Abstract
Mendelian susceptibility to mycobacterial diseases (MSMD) is a rare syndrome, the known genetic etiologies of which impair the production of, or the response to interferon-gamma (IFN-γ). We report here a patient (P1) with MSMD whose cells display mildly impaired responses to IFN-γ, at levels, however, similar to those from MSMD patients with autosomal recessive (AR) partial IFN-γR2 or STAT1 deficiency. Whole-exome sequencing (WES) and Sanger sequencing revealed only one candidate variation for both MSMD-causing and IFN-γ-related genes. P1 carried a heterozygous frame-shift IFNGR2 mutation inherited from her father. We show that the mutant allele is intrinsically loss-of-function and not dominant-negative, suggesting haploinsufficiency at the IFNGR2 locus. We also show that Epstein-Barr virus transformed B lymphocyte cells from 10 heterozygous relatives of patients with AR complete IFN-γR2 deficiency respond poorly to IFN-γ, in some cases as poorly as the cells of P1. Naive CD4+ T cells and memory IL-4-producing T cells from these individuals also responded poorly to IFN-γ, whereas monocytes and monocyte-derived macrophages (MDMs) did not. This is consistent with the lower levels of expression of IFN-γR2 in lymphoid than in myeloid cells. Overall, MSMD in this patient is probably due to autosomal dominant (AD) IFN-γR2 deficiency, resulting from haploinsufficiency, at least in lymphoid cells. The clinical penetrance of AD IFN-γR2 deficiency is incomplete, possibly due, at least partly, to the variability of cellular responses to IFN-γ in these individuals.
INTRODUCTION
Mendelian susceptibility to mycobacterial diseases (MSMD, MIM209950) is a rare condition characterized by the occurrence of clinical disease caused by poorly virulent mycobacteria, such as BCG vaccines or environmental mycobacteria, in otherwise healthy children (1–4). The patients are also vulnerable to the more virulent Mycobacterium tuberculosis (5,6). Non-typhoidal Salmonella infections are also seen in MSMD patients, but infections due to other intra-macrophagic pathogens are less common (3,7). Mendelian mutations in six autosomal (IFNGR1, IFNGR2, STAT1, IRF8, IL12B and IL12RB1) and two X-linked (CYBB and NEMO) genes have been discovered which account for about half of the patients with MSMD (7–9). They collectively define up to 15 different genetic etiologies, based on the mode of inheritance (dominant or recessive), functional impairment (complete or partial defect) and biochemical features (depending on both the mutation and the protein domain affected). We also recently identified, by genome-wide linkage analysis combined with whole-exome sequencing (WES), patients with MSMD and AR ISG15 deficiency, thereby revealing an unexpected key role of ISG15 as an interferon-gamma (IFN-γ)-inducing cytokine (10). All these defects impair the production of or the response to IFN-γ directly or indirectly, indicating that IFN-γ is essential for the confinement of most mycobacterial species in natura (11).
The inherited defects of IL12RB1 (12,13), IL12B (14) and ISG15 (10) in patients with MSMD are autosomal recessive (AR) traits characterized by an impairment of IFN-γ production by T or natural killer (NK) cells. The encoded proteins of all three genes are indispensable for an adequate IL-12- (IL12B, IL12RB1) or ISG15-dependent (ISG15) induction of IFN-γ. An X-linked NEMO defect can lead to weak IL-12 induction, with indirect effects on IFN-γ production (15). There are 11 types of disorders in which responses to IFN-γ are impaired, including four different IFN-γR1 defects (16–20), leading to clinical MSMD of different severities. Dominant partial STAT1 defects result in the impairment of the IFN-γ response through various mechanisms, including phosphorylation and DNA binding (21,22). Autosomal dominant (AD) IRF-8 defects probably confer susceptibility to MSMD through low CD11c+CD1c+ dendritic cell count and an IFN-γ-induced pathway (9). MSMD-CYBB defects are also linked to the IFN-γ pathway as CYBB is an IFN-γ-inducible component of the oxidative burst (8). IFN-γR2 plays an important role in mediating the IFN-γ response and both recessive complete and partial IFN-γR2 defects have been reported to cause MSMD (23–27).
