Abstract
An immune correlates analysis of the RV144 HIV-1 vaccine trial revealed that antibody responses to the gp120 V1/V2 region correlated inversely with infection risk. The RV144 protein immunogens (A244-rp120 and MN-rgp120) were modified by an N-terminal 11-amino-acid deletion (Δ11) and addition of a herpes simplex virus (HSV) gD protein-derived tag (gD). We investigated the effects of these modifications on gp120 expression, antigenicity, and immunogenicity by comparing unmodified A244 gp120 with both Δ11 deletion and gD tag and with Δ11 only. Analysis of A244 gp120, with or without Δ11 or gD, demonstrated that the Δ11 deletion, without the addition of gD, was sufficient for enhanced antigenicity to gp120 C1 region, conformational V2, and V1/V2 gp120 conformational epitopes. RV144 vaccinee serum IgGs bound more avidly to A244 gp120 Δ11 than to the unmodified gp120, and their binding was blocked by C1, V2, and V1/V2 antibodies. Rhesus macaques immunized with the three different forms of A244 gp120 proteins gave similar levels of gp120 antibody titers, although higher antibody titers developed earlier in A244 Δ11 gp120-immunized animals. Conformational V1/V2 monoclonal antibodies (MAbs) gave significantly higher levels of blocking of plasma IgG from A244 Δ11 gp120-immunized animals than IgG from animals immunized with unmodified A244 gp120, thus indicating a qualitative difference in the V1/V2 antibodies induced by A244 Δ11 gp120. These results demonstrate that deletion of N-terminal residues in the RV144 A244 gp120 immunogen improves both envelope antigenicity and immunogenicity.
INTRODUCTION
The RV144 vaccine trial in Thailand demonstrated an estimated vaccine efficacy of 31.2% in preventing HIV-1 acquisition in a heterosexual population (1). A previous trial involving high-risk intravenous drug users (IVDU) using AIDSVAX B/E (2–5) had not shown protection (6, 7). The RV144 vaccine comprises a canarypox ALVAC prime with the E.92TH023 gp120 membrane-anchored insert and an AIDSVAX B/E gp120 boost. This vaccine regimen induced Env antibody responses in 98.6% and CD4 T cell responses in 90.1% of vaccinated subjects (6) and induced tier 1 virus- but not tier 2 virus-neutralizing antibodies (1). The majority of breakthrough infections in RV144 trial were subtype CRF01_AE, (89% and 91.7% in the infected and placebo groups, respectively) (6), suggesting that the immune responses elicited against the clade E gp120 A244 Env protein were involved in lowering infection risk of HIV-1 acquisition.
The target of potentially protective or neutralizing antibodies is the trimeric Env spike, which is sparsely present on HIV-1 virions (8, 9). Neutralizing epitopes presented on gp120 may be masked by glycans, may be exposed only transiently following receptor/coreceptor engagement, or may depend strongly on intact quaternary structures (10–12). A major hurdle in HIV-1 Env protein vaccine design is the preservation of the structural properties in soluble versions of Env proteins that mimic those on intact viruses (13), particularly when the Env gp120 proteins are expressed as monomers. Furthermore, the gp120 inner domains and the coreceptor binding epitopes can be occluded in dimeric (and probably misfolded) forms of recombinant gp120, which are often produced by mammalian cells together with gp120 monomers (14). Thus, optimal presentation of neutralizing epitopes on gp120 depends critically on its conformational state.
A number of conformational V2 antibodies that bind well to epitopes on scaffolded murine leukemia viruses (gp70-V1/V2) and to other recently described V1/V2 scaffold proteins have been described (15–19). A clonal lineage of V1/V2 conformational gp120 broadly neutralizing antibodies (bnAbs) CH01 to CH04, which show blocking by the prototype V1/V2 conformational gp120 monoclonal antibodies (MAbs) PG9 and PG16, bind to only a subset of gp120 monomers, including clade E.A244 gp120 (20). Although previously described as quaternary-structure-specific MAbs, with preferential binding to membrane-anchored trimeric HIV Env (21), PG9 and PG16 bnAbs can bind to monomeric and trimeric forms of some gp140 (22) and to rare monomeric gp120 as well (20). The PG9 bnAb has been crystallized bound to a V1/V2 scaffold protein and shown to bind primarily to the V1/V2 C-β strand and to adjacent glycans (17). Thus, those V1/V2 conformational bnAbs for which PG9 is a prototype bind to a conformational peptidoglycan epitope of gp120 V1/V2 (17). The RV144 Env, A244-rgp120 (20), a component of AIDSVAX B/E (2, 5) is among the rare monomeric gp120s to which the CH01-to-CH04 and PG9 antibodies bind. The unmutated ancestor antibodies of the CH01-to-CH04 clonal lineage also bind A244 gp120 monomers, with an affinity within the range appropriate for B-cell receptor triggering (20).
One unusual feature of the RV144 protein gp120 design was that the proteins were constructed with a herpes simplex virus (HSV) gD peptide tag and an 11-amino-acid (aa) deletion at the gp120 N terminus (2, 5). Could features of the A244-rgp120 design have contributed to enhanced exposure of V1/V2 and V2 conformational epitopes on the vaccine proteins? If so, induction of antibodies with specificity for the more prominently exposed epitopes might be observed in RV144 vaccinees. A recently conducted analysis of the RV144 case-control study showed that antibody responses were to the C1, V2, V3, and C5 gp120 regions and that high levels of IgG antibodies to a V1/V2 scaffold protein correlated inversely with HIV-1 infection rate in vaccinees (23). Thus, one hypothesis is that addition of the gD tag and/or the Δ11 mutation provided enhanced presentation of certain gp120 epitopes and contributed to the induction of V1/V2 antibody responses in RV144-vaccinated subjects.
We report here that the RV144 gp120 protein immunogen, A244-rgp120, was associated with enhanced antigenicity for C1, V2, and V1/V2 conformational epitopes and that the gp120 N-terminal deletion (Δ11) without inclusion of the HSV gD tag is sufficient for enhanced antigenicity and immunogenicity in both humans and rhesus macaques.
MATERIALS AND METHODS
Proteins and antibodies.
RV144 vaccine immunogen proteins (Table 1) A244-rgp120 and MN-rgp120 were produced originally by Genentech, Inc., further developed by VaxGen Inc., and supplied for this study by GSID (Global Solutions for Infectious Diseases, South San Francisco, CA). A244 gp120, A244 gDΔ11, A244 Δ11, A244 gD N160K, MN gDΔ11, and MN gp120 were expressed in 293T cells (Table 1; Fig. 1) and lectin affinity purified (24) followed by size exclusion chromatography (SEC) on a Superdex 200 FPLC (GE Healthcare) to homogeneity for monomeric gp120. Expression of additional gp120 proteins with N-terminal deletion included the subtype B (63521 and 6240) and subtype C (C.089C and C.1086) Env proteins described earlier (20, 25, 26). N-terminal deletion for all Env gp120 involved 11 aa, except for C.1086, in which the corresponding shorter N-terminal segment (7 aa) of the mature Env protein was deleted. AE.A244 V1/V2 Tags and B.Case A2 V1/V2 Tags proteins were constructed with an Ig leader sequence (METDTLLLWVLLLWVPGSTGD) as a protein cleavage and secretion signal at the N terminus and Avi-tag followed by a His6 tag and were produced in 293F cells by transfection and purified by nickel columns (H.-X. Liao, M. Bonsignori, S. M. Alam, J. S. McLellan, G. D. Tomaras, and B. F. Haynes, unpublished data). Synagis (palivizumab; MedImmune LLC, Gaithersburg, MD), a human respiratory syncytial virus (RSV) MAb, was used as a negative control. The C1 MAb A32, the V3 MAb 19b, and gp41 immunodominant MAb 7B2 were supplied by James Robinson (Tulane University, New Orleans, LA). CH01 MAb as previously described was isolated, and its unmutated ancestor antibodies were inferred, from IgG+ memory B cells of a broad neutralizer subject (20). Conformational V2 MAbs 697-D, 830A, 2158, and 697-D Fab were provided by S.Z.-P. (New York University, NY) and described previously (15, 27). V1/V2 conformational/quaternary MAbs PG9 and PG16 were provided by Dennis Burton (IAVI and Scripps Research Institute, La Jolla, CA), and the CD4 binding site MAb VRC01 was from the Vaccine Research Institute, NIH (Bethesda, MD). CH58 MAb, recently isolated from the plasma of an RV144 vaccinee by culturing and screening of memory B cells, binds to the V1/V2 region of gp120 (Liao et al., unpublished).
Table 1.
Env gp120 protein constructs used in the study
| Env protein | gD peptide | N-terminal deletion size (aa) |
|---|---|---|
| A244-rgp120a | + | 11 |
| MN-rgp120a | + | 11 |
| A244 gp120 | − | |
| A244 gD Δ11 gp120 | + | 11 |
| A244 Δ11 gp120 | − | 11 |
| MN gp120 | − | |
| MN gD Δ11 gp120 | + | 11 |
| MN Δ11 gp120 | − | 11 |
| 92TH023 gp120 | − | |
| 92TH023 gD Δ11 gp120 | + | 11 |
| 63521 Δ11 gp120 | − | 11 |
| 6240 Δ11 gp120 | − | 11 |
| 1086 Δ7 gp120b | − | 7 |
RV144 vaccine immunogen proteins A244-rgp120 and MN-rgp120 were produced by Genentech Inc., developed by VaxGen Inc., and supplied by GSID.
1086 Env, in which the corresponding N-terminal segment (7 aa) is shorter, was designed with Δ7 deletion.
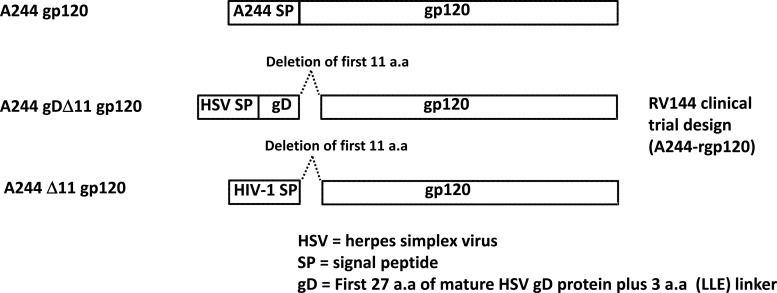
Fig 1.
