Abstract
Rift Valley fever virus (RVFV) is a Phlebovirus (Bunyaviridae family) transmitted by mosquitoes. It infects humans and ruminants, causing dramatic epidemics and epizootics in Africa, Yemen, and Saudi Arabia. While recent studies demonstrated the importance of the nonstructural protein NSs as a major component of virulence in vertebrates, little is known about infection of mosquito vectors. Here we studied RVFV infection in three different mosquito cell lines, Aag2 cells from Aedes aegypti and U4.4 and C6/36 cells from Aedes albopictus. In contrast with mammalian cells, where NSs forms nuclear filaments, U4.4 and Aag2 cells downregulated NSs expression such that NSs filaments were never formed in nuclei of U4.4 cells and disappeared at an early time postinfection in the case of Aag2 cells. On the contrary, in C6/36 cells, NSs nuclear filaments were visible during the entire time course of infection. Analysis of virus-derived small interfering RNAs (viRNAs) by deep sequencing indicated that production of viRNAs was very low in C6/36 cells, which are known to be Dicer-2 deficient but expressed some viRNAs presenting a Piwi signature. In contrast, Aag2 and U4.4 cells produced large amounts of viRNAs predominantly matching the S segment and displaying Dicer-2 and Piwi signatures. Whereas 21-nucleotide (nt) Dicer-2 viRNAs were prominent during early infection, the population of 24- to 27-nt Piwi RNAs (piRNAs) increased progressively and became predominant later during the acute infection and during persistence. In Aag2 and U4.4 cells, the combined actions of the Dicer-2 and Piwi pathways triggered an efficient antiviral response permitting, among other actions, suppression of NSs filament formation and allowing establishment of persistence. In C6/36 cells, Piwi-mediated RNA interference (RNAi) appeared to be sufficient to mount an antiviral response against a secondary infection with a superinfecting virus. This study provides new insights into the role of Dicer and Piwi in mosquito antiviral defense and the development of the antiviral response in mosquitoes.
INTRODUCTION
Flaviviridae, Togaviridae, and Bunyaviridae, which constitute the main group of arthropod-borne viruses (arbovirus), have an important impact on human and animal health worldwide. Many arboviruses are transmitted by mosquitoes which become infected when taking a blood meal from an infected vertebrate. After infecting the mosquito midgut, the virus disseminates through the whole body, including the salivary glands and the genital tract. When present in the salivary glands, the virus can be further propagated from an infected mosquito to a vertebrate through the mosquito saliva during biting. In addition, infection of the genital tract allows vertical transmission from the adult to the offspring. In contrast to the case with vertebrates, arbovirus infection is usually not detrimental to mosquitoes, which remain infectious during their entire lifetime. Although they lack an adaptive immune response, arthropods have an innate immune defense against microbes based mainly on RNA interference (RNAi), a conserved mechanism found in almost all eukaryotes and serving as an antiviral defense mechanism (1, 2; reviewed in references 3, 4, and 1).
Activation of RNAi-based immunity has been found in many mosquitoes and mosquito cells infected by different arboviruses, such as Sindbis virus, dengue virus, West Nile virus, O'nyong-nyong virus, and La Crosse virus (5–11). Essentially, it was shown that knocking down the components of the RNAi pathway leads to an increase in viral replication, thus indicating the key role played by RNAi in establishing virus protection in mosquitoes. RNAi relies on the presence of intracellular double-stranded RNAs (dsRNAs) generated either by RNA secondary structures in the viral genome or from viral replicative intermediates which are processed into 21-nucleotide (nt) small virus-derived RNA duplexes (viRNAs) by an RNase III enzyme, Dicer-2 (12). The RNA duplexes contain a guide strand that provides sequence specificity for RISC (13) and a passenger strand that is removed from the activated RISC by the combined endonuclease activities of Argonaute-2 (Ago-2) and C3PO (13–15). The remaining guide directs RISC to cognate RNAs in the cells and induces Ago-2-mediated cleavage and sequence-specific degradation of the target RNA molecules.
Besides the Dicer-2 pathway, which relies on small interfering RNAs (siRNAs), other RNAi silencing pathways involving microRNAs (miRNAs) or Piwi RNAs (piRNAs) participate in the innate immune response of mosquitoes (reviewed in reference 16). In general, the size of the viRNAs reveals the pathway utilized (17). In Drosophila melanogaster, siRNAs and miRNAs depend, respectively, on Dicer-2 and Dicer-1 and are 21 and 22 nt long. In contrast, piRNAs are 24 to 30 nt long, are generated in a Dicer-independent manner, and require Ago-3, Aubergine (Aub), and Piwi for biogenesis (18). Originally, piRNAs were thought to be specific to germ line cells and essential for silencing retrotransposons, but since then, they have also been detected in mosquito and vertebrate somatic cells (19–22). The biogenesis of piRNAs has been studied mostly in Drosophila. It involves synthesis of primary piRNAs from single-stranded precursors, which then initiate an amplification cycle known as the ping-pong amplification cycle (23). Primary piRNAs are associated with Piwi and Aub; they are typically generated from one polarity and possess predominantly a 5′-uridine bias (U1). Cleavage of target RNA by primary piRNAs initiates the “ping-pong” amplification process, which involves Ago-3 and Aub activities. In contrast to the siRNA duplexes generated by Dicer, complementarity between sense and antisense piRNAs is generally limited to 10 nt; they contain 2′-O-methyl groups at their 3′ ends. The secondary piRNAs possess a specific signature with a strong U1 bias for the Aub-associated piRNAs and an adenosine bias at position 10 (A10) for the Ago3-associated piRNAs (24, 25).
Rift Valley fever (RVF) is a mosquito-borne zoonotic viral disease affecting livestock and humans (26). The causative agent, the RVF virus (RVFV), a Phlebovirus of the Bunyaviridae family (27), is transmitted to vertebrates mainly by mosquitoes, but aerosols and direct contact with infected biological material are also involved in transmission during outbreaks. RVFV was first isolated in the 1930s during a severe outbreak affecting sheep that caused many animal deaths and abortions (28). RVFV is responsible for high fatality rates in sheep and cattle. During outbreaks, various symptoms are observed in humans, from mild febrile illness to fatal hemorrhagic fever (for a recent review, see reference 29). Initially confined to sub-Saharan regions of Africa where periodic epidemics and epizootics had occurred, RVF spread to Egypt in 1977 and to the Middle East in 2000 (reviewed in references 30, 29, and 31). Like all the members of the family, RVFV possesses a single-stranded tripartite RNA genome of negative/ambisense polarity (32, 33). The L and M segments code, respectively, for the L RNA-dependent RNA polymerase protein and for the precursor to the glycoproteins Gn and Gc, which also generates two nonstructural proteins during processing. The S segment utilizes an ambisense strategy and codes for two proteins in opposite polarities, the nucleoprotein N and the nonstructural NSs protein (34). The two open reading frames (ORFs) encoded by the S segment are separated by a highly conserved intergenic region (IGR) which possesses signals for transcription termination of the N and NSs mRNAs (35–37).
During the last decade, efforts have been made to better understand the pathogenesis in vertebrates. Recent studies have established the fundamental role of the NSs protein in the mechanisms of pathogenicity. This viral protein forms filaments in nuclei of infected mammalian cells (38–40) and inhibits basal cellular transcription and interferon-β gene transcription via the interaction of NSs with, respectively, the p44 and p62 subunits of the TFIIH general transcription factor (41, 42) and the SAP30 subunit of the Sin3A repressor complex (43). NSs was also shown to downregulate the expression of PKR by degrading the protein through the proteasome pathway (44–46). In addition, NSs interacts with gamma satellite pericentromeric sequences, provoking abnormal nuclei during cell division (38).
In contrast to the case with vertebrates, very little is known about the effect of RVFV infection in mosquitoes and the response to RVFV infection developed by these arthropods (47, 48). In this work, we analyzed infection in three mosquito cell lines originating with Aedes aegypti (Aag2) and A. albopictus (U4.4 and C6/36). We characterized RVFV-mosquito interactions using immunofluorescence, biochemical analysis, and deep RNA sequencing approaches. We showed that all these cell lines were sensitive to RVFV and produced virus but responded differently to RVFV infection. In the case of Aag2 cells, the most obvious manifestation was the rapid disappearance of the NSs filaments and the clearance of the protein from both the nuclear and cytoplasmic compartments. This phenomenon seemed to be exacerbated in U4.4 cells, where NSs filaments were never observed. On the contrary, in C6/36 cells, the NSs filaments that were observed in nuclei early after infection remained visible throughout the time course of infection. Deep-sequencing analysis of viRNAs produced in these cell lines after RVFV infection revealed the presence of viRNAs in the three cell types. The viRNAs increased in number during infection and targeted the three segments of the RVFV genome, with a preference for the S and M segments. The production of viRNAs was extremely low in C6/36 cells, indicating an inefficient RNAi system, in agreement with recent reports showing that the Dicer RNAi pathway is disabled in these cells (49). In Aag2 cells, the average size and the pattern of the viRNA population evolved as infection progressed. The Dicer-2-mediated RNAi that appeared to be preponderant in the early phase of infection was progressively overtaken by Piwi-mediated RNAi in the later phases of infection and during persistence. In the Dicer-2-incompetent C6/36 cells, even though the Piwi-mediated RNAi pathway failed to mount a primary antiviral response strong enough to allow the establishment of persistence, it was sufficient to silence viral replication during secondary infection with a superinfecting virus. In Aag2 and U4.4 cells, an efficient RNAi response implicating both Dicer2 and Piwi was able to control RVFV replication, eliminate the NSs protein, and establish persistence.
MATERIALS AND METHODS
Cells and viruses.
Aag2 mosquito cells (a kind gift of the Lan Department of Entomology, University of Wisconsin—Madison), which were derived from Aedes aegypti larvae, were cultured and maintained in Schneider's Drosophila medium (21720; Gibco, Invitrogen) containing 10% fetal calf serum (FCS) (Biowest-S1810-500). Aedes albopictus-derived C6/36 and U4.4 cells were grown in Leibowitz's L-15 medium (Gibco, Invitrogen-BRL) supplemented with 10% FCS (Lonza-DE14-801-F)–2% tryptose phosphate broth (Sigma). Mosquito cell lines were incubated at 28°C without CO2. Murine fibroblastic L929 cells were cultured and maintained Dulbecco's modified Eagle's medium (DMEM) (31966; Gibco, Invitrogen) supplemented with l-glutamine and 10% of FCS. Penicillin (5 units/ml) and streptomycin (5 μg/ml) were added to the medium.
RVFV strain ZH548 was isolated from a human case in Egypt (50). The rZHNSs and rZHDelNSs-RLuc RVFV recombinant strains were produced using the reverse-genetics system with the ZH548 genome (51, 52). Virus stocks were produced by infecting Vero cells at multiplicity of infection (MOI) of 10−3 and by harvesting the medium 72 h postinfection (p.i.).
For most of the experiments, Aag2, C6/36, and L929 cells were infected with ZH548 strain at an MOI of 5 PFU per cell and incubated in maintenance medium supplemented with only 2% FCS during various periods of time.
Virus titration.
Virus was titrated by plaque assay by infecting confluent monolayers of Vero E6 cells with serial viral dilutions (10−1 to 10−7). Cells were incubated under an agarose layer for 4 to 5 days at 37°C before being fixed and stained with crystal violet solution (0.2% crystal violet, 3.7% formaldehyde, and 20% ethanol). Viral titers were calculated, taking into account plaque numbers and the dilution factor. All the manipulations involving RVFV were performed in biosafety level 3 (BSL-3) containment.
Immunostaining and confocal microscopy analysis.
