Abstract
Copy number variations (CNVs) in the human genome contribute significantly to disease. De novo CNV mutations arise via genomic rearrangements, which can occur in ‘trans’, i.e. via interchromosomal events, or in ‘cis’, i.e. via intrachromosomal events. However, what molecular mechanisms occur between chromosomes versus between or within chromatids has not been systematically investigated. We hypothesized that distinct CNV mutational mechanisms, based on their intrinsic properties, may occur in a biased intrachromosomal versus interchromosomal manner. Here, we studied 62 genomic duplications observed in association with sporadic Potocki–Lupski syndrome (PTLS), in which multiple mutational mechanisms appear to be operative. Intriguingly, more interchromosomal than intrachromosomal events were identified in recurrent PTLS duplications mediated by non-allelic homologous recombination, whereas the reciprocal distribution was found for replicative mechanisms and non-homologous end-joining, likely reflecting the differences in spacial proximity of homologous chromosomes during different mutational processes.
INTRODUCTION
Copy number variations (CNVs), including deletions and duplications resulting from genomic rearrangements, are a major type of genetic variation between human individuals (1). Whereas many CNVs represent benign polymorphisms, others are associated with human diseases, including chromosomal syndromes, genomic disorders, Mendelian traits and common diseases (1,2).
A fundamental question in genetics and evolution is: What is the origin of DNA mutations? Furthermore, determining mutational origins may also facilitate genetic diagnostics of human diseases. De novo CNV mutations can occur as germ line events during either mitotic cycle of germ stem cells or subsequent meiosis, but may also occur post-zygotically in early stages of embryo development (3,4). Notably, paternally versus maternally derived mutations may have different contributions to certain diseases due to sex-specific germ cell processes, or even convey distinct phenotypes due to imprinting (5). A greater contribution of paternal mutations than maternal mutations has been suggested for de novo single nucleotide variants and CNV mutations at some specific loci in the human genome (6,7). Besides parental origins, CNVs can arise in different ways based on the chromosomal rearrangement patterns (8): intrachromatidal (within the same chromatid), interchromatidal (between sister chromatids) and interchromosomal (between homologous chromosomes) events. The first two patterns represent intrachromosomal events. Notably, the variable ratios of interchromosomal versus intrachromosomal rearrangements have been identified across different CNV loci (6,9,10), but the underlying potential reasons remain obscure.
Various molecular mechanisms have been proposed for human CNV formation. Three major mechanisms delineated include non-allelic homologous recombination (NAHR) (8), non-homologous end-joining (NHEJ) (11) and the replicative mechanisms (RMs) (12–14). Considering the differences in intrinsic properties between CNV mutations, we hypothesized that various mutational mechanisms may differentially display parental origin biases and/or have a preferential involvement of interchromosomal versus intrachromosomal CNV mutations.
Since the majority of the CNVs identified in human populations represent inherited genetic polymorphisms, they do not directly reflect the process of de novo CNV formation, and therefore, are not suitable for investigation into CNV origin. In this study, in order to specifically examine de novo CNV events, we took advantage of genomic disorders, many of which present as sporadic conditions manifesting developmental delays and intellectual disabilities for which families seek medical consultation (15).
Potocki–Lupski syndrome (PTLS, [MIM 610883]) is associated with 17p11.2 duplications (Fig. 1); clinical manifestations include infantile hypotonia, failure to thrive, mental retardation, autism, behavioral abnormalities and structural cardiovascular anomalies (10,16). We utilized PTLS as a model to study the potential roles of molecular mechanisms in CNV origins based on the following attributes that provide experimental advantages: (a) PTLS is a duplication syndrome, which has extra genomic copies and provides more genetic and molecular information than deletion syndromes to trace CNV origins; (b) essentially all PTLS-associated duplications are de novo mutations and associated with this sporadic trait and (c) various molecular mechanisms have been revealed and comprehensively investigated in our previous studies on PTLS duplications (10,13,17).
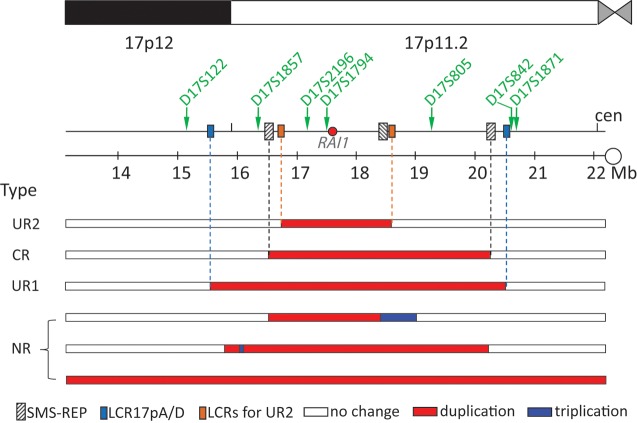
Figure 1.
PTLS-associated duplication types and the STR markers used for the parent-of-origin study. Schematic representation of 17p11.2 and partial 17p12 (centromere-cen, to the right) with LCRs (not to scale). The red horizontal bars depict the portions of the duplicated genomic intervals. LCR-mediated recurrent PTLS duplications: CR duplication between SMS-REPs, uncommon recurrent (Type I, UR1) duplications between LCR17pA and LCR17pD, uncommon recurrent (Type II, UR2) duplications between two LCRs of 24 kb in length. Nonrecurrent (NR) duplications have variable sizes and locations, some of which can have complex structures, for example, the triplicated intervals shown by blue bars. Three nonrecurrent duplications are chosen arbitrarily to represent the 16 NR duplications. The positions of seven STR markers are shown by green arrows.