Diagram of HIV-1 gp120 proteins constructs with modifications. Designs for each of the A244 proteins either with no modifications (gp120), with both the HSV gD tag (first 27 aa of mature HSV gD protein) and N-terminal 11-aa deletion (gDΔ11 gp120), or with only the 11-aa deletion (Δ11 gp120) are outlined. The gDΔ11 gp120 constructs for all three proteins are similar with respect to modifications to the design used in the RV144 vaccine trial for A244-rgp120 and MN-rgp120. The Env gp120 proteins were expressed and purified as described in Materials and Methods. A similar design was used to construct gp120 proteins with or without N-terminal deletion and included the clade B Env MN, 63521, and 6240, the clade C 1086, and the E clade 92TH023. LLE, linear linkage encoding.
Surface plasmon resonance (SPR) kinetics and dissociation constant (Kd) measurements.
Env gp120 binding Kd and rate constant measurements were carried out on BIAcore 3000 instruments using an anti-human Ig Fc capture assay as described earlier (28–30). Anti-RSV Synagis MAb was captured on the same sensor chip as a control surface. Nonspecific binding of Env gp120 to the control surface and/or blank buffer flow was subtracted for each MAb-gp120 binding interactions (see Fig. S1 in the supplemental material). 697D Fab was directly coupled via amine coupling chemistry to the sensor surfaces, Env gp120 was flowed, and data were collected as above. All curve fitting analyses were performed using global fit of multiple titrations to the 1:1 Langmuir model. Binding of 697D and CH01 MAbs to A244 gp120 (unmodified) gave biphasic dissociations, and the reported kd values are for the faster components of the fit. Means and standard deviations (SD) of rate constants and Kd values were calculated from at least three measurements on individual sensor surfaces with equivalent amounts of captured antibody. All data analysis was performed using the BIAevaluation 4.1 analysis software (GE Healthcare).
Isolation and purification of IgG from plasma.
Total IgG was isolated from individual RV144 vaccine recipient plasma samples using protein G resin prepacked into 96-well depletion plates (GE Healthcare) as previously described (31). The sample volume was reduced to 50 μl by centrifugation at 14,000 × g in a microcentrifuge precooled to 4°C. A buffer exchange was then performed using 2.5 volumes of phosphate-buffered saline (PBS), pH 7.5. The concentrated IgG was diluted to the desired volume with PBS and assayed for protein concentration using a NanoDrop 8000 Spectrophotometer (Thermo Fisher Scientific) using the IgG reference setting.
Binding antibody multiplex assays for anti-Env IgG were performed as previously described (32). Briefly, antibody measurements from vaccine plasma (1:200 dilution) were acquired on a Bio-Plex instrument (Bio-Rad), and the readout was expressed as mean fluorescent intensity (MFI) and concentration (μg/ml) based on an HIVIG standard curve. Positive and negative controls were included in each assay to ensure specificity and for maintaining consistency and reproducibility between assays. To control for Env protein performance, the positive-control titer (HIVIG) included on each assay had to be within ±3 SD of the mean for each antigen (tracked with a Levy-Jennings plot with preset acceptance of titer (calculated with a four-parameter logistic equation; SigmaPlot; Systat Software).
SPR measurements of plasma IgG avidity.
RV144 vaccine recipient IgG avidity was measured on a BIAcore 4000 instrument (BIAcore/GE Healthcare) using the multiplex array format (1 by 16) in which each IgG sample was flowed over duplicate spots of 8 different Env gp120 antigen surfaces, as described in the recent RV144 correlate analysis studies (23). Antigen surface activity was monitored using the C1 MAb A32 as a positive control and an irrelevant anti-RSV (Synagis) MAb as a negative control (see Fig. S1 in the supplemental material). V1/V2 MAb CH01, which is sensitive to N160K substitution, was used as a negative control for antigen spots with A244gD/N160K gp120. An anti-gD Fab was used to monitor binding to the gD peptide tag in Env gp120 with gD and to select IgG samples with low gD reactivity for MAb blocking studies. The IgG samples (n = 97) from vaccinee plasma at week 26 (2 weeks following the final immunization) and week 0 were diluted in PBS to 200 μg/ml and injected over each of the flow cells with replicate spots (2×) at 10 μl/min for an association time of 120 s and a dissociation time of 600 s. A random selection of IgG samples collected at visit 0 from 20 vaccinees was also included. Each surface activity was monitored by including A32 MAb (20 μg/ml) injection every 20 cycles of IgG samples, and surface decay of A32 binding over the entire experimental run was used to normalize the binding signal of plasma IgG samples. Nonspecific binding of the negative-control MAb was subtracted from each IgG sample binding data point. Data analyses were performed with BIAevaluation 4000 and BIAevaluation 4.1 software (BIAcore/GE Healthcare) as described earlier for Biacore 3000 (29) and Biacore A100 (33) data analysis, respectively. Kinetic binding responses and dissociation rate constant (Kd) values (s−1) were measured using methods described earlier (23, 33) The majority of IgG bound with a relatively slow dissociation rate (<10−3 s−1), and the previously described method for BIAcore A100 ranking of dissociation rates in complex or polyclonal samples as a ratio of response units measured as binding late and stability late (33, 34) was modified to include binding response and dissociation rate constant measurements as described earlier (23, 35). A relative avidity binding score was calculated for each IgG sample as follows: avidity score = binding response/Kd, where the avidity score is in RU · s, the binding response is in RU, and Kd is in s−1, with higher binding responses and slower Kd being indicators of a higher-affinity interaction (23, 35).
Antibody blocking assay.
Antibody blocking using an enzyme-linked immunosorbent assay (ELISA) was carried out as described earlier (30). ELISA plates (384-well plates; Costar 3700) were coated with 30 ng/well Env gp120 overnight at 4°C and blocked with assay diluent (PBS containing 4% [wt/vol] whey protein–15% normal goat serum–0.5% Tween 20–0.05% sodium azide) for 1 h at room temperature. All assay steps were conducted in assay diluent (except the substrate step) and incubated for 1 h at room temperature followed by washing with PBS–0.1% Tween 20. Sera were diluted 1:50 and incubated in quadruplicate wells. For CD4 (binding site) blocking assays, 10 μl of a saturating concentration of soluble CD4 (Progenics Pharm Inc.) was added following the serum incubation step. Biotinylated target MAb (10 μl) was added at the 50% effective concentration (EC50) (determined by a direct binding of biotinylated MAb to JRFL gp140). Values from quadruplicate wells were background subtracted and averaged. Percent inhibition was calculated as follows: 100 − (sera triplicate mean/no inhibition control mean) ×100.
SPR antibody blocking using RV144 vaccinee IgG samples was measured on BIAcore 3000 instruments. Env immunogen A244 Δ11 gp120 was immobilized on all flow cells of a CM5 sensor chip to about 5,000 to 6,000 RU using standard amine coupling chemistry. Blocking antibodies were sequentially injected at predetermined concentrations to capture near saturation. A zero baseline was set, and RV144 patient IgGs were injected at 10 μl/min for an association time of 180 s and a dissociation time of 600 s. Kinetic binding responses were measured 15 s after the end of the injection. The IgG samples (n = 119) with high and midlevel binding (>80 RU) to A244 Δ11 gp120 were selected from a panel of week 26 (2 weeks following the final immunization) plasma samples that included infected vaccinee (n = 41) and uninfected vaccinee (n = 205) groups. Randomly selected visit 0 vaccinee IgG samples (n = 19) with no binding to A244 Δ11 gp120 were included to assess nonspecific interactions. Anti-gp41 MAb 7B2 was used as a negative-control blocking antibody. Test antibodies included A32 (C1 region), PG9 and CH01 (V1/V2 conformational/quaternary), 2158, 697-30D and 830A (conformational V2), and19b (V3) MAbs and the V2 MAb CH58. Following each binding cycle, surfaces were regenerated with a short injection (10 to 15 s) of either glycine-HCl, pH 2.0, or 100 mM phosphoric acid. Blocking percentages were calculated from the ratio of binding response after negative-control 7B2 MAb blocking to the binding response after test antibody blocking: % blocking = [1 − (RU after test MAb blocking/RU after control MAb blocking)*100].
Isolation of antibodies from RV144 vaccinee plasma memory B cell.
Monoclonal antibodies CH51 and CH54 were isolated from circulating IgG+ memory B cells obtained from a vaccine recipient (subject 210884) as previously described ((20). Briefly, CD2−, CD14−, CD16−, CD235a−, IgD−, and IgG+ cells were isolated from frozen peripheral blood mononuclear cells (PBMCs) using magnetic activated cell sorting (Miltenyi Biotec, Auburn, CA), and cultures were screened for binding to HIV-1 gp120 envelope glycoproteins contained in the vaccine formulation (36). Cells from positive cultures were single-cell sorted, and PCR was performed as previously described (37, 38). The supernatants were harvested from the PCR-produced IgH and IgL gene expression cassette-transfected 293T cells after 3 days of incubation at 37°C in 5% CO2, and the monoclonal antibodies were purified as previously described (37).
NHP immunization.
Two groups of three rhesus macaques were immunized by both intramuscular and nasal routes with either A244 gp120 (no modification) or A244 gDΔ11 gp120 proteins. A third group of four animals were similarly immunized with A244 Δ11 gp120 proteins (without gD). Animals in all three groups were immunized with 100 μg of gp120 monomeric proteins formulated in squalene and adjuvanted with oCpG, R848, and MPL-A as described earlier (39, 40). The animals were immunized at weeks 0, 4, 8, and 16, and plasma samples were collected at time points corresponding to preimmunization (week 0) and at 2 weeks following each immunization. Plasma antibody titers were measured by binding ELISA to A244 gp120 and SPR blocking assay with conformational V2 and V1/V2 MAbs as described above. For SPR blocking of nonhuman primate (NHP) plasma IgG, biotin-labeled A244 V1/V2 Tags protein was captured on a streptavidin-coated CM5 (GE Healthcare) sensor chip. As described above, each of the blocking MAbs and control MAbs was bound to near saturation. Purified NHP plasma IgG (200 μg/ml) samples were then injected, and the binding levels of NHP IgG in the presence and absence of blocking MAbs were calculated; the percent blocking of NHP IgG binding by each of the V1/V2 (CH01 and CH58) and V2 (697D) MAbs was calculated as described above for RV144 plasma IgG, using the same formula, % blocking = [1 − (RU after test MAb blocking/RU after control MAb blocking)*100].
