (0.005–0.007) | 0.6 |
| L1 | 0.028 (0.015–0.034) | 0.031 (0.022–0.036) | 0.4 | 0.012 (0.009–0.018) | 0.017 (0.014–0.021) | 0.04 |
| R1 | 0.016 (0.014–0.017) | 0.018 (0.015–0.022) | 0.02 | 0.009 (0.007–0.011) | 0.011 (0.007–0.013) | 0.09 |
| V1 | 0.050 (0.031–0.054) | 0.045 (0.030–0.055) | 0.5 | 0.005 (0.002–0.006) | 0.008 (0.005–0.010) | 0.008 |
IQR, interquartile ratio.
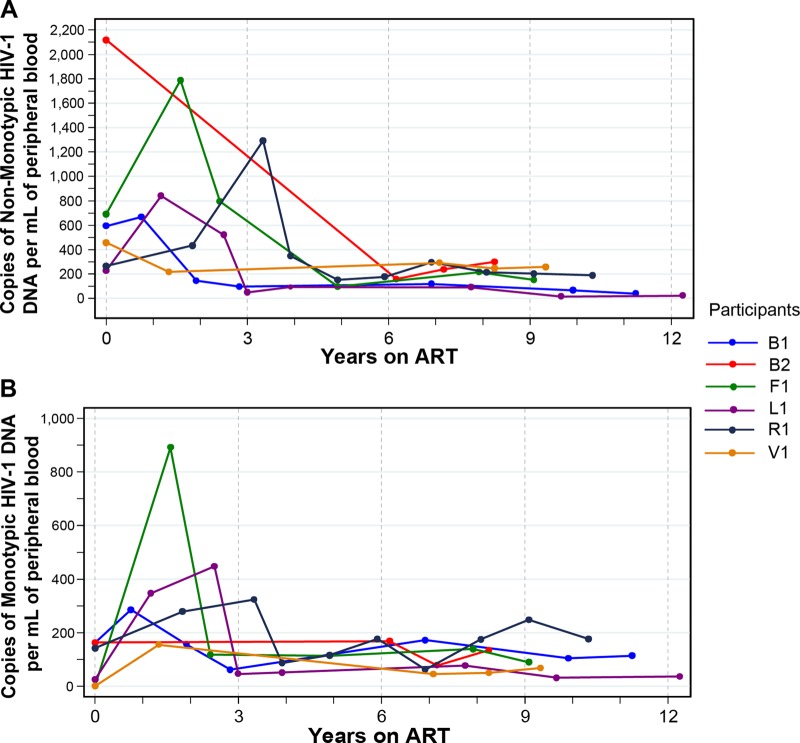
HIV-1-infected CD4 lymphocytes that produce virus likely undergo cell lysis and decay faster than cells that produce defective variants. Over time this could result in the relative predominance of defective sequences that may appear to be monotypic. In the absence of cellular proliferation, the populations of both unique and monotypic sequences would be expected to decrease, although if defective, the monotypic variants would decrease at a lower rate. This possibility was explored by estimating the change in the concentration of unique and monotypic HIV DNA sequences in the peripheral blood over time. All participants had a decrease in the estimated concentration of HIV-1 DNA per ml of blood over the time period analyzed (median decrease, 320 copies/ml; range, 42 to 1,850 copies/ml). The nonmonotypic sequences decreased in all six participants, with an estimated linear decrease of −6.2% per year (95% CI, −9.5% to −3.0; P < 0.0001) (Fig. 4A). In contrast, while the monotypic sequences decreased slightly in two participants and increased slightly in the other four, these changes were small and the overall change was not statistically significant (estimated linear decrease, −1.0% per year; 95% CI, −2.3 to −0.2% per year; P > 0.10) (Fig. 4B). The apparent stability of the concentration of monotypic sequences occurred despite a weight gain of 186% (the median weight increased from 20.4 to 58.3 kg) in these pediatric patients, which would be expected to dilute the concentration of monotypic HIV sequences without replenishment of infected cells.