In this study, we observed at the PTLS locus that interchromosomal rearrangements predominate in recurrent duplications mediated by NAHR, whereas intrachromosomal duplications seem to be preferred by RMs. This significantly biased distribution of interchromosomal versus intrachromosomal origins is consistent with the spacial proximity of chromosome homologs and substrate sequences utilized by different molecular mechanisms for CNV formation during meiotic versus mitotic events.
RESULTS
CNV types, mechanisms and origins in 62 PTLS duplications
The 17p11.2 genomic duplications associated with PTLS were investigated using two custom-designed comparative genomic hybridization (CGH) microarrays in this study and our previous studies (10,13,17–19). One utilized bacterial artificial chromosome (BAC) clones as interrogating probes (10). The other employed high-density oligonucleotide probes (13), which resolved copy number changes as small as ∼ 500 bp and achieved a higher resolution than the BAC CGH microarray. We summarized the duplication information (sizes, types and breakpoints), investigative methods, inferred mutational mechanisms and CNV origins of a total of 62 duplications associated with PTLS in Table 1. These duplications include 46 recurrent and 16 nonrecurrent duplications. Notably, all nonrecurrent PTLS duplications were assayed using the high-density oligonucleotide CGH microarray for fine mapping of breakpoint junctions. In addition, four nonrecurrent duplications (subjects 1229, 2695, 2711, and 2965) were also investigated using high-resolution single nucleotide polymorphism (SNP) microarrays (see Methods and Supplementary Material, Fig. S1).
Table 1.
Summary of 62 PTLS-associated duplications with duplication information, mutational mechanisms, parent-of-origins and rearrangement patterns
| Subject | Size (Mb)a | CGH/SNP array | Type | Breakpoint | Reference | Mechanism | Reference | P/M, inter/intra | Reference |
|---|---|---|---|---|---|---|---|---|---|
| 504 | 3.7 | BAC | CR | Paired LCRs | (10) | NAHR | (10) | P, inter | (10) |
| 527 | 10.4 | BAC, Oligo | SNR | One LCR | (10,17) | NHEJ/RMs | (17) | M, intra | (10) |
| 563 | 5.6 | BAC, Oligo | CNR | Microhomologies CCTC, CTCCC | (10,13) | RMs | (13) | P | (10) |
| 621 | 15.5 | BAC, Oligo | CNR | Microhomologies CC, TTGGT | (10,13) | RMs | (13) | M | (10) |
| 990 | 3.7 | BAC | CR | Paired LCRs | (10) | NAHR | (10) | P, inter | (10) |
| 1006 | 3.7 | BAC | CR | Paired LCRs | (10) | NAHR | (10) | P, intra | (10) |
| 1192 | 3.7 | BAC | CR | Paired LCRs | (10) | NAHR | (10) | P. inter | (10) |
| 1229 | 6.4 | BAC, Oligo, Omni-1 | CNR | Microhomology ACCTTC, one LCR | (10,13), TS | RMs | (13) | P, intra | (10) |
| 1251 | 3.7 | BAC | CR | Paired LCRs | (10) | NAHR | (10) | P, intra | (10) |
| 1353 | 3.7 | BAC | CR | Paired LCRs | (10) | NAHR | (10) | M, inter | (10) |
| 1364 | 3.7 | BAC | CR | Paired LCRs | (10) | NAHR | (10) | M, intra | (10) |
| 1458 | 7.6 | BAC, Oligo | CNR | One LCR | (10,13) | RMs | (13) | M, intra | (10) |
| 1579 | 3.7 | BAC | CR | Paired LCRs | (10) | NAHR | (10) | M, inter | (10) |
| 1618 | 3.7 | BAC | CR | Paired LCRs | (10) | NAHR | (10) | P, inter | (10) |
| 1632 | 3.7 | BAC | CR | Paired LCRs | (10) | NAHR | (10) | M, inter | (10) |
| 1671 | 3.7 | BAC | CR | Paired LCRs | (10) | NAHR | (10) | M, intra | (10) |
| 1786 | 3.7 | BAC | CR | Paired LCRs | (10) | NAHR | (10) | P, intra | (10) |
| 1789 | 3.7 | BAC | CR | Paired LCRs | (10) | NAHR | (10) | P, intra | (10) |
| 1838 | 3.7 | BAC | CR | Paired LCRs | (10) | NAHR | (10) | P, intra | (10) |
| 1861 | 7.7 | BAC, Oligo | SNR | One LCR | (10,13) | NHEJ/RMs | (13) | M, intra | (10) |
| 1913 | 3.7 | BAC | CR | Paired LCRs | (10) | NAHR | (10) | P, intra | (10) |
| 2153 | 3.7 | BAC, Oligo | CR | Paired LCRs | (10,13) | NAHR | (13) | P, inter | (10) |