Aedes mosquito cells and L929 cells grown on coverslips to 80% confluence were infected with ZH548 at a MOI. of 5 PFU per cell. Cells were fixed with 3.7% formaldehyde and permeabilized 10 min at room temperature with 0.25% Triton X-100 in phosphate-buffered saline (PBS). After 30 min of incubation with PBS–5% BSA, the cells were incubated with the primary antibodies, rabbit anti-N and mouse polyclonal anti-NSs, both diluted 1:500 in PBS–5% BSA. After 1 h of incubation at room temperature, the coverslips were washed four times in PBS–5% BSA and then treated with the corresponding secondary antibodies (Alexa Fluor goat antibodies; Invitrogen) at a dilution of 1:800. Then, cell nuclei were stained with Hoechst stain (1 μg/ml; Invitrogen). Cells were washed four times in PBS–5% BSA and mounted by using Fluoromount solution (SouthernBiotech). Stained cell samples were examined under a confocal microscope (Leica SP5), and images were analyzed by the ImageJ software program (W. S. Rasband, ImageJ, National Institutes of Health, Bethesda, MD [http://imagej.nih.gov/ij/], 2011).
Western blot.
Subcellular fractionation of insect and mouse cells was performed as follows. Infected cells were pelleted by centrifugation at 2,000 rpm for 5 min, resuspended in cytoplasmic lysis buffer (25 mM Tris HCl [pH 7.5], 50 mM NaCl, 2 mM EDTA, and 0.6% Nonidet P-40) containing protease inhibitors (Complete protease inhibitor; Roche) and incubated for 5 min on ice. The lysate was pelleted at 3,600 rpm for 2 min; the supernatant (cytoplasmic fraction) was collected. The pellet was washed gently and lysed in nuclear lysis buffer (25 mM Tris-HCl [pH 7.5], 50 mM NaCl, 2 mM EDTA, 0.6% Nonidet P-40, and 0.3% SDS) containing protease inhibitors (Complete protease inhibitor; Roche). After 5 min of incubation on ice, the nuclear fraction was centrifuged at 10,000 rpm for 10 min before being treated with DNase I (Roche) for 30 min at 37°C. Whole-cell lysates were obtained by lysing pelleted cells directly in the nuclear lysis buffer and treated as described above.
Lysates were run on a 12% polyacrylamide-SDS gel and blotted onto nitrocellulose. Proteins were detected by using rabbit polyclonal antibodies against NSs (a kind gift from J. M. Egly, IGBMC, Strasbourg, France) (43) or mouse polyclonal antibodies against N.
Northern blots.
Mosquito cells were infected with ZH548 at a MOI of 3 to 5 and incubated for various times at 28°C, and total cellular RNAs were extracted by TRIzol reagent (Invitrogen) according to the manufacturer's instruction. The aqueous phase was precipitated with propan-2-ol. The precipitate was centrifuged for 30 min at 17,000 × g, and the pellet was washed with 70% ethanol, dried, and dissolved in 30 μl of RNase-free distilled water. Eight micrograms of total RNA was denatured in 10 mM MOPS buffer containing 1 M glyoxal (Merck) and 50% of dimethyl sulfoxide (DMSO). The MOPS buffer contains 20 mM 3-[N-morpholino]-propanesulfonic acid, 5 mM sodium acetate, and 1 mM EDTA at pH 7. The RNA samples were separated by electrophoresis at 50 V in 1.5% agarose gels in MOPS buffer. RNAs were transferred onto Hybond-N membranes (Amersham Pharmacia Biotech). Blotting was performed as already described (37). Briefly, the N or NSs probe was synthesized in vitro from linearized plasmids using either T7 or SP6 RNA polymerase in the presence of [α-32P]CTP (3,000 Ci/mmol; PerkinElmer). After TRIzol extraction, the probes were purified by filtration through Sephadex Quick spin columns (Roche).
Luciferase reporter assay.
Monolayers of mosquito cells grown in 12-well tissue culture plates were either left uninfected or infected with ZH548 or rZHDelNSs at a MOI of 2 PFU per cell. After incubation for 24 h, 48 h, or 72 h, the cells were infected with the recombinant virus rZHDelNSs-RLuc at a MOI of 2 PFU per cell and incubated for 24 h. Cells were lysed in passive lysis buffer (Promega), and Renilla luciferase activity was determined using a luciferase reporter assay system (Promega). The results were normalized to those for mock-infected cells which were infected with rZHDelNSs-RLuc.
Small-RNA isolation and deep sequencing.
Approximately 3 × 106 Aag2, U4.4, and C6/36 cell grown in 25-cm2 flasks were infected with the ZH548 RVFV strain with a MOI of 5 PFU per cell. Total RNA from mosquito cells was extracted using the TRIzol reagent (Invitrogen) at 24 h, 48 h, or 72 h postinfection. The concentration, purity, and integrity of the RNA samples were determined using the NanoDrop ND-1000 (Thermo Scientific) and BioAnalyzer 2100 (Agilent) instruments.
Small-RNA libraries were prepared with the “small RNA sample prep v1.5 conversion kit,” following the manufacturer's instructions (Illumina). Briefly, the 10- to 30-bp RNA fragments were purified from total RNA on a 15% urea PAGE gel (Bio-Rad). The purified small RNAs were ligated with 3′ RNA adaptor (5′-/5rApp/ATCTCGTATGCCGTCTTCTGCTTG/3ddC/), which is specifically modified to target miRNAs and other small RNAs that have a 3′ hydroxyl group resulting from cleavage by Dicer and other RNA processing enzymes. The 5′ RNA adaptor (5′-GUUCAGAGUUCUACAGUCCGACGAUC) was then ligated to the 5′ phosphate end of small RNA. Reverse transcription was done to convert the RNA to cDNA, which was then selectively amplified by PCR. The PCR products were purified by 10% PAGE (Bio-Rad) in a size range between 90 to 130 nt and checked on a high-sensitivity DNA chip (Agilent). Libraries were sequenced using the Illumina genome analyzer II to generate single-end 36-bp reads. Adapter sequences and sequences of low quality were removed using a KNIME workflow (53–55). The sequences were then aligned to the RVFV reference genome (GenBank accession number DQ380206) using the software program Bowtie (56) (version 0.12.7 and the following parameters: -a/–best/-q/-m50/-e50/-solexa1.3-quals). Reads conforming to specific length constraints (18 to 30, 21, 24 to 25, or 27 to 28) were subselected and further partitioned according to the strand they aligned to. Counts per position for statistical analysis were calculated using the pileup command from the samtools software package (57). The logos sequences were generated using a local version of the software program Seqlogo (53–55) from weblogo.berkeley.edu.
RESULTS
Mosquito cells are susceptible to infection by the ZH548 strain of RVFV.
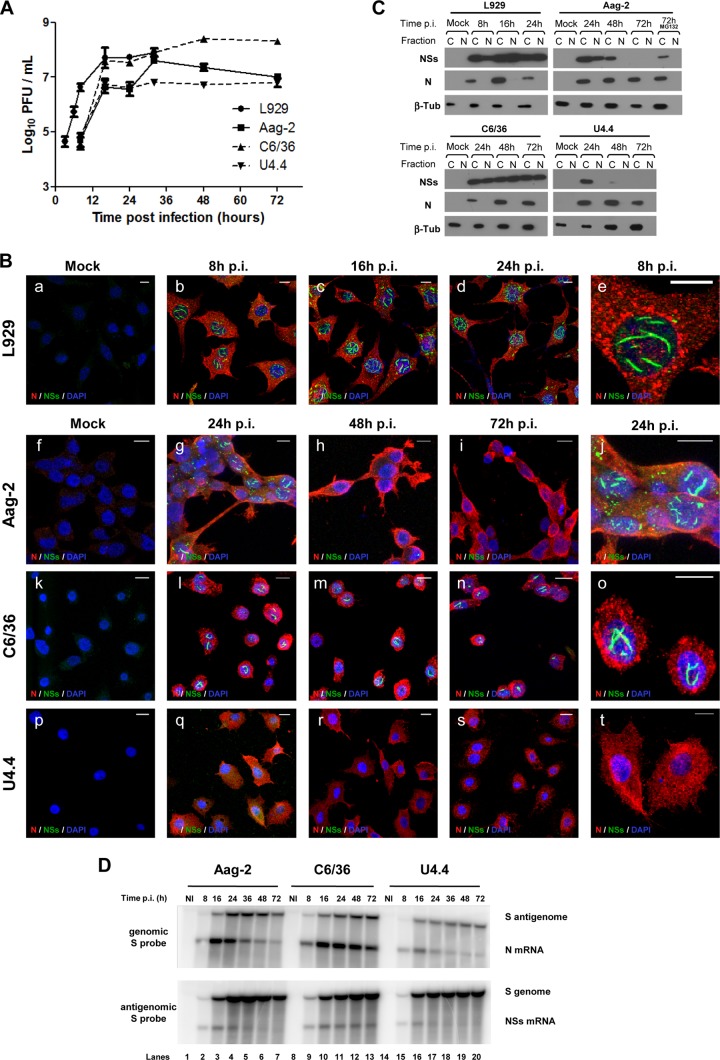
To examine the impact of RVFV infection on mosquito cells, we used continuous cell lines derived from Aedes mosquitoes, which are a competent vector for RVFV, the C6/36 and U4.4 cells subcloned from the Singh's original Aedes albopictus cultures (58, 59), and the Aag2 cells originally derived from Aedes aegypti by Peleg (60) and further characterized by Lan and Fallon (61). The three mosquito cell lines were infected at a MOI of 2 with the naturally virulent ZH548 strain of RVFV isolated from a human case during the Egyptian outbreak in 1977-1978. After infection, cells were incubated at 28°C for 24, 48, and 72 h p.i., and the presence of infectious viral particles in the medium was determined by plaque assay (Fig. 1A). For comparison, the L929 murine fibroblasts were infected and analyzed in parallel. The best viral yield (108.4 PFU/ml) was obtained in C6/36 cells; it was slightly higher than the titer produced in L929 cells (107.8 PFU/ml). The most restricted viral growth was observed in Aag2 or U4.4 cells, with a maximal titer of 106.7 PFU/ml. In mosquito cells, the complete virus cycle lasted longer than that in mammalian L929 cells due to the lower incubation temperature: a plateau was reached at approximately 24 h p.i., compared to 16 h p.i. in L929 cells. Repeatedly, C6/36 cells produced the highest viral titers compared to Aag2 or U4.4 cells, which is in agreement with the sensitivity of C6/36 cells to arbovirus growth. It is noteworthy that the infected L929 cells were all dead within 48 h, whereas none of the RVFV-infected mosquito cells exhibited any cytopathic effect after 6 or 7 days p.i. (data not shown).
Fig 1.
RVFV replicates in mosquito cells but downregulates expression of its NSs protein. (A) Cell cultures were infected with ZH548 at a MOI of 3 to 5. Maintenance medium was collected, and virus was titrated by PFU. (B) Cells stained with 4′,6-diamidino-2-phenylindole (DAPI) and using antibodies against N (red) NSs (green) were examined by immunofluorescence. (C) Western blot of cytoplasmic (C) and nuclear (N) extracts using antibodies against RVFV N and NSs and beta-tubulin. MG132 was added for 6 h, from 66 to 72 h p.i. (+MG132) (D) Total RNAs analyzed by Northern blotting using probes hybridizing with the S antigenome and N mRNA (upper panels) and genome and NSs mRNA (lower panels).
RVFV NSs protein is differentially expressed in C6/36, U4.4, and Aag2 cells.