RESULTS
Expression of gp120 variants of the RV144 trial protein immunogens.
To address the effect of the modifications on the RV144 clinical trial protein Env gp120 antigenicity, we expressed the immunogen proteins A244-rgp120 and MN-rgp120 in 293T cells, with no modifications (A244 gp120 and MN gp120); with only the 11-aa N-terminal deletion (A244 Δ11 gp120); or, as in the RV144 Env immunogens, with both the HSV gD peptide tag and the N-terminal deletion (A244 gDΔ11 gp120 and MN gDΔ11 gp120) (Fig. 1; Table 1). The E clade 92TH023 gp120 was also expressed either with no modification (92TH023 gp120) or with both the Δ11 deletion and the gD tag (92TH023 gDΔ11 gp 120). These Env proteins were compared for gp120 monomer expression and for their binding to MAbs that recognize conformational epitopes on gp120.
Presentation of gp120 conformational epitopes on RV144 vaccine Env gp120 proteins with the gD tag and Δ11 deletion.
It has been reported previously that one component of the RV144 Env immunogen, A244-rgp120, binds to MAbs with specificity for the gp120 conformational V1/V2 epitopes (17, 20). The CH01 to CH04 lineage V1/V2 bnAbs and PG9 bnAb bound to A244 gp120 with Kd values ranging from 100 to 300 nM (20). Since PG9/PG16 MAbs bind preferentially to native trimers (21) and only to rare gp120 monomers (20), the binding of PG9 and CH01 MAbs suggests that the RV144 Env gp120 might show enhanced expression of conformational epitopes in the V1/V2 loops.
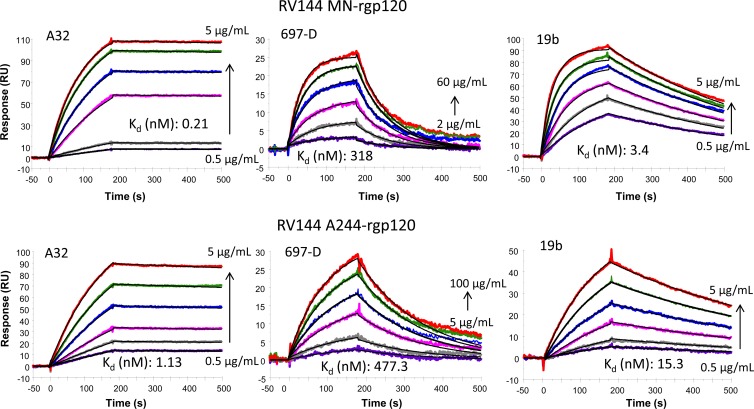
We found that other conformational epitopes were also presented on both RV144 Env A244-rgp120 and MN-rgp120. In particular, the conformational V2 MAb 697-D, which fails to bind linear V2 peptides (15), bound to MN- and A244-rgp120 with Kd values of 477 nM and 318 nM, respectively (Fig. 2) The gp120 C1 MAb A32, which binds to the surface of transmitted/founder infected CD4 T cells and mediates antibody-dependent cellular cytotoxicity (ADCC) (41), also bound strongly to the two RV144 Env gp120 proteins, with a relatively higher affinity for MN-rgp120 (Fig. 2). The Kd of the V3 MAb 19b for MN-rgp120 was about 5-fold lower than that of A244-rgp120 but was within the range reported for other V3 MAb binding to Env gp120 proteins (42). Thus, both of the RV144 vaccine gp120 immunogens expressed conformational epitopes within the C1, V2, and V1/V2 regions of gp120. The presentation of gp120 variable loop conformational epitopes and the recent association of conformational V2 antibodies with a lower rate of HIV-1 infection in RV144 (23) raised the question as to whether one or both of the two RV144 vaccine Env modifications—inclusion of the HSV gD peptide tag and/or the N-terminal Δ11 deletion—might have led to enhanced exposure of conformational epitopes within the C1 and V1/V2 regions.
Fig 2.
Binding of C1, V2, and V3 antibodies to RV144 immunogen gp120 proteins. RV144 Env proteins MN-rgp120 and A244-rgp120 binding at various concentrations (0.5, 1.0, 2.0, 3.0, 4.0, and 5 μg/ml for A32; 2.0, 5.0, 10, 20, 40, and 60 μg/ml and 5.0, 10.0, 25.0, 50.0, 75.0, and 100 μg/ml for 697D MAb binding to MN gp120 and A244 gp120, respectively; and 0.5, 1.0, 2.0, 3.0, 4.0, and 5 μg/ml for 19b) to A32, 697D, and 19b are shown. The calculated Kd values and fitted curves (black line) for each of the binding interactions are shown. Each MAb was captured on anti-Fc antibody immobilized surfaces, and gp120 monomeric proteins were injected as analytes, as described in Materials and Methods. The data are representative of three measurements.
The N-terminal 11-aa deletion (Δ11) in A244 gp120 reduces dimer formation.
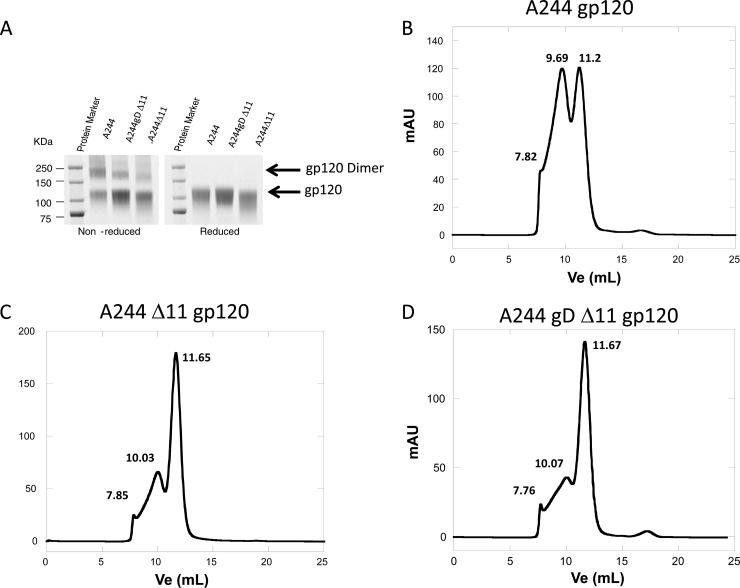
Expression of recombinant gp120 produces a substantial amount of disulfide-linked gp120 dimer, in which gp120 inner domain epitopes and the coreceptor binding surface are occluded (14). To determine the effect the two modifications on A244 gp120 might have on protein expression, we first compared the oligomerization states of the three different A244 gp120 proteins. In reducing SDS-PAGE gel, A244 gp120 proteins migrated as single bands of the expected size; under nonreducing conditions, they gave a mixture of bands that corresponded to monomers and dimers of gp120 (Fig. 3A). Size exclusion chromatography showed that A244 gp120 (Fig. 3B) had more dimer (58% ± 1%) than monomer (38% ± 1%). In contrast, the monomer fraction of Env A244 Δ11 gp120 was enriched almost 2-fold (66% ± 1%), and the dimer fraction was correspondingly reduced (30% ± 1%; monomer-to-dimer ratio, 2.2:1 (Student's t test, P < 0.001 for monomer fractions in A244 gp120 and A244 Δ11 gp120) (Fig. 3C). The inclusion of the HSV gD peptide, in addition to the Δ11 modification, in A244 gD Δ11 (Fig. 3D) did not further improve monomer enrichment, with a ratio of monomer to dimer similar to that of A244 Δ11 (63% ± 1% and 33% ± 1%, respectively) (Student's t test, P = 0.11 for monomer fractions in A244 gD Δ11 and A244 Δ11 gp120). The amount of higher-order oligomers or aggregates was the same for all three expressed proteins (about 3 to 4%) (Fig. 3). We observed a similar profile for the 92TH023 gp120 proteins, with a higher proportion of monomers in 92TH023 gp120s with Δ11 and gD tag (∼65%) than without any modifications (∼38%). MN gp120 expressed with Δ11 and gD (MN gD gp120) or with no modifications (MN gp120) gave similar proportions of dimers (34% and 31%, respectively). Additional gp120 constructs derived from different clades including clade B (63521 and 6240) and clade C (C.1086), designed to contain their original signal peptide and deletion of the first 11 or 7 (C.1086) amino acid residues, were produced in 293 cells by transient transfection. For each of the above proteins, we found that inclusion of Δ11 alone yielded predominantly monomers, as detected by SDS-PAGE under nonreducing conditions (data not shown), and greater than 90% monomers in gel filtration chromatography analysis. Thus, the N-terminal Δ11 modification alone resulted in markedly smaller amounts of gp120 dimer formation when Env proteins were expressed in mammalian cells.
Fig 3.
Relative proportions of monomer and dimer in A244 gp120 proteins. (A) SDS-PAGE analysis (under reduced and nonreduced conditions) of A244, A244 gD Δ11, and A244 Δ11 gp120 proteins showing the presence of disulfide-linked dimers in various proportions. Each of the gp120 preparations was analyzed by size exclusion chromatography (SEC), which showed a relatively larger proportion of monomer in A244 gp120 (B) than in either A244 Δ11 (C) or A244 gD Δ11 (D). Peak volume analysis of the monomer and dimer fractions gave the following proportions of monomer and dimer: in A244 gp120-A244 gp120, dimer, 58%, and monomer, 38%; in A244 Δ11 gp120, dimer, 30%, and monomer, 66%; in A244 gD Δ11 gp120, dimer, 33%, and monomer, 63%. Similarly, for B.63521 gp120, the proportion of monomer was improved by N-terminal deletion: for 63521 gp120, dimer, 23%, and monomer, 77%; and for B.63251 Δ11 gp120, there was no resolved peak for the dimer and the result for monomer was >95%. For C.1086, a modest improvement was observed with C.1086 gp120 (dimer, 33%; monomer, 67%) and C.1086Δ7 gp120 (dimer, 27%; monomer, 73%). The standard deviation of the measurements of percentage of monomer and dimer was within 1%.
Enhanced binding of conformational V2 antibodies and V1/V2 bnAbs to A244 Δ11 gp120 monomers.
