Abstract
Porcine circovirus type 2 (PCV2) is the primary causative agent of porcine circovirus-associated diseases in pigs. To date, viral proteins Cap, Rep, Rep′, and ORF3, encoded by the PCV2 genome, have been described. Here, transcription and translation of a novel viral gene within the PCV2 genome (designated ORF4) was determined and functionally analyzed in vitro and in vivo. Northern blot analysis indicated that the RNA transcribed from the ORF4 gene is about 180 bp in length and overlaps ORF3 in the same direction. Site-directed mutagenesis confirmed that the viral ORF4 protein is not essential for virus replication in PK-15 cells and in mice infected with an ORF4-deficient PCV2 (PCV2Δ). PCV2Δ triggered higher activity levels of caspase-3 and -8 than wild-type PCV2 (wPCV2) in PK-15 cells. The antigenic epitopes of two mouse monoclonal antibodies (MAbs) raised against the viral ORF4 protein were mapped to the same 19KSSASPR25 peptide. Expression of ORF4 was confirmed using the specific MAbs in wPCV2-infected PK-15 cells and mice. Mice infected with PCV2Δ had a higher serum viral load (genomic copies) and more severe lymphoid tissue damage in the spleen than those infected with wPCV2. Meanwhile, flow-cytometric analysis indicated that the PCV2Δ infection caused a significant decrease of CD4+ and CD8+ T lymphocytes. Our results demonstrate that ORF4 is a newly discovered viral protein that is not essential for PCV2 replication but plays a role in suppressing caspase activity and regulating CD4+ and CD8+ T lymphocytes during PCV2 infection.
INTRODUCTION
Circoviruses are a family of small nonenveloped icosahedral viruses infecting pigs, geese, canaries, pigeons, and parrots (1–3). Porcine circovirus type 1 (PCV1) was initially isolated as a persistent contaminant from a porcine kidney cell line and is nonpathogenic for experimentally infected pigs (2, 4). In 1991, a new disease, postweaning multisystemic wasting syndrome (PMWS), was found in pigs (5). A novel strain of PCV isolated from pigs with PMWS was named PCV2 (6–9). PMWS primarily affects pigs between 5 and 18 weeks of age. Signs of PMWS include progressive weight loss, tachypnea, anemia, diarrhea, and jaundice. In PMWS-affected pigs, microscopic lesions of lymphoid tissues are present, characterized by lymphocyte depletion of follicular and interfollicular areas together with macrophage infiltration. As an emerging infectious disease, PMWS is now considered an epidemic in swine-producing countries worldwide and a major concern in the pig industry.
PCV1 contains an ambisense, single-stranded, closed-circular genome of 1,759 bp, while the genomes of PCV2 isolates range from 1,766 to 1,768 bp (9–13). The overall DNA sequence homology within PCV1 or PCV2 isolates is greater than 90%, while the homology between PCV1 and PCV2 is only about 68 to 76% (9, 10). Phylogenetic studies have shown that PCV1 and PCV2 belong to two different genotypes (12). The organizational structures of the PCV1 and PCV2 genomic DNA are similar, both containing 11 predicted open reading frames (ORFs). ORF1, ORF5, ORF7, and ORF10 are located on the virus (positive) strand and are transcribed clockwise, while ORF2, ORF3, ORF4, ORF6, ORF8, ORF9, and ORF11 are encoded by the complementary (negative) strand and are transcribed counterclockwise (10). ORF1 and ORF2, oriented in opposite directions, are the two major ORFs in PCV1 and PCV2. To date, four virally encoded proteins have been described in PCV2-infected cells. Rep and Rep′, both encoded by ORF1, are considered essential proteins for viral replication (14, 15), while ORF2 encodes a 27.8-kDa immunogenic capsid protein which is the only structural protein of PCV2 (16). Identified in 2005, the ORF3 gene of PCV2 is embedded within ORF1, oriented in the opposite direction, and encodes an 11.9-kDa protein. Recent studies have suggested that ORF2- and ORF3-related proteins play important roles in the pathogenesis of PCV2. A two-amino-acid (aa) mutation or nucleotide deficiency in the capsid protein can alter the virulence of PCV2 (17, 18), while ORF3-deficient mutant PCV2 may be attenuated in vivo (19, 20). However, contradictory results have also been found. For example, the chimeric PCV1-2 DNA clone, which contains the PCV2 capsid gene in the backbone of the nonpathogenic PCV1, was found to be attenuated in pigs (21), while no remarkable difference was observed in pathogenicity between wild-type PCV2-inoculated groups and ORF3-deficient PCV2-inoculated groups (22). All of these observations indicate that viral proteins encoded by ORF2 and ORF3 genes do not solely determine the pathogenicity of PCV2. Until now, the total number of viral proteins encoded by PCV2 that control pathogenicity is not clear.
The putative ORF4 of porcine circovirus is located within ORF3 and is oriented in the same direction. The ORF4-encoded peptide is predicted to be 59 aa long with a molecular mass of 6.5 kDa in PCV2, while the corresponding region of PCV1 would produce a 115-aa peptide of 13.3 kDa. The putative ORF4 regions of PCV2 and PCV1 are 83% similar at the amino acid level (10), but it has not been determined whether this high sequence homology is indicative of a role in pathogenicity. Until now, the ORF4-encoded protein of PCV has not been experimentally confirmed.
In the present study, the ORF4 protein was identified in replicating PCV2. In addition to the preparation of monoclonal antibodies (MAbs) raised against the recombinant ORF4 protein, ORF-specific DNA/RNA probes were synthesized, and an ORF4-deficient PCV2 infectious DNA clone (PCV2Δ) was constructed to aid in the functional analysis of this newly identified protein both in vitro and in vivo.
MATERIALS AND METHODS
Virus, cells, sera, plasmids, and animals.
The PCV2 strain HZ0201 (AY188355), isolated from a pig with naturally occurring PMWS, was propagated in PK-15 cells (13). PCV-free PK-15 cells were maintained in minimal essential medium supplemented with 10% heat-inactivated fetal bovine serum (GibcoBRL Life Technologies, Grand Island, NY) at 37°C with 5% CO2. The MAbs against PCV2 Rep and Cap proteins were kept in our laboratory (23, 24). The PCV2-ORF3 polyclonal antibody (PAb) was a kind gift from Jimmy Kwang of the Animal Health Biotechnology Group, Temasek Life Sciences Laboratory, National University of Singapore (25). Eukaryotic expression plasmids pCI-neo-PCV2-ORF1, -ORF2, -ORF3 (23, 26), and -ORF4, as well as the plasmids pEGFP-PCV2-ORF1, -ORF2, -ORF3, -ORF4, and pEGFP-PCV1-ORF4 (our unpublished constructs), were maintained in our laboratory. Seven-week-old specific-pathogen-free (SPF) BALB/c mice were purchased from the Shanghai Laboratory Animal Center, Chinese Academy of Sciences, Shanghai, China.
Northern blotting.
The ORF4 transcript (Fig. 1A) was detected with a DNA probe and RNA probes. The digoxigenin (DIG)-labeled ORF4 DNA probe was generated using the DIG labeling mix (Roche, Basel, Switzerland) with a primer pair (forward primer, 5′ATGACGTGTACATTAGTCTT3′; reverse primer, 5′TCAGGGACAACGGAGTGA-3′) and pCI-neo-ORF4 as the PCR template. After purification using a PCR cleanup kit (Axygen Biosciences, Union City, CA), the PCR fragment was used as a DNA probe (probe D). To synthesize the ORF4 RNA probes, linearized pCI-neo-ORF4 digested with SalI or EcoRI was purified with the PCR cleanup kit (Axygen Biosciences) and then used as the template for synthesis of DIG-labeled ORF4-specific RNA sequence probes by in vitro transcription with T7 or T3 RNA polymerase and an RNA-DIG labeling mix (Roche) according to the manufacturer's protocol. The synthesized RNA probes were designated probes S1 and S2.
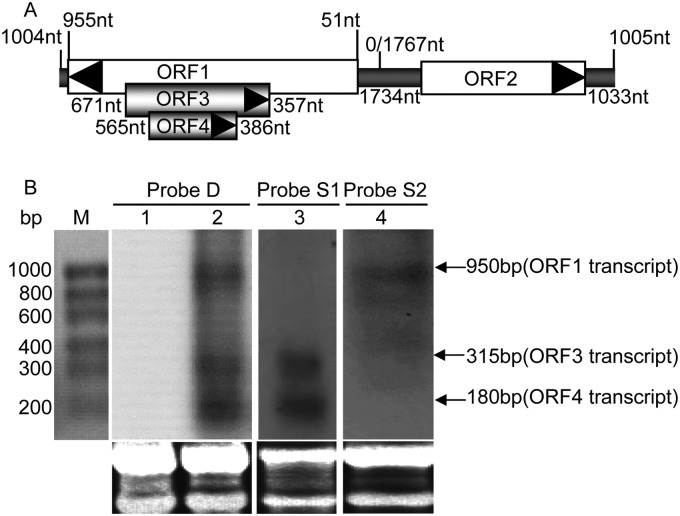
Fig 1.
Genomic location of ORF4 gene and Northern blot detection of ORF4 transcripts within PCV2-infected cells. (A) Genomic schematic of PCV2 strain HZ0201 (AY188355). Coding sequences of the four ORFs are annotated with the nucleotide coordinates of each gene. ORF4, ORF3, and ORF2 are transcribed from the positive strand (left to right), while ORF1 is transcribed from the negative strand (right to left). (B) Northern blot identification of the ORF4 transcript in PK-15 cells infected with wPCV2. Total RNA samples were isolated from mock- and wPCV2-infected PK-15 cells. Lane M, molecular size marker; lane 1, no RNA signal was detected in mock-infected PK-15 cells after hybridization with a DIG-labeled ORF4 DNA probe (probe D); lane 2, ORF4, ORF3, and ORF1 transcripts were hybridized with probe D in wPCV2-infected PK-15 cells; lane 3, ORF4 and ORF3 RNA transcripts were detected in wPCV2-infected PK-15 cells with the DIG-labeled ORF4 RNA probe (probe S1); lane 4, the ORF1 RNA signal was hybridized in wPCV2-infected PK-15 cells with the DIG-labeled RNA probe complementary to ORF4 (probe S2).