| 2167 | 3.7 | BAC | CR | Paired LCRs | (10) | NAHR | (10) | P | (10) |
| 2211 | 8.7 | BAC, Oligo | CNR | One LCR | (10,13) | RMs | (13) | M, intra | (10) |
| 2306 | 3.7 | BAC | CR | Paired LCRs | (10) | NAHR | (10) | M, inter | (10) |
| 2337 | 5.9 | BAC, Oligo | CNR | NA | (10,13) | RMs | (13) | M, intra | (10) |
| 2440 | 4.9 | BAC, Oligo | SNR | One LCR | (10,13) | NHEJ/RMs | (13) | P, inter | (10) |
| 2488 | 7.5 | BAC, Oligo | SNR | One LCR | (10,13) | NHEJ/RMs | (13) | M, intra | (10) |
| 2543 | 1.5 | BAC, Oligo | SNR | One LCR | (10,13) | NHEJ/RMs | (13) | M, inter | (10) |
| 2555 | 3.7 | BAC | CR | Paired LCRs | (10) | NAHR | (10) | M, inter | (10) |
| 2571 | 3.7 | BAC | CR | Paired LCRs | (10) | NAHR | (10) | P, inter | (10) |
| 2578 | 3.7 | Oligo | CR | Paired LCRs | (17) | NAHR | (17) | P, inter | TS |
| 2581 | 3.7 | Oligo | CR | Paired LCRs | (17) | NAHR | (17) | P, inter | TS |
| 2592 | 3.7 | Oligo | CR | Paired LCRs | (17) | NAHR | (17) | M, inter | TS |
| 2597 | 3.7 | Oligo | CR | Paired LCRs | (17) | NAHR | (17) | Inter | TS |
| 2634 | 3.7 | Oligo | CR | Paired LCRs | (17) | NAHR | (17) | Inter | TS |
| 2659 | 3.7 | Oligo | CR | Paired LCRs | (18) | NAHR | (18) | M, inter | (18) |
| 2660 | 3.7 | Oligo | CR | Paired LCRs | (18) | NAHR | (18) | P, inter | (18) |
| 2661 | 11.1 | Oligo | SNR | Microhomology AT | (13) | NHEJ/RMs | (13) | M, intra | (18) |
| 2671 | 3.7 | Oligo | CR | Paired LCRs | (17) | NAHR | (17) | Inter | TS |
| 2692 | 3.7 | Oligo | CR | Paired LCRs | (17) | NAHR | (17) | Inter | TS |
| 2695 | 4.4 | Oligo, Omni-1 | CNR | Two AluYs, one LCR | (13), TS | RMs | (13) | P, inter | TS |
| 2708 | 3.7 | Oligo | CR | Paired LCRs | (17) | NAHR | (17) | M, inter | TS |
| 2711 | 11.3 | Oligo, Omni-1 | CNR | One LCR | (13), TS | RMs | (13) | M, intra | TS |
| 2724 | 3.7 | Oligo | CR | Paired LCRs | (17) | NAHR | (17) | P, intra | TS |
| 2728 | 3.7 | Oligo | CR | Paired LCRs | (17) | NAHR | (17) | M, inter | TS |
| 2745 | 3.7 | Oligo | CR | Paired LCRs | (17) | NAHR | (17) | M, intra | TS |
| 2748 | 3.7 | Oligo | CR | Paired LCRs | (17) | NAHR | (17) | Inter | TS |
| 2758 | 3.7 | Oligo | CR | Paired LCRs | (17) | NAHR | (17) | P, inter | TS |
| 2767 | 3.7 | Oligo | CR | Paired LCRs | (17) | NAHR | (17) | P, inter | TS |
| 2808 | 5.0 | Oligo | UR1 | Paired LCRs | (17) | NAHR | (17) | P, inter | TS |
| 2946 | 2.8 | Oligo | CNR | Two LCRs | (17) | RMs | (17) | M, intra | (35) |
| 2948 | 3.7 | Oligo | CR | Paired LCRs | (17) | NAHR | (17) | P, inter | TS |
| 2953 | 3.7 | Oligo | CR | Paired LCRs | (17) | NAHR | (17) | Inter | TS |
| 2956 | 3.7 | Oligo | CR | Paired LCRs | (17) | NAHR | (17) | M, inter | TS |
| 2959 | 5.0 | Oligo | UR1 | Paired LCRs | (17) | NAHR | (17) | M, inter | TS |
| 2962 | 3.7 | Oligo | CR | Paired LCRs | (17) | NAHR | (17) | M, inter | TS |
| 2965 | 2.5 | Oligo, Omni-1 | CNR | Four LCRs | (17), TS | RMs | (17) | P, inter | TS |
| 2995 | 3.7 | Oligo | CR | Paired LCRs | (17) | NAHR | (17) | M, inter | TS |
| 2998 | 3.7 | Oligo | CR | Paired LCRs | (17) | NAHR | (17) | M, intra | TS |
| 3028 | 3.7 | Oligo | CR | Paired LCRs | TS | NAHR | TS | M, inter | TS |
| 3142 | 2.1 | Oligo | UR2 | Paired LCRs | (19) | NAHR | (19) | Inter | TS |
BAC, bacterial artificial chromosome; CGH, comparative genomic hybridization; CNR, complex nonrecurrent duplication; CR, common recurrent duplication; inter, interchromosomal rearrangement; intra, intrachromosomal rearrangement; LCR, low-copy repeat; M, maternal origin; NA, not available; NAHR, non-allelic homologous recombination; NHEJ, non-homologous end-joining; oligo, oligonucleotide; P, paternal origin; RMs, replicative mechanisms; SNR, simple nonrecurrent duplication; TS, this study; UR1, uncommon recurrent type I duplication; UR2, uncommon recurrent type II duplication.
aFor complex nonrecurrent duplications, the sizes of continuous RAI1 gene duplications were given.