Inherited IFN-γR2 deficiency was first identified in 1998 in a child with MSMD (24). Eleven patients from nine kindreds and eight IFNGR2 mutations have now been reported (Fig. 1A), with two forms of IFN-γR2 deficiency, defined as partial and complete (23–28). Nine patients from seven kindreds display complete IFN-γR2 deficiency with abolished cellular responses to IFN-γ, in terms of STAT1 phosphorylation, gamma-activated factor (GAF) activation and the induction of GAF-dependent target genes (24–28). Two forms of complete IFN-γR2 deficiency have been reported on the basis of protein expression at the cell surface. In some patients, a premature stop codon in the region encoding the extracellular domain results in a lack of receptor expression at the cell surface (24). In other patients, the defect is characterized by the detectable protein expression of non-functional receptors at the cell surface (26,27). Finally, two patients have been shown to display partial IFN-γR2 deficiency, with impaired but not abolished responses to IFN-γ (23, Moncada-Vélez, M. et al., submitted). All these patients display AR IFN-γR2 deficiency. However, an IFNGR2 null allele has been shown to exert negative dominance at the cellular level in a healthy individual (25), raising the possibility that there may be symptomatic patients with AD IFN-γR2 deficiency. We report here the first MSMD patient with AD IFN-γR2 deficiency. The underlying mechanism involves haploinsufficiency at the IFNGR2 locus, as all heterozygous relatives of patients with complete IFN-γR2 deficiency respond poorly to IFN-γ. z
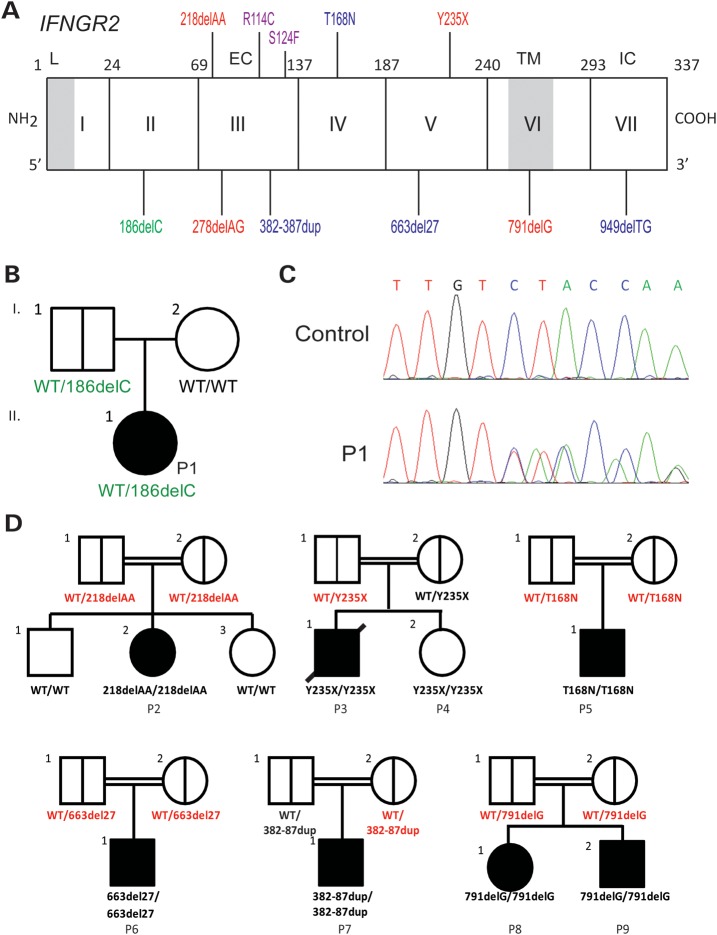
Figure 1.
Pedigree and mutations of IFNGR2. (A) Schematic presentation of the mutations identified in the IFNGR2 gene. The leader sequence (L, 1–22), extracellular domain (EC, 23–248), transmembrane domain (TM, 249–272) and intracellular domain (IC, 273–337) are indicated. Mutations marked in red cause complete AR IFN-γR2 deficiency with no detectable expression of IFN-γR2 at the cell surface. The mutations marked in blue cause complete AR IFN-γR2 deficiency with detectable surface expression of a non-functional IFN-γR2. With the antibody now available, the cells of patients carrying the 382–387dup and 663del27 mutations (26) were shown to have impaired, but detectable IFN-γR2 expression at the surface of Epstein-Barr virus transformed B lymphocyte (EBV-B) cells. The mutation marked in purple cause partial AR IFN-γR2 deficiency. The mutation marked in green is the heterozygous mutation reported here. (B) Family pedigree of P1. (C) Electropherogram showing the heterozygous IFNGR2 186delC mutation found in P1. (D) Pedigree of the families with IFN-γR2 deficiency recruited for this study. The IFNGR2 genotype of the heterozygous individuals included is shown in red.