To identify the ORF4 transcript, total RNA from PK-15 cells infected with the wild-type PCV2 strain HZ0201 (wPCV2) was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) and treated with DNase I. The total RNA was then separated by electrophoresis in a 2.2% agarose-formaldehyde gel and transferred to a nylon membrane (Amersham Pharmacia Biotech AB, Uppsala, Sweden). The transferred RNA was hybridized with DIG-labeled ORF4 DNA or each of the RNA probes by following the manufacturer's procedures (Roche). The anti-DIG antibody conjugated with alkaline phosphatase (1:10,000 dilution; Roche) was applied to the membrane at room temperature for 1 h. The hybridized bands were visualized using CDP-Star (Roche) with the FluorChem E system image analyzer (Cell Biosciences, Santa Clara, CA) and compared to immobilized molecular size standards stained with methylene blue. Mock-infected PK-15 cells were used as a control.
Generation of MAbs against the PCV2 ORF4 protein.
The full 180-bp ORF4 fragment (nucleotide [nt] 386 to 565) was PCR amplified from PCV2 genomic DNA (Fig. 1A) using the following primers: forward, 5′-GCGAATTCATGACGTGTACATTAGTCTT-3′, and reverse, 5′-GGCCTCGAGTCAGGGACAACGGAGTGA-3′. The purified PCR product, digested with EcoRI and XhoI, was cloned into the vector pGEX-4T-1 (Amersham). The recombinant plasmid (pGEX-ORF4) was transformed into Escherichia coli BL21 (Invitrogen) and sequenced. The resulting cells were induced to express ORF4 with isopropyl-β-d-thiogalactopyranoside according to the manufacturer's protocols (Amersham). The purified GST-ORF4 protein was obtained after separation by 15% SDS-PAGE, excising the specific gel band of interest and releasing the protein from the gel in a dialyzer (Serva, Heidelberg, Germany) by electrophoresis in protein electrophoresis buffer (25 mM Tris base, 192 mM glycine, 3.5 mM SDS). The preparation was then injected into SPF BALB/c mice to raise an anti-ORF4 MAb as described previously (24). Reactivity of the anti-ORF4 MAb was analyzed by indirect immunofluorescence assay (IFA), immunoperoxidase monolayer assay (IPMA), and Western blotting in wPCV2-infected or ORF4-transfected PK-15 cells.
Peptide dot-ELISA for fine mapping epitopes with overlapping peptides.
Three 28-mer peptides, overlapping by 15 amino acids and spanning residues 1 to 59 of the PCV2 ORF4 protein, were initially synthesized by the solid-phase peptide synthesis method using a Symphony multiplex peptide synthesizer (Protein Technologies, Inc., Tuscson, AZ). During synthesis, a cysteine residue was added at the N terminal of all peptides except those already containing a cysteine residue in that position for conjugation. Peptides were conjugated to a bovine serum albumin (BSA) carrier protein using the heterobifunctional cross-linker Sulfo-SMCC (Sigma-Aldrich, St. Louis, MO). These BSA-conjugated peptides were tested for their reactivity with MAbs by peptide dot-enzyme-linked immunosorbent assay (ELISA) as described by Shang et al. (18) with some modifications. In this assay, 1.5 μg of each BSA-conjugated peptide was dotted onto a nitrocellulose membrane. The membrane was then blocked with 5% skim milk in phosphate-buffered saline (PBS) and incubated with the 6A5 or 2D3 MAb (diluted 1:100) at 37°C for 2 h, followed by incubation with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (Sigma) at 37°C for 1 h. The membrane was washed after each incubation step with PBS-Tween (PBST). Immunoreactive signals were visualized by an ECL Western blotting system (Thermo Scientific, Waltham, MA) using the FluorChem E system image analyzer. The carrier protein BSA was also included as a negative control.
Construction of a mutant PCV2 infectious clone.
Using the PCV2 strain HZ0201 as a genomic template (Fig. 1A), the infectious ORF4-deficient PCV2 mutant clone (pSK-PCV2Δ) was constructed by fusion PCR with two sets of primers, which introduced a specific mutation to change the initiation codon of ORF4 from ATG to GTG in the cloned PCV2 genome. Briefly, using extracted cellular DNA as the template, mutagenesis was performed in a two-step PCR. The first PCR was performed in two reactions with primer pairs F-SacII (5′-GAACCGCGGGCTGGCTGAACTTTTGAAAGT-3′)/R580 (5′-TAATGTACACGTCACTGTGGGGCCACCTGG-3′) and F551 (5′-CCAGGTGGCCCCACAGTGACGTGTACATTA-3′)/R-SacII (5′-GCACCGCGGAAATTTCTGACAAACGTTACA-3′) to generate two modified subgenomic DNA fragments of 88 bp (nt 491 to 580) and 1,711 bp (nt 551 to 496). After purification by using the AxyPrep DNA gel extraction kit (Axygen Biosciences), the mixed PCR segments were used as the template for amplification of the full-length mutant genome with the primers F-SacII and R-SacII. The genomic PCR fragment containing the mutation site was then purified, digested with SacII, and cloned into the vector pBluescript SK+ (Stratagene, La Jolla, CA). Finally, the resultant plasmid, pSK-PCV2Δ, was transformed into E. coli Top10 cells (Amersham), and the target sequence of the clones was confirmed by PCR, restriction analysis, and DNA sequencing.
Transfection.
In vitro expression of PCV2 ORF1, ORF2, ORF3, and ORF4 as well as ORF4 of PCV1 was conducted using PK-15 cells. Cells grown in 96-well plates were separately transfected with the following plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer's guidelines: pEGFP-PCV2-ORF1, pEGFP-PCV2-ORF2, pEGFP-PCV2-ORF3, pEGFP-PCV2-ORF4, pEGFP-PCV1-ORF4, pCI-neo-PCV2-ORF1, pCI-neo-PCV2-ORF2, pCI-neo-PCV2-ORF3, and pCI-neo-PCV2-ORF4. At 60 h posttransfection, protein expression was determined by IPMA using mouse MAbs to Rep, Cap, and ORF4, as well as anti-ORF3 PAb.
Generation and determination of titers of infectious mutant PCV2.
The pSK-PCV2Δ plasmid extracted from transformed E. coli (Top10 strain) was digested by SacII and circularized by ligation with T4 DNA ligase at 16°C overnight. The ligated DNA mixture was transfected into approximately 60 to 80% confluent PK-15 cells in a 24-well plate with Lipofectamine 2000 by following the manufacturer's guidance. At 72 h posttransfection, genome-transfected cells were subjected to three successive freeze-thaw cycles to harvest the virus stock. The infectivity of recovered virus was analyzed by IFA with an anti-ORF4 MAb or anti-Cap MAb as described previously (24).
To analyze the replicative ability of the rescued virus, PK-15 cells were inoculated with wPCV2 or PCV2Δ, and the cell monolayers were harvested at 18, 24, 36, 48, 72, and 96 h postinoculation. The titers of viruses were determined by calculating the 50% tissue culture infective doses (TCID50) per 0.1 ml according to the method of Reed and Muench (27). The above-mentioned assay was conducted three times in separate experiments. Results are presented as means ± standard deviations (SD).
Mouse inoculation and pathological analysis.
To study the pathogenicity of ORF4-deficient PCV2, 60 8-week-old SPF BALB/c mice were randomly separated into three groups of 20 mice per group. The members of the first group of mice were sham inoculated on day 0 with 0.2 ml intraperitoneally and 0.03 ml intranasally of cell minimal essential medium to serve as a negative control. Each mouse in groups 2 and 3 was inoculated in the same manner with 105.5 TCID50 of wPCV2 and PCV2Δ, respectively. Treated mice were housed in isolation filter-top cages and monitored daily for clinical signs. At 14, 21, and 42 days postinoculation (dpi), five mice were sacrificed with phenobarbital anesthesia at each time point in each group. At necropsy, sera were collected for detection of viral DNA, virus reisolation, and virus genome sequencing as we have described previously (28). The spleen was removed and divided into three parts. One part, fixed in 10% neutral buffered formalin, was used for pathological and immunohistochemical (IHC) analyses. The pathological analysis was performed according to a previously described method (26). Briefly, formalin-fixed spleens were sectioned and stained with hematoxylin and eosin (H&E). Microscopic changes in splenic tissues of challenged mice were determined by comparing their tissues to those of control mice. The severity of lesions was estimated as the frequency of abnormal spleen follicles, which is the ratio of the number of the follicles with lymphoid depletion and histiocytic infiltration to the total number of follicles counted in the spleen section of each mouse. The second part, frozen in liquid nitrogen, was used for IFA analysis. IHC and IFA were performed using anti-Cap and anti-ORF4 MAbs as primary antibodies as stated previously (28). The final portion of the spleen was prepared for flow-cytometric analysis to determine T-lymphocyte subsets.
IFA and IPMA.