The 46 recurrent duplications can be further divided into 43 common recurrent (CR) duplications (16), two uncommon recurrent type I (UR1) duplications (17) and one uncommon recurrent type II (UR2) duplication (19), all of which were mediated by NAHR between various low-copy repeats (LCRs, also known as segmental duplications) in 17p (Fig. 1) (8,20). The 16 nonrecurrent PTLS duplications were of variable sizes and genomic locations, but can be categorized based on structural complexity into six simple nonrecurrent (SNR) duplications and ten complex nonrecurrent (CNR) duplications (Fig. 1). Both NHEJ and RMs have been proposed to cause SNR duplications, whereas RMs have been proposed to play a prominent role in mediating CNR rearrangements (1,11,21).
PTLS duplication origins
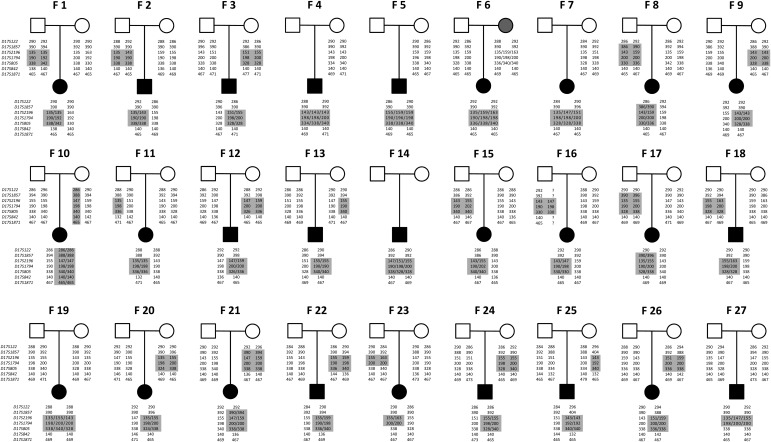
PTLS duplications from 27 subjects (2578, 2581, 2592, 2597, 2634, 2672, 2692, 2695, 2708, 2711, 2724, 2728, 2745, 2748, 2758, 2767, 2808, 2948, 2953, 2956, 2959, 2962, 2965, 2995, 2998, 3028 and 3142) are newly investigated for duplication origins using short tandem repeat (STR) genotyping analysis (Supplementary Material, Table S1). The copy number status of the seven STR markers (Fig. 1; Supplementary Material, Table S2) was confirmed in these PTLS subjects before inferring their duplication origins using STR genotypes in each core family; that is trios of subject plus biological parents. Interchromosomal rearrangements were identified in 23 PTLS duplications and intrachromosomal rearrangements were found in the four remaining duplications (Fig. 2; Supplementary Material, Table S1). Furthermore, paternal and maternal origins were determined in nine and eleven PTLS duplications respectively, whereas the remaining seven duplications lack sufficient information to infer parental origin (Fig. 2; Supplementary Material, Table S1).
Figure 2.
STR genotyping revealed duplication origins in 27 newly investigated families (F1–F27). Seven STR markers were listed in order from distal (top) to proximal positions (bottom) of human chromosome 17p. The duplicated alleles in patients and their origins in parental alleles were shadowed. The mother in F6 was a carrier of mosaic PTLS duplication.
In the entire dataset of 62 PTLS duplications, 26 were paternal in origin and 29 were maternal in origin (Table 1). The remaining seven duplications were not informative. No evidence was found for a predominant paternal origin. These 62 independent PTLS associated duplication events revealed a distribution of chromosomal rearrangement patterns: 38 interchromosomal rearrangements, 21 intrachromosomal rearrangements and three with uninformative patterns (Table 1).
Biased distribution of interchromosomal versus intrachromosomal origins with different duplication mechanisms
Interestingly, a high frequency (75.6%) of interchromosomal duplications was found in NAHR-mediated PTLS duplications (34 interchromosomal and 11 intrachromosomal duplications; uninformative for one NAHR-mediated duplication) (Table 1). However, only 33.3% (two of six) of the PTLS duplications mediated by NHEJ and/or RMs and 25.0% (two out of eight informative duplications) of the complex duplications mediated by RMs were interchromosomal (Table 1). This biased distribution of interchromosomal versus intrachromosomal PTLS duplications between various mutational mechanisms is significant (Table 2).
Table 2.
Biased distribution of inter-/intra-chromosomal rearrangements in PTLS duplications with different mutational mechanisms
| Mechanism | Inter | Intra | Mechanism | Inter | Intra | Mechanism | Inter | Intra |
|---|---|---|---|---|---|---|---|---|
| NAHR | 34 | 11 | NAHR | 34 | 11 | NAHR | 34 | 11 |
| NHEJ | 2 | 4 | RMs | 2 | 6 | NHEJ+RMs | 4 | 10 |
| P = 0.054a | P = 0.0096a | P = 0.0029a | ||||||
aFisher's exact test, two-sided.
In this study, a potential correlation was also examined between parental origins and mutational mechanisms, and between parental origins and rearrangement patterns (interchromosomal versus intrachromosomal). More duplications were found to be mediated by NHEJ or RMs in the group of maternal origin (11 of 29) than those in paternal origins (5 of 26) (Supplementary Material, Table S3). However, no significantly biased distribution was identified.