Monomers of each of the A244 gp120 proteins (A224, A244 Δ11, and A244 gD Δ11) were purified to homogeneity by removal of dimeric and aggregate fractions with size exclusion chromatography (SEC) (see Fig. S2 in the supplemental material). Following SEC fractionation, the monomeric gp120 proteins were stable and did not redistribute into dimer or aggregate fractions (see Fig. S2 in the supplemental material). Each of the three purified gp120 monomers bound to CD4 and showed CD4-induced (CD4i) epitope upregulation as assessed by 17b MAb binding (data not shown). A comparison of the binding of the size-fractionated monomeric and dimeric A244 gp120 (data not shown) showed that the dimer fraction had markedly reduced binding of the C1 MAb A32 and little or no binding of the conformational V2 MAb 697-D. This result is consistent with previous reports (14) that the V1/V2 loop and the N and C termini are involved in gp120 dimer formation and that the epitopes on the Env inner domain are occluded in gp120 dimers.
Using a panel of antibodies with specificities that included conformational C1, V2, and V1/V2 epitopes, we compared MAb binding Kd and rate constants for each of the monomeric clade E A244 gp120 proteins, to assess whether the Δ11 and/or gD tag had any effect on Env antigenicity (Fig. 4; Table 2). We observed that inclusion of Δ11 had no effect on exposure of the V3 loop, since the V3 MAb 19b bound with similar Kd and kinetic rate constants to each of A244 gp120 proteins (Table 2; Fig. 4A). The ADCC-mediating, C1 MAb A32 (41), however, had a 9-fold- and 6-fold-higher affinity for A244 Δ11 and A244 gD Δ11, respectively, than for A244 gp120 (Table 2; Fig. 4B). Similarly, the conformational V2 MAb 697-D (15) had a nearly 10-fold-higher affinity for A244 gD Δ11 and A244 Δ11 than for A244 gp120 (Kd = 218, 157 and 1465 nM, respectively) (Fig. 4D; Table 2). We also observed these differences using the Fab fragment of the V2 conformational MAb 697-D, which bound to A244 gD Δ11 with 8-fold-higher affinity than to unmodified A244 gp120 (see Fig. S3 in the supplemental material; Kd = 690 and 5,700 nM, respectively). While the dissociation rate constants were similar, the ka (on-rate or association rate constant) was nearly 10-fold higher for binding of 697-D to A244 gD Δ11 than for binding to A244 gp120 (see Fig. S3 in the supplemental material). We also studied two other conformation-dependent V2 MAbs, 2158 and 830A, which had higher affinities for A244 gp120 than did 697-D (Table 2). Both had between 3- and 5-fold-higher affinities for A244 gD Δ11 and A244 Δ11 gp120 than for unmodified A244 gp120. Conformational V1/V2 bNab CH01 showed a similar differential, while relatively smaller differences were observed with PG9 (Table 2). We conclude that the epitopes recognized by conformational V2 and V1/V2 are well exposed on A244 Δ11 gp120.
Fig 4.
Enhanced binding of C1, V2, and V1/V2 antibodies to E.A244gp120 proteins with Δ11 deletion. Each of the analyte gp120 proteins (left panel, A244 gp120; middle panel, A244gDΔ11; right panel, A244Δ11 gp120) was injected over the listed antibodies captured on an anti-Fc immobilized surface. Each gp120 protein was titrated at 0.5, 1.0, 2.0, 3.0, 4.0, and 5.0 μg/ml for 19b (A) and A32 (B); A244 gp120 at 10, 20, 30, 40, and 50 μg/ml for 697D (C); 10, 25, 50, 75, and 100 μg/ml on CH01(D) and PG9 (E); A244gDΔ11 at 5, 10, 20, 30, and 40 μg/ml for 697-D and 10, 25, 50, 75, and 100 μg/ml for CH01 and at 10, 20, 30, and 40 μg/ml for PG9; A244Δ11 at 2, 4, 6, 10, 25, and 50 μg/ml for 697-D, 10, 25, 50, 75, and 100 μg/ml for CH01 and 10, 25, 50, 75, and 100 μg/ml for PG9 MAb captured surfaces. Data are representative of at least 3 measurements made on individual flow cells with equivalent amounts of captured antibodies. All SPR binding experiments were carried out using purified monomeric gp120 as assessed by size exclusion chromatography.
Table 2.
Dissociation and kinetic rate constants of antibody binding to A244 gp120 proteinsa
| Antibodyb | Rate constant and/or Kd | A244 gp120 | A244 gD Δ11 gp120 | E.A244 Δ11 gp120 |
|---|---|---|---|---|
| A32 | ka (× 103 M−1 s−1) | 76.8 ± 11.4 | 134 ± 14.0 | 222.6 ± 20.4 |
| (C1) | kd (× 103 s−1) | 0.47 ± 0.05 | 0.13 ± 0.017 | 0.15 ± 0.03 |
| Kd (nM) | 6.25 ± 1.4 | 1.0 ± 0.22 | 0.67 ± 0.13 | |
| 19b | ka (× 103 M−1 s−1) | 130.3 ± 10.5 | 170.3 ± 8.5 | 239.3 ± 19.8 |
| (V3) | kd (× 103 s−1) | 1.54 ± 0.095 | 1.4 ± 0.08 | 1.56 ± 0.08 |
| Kd (nM) | 11.8 ± 0.21 | 8.24 ± 0.23 | 6.54 ± 0.38 | |
| 697-D | ka (× 103 M−1s−1) | 4.9 ± 1.1 | 24.9 ± 5.6 | 26.75 ± 0.71 |
| (V2) | kd (× 103 s−1) | 7.0 ± 1.98 | 5.24 ± 0.54 | 5.18 ± 0.6 |
| Kd (nM) | 1,465.3 ± 317 | 217.6 ± 45.7 | 156.7 ± 34.0 | |
| 830A | ka (× 103 M−1 s−1) | 21.8 ± 3.1 | 41.1 ± 1.9 | 59.9 ± 4.6 |
| (V2) | kd (× 103 s−1) | 0.22 ± 0.06 | 0.07 ± 0.003 | 0.088 ± 0.01 |
| Kd (nM) | 10.2 ± 3.6 | 1.7 ± 0.16 | 1.56 ± 0.09 | |
| 2158 | ka (× 103 M−1 s−1) | 16.4 ± 0.98 | 28.7 ± 1.0 | 36.5 ± 1.8 |
| (V2) | kd (× 103 s−1) | 0.19 ± 0.04 | 0.10 ± 0.03 | 0.13 ± 0.04 |
| Kd (nM) | 11.2 ± 1.6 | 3.7 ± 0.9 | 3.68 ± 1.1 | |
| CH01 | ka (× 103 M−1 s−1) | 3.73 ± 1.6 | 37.2 ± 15.1 | 49.0 ± 5.4 |
| (V1/V2) | kd (× 103 s−1) | 4.38 ± 0.52 | 9.9 ± 2.8 | 15.6 ± 1.5 |
| Kd (nM) | 1,639 ± 601 | 277.8 ± 42 | 317 ± 31.9 | |
| CH01-04 UA1 (V1/V2) | ka (× 103 M−1 s−1) | 6.7 ± 2.5 | 16.7 ± 6.3 | 17.6 ± 0.5 |
| kd (× 103 s−1) | 21.5 ± 0.6 | 19.2 ± 1.3 | 18.8 ± 0.5 | |
| Kd (nM) | 3,500 ± 1200 | 1,100 ± 250 | 1,050 ± 220 | |
| CH01-04 UA2 (V1/V2) | ka (× 103 M−1 s−1) | 4.4 ± 2.3 | 20.8 ± 10.1 | 27.3 ± 11.4 |
| kd (× 103 s−1) | 13.9 ± 0.2 | 17.7 ± 0.6 | 26.2 ± 1.0 | |
| Kd (nM) | 3,200 ± 950 | 870 ± 200 | 940 ± 50 | |
| PG9 | ka (× 103 M−1 s−1) | 5.0 ± 3.5 | 11.5 ± 0.6 | 10.9 ± 0.9 |
| (V1/V2) | kd (× 103 s−1) | 1.1 ± 0.4 | 0.55 ± 0.03 | 0.57 ± 0.06 |
| Kd (nM) | 183 ± 44.0 | 48.1 ± 0.15 | 52.6 ± 2.9 | |
| VRC01 | ka (× 103 M−1 s−1) | 17.6 ± 0.52 | 13.3 ± 0.57 | 9.7 ± 0.43 |
| (CD4 bs) | kd (× 103 s−1) | 0.28 ± 0.02 | 0.21 ± 0.06 | 0.39 ± 0.03 |
| Kd (nM) | 15.7 ± 1.7 | 15.8 ± 3.9 | 36.7 ± 1.4 |
Each of the rate constants and Kd values was derived from at least three measurements on individual flow cells of the same sensor chip or from binding data collected on different sensor chips. The mean and SD of rate constants (ka and kd) and Kd values are reported for each antibody binding to the three different forms of monomeric E.A244 gp120 proteins.
Parentheses indicate gp120 region recognized by the MAbs.
These results suggest that the conformational V2 and V1/V2 epitopes recognized by MAbs 697-D and CH01 are better exposed or conformationally more stable on A244 gp120 proteins with the Δ11 modification, with or without the inclusion of HSV gD, than on the unmodified protein. In most cases, the differences in Kd were due to differences in the association rates, ka, with roughly 5- to 10-fold-higher rates of association with gp120 bearing a Δ11 modification for both 697-D Fab and CH01 MAb (Table 2; see Fig. S3 in the supplemental material). The contribution of ka to the differences in Kd supports the notion that exposure of particular conformational epitopes is a critical factor in the enhanced antigenicity.
RV144 A244 Δ11 Env binds well to CH01-CH04 clonal lineage unmutated antibodies.
Two unmutated ancestor antibodies (UAs) of the MAb CH01 to CH04 clonal lineage, CH01_RUA1 and CH01_RUA2, have recently been shown to bind to the RV144 vaccine trial immunogen A244-rgp120 (20, 43). We compared binding of the CH01-04 UAs to A244 Δ11 gp120 with their binding to unmodified A244 gp120. CH01_RUA1 and CH01_RUA2 bound to A244 Δ11 and A244 gD Δ11 gp120 with about 3- to 4-fold-higher affinity than to A244 gp120 (Fig. 5); the two CH01 UAs had roughly equivalent Kd values for the A244 gp120Δ11 proteins. As observed for CH01 MAb, the binding association rates of the UAs to A244 Δ11 were greater than binding to unmodified gp120; the dissociation rates were similar (Fig. 5). The affinity of the CH01-04 UAs for A244 Δ11 gp120 was lower, about 3-fold, than was the affinity of the mature CH01 MAb; the effects of the Δ11 deletion were less pronounced for the UAs than for the CH01 mature antibody.