The inoculation procedure and IFA protocol were performed as previously described (24). Briefly, PK-15 cells were mixed with wPCV2 or PCV2Δ (at a ratio of 10:1, vol/vol) and seeded in 96-well flat-bottom microtiter plates at 100 μl/well. After incubation at 37°C for 72 h, the cells were washed twice with PBS and fixed with a methanol-acetone mixture (1:1, vol/vol) at −20°C for 20 min. The cells were blocked by PBS with 5% skim milk at 37°C for 1 h. For IFA, the cells were incubated with a primary anti-Cap or anti-ORF4 2D3 MAb for 60 min at 37°C, followed by fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG (Kirkegaard & Perry Laboratories Inc. [KPL], Gaithersburg, MD) for 60 min at 37°C. Finally, the cells were stained with 4,6-diamidino-2-phenylindole (DAPI) for 5 min at room temperature. The images were photographed under a laser confocal microscope (Zeiss, Oberkochen, Germany). For IPMA, the transfected cells were separately incubated with mouse MAbs to Rep, Cap, and ORF4, as well as an anti-ORF3 PAb in PBST containing 2% skim milk for 1 h at 37°C. After three washes with PBST, the cells were incubated for 1 h at 37°C with HRP-conjugated goat anti-guinea pig IgG (KPL) or HRP-conjugated goat anti-mouse IgG (KPL) in PBST containing 2% skim milk and washed again. Staining was developed in the presence of the peroxidase substrate 3-amino-9-ethylcarbazol (AEC; Solarbio, Beijing, China) for 10 to 20 min at room temperature and was stopped by substrate removal. The stained cells were observed using an IX71 inverted fluorescence microscope (Olympus, Tokyo, Japan).
Immunoblot analysis.
To detect the ORF4 protein expressed in PK-15 cells after transfection with the pEGFP-ORF4 vector or infection with wPCV2 virions as well as in wPCV2-infected mouse splenocytes, Western blot analysis was performed as described elsewhere, with slight modifications (24). pEGFP-C-transfected or mock-infected PK-15 cells and mock-infected mice were used as controls. Briefly, transfected PK-15 cells were cultured for 60 h, while mouse splenocytes were infected for 14, 21, and 42 days before harvesting. The harvested cells were lysed using the CytoBuster protein extraction reagent (Merck, Darmstadt, Germany) according to the manufacturer's instructions. Proteins in all of the supernatants collected as described above were separated both by 15% SDS-PAGE and tricine SDS-PAGE. The separated proteins were blotted onto nitrocellulose membranes, which were blocked with 5% skim milk in PBST at 37°C for 1 h and then reacted with mouse anti-ORF4 2D3 MAb at 37°C for 2 h. After three washes in PBST, the membranes were incubated with HRP-conjugated goat anti-mouse IgG (KPL) for 1 h. Immunoreactive bands were visualized using an ECL Western blotting system (Thermo Scientific) and the FluorChem E system image analyzer (Cell Biosciences). Meanwhile, lysed samples were dotted onto nitrocellulose membranes for dot-ELISA, which was performed by following the procedures described above using the anti-ORF4 2D3 MAb as the primary antibody.
Colorimetric caspase activity assay.
Proteolytic caspase activities induced by wPCV2 and PCV2Δ were detected using ApoAlert caspase-3 and caspase-8 colorimetric assay kits (Clontech, Franklin Lakes, NJ) and a caspase-9 colorimetric assay kit (Millipore, Bedford, MA), with some modifications. Briefly, PK-15 cells were infected with PCV2Δ and wPCV2 at a multiplicity of infection (MOI) of 1 TCID50 as mentioned above. Mock-infected PK-15 cells were processed in parallel as a control. At 24 and 72 h postinfection (hpi), cells were collected and counted, and 2 × 106 cells were prepared per sample. After lysing each sample in 50 μl of lysis buffer on ice for 10 min followed by centrifugation, the supernatant was collected and 3 μl of dimethyl sulfoxide or an inhibitor for caspase-3 (DEVD-fmk), caspase-8 (IETD-fmk), or caspase-9 (Ac-LEHD-CHO) was added. The sample was then divided and incubated in separate reaction buffers containing the p-nitroaniline (pNA)-labeled substrate for caspase-3 (DEVD-pNA), caspase-8 (IETD-pNA), or caspase-9 (LEHD-pNA) at 37°C for 2 h. Finally, caspase activity was assessed by quantification of released pNA resulting from substrate cleavage using a microplate reader at 405 nm and a pNA calibration curve.
Flow cytometry.
Each spleen was minced to prepare a cell suspension. Briefly, after removing red blood cells using 0.83% NH4Cl-Tris buffer (pH 7.4), splenocytes were washed with PBS three times and resuspended at 107 cells/ml in PBS. For T-lymphocyte subset analysis, this splenocyte suspension was aliquoted at 100 μl/tube, placed on ice, and mixed with an antibody mixture containing 1 μl PE anti-mouse CD3, 1 μl PE/Cy5 anti-mouse CD4, and 1 μl FITC anti-mouse CD8 (all from Biolegend, San Diego, CA). Meanwhile, irrelevant antibodies served as background controls, and single fluorescence-stained samples were used to optimize the compensation of signals. All samples were placed on ice for 45 min on a shaker and protected from light. Thereafter, the antibody-labeled cells were washed three times with PBS and analyzed with an FC500 flow cytometer (Beckman, Brea, CA). Data analysis was performed with CXP software (Beckman).
Quantitative real-time PCR.
Viral DNA levels in the mouse serum samples collected after wPCV2 or PCV2Δ challenge were determined using quantitative real-time PCR as previously described (26). Briefly, total DNA was extracted from 100 μl of serum by using a UNlQ-10 column virus genomic DNA isolation kit (Sangon Biotech Co., Ltd., Shanghai, China) by following the manufacturer's instructions. Forward (5′-ACTACTCCTCCCGCCATACC-3′) and reverse (5′-GGTCCACATTTCCAGCAGTT-3′) primers were used to amplify a 155-bp fragment from the ORF2 gene of PCV2. The PCR mixture contained a final concentration of 1× SYBR Premix Ex Taq (TaKaRa, Dalian, China), 0.2 mM each primer, and DNA equivalent to that of 1 ml serum as the template. All reactions were carried out in duplicate on a RealPlex4 real-time PCR system (Eppendorf, Hamburg, Germany). The program consisted of one cycle at 95°C for 2 min and 40 cycles at 95°C for 5 s, 57°C for 5 s, and 72°C 15 s, followed by melting curve analysis to determine specificity. Serial dilutions of the pCI-neo-ORF2 plasmid were used to obtain a standard curve. The number of virus copies for each sample was calculated as the mean value of duplicate reactions.
Statistical analysis.
The statistically significant differences between groups were determined using the Student t test. A P value of less than 0.05 was considered significant.
RESULTS
Transcription of ORF4 within PCV2-infected cells.
To confirm the transcription of ORF4 from the PCV2 genome, total cellular RNA extracts from wPCV2-infected PK-15 cells were hybridized with DIG-labeled DNA and RNA probes. After hybridization with DNA probe D, three RNA bands of approximately 950, 300, and 180 bp were found in total cellular RNA of wPCV2-infected PK-15 cells (Fig. 1B, lane 2), which were identical to the lengths of the ORF1, ORF3, and putative ORF4 transcripts, while no hybridized band was detected in total cellular RNA of mock-infected PK-15 cells (Fig. 1B, lane 1). To determine the polarity of these transcripts, RNA probes were used for hybridization with total RNA extracted from wPCV2-infected PK-15 cells. Hybridization with probe S1 revealed two transcripts (300 and 180 bp in length) in total cellular RNA of wPCV2-infected PK-15 cells (Fig. 1B, lane 3). The 300-bp transcript was determined to have the same size and orientation as those previously reported for the PCV2 ORF3 gene (14), and the 180-bp band was identical to the size of the putative ORF4 transcript. Using the cRNA probe S2 (oriented in the negative direction) for hybridization, a 950-bp RNA band was detected, consistent with that reported previously for the Rep transcript (14) (Fig. 1B, lane 4). The results indicated that the ORF4 gene was transcribed from the viral minus strand and in the same orientation as ORF3.
Generation and characterization of MAbs to ORF4 protein of PCV2.
The genomic ORF4 sequence consisting of 180 bp was cloned into the vector pGEX-4T-1-F4, which produced a GST-fused ORF4 protein with an approximate molecular mass of 32.5 kDa, as confirmed by Western blotting using a rabbit anti-GST polyclonal antibody (Fig. 2A, lane 2). Subsequently, the specific protein band was gel purified after separation by 15% SDS-PAGE. To obtain specific MAbs, PCV2 ORF4 proteins expressed and purified from E. coli were used to immunize SPF BALB/c mice. Ultimately, two hybridoma cell lines, 2D3 and 6A5, secreting MAbs against the PCV2 ORF4 protein, were generated. Western blot analysis showed that the generated MAbs could recognize the viral ORF4 protein expressed in E. coli transformed with the pGEX-ORF4 vector (Fig. 2A, lane 3) and in PK-15 cells transfected with the pEGFP-ORF4 plasmid (Fig. 2A, lane 5). IPMA analysis also demonstrated that the generated MAbs could specifically recognize ORF4-encoded proteins in PK-15 cells transfected with the pEGFP-ORF4 vector (Fig. 2B). In both Western blot and IPMA analyses, the generated MAbs could not react with PCV2 ORF1-, ORF2-, and ORF3-encoded proteins expressed in E. coli (data not shown) or PK-15 cells nor the eukaryotically expressed enhanced green fluorescent protein (Fig. 2A, lane 4) and ORF4 protein of PCV1 in PK-15 cells (data not shown), indicating that they were specific for the ORF4 protein of PCV2.
Fig 2.