DISCUSSION
NAHR events are found to be frequent in meiosis, although NAHR can also occur in mitosis (22,23). NAHR can mediate recurrent PTLS duplications (16,17,19), the majority (75.6%) of which were found to originate from interchromosomal rearrangements in this study (Table 2). This observation is consistent with the molecular evidence for how NAHR or ectopic recombination appears to occur. In meiosis, NAHR occurs via the alignment of paralogous genomic segments and the subsequent ectopic crossover (8). These alignments can take place between LCRs on the same chromatid (intrachromatid), from sister chromatids (interchromatid) and from homologous chromosomes (interchromosomal). Interestingly, homologous chromosomes undergo synapsis during metaphase, which can facilitate interchromosomal crossing over. Additional consistent findings were obtained from four NAHR hotspots (CMT1A-REP, AZFa HERV, WBS LCR and LCR17p) that were previously investigated using pooled sperm assays and were shown to be meiosis-specific (24). In that study, the rate of interchromosomal CMT1A duplications is estimated to be 50-fold higher than the rate of interchromatid events (i.e. the only intrachromosomal pattern that can result in duplications) (24). Preliminary estimates for the WBS locus also suggest that interchromatid NAHR is much less frequent than the interchromosomal event (24). These findings are consistent with ectopic synapsis more readily occurring between homologous chromosomes and thereby setting the stage for ectopic crossovers to occur (19).
NHEJ and RMs occur in both meiosis and mitosis; however, homologous chromosomes are paired in meiosis but not in mitosis. Therefore, interchromosomal rearrangements may not be as topologically favored as the intrachromosomal ones, at least during mitosis.
In eukaryotic cells, NHEJ is routinely utilized by human cells to repair DNA double strand breaks (DSBs), such as those caused by ionizing radiation, reactive oxygen species and physiological V(D)J recombination (11). The four steps of NHEJ include the detection of DSB, the molecular bridging, the modification of the broken DNA ends and the final ligation step (25). In meiosis, NHEJ can mediate both intrachromosomal and interchromosomal events. However, in mitosis, the probability of homologous chromosomes becoming physically associated in the three-dimensional space is very small, which potentially restricts interchromosomal rearrangements. According to our observations in NHEJ-mediated PTLS duplications, intrachromosomal duplications were generated at a higher rate than interchromosomal duplications (Table 2). This phenomenon is consistent with the findings in PLP1 duplications, for which it was proposed that a single DSB occurred in one strand, then one of the broken ends invaded and copied from the sister chromatid and caused duplication by rejoining the ends via NHEJ (26,27).
RMs have been found to be involved in generating complex CNVs (21). In our previous study, a DNA replication-based mechanism of fork stalling and template switching (FoSTeS) was proposed to explain human complex CNVs (12). According to the FoSTeS model, during DNA replication, the replication fork stalls at one position, then the lagging strand disengages from the original template, switches to another fork in physical proximity by the virtue of microhomology and restarts the DNA synthesis. Therefore, intrachromosomal rearrangements occurring between genomic regions that are topologically constrained may be in three-dimensional spatial proximity and potentially occur more frequently than interchromosomal rearrangements (28). According to our results of complex PTLS duplications mediated by RMs, intrachromosomal duplications are generated at a higher rate than interchromosomal ones, which showed a similar propensity to that which we observed in NHEJ (Table 2).
Additionally, no parental origin bias was identified for PTLS duplications, even after dividing PTLS duplications into subgroups based on the mutational mechanisms or rearrangement patterns (Supplementary Material, Tables S3 and S4). Biased paternal origins of CNV mutations have been observed for some specific disease-associated loci in the human genome (29,30), which was previously hypothesized to result from the more DNA replication cycles in spermatogenesis than in oogenesis (6,7). However, no biased distribution was observed for PTLS duplications in this study (Supplementary Material, Table S3). Interestingly, a bias of maternal origin was observed for the deletion of CNVs associated with the Duchenne muscular dystrophy (DMD) gene (31), a locus at which RMs have been implicated in CNV instability (32,33). Therefore, variable contributions of paternal versus maternal origins were identified across different CNV loci. We hypothesize that additional factors other than excess DNA replication events in sperm production may play roles in such phenomena. Since most of the replicative errors involved in CNV formation are induced by DNA strand breaks (21), the load of DNA damage may be another factor affecting the incidence of CNV mutations mediated by RMs. Considering the fact that meiosis is arrested after diplotene of prophase I for decades during human oogenesis (34), it is plausible that DNA DSBs accumulating in oocytes during such a long arrest period could be potentially transmitted to zygotes and induce CNV formation via break-induced replication mechanisms in zygotes.
In summary, our study showed that the NAHR mechanism favors interchromosomal PTLS duplications, whereas RMs and NHEJ were prone to mediate intrachromosomal PTLS duplications. Since NAHR is frequent (but not specific) in meiosis, the pairing between homologous chromosomes during synapsis provides one potential explanation for the bias towards interchromosomal rearrangements. On the other hand, the probability of successful repair of DSBs via NHEJ or break-induced replication will be increased if the interacting genomic regions are in spacial proximity, i.e. intrachromosomal rearrangements within the same chromosome. The correlation between mutational mechanisms and duplication patterns suggests that different spacial proximity for chromosome homologs could influence CNV formation via different mutational mechanisms. This contention may also explain the reasons for inconsistent interchromosomal versus intrachromosomal ratios across different CNV loci in the human genome. Notably, our findings are based on the observations at the PTLS locus. We anticipate that further studies on the possible relationship between CNV origins and mutational mechanisms at additional loci can further our understanding of CNV formation in the human genome.