Because NSs plays a major role in virulence and cytotoxicity in vertebrates (62, 63), its expression was assessed by immunofluorescence and confocal microscopy (Fig. 1B), as well as by Western blotting (Fig. 1C), for all three mosquito cell lines infected with the RVFV ZH548 strain. Characteristic filaments formed by NSs, which have been extensively described for mammalian cells (38–40), were observed in L929 cells at 8 h p.i. run as a control (Fig. 1B). In Aag2 cells, nuclear NSs filaments that were observed at early times (24 h) p.i. almost totally disappeared as infection progressed (Fig. 1B). In C6/36 cells, NSs formed nuclear filaments, as in the case of L929 cells, starting at a relatively early time p.i. (16 h p.i.) and remaining visible during the entire time course of infection. In contrast, formation of NSs filaments was never observed in U4.4 cells. As in the mammalian cells, the viral N protein accumulated in the cytoplasm of all cell lines tested (Fig. 1B), confirming the sensitivity of the mosquito cells to RVFV infection.
To ascertain these results, the presence of the viral proteins N and NSs was also analyzed by Western blotting after fractionating RVFV-infected cells into cytoplasmic and nuclear extracts (Fig. 1C). As expected, the N protein was detected only in the cytoplasmic fraction at all collection times and in all the cell lines, indicating that the fractionation was performed adequately since this virus replicates only in the cytoplasm. In agreement with immunofluorescence data, the NSs protein was present in both the cytoplasmic and nuclear fractions at all the collection time points in L929 and C6/36 cells. In Aag2 cells, NSs was detected in both the cytoplasm and nuclear fractions at 24 h p.i. but disappeared progressively, first from the nuclear fraction at 48 h p.i. and finally from both compartments, so that at 72 h p.i., no NSs band was visible in either of these fractions (Fig. 1C). Since the NSs filaments are stable, at least in mammalian cells, their disappearance suggested a possible degradation by the proteasome. Treatment of the infected cells with the proteasome inhibitor MG132 for 6 h from 66 to 72 h p.i. restored NSs only in the cytoplasmic fraction (Fig. 1C), but no filament could be recovered in nuclei after the treatment (not shown). This indicated that the filament formed in the early phase of infection in Aag2 cells was likely eliminated through proteasome degradation. In the case of U4.4 cells, the NSs protein was never detected in the nuclear fraction and was only visible in the cytoplasmic fraction at 24 h p.i. (Fig. 1C). It is noteworthy that RVFV produced in mosquito cells was genetically stable since no mutation in the viral genome was observed during sequencing even after 12 passages in Aag2 cells, with NSs still forming nuclear filaments after infection of L929 cells (not shown). The disappearance of the NSs protein in Aag2 cells and its absence in U4.4 cells suggested that expression of NSs could be inhibited at the transcriptional level in these mosquito cells.
NSs expression is downregulated at the transcriptional level in Aag2 and U4.4 cells but not in C6/36 cells.
To determine whether the expression of NSs was regulated at the transcriptional level, total cellular RNAs from the RVFV-infected cells were extracted and analyzed by Northern blotting using radioactive probes allowing detection of the S genome or antigenome and of the NSs and N mRNAs (Fig. 1D). At early time points, the four RNA molecules were easily observed in the three cell lines, the N and NSs mRNAs being at optimal levels at 16 h p.i. However, at later time points, the synthesis of N and NSs mRNAs was regulated differently depending on the cell line. In C6/36 cells, the two mRNAs remained at a high and constant steady-state level all through infection, whereas in Aag2 cells, both mRNAs declined rapidly starting at 24 h p.i. This phenomenon was more pronounced in U4.4 cells than in Aag2 cells. Concerning the synthesis of the RVFV genome/antigenome, both RNA molecules accumulated in C6/36 cells through the course of infection, while the amounts of genome and antigenome were significantly reduced at late times in Aag2 cells. Slightly smaller amounts of antigenome accumulated in U4.4 cells compared to those on C6/36 or Aag2 cells (Fig. 1D).
The viRNA population in Aag2 cells evolves during the course of infection.
In insects, RNAi is a major antiviral defense mechanism that activates the cleavage of viral double-stranded RNAs into 21-nt siRNAs by Dicer-2 (reviewed in reference 16). To determine if RVFV infection differentially induced an RNAi response in the three mosquito cell lines used in this work, the presence of viRNAs was investigated by deep RNA sequencing and mapping to the RVFV genome and antigenome. Because Aag2 cells constitute a good model when analyzing RVFV infection in mosquito cells, the presence of viRNAs was first analyzed in this cell type. Thus, Aag2 cells were infected with ZH548 RVFV at a MOI of 5, and total cellular RNAs were extracted at 24 h, 48 h, and 72 h p.i. (Table 1). The sizes of RNAs selected for sequencing ranged from 10 to 36 nt. Each sample generated similar numbers of reads, i.e., approximately 20 × 106 reads. Most of these siRNAs aligned to the RVFV genome or antigenome and were therefore considered viRNAs. Their population increased during the time course of infection, representing 4%, 10%, and 24% of total siRNAs at 24 h, 48 h, and 72 h p.i., respectively. More than 95% of the viRNAs had sequences strictly identical to the viral sequence, with only 4 to 7% of viRNAs carrying one mismatch compared to the RVFV ZH548 reference sequence.
Table 1.
Characteristics of viRNAs expressed in RVFV-infected mosquito cells; evolution during acute and persistent infection in Aag2 cells
| Small-RNA library | RVFV RNA segment | Time p.i. (h) | Reads aligning to RVFV genome |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Total reads |
Reads with mismatches (%) |
||||||||
| No. total (unique) | Avg length (nt) | Avg quality scorea | No. (%): |
0 | ≥1 | ||||
| Genomic sense | Antigenomic sense | ||||||||
| Aag-2 | L segment | Mock infection | 21,888 (16,682) | 10.63 | 35.44 | 7,274 (33.2) | 15,624 (71.3) | 528 (2.41) | 21,360 (97.6) |
| 24 | 54,195 (53,682) | 19.11 | 36.23 | 23,308 (43) | 31,092 (57.3) | 41,911 (77.3) | 12,284 (22.6) | ||
| 48 | 124,516 (119,998) | 18.66 | 36.53 | 61,825 (49.6) | 63,929 (51.3) | 92,358 (74.2) | 32,158 (25.8) | ||
| 72 | 312,657 (306,790) | 23.68 | 36.67 | 112,001 (35.8) | 201,910 (64.6) | 282,132 (90.2) | 30,525 (9.8) | ||
| Passage 3 | 609,884 (593,874) | 23.01 | 36.69 | 336,885 (55.2) | 281,208 (46.1) | 553,914 (90.8) | 55,970 (9.2) | ||
| M segment | Mock infection | 10,352 (9,450) | 11.33 | 34.75 | 6,625 (64) | 4,228 (40.8) | 527 (5.1) | 9,825 (94.9) | |
| 24 | 355,721 (355,353) | 22.77 | 36.37 | 190,798 (53.6) | 165,063 (46.4) | 341,638 (96) | 14,083 (4) | ||
| 48 | 678,728 (675,733) | 21.51 | 36.6 | 349,433 (51.5) | 330,379 (48.7) | 639,250 (94.2) | 39,478 (5.8) | ||
| 72 | 2,265,853 (2,249,092) | 24.34 | 36.63 | 1,045,619 (46.1) | 1,222,596 (53.9) | 2,185,093 (96.4) | 80,760 (3.6) | ||
| Passage 3 | 1,469,089 (1,458,095) | 24.05 | 36.58 | 936,518 (63.7) | 535,245 (36.4) | 1,408,329 (95.9) | 60,760 (4.1) | ||
| S segment | Mock infection | 4,338 (4,139) | 11.78 | 35.08 | 1,435 (33.1) | 2,975 (68.6) | 446 (10.3) | 3,892 (89.7) | |
| 24 | 472,666 (464,823) | 19.61 | 35.92 | 56,205 (11,9) | 416,499 (88.1) | 454,575 (96.2) | 18,091 (3.8) | ||
| 48 | 1,147,187 (1,136,372) | 18.83 | 36.07 | 81,385 (7.1) | 1,065,959 (92.9) | 1,102,611 (96.1) | 44,576 (3.9) | ||
| 72 | 2,412,216 (2,407,348) | 23.12 | 36.37 | 164,089 (6.8) | 2,248,302 (93.2) | 2,340,636 (97) | 71,580 (3) | ||
| Passage 3 | 2,828,925 (2,824,469) | 22.93 | 36.5 | 187,089 (6.6) | 2,641,998 (93.4) | 2,742,861 (97) | 86,064 (3) | ||
| U4.4 | L segment | Mock infection | 23,802 (16,542) | 10.7 | 35.22 | 19,110 (80.3) | 7,281 (30.6) | 627 (2.6) | 23,175 (97.3) |
| 48 | 153,413 (151,045) | 23.2 | 36.14 | 73,141 (47.7) | 81,092 (52.8) | 138,236 (90.1) | 15,177 (9.9) | ||
| M segment | Mock infection | 17,856 (14,417) | 10.54 | 34.71 | 8,646 (48.4) | 9,616 (53.8) | 579 (3.2) | 17,277 (96.8) | |
| 48 | 1,967,446 (1,966,524) | 24.95 | 36.28 | 1,300,017 (66) | 667,784 (33.9) | 1,855,337 (94.3) | 112,109 (5.7) | ||
| S segment | Mock infection | 6,386 (6,120) | 11.48 | 34.72 | 1,485 (23.2) | 4,955 (77.6) | 890 (13.9) | 5,496 (86.1) | |
| 48 | 2,114,814 (2,113,941) | 25.06 | 36.1 | 257,160 (12.1) | 1,857,749 (87.8) | 2,020,445 (95.5) | 94,369 (4.5) | ||
| C6/36 | L segment | Mock infection | 17,784 (11,576) | 10.6 | 35.4 | 13,011 (73.1) | 7,515 (42.3) | 787 (4.4) | 16,997 (95.6) |
| 48 | 47,428 (44,986) | 15.86 | 32.9 | 24,092 (50.8) | 24,541 (51.7) | 30,441 (64.2) | 16,987 (35.8) | ||
| M segment | Mock infection | 13,259 (12,627) | 10.96 | 35.1 | 5,117 (38.6) | 8,531 (64.3) | 601 (4.5) | 12,658 (95.5) | |
| 48 | 198,265 (196,710) | 17.28 | 33.45 | 133,373 (67.2) | 66,138 (33.3) | 176,283 (88.9) | 21,982 (11.1) | ||
| S segment | Mock infection | 9,343 (9,148) | 11.66 | 35.72 | 3,064 (32.8) | 6,318 (67.6) | 882 (9.4) | 8,461 (90.6) | |
| 48 | 134,035 (133,678) | 17.27 | 32.99 | 25,488 (19) | 108,613 (81) | 118,869 (88.7) | 15,166 (11.4) | ||
Quality score represents an average of the confidence in each sequenced nucleotide in each viRNA.
The viRNAs were distributed according to each segment of the viral genome (Table 1). Considering the number of reads normalized per segment length, the S segment (1,690 nt in length) was the most intensely targeted through the entire time course of infection, with 280 × 103, 680 × 103, and 1,400 × 103 reads per kb, respectively, at 24, 48, and 72 h p.i., followed by the M segment (3,885 nt in length) with 90 × 103, 170 × 103, and 580 × 103 reads per kb, respectively, at 24, 48, and 72 h p.i. The L segment (6,404 nt in length) was the least targeted, with 8.4 × 103, 19.4 × 103, and 48.8 × 103 reads per kb, respectively, at the three selected time points. It can be noticed that the number of reads mapping to each segment increased similarly for the L, M, and S segments during the time course of infection, since the increase for each segment at 72 h compared to the result at 24 h p.i. was, respectively, 5.7-, 6.3-, and 5.1-fold. A small population of siRNAs from noninfected cells mapped to each of the segments of the viral genome, but their average length of 11 nt differed significantly from that of the viRNAs present in infected cells (Table 1).