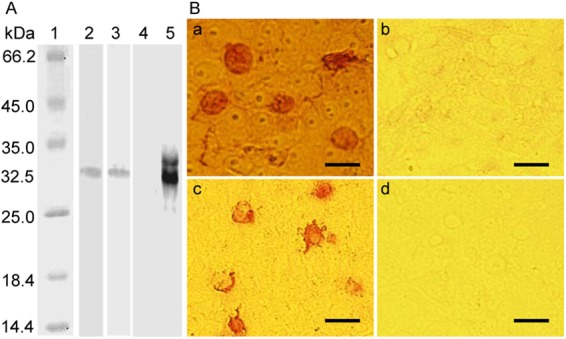
Reactivity and specificity of MAbs raised against viral ORF4 protein. (A) Western blot analysis of ORF4 protein expressed in bacterial cells or PK-15 cells. Lysed samples were electrophoresed on 15% SDS-PAGE gels, transferred onto nitrocellulose membranes, and probed with specific antibodies. Lane 1, protein molecular size marker; lane 2, the GST-ORF4 protein purified from E. coli was detected by a rabbit anti-GST PAb; lane 3, the lysate of GST-ORF4-expressing E. coli was detected by the 2D3 MAb against PCV2 ORF4 protein; lane 4, pEGFP-C2-transfected PK-15 cells were probed with the 2D3 MAb; lane 5, pEGFP-ORF4-transfected PK-15 cells were probed with the 2D3 MAb. (B) IPMA analysis of PK-15 cells transfected with pEGFP-ORF4 or pEGFP-C2. At 48 h after transfection, the cells were fixed and labeled with the 6A5 (a) and 2D3 (c) mouse MAbs raised against the PCV2 ORF4 protein. As negative controls, pEGFP-C2-transfected cells were probed with the 6A5 (b) and 2D3 (d) MAbs. Bars, 25 μm.
Identification of antigenic epitopes on PCV2 ORF4 protein.
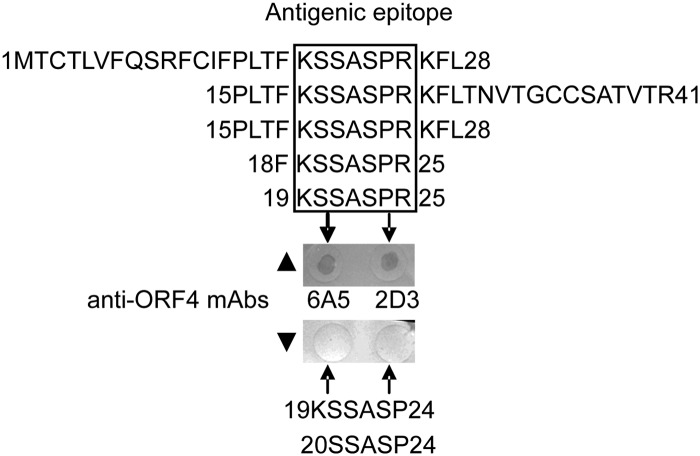
For mapping and localization of linear B cell epitopes recognized by the MAbs 2D3 and 6A5, overlapping linear peptides of the ORF4 protein were synthesized by the PEPSCAN technique. As shown in Fig. 3, peptides corresponding to aa 1 to 28 and 15 to 41 were recognized by both 2D3 and 6A5 MAbs to the PCV2 ORF4 protein, while the peptide matching aa 29 to 59 (data not shown) could not be detected by either MAb. Subsequently, by using shorter N- and C-terminally truncated peptides derived from the two recognized peptides to fine map the epitope, both anti-ORF4 protein MAbs were found to react with the 15PLTFKSSASPRKFL28 peptide. After shortening this sequence further, the 19KSSASPR25 peptide was still recognized by both 2D3 and 6A5 MAbs (Fig. 3), indicating that this peptide sequence is the common antigenic epitope and core motif for the ORF4-specific MAbs.
Fig 3.
Fine mapping of linear B cell epitopes on the PCV2 ORF4 protein using peptide dot-ELISA. Eight BSA-conjugated peptides spanning residues 1 to 28, 15 to 41, 15 to 28, 18 to 25, 19 to 25, 19 to 24, and 20 to 24 were dotted on a nitrocellulose membrane in a peptide dot-ELISA and probed with MAbs 6A5 and 2D3 to the PCV2 ORF4 protein. ▲, two representative positive peptide dot-ELISA wells. Peptides corresponding to residues 1 to 28, 15 to 41, 15 to 28, 18 to 25, and 19 to 25 were recognized by the 6A5 and 2D3 MAbs. ▼, two representative non-positive peptide dot-ELISA wells. Peptides corresponding to residues 19 to 24 and 20 to 24 did not react with the 6A5 and 2D3 MAbs.
PCV2-infected cells express the ORF4-encoded protein in vitro and in vivo.
To verify the expression of the ORF4 protein in replicating PCV2, PK-15 cells were infected with wPCV2 or PCV2Δ. Expression of the PCV2 ORF4 protein in wPCV2-infected PK-15 cells was observed using the ORF4-specific 2D3 MAb by IFA and confocal microscopy as early as 24 hpi (data not shown) and peaked at 60 hpi (Fig. 4B). The PCV2 Cap protein was also detected as a positive control in these cells by a specific MAb at 48 hpi (Fig. 4A). As expected, no PCV2 ORF4 protein was detected with the ORF4-specific MAb in PCV2Δ-infected PK-15 cells (data not shown).
Fig 4.
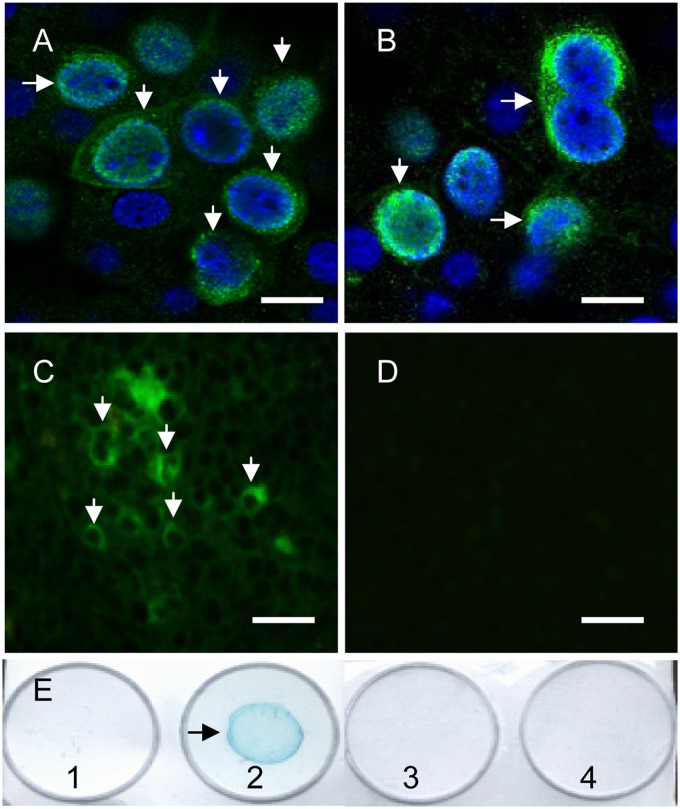
Expression of viral ORF4 protein in wPCV2-infected PK-15 cells and mice. At 60 hpi with wPCV2, PK-15 cells (arrow) were fixed and probed with an anti-Cap MAb (A) and anti-ORF4 2D3 MAb (B) for confocal microscopic analysis. Nuclei were labeled with DAPI (bars, 25 μm). At 21 dpi, the 2D3 MAb to the PCV2 ORF4 protein was used to stain (green) cryosectioned spleens of wPCV2-inoculated mice (C) or PCV2Δ-inoculated mice (D) for IFA analysis (bars, 50 μm). (E) Dot blot analysis in which mock- and wPCV2-infected PK-15 cells were lysed with the CytoBuster protein extraction reagent. After treatment with or without reducing loading buffer, the lysates were dotted onto a nitrocellulose membrane, which was incubated with the anti-ORF4 2D3 MAb followed by HRP-labeled goat anti-mouse IgG. Well 1, nonreduced mock-infected PK-15 cell lysate; well 2, nonreduced PCV2-infected PK-15 cell lysate; well 3, reduced wPCV2-infected PK-15 cell lysate; well 4, reduced mock-infected PK-15 cell lysate.
In addition, mice were inoculated with PCV2Δ or wPCV2, and their spleens were obtained at different time points postinoculation and cryosectioned for analysis by IFA using anti-Cap and ORF4-specific 2D3 MAb as primary antibodies. The expressed ORF4 protein was detected in the spleen of wPCV2-infected mice from 14 to 42 dpi (only 21 dpi is shown in Fig. 4C), while no signal was detected in the spleen of PCV2Δ-infected mice (Fig. 4D), demonstrating that PCV2Δ indeed failed to express the ORF4 protein in vivo. By dot-ELISA, the ORF4 protein signal could be visualized only by using the ORF4-specific 2D3 MAb in wPCV2-infected PK-15 cell lysates that were not mixed with 4× reducing loading buffer (Fig. 4E, well 2) or boiled (data not shown). No ORF4 protein signal was detected in mock-infected PK-15 cell lysates serving as a negative control (Fig. 4E, well 1). However, when the samples were treated with 4× reducing loading buffer and boiled, the viral ORF4 protein could no longer be detected in wPCV2-infected PK-15 cells (Fig. 4E, well 3) or in the corresponding mock-infected PK-15 cells (Fig. 4E, well 4) by dot-ELISA. In addition, no ORF4 protein signal was visualized by Western blotting using the ORF4-specific 2D3 MAb in lysates of wPCV2-infected PK-15 cells or mouse splenocytes (data not shown). Together, these results indicated that the antigenic epitope of the PCV2 ORF4 protein was disrupted in the reduced or boiled samples.
PCV2 ORF4 protein is not essential for viral replication.
To determine the influence of the PCV2 ORF4 protein on viral replication, the ORF4-deficient PCV2 infectious DNA clone was constructed and used to infect PK-15 cells. As shown in Fig. 5A and B, ORF4-deficient PCV2 virions were recovered from PK-15 cells, and only the Cap protein could be detected when PCV2Δ-infected PK-15 cells were immunostained with the anti-Cap MAb or anti-ORF4 2D3 MAb. Subsequently, the replicative abilities of wPCV2 and PCV2Δ were determined in PK-15 cells. As shown in Fig. 5C, the titer of PCV2Δ was similar to that of wPCV2 over the course of the infection. At 36 hpi, the titers of PCV2Δ and wPCV2 reached 105.3 ± 100.18 TCID50/0.1 ml and 105.5 ± 100.2 TCID50/0.1 ml, respectively. These results indicated that the viral protein ORF4 was not essential for wPCV2 replication in PK-15 cells.