Fig 5.
Binding of CH01_RUA1 and CH01_RUA2 to A244Δ11 gp120. Each of the CH01 RUAs (RUA1 and RUA2 data are shown in top and bottom panels, respectively) was captured as described in the legend for Fig. 4. Each of the three forms of A244 gp120 was flowed at concentrations of 10, 25, 50, 75, and 100 μg/ml. Kinetic rate constants and Kd were derived as described in Materials and Methods. Data are representative of at least 3 measurements made on individual flow cells with equivalent amounts of captured antibodies.
HSV gD and Δ11 modifications have smaller effects on the antigenicity of B.MN gp120 and AE.92TH023 gp120.
We next compared the antigenicity of N-terminally deleted B.MN and AE.92TH023 gp120 and a clade C gp120, C.1086, with their unmodified counterparts (see Tables S1 and S2 in the supplemental material). The N-terminal deletions had weaker effects on the antigenicity of B.MN, AE.92TH023, and C.1086 gp120s than they did for A244gp120 in particular, showing little or no effect on the binding of C1 or V1/V2 MAbs. However, there was 1 log more avid binding of the CD4 binding site bnAb, VRC01, to MNΔ11 gp120 than to MN gp120 (see Table S1 in the supplemental material). Thus, the effects of N-terminal deletions on gp120 immunogens depend on the particular gp120 tested.
Plasma IgG from RV144 vaccinees binds with higher avidity to A244 gD Δ11 gp120.
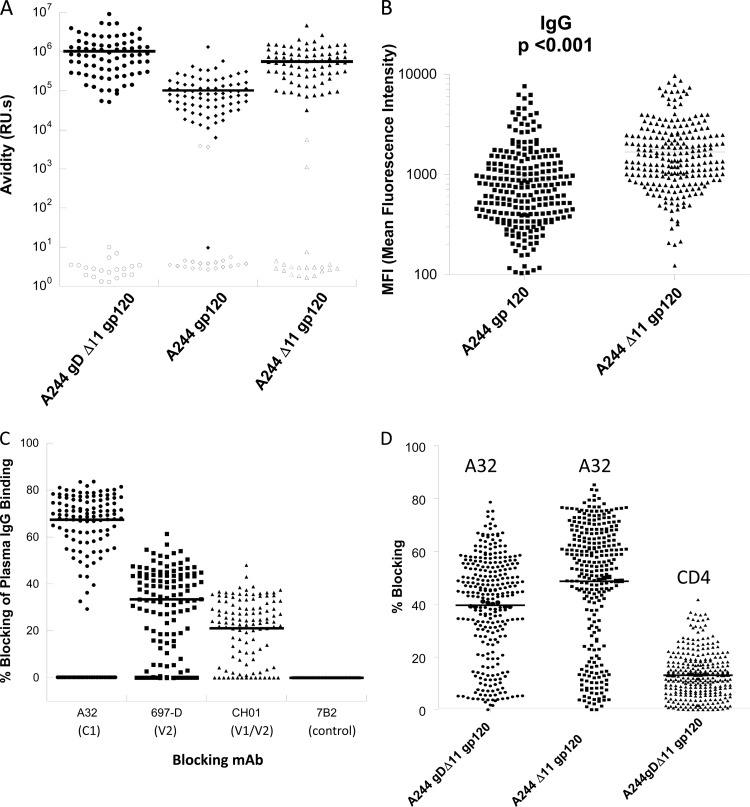
The effect of Δ11 modification in enhancing the antigenicity of gp120 conformational epitopes to C1, V2, and V1/V2 raises the question as to whether antibodies induced by the RV144 immunogen (A244 gD Δ11) also bind more avidly to A244 Δ11 than to A244 gp120. We purified IgGs from RV144 vaccinee plasma taken 2 weeks after the final ALVAC/AIDSVAX B/E immunization (week 26) and measured their relative avidity scores for each of the A244 gp120 proteins in an SPR binding assay. Compared to placebo and prevaccination visit 1 IgG samples (no binding), IgG samples from the week 26 vaccinee group bound A244 gD Δ11 gp120 with avidity scores that ranged over 2 orders of magnitude (Fig. 6A). The mean avidity for both A244 Δ11 and A244 gD Δ11 were both significantly higher (P < 0.001) than that for A244 gp120 (Fig. 6A). In the HIV-1 binding antibody multiplex assay, we also found a significant difference; RV144 plasma IgG showed more avid binding to A244 Δ11 gp120 than did A244 gp120 (P < 0.001; Fig. 6B). Thus, the RV144 vaccine gave rise to antibodies with higher avidity for A244 gp120 with the Δ11 modification than for unmodified A244 gp120.
Fig 6.
RV144 vaccinee sera antibody responses. (A) RV144 vaccinee IgG binding to A244 gp120 proteins show higher avidity for A244 gp120 with Δ11. RV144 visit 8 (week 26, 2 weeks following the final immunization) IgG (n = 97) binding was measured for A244 gp120, A244 gD Δ11, and A244 Δ11 gp120 proteins. Binding responses and dissociation rate constants for avidity score measurements were calculated as described in Materials and Methods. The mean avidity of binding to A244 gp120, A244 gD Δ11, and A244 Δ11 gp120 were 1.0 ± 1.5, 10.0 ± 0.5, and 5.7 ± 0.7 RU · s (× 105), respectively. The differences in avidity were significant for A244 gp120 versus A244gDΔ11 (Student's t test, P < 0.001) and A244 gp120 versus A244Δ11 gp120 (Student's t test, P < 0.001). The binding of plasma IgG samples from the placebo group is shown as open circles. (B) RV144 vaccinee plasma IgG binding to A244 gp120 proteins show higher relative binding to A244 gp120 with Δ11. RV144 visit 8 (week 26, 2 weeks following the final immunization) plasma antibody was measured against A244 gp120, A244 gD Δ11, and A244 Δ11 gp120 proteins in a binding antibody multiplex assay, and the mean fluorescence intensity (MFI) values were plotted. The differences in binding responses were significant for A244 gp120 versus A244 Δ11 gp120 (Student's t test, P < 0.001). (C) Blocking of RV144 induced IgG binding to A244 gD Δ11 gp120 by conformational C1 (A32), V2 (697D), and V1/V2 (CH01) antibodies. RV144 IgG samples (n = 109) with high levels (>80 RU measured at 200 μg/ml) of binding to A244 Δ11 gp120 were selected for antibody blocking studies. A control group (n = 19, open circles) showing no binding to A244 Δ11 gp120 was included to assess nonspecific signal in IgG samples. The mean blocking rates of RV144 plasma IgG by MAbs A32, 697-D, and CH01 were 67.4% ± 11.4%, 34.1% ± 14.5%, and 22.1% ± 12.5%, respectively. (D) ELISAs showing high levels of A32 blocking (mean = 39.6% ± 19.2%) by RV144 IgG and low levels of CD4 blocking antibodies (mean = 13% ± 8.9%). Blocking of IgG from visit 1 were 6.7% ± 4.2% and 8.9% ± 7.6% for A32 and CD4, respectively.
Conformational antibodies to C1, V2, and V1/V2 block RV144-induced IgG binding to A244 Δ11 gp120.
To assess the specificity of the antibodies induced by RV144 vaccine gp120 immunogens, we measured the relative level of blocking of vaccinee IgG binding by a panel of MAbs, including those that showed higher affinity for A244 Δ11 gp120. As shown in Fig. 6C, the binding of vaccinee IgG was blocked by each of the C1 (A32), V2 (697-D), and V1/V2 (CH01) bnAbs tested, with the strongest blocking observed with A32 (67.4%). For the V2 epitope, we also used two additional V2 MAbs, 2158 and 830A, which show various levels of overlap; 830A strongly blocks all other V2 MAbs (27) (data not shown). Among the V2 MAbs, blocking of RV144 IgG was stronger with 697-D and 830A (see Fig. S4 in the supplemental material), both of which had enhanced binding to A244 with Δ11 modifications (Table 2).
Of the two conformational V1/V2 gp120 bnAbs, we found no blocking of RV144 IgG binding by PG9 (data not shown) but detectable blocking (22.1%) by CH01 (Fig. 6C). We also determined blocking of plasma antibodies in ELISAs for antibodies that inhibit binding of biotinylated MAb A32 and of soluble CD4. In these assays, the mean blocking rates of A32 and sCD4 binding were 39.6% and 13%, respectively (Fig. 6D).
These results suggest that the RV144 vaccine induced a relatively larger proportion of antibodies directed against the conformational C1 (A32) epitope, which is the target for ADCC in the trial vaccine plasma (36). The next-largest population of antibodies tested targeted the conformational V2 epitopes recognized by the conformational MAb 697-D, followed by CH01-like bnAbs. No PG9-like antibodies were detected.
Monoclonal antibodies from RV144 vaccinees recognize epitopes enhanced on A244 Δ11 gp120 monomers.
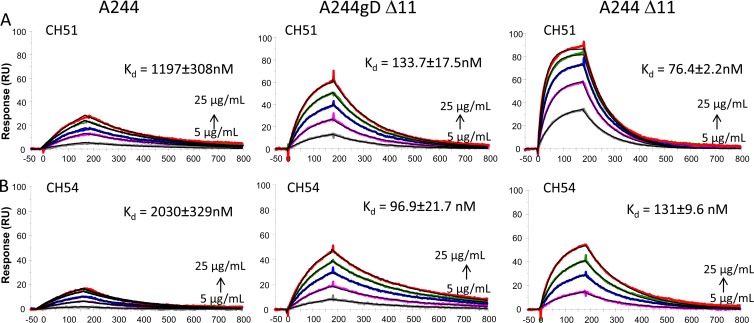
Using previously described methodologies for isolating antibodies from memory B cells (20), two IgG antibodies, CH51 and CH54, were isolated from circulating IgG+ memory B cells of an RV144 vaccine recipient (subject 210884) (36). A32 blocked binding of both CH51 and CH54, suggesting that RV144-derived MAbs bound to epitopes that overlap the C1 conformational epitope of A32. The overall Kd values of CH51 and CH54 MAbs for binding to A244gDΔ11 were higher than that of the C1 MAb A32 (Fig. 4 and 7), but both of these RV144 MAbs bound to A244 gD Δ11 and A244 Δ11 with a 1-order-of-magnitude-lower Kd than they did to A244 gp120 (Fig. 7A and B). Thus, the RV144 vaccinee-derived antibodies (CH51 and CH54) with A32-like specificity showed tighter binding to A244 gp120 with the N-terminal deletion and mirrored the overall enhanced avidity detected in RV144 plasma samples with A244 gD Δ11.