Fig 4.
Change in HIV-1 DNA copy numbers per volume of peripheral blood over time of suppressive ART. The estimated numbers of nonmonotypic (A) and monotypic (B) sequences per ml are shown. The numbers of HIV-1 DNA copies per ml were estimated from the HIV-1 DNA load per million PBMCs and the CD4 count, assuming that all the HIV-1 DNA came from CD4 cells. Each colored line represents an individual participant.
DISCUSSION
This study of HIV-1 DNA sequences that persist over a decade of effective ART demonstrated a significant increase in the proportion of monotypic HIV-1 env and pol sequences. Four alternative hypotheses were considered to explain these findings. Specifically, no identifiable genotypic features of the monotypic sequences suggested ongoing viral replication. Fewer N-linked glycosylation sites in the monotypic env clusters than in nonmonotypic sequences is consistent with proliferation of cells with HIV-1 proviruses rather than ongoing viral replication with humoral immune pressure selecting glycosylated sites. The observation that monotypic sequences were not more closely related to the founder sequence or to those replicating just prior to the initiation of ART indicates that relatively more homogeneous genotypes from these stages of infection were not favored to persist. Because there was no decay in the concentration of monotypic sequences over approximately 10 years, we suspect that proliferation of HIV-1-infected cells contributes to maintaining the HIV-1 population during suppressive ART.
Several features of this study facilitated our observations of monotypic sequences during ART. First, a relatively large number of single-genome HIV-1 DNA sequences were derived from each participant, increasing the likelihood that monotypic sequences would be detected. Second, sequences were derived from longitudinal specimens collected over a decade, which increased our ability to measure changes in the prevalence of monotypic sequences. Third, the analysis of HIV-1 DNA from both PBMCs and sputa demonstrated increasing monotypic sequences in both specimen types, suggesting that this phenomenon occurs in tissues other than blood. Fourth, our primary analyses are based on the proportion of monotypic sequences and so are independent of potential errors introduced by sequence alignments or assumptions made in the phylogenetic analyses, excluding the possibility that our findings are an artifact of phylogenetic reconstruction.
Our analysis has several limitations. First, we did not investigate whether the monotypic sequences were from replication-competent viruses. The monotypic HIV-1 templates that we sequenced likely include defective proviruses that were less likely to undergo lytic infection and less likely to be cleared by cellular immune responses and therefore were enriched over time on ART. Persistence of defective forms has been described previously (44, 45). However, PBMCs harboring replication-competent HIV-1 that is genetically identical to persistent and predominant monotypic plasma variants have also been identified (28), indicating that not all identical sequences are defective. As expression of HIV-1 proteins does not necessarily result in cell death even in the presence of autologous lymphocytes (46), cells infected with replication-competent HIV-1 could persist for a prolonged period of time. Second, while our genotypic analyses did not detect evidence of ongoing viral replication, they were restricted to two gene regions and may not have had sufficient power to detect very low rates of viral replication. Cell-to-cell transmission of HIV-1 has been hypothesized to occur during effective ART by transfer of numerous virions, overwhelming intracellular ARV concentrations (47). Cells infected in this manner could appear to harbor monotypic viruses. Third, selection of cytotoxic T lymphocyte (CTL)-restricted epitopes could not be fully evaluated in this study because HLA typing of the participants was not performed. CTL-mediated clearance of HIV-1-infected cells could result in a relative increase in the proportion of monotypic sequences with CTL-escape mutations, even in the absence of viral replication or cellular proliferation. Fourth, as the PBMC and sputum cell subsets were not separated, we cannot determine whether the monotypic sequences were derived from a specific cell type. Lastly, our calculation of the HIV-1 DNA concentration in blood is only an indirect estimation of total HIV-1 and may not accurately reflect changes in the amount of HIV-1 in other lymphoid tissues, where the majority of HIV-1-infected cells are thought to reside. However, it still seems unlikely that a single replication-defective virus would infect a large number of cells, and we think that proliferation of infected cells is a more likely explanation.