Fig 5.
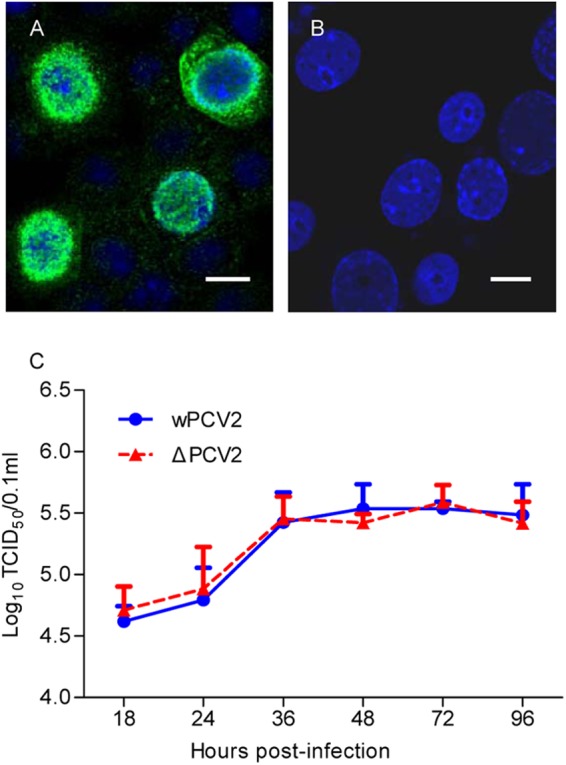
Recovery and replication kinetics of ORF4-deficient PCV2. PK-15 cells were transfected with T4 DNA ligase-circularized, ORF4-deficient PCV2 infectious DNA for rescuing PCV2Δ. (A and B) Recovered PCV2Δ was detected by an anti-Cap protein MAb (A) and anti-ORF4 protein 2D3 MAb (B), respectively. Nuclei were stained with DAPI (bars, 25 μm). (C) One-step growth curves of PCV2Δ and wPCV2 in PK-15 cells. PK-15 cells were infected by PCV2Δ or wPCV2 at an MOI of 1. Virus stocks were harvested from 18 to 96 hpi at 6-h intervals, and titers were determined as described in Materials and Methods. Results are presented as the mean TCID50 ± SD (n = 3).
Profiles of caspases and T cell subsets during ORF4-deficient PCV2 infection.
Infected host cells undergo apoptosis to limit viral propagation (25). Therefore, many viruses carry antiapoptotic factors to facilitate replication, such as poliovirus (29), rotavirus (30), influenza virus (31, 32), dengue virus (33), infectious bursal disease virus (34), and cowpox virus (35). To determine whether the ORF4 protein affects apoptosis in host cells, caspase-3, -8, and -9 activities were measured in PCV2Δ-infected PK-15 cells and compared to those of wPCV2-infected PK-15 cells, which served as positive controls. After infection, the proteolytic activities of caspase-3 (Fig. 6A), caspase-8 (Fig. 6B), and caspase-9 (Fig. 6C) were induced by both PCV2Δ and wPCV2. The activities of caspase-3 and -8 from PCV2Δ- and wPCV2-infected PK-15 cells were significantly higher than those from mock-infected PK-15 cells (P < 0.05), indicating that caspase-3 and -8 were involved in PCV2-induced apoptosis. At 24 and 72 hpi, the activities of caspase-3 and -8 induced by PCV2Δ were higher than those of wPCV2 (P < 0.05), suggesting that the ORF4 protein plays an important role in suppressing apoptosis in PCV2 infection. In addition, DEVD-fmk, IETD-fmk, and Ac-LEHD-CHO peptide inhibitors to caspase-3, -8, and -9, respectively, could inhibit the induction of these enzymes, decreasing their activities to below basal levels in PK-15 cells (Fig. 6).
Fig 6.
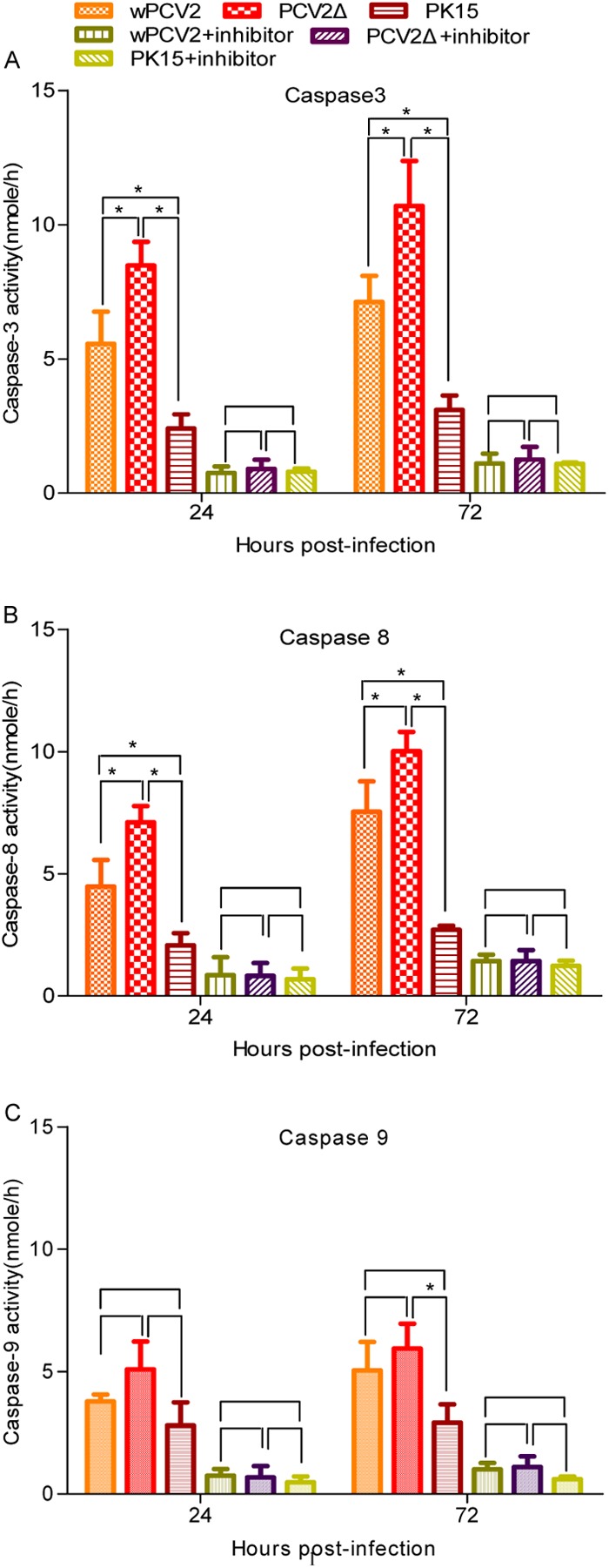
Detection of caspase-3, -8, and -9 activities during wPCV2 and PCV2Δ infection. Caspase activity was detected in PK-15 cell lysates harvested at 24 and 72 hpi with PCV2Δ or wPCV2. DEVD-pNA, IETD-pNA, and LEHD-pNA were used as substrates of caspase-3, caspase-8, and caspase-9, respectively, in the colorimetric detection assays. Mock-, wPCV2-, and PCV2Δ-infected PK-15 cells treated with inhibitors to caspase-3 (DEVD-fmk), caspase-8 (IETD-fmk), and caspase-9 (Ac-LEHD-CHO) were used as controls. Results are presented as mean values of caspase activity ± SD (n = 3) (*, P < 0.05). Significantly higher activities of caspase-3 (A) and caspase-8 (B) were induced in PCV2Δ-infected cells than in wPCV2- or mock-infected cells (P < 0.05). (C) The caspase-9 activity of PCV2Δ-infected cells was not significantly increased compared to that in PCV2Δ- or mock-infected cells (P > 0.05).
The percentages of spleen T-lymphocyte subsets were analyzed using flow cytometry at 14, 21, and 42 dpi in mice. In cross comparisons among groups, the changes in relative proportions of splenic CD3+ CD8+, CD3+ CD4+ CD8+, and CD3+ CD4+ cell subsets showed different trends among different groups. The proportion of the CD3+ CD8+ cell subset of PCV2Δ-infected mice was significantly lower (P < 0.05) at 42 dpi than that of wPCV2-inoculated mice (Fig. 7A). As shown in Fig. 7B, the proportion of the CD3+ CD4+ cell subset of PCV2Δ-inoculated mice was significantly lower (P < 0.05) than that of wPCV2- or mock-inoculated mice, except at 21 dpi (P > 0.05). Meanwhile, the relative proportion of the CD3+ CD4+ CD8+ cell subset was lower in PCV2Δ-infected mice than in wPCV2- and mock-infected mice, but not significantly (P > 0.05) (Fig. 7C). To some degree, these results imply that the ORF4 protein plays a regulatory role in the decrease of CD4+ and CD8+ T cells during PCV2 infection.
Fig 7.