Fig 7.
RV144 MAbs CH51 and CH54 show higher-affinity binding to A244gp120 with Δ11 modification. A244, A244 gD Δ11, and A244 Δ11 gp120 were each injected at increasing concentrations of 5, 10, 15, 20, and 25 μg/ml over either CH51 (A) or CH54 (B) captured on anti-Fc immobilized surfaces. Binding of both CH51 and CH54 was 1 order of magnitude higher for both A244 gp120 with the Δ11 modification than for A244 gp120 (left panel) with no modification.
A244 Δ11 gp120 induces antibodies blocked by conformational V2 and V1/V2 MAbs in nonhuman primates.
One prediction of the observed enhanced antigenicity of N-terminal deletion in A244 Δ11 gp120 would be that there would be qualitative differences in the induced V2 or V1/V2 antibodies. This prediction is based on the results described above that gp120 N-terminal deletion alone enhances the affinity of selected conformational V2 (697-D) and V1/V2 (CH01) MAbs to recombinant A244 gp120 proteins. Moreover, since the RV144 immune correlates study showed that conformational V2 antibody responses were associated with a lower rate of HIV-1 infection (23), we were interested to determine whether the A244 Δ11 gp120 would induce higher levels of conformational V2 and V1/V2 antibodies than those induced by A244 gp120 with no modification. Thus, to assess the immunogenicity of each of the different forms of the A244 gp120 proteins, we immunized three groups of rhesus macaques with either A244 gp120, A244 gD Δ11, or A244 Δ11 gp120 proteins.
We first tested the magnitude of the antibody responses in each of the immunogen groups against various proteins. For each of the postimmunization plasma sample groups PI-2, PI-3, and PI-4 (samples harvested 2 weeks after immunization time points of 4, 8, and 16 weeks, respectively), the reactivity levels (50% effective dilution [ED50]) against the RV144 immunogen protein A244 gD Δ11 were similar in all three immunogen groups (Fig. 8A). The ED50 dilution titers were also similar for all three groups of NHP plasma when binding to A244 gp120 either with or without N-terminal deletion (data not shown). The NHP plasma samples gave high background to the gp70 carrier protein when reactivity to gp70 Case A2 V1/V2 protein (18), which was used in the RV144 correlate analysis (23), was studied. Therefore, to assess the reactivity of NHP plasma antibodies to the gp120 Case A2 V1/V2 region, we used an alternate antigen design lacking the gp70 carrier, the B. Case A V1/V2 Tags protein. Using the B. Case A V1/V2 Tags protein, 5-fold-higher mean plasma antibody ED50 binding values were observed at PI-2 in A244 Δ11-immunized macaques than in macaques immunized with A244 gp120 with no N-terminal deletion (Fig. 8B). Thus, while the overall magnitude of the antibody responses to gp120 was similar in animals immunized with A244 gp120 with either no modification or with N-terminal deletion, binding to the clade B Case A2 V1/V2 loop region was enhanced 5-fold after two immunizations with A244 Δ11 gp120.
Fig 8.
Higher levels of V1/V2 MAb blocking antibodies induced in rhesus macaques immunized with A244 Δ11 gp120 protein. Binding ELISA titers (ED50, half-maximal dilution titer) of NHP plasma antibodies to RV144 immunogen protein A244 gDΔ11 gp120 (A) and the V1/V2 protein B. Case A V1/V2 Tags (B) were measured for each animal in the three immunogen groups. Data from A244 gp120-, A244 gD Δ11 gp120-, and A244 Δ11 gp120-immunized animals are shown in open circles, filled circles, and filled diamonds, respectively. Bars in each plot indicate mean values for each of the indicated immunogen group. PI-2, PI-3, and PI-4 indicate postimmunization plasma samples harvested 2 weeks after immunization time points of 4, 8, and 16 weeks, respectively. The difference in titers between animal groups immunized with A244 gp120 (median ED50 = 342 ± 125) versus A244 Δ11 gp120 (median ED50 = 1,069 ± 583) for binding to B. Case V1/V2 Tags did not reach statistical significance (P = 0.09). (C) Blocking of NHP IgG with conformational V2 and V1/V2 MAbs. For 697-D blocking, the median values (± SD) for A244 gp120-, A244 gD Δ11-, and A244 Δ11 gp120-immunized NHP groups were 30.9 ± 4.2 (28.7), 37.0 ± 9.8 (33.6), and 56.6 ± 9.8 (51.7), respectively (*, P = 0.013 for A244 Δ11 versus A244 gp120). For CH01 blocking, the blocking rates were 20.3% ± 3.8% (24.1%), 18.6% ± 8.1% (21.8%), and 45.2% ± 7.4% (51.7%), respectively (*, P = 0.006 for A244 Δ11 versus A244 gp120; **, P = 0.026 for A244 Δ11 versus A244 gD gp120). For CH58, the blocking rates were 77.5% ± 9.4%, 90.5% ± 4.6%, and 74.1% ± 24.3%, respectively, for A244 gp120-, A244 gDΔ11-, and A244 Δ11 gp120-immunized groups. The median values in a second independent experiment were similar and are given in parentheses above. The measurement of blocking antibodies was carried out on NHP IgG samples harvested from PI-3 plasma of each immunogen group and was performed as described for data in Fig. 6 and in Materials and Methods.
We had earlier observed in RV144 IgG samples that the induced antibodies were blocked by certain conformational V2 and V1/V2 antibodies (Fig. 6C). Thus, we reasoned that while the overall magnitude of gp120 antibody responses was similar in the three NHP immunogen groups, the A244 Δ11 gp120 immunogen might have induced qualitatively different antibodies in rhesus macaques compared to those immunized with A244 gp120 with no modification. Therefore, we compared the levels of blocking by conformational V2 and V1/V2 MAbs of purified plasma IgG harvested from animals in each of the NHP immunogen groups. Blocking of binding to A244 V1/V2 protein was tested with V2 and V1/V2 mabs (Fig. 8C) at the time of peak gp120 responses (PI-3 IgG samples). Blocking by the conformational V1/V2 bNAb CH01 was significantly higher (>2-fold-higher median values) in the A244 Δ11 animals (45.2% ± 7.2%) than in either those immunized with A244 gp120 with no modification (20.3% ± 3.8%; P = 0.006) or those with A244 gp120 with both Δ11 and gD tag (18.7% ± 8.1%; P = 0.026) (Fig. 8C). Similarly, for the conformational V2 MAb 697-D, the observed blocking was higher for the A244 Δ11 gp120 immunogen group (56.6% ± 9.8%) than for the A244 gp120 (30.9% ± 4.2%; P = 0.013 for A244 Δ11 versus A244 gp120) immunogen group. Blocking with the RV144 CH58 MAb, which binds a distinct V2 linear epitope (Liao et al., unpublished), gave much higher blocking overall, but there were no significant differences between the IgG samples from the three immunogen groups (Fig. 8C).
These results suggest that while all three forms of A244 gp120 proteins induce an overall similar magnitude of gp120 antibodies, inclusion of N-terminal deletion alone allowed the induction of antibodies that were qualitatively different; a higher proportion of V1/V2 antibody responses that were specifically blocked by the conformational V2 and V1/V2 MAbs were induced.
DISCUSSION
The RV144 trial showed the estimated vaccine efficacy to be 31.2%. Future HIV-1 vaccine efficacy trials will therefore require an improved immunogen design, and analysis of the RV144 immunogens is an important first step. In this work, we have probed the effects of gp120 design on antigenicity and immunogenicity of the immunogens used in the RV144 HIV-1 vaccine efficacy trial. We have demonstrated that deletion of the N-terminal amino acid residues of A244 gp120, which was used as boosts in the trial, enhanced the antigenicity of gp120 conformational epitopes. The enhanced epitopes were immunogenic in RV144 vaccinees, and they induced immune responses with higher avidity for these conformational epitopes than for the same epitopes on the unmodified immunogen A244 gp120. We have also shown that an HSV gD tag, introduced into the AIDSVAX B/E rgp120 as part of early expression and purification strategies (5), did not contribute to the antigenicity enhancement. Antigenic enhancement by the N-terminal deletion Δ11 was more pronounced with A244 (clade E) than with MN (clade B), 92TH023 (clade E), or 1086 (clade C) gp120 proteins, suggesting that these effects may depend on the particular gp120 vaccine immunogen. Finally, we demonstrated that the N-terminal deleted gp120 A244 protein induced significantly higher levels of macaque plasma antibodies that could be blocked by conformational V2 MAbs compared to intact A244 gp120.
The higher proportion of disulfide-linked dimers in the preparations of unmodified A244 rgp120 than in those with a Δ11 deletion (with or without the gD tag replacement) suggested that the principal effect of removing the N-terminal residues may be to enhance the reliability of folding in the endoplasmic reticulum. Correctly folded gp120 has no unpaired cysteines, and any interchain disulfides must form at the expense of correct intrachain pairings. That is, at least part of the protein must be misfolded for disulfide-linked dimers to form at all. Even the monomeric protein in any preparation may be conformationally heterogeneous; the proportion of dimer will tend to reflect the degree of misfolding within the monomer population. Because the consequences of the N-terminal deletion depend on the rest of the gp120 sequence—it had a less marked effect on rgp120 from other isolates—its influence on folding is Env dependent. Thus, we observed marked improvement in purification of gp120 monomers on certain Envs, like E.A244 gp120 and B.63521, and a more modest improvement in the case of C.1086 gp120.