Multiple questions that warrant further study are raised by this project. What proportion of monotypic HIV-1 DNA sequences represents replication-competent proviruses? Are our findings in blood and sputum representative of those in lymphoid tissues throughout the body? Are our findings unique to children/adolescents, due perhaps to greater thymic reserve and potential for immune proliferation? If proliferation of HIV-1-infected cells sustains the infections, are there interventions that can safely curtail or circumvent the proliferative response?
In addition to the proliferation of infected cells, their long half-life and low-level viral replication likely contribute to HIV-1 persistence. Each of these three mechanisms presents distinct challenges to the development of therapeutic strategies to eradicate HIV-1 infection. Extending the duration or intensity of ART will not target viruses that persist due to proliferation of cells with integrated provirus. Strategies designed to engineer CD4+ cells that resist HIV-1 infection, such as zinc finger endonuclease modification of HIV-1 coreceptors (48), may produce a functional cure. However, strategies that specifically target HIV-1-infected cells, such as therapeutic vaccines (49), will likely be needed to eliminate infection that persists due to cellular proliferation.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Foster Foundation (to L.M.F. and T.A.W.) and the National Institutes of Health, including R21 AI058723 (to L.M.F.), R01 AI091550 (to L.M.F.), K23 AI058683 (to N.H.T.), and K23 AI077357 (to T.A.W.); the University of Washington CFAR P30 AI27757; and the General Clinical Research Center at the University of Washington M01RR-00037.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 21 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.01985-12.
REFERENCES
- 1. Ho DD. 1995. Time to hit HIV, early and hard. N. Engl. J. Med. 333:450–451 [DOI] [PubMed] [Google Scholar]
- 2. Chun T-W, Justement JS, Moir S, Hallahan CW, Maenza J, Mullins JI, Collier AC, Corey L, Fauci AS. 2007. Decay of the HIV reservoir in patients receiving antiretroviral therapy for extended periods: implications for eradication of virus. J. Infect. Dis. 195:1762–1764 [DOI] [PubMed] [Google Scholar]
- 3. Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. 2003. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat. Med. 9:727–728 [DOI] [PubMed] [Google Scholar]
- 4. Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel Ghattas M-RG, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy Haddad J-PEK, Sékaly R-P. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 15:893–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183–188 [DOI] [PubMed] [Google Scholar]
- 6. Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. 1995. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat. Med. 1:1284–1290 [DOI] [PubMed] [Google Scholar]
- 7. Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A. 94:13193–13197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300 [DOI] [PubMed] [Google Scholar]
- 9. Wong JK, Hezareh M, Günthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291–1295 [DOI] [PubMed] [Google Scholar]
- 10. Chun TW, Davey RT, Jr, Ostrowski M, Shawn Justement J, Engel D, Mullins JI, Fauci AS. 2000. Relationship between pre-existing viral reservoirs and the re-emergence of plasma viremia after discontinuation of highly active anti-retroviral therapy. Nat. Med. 6:757–761 [DOI] [PubMed] [Google Scholar]