MATERIALS AND METHODS
Subjects
A total of 27 core families (subject and biological parents) associated with PTLS were newly investigated for duplication origins (Supplementary Material, Table S1). Our previously published data of PTLS duplication types, mutational mechanisms, duplication origins were also summarized for re-analyses in this study (10,13,17–19,35). Blood samples from the PTLS subjects and their parents were obtained with informed consent approved by the Institutional Review Board for Human Subject Research at Baylor College of Medicine. The experiments conducted in Shanghai, China, were also approved by the Institutional Review Board at School of Life Sciences in Fudan University.
CGH microarray analysis
We designed high-density oligonucleotide-based CGH microarrays to finely examine the location, size, genomic content and breakpoint interval of 43 PTLS-associated duplications in this study and our previous ones (Table 1) (13,17–19). These microarrays are based on the Agilent 4 × 44 K format. Approximately 44 000 oligonucleotide probes were used to interrogate the entire 17p with a genome resolution of ∼500 bp. Probes having similar sequences in other genomic loci had been filtered out, and only unique probes were employed. After digestion with AluI and RsaI, the test DNAs were labeled with Cy5-dCTP and the control DNA was labeled with Cy3-dCTP using the BioPrime Array CGH genomic labeling kit (Invitrogen Corporation, Carlsbad, CA, USA). Purification of labeling products, array hybridization, washing, scanning and data analysis were conducted by following the Agilent oligonucleotide CGH protocol (version 5.0).
A total of 31 PTLS duplications were previously assayed using a custom human 17p CGH microarray (Table 1) (10), which was developed with 83 human BAC clones tiling an 11 Mb region from clone RP11-462C21 in 17p13.1 to clone RP11-1109M24 in 17p11.1. We found 12 nonrecurrent PTLS duplications and repeatedly assayed them using high-density oligonucleotide CGH microarrays for fine mapping. Common and recurrent PTLS duplications were identified in the remaining 19 subjects; therefore, they were not further investigated on oligonucleotide CGH microarrays.
SNP microarray analysis
Four PTLS duplications (subjects 1229, 2695, 2711 and 2965) identified by high-density CGH microarrays were repeatedly analyzed using the Human Omni-1 Quad BeadChip (Illumina, USA). DNA sample processing and genotyping were performed by following the Infinium HD assay super manual protocol (Illumina). In brief, genomic DNA was amplified, fragmented and labeled prior to hybridization to the genotyping chip. The resulting microarray was imaged with the iScan reader (Illumina). After allele detection and genotype calling were performed with Genome Studio software (Illumina), B allele frequencies and logR ratios were obtained. The CNV Partition algorithm was run as a plug-in within the Genome Studio browser to obtain the CNV values. The detailed information of microarray data analysis was previously described (36).
Short tandem repeat analysis
Seven STR markers in the 3.7 Mb common PTLS duplication interval and its flanking regions were chosen for the duplication origin study (Fig. 1). The primer information is provided in Supplementary Material, Table S2. FAMTM (Blue) fluorescent dye was used to label the 5′ end of the forward primers. PCR amplifications were conducted with TaKaRa Hot start Taq polymerase. A 10 μl PCR reaction was performed with 0.1 μl of TaKaRa Hot start Taq polymerase with 1 μl of 10 × PCR buffer, 1.2 μl of dNTPs, 2 pmol of each primer and 10 ng DNA template. The PCR conditions were as follows: 95°C for 1 min, 30 cycles at 95°C for 30 s, 61°C for 30 s and 72°C for 30 s, followed by 72°C for 10 min. The PCR products were analyzed by capillary electrophoresis using an ABI 3130 genetic analyzer, and the STR alleles in each sample were genotyped using GeneMapper V3.2 software (Applied Biosystems, USA).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by National Basic Research Program of China (2012CB944600 and 2011CBA00401), National S&T Major Special Project (2011ZX09102-010-01), National Natural Science Foundation of China (31000552 and 31171210) and National Institute of Neurological Disorders and Stroke, National Institutes of Health (R01NS058529).
Supplementary Material
ACKNOWLEDGEMENTS
We thank the participating subjects and families for their collaboration and the referring physicians and genetic counselors for sample acquisition. We also thank Dr CMB Carvalho for her help with SNP microarray analysis.
Conflict of Interest statement. J.R.L. is a consultant for Athena Diagnostics, has stock ownership in 23andMe and Ion Torrent Systems and is a coinventor on multiple United States and European patents for DNA diagnostics. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from clinical testing by high-resolution human genome analysis.