RESULTS
Impaired response to IFN-γ in EBV-B cells from a patient with MSMD
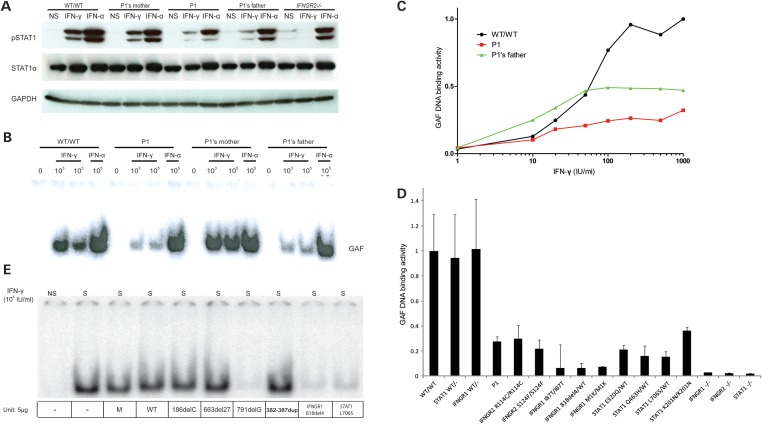
We investigated a 15-year-old Polish girl (P1) with MSMD by first studying the response of her Epstein-Barr virus transformed B lymphocyte (EBV-B) cells to IFN-γ. Upon stimulation with IFN-γ, the patient's EBV-B cells displayed weaker STAT1 phosphorylation than a healthy control as measured by western blotting with a specific phosphotyrosine-701 STAT1 antibody (Fig. 2A). GAF DNA-binding activity was also impaired, as shown by electrophoretic mobility shift assay (EMSA) (Fig. 2B and Supplementary Material, Fig. S1), but this activity was not entirely abolished. In contrast, the patient's cells responded normally to IFN-α (as shown by western blotting and EMSA). Dose–response experiments further showed that the defect was more pronounced at higher than at lower concentrations of IFN-γ (Fig. 2C). We then compared IFN-γ-induced GAF DNA-binding activities in P1 with those in other reported human partial and complete defects in the IFN-γ pathway, due to mono- or bi-allelic mutations in IFNGR1 (16–20), IFNGR2 (24–27, Moncada-Vélez, M. et al., submitted) and STAT1 (21,22,29,30). P1's IFN-γ response was similar to that for cells from patients with partial recessive IFN-γR2 deficiency (23) and partial recessive STAT1 deficiency (particularly, the K201N/K201N mutant) (30) (Fig. 2D and Supplementary Material, Fig. S2). Collectively, these results suggest that the impaired GAF-DNA-binding activity observed in EBV-B cells stimulated by IFN-γ is responsible for MSMD in P1.
Figure 2.
P1's IFN-γ response and exploration of a possible dominant-negative effect. EBV-B cells from a healthy control, P1, P1's father and mother were stimulated with IFN-γ or IFN-α and subjected to western blot analysis for STAT1, Tyr701 phosphorylated STAT1 (A) or to EMSA for the assessment of GAF DNA-binding activities (B). (C) The GAF DNA-binding activities of EBV-B cells stimulated with various concentrations of IFN-γ were compared between a healthy control, P1 and P1's father. (D) GAF DNA-binding activities in the EBV-B cells were compared between a healthy control, individuals heterozygous for IFNGR1 or STAT1, P1 and patients with various IFNGR1, IFNGR2 and STAT1 mutations resulting in partial or complete deficiency, after stimulation with IFN-γ. The genotype of each cell line is written for the corresponding gene. (E) SV40-transformed fibroblasts from a healthy control were transfected with various plasmids encoding the WT or a mutant form of IFN-γR2. The various mutations included 186delC, 663del27 and 791delG, 382–87dup. Cells were also transfected with an IFNGR1 818del4 or STAT1 L706S expression plasmid as a positive control, for the detection of dominant-negative effects on IFN-γ signaling.
A heterozygous IFNGR2 mutation in P1
We, therefore, sequenced the exons and flanking intron regions of IFNGR1, IFNGR2 and STAT1 by Sanger sequencing. We identified a heterozygous 186delC mutation in IFNGR2 exon 2 (Fig. 1C). No other mutation was found in the three genes. We nevertheless subjected the genomic DNA of P1 and P1's father to WES (Supplementary Material, Table S1), but did not identify any other new variation particularly in the 15 genes known to be involved in the IFN-γ signaling pathway or the nine genes known to cause MSMD (Supplementary Material, Tables S2 and 3). The patient was heterozygous for IFNGR2, having a mutant allele and a WT allele. The mutant allele was not found, by sequencing, in 200 controls from Poland and was also absent from public databases (dbSNP, 1000genome) and our own in-house database of 700 exomes. The mutation is predicted to generate a premature stop codon at position 82, upstream from the segment encoding the transmembrane domain. The patient's father carried the same mutation in IFNGR2, whereas her mother had two wild-type (WT) alleles (Fig. 1B). Since causal genetic lesions have been identified as far as 500 kb upstream or downstream from other morbid genes (31), we assessed the expression of the two IFNGR2 alleles by RT-PCR. Both the alleles appeared to be spliced and expressed with a similar efficiency in P1 and her father (data not shown), arguing against compound heterozygosity for two morbid alleles at the IFNGR2 locus in P1. However, these data from EBV-B cells do not rule out the remote but finite possibility of a second, subtle regulatory mutation affecting IFNGR2 expression in vivo in relevant primary cells, such as T cells and macrophages, which express IFN-γR, and are critical for immunity against mycobacteria.