- 11. Zhu T, Muthui D, Holte S, Nickle D, Feng F, Brodie S, Hwangbo Y, Mullins JI, Corey L. 2002. Evidence for human immunodeficiency virus type 1 replication in vivo in CD14(+) monocytes and its potential role as a source of virus in patients on highly active antiretroviral therapy. J. Virol. 76:707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Edén A, Fuchs D, Hagberg L, Nilsson S, Spudich S, Svennerholm B, Price RW, Gisslén M. 2010. HIV-1 viral escape in cerebrospinal fluid of subjects on suppressive antiretroviral treatment. J. Infect. Dis. 202:1819–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Guadalupe M, Sankaran S, George MD, Reay E, Verhoeven D, Shacklett BL, Flamm J, Wegelin J, Prindiville T, Dandekar S. 2006. Viral suppression and immune restoration in the gastrointestinal mucosa of human immunodeficiency virus type 1-infected patients initiating therapy during primary or chronic infection. J. Virol. 80:8236–8247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. North TW, Higgins J, Deere JD, Hayes TL, Villalobos A, Adamson L, Shacklett BL, Schinazi RF, Luciw PA. 2010. Viral sanctuaries during highly active antiretroviral therapy in a nonhuman primate model for AIDS. J. Virol. 84:2913–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bailey JR, Sedaghat AR, Kieffer T, Brennan T, Lee PK, Wind-Rotolo M, Haggerty CM, Kamireddi AR, Liu Y, Lee J, Persaud D, Gallant JE, Cofrancesco J, Quinn TC, Wilke CO, Ray SC, Siliciano JD, Nettles RE, Siliciano RF. 2006. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J. Virol. 80:6441–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dinoso JB, Kim SY, Wiegand AM, Palmer SE, Gange SJ, Cranmer L, O'Shea A, Callender M, Spivak A, Brennan T, Kearney MF, Proschan MA, Mican JM, Rehm CA, Coffin JM, Mellors JW, Siliciano RF, Maldarelli F. 2009. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A. 106:9403–9408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hatano H, Delwart EL, Norris PJ, Lee Dunn-Williams T-HJ, Hunt PW, Hoh R, Stramer SL, Linnen JM, McCune JM, Martin JN, Busch MP, Deeks SG. 2009. Evidence for persistent low-level viremia in individuals who control human immunodeficiency virus in the absence of antiretroviral therapy. J. Virol. 83:329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maldarelli F, Palmer S, King MS, Wiegand A, Polis MA, Mican J, Kovacs JA, Davey RT, Rock-Kress D, Dewar R, Liu S, Metcalf JA, Rehm C, Brun SC, Hanna GJ, Kempf DJ, Coffin JM, Mellors JW. 2007. ART suppresses plasma HIV-1 RNA to a stable set point predicted by pretherapy viremia. PLoS Pathog. 3:e46 doi:10.1371/journal.ppat.0030046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tobin NH, Learn GH, Holte SE, Wang Y, Melvin AJ, McKernan JL, Pawluk DM, Mohan KM, Lewis PF, Mullins JI, Frenkel LM. 2005. Evidence that low-level viremias during effective highly active antiretroviral therapy result from two processes: expression of archival virus and replication of virus. J. Virol. 79:9625–9634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Doyle T, Smith C, Vitiello P, Cambiano V, Johnson M, Owen A, Phillips AN, Geretti AM. 2012. Plasma HIV-1 RNA detection below 50 copies/ml and risk of virologic rebound in patients receiving highly active antiretroviral therapy. Clin. Infect. Dis. 54:724–732 [DOI] [PubMed] [Google Scholar]
- 21. Frenkel LM, Wang Y, Learn GH, McKernan JL, Ellis GM, Mohan KM, Holte SE, De Vange SM, Pawluk DM, Melvin AJ, Lewis PF, Heath LM, Beck IA, Mahalanabis M, Naugler WE, Tobin NH, Mullins JI. 2003. Multiple viral genetic analyses detecting low-level human immunodeficiency virus type 1 replication during effective highly active antiretroviral therapy. J. Virol. 77:5721–5730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Günthard HF, Frost SD, Leigh-Brown AJ, Ignacio CC, Kee K, Perelson AS, Spina CA, Havlir DV, Hezareh M, Looney DJ, Richman DD, Wong JK. 1999. Evolution of envelope sequences of human immunodeficiency virus type 1 in cellular reservoirs in the setting of potent antiviral therapy. J. Virol. 