REFERENCES
- 1.Zhang F., Gu W., Hurles M.E., Lupski J.R. Copy number variation in human health, disease, and evolution. Annu. Rev. Genomics Hum. Genet. 2009;10:451–481. doi: 10.1146/annurev.genom.9.081307.164217. doi:10.1146/annurev.genom.9.081307.164217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lupski J.R., Belmont J.W., Boerwinkle E., Gibbs R.A. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. doi:10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lupski J.R. New mutations and intellectual function. Nat. Genet. 2010;42:1036–1038. doi: 10.1038/ng1210-1036. doi:10.1038/ng1210-1036. [DOI] [PubMed] [Google Scholar]
- 4.Boone P.M., Wiszniewski W., Lupski J.R. Genomic medicine and neurological disease. Hum. Genet. 2011;130:103–121. doi: 10.1007/s00439-011-1001-1. doi:10.1007/s00439-011-1001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reik W., Walter J. Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2001;2:21–32. doi: 10.1038/35047554. doi:10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 6.Arnheim N., Calabrese P. Understanding what determines the frequency and pattern of human germline mutations. Nat. Rev. Genet. 2009;10:478–488. doi: 10.1038/nrg2529. doi:10.1038/nrg2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arlt M.F., Wilson T.E., Glover T.W. Replication stress and mechanisms of CNV formation. Curr. Opin. Genet. Dev. 2012;22:204–210. doi: 10.1016/j.gde.2012.01.009. doi:10.1016/j.gde.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stankiewicz P., Lupski J.R. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. doi:10.1016/S0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 9.Lopes J., Tardieu S., Silander K., Blair I., Vandenberghe A., Palau F., Ruberg M., Brice A., LeGuern E. Homologous DNA exchanges in humans can be explained by the yeast double-strand break repair model: a study of 17p11.2 rearrangements associated with CMT1A and HNPP. Hum. Mol. Genet. 1999;8:2285–2292. doi: 10.1093/hmg/8.12.2285. doi:10.1093/hmg/8.12.2285. [DOI] [PubMed] [Google Scholar]
- 10.Potocki L., Bi W., Treadwell-Deering D., Carvalho C.M., Eifert A., Friedman E.M., Glaze D., Krull K., Lee J.A., Lewis R.A., et al. Characterization of Potocki–Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. Am. J. Hum. Genet. 2007;80:633–649. doi: 10.1086/512864. doi:10.1086/512864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lieber M.R., Ma Y., Pannicke U., Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat. Rev. Mol. Cell Biol. 2003;4:712–720. doi: 10.1038/nrm1202. doi:10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 12.Lee J.A., Carvalho C.M., Lupski J.R. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131:1235–1247. doi: 10.1016/j.cell.2007.11.037. doi:10.1016/j.cell.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 13.Zhang F., Khajavi M., Connolly A.M., Towne C.F., Batish S.D., Lupski J.R. The DNA replication FoSTeS/MMBIR mechanism can generate human genomic, genic, and exonic complex rearrangements. Nat. Genet. 2009;41:849–853. doi: 10.1038/ng.399. doi:10.1038/ng.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu P., Carvalho C.M., Hastings P., Lupski J.R. Mechanisms for recurrent and complex human genomic rearrangements. Curr. Opin. Genet. Dev. 2012;22:211–220. doi: 10.1016/j.gde.2012.02.012. doi:10.1016/j.gde.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lupski J.R., Stankiewicz P. Genomic disorders: molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. doi:10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potocki L., Chen K.S., Park S.S., Osterholm D.E., Withers M.A., Kimonis V., Summers A.M., Meschino W.S., Anyane-Yeboa K., Kashork C.D., et al. Molecular mechanism for duplication 17p11.2—the homologous recombination reciprocal of the Smith-Magenis microdeletion. Nat. Genet. 2000;24:84–87. doi: 10.1038/71743. doi:10.1038/71743. [DOI] [PubMed] [Google Scholar]
- 17.Zhang F., Potocki L., Sampson J.B., Liu P., Sanchez-Valle A., Robbins-Furman P., Navarro A.D., Wheeler P.G., Spence J.E., Brasington C.K., et al. Identification of uncommon recurrent Potocki-Lupski syndrome-associated duplications and the distribution of rearrangement types and mechanisms in PTLS. Am. J. Hum. Genet. 2010;86:462–470. doi: 10.1016/j.ajhg.2010.02.001. doi:10.1016/j.ajhg.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doco-Fenzy M., Holder-Espinasse M., Bieth E., Magdelaine C., Vincent M.C., Khoury M., Andrieux J., Zhang F., Lupski J.R., Klink R., et al. The clinical spectrum associated with a chromosome 17 short arm proximal duplication (dup 17p11.2) in three patients. Am. J. Med. Genet. A. 2008;146:917–924. doi: 10.1002/ajmg.a.32195. [DOI] [PubMed] [Google Scholar]