The patient displays AD IFN-γR2 deficiency but the IFNGR2 allele is not dominant-negative
We tested the hypothesis of AD IFN-γR2 deficiency in this patient further, by assessing the response to IFN-γ of EBV-B cells from P1 in more depth and comparing this response with that of her parents. EBV-B cells from P1 and her father displayed similar low levels of GAF DNA-binding activity and STAT1 Tyr701 phosphorylation upon IFN-γ stimulation, whereas such activity was normal in EBV-B cells from the patient's mother (Fig. 2A and B). In contrast, the IFN-α response was similar in all three cell lines, strongly suggesting that the heterozygous IFNGR2 mutation is exclusively responsible for the inherited cellular phenotype of impaired response to IFN-γ in P1 and her father. We then investigated whether this heterozygous IFNGR2 allele caused AD IFN-γR2 deficiency by negative dominance or haploinsufficiency. We first tested the hypothesis of a dominant-negative effect accounting for the cellular phenotype in cells heterozygous for the 186delC IFNGR2 allele, consistent with a previous report showing that the 791delG mutation was dominant-negative at the cellular level in vitro (the two heterozygous individuals were clinically healthy) (25). SV40-transformed fibroblasts from a healthy control were transfected with either a WT or a mutant IFNGR2 allele (the mutants tested were 186delC, 663del27 (27), 791delG (25) and 382–387dup (26)), and two previously reported dominant-negative alleles of IFNGR1 818del4 (19) and STAT1 L706S (22). They were then stimulated with IFN-γ. Consistent with previous reports, a dominant-negative effect was observed in fibroblasts transfected with IFNGR1 818del4, IFNGR2 791delG and STAT1 L706S alleles; however, we observed no dominant-negative effect on fibroblasts transfected with the other IFNGR2 mutant alleles, including 186delC (this report), 663del27 (27) and 382–387dup (26) (Fig. 2E). Thus, the heterozygous IFNGR2 186delC allele in P1 and her father is responsible for the impaired response to IFN-γ, as suggested by the strict familial cosegregation of the IFNGR2 genotype and IFN-γ responsiveness. However, this impairment is not caused by a dominant-negative effect of the mutant allele. Instead, it probably results from haploinsufficiency.
Levels of IFN-γR2 expression at the cell surface are low in EBV-B cells
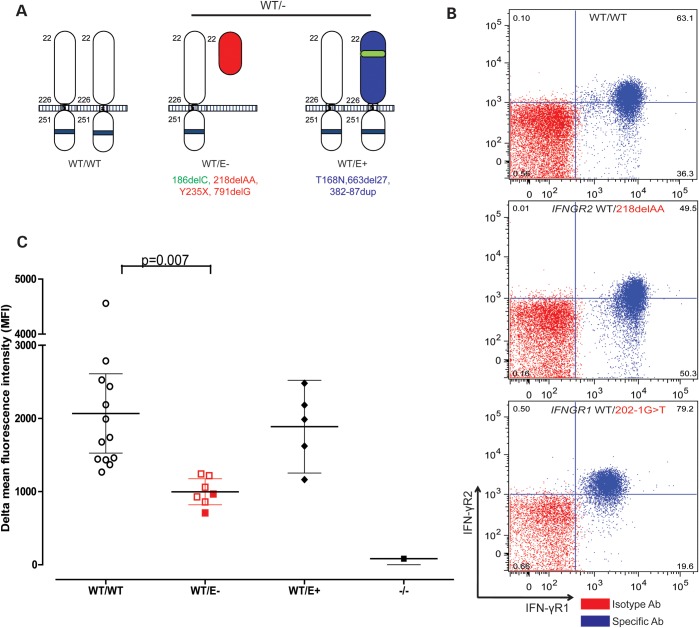
We tested the hypothesis that the IFNGR2 mutation is acting by haploinsufficiency, by expanding our studies to include the heterozygous relatives of six patients with complete IFN-γR2 deficiency (24–27), with the heterozygous relatives of patients with complete IFN-γR1 deficiency serving as controls (16,33). We first investigated the cell-surface expression of IFN-γR2 in the EBV-B cells heterozygous for a loss-of-function IFNGR2 allele, referred to below as IFNGR2 WT/−. We designated the heterozygous individuals WT/E+ or WT/E−, on the basis of the intrinsic expression (E+) or lack of expression (E−) of the mutant IFN-γR2 protein at the cell surface. Different types of mutations have different consequences for the expression of the encoded receptor at the cell surface. Some mutations prevent the cell-surface expression of the mutated receptor (186delC, 218delAA, Y235X and 791delG, Fig. 3A) (25), whereas others allow the expression of a non-functional receptor at the cell surface (T168N, 663del27 and 382–87dup, Fig. 3A) (26,27). We showed, by flow cytometry, that EBV-B cells from WT/E− individuals had significantly lower levels of IFN-γR2 expression at the cell surface than EBV-B cells from WT/WT controls (P = 0.007, Fig. 3C). In contrast, no significant difference was observed between the WT/WT and WT/E+ groups. Moreover, the levels of IFN-γR1 expression were similar between heterozygous IFNGR1 WT/− and heterozygous IFNGR2 WT/− EBV-B cells, in which they were normal (data not shown). We analyzed 12 cell lines carrying seven different IFNGR2 mutations and obtained results that were both reproducible and consistent with no outliers (Fig. 3C). The levels of IFN-γR2 expression were consistently lower in the WT/E− EBV-B cells, as shown by the mean, median, delta mean or delta median fluorescence intensity of allophycocyanin (APC)-labeled IFN-γR2 antibody. These data indicate that heterozygosity for null IFNGR2 alleles is associated with lower than normal levels of expression of the encoded receptor on the surface of EBV-B cells. These data further suggest that there may be haploinsufficiency at the IFNGR2 locus, in terms of IFN-γ responsiveness, for heterozygous cells other than those of P1 and her father.