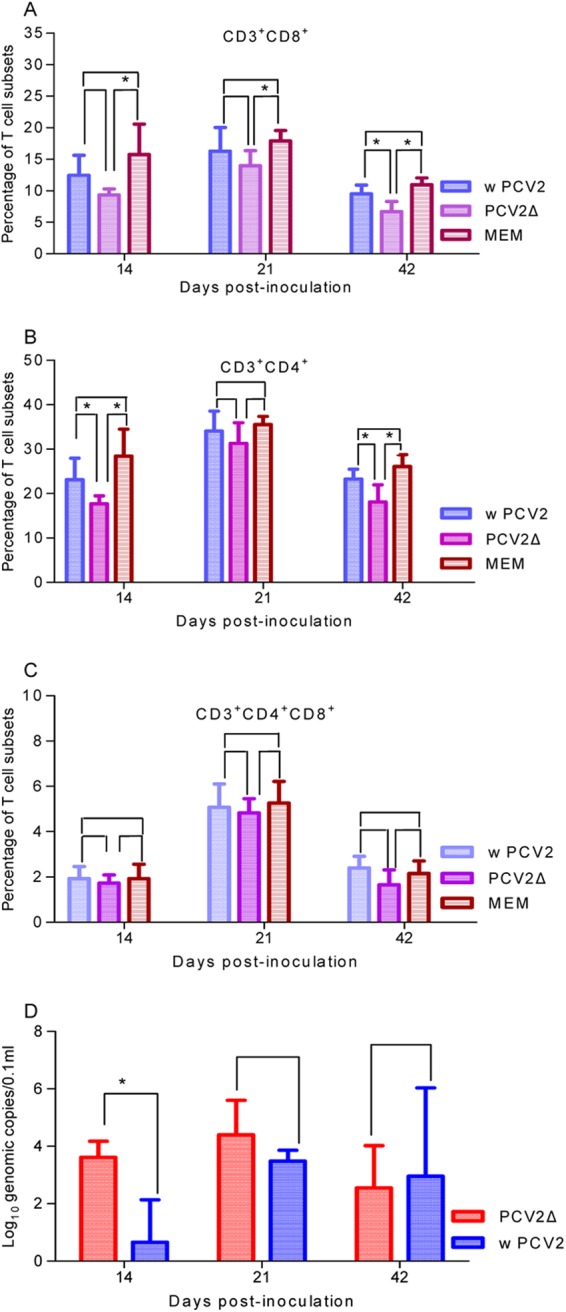
Kinetics of serum viral load and splenic T-lymphocyte subsets in mice inoculated with ORF4-deficient PCV2. Spleens were collected at different time points from PCV2Δ-, wPCV2-, and mock-inoculated mice and minced to prepare splenocyte suspensions at a concentration of 1 × 107 cells/ml. Spleen cell suspensions of PCV2Δ-, wPCV2-, and mock-inoculated mice were labeled with an antibody mixture containing PE anti-mouse CD3, PE/Cy5 anti-mouse CD4, and FITC anti-mouse CD8 for flow-cytometric analysis. CD3+ cells were gated, and average percentages of T cell subsets for each group are shown. (A) CD3+ CD8+ T lymphocytes in spleen. (B) CD3+ CD4+ T lymphocytes in spleen. (C) CD3+ CD4+ CD8+ T lymphocytes in spleen. (D) Viral genomic copies in sera of BALB/c mice inoculated with PCV2Δ or wPCV2 at the same dose. Results are presented as mean percentages of T lymphocytes or serum genomic copies ± SD (n = 5) (*, P < 0.05).
Viremia and virus reisolation from mice inoculated with ORF4-deficient PCV2.
PCV2 viremia was analyzed in serum of inoculated mice by quantitative real-time PCR. As shown in Fig. 7D, viral DNA from five PCV2Δ-inoculated mice reached, on average, 103.61 ± 100.56, 104.40 ± 101.21, and 102.55 ± 101.47 copies/μl at 14, 21, and 42 hpi, respectively. In mice inoculated with wPCV2, the average abundance of the viral genome was 100.66 ± 101.48, 103.48 ± 100.38, and 102.95 ± 103.09 copies/μl in serum at 14, 21, and 42 dpi, respectively. The results suggest that the replicative ability of PCV2Δ was enhanced significantly in mice at 14 dpi (P < 0.05). To confirm the genetic stability in PCV2Δ-infected mice, virus was reisolated from the serum of inoculated mice and sequenced. PCV2Δ was detected from the serum of all inoculated mice, confirming that they were all infected, and the reisolated PCV2Δ showed no reversal of the nucleotide mutation.
Detection of microscopic lesions and viral antigens in mice infected with ORF4-deficient PCV2.
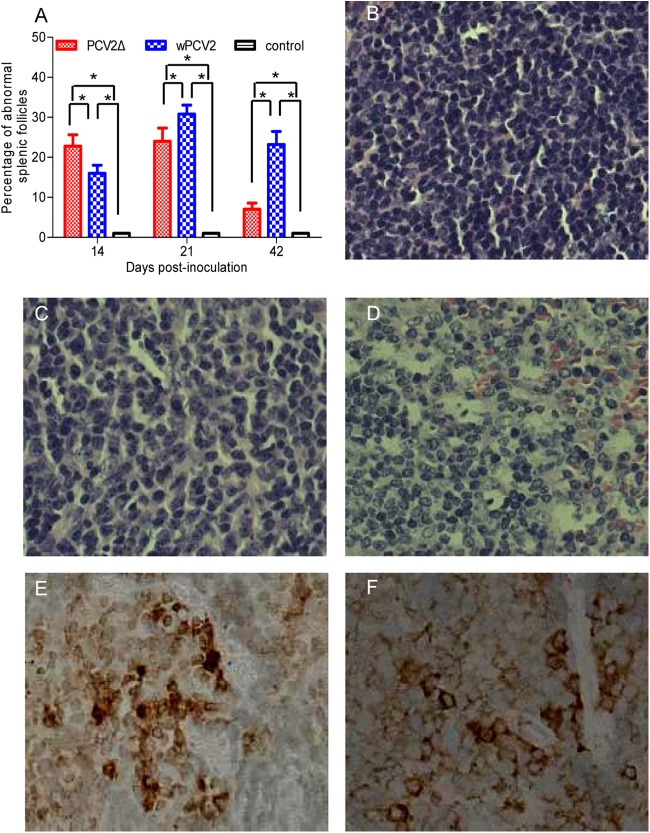
To determine the effect of ORF4 on pathogenicity of the virus, BALB/c mice were inoculated with PCV2Δ or wPCV2. No clinical signs (i.e., weight loss, wasting, or tachypnea) were observed in any of the PCV2Δ- or wPCV2-inoculated mice. Compared to the hematoxylin and eosin (H&E)-stained spleen sections of mock-inoculated mice (Fig. 8B), those obtained from PCV2Δ- or wPCV2-inoculated mice at necropsy at 14, 21, and 42 dpi showed microscopic alterations characterized by lymphocyte depletion, histiocytic infiltration, and apoptotic cells in the marginal zone of the splenic follicles (Fig. 8C and D). In comparing the degree of pathological changes, the PCV2Δ-infected mice revealed a higher frequency of lesions in splenic follicles (P < 0.05) (Fig. 8A) and more severe microscopic lesions (Fig. 8D) than wPCV2-inoculated mice (Fig. 8C) at 14 dpi. In contrast, at 21 and 42 dpi, the PCV2Δ-infected mice had a lower frequency of lesions in splenic follicles than the wPCV2-inoculated mice (P < 0.05) (Fig. 8A).
Fig 8.
Splenic microscopic lesions and antigen detection in mice inoculated with ORF4-deficient PCV2. (A to D) Pathological analysis of H&E-stained splenic sections. (A) Percentages of abnormal splenic follicles relative to total follicles in spleens of PCV2Δ- or wPCV2-inoculated mice. Results are means ± SD (n = 5) (*, P < 0.05). (B) Spleen from a mock-infected mouse. (C and D) Spleens from wPCV2-infected (C) and PCV2Δ-infected (D) mice showing lymphocyte depletion and histiocytic infiltration. (E and F) IHC staining with the anti-Cap MAb (brown color) of paraffin-embedded spleen sections of wPCV2-inoculated (C) and PCV2Δ-inoculated (D) mice. Magnification, ×100.
IHC staining of the PCV2 Cap antigen was also performed on the paraffin-embedded mouse spleen sections. In PCV2Δ-inoculated mice (Fig. 8E), the PCV2 Cap antigen was detected in the spleen of 2/5 mice at 14 dpi, 4/5 mice at 21 dpi, and 2/5 mice at 42 dpi. In wPCV2-inoculated mice (Fig. 8F), the PCV2 Cap antigen was detected in the spleen of 1/5 mice at 14 dpi, 2/5 mice at 21 dpi, and 3/5 five mice at 42 dpi. The anti-Cap-labeled cells in the spleen were mainly histiocytic cells and macrophages in the marginal zone of splenic follicles. However, no ORF4 antigen signal could be detected by IHC staining of the spleen sections from either PCV2Δ- or wPCV2-inoculated mice (data not shown), possibly due to disruption of the antigenic epitope of the viral ORF4 protein by paraffin embedding.
DISCUSSION
With an ambisense, single-stranded, closed-circular genome, PCV2 is estimated to have 11 ORFs (10), while 10 RNA transcripts have been detected during viral replication (14, 25). The viral RNAs are oriented in two directions. RNA of the ORF2 and ORF3 genes are transcribed from the cDNA strand, while a cluster of five Rep-associated RNAs (Rep, Rep′, Rep3a, Rep3b, and Rep3c) and three non-structure (NS)-associated RNAs (NS515, NS672, and NS0) are transcribed in the opposite orientation. Until now, only viral proteins encoded by ORF1, ORF2, and ORF3 have been found in replicating PCV2. ORF1 encodes two viral replication-associated proteins, Rep and Rep′ (36). The ORF2-encoded protein Cap is an immune-associated protein (16), while the ORF3 protein has been characterized as an inducer of apoptosis (25). Whether ORF4 is transcribed in PCV2 infection was previously unclear. In our study, a hybridization band of approximately 180 bp in length was detected in a Northern blot assay of PCV2-infected cells (Fig. 1B). This transcript length was shorter than those of all confirmed viral RNAs but similar to that of the putative ORF4 previously predicted to completely overlap ORF3 and orient in the same direction (10). The results demonstrated that ORF4 was transcribed during wPCV2 replication.