For each segment, viRNAs were mapped to the genome or the antigenome: for the L and M segments, viRNAs were almost evenly distributed among the genome and antigenome, but for the S segment, a strong bias was observed at each time point, since approximately 90% of the viRNAs specific for the S segment aligned to the antigenome (Table 1).
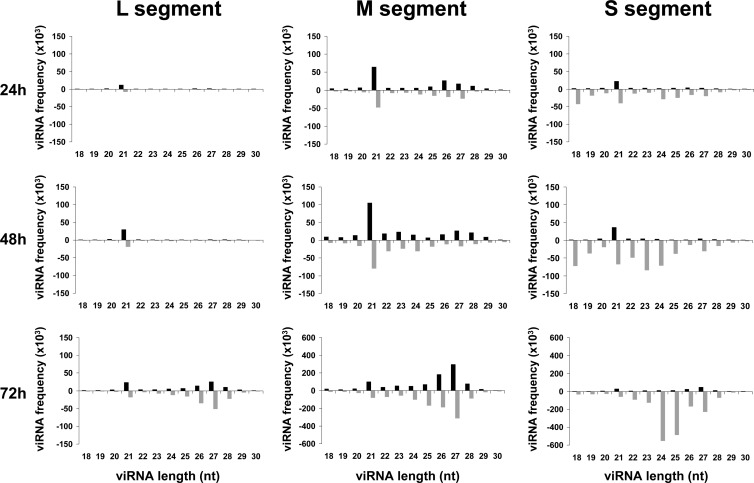
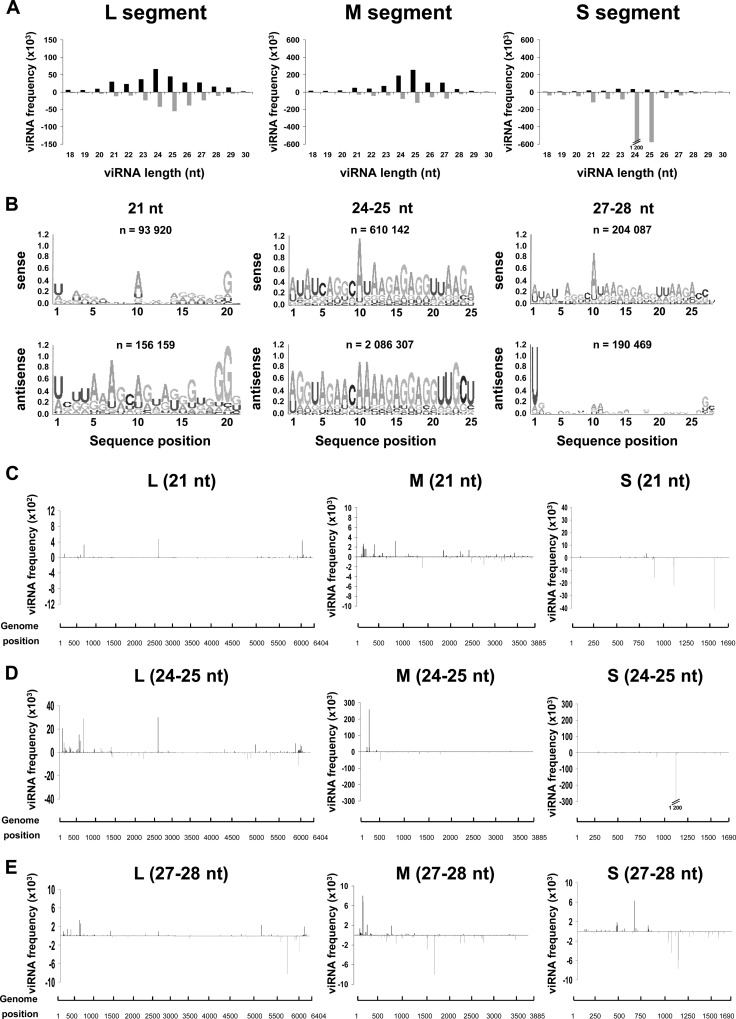
The size distribution of the viRNAs, ranging from 18 to 30 nt and aligning to the whole genome/antigenome, is shown in Fig. 2. At each time point, the pattern was different between those mapping to the S segment and those mapping to the L and M segments. For the L and M segments, the class of 21-nt viRNAs was predominant in samples collected at 24 h and 48 h p.i., and each size class of viRNAs was relatively homogenously distributed between the genome and antigenome. While the total amount of viRNAs derived from the L and M segments increased between 24, 48, and 72 h p.i. (note the different scales on the histograms), an important change in size distribution occurred at 72 h p.i., with a broad class of 24- to 28-nt viRNAs becoming predominant at this time p.i. (Fig. 2). In the case of the S segment, the 21-nt-long viRNAs of both polarities were present at 24 and 48 h p.i., but additional classes smaller (18 to 20 nt) and larger (24 to 28 nt) in size, mainly aligning to the antigenome, also contributed to the population of viRNAs. Between 24 h and 48 h p.i., the number of reads in each size class increased by at least a factor of 2. At 72 h p.i., the profile of the size distribution changed significantly as the class of 21 nt became minor, giving the impression of disappearing even though they remained constant or increased slightly over time compared to the class of 24 to 28 nt. In addition, the antigenomic viRNAs were strikingly more abundant than the genomic ones; this was also true at 24 h and 48 h p.i. but was exacerbated at 72 h as the genome-derived viRNAs became a minority. At 72 h p.i., while viRNAs derived from the L and M segments had a predominant size of 27 to 28 nt, those derived from the S segment were predominantly 24 or 25 nt long.
Fig 2.
Kinetics of viRNA profiles in Aag2 cells during RVFV infection. Size distribution and density plots of viRNAs mapping to the L, M, and S segments, with the upper and lower histograms representing the genome and antigenome sense, respectively.
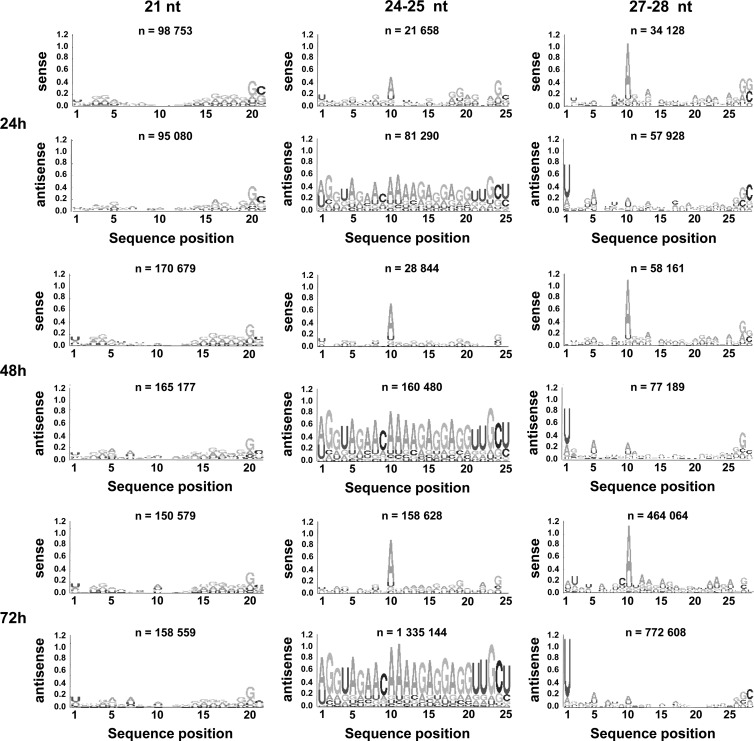
According to current knowledge, the 21-nt-long viRNAs are typical siRNAs generated by the Dicer-2-dependent RNAi pathway, whereas RNAs of 24 to 30 nt are processed through the Dicer-independent Piwi (piRNA) pathway (64). In Drosophila, the piRNAs were reported to possess a 5′ U bias, with the ping-pong piRNAs enriched in A at position 10 (24). Although little is known about the mosquito Piwi pathway, a similar sequence signature was observed in arbovirus-infected mosquitoes (49, 65). Therefore, based on the size range of the viRNAs produced in Aag2 cells after RVFV infection, we assumed that the 21-nt viRNAs, produced predominantly during the early phase of infection, were generated by the Dicer-2-dependent pathway while the 24- to 30-nt viRNAs, which became predominant later, were produced by the Dicer-2-independent Piwi-derived pathway. This assumption was clearly substantiated by the Logo analysis (Fig. 3): none of the 21-nt viRNAs in the genomic or antigenomic polarity displayed any signature of piRNAs. In contrast, an A at position 10 was predominant among the 27- to 28-nt viRNAs of genomic polarity, and a U at the 5′ end was often observed for those of opposite polarity, indicating a ping-pong signature. It should be noted that at the early time point (24 h p.i.), viRNAs with a Piwi signature were already visible, even though they were much less abundant than later after infection (Fig. 3). Because the class of 24- to 25-nt viRNAs became important at later times p.i., particularly for the S segment, the sequences of these viRNAs were also analyzed: an A at position 10 was predominant in viRNAs aligning to the genome, while those which aligned to the antigenome were rather heterogeneous in sequence (Fig. 3), suggesting that they were not ping-ping piRNAs like the 27- to 28-nt viRNAs but still derived from the Piwi pathway (see Discussion).
Fig 3.
Sequence analysis of viRNAs. Logo analysis of 21-, 24/25-, and 27/28-nt viRNA of the genomic (sense) or antigenomic (antisense) polarity at each time point.
Hot spots targeted by the 21- and 27-nt viRNAs in Aag2 cells localized at different positions in the RVFV genome.
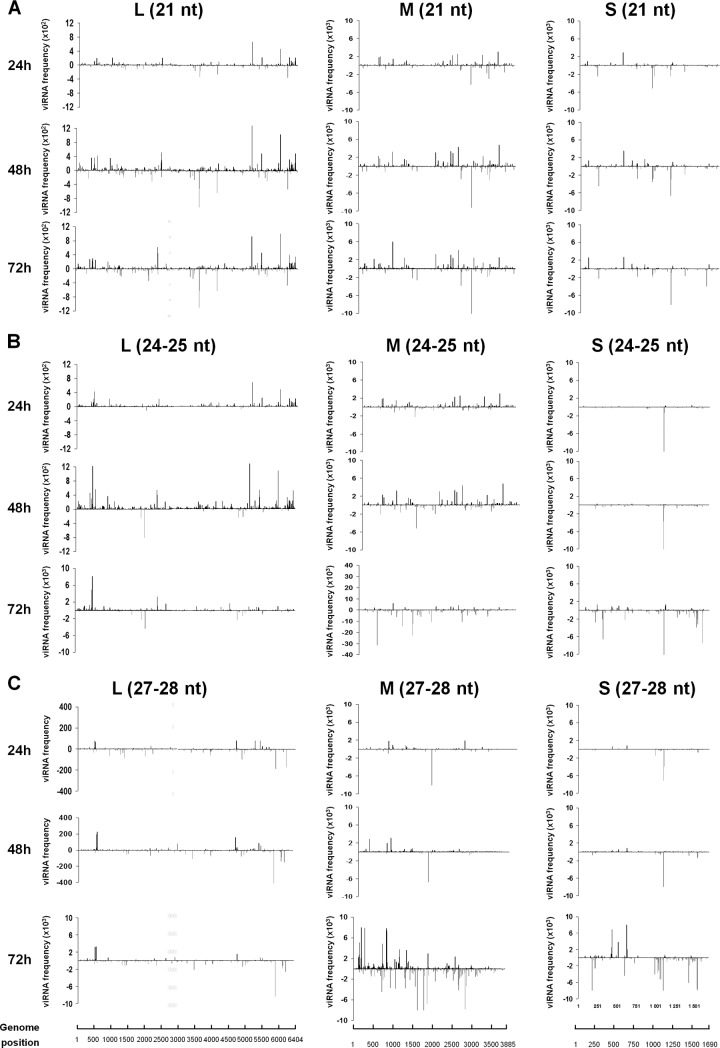
To characterize the viRNA populations more accurately, we aligned them to the viral genome sequences, and their matching frequency was determined for each nucleotide across the length of each segment in both orientations. The three classes, 21, 24/25, and 27/28 nt, were analyzed separately (Fig. 4). Figure 4A shows that the 21-nt-long viRNAs mapped to the three segments of the genome at 24, 48, and 72 h p.i. Regions intensely targeted, defined as “hot spots,” were scattered over each segment in both the genome and antigenome strands. These hot spots increased in intensity at 48 and 72 h, but their position in each segment remained exactly the same. Targeting on both strands suggests that the viral replicative forms, which might be transiently in double-stranded form at the fork of replication in this negative-stranded RNA virus, triggered the RNAi response.