Figure 3.
IFN-γR2 expression in heterozygous IFNGR2 cells. (A) Schematic diagram of IFN-γR2 structures. In WT/WT healthy controls, the protein is encoded by two WT alleles. WT/E− individuals have a heterozygous loss-of-function mutation preventing the expression of the receptor on the cell surface. WT/E+ individuals have a loss-of-function mutation changing the structure of IFN-γR2, but with protein detectable on the cell surface. (B) Monocytes from a healthy control, an individual carrying a heterozygous IFNGR2 218delAA mutation and an individual carrying a heterozygous IFNGR1 202–1G>T mutation were incubated with an antibody recognizing the extracellular domain of IFN-γR1 or IFN-γR2. The red plot corresponds to the isotype control. The blue plot corresponds to the specific antibody for IFN-γR1 and IFN-γR2. (C) Delta mean fluorescence intensity (ΔMFI) of IFN-γR2 in EBV-B cells was compared between healthy controls, WT/E−, WT/E+ and 218delAA/218delAA patients. The solid red circles indicate the ΔMFI of the parent with a heterozygous 791delG mutation. Student's t-test was used to assess the significance of differences between two groups. Each experiment was carried out at least three times.
Dominance due to IFNGR2 haploinsufficiency in EBV-B cells
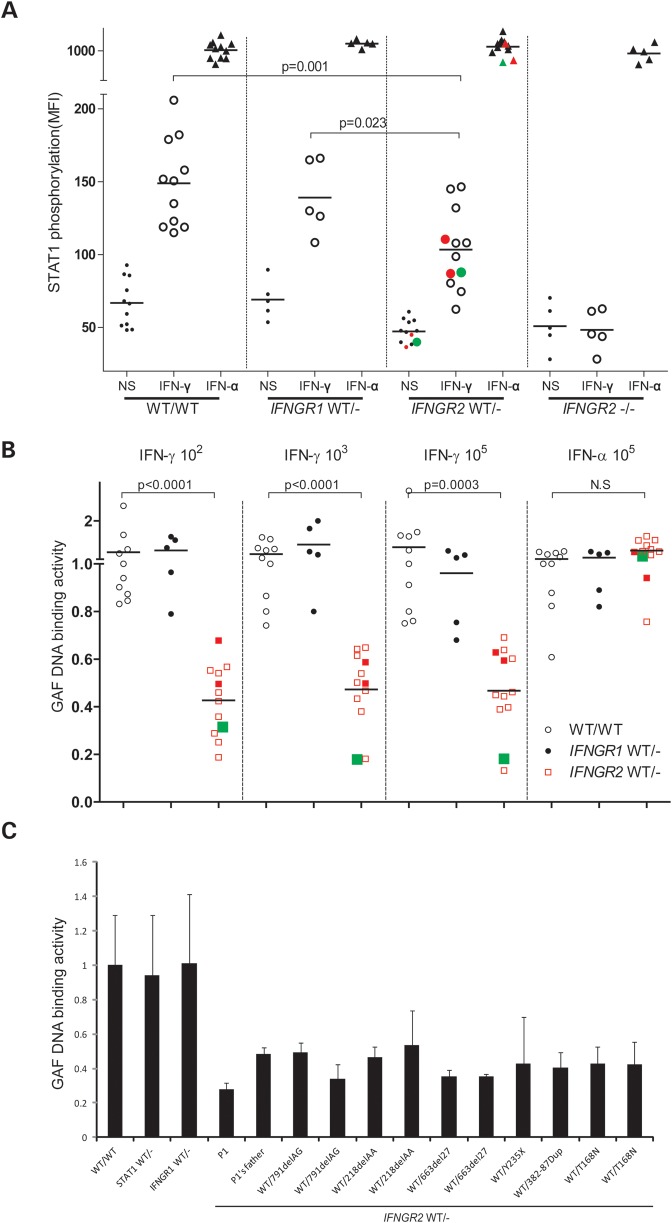
We tested the hypothesis of haploinsufficiency at the IFNGR2 locus, by evaluating Tyr701 STAT1 phosphorylation by flow cytometry in EBV-B cells from 12 individuals heterozygous for null IFNGR2 alleles (186delC, 218delAA, Y235X, 791delG, T168N, 663del27 and 382–87dup) (25–27). STAT1 Tyr701 phosphorylation levels were significantly lower in all heterozygous IFNGR2 individuals (WT/−) than in WT individuals, after stimulation with IFN-γ, but not IFN-α (Fig. 4A). We also found that GAF DNA-binding activity levels were significantly lower in heterozygous cells than in cells from healthy controls (Fig. 4B). The mean GAF activity in heterozygotes was ∼40% that of controls. In contrast, EBV-B cells with a heterozygous loss-of-function mutation in either STAT1 or IFNGR1 had normal responses, suggesting a gene-specific gene dosage effect for IFNGR2, but not for IFNGR1 and STAT1 (21,30) (Fig. 4A and B, data not shown). Consistent with haploinsufficiency, we found that EBV-B cells from P1 displayed a more pronounced defect at high concentrations of IFN-γ, requiring the presence of larger numbers of functional receptors (Fig. 2C). We also compared the responses of heterozygous mutant cells. There were some inter-individual and inter-experimental differences, but we consistently found that all cell lines heterozygous for a null IFNGR2 allele had an impaired response to IFN-γ (Fig. 4C and Supplementary Material, Fig. S2), and that P1's response was in the lower range for this group. Finally, as