In order to investigate whether PCV2 translates the ORF4 transcript into a protein during replication, we generated MAbs from mice immunized with an E. coli-expressed recombinant protein derived from the putative ORF4 sequence of wPCV2. By IFA analysis, the ORF4-specific 2D3 MAb could specifically recognize productive infection of wPCV2 in PK-15 cells (Fig. 4B) and mouse splenocytes (Fig. 4C) but not in PCV2Δ-infected PK-15 cells (Fig. 5B) and mouse splenocytes (Fig. 4D). Furthermore, the PCV2 ORF4-specific MAb did not recognize PCV1 antigens in IFA analysis. These results demonstrate that the newly identified ORF4 protein of wPCV2 was expressed during infection, and the anti-PCV2 ORF4 MAbs generated in this study were type specific. Fine epitope mapping defined the core amino acid sequence recognized by the PCV2 ORF4-specific MAbs to be 19KSSASPR25 (Fig. 3). Interestingly, the ORF4 protein signal could only be detected in lysates of wPCV2-infected PK-15 cells not reduced by loading buffer or boiling in dot-ELISA and IFA analysis (Fig. 4E, well 2) and in cryosections of mouse spleens infected with wPCV2 (Fig. 4C). In analyses by Western blotting and dot-ELISA or IHC, no ORF4 protein signal (Fig. 4E, wells 3 and 4) could be detected when the protein samples were treated with loading buffer and boiled or when the wPCV2-infected tissue was embedded in paraffin (data not shown). Hence, these observations suggest that the antigenic epitope recognized by the MAbs to the PCV2 ORF4 protein is a conformational epitope.
Apoptosis is a genetically programmed cell death process (37–39) that allows infected host cells to sacrifice themselves in response to infection in order to limit viral replication (40). However, viruses have developed a variety of strategies to prevent apoptosis through activating endogenous antiapoptotic processes and genes or by expressing their own antiapoptotic genes to complete its propagation (41, 42). On the other hand, viruses may benefit from stimulating apoptosis, either to kill uninfected immune cells or to induce the breakdown of infected cells in the host, thereby favoring viral dissemination (43). The two major apoptotic pathways, extrinsic and intrinsic, have been well studied (37, 44). The extrinsic pathway occurs by activation of tumor necrosis factor receptors to recruit and activate initiator caspases such as caspase-8. Caspase-8 directly or indirectly initiates the proteolytic activities of downstream effector caspases, typically caspase-3. The intrinsic pathway is largely centered around and/or regulated by the mitochondria (44–46). Cytochrome c released from the mitochondria leads to apoptosome formation, which can activate initiator caspases, typically caspase-9. Activation of caspase-9 leads to the activation of the executioner caspase-3, which results in the same type of apoptotic response as that observed for the extrinsic pathway. The apoptosis induced by wPCV2 is known to be caspase dependent, involving significant activation of both the effector caspase-3 and the initiator caspase-8 (25). The PCV2 ORF3 protein was reported to be an apoptosis inducer in vivo and in vitro (20) and found to initiate apoptosis by interacting with pPirh2 and upregulating p53 expression (47). In the current study, progeny of the ORF4-deficient PCV2 infectious clone could be recovered from inoculated PK-15 cells (Fig. 5A), and its replicative capacity was similar to that of wPCV2 (Fig. 5C). However, compared to wPCV2, the ORF4-deficient PCV2 induced higher caspase-3 and -8 activities in PK-15 cells (Fig. 6), a higher viral load at 14 dpi in mouse sera (Fig. 7D), and more severe microscopic lesions in the spleen at the early stage of infection (Fig. 8). Therefore, these results suggest that the ORF4 protein is not essential for wPCV2 replication. Based on previous studies (20, 25), we can also hypothesize that the ORF3 and ORF4 proteins interact with each other in regulating apoptosis. However, whether the ORF4 protein is truly involved in an antiapoptosis mechanism requires further investigation.
Lymphocyte depletion and histiocytic infiltration are the most characteristic features of PMWS (8, 48–50). In previous studies, BALB/c mice inoculated with wPCV2 showed microscopic lesions similar to those found in wPCV2-infected pigs (20, 26, 51). Therefore, we used the mouse model in the present study to compare PCV2Δ and wPCV2 infection in vivo. Obvious pathological splenic lesions and higher serum virus loads (P < 0.05) were detected in the earlier stages of infection in ORF4-deficient PCV2-infected mice (Fig. 7D and 8A) compared to the wPCV2-infected mice inoculated at the same dose. Our results suggest that the ORF4-deficient PCV2 is involved in virulence and replication in vivo. We also found that the relative proportions of CD4+, CD8+, and CD4+ CD8+ T cells were downregulated in PCV2Δ- and wPCV2-infected mice, similar to previous findings for PCV2-infected pigs (52). Furthermore, compared to wPCV2-infected mice, the mutant virus exhibited a greater decrease of CD4+ and CD8+ T cells in infected mice (Fig. 7A to C). A previous study also found that cell depletion in PMWS lymphoid tissues is mainly related to the decreased cytokine secretion (53). Taken together, these findings implicate a role for the PCV2 ORF4 protein in modulating the host immune system via regulation of CD4+ and CD8+ T lymphocytes during infection.
ACKNOWLEDGMENTS
This work was supported by grants from National Natural Science Foundation of China (project no. 31230072), National High-Tech Program of China (project no. 2011AA10A208), and the Zhejiang provincial Sci & Tech Department (project no. 2011C12007).
Footnotes
Published ahead of print 14 November 2012
REFERENCES
- 1. Fauquet CM, Maniloff MMJ, Desselberger U, Ball LA. 2005. Virus taxonomy: 8th report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, Amsterdam, the Netherlands [Google Scholar]
- 2. Tischer I, Gelderblom H, Vettermann W, Koch MA. 1982. A very small porcine virus with circular single-stranded DNA. Nature 295:64–66 [DOI] [PubMed] [Google Scholar]
- 3. Woods LW, Latimer KS, Barr BC, Niagro FD, Campagnoli RP, Nordhausen RW, Castro AE. 1993. Circovirus-like infection in a pigeon. J. Vet. Diagn. Investig. 5:609–612 [DOI] [PubMed] [Google Scholar]
- 4. Tischer I, Rasch R, Tochtermann G. 1974. Characterization of papovavirus- and picornavirus-like particles in permanent pig kidney cell lines. Zentralbl. Bakteriol. Orig. A 226:153–167 [PubMed] [Google Scholar]
- 5. Harding JCS, Clark EG. 1997. Recognizing and diagnosing postweaning multisystemic wasting syndrome (PMWS). J. Swine Health Prod. 5:201–203 [Google Scholar]
- 6. Allan G, Meehan B, Todd D, Kennedy S, McNeilly F, Ellis J, Clark EG, Harding J, Espuna E, Botner A, Charreyre C. 1998. Novel porcine circoviruses from pigs with wasting disease syndromes. Vet. Rec. 142:467–468 [PubMed] [Google Scholar]
- 7. Allan GM, McNeilly F, Meehan BM, Kennedy S, Mackie DP, Ellis JA, Clark EG, Espuna E, Saubi N, Riera P, Botner A, Charreyre CE. 1999. Isolation and characterisation of circoviruses from pigs with wasting syndromes in Spain, Denmark and Northern Ireland. Vet. Microbiol. 66:115–123 [DOI] [PubMed] [Google Scholar]
- 8. Ellis J, Hassard L, Clark E, Harding J, Allan G, Willson P, Strokappe J, Martin K, McNeilly F, Meehan B, Todd D, Haines D. 1998. Isolation of circovirus from lesions of pigs with postweaning multisystemic wasting syndrome. Can. Vet. J. 39:44–51 [PMC free article] [PubMed] [Google Scholar]
- 9. Morozov I, Sirinarumitr T, Sorden SD, Halbur PG, Morgan MK, Yoon KJ, Paul PS. 1998. Detection of a novel strain of porcine circovirus in pigs with postweaning multisystemic wasting syndrome. J. Clin. Microbiol. 36:2535–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hamel AL, Lin LL, Nayar GPS. 1998. Nucleotide sequence of porcine circovirus associated with postweaning multisystemic wasting syndrome in pigs. J. Virol. 72:5262–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meehan BM, Creelan JL, McNulty MS, Todd D. 1997. Sequence of porcine circovirus DNA: affinities with plant circoviruses. J. Gen. Virol. 78:221–227 [DOI] [PubMed] [Google Scholar]
- 12. Meehan BM, McNeilly F, Todd D, Kennedy S, Jewhurst VA, Ellis JA, Hassard LE, Clark EG, Haines DM, Allan GM. 1998. Characterization of novel circovirus DNAs associated with wasting syndromes in pigs. J. Gen. Virol. 79:2171–2179 [DOI] [PubMed] [Google Scholar]
- 13. Zhou JY, Chen QX, Ye JX, Shen HG, Chen TF, Shang SB. 2006. Serological investigation and genomic characterization of PCV2 isolates from different geographic regions of Zhejiang province in China. Vet. Res. Commun. 30:205–220 [DOI] [PubMed] [Google Scholar]
- 14. Cheung AK. 2003. Transcriptional analysis of porcine circovirus type 2. Virology 305:168–180 [DOI] [PubMed] [Google Scholar]
- 15. Mankertz A, Hillenbrand B. 2001. Replication of porcine circovirus type 1 requires two proteins encoded by the viral rep gene. Virology 279:429–438 [DOI] [PubMed] [Google Scholar]
- 16. Nawagitgul P, Morozov I, Bolin SR, Harms PA, Sorden SD, Paul PS. 2000. Open reading frame 2 of porcine circovirus type 2 encodes a major capsid protein. J. Gen. Virol. 81:2281–2287 [DOI] [PubMed] [Google Scholar]