Two observations show that the upregulation of conformational C1, V2, and V1/V2 epitopes on A244 gp120 was relevant to the antibody responses induced in the RV144 trial. First, using ELISA and SPR blocking assays, we have identified RV144 vaccinee antibodies that react with epitopes related to those recognized by MAbs A32 (C1), 697-D (conformational V2), and CH01 (conformational V1/V2), and we have isolated from RV144 vaccinees human MAbs that are blocked in their binding to A244 gp120 by MAb A32 (44, 45). Moreover, MAb 697-D, which binds to A244 gp120 and the gp70V1/V2 Case A2 clade B scaffolded protein, binds to A244 Δ11 gp120 nearly 10-fold more avidly than it does to A244 gp120 with no modifications. Second, the RV144-induced plasma antibody response had a higher avidity for A244 Envs with gD Δ11 or with Δ11 alone than for their unmodified counterparts (Fig. 6). The conformational V2 and V1/V2 epitope specificities induced by the vaccine included those that could be blocked by MAbs CH01 and 697D, but not by MAb PG9 (Fig. 6C; see also Fig. S4 in the supplemental material). Although we have not yet been able to rescue a V2 MAb against the conformational V2 or V1/V2 epitopes selectively recognized by 697-D or CH01, we have demonstrated the presence of plasma antibodies with specificities capable of blocking the binding of these MAbs to A244 gp120 (Fig. 6C). Both the A244 gp120 MAbs isolated from RV144 vaccinees (CH51 and CH54) bound A244 and MN gp120s, their binding was blocked by A32, and both mediated antibody-dependent cellular cytotoxicity (ADCC) to HIV-1 AE_01-infected CD4 T cell targets (36). Binding of both of these A32-like MAbs (CH51 and CH54) to A244 gp120 was enhanced when the Δ11 deletion was introduced. These data strongly suggest that the observed Δ11-enhanced gp120 antigenicity of RV144 gp120 immunogens played a role in the induction of certain antibody types (C1, V2, and V1/V2) in the RV144 vaccinees.
Finally, the effect of Δ11 modification in enhancing the immunogenicity of E.A244 gp120 was also supported by immunization data from rhesus macaques showing a significantly higher level of plasma antibodies that could be blocked by conformational V2 MAbs induced in animals immunized with A244 gp120Δ11 than the level found in the group immunized with A244 gp120 with no modification (Fig. 8).
The underlying mechanism of protection in the RV144 trial has yet to be elucidated—the immune correlates study has so far identified only antibody responses that correlate directly (plasma HIV-1 Env IgA) or inversely (plasma Abs binding to gp70-V1/V2) with infection risk (23). Subsequent studies are required to determine if these antibody types are causal correlates or are surrogate markers of other factors.
In summary, we have shown that the Δ11 N-terminal deletion on the gp120 Envs used in the AIDSVAX B/E boost of the RV144 HIV-1 vaccine trial enhanced gp120 epitope expression and augmented both antigenicity and immunogenicity for the V2 and V1/V2 gp120 regions. The Δ11 deletion (with or without gD) in A244 gp120 leads to expression of a higher proportion of correctly folded recombinant protein, and the stability and conformational homogeneity of the immunogen are likely to have contributed substantially to its properties. Our data suggest that careful attention to Env conformations and antigenicity will be critical when designing immunogens in future trials.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dennis Burton (Scripps Research Institute, La Jolla, CA) and Peter Kwong (VRC, NIH, Bethesda, MD) for provision of MAb PG9. We thank Amanda Lilley for SEC and Judith T. Lucas, Vicki Ashley, and Michele Donathan for technical assistance.
This study was supported in part by the Bill & Melinda Gates Foundation and Collaboration for AIDS Vaccine Discovery (CAVD) and CAVD-VIMC grants, by the National Institutes of Allergy and Infectious Diseases, Division of AIDS, Department of Health and Human Sciences, through a U01-grant from the NIH, NIAID, AI067854 (the Center for HIV/AIDS Vaccine Immunology), by NIH, NHLBI, HL59725, and by research funds from the Department of Veterans Affairs. This work was also supported in part by an Interagency Agreement (Y1-AI-2642-12) between the U.S. Army Medical Research and Materiel Command and the National Institute of Allergy and Infectious Diseases, by a cooperative agreement (W81XWH-07-2-0067) between the Henry M. Jackson Foundation for the Advancement of Military Medicine and the U.S. Department of Defense, and by a contract between Fisher BioServices and NYU (FBS50035-15).
Author contributions: B.F.H., S.M.A., S.C.H., G.D.T., and H.-X.L. designed the research and experiments and reviewed the data; Shelley Stewart, K.A., F.H.J., L.S., K.E.L., C.B., K.A., R.P., R.G.O., M.B., T.L.J., K.-K.H., and Sampa Santra performed research and data analysis; S.Z.-P., F.S., P.W.B., J.K., N.M., N.K., M.R., N.L.L., Sampa Santra, N.L.L., S.C.H., and S.Z.-P. provided support in research, reagents and comments; B.F.H. and S.M.A. wrote the paper, with support from S.C.H., G.D.T., H.-X.L., S.Z.-P., P.B., N.L.M., and J.H.K. providing comments and revisions.
The views expressed are those of the authors and should not be construed to represent the positions of the U.S. Army, Department of Defense, or Department of Veterans Affairs.
Footnotes
Published ahead of print 21 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00718-12.
REFERENCES
- 1. Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH, Investigators MOPH-TAVEG. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209–2220 [DOI] [PubMed] [Google Scholar]
- 2. Berman PW. 1998. Development of bivalent rgp120 vaccines to prevent HIV type 1 infection. AIDS Res. Hum. Retroviruses 14:S277–S289 [PubMed] [Google Scholar]
- 3. Berman PW, Huang W, Riddle L, Gray AM, Wrin T, Vennari J, Johnson A, Klaussen M, Prashad H, Kohne deWit CC, Gregory TJ. 1999. Development of bivalent (B/E) vaccines able to neutralize CCR5-dependent viruses from the United States and Thailand. Virology 265:1–9 [DOI] [PubMed] [Google Scholar]
- 4. Francis DP, Gregory T, McElrath MJ, Belshe RB, Gorse GJ, Migasena S, Kitayaporn D, Pitisuttitham P, Matthews T, Schwartz DH, Berman PW. 1998. Advancing AIDSVAX to phase 3: safety, immunogenicity, and plans for phase 3. AIDS Res. Hum. Retroviruses 14:325–331 [PubMed] [Google Scholar]
- 5. Lasky LA, Groopman JE, Fennie CW, Benz PM, Capon DJ, Dowbenko DJ, Nakamura GR, Nunes WM, Renz ME, Berman PW. 1986. Neutralization of the AIDS retrovirus by antibodies to a recombinant envelope glycoprotein. Science 233:209–212 [DOI] [PubMed] [Google Scholar]
- 6. Flynn NM, Fortahl DN, Harro CD, Judson FN, Mayer KH, Para MF, rgp120 HIV Vaccine Study Group 2005. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J. Infect. Dis. 191:654–665 [DOI] [PubMed] [Google Scholar]
- 7. Pitisuttithum P. 2008. HIV vaccine research in Thailand: lessons learned. Expert Rev. Vaccines 7:311–317 [DOI] [PubMed] [Google Scholar]
- 8. Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhu P, Liu J, Bess J, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. 2006. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441:847–852 [DOI] [PubMed] [Google Scholar]
- 10. Gorny MK, Stamatatos L, Volsky B, Revesz K, Williams C, Wang XH, Cohen S, Staudinger R, Zolla-Pazner S. 2005. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. J. Virol. 79:5232–5237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Honnen WJ, Krachmarov C, Kayman SC, Gorny MK, Zolla-Pazner S, Pinter A. 2007. Type-specific epitopes targeted by monoclonal antibodies with exceptionally potent neutralizing activities for selected strains of human immunodeficiency virus type 1 map to a common region of the V2 domain of gp120 and differ only at single positions from the clade B consensus sequence. J. Virol. 81:1424–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kwong PD, Doyle ML, Casper DJ, Cicala C, SALeavitt Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinge H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678–682 [DOI] [PubMed] [Google Scholar]
- 13. Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, Klasse PJ, Milne JL, Wilson IA, Olson WC, Moore JP, Subramaniam S. 2011. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glysoproteins display the same and open quaternary molecular architectures. Proc. Natl. Acad. Sci. U. S. A. 108:11440–11445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Finzi A, Pacheco B, Zeng X, Do Kwon Y, Kwong PD, Sodroski J. 2010. Conformational characterization of aberrant disulfide-linked HIV-1 gp120 dimers secreted from overexpressing cells. J. Virol. Methods 168:155–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gorny MK, Moore JP, Conley AJ, Karwowska S, Sodroski J, Williams C, Burda S, Boots LJ, Zolla-Pazner S. 1994. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of human immunodeficiency virus type 1. J. Virol. 68:8312–8320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kayman SC, Wu Z, Revesz K, Chen H, Kopelman R, Pinter A. 1994. Presentation of native epitopes in the V1V2 and V3 regions of human immunodeficiency virus type I gp120 by fusion glycoproteins containing isolated gp120 domains. J. Virol. 68:400–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McLellan J, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O'Dell S, Patel N, Shahzad-ul Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang GY, Diwanji D, Georgiev I, Kwon YD, Lee D, Louder MK, Moquin S, Schmidt SD, Yang ZY, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang LX, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pinter A, Honnen WJ, He Y, Gorny MK, Zolla-Pazner S, Kayman SC. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 78:5205–5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pinter A, Honnen WJ, Kayman SC, Trochev Wu OZ. 1998. Potent neutralization of primary HIV-1 isolates by antibodies directed against epitopes present in the V1/V2 domain of HIV-1 gp120. Vaccine 16:1803–1811 [DOI] [PubMed] [Google Scholar]