- 19.Liu P., Lacaria M., Zhang F., Withers M., Hastings P.J., Lupski J.R. Frequency of nonallelic homologous recombination is correlated with length of homology: evidence that ectopic synapsis precedes ectopic crossing-over. Am. J. Hum. Genet. 2011;89:580–588. doi: 10.1016/j.ajhg.2011.09.009. doi:10.1016/j.ajhg.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. doi:10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- 21.Hastings P.J., Lupski J.R., Rosenberg S.M., Ira G. Mechanisms of change in gene copy number. Nat. Rev. Genet. 2009;10:551–564. doi: 10.1038/nrg2593. doi:10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam K.W., Jeffreys A.J. Processes of de novo duplication of human alpha-globin genes. Proc. Natl Acad. Sci. USA. 2007;104:10950–10955. doi: 10.1073/pnas.0703856104. doi:10.1073/pnas.0703856104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flores M., Morales L., Gonzaga-Jauregui C., Dominguez-Vidana R., Zepeda C., Yanez O., Gutierrez M., Lemus T., Valle D., Avila M.C., et al. Recurrent DNA inversion rearrangements in the human genome. Proc. Natl Acad. Sci. USA. 2007;104:6099–6106. doi: 10.1073/pnas.0701631104. doi:10.1073/pnas.0701631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turner D.J., Miretti M., Rajan D., Fiegler H., Carter N.P., Blayney M.L., Beck S., Hurles M.E. Germline rates of de novo meiotic deletions and duplications causing several genomic disorders. Nat. Genet. 2008;40:90–95. doi: 10.1038/ng.2007.40. doi:10.1038/ng.2007.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weterings E., van Gent D.C. The mechanism of non-homologous end-joining: a synopsis of synapsis. DNA Repair (Amst) 2004;3:1425–1435. doi: 10.1016/j.dnarep.2004.06.003. doi:10.1016/j.dnarep.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 26.Woodward K.J., Cundall M., Sperle K., Sistermans E.A., Ross M., Howell G., Gribble S.M., Burford D.C., Carter N.P., Hobson D.L., et al. Heterogeneous duplications in patients with Pelizaeus–Merzbacher disease suggest a mechanism of coupled homologous and nonhomologous recombination. Am. J. Hum. Genet. 2005;77:966–987. doi: 10.1086/498048. doi:10.1086/498048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee J.A., Inoue K., Cheung S.W., Shaw C.A., Stankiewicz P., Lupski J.R. Role of genomic architecture in PLP1 duplication causing Pelizaeus–Merzbacher disease. Hum. Mol. Genet. 2006;15:2250–2265. doi: 10.1093/hmg/ddl150. doi:10.1093/hmg/ddl150. [DOI] [PubMed] [Google Scholar]
- 28.Carvalho C.M., Ramocki M.B., Pehlivan D., Franco L.M., Gonzaga-Jauregui C., Fang P., McCall A., Pivnick E.K., Hines-Dowell S., Seaver L.H., et al. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat. Genet. 2011;43:1074–1081. doi: 10.1038/ng.944. doi:10.1038/ng.944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hehir-Kwa J.Y., Rodriguez-Santiago B., Vissers L.E., de Leeuw N., Pfundt R., Buitelaar J.K., Perez-Jurado L.A., Veltman J.A. De novo copy number variants associated with intellectual disability have a paternal origin and age bias. J. Med. Genet. 2011;48:776–778. doi: 10.1136/jmedgenet-2011-100147. doi:10.1136/jmedgenet-2011-100147. [DOI] [PubMed] [Google Scholar]
- 30.Sibbons C., Morris J.K., Crolla J.A., Jacobs P.A., Thomas N.S. De novo deletions and duplications detected by array CGH: a study of parental origin in relation to mechanisms of formation and size of imbalance. Eur. J. Hum. Genet. 2012;20:155–160. doi: 10.1038/ejhg.2011.182. doi:10.1038/ejhg.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimm T., Meng G., Liechti-Gallati S., Bettecken T., Muller C.R., Muller B. On the origin of deletions and point mutations in Duchenne muscular dystrophy: most deletions arise in oogenesis and most point mutations result from events in spermatogenesis. J. Med. Genet. 1994;31:183–186. doi: 10.1136/jmg.31.3.183. doi:10.1136/jmg.31.3.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitsui J., Takahashi Y., Goto J., Tomiyama H., Ishikawa S., Yoshino H., Minami N., Smith D.I., Lesage S., Aburatani H., et al. Mechanisms of genomic instabilities underlying two common fragile-site-associated loci, PARK2 and DMD, in germ cell and cancer cell lines. Am. J. Hum. Genet. 2012;87:75–89. doi: 10.1016/j.ajhg.2010.06.006. doi:10.1016/j.ajhg.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ankala A., Kohn J.N., Hegde A., Meka A., Ephrem C.L., Askree S.H., Bhide S., Hegde M.R. Aberrant firing of replication origins potentially explains intragenic nonrecurrent rearrangements within genes, including the human DMD gene. Genome Res. 2012;22:25–34. doi: 10.1101/gr.123463.111. doi:10.1101/gr.123463.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Handel M.A., Schimenti J.C. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat. Rev. Genet. 2012;11:124–136. doi: 10.1038/nrg2723. [DOI] [PubMed] [Google Scholar]
- 35.Stankiewicz P., Parka S.S., Holder S.E., Waters C.S., Palmer R.W., Berend S.A., Shaffer L.G., Potocki L., Lupski J.R. Trisomy 17p10-p12 resulting from a supernumerary marker chromosome derived from chromosome 17: molecular analysis and delineation of the phenotype. Clin. Genet. 2001;60:336–344. doi: 10.1034/j.1399-0004.2001.600503.x. doi:10.1034/j.1399-0004.2001.600503.x. [DOI] [PubMed] [Google Scholar]
- 36.Prakash S.K., LeMaire S.A., Guo D.C., Russell L., Regalado E.S., Golabbakhsh H., Johnson R.J., Safi H.J., Estrera A.L., Coselli J.S., et al. Rare copy number variants disrupt genes regulating vascular smooth muscle cell adhesion and contractility in sporadic thoracic aortic aneurysms and dissections. Am. J. Hum. Genet. 2010;87:743–756. doi: 10.1016/j.ajhg.2010.09.015. doi:10.1016/j.ajhg.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.