Fig 4.
Mapping of viRNAs produced in Aag2 cells on the RVFV genome. Histograms represent the distribution of the 21-nt (A), 24/25-nt (B), or 27/28-nt (C) viRNAs, with the upper and lower histograms representing the genome and antigenome sense, respectively.
The 24/25-nt and 27/28-nt viRNAs also mapped to the three segments of the RVFV genome/antigenome, with some intensely targeted regions (Fig. 4B and C). For the S segment in particular, these viRNAs generally targeted the antigenome more abundantly than the genome. Generally, the hot spots corresponding to the three size classes of viRNAs did not target the same positions, strongly suggesting that they were produced by different mechanisms. There is one noticeable exception, however: the hot spot near position 1100 in the S segment was observed in the profile of the three size classes of viRNAs. In general, new hot spots appeared at 72 h p.i., compared to 24 and 48 h p.i., especially on the M and S segments, and the viRNA frequencies increased significantly during the course of infection.
Altogether, these results indicated that the RNAi pathway induced in Aag2 cells after RVFV infection changed during the course of infection, with the Dicer-2 and Piwi pathways dominating, respectively, the early and late phases of infection.
The Piwi RNA pathway becomes preponderant in persistently RVFV-infected Aag2 cells.
Infection of mosquito cell lines by arboviruses usually leads to persistent infection with no cytopathic effect, survival of the culture, which can be passaged many times, and production of infectious virus, though viral titers may vary with passages. When choosing the different time points, 24 h, 48 h, and 72 h p.i., we selected early, medium, and late times of the acute phase. However, it is difficult to determine when the acute phase ends and when the persistent/chronic infection starts. Considering that the viral cycle lasts approximately 20 to 24 h, it is likely that at 48 h and 72 h p.i., the cells have already entered the persistent state. To investigate RNAi during persistence, ZH548-infected Aag2 cells were passaged at regular intervals (4 to 6 days). As already described for other arboviruses, RVFV titers were found to fluctuate from approximately 105 to 102 PFU/ml, depending on the passage. The NSs filaments were never observed in any of the 12 to 15 passages, while immunostaining showed that N was expressed (not shown).
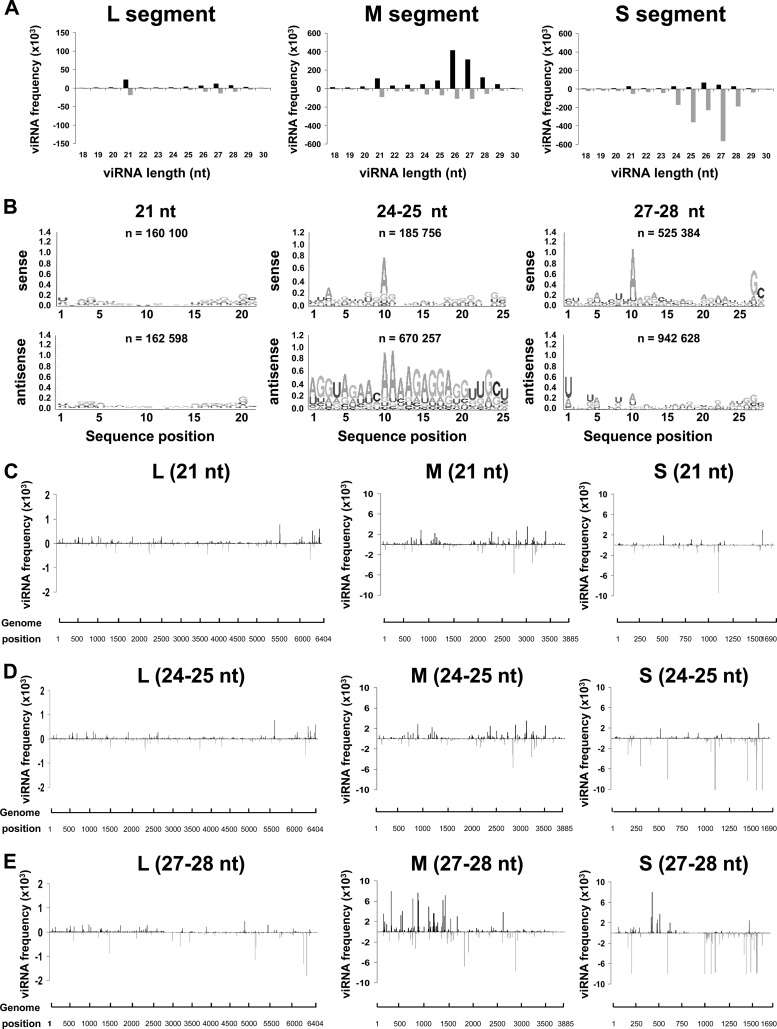
To determine how RNAi was regulated during persistence, we selected ZH548-infected Aag2 cells at passage 3 and subcultured them for 24 h after the third passage, extracted their RNAs, and sequenced their small RNAs. The number of reads and the proportion of viRNAs aligning to the RVFV genome were similar to those obtained during acute infection, with a bias again toward viRNAs of antigenomic sense for the S segment (Table 1). Figure 5A shows the distribution of these viRNAs among the three segments according to their sizes. Although the presence of some 21-nt viRNAs was detected, they represented a minor class, especially among those mapping to the S and M segments. The majority of the viRNA population ranged from 23 to 28 nt in size, with a peak at 24 to 25 nt. Similarly to what was observed during the acute infection, the viRNA aligning to the L and M segments mapped almost equally to the genome and antigenome, whereas the viRNAs aligning to the S segment mapped almost exclusively to the antigenome. The sequence of the viRNAs was analyzed: the Piwi ping-pong signature was clearly visible for the 27- to 28-nt viRNAs, which were enriched in 5′-terminal uridine and an adenosine at position 10, respectively, in the viRNA aligning to the antigenome or the genome (Fig. 5B). In the case of the 24/25-nt viRNAs, a significant enrichment in A10 and U1 for the genome and antigenome viRNAs could be observed (Fig. 5B), suggesting a Piwi origin. The origin of the 21-nt viRNAs was not clear, since they were enriched in A10 and U1. The distribution of the hot spots (21-nt, 24/25-nt, and 27/28-nt viRNAs) on the three segments was also analyzed (Fig. 5C to E). The localization of the major hot spots was similar to that found during the acute infection. Altogether, these results indicate that the RNAi response mounted by the infected cells at passage 3 had the same characteristics as the one observed in cells infected for 72 h, with a clear preponderance of piRNAs.
Fig 5.
Production of viRNAs in RVFV-persistently infected Aag2 cells at passage 3. Size distribution and density of the viRNAs aligning to each segment of RVFV genomic (upper) and antigenomic (lower) strand in ZH548-persistently infected Aag2 cells which underwent 3 passages are shown. Total RNAs were collected at 24 h after the third subculture (A). (B) Logo analysis of 21-, 24/25-, and 27/28-nt viRNA of the genomic (sense) or antigenomic (antisense) polarity. (C to E) Mapping of the 21-nt (C), 24/25-nt (D), or 27/28-nt (E) viRNAs on the RVFV genome.
Dicer-2- and Piwi-mediated RNAi responses are expressed in U4.4 cells.
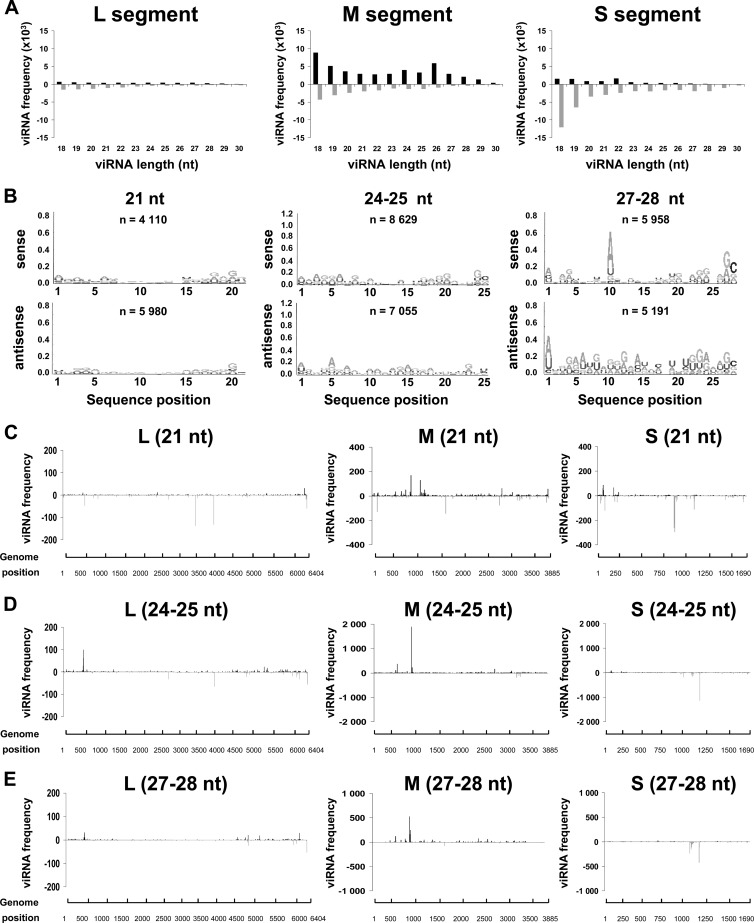
To determine if the production of viRNAs was dependent on cell lines and/or on the mosquito species, we carried out deep RNA sequencing on ZH458 RVFV-infected U4.4 and C6/36 cells, which are both derived from A. albopictus mosquitoes. For logistic reasons, only the 48-h time point was analyzed. Table 1 indicates that the total number of reads obtained with these samples was similar to that obtained from infected Aag2 cells. Concerning the reads aligning to the RVFV genome, a strong difference was observed between U4.4 (or Aag2) and C6/36 cells. In C6/36 cells, a much smaller percentage (2.1%) of reads aligned to the RVFV genome than in U4.4 (20% of the total number of reads) or Aag2 (10.3%) cells. Interestingly, the number of viRNAs observed at 48 h in U4.4 cells (4.2 × 106) was approximately 2-fold higher than that in Aag2 cells at the same time point and was close to that observed at 72 h in Aag2 cells (4.98 × 106) (see Table 1), suggesting that the RNAi activity was very efficient in U4.4 cells, with an accelerated and probably exacerbated process. This is in agreement with the results shown in Fig. 1, showing the downregulation of the viral mRNAs.