- 20. Bonsignori M, Hwang KK, Chen X, Tsao CY, Morris L, Gray E, Marshall DJ, Crump JA, Kapiga SH, Sam NE, Sinangil F, Pancera M, Ypongpimg Y, Zhang B, Zhu J, Kwong PD, O'Dell S, Mascola JR, Wu Nabel L, Phogat GJS, Seaman MS, Seaman MS, Whitesides JF, Moody MA, Kelsoe G, Yang X, Sodroski J, Shaw GM, Montefiori DC, Kepler TB, Tomaras GD, Alam SM, Liao H-X, Haynes BF. 2011. Analysis of a clonal lineage of HIV-1 Envelope V2/V3 conformational epitope-specific broadly neutralizing antibodies and their inferred unmutated common ancestors. J. Virol. 85:9998–10009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, Lehrman JK, Priddy FH, Olsen OA, Frey SM, Hammond PW, Kaminsky S, Zamb T, Moyle M, Koff WC, Poignard P, Burton DR. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Davenport T, Friend D, Ellingson K, Xu H, Caldwell Z, Sellhorn G, Kraft Z, Strong RK, Stamatatos L. 2011. Binding interactions between soluble HIV envelope glycoproteins and quaternary-structure-specific monoclonal antibodies PG9 and PG16. J. Virol. 85:7095–7107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haynes BF, McElrath GPMJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N. Engl. J. Med. 366:1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liao H-X, Sutherland LL, Xia S-M, Brock ME, Scearce RM, Vanleeuwen S, Alam SM, McAdams M, Weaver EA, Camacho ZT, Ma B-J, Li Y, Decker JM, Nabel GJ, Montefiori DC, Hahn BH, Korber BT, Gao F, Haynes BF. 2006. A group M consensus envelope glycoprotein induces antibodies that neutralize subsets of subtype B and C HIV-1 primary viruses. Virology 353:268–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, Wei X, Jiang C, Kirchherr JL, Gao F, Anderson JA, Ping LH, Swanstrom R, Tomaras GD, Blattner WA, Goepfert PA, Kilby JM, Saag MS, Delwart EL, Busch MP, Cohen MS, Montefiori DC, Haynes BF, Gaschen B, Athreya GS, Lee HY, Wood N, Seoighe C, Perelson AS, Bhattacharya T, Korber BT, Hahn BH, Shaw GM. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 105(21):7552–7557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tsao C, Liao H, Ma B, Chen H, Foulger AS, Lu X, Jaeger F, Hahn B, Shaw G, Swanstrom R, Cohen M, Kamanga G, Mascola J, Montefiori D, Alam MS, Haynes BF. 2010. Antigenicity and immunogenicity of transmitted/founder HIV envelope oligomers compared to chronic HIV envelopes. AIDS Res. Hum. Retroviruses 10(01):A27 [Google Scholar]
- 27. Gorny MK, Williams C, Wang X, Volsky B, O'Neal T, Li L, Seaman MS, Zolla-Pazner S. 2012. Functional and immunochemical cross-reactivity of V2-specific monoclonal antibodies from human immunodeficiency virus type 1-infected individuals. Virology 427:198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Alam SM, McAdams M, Boren D, Rak M, Scearce RM, Gao F, Camacho ZT, Gewirth D, Kelsoe G, Chen P, Haynes BF. 2007. The role of antibody polyspecificity and lipid reactivity in binding of broadly neutralizing anti-HIV-1 envelope human monoclonal antibodies 2F5 and 4E10 to glycoprotein 41 membrane proximal envelope epitopes. J. Immunol. 178:4424–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alam SM, Morelli M, Dennison SM, Liao HX, Zhang R, Xia SM, Rits-Volloch S, Sun L, Harrison SC, Haynes BF, Chen B. 2009. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc. Natl. Acad. Sci. U. S. A. 106:20234–20239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alam SM, Scearce RM, Parks RJ, Plonk K, Plonk SG, Sutherland LL, Gorny MK, Zolla-Pazner S, Vanleeuwen S, Moody MA, Xia SM, Montefiori DC, Tomaras GD, Weinhold KJ, Karim SA, Hicks CB, Liao HX, Robinson J, Shaw GM, Haynes BF. 2008. Human immunodeficiency virus type 1 gp41 antibodies that mask membrane proximal region epitopes: antibody binding kinetics, induction, and potential for regulation in acute infection. J. Virol. 82:115–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu P, Overman RG, Yates NL, Alam SM, Vandergrift N, Chen Y, Graw F, Freel SA, Kappes JC, Ochsenbauer C, Montefiori DC, Gao F, Perelson AS, Cohen MS, Haynes BF, Tomaras GD. 2011. Dynamic antibody specificities and virion concentrations in circulating immune complexes in acute to chronic HIV-1 infection. J. Virol. 85:11196–11207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tomaras GD, Yates NL, Liu P, Qin L, Fouda GG, Chavez LL, Decamp AC, Parks RJ, Ashley VC, Lucas JT, Cohen M, Eron J, Hicks CB, Liao HX, Self SG, Landucci G, Forthal DN, Weinhold KJ, Keele BF, Hahn BH, Greenberg ML, Morris L, Karim SS, Blattner WA, Montefiori DC, Shaw GM, Perelson AS, Haynes BF. 2008. Initial B-cell responses to transmitted human immunodeficiency virus type 1: virion-binding immunoglobulin M (IgM) and IgG antibodies followed by plasma anti-gp41 antibodies with ineffective control of initial viremia. J. Virol. 82:12449–12463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Safsten P, Klakamp SL, Drake AW, Karlsson R, Myszka DG. 2006. Screening antibody-antigen interactions in parallel using BIAcore A100. Anal. Biochem. 353:181–190 [DOI] [PubMed] [Google Scholar]
- 34. Kasturi SP, Skountozi I, Albrecht RA, Koutsonanos D, Hua T, Nakaya HI, Ravindran R, Stewart S, Alam M, Kwissa M, Villinger F, Murthy N, Steel J, Jacob J, Hogan RJ, GarcíA-Sastre A, Compans R, Pulendran B. 2011. Programming the magnitude and persistence of antibody responses with innate immunity. Nature 470:543–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Flynn BJ, Kastenmuller K, Willie-Reece U, Tomaras GD, Alam SM, Lindsay RW, Salazar AM, Perdiguero B, Gomez CE, Wagner R, Esteban M, Park CG, Trumpfheller C, Keler T, Panteleo G, Steinman RM, Seder R. 2011. Immunization with HIV Gag targeted to dendritic cells followed by recombinant New York Vaccinia virus induces robust T-cell immunity in nonhuman primates. Proc. Natl. Acad. Sci. U. S. A. 108:7131–7136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bonsignori M, Moody JPMA, Alpert MD, Chen X, Hwang K-K, Gilbert PB, Huang YT, Gurley C, Kozink D, MArshall DJ, Whitesides JF, Tsao C-Y, Kaewkungwal J, Nitayaphan S, Pitisuttithum P, Rerks-Ngam S, Kim JH, Michael NL, Tomaras GD, Lewis GK, Devico A, Evans DT, Ferrari G, Liao HX, Haynes BF. 2012. ADCC-mediating antibodies from an HIV-1 vaccine efficacy trial target multiple epitopes and preferentially use the VH1 gene family. J. Virol. 86:11521–11532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liao HX, Levesque MC, Nagel A, Dixon A, Zhang R, Walter E, Parks R, Whitesides J, Marshall DJ, Hwang KK, Yang Y, Chen X, Gao F, Munshaw S, Kepler TB, Denny T, Moody MA, Haynes BF. 2009. High-throughput isolation of immunoglobulin genes from single human B cells and expression as monoclonal antibodies. J. Virol. Methods 158:171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng NY, Mays I, Garman L, Helms C, James J, Air GM, Capra JD, Ahmed R, Wilson PC. 2008. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 453:667–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dennison SM, Sutherland LL, Jaeger FH, Anasti KM, Parks R, Stewart S, Bowman C, Xia SM, Zhang R, Shen X, Scearce RM, Ofek G, Yang Y, Kwong PD, Santra S, Liao HX, Tomaras G, Letvin NL, Chen B, Alam SM, Haynes BF. 2011. Induction of antibodies in rhesus macaques that recognize a fusion-intermediate conformation of HIV-1 gp41. PLoS One 6:e27824 doi:10.1371/journal.pone.0027824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ma BJ, Alam SM, Go EP, Lu X, Desaire H, Tomaras GD, Bowman C, Sutherland LL, Scearce RM, Santra S, Letvin NL, Kepler TB, Liao HX, Haynes BF. 2011. Envelope deglycosylation enhances antigenicity of HIV-1 gp41 epitopes for both broad neutralizing antibodies and their unmutated ancestor antibodies. PLoS Pathog. 7:e1002200 doi:10.1371/journal.ppat.1002200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ferrari G, Pollara J, Kozink D, Harms T, Drinker M, Freel S, Moody MA, Alam SM, Tomaras GD, Ochsenbauer C, Kappes JC, Shaw GM, Hoxie JA, Robinson JE, Haynes BF. 2011. An HIV-1 gp120 envelope human monoclonal antibody that recognizes a C1 conformational epitope mediates potent antibody-dependent cellular cytotoxicity (ADCC) activity and defines a common ADCC epitope in human HIV-1 serum. J. Virol. 85:7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. VanCott TC, Bethke FR, Polonis VR, Gorny MK, Zolla-Pazner S, Redfield RR, Birx DL. 1994. Dissociation rate of antibody-gp120 binding interactions is predictive of V3-mediated neutralization of HIV-1. J. Immunol. 153:449. [PubMed] [Google Scholar]
- 43. Bonsignori M, Wu X, Moody MA, Liao H, Hwang K, Crump JA, Capiga SH, Sam NE, Tomaras GD, Chen X, Tsao C, Alam SM, Nabel GJ, Kwong PD, Morris L, Montefiori D, Mascola JR, Haynes BF. 2011. Isolation of CD4-binding site and V2/V3 conformational (quaternary) broadly neutralizing antibodies from the same HIV-1 infected African subject. AIDS Res. Hum. Retroviruses 27:A120 [Google Scholar]
- 44. Billings EA, Karasavvas N, de Souza MS, Currier J, Pitisuttithum P, Kaewkunwal J, Nitayaphan S, Gilbert PB, Tomaras GD, Zolla-Pazner SB, Haynes BF, Michael NL, Rerks-Ngarm S, Kim JH, Rao M. 2011. Surface plasmon resonance analysis of anti-gp120 V2-specific IgG antibodies generated in the RV144 Thai Trial. AIDS Res. Hum. Retroviruses 27:21 [Google Scholar]
- 45. Zolla-Pazner S, Cardozo T, Decamp A, Haynes B, Kim J, Kong X, Michael N, Rerks-Ngam S, Williams C. 2011. V2-reactive antibodies in RV144 vaccinees' plasma. AIDS Res. Hum. Retroviruses 27:A21 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.