73:9404–9412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sharkey ME, Teo I, Greenough T, Sharova N, Luzuriaga K, Sullivan JL, Bucy RP, Kostrikis LG, Haase A, Veryard C, Davaro RE, Cheeseman SH, Daly JS, Bova C, Ellison RT, Mady B, Lai KK, Moyle G, Nelson M, Gazzard B, Shaunak S, Stevenson M. 2000. Persistence of episomal HIV-1 infection intermediates in patients on highly active anti-retroviral therapy. Nat. Med. 6:76–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shiu C, Cunningham CK, Greenough T, Muresan P, Sanchez-Merino V, Carey V, Jackson JB, Ziemniak C, Fox L, Belzer M, Ray SC, Luzuriaga K, Persaud D. 2009. Identification of ongoing human immunodeficiency virus type 1 (HIV-1) replication in residual viremia during recombinant HIV-1 poxvirus immunizations in patients with clinically undetectable viral loads on durable suppressive highly active antiretroviral therapy. J. Virol. 83:9731–9742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang L, Ramratnam B, Tenner-Racz K, He Y, Vesanen M, Lewin S, Talal A, Racz P, Perelson AS, Korber BT, Markowitz M, Ho DD. 1999. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N. Engl. J. Med. 340:1605–1613 [DOI] [PubMed] [Google Scholar]
- 26. Persaud D, Ray SC, Kajdas J, Ahonkhai A, Siberry GK, Ferguson K, Ziemniak C, Quinn TC, Casazza JP, Zeichner S, Gange SJ, Watson DC. 2007. Slow human immunodeficiency virus type 1 evolution in viral reservoirs in infants treated with effective antiretroviral therapy. AIDS Res. Hum. Retroviruses 23:381–390 [DOI] [PubMed] [Google Scholar]
- 27. Persaud D, Siberry GK, Ahonkhai A, Kajdas J, Monie D, Hutton N, Watson DC, Quinn TC, Ray SC, Siliciano RF. 2004. Continued production of drug-sensitive human immunodeficiency virus type 1 in children on combination antiretroviral therapy who have undetectable viral loads. J. Virol. 78:968–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Anderson JA, Archin NM, Ince W, Parker D, Wiegand A, Coffin JM, Kuruc J, Eron J, Swanstrom R, Margolis DM. 2011. Clonal sequences recovered from plasma from patients with residual HIV-1 viremia and on intensified antiretroviral therapy are identical to replicating viral RNAs recovered from circulating resting CD4+ T cells. J. Virol. 85:5220–5223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brennan TP, Woods JO, Sedaghat AR, Siliciano JD, Siliciano RF, Wilke CO. 2009. Analysis of human immunodeficiency virus type 1 viremia and provirus in resting CD4+ T cells reveals a novel source of residual viremia in patients on antiretroviral therapy. J. Virol. 83:8470–8481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Buzón MJ, Codoñer FM, Frost SDW, Pou C, Puertas MC, Massanella M, Dalmau J, Llibre JM, Stevenson M, Blanco J, Clotet B, Paredes R, Martinez-Picado J. 2011. Deep molecular characterization of HIV-1 dynamics under suppressive HAART. PLoS Pathog. 7:e1002314 doi:10.1371/journal.ppat.1002314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Evering TH, Mehandru S, Racz P, Tenner-Racz K, Poles MA, Figueroa A, Mohri H, Markowitz M. 2012. Absence of HIV-1 evolution in the gut-associated lymphoid tissue from patients on combination antiviral therapy initiated during primary infection. PLoS Pathog. 8:e1002506 doi:10.1371/journal.ppat.1002506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rong L, Perelson AS. 2009. Modeling HIV persistence, the latent reservoir, and viral blips. J. Theor. Biol. 260:308–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bull ME, Learn GH, McElhone S, Hitti J, Lockhart D, Holte S, Dragavon J, Coombs RW, Mullins JI, Frenkel LM. 2009. Monotypic human immunodeficiency virus type 1 genotypes across the uterine cervix and in blood suggest proliferation of cells with provirus. J. Virol. 83:6020–6028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, Longworth SA, Vinup KE, Mrass P, Oliaro J, Killeen N, Orange JS, Russell SM, Weninger W, Reiner SL. 2007. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science 315:1687–1691 [DOI] [PubMed] [Google Scholar]