The distribution of the viRNAs in U4.4 cells is shown in Fig. 6A. The overall profile resembles the one shown in Fig. 2 for Aag2 cells. We aligned the viRNAs to the viral sequence and determined their frequency per nucleotide across the length of each segment in both orientations. Similarly to Aag2 cells, the viRNAs matched to the S segment more abundantly than to the M segment, which in turn was more targeted than the L segment. The size distribution of viRNAs was also similar to that observed in Aag2 cells. However, the profile observed in U4.4 cells at 48 h p.i. was closer to that found in Aag2 cells at 72 h p.i. For the L and M segments, two preponderant classes of 21 and 27 nt aligned almost equally to the genome and antigenome. For the S segment, the dominant class was 24- to 28-nt viRNAs, aligning almost exclusively to the antigenome. The Logo analysis of each size class of viRNAs indicated that the 21-nt viRNAs had no Piwi signature, but the 24- or 25-nt and 27- or 28-nt viRNAs did, even though only the 27- or 28-nt ones had a clear ping-pong signature (Fig. 6B), also as previously observed in Aag2 cells (Fig. 2).
Fig 6.
Production of viRNAs in U4.4 cells infected with RVFV. (A) Size distribution and density of the viRNAs aligning to each segment of RVFV genomic (upper) and antigenomic (lower) polarity in U4.4 cells infected by RVFV ZH548 collected at 48 h p.i.. (B) Logo analysis of 21-, 24/25-, and 27/28-nt viRNA of the genomic (sense) or antigenomic (antisense) polarity. (C to E) Mapping of the 21-nt (C), 24/25-nt (D), or 27/28-nt (E) viRNAs on the RVFV genome.
Again as in Aag2 cells, the 21-, 24/25-, and 27/28-nt viRNAs observed in U4.4 cells revealed significant differences in intensity and localization (compare Fig. 4A to C to Fig. 6C to E). In U4.4 cells, the positions of the hot spots in the three segments were the same as those found in Aag2 cells, suggesting the existence of conserved features for the generation of hot spots. These data indicated that the two different species of mosquitoes, A. aegypti and A. albopictus, displayed similar antiviral responses after RVFV infection.
viRNAs in C6/36 cells possess a Piwi signature.
In C6/36 cells, not only was the number of reads aligning to the RVFV genome much lower (approximately 10-fold) than that in U4.4 or Aag2 cells (Table 1), but also the profile of the viRNAs was quite different from that observed for U4.4 or Aag2 cells (Fig. 7A). Every size class, including 21- or 27-nt-long viRNAs, was represented, but the distribution was heterogeneous. It is noteworthy that the class of 18- to 20-nt viRNAs was abnormally abundant compared to those observed in Aag2 and U4.4 cells, suggesting that they were degraded by a nuclease activity, similar to what has been observed in West Nile virus-infected C6/36 cells (66). However, this was unlikely to be due to inappropriate treatments, since ribosomal (not shown) and viral RNAs (Fig. 1) did not exhibit any degradation. The Logo analysis showed that the 27- or 28-nt viRNAs presented the A10/U1 ping-pong signature; no sequence preference was visible in the 24/25- and 21-nt viRNAs (Fig. 7B). The location of hot spots corresponding to the 21-, 24/25-, or 27- /28-nt viRNAs was determined for each segment (Fig. 7C to E). Many hot spots were found at the same position as in Aag2 or U4.4 cells.
Fig 7.
Production of viRNAs produced in C6/36 cells infected with RVFV. (A) Size distribution of the viRNAs of genomic (upper) and antigenomic (lower) polarity in C6/36 cells infected by RVFV ZH548 collected at 48 h p.i. (B) Logo analysis of 21-, 24/25-, and 27/28-nt viRNA of the genomic (sense) or antigenomic (antisense) polarity. (C to E) Mapping of the 21-nt (C), 24/25-nt (D), or 27/28-nt (E) viRNAs on the RVFV genome.
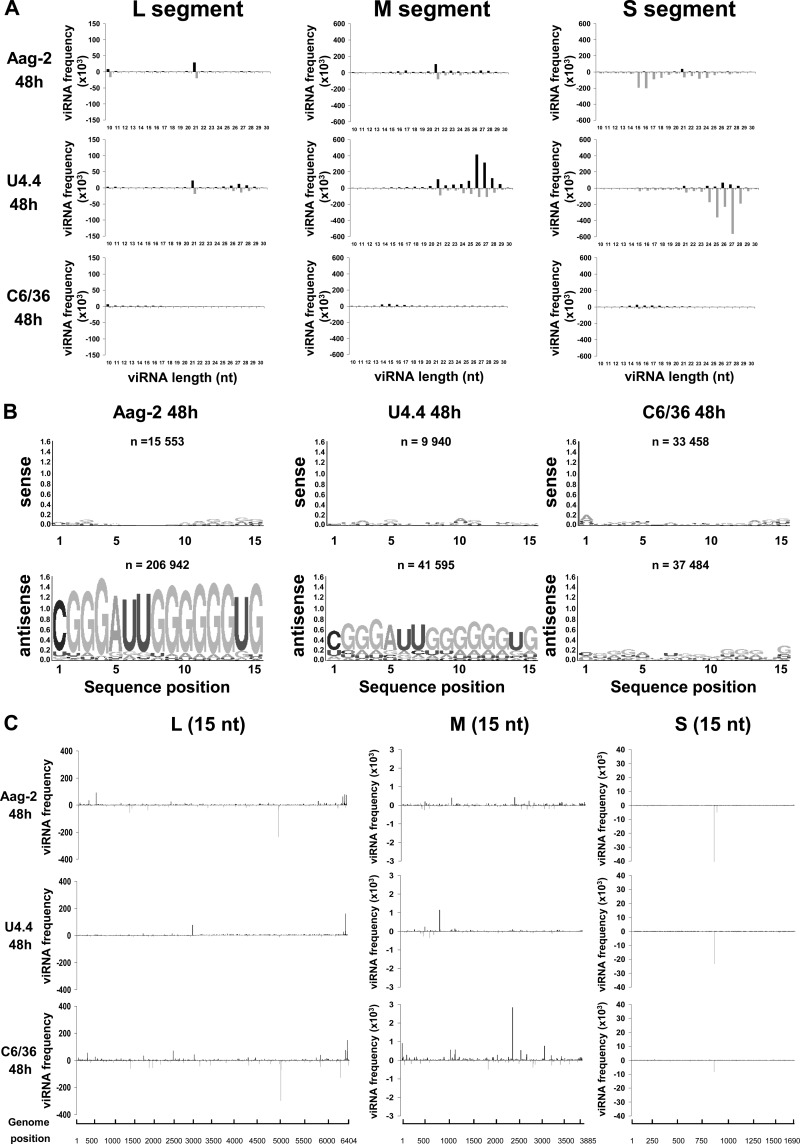
Because of the unexpected size profile of the viRNAs in C6/36 cells, we extended the analysis to a broader range of 10 to 30 nt instead of the usual 18 to 30 nt (Fig. 8), and we compared the viRNA profiles obtained at 48 h p.i. in the three cell lines. In C6/36 cells, we observed a prominent peak of 14 to 19 nt aligning to the three segments of the genome and antigenome in addition to the 21- and 23- to 28-nt viRNAs already described (Fig. 8A). In Aag2 cells, viRNAs of 14 to 19 nt were also visible in the profile of the S segment and to a lesser extent of the M segment (Fig. 8A). In U4.4 cells, the small class of viRNAs was detected but represented only a minor population. To determine if these viRNAs were derived from longer ones, such as the 21-, 24/25-, or 27/28-nt viRNAs, we mapped them across the three segments and compared the locations of the hot spots (Fig. 8C). The profile was not comparable either to the 27/28-nt piRNAs or to the 21-nt viRNAs, suggesting that they were not generated by the same process. To further characterize these small viRNAs, we analyzed their sequences: no canonical signature was observed in any strand, but a predominant sequence enriched in Gs in the antigenomic strand appeared to be conserved in the three cell lines (Fig. 8B). Such a sequence was not detected in noninfected cells (not shown).
Fig 8.
Analysis of unusually small viRNAs produced in mosquito cells infected with RVFV. (A) Size distribution of the viRNAs of genomic (upper) and antigenomic (lower) polarity in Aag2, U4.4, and C6/36 cells infected by RVFV ZH548, collected at 48 h p.i. (B) Logo analysis of 15-nt viRNA of the genomic (sense) or antigenomic (antisense) polarity. (C) Mapping of the 15-nt-long viRNAs on the RVFV genome in the three cell lines.
Altogether, these results suggest that upon RVFV infection, the RNAi responses are similar in Aag2 and U4.4 cells but different in C6/36 cells. Concerning C6/36 cells, our data are in accordance with the recent reports (49, 66, 67) which indicated that C6/36 cells are defective in RNAi due to a homozygous single nucleotide deletion in the Dicer-2 gene which leads to a premature termination codon and a truncated inactive protein (49). Unexpectedly, we found a significant number of very small viRNAs (13 to 19 nt) in these cells and in Aag2 cells. Similarly, unusually small viRNAs were observed in dengue-2 virus-infected Aedes aegypti mosquitoes and in mammalian cells (68, 69). Their origin remained undetermined (see Discussion).
RVFV-infected C6/36 cells are able to mount an efficient PDR in spite of the absence of a functional Dicer-2 pathway.
In addition to a defense mechanism against viruses, RNAi is involved in RNA silencing implicated in several phenomena, such as pathogen-derived-resistance (PDR) or posttranscriptional gene silencing. However, the importance of the siRNA and piRNA pathways in these phenomena has not yet been characterized. PDR is developed by insects and plants to prevent superinfection with a homologous virus (70). Therefore, we took advantage of the dysfunctional Dicer pathway in C6/36 cells to determine whether this pathway is required for PDR. In order to do this, the C6/36 cells were primarily infected with RVFV ZH548 (or rZHDelNSs, in case NSs would have a specific detrimental effect) and then superinfected with the homologous RVFV rZHDelNSs-RLuc, expressing Renilla luciferase in place of NSs (51, 52). The experiments designed to assess PDR in C6/36 cells were also performed with U4.4 and Aag2 cells. Cells were superinfected either at 24 h, 48 h, or 72 h after primary infection, and the ability of the primarily infected cells to replicate the superinfecting virus was assessed through the expression of Renilla luciferase. Cells which were not infected with the primary RVFV were used to evaluate the 100% replication activity of rZHDelNSs-RLuc. In C6/36 cells infected by ZH458, replication of the superinfecting virus was inhibited drastically (approximately 99%) at the three time points, and the level of inhibition was similar to that observed in ZH548-infected Aag2 cells (approximately 98 to 99%). The restriction observed in C6/36 cells was even more stringent than that in U4.4 cells, where viral replication was less inhibited, particularly in the 24-h-p.i. sample (88%). Interestingly, replication of the superinfecting virus was also drastically inhibited in the three cell lines infected primarily with rZHDelNSs, indicating that NSs had no significant impact on the phenomenon. Altogether, these results demonstrated that the Dicer-2-defective C6/36 cells were able to mount an efficient PDR against the superinfecting virus at least as efficiently as Aag2 or U4.4 cells, indicating that a functional Dicer-2 pathway was not required for PDR and that most likely the Piwi pathway alone was able to carry out this activity.
DISCUSSION
Many species of mosquitoes infected by RVFV have been found in nature, suggesting that they are competent vectors for transmitting the virus. Once infected, females of competent vectors can transmit RVFV to their eggs by vertical transmission and infect vertebrates via saliva when taking a blood meal. In order to avoid any detrimental effect in the infected mosquito cells, viral replication must be tightly controlled. To this end, mosquitoes have developed RNAi mechanisms which control virus replication. Contrary to the case for RVFV infection in vertebrates, little is known concerning RVFV infection in mosquito cells. In this work, we have analyzed the mosquito-RVFV interactions at the cellular and molecular levels with a particular emphasis on the RNAi response, which was characterized using a large-scale deep-sequencing approach. This analysis was carried out with cell cultures from A. albopictus and A. aegypti mosquitoes known to be competent vectors.