expected, the GAF DNA-binding activity in response to IFN-α was normal in IFNGR2 heterozygotes, suggesting that the impairment of GAF DNA-binding activity is specific to the IFN-γ pathway. Overall, these results strongly suggest that haploinsufficiency at the IFNGR2 locus in EBV-B cells constitutes a general mechanism for impaired IFN-γ signaling in the cells heterozygous for a null allele and is not specific to P1 and her father.
Figure 4.
Haploinsufficiency in EBV-B cells from heterozygous IFNGR2 individuals. (A) Tyr701 STAT1 phosphorylation was quantified by FACS in EBV-B cells after stimulation with 105 IU/ml IFN-γ or IFN-α for 30 min. These experiments were carried out twice. (B) GAF DNA-binding activities in EBV-B cells were compared between healthy controls, heterozygous IFNGR1 individuals and heterozygous IFNGR2 individuals following stimulation with various concentrations of IFN-γ or IFN-α, as indicated. The dot indicates the relative value of GAF DNA-binding activity for a given cell with respect to the same healthy control. The solid red circles and squares indicate the values for the parent with a heterozygous 791delG mutation. The solid green circles and squares indicate the values for P1. (C) P1's GAF DNA-binding activity was compared with that of the other individuals heterozygous for IFNGR2.
Impaired ISGs induction due to IFNGR2 haploinsufficiency
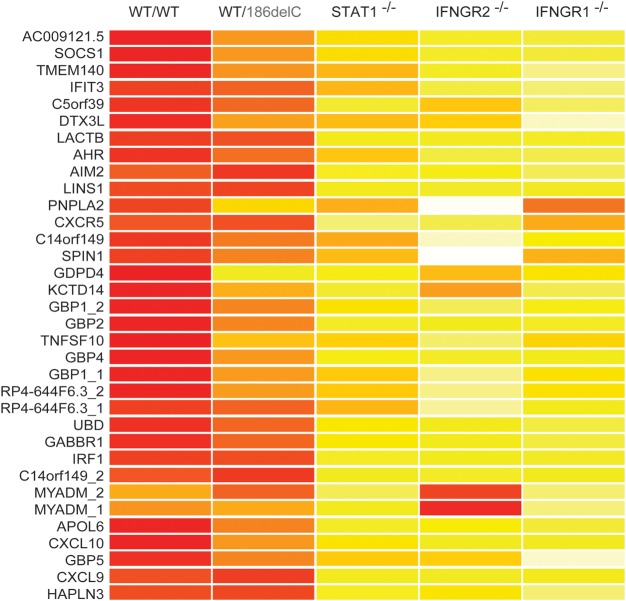
We defined the molecular consequences of AD IFN-γR2 deficiency, more precisely, by carrying out a whole-transcriptome microarray analysis of EBV-B cells from three healthy controls, three patients with AR complete IFN-γR2, IFN-γR1 or STAT1 deficiency and one individual heterozygous for an IFNGR2 mutation (P1). These cells were either being left unstimulated or stimulated with IFN-γ for 2 h. We detected no difference between the EBV-B cells of any of these groups of individuals in the absence of stimulation. However, we defined a set of IFN-stimulated genes (ISGs), induced or inhibited by IFN-γ, in the healthy controls. These genes were defined as displaying an at least 2-fold change in expression in response to stimulation. We identified 31 such genes, which were highly induced in EBV-B cells from healthy controls. The cells with complete IFN-γR1, IFN-γR2 or STAT1 deficiency showed no upregulation of these genes in response to stimulation with IFN-γ. Interestingly, the level of induction of most of these genes was intermediate in cells heterozygous for IFNGR2 alleles. We observed only weak induction of various target genes, including CXCL10, GBP1 and SOCS1 (Fig. 5). Collectively, these data clearly indicate that haploinsufficiency at the IFNGR2 locus is of physiological relevance, because of its broad impact on the transcription of IFN-γ-inducible genes.