- 35. Wagner TA, Tobin NH, McKernan JL, Xu M, Melvin AJ, Mohan KM, Learn GH, Mullins JI, Frenkel LM. 2009. Increased mutations in Env and Pol suggest greater HIV-1 replication in sputum-derived viruses compared with blood-derived viruses. AIDS 23:923–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rodrigo AG, Goracke PC, Rowhanian K, Mullins JI. 1997. Quantitation of target molecules from polymerase chain reaction-based limiting dilution assays. AIDS Res. Hum. Retroviruses 13:737–742 [DOI] [PubMed] [Google Scholar]
- 37. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 38. Deng W, Maust BS, Nickle DC, Learn GH, Liu Y, Heath L, Kosakovsky Pond SL, Mullins JI. 2010. DIVEIN: a web server to analyze phylogenies, sequence divergence, diversity, and informative sites. Biotechniques 48:405–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gouy M, Guindon S, Gascuel O. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27:221–224 [DOI] [PubMed] [Google Scholar]
- 40. Jensen MA, Li F-S, van't Wout AB, Nickle DC, Shriner D, He H-X, McLaughlin S, Shankarappa R, Margolick JB, Mullins JI. 2003. Improved coreceptor usage prediction and genotypic monitoring of R5-to-X4 transition by motif analysis of human immunodeficiency virus type 1 env V3 loop sequences. J. Virol. 77:13376–13388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Korber B, Myers G. 1992. Signature pattern analysis: a method for assessing viral sequence relatedness. AIDS Res. Hum. Retroviruses 8:1549–1560 [DOI] [PubMed] [Google Scholar]
- 42. Gupta R, Jung E, Gooley AA, Williams KL, Brunak S, Hansen J. 1999. Scanning the available Dictyostelium discoideum proteome for O-linked GlcNAc glycosylation sites using neural networks. Glycobiology 9:1009–1022 [DOI] [PubMed] [Google Scholar]
- 43. Julenius K, Mølgaard A, Gupta R, Brunak S. 2005. Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15:153–164 [DOI] [PubMed] [Google Scholar]
- 44. Josefsson L, Eriksson S, Sinclair E, Ho T, Killian M, Epling L, Shao W, Lewis B, Bacchetti P, Loeb L, Custer J, Poole L, Hecht FM, Palmer S. 2012. Hematopoietic precursor cells isolated from patients on long-term suppressive HIV therapy did not contain HIV-1 DNA. J. Infect. Dis. 206:28–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kearney, et al. Abstr. 17th Conf. Retroviruses Opportunistic Infect., abstr. 98.2010. [Google Scholar]
- 46. Shan L, Deng K, Shroff NS, Durand CM, Rabi SA, Yang H-C, Zhang H, Margolick JB, Blankson JN, Siliciano RF. 2012. Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity 36:491–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sigal A, Kim JT, Balazs AB, Dekel E, Mayo A, Milo R, Baltimore D. 2011. Cell-to-cell spread of HIV permits ongoing replication despite antiretroviral therapy. Nature 477:95–98 [DOI] [PubMed] [Google Scholar]
- 48. Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee Y-L, Guschin DY, Rupniewski I, Waite AJ, Carpenito C, Carroll RG, Orange JS, Urnov FD, Rebar EJ, Ando D, Gregory PD, Riley JL, Holmes MC, June CH. 2008. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat. Biotechnol. 26:808–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fuller DH, Rajakumar P, Che JW, Narendran A, Nyaundi J, Michael H, Yager EJ, Stagnar C, Wahlberg B, Taber R, Haynes JR, Cook FC, Ertl P, Tite J, Amedee AM, Murphey-Corb M. 2012. Therapeutic DNA vaccine induces broad T cell responses in the gut and sustained protection from viral rebound and AIDS in SIV-infected rhesus macaques. PLoS One 7:e33715 doi:10.1371/journal.pone.0033715 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.