- 17. Fenaux M, Opriessnig T, Halbur PG, Elvinger F, Meng XJ. 2004. Two amino acid mutations in the capsid protein of type 2 porcine circovirus (PCV2) enhanced PCV2 replication in vitro and attenuated the virus in vivo. J. Virol. 78:13440–13446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shang SB, Jin YL, Jiang XT, Zhou JY, Zhang X, Xing G, He JL, Yan Y. 2009. Fine mapping of antigenic epitopes on capsid proteins of porcine circovirus, and antigenic phenotype of porcine circovirus type 2. Mol. Immunol. 46:327–334 [DOI] [PubMed] [Google Scholar]
- 19. Karuppannan AK, Jong MH, Lee SH, Zhu Y, Selvaraj M, Lau J, Jia Q, Kwang J. 2009. Attenuation of porcine circovirus 2 in SPF piglets by abrogation of ORF3 function. Virology 383:338–347 [DOI] [PubMed] [Google Scholar]
- 20. Liu J, Chen I, Du Q, Chua H, Kwang J. 2006. The ORF3 protein of porcine circovirus type 2 is involved in viral pathogenesis in vivo. J. Virol. 80:5065–5073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fenaux M, Opriessnig T, Halbur PG, Elvinger F, Meng XJ. 2004. A chimeric porcine circovirus (PCV) with the immunogenic capsid gene of the pathogenic PCV type 2 (PCV2) cloned into the genomic backbone of the nonpathogenic PCV1 induces protective immunity against PCV2 infection in pigs. J. Virol. 78:6297–6303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Juhan NM, LeRoith T, Opriessnig T, Meng XJ. 2010. The open reading frame 3 (ORF3) of porcine circovirus type 2 (PCV2) is dispensable for virus infection but evidence of reduced pathogenicity is limited in pigs infected by an ORF3-null PCV2 mutant. Virus Res. 147:60–66 [DOI] [PubMed] [Google Scholar]
- 23. Zhang X, Ma G, Li Y, Jiang X, He J, Zhou J. 2009. Characterization of monoclonal antibody against replication-associated protein of porcine circovirus. DNA Cell Biol. 28:23–29 [DOI] [PubMed] [Google Scholar]
- 24. Zhou JY, Shang SB, Gong H, Chen QX, Wu JX, Shen HG, Chen TF, Guo JQ. 2005. In vitro expression, monoclonal antibody and bioactivity for capsid protein of porcine circovirus type II without nuclear localization signal. J. Biotechnol. 118:201–211 [DOI] [PubMed] [Google Scholar]
- 25. Liu J, Chen I, Kwang J. 2005. Characterization of a previously unidentified viral protein in porcine circovirus type 2-infected cells and its role in virus-induced apoptosis. J. Virol. 79:8262–8274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shen HG, Zhou JY, Huang ZY, Guo JQ, Xing G, He JL, Yan Y, Gong LY. 2008. Protective immunity against porcine circovirus 2 by vaccination with ORF2-based DNA and subunit vaccines in mice. J. Gen. Virol. 89:1857–1865 [DOI] [PubMed] [Google Scholar]
- 27. Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493–497 [Google Scholar]
- 28. Sorden SD, Harms PA, Nawagitgul P, Cavanaugh D, Paul PS. 1999. Development of a polyclonal-antibody-based immunohistochemical method for the detection of type 2 porcine circovirus in formalin-fixed, paraffin-embedded tissue. J. Vet. Diagn. Investig. 11:528–530 [DOI] [PubMed] [Google Scholar]
- 29. Autret A, Martin-Latil S, Brisac C, Mousson L, Colbere-Garapin F, Blondel B. 2008. Early phosphatidylinositol 3-kinase/Akt pathway activation limits poliovirus-induced JNK-mediated cell death. J. Virol. 82:3796–3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bagchi P, Dutta D, Chattopadhyay S, Mukherjee A, Halder UC, Sarkar S, Kobayashi N, Komoto S, Taniguchi K, Chawla-Sarkar M. 2010. Rotavirus nonstructural protein 1 suppresses virus-induced cellular apoptosis to facilitate viral growth by activating the cell survival pathways during early stages of infection. J. Virol. 84:6834–6845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ehrhardt C, Marjuki H, Wolff T, Nurnberg B, Planz O, Pleschka S, Ludwig S. 2006. Bivalent role of the phosphatidylinositol-3-kinase (PI3K) during influenza virus infection and host cell defence. Cell. Microbiol. 8:1336–1348 [DOI] [PubMed] [Google Scholar]
- 32. Ehrhardt C, Wolff T, Pleschka S, Planz O, Beermann W, Bode JG, Schmolke M, Ludwig S. 2007. Influenza A virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J. Virol. 81:3058–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee CJ, Liao CL, Lin YL. 2005. Flavivirus activates phosphatidylinositol 3-kinase signaling to block caspase-dependent apoptotic cell death at the early stage of virus infection. J. Virol. 79:8388–8399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu M, Vakharia VN. 2006. Nonstructural protein of infectious bursal disease virus inhibits apoptosis at the early stage of virus infection. J. Virol. 80:3369–3377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Soares JA, Leite FG, Andrade LG, Torres AA, De Sousa LP, Barcelos LS, Teixeira MM, Ferreira PC, Kroon EG, Souto-Padron T, Bonjardim CA. 2009. Activation of the PI3K/Akt pathway early during vaccinia and cowpox virus infections is required for both host survival and viral replication. J. Virol. 83:6883–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cheung AK. 2003. The essential and nonessential transcription units for viral protein synthesis and DNA replication of porcine circovirus type 2. Virology 313:452–459 [DOI] [PubMed] [Google Scholar]
- 37. Duprez L, Wirawan E, Vanden Berghe T, Vandenabeele P. 2009. Major cell death pathways at a glance. Microbes Infect. 11:1050–1062 [DOI] [PubMed] [Google Scholar]
- 38. Fulda S, Gorman AM, Hori O, Samali A. 2010. Cellular stress responses: cell survival and cell death. Int. J. Cell Biol. 2010:214074 doi:10.1155/2010/214074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hotchkiss RS, Strasser A, McDunn JE, Swanson PE. 2009. Mechanisms of disease cell death. N. Engl. J. Med. 361:1570–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Labbe K, Saleh M. 2008. Cell death in the host response to infection. Cell Death Differ. 15:1339–1349 [DOI] [PubMed] [Google Scholar]
- 41. Busca A, Saxena M, Kryworuchko M, Kumar A. 2009. Anti-apoptotic genes in the survival of monocytic cells during infection. Curr. Genomics 10:306–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Galluzzi L, Kepp O, Morselli E, Vitale I, Senovilla L, Pinti M, Zitvogel L, Kroemer G. 2010. Viral strategies for the evasion of immunogenic cell death. J. Intern. Med. 267:526–542 [DOI] [PubMed] [Google Scholar]
- 43. Galluzzi L, Brenner C, Morselli E, Touat Z, Kroemer G. 2008. Viral control of mitochondrial apoptosis. PLoS Pathog. 4:e1000018 doi:10.1371/journal.ppat.1000018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sprick MR, Walczak H. 2004. The interplay between the Bcl-2 family and death receptor-mediated apoptosis. Biochim. Biophys. Acta 1644:125–132 [DOI] [PubMed] [Google Scholar]
- 45. Brunelle JK, Letai A. 2009. Control of mitochondrial apoptosis by the Bcl-2 family. J. Cell Sci. 122:437–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Galluzzi L, Morselli E, Kepp O, Vitale I, Rigoni A, Vacchelli E, Michaud M, Zischka H, Castedo M, Kroemer G. 2010. Mitochondrial gateways to cancer. Mol. Aspects Med. 31:1–20 [DOI] [PubMed] [Google Scholar]
- 47. Liu J, Zhu Y, Chen I, Lau J, He F, Lau A, Wang ZL, Karuppannan AK, Kwang J. 2007. The ORF3 protein of porcine circovirus type 2 interacts with porcine ubiquitin E3 ligase Pirh2 and facilitates p53 expression in viral infection. J. Virol. 81:9560–9567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Allan GM, McNeilly F, Kennedy S, Daft B, Clarke EG, Ellis JA, Haines DM, Meehan BM, Adair BM. 1998. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the U. S. A. and Europe. J. Vet. Diagn. Investig. 10:3–10 [DOI] [PubMed] [Google Scholar]
- 49. Kiupel M, Stevenson GW, Mittal SK, Clark EG, Haines DM. 1998. Circovirus-like viral associated disease in weaned pigs in Indiana. Vet. Pathol. 35:303–307 [DOI] [PubMed] [Google Scholar]
- 50. Rosell C, Segales J, Plana-Duran J, Balasch M, Rodriguez-Arrioja GM, Kennedy S, Allan GM, McNeilly F, Latimer KS, Domingo M. 1999. Pathological, immunohistochemical, and in-situ hybridization studies of natural cases of postweaning multisystemic wasting syndrome (PMWS) in pigs. J. Comp. Pathol. 120:59–78 [DOI] [PubMed] [Google Scholar]
- 51. Kiupel M, Stevenson GW, Choi J, Latimer KS, Kanitz CL, Mittal SK. 2001. Viral replication and lesions in BALB/c mice experimentally inoculated with porcine circovirus isolated from a pig with postweaning multisystemic wasting disease. Vet. Pathol. 38:74–82 [DOI] [PubMed] [Google Scholar]
- 52. Darwich L, Segales JS, Domingo M, Mateu E. 2002. Changes in CD4+, CD8+, CD4+ CD8+, and immunoglobulin M-positive peripheral blood mononuclear cells of postweaning multisystemic wasting syndrome-affected pigs and age-matched uninfected wasted and healthy pigs correlate with lesions and porcine circovirus type 2 load in lymphoid tissues. Clin. Diagn. Lab. Immunol. 9:236–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mandrioli L, Sarli G, Panarese S, Baldoni S, Marcato PS. 2004. Apoptosis and proliferative activity in lymph node reaction in postweaning multisystemic wasting syndrome (PMWS). Vet. Immunol. Immunopathol. 97:25–37 [DOI] [PubMed] [Google Scholar]