Altogether, our results point out the major role of the Dicer-2 pathway for restriction of viral growth during RVFV infection. C6/36 cells from A. albopictus, which have been recently reported to be deficient in the Dicer-2 pathway (49, 66, 67), were able to replicate RVFV to high titers, while the fully competent Dicer-2 U4.4 cells, also derived from A. albopictus, were more efficient in restricting RVFV growth. The restriction was particularly evident when examining the NSs filament: U4.4 cells were able to extinguish the NSs expression and the filament formation, while C6/36 cells did not. However, the C6/36 cells, which possess a functional Piwi pathway, were able to mount an antiviral response against a secondary infection with a related virus during PDR. Although it cannot be excluded that mechanisms other than RNAi may be involved in PDR (71–73), these data strongly suggest that the Dicer-2 pathway is not necessary for the establishment of PDR and that the Piwi pathway alone can support the antiviral response implicated in PDR. The observation that PDR is efficient in C6/36 cells is not unique to RVFV, since it was also found for C6/36 cells infected by other arboviruses or expressing dengue-2 virus-specific sequences (74–77), but the role of Piwi could not be discussed at that time because it was not known that these cells were defective for Dicer-2.
In several recent studies, piRNAs as well as siRNAs have been detected in mosquitoes or mosquito cells infected by several arboviruses, such as dengue virus (68), West Nile virus (68), chikungunya virus, Sindbis virus, and La Crosse virus (65), indicating that the Piwi pathway could be implicated in the antiviral defense of arthropods. In this study, we also present evidence that Dicer-2-mediated 21-nt siRNAs, as well as 24- to 28-nt piRNAs, were generated during RVFV infection. In addition, we demonstrated by a kinetics study that in RVFV-infected Aag2 cells, the two different pathways are regulated during the time course of infection, with a siRNA Dicer-2-mediated response taking a preponderant place early during infection, followed by a Piwi-mediated RNAi response dominant at late times after infection and during persistence. Thus, our results demonstrate, for the first time to our knowledge, that the two types of RNAi response were differentially controlled during the course of infection.
In the three cell lines, including Aag2 cells at each time point, the 27/28-nt viRNAs presented the clear characteristic of RNAs produced by the Piwi ping-pong pathway. These ping-pong 27/28-nt viRNAs were generated from the three segments, but for each segment, differences were observed with regard to the predominant species: viRNAs derived from the S segment were mostly 24 or 25 nt long, whereas those derived from the M and L segments were 27 or 28 nt long. Unlike the 27- or 28-nt viRNAs, which possess the ping-pong signature, the 24- or 25-nt viRNAs had no U1 preference but only an A at position 10 in the genomic strand, strongly suggesting that they were also mediated by the Piwi pathway. A similar trend could be observed for the L and M segments, which generated a majority of 27/28-nt viRNAs at 72 h p.i. but shifted to 24 or 25 nt at passage 3, suggesting that the size of the dominant population of viRNAs might change as infection progresses. The absence of U1 in piRNAs generated in dengue-2 virus-infected mosquitoes has also been reported; it appeared to be specific to this virus (67, 68, 78). In Drosophila, piRNAs that associate with each member of the Piwi protein family have a distinct size: 25, 24, and 23 nt for Piwi, Aub, and Ago3, respectively (23). In addition, piRNAs associated with Ago3 have a strong preference for adenosine at position 10 but no U1 bias, while those associated with Aub have a U1 bias. Assuming that mosquitoes and flies have similar Piwi pathways, our observations suggest that various Piwi proteins were implicated in their biogenesis mechanisms and evolved during RVFV infection.
Several articles describing RNAi in infected C6/36 cells (65–67) reported the existence of piRNAs. Interestingly, the sizes of these small RNAs seem to vary: in many cases they were 26 and 27 nt long, but smaller classes (19 to 20 nt long) were found in West Nile virus-infected C6/36 cells (23). Our data show that a population of unusually small viRNAs was particularly evident in C6/36 cells but was also detected in Aag2 and U.4 cells. Small RNAs (13 to 19 nt) below the canonical size were also reported in Aedes aegypti infected with dengue-2 virus (68) and in human cells associated with Kaposi sarcoma-associated herpesvirus K12-1 miRNA (69). The existence of these unusually small siRNAs adds to the complexity of the mosquito response, since they might participate in biological process.
In the Dicer-2 competent U4.4 and Aag2 cells, the major manifestation of the functional RNAi activity capable of setting up an efficient antiviral response resided in the extinction of the NSs filament. Although we mainly focused here on acute infection, we should mention that RVFV-infected C6/36 cells could not be passaged, so that persistent infection could not be established, while Aag2 and U4.4 cells underwent more than 12 passages of persistent infection. In mammalian cells, NSs inhibits cellular transcription through its interaction with p44 and p62 of TFIIH (41, 42). Preliminary data obtained by the two-hybrid system indicated that Aedes aegypti p44 is also able to interact with NSs in Aag2 cells (not shown). Knowing that NSs is a strong inhibitor of general transcription and that it prevents normal chromosome segregation through its interaction with pericentromeric sequences at least in vertebrate cells (38, 42), it seems logical that persistence could be established only if the formation of the NSs filament was prevented during the acute phase of infection, which was achieved in U4.4 and Aag2 cells by downregulating NSs synthesis. Interestingly, the NSs protein of Bunyamwera orthobunyavirus, which also inhibits cellular transcription, was shown to be required for efficient viral replication in U4.4 cells (79).
In conclusion, our results indicated that RVFV infection in mosquito cells induced both Dicer-2- and Piwi-mediated RNAi, which manifested itself with the extinction of the characteristic filaments formed by NSs. The elimination of NSs, which otherwise would have a detrimental effect on cell transcription, allows the cells to survive and establish persistent infections. Presently, most of what is known about the Piwi pathway concerns cells of the germ line. Further studies will be needed to determine precisely the components involved in Piwi-mediated regulation in mosquitoes. Similar to the case with Dicer-2-deficient C6/36 cells, it will be important to produce Piwi-deficient cells to further investigate and confirm the role of this pathway in the natural maintenance of arboviruses in mosquitoes.
ACKNOWLEDGMENTS
We thank M. Flamand and A. Billecocq for their support and helpful discussions, C. Tamietti for her excellent technical assistance, and M. A. Dillies for her valuable help.
This work was supported in part by grants from ANR MIE-08-022, from the EU FP7 project Arbo-zoonet, and from InterVet.
Footnotes
Published ahead of print 21 November 2012
REFERENCES
- 1. Ghildiyal M, Zamore PD. 2009. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10:94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hammond SM, Bernstein E, Beach D, Hannon GJ. 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293–296 [DOI] [PubMed] [Google Scholar]
- 3. Aliyari R, Ding SW. 2009. RNA-based viral immunity initiated by the Dicer family of host immune receptors. Immunol. Rev. 227:176–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ding SW. 2010. RNA-based antiviral immunity. Nat. Rev. Immunol. 10:632–644 [DOI] [PubMed] [Google Scholar]
- 5. Adelman ZN, Blair CD, Carlson JO, Beaty BJ, Olson KE. 2001. Sindbis virus-induced silencing of dengue viruses in mosquitoes. Insect Mol. Biol. 10:265–273 [DOI] [PubMed] [Google Scholar]
- 6. Blair CD, Adelman ZN, Olson KE. 2000. Molecular strategies for interrupting arthropod-borne virus transmission by mosquitoes. Clin. Microbiol. Rev. 13:651–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blakqori G, Delhaye S, Habjan M, Blair CD, Sanchez-Vargas I, Olson KE, Attarzadeh-Yazdi G, Fragkoudis R, Kohl A, Kalinke U, Weiss S, Michiels T, Staeheli P, Weber F. 2007. La Crosse bunyavirus nonstructural protein NSs serves to suppress the type I interferon system of mammalian hosts. J. Virol. 81:4991–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cirimotich CM, Scott JC, Phillips AT, Geiss BJ, Olson KE. 2009. Suppression of RNA interference increases alphavirus replication and virus-associated mortality in Aedes aegypti mosquitoes. BMC Microbiol. 9:49 doi:10.1186/1471-2180-9-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Keene KM, Foy BD, Sanchez-Vargas I, Beaty BJ, Blair CD, Olson KE. 2004. RNA interference acts as a natural antiviral response to O'nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proc. Natl. Acad. Sci. U. S. A. 101:17240–17245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Powers AM, Olson KE, Higgs S, Carlson JO, Beaty BJ. 1994. Intracellular immunization of mosquito cells to LaCrosse virus using a recombinant Sindbis virus vector. Virus Res. 32:57–67 [DOI] [PubMed] [Google Scholar]
- 11. Sanchez-Vargas I, Scott JC, Poole-Smith BK, Franz AW, Barbosa-Solomieu V, Wilusz J, Olson KE, Blair CD. 2009. Dengue virus type 2 infections of Aedes aegypti are modulated by the mosquito's RNA interference pathway. PLoS Pathog. 5:e1000299 doi:10.1371/journal.ppat.1000299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bernstein E, Caudy AA, Hammond SM, Hannon GJ. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363–366 [DOI] [PubMed] [Google Scholar]
- 13. Tomari Y, Du T, Zamore PD. 2007. Sorting of Drosophila small silencing RNAs. Cell 130:299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. 2005. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123:607–620 [DOI] [PubMed] [Google Scholar]
- 15. Rand TA, Petersen S, Du F, Wang X. 2005. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123:621–629 [DOI] [PubMed] [Google Scholar]
- 16. Ding SW, Voinnet O. 2007. Antiviral immunity directed by small RNAs. Cell 130:413–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Farazi TA, Juranek SA, Tuschl T. 2008. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development 135:1201–1214 [DOI] [PubMed] [Google Scholar]
- 18. Sarot E, Payen-Groschene G, Bucheton A, Pelisson A. 2004. Evidence for a piwi-dependent RNA silencing of the gypsy endogenous retrovirus by the Drosophila melanogaster flamenco gene. Genetics 166:1313–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, Kittler EL, Zapp ML, Klattenhoff C, Schulz N, Theurkauf WE, Weng Z, Zamore PD. 2009. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137:509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137:522–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reynolds SH, Ruohola-Baker H. 2009. PIWI goes solo in the soma. Dev. Cell 16:627–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tchurikov NA, Kretova OV. 2011. Both piRNA and siRNA pathways are silencing transcripts of the suffix element in the Drosophila melanogaster germline and somatic cells. PLoS One 6:e21882 doi:10.1371/journal.pone.0021882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Siomi MC, Sato K, Pezic D, Aravin AA. 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat. Rev. Mol. Cell Biol. 12:246–258 [DOI] [PubMed] [Google Scholar]
- 24. Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128:1089–1103 [DOI] [PubMed] [Google Scholar]
- 25. Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC. 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315:1587–1590 [DOI] [PubMed] [Google Scholar]
- 26. Gerdes GH. 2004. Rift Valley fever. Rev. Sci. Tech. 23:613–623 [DOI] [PubMed] [Google Scholar]
- 27. Nichol ST, Beaty BJ, Elliott RM, Goldbach RW, Plyusnin A, Tesh RB. 2005. The Bunyaviridae, p 695–716 In Fauquet C, Mayo MA, Maniloff LSM, J, Desselberger U, Ball LA. (ed), Virus taxonomy. Classification and nomenclature of viruses. Eighth report of the International Committee on Taxonomy of Viruses Elsevier Academic Press, London, United Kingdom [Google Scholar]
- 28. Daubney RJ, Hudson JR, Garnham PC. 1931. Enzootic hepatitis of Rift Valley fever. An underdescribed virus disease of sheep, cattle and man from East Africa. J. Pathol. Bacteriol. 34:545–579 [Google Scholar]
- 29. Pepin M, Bouloy M, Bird BH, Kemp A, Paweska J. 2010. Rift Valley fever virus (Bunyaviridae: Phlebovirus): an update on pathogenesis, molecular epidemiology, vectors, diagnostics and prevention. Vet. Res. 41:61 doi:10.1051/vetres/2010033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Balkhy HH, Memish ZA. 2003. Rift Valley fever: an uninvited zoonosis in the Arabian peninsula. Int. J. Antimicrob. Agents 21:153–157 [DOI] [PubMed] [Google Scholar]
- 31. Peters C, Linthicum K. 1994. Rift Valley fever. CRC Press, Boca Raton, FL [Google Scholar]
- 32. Plyusnin A, Elliott RM. (ed). 2011. Bunyaviridae. Molecular and cellular biology. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 33. Schmaljohn CS, Nichol ST. 2007. Bunyaviridae, p 1741–1789 In Knipe DM, Howley PM. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 34. Giorgi C, Accardi L, Nicoletti L, Gro MC, Takehara K, Hilditch C, Morikawa S, Bishop DH. 1991. Sequences and coding strategies of the S RNAs of Toscana and Rift Valley fever viruses compared to those of Punta Toro, Sicilian Sandfly fever, and Uukuniemi viruses. Virology 180:738–753 [DOI] [PubMed] [Google Scholar]
- 35. Albarino CG, Bird BH, Nichol ST. 2007. A shared transcription termination signal on negative and ambisense RNA genome segments of Rift Valley fever, sandfly fever Sicilian, and Toscana viruses. J. Virol. 81:5246–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ikegami T, Won S, Peters CJ, Makino S. 2007. Characterization of Rift Valley fever virus transcriptional terminations. J. Virol. 81:8421–8438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lara E, Billecocq A, Leger P, Bouloy M. 2011. Characterization of wild-type and alternate transcription termination signals in the Rift Valley fever virus genome. J. Virol. 85:12134–12145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mansuroglu Z, Josse T, Gilleron J, Billecocq A, Leger P, Bouloy M, Bonnefoy E. 2010. Nonstructural NSs protein of Rift valley fever virus interacts with pericentromeric DNA sequences of the host cell, inducing chromosome cohesion and segregation defects. J. Virol. 84:928–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Swanepoel R, Blackburn NK. 1977. Demonstration of nuclear immunofluorescence in Rift Valley fever infected cells. J. Gen. Virol. 34:557–561 [DOI] [PubMed] [Google Scholar]
- 40. Yadani FZ, Kohl A, Prehaud C, Billecocq A, Bouloy M. 1999. The carboxy-terminal acidic domain of Rift Valley fever virus NSs protein is essential for the formation of filamentous structures but not for the nuclear localization of the protein. J. Virol. 73:5018–5025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kalveram B, Lihoradova O, Ikegami T. 2011. NSs protein of rift valley fever virus promotes posttranslational downregulation of the TFIIH subunit p62. J. Virol. 85:6234–6243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Le May N, Dubaele S, Proietti De Santis L, Billecocq A, Bouloy M, Egly JM. 2004. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell 116:541–550 [DOI] [PubMed] [Google Scholar]
- 43. Le May N, Mansuroglu Z, Leger P, Josse T, Blot G, Billecocq A, Flick R, Jacob Y, Bonnefoy E, Bouloy M. 2008. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS Pathog. 4:e13 doi:10.1371/journal.ppat.0040013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Habjan M, Pichlmair A, Elliott RM, Overby AK, Glatter T, Gstaiger M, Superti-Furga G, Unger H, Weber F. 2009. NSs protein of rift valley fever virus induces the specific degradation of the double-stranded RNA-dependent protein kinase. J. Virol. 83:4365–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ikegami T, Narayanan K, Won S, Kamitani W, Peters CJ, Makino S. 2009. Dual functions of Rift Valley fever virus NSs protein: inhibition of host mRNA transcription and post-transcriptional downregulation of protein kinase PKR. Ann. N. Y. Acad. Sci. 1171(Suppl. 1):E75–E85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ikegami T, Narayanan K, Won S, Kamitani W, Peters CJ, Makino S. 2009. Rift Valley fever virus NSs protein promotes post-transcriptional downregulation of protein kinase PKR and inhibits eIF2alpha phosphorylation. PLoS Pathog. 5:e1000287 doi:10.1371/journal.ppat.1000287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Crabtree MB, Kent Crockett RJ, Bird BH, Nichol ST, Erickson BR, Biggerstaff BJ, Horiuchi K, Miller BR. 2012. Infection and transmission of Rift Valley fever viruses lacking the NSs and/or NSm genes in mosquitoes: potential role for NSm in mosquito infection. PLoS Negl. Trop. Dis. 6:e1639 doi:10.1371/journal.pntd.0001421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vaughn VM, Streeter CC, Miller DJ, Gerrard SR. 2010. Restriction of Rift valley Fever virus virulence in mosquito cells. Viruses 2:655–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Morazzani EM, Wiley MR, Murreddu MG, Adelman ZN, Myles KM. 2012. Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PLoS Pathog. 8:e1002470 doi:10.1371/journal.ppat.1002470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Laughlin LW, Meegan JM, Strausbaugh LJ, Morens DM, Watten RH. 1979. Epidemic Rift Valley fever in Egypt: observations of the spectrum of human illness. Trans. R. Soc. Trop. Med. Hyg. 73:630–633 [DOI] [PubMed] [Google Scholar]
- 51. Billecocq A, Gauliard N, Le May N, Elliott RM, Flick R, Bouloy M. 2008. RNA polymerase I-mediated expression of viral RNA for the rescue of infectious virulent and avirulent Rift Valley fever viruses. Virology 378:377–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gommet C, Billecocq A, Jouvion G, Hasan M, Zaverucha do Valle T, Guillemot L, Blanchet C, van Rooijen N, Montagutelli X, Bouloy M, Panthier JJ. 2011. Tissue tropism and target cells of NSs-deleted Rift Valley fever virus in live immunodeficient mice. PLoS Negl. Trop. Dis. 5:e1421 doi:10.1371/journal.pntd.0001421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jagla B, Wiswedel B, Coppee JY. 2011. Extending KNIME for next-generation sequencing data analysis. Bioinformatics 27:2907–2909 [DOI] [PubMed] [Google Scholar]
- 55. Schneider TD, Stephens RM. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 18:6097–6100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25 doi:10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Miller ML, Brown DT. 1992. Morphogenesis of Sindbis virus in three subclones of Aedes albopictus (mosquito) cells. J. Virol. 66:4180–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Singh KRP. 1967. Cell cultures derived from larvae of Aedes albopictus (Skuse) and Aedes aegypti (L.). Curr. Sci. 36:506–508 [Google Scholar]
- 60. Peleg J. 1968. Growth of arboviruses in primary tissue culture of Aedes aegypti embryos. Am. J. Trop. Med. Hyg. 17:219–223 [DOI] [PubMed] [Google Scholar]
- 61. Lan Q, Fallon AM. 1990. Small heat shock proteins distinguish between two mosquito species and confirm identity of their cell lines. Am. J. Trop. Med. Hyg. 43:669–676 [DOI] [PubMed] [Google Scholar]
- 62. Bouloy M, Janzen C, Vialat P, Khun H, Pavlovic J, Huerre M, Haller O. 2001. Genetic evidence for an interferon-antagonistic function of Rift Valley fever virus nonstructural protein NSs. J. Virol. 75:1371–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Vialat P, Billecocq A, Kohl A, Bouloy M. 2000. The S segment of rift valley fever phlebovirus (Bunyaviridae) carries determinants for attenuation and virulence in mice. J. Virol. 74:1538–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313:320–324 [DOI] [PubMed] [Google Scholar]
- 65. Vodovar N, Bronkhorst AW, van Cleef KW, Miesen P, Blanc H, van Rij RP, Saleh MC. 2012. Arbovirus-derived piRNAs exhibit a ping-pong signature in mosquito cells. PLoS One 7:e30861 doi:10.1371/journal.pone.0030861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Brackney DE, Scott JC, Sagawa F, Woodward JE, Miller NA, Schilkey FD, Mudge J, Wilusz J, Olson KE, Blair CD, Ebel GD. 2010. C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Negl. Trop. Dis. 4:e856 doi:10.1371/journal.pntd.0000856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Scott JC, Brackney DE, Campbell CL, Bondu-Hawkins V, Hjelle B, Ebel GD, Olson KE, Blair CD. 2010. Comparison of dengue virus type 2-specific small RNAs from RNA interference-competent and -incompetent mosquito cells. PLoS Negl. Trop. Dis. 4:e848 doi:10.1371/journal.pntd.0000848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hess AM, Prasad AN, Ptitsyn A, Ebel GD, Olson KE, Barbacioru C, Monighetti C, Campbell CL. 2011. Small RNA profiling of Dengue virus-mosquito interactions implicates the PIWI RNA pathway in anti-viral defense. BMC Microbiol. 11:45 doi:10.1186/1471-2180-11-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Li Z, Kim SW, Lin Y, Moore PS, Chang Y, John B. 2009. Characterization of viral and human RNAs smaller than canonical MicroRNAs. J. Virol. 83:12751–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Grumet R, Sanford JC, Johnston SA. 1987. Pathogen-derived resistance to viral infection using a negative regulatory molecule. Virology 161:561–569 [DOI] [PubMed] [Google Scholar]
- 71. Ellenberg P, Linero FN, Scolaro LA. 2007. Superinfection exclusion in BHK-21 cells persistently infected with Junin virus. J. Gen. Virol. 88:2730–2739 [DOI] [PubMed] [Google Scholar]
- 72. Lee YM, Tscherne DM, Yun SI, Frolov I, Rice CM. 2005. Dual mechanisms of pestiviral superinfection exclusion at entry and RNA replication. J. Virol. 79:3231–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zou G, Zhang B, Lim PY, Yuan Z, Bernard KA, Shi PY. 2009. Exclusion of West Nile virus superinfection through RNA replication. J. Virol. 83:11765–11776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Adelman ZN, Sanchez-Vargas I, Travanty EA, Carlson JO, Beaty BJ, Blair CD, Olson KE. 2002. RNA silencing of dengue virus type 2 replication in transformed C6/36 mosquito cells transcribing an inverted-repeat RNA derived from the virus genome. J. Virol. 76:12925–12933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gaines PJ, Olson KE, Higgs S, Powers AM, Beaty BJ, Blair CD. 1996. Pathogen-derived resistance to dengue type 2 virus in mosquito cells by expression of the premembrane coding region of the viral genome. J. Virol. 70:2132–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Karpf AR, Lenches E, Strauss EG, Strauss JH, Brown DT. 1997. Superinfection exclusion of alphaviruses in three mosquito cell lines persistently infected with Sindbis virus. J. Virol. 71:7119–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sanchez-Vargas I, Travanty EA, Keene KM, Franz AW, Beaty BJ, Blair CD, Olson KE. 2004. RNA interference, arthropod-borne viruses, and mosquitoes. Virus Res. 102:65–74 [DOI] [PubMed] [Google Scholar]
- 78. Wu Q, Luo Y, Lu R, Lau N, Lai EC, Li WX, Ding SW. 2010. Virus discovery by deep sequencing and assembly of virus-derived small silencing RNAs. Proc. Natl. Acad. Sci. U. S. A. 107:1606–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Szemiel AM, Failloux AB, Elliott RM. 2012. Role of Bunyamwera orthobunyavirus NSs protein in infection of mosquito cells. PLoS Negl. Trop. Dis. 6:e1823 doi:10.1371/journal.pntd.0001823 [DOI] [PMC free article] [PubMed] [Google Scholar]