Figure 5.
Microarray analysis of ISGs in EBV-B cells. EBV-B cells from three healthy controls, P1 and patients with complete STAT1, IFN-γR1 and IFN-γR2 deficiencies were stimulated by incubation with 104 IU/ml IFN-γ for 2 h. Red colors represent high stimulated/basal expression fold induction level. Whole-transcriptome analysis was then carried out on mRNA extracted from EBV-B cells.
IFN-γR2 expression in fresh leukocyte subsets
We then investigated whether the haploinsufficiency observed in EBV-B cells may also occur in more relevant cells: T cells and macrophages, both of which normally express IFN-γR2 and are critical for immunity to mycobacteria. We assessed the IFN-γR2 expression in fresh leukocyte subsets ex vivo, including CD3+ T cells, CD14+ monocytes, CD19+ B cells and CD56+ NK cells. IFN-γR2 surface expression was strongest on control monocytes, intermediate in B cells and weakest in T and NK cells. In contrast, IFN-γR1 was expressed to similar levels on the surfaces of all four cell types tested (Supplementary Material, Fig. S3). These results are consistent with previous reports (33–35), suggesting a cell-specific pattern of IFN-γR2 expression governing IFN-γ responsiveness. Levels of IFN-γR2 expression at the cell surface were lower in monocytes from heterozygous WT/E− IFNGR2 individuals, but IFN- γR1 expression was normal in these cells (Fig. 3B). IFN-γR1 expression levels were also low in heterozygous IFNGR1 monocytes, which had normal levels of IFN-γR2 expression (Fig. 3B). IFN-γR2 expression levels in both controls and WT/E− individuals were lower if the collection of blood samples were delayed. However, our results showed that IFN-γR2 surface expression was consistently weaker on the monocytes from all WT/E− individuals tested than in WT/WT controls (Supplementary Material, Fig. S4).
IFNGR2 haploinsufficiency in fresh leukocyte subsets
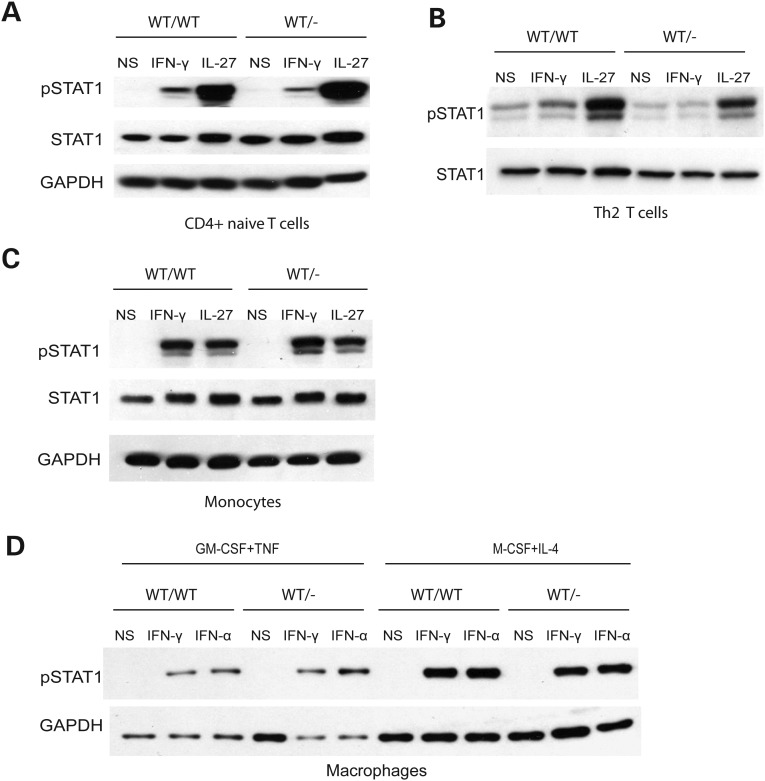
Activated pSTAT1 levels in control cells were the highest in monocytes, intermediate in B cells and lowest (barely detectable) in T and NK cells following IFN-γ stimulation (Supplementary Material, Fig. S5), consistent with previous reports (34,35). We then investigated the occurrence of haploinsufficiency in T cells and monocytes from IFNGR2 heterozygous individuals. We studied naive CD4+ T cells and differentiated IFN-γ-producing (‘Th1’) and IL-4-producing (‘Th2’) CD4+ T cells. Naive T cells and IL-4-producing T cells responded to IFN-γ in terms of STAT1 phosphorylation, whereas IFN-γ-producing T cells did not (data not shown), consistent with previous reports that IFN-γ-producing T cells lack IFN-γR2 expression and are unresponsive to IFN-γ (36–39). The levels of pSTAT1 in IFNGR2 heterozygous naive and IL-4-producing CD4+ T cells were lower than those in healthy controls (Fig. 6A and B). We then tested monocytes and monocyte-derived macrophages (MDMs) by stimulation with macrophage colony-stimulating factor + interleukins-4 (M-CSF + IL-4) or granulocyte colony-stimulating factor + tumor necrosis factor-α (GM-CSF + TNF-α). Control monocytes generated similar amounts of pSTAT1 in response to stimulation with IFN-α and IFN-γ. Control MDMs induced to differentiate with M-CSF + IL-4 responded more strongly to IFN-γ and produced larger amounts of IFN-γR2 than macrophages induced to differentiate with GM-CSF + TNF-α (data not shown). Following stimulation with IFN-γ, pSTAT1 levels in monocytes and MDMs were no lower in IFNGR2 heterozygotes than in controls (Fig. 6C and D). Thus, IFNGR2 haploinsufficiency seems to be restricted to EBV-B cell lines and freshly analyzed T cells and does not affect monocytes and MDMs, at least as assessed by STAT1 phosphorylation in response to stimulation with IFN-γ. Unsurprisingly, IFNGR2 haploinsufficiency seems to affect principally the cell types that normally express low levels of IFN-γR2 at their surface.
Figure 6.
Haploinsufficiency in the primary immune cells from heterozygous IFNGR2 individuals. (A) Naive CD4+ T cells, (B) IL-4-producing T helper cells, (C) monocytes (D) MDMs generated in the presence of either M-CSF + IL-4 or GM-CSF + TNF-α were stimulated by incubation with IFN-γ, IFN-α or IL-27 for 15 min. Western blots were carried out to evaluate STAT1 phosphorylation. These experiments were carried out at least twice.
DISCUSSION
This report identifies AD IFN-γR2 deficiency as a new genetic etiology of MSMD. We observed haploinsufficiency at the human IFNGR2 locus, at least in some lymphoid cells (both EBV-B cell lines and primary T cells), and in all heterozygous individuals tested. Under the conditions tested, this finding is specific to IFNGR2, as there was no detectable haploinsufficiency for the IFNGR1 and STAT1 loci. Haploinsufficiency and dominant negativity are the two independent, mutually exclusive mechanisms that can account for dominant phenotypes in diploid cells heterozygous for a mutant allele. Haploinsufficiency at the IFNGR2 locus can be explained biochemically, because the assembly and expression of the membrane-bound IFN-γ receptor requires IFN-γR2 expression which is a limiting factor. Unlike IFN-γR1, which is ubiquitously and uniformly expressed, IFN-γR2 is expressed under tight control and serves as a key regulator of IFN-γ responsiveness and signaling in the mouse model and in human subjects (36,39,40). IFN-γR2 had been described as a tuning factor preventing the antiproliferative effect of IFN-γ in subsets of T helper cells (36). Recent biochemical studies of IFN-α receptors have also shown that different levels of cell-surface receptors are required for different biological functions (41). For IFN-α, small numbers of receptor binding sites are sufficient to mediate antiviral responses, but not to mediate antiproliferative effects (41,42). Consistent with this, our results show that the cellular penetrance of IFNGR2 haploinsufficiency depends on the level of receptor expression at the cell surface. Cellular penetrance is complete in lymphocytes, including T cells, which normally express small numbers of IFN-γR2. Conversely, heterozygosity for IFNGR2 null alleles does not seem to affect the response of cells that normally express larger numbers of these receptors, such as phagocytes. This finding suggests that T cells play a role in the pathogenesis of MSMD in patients with various mutations affecting the IFN-γ response pathway, including patients with AD IFN-γR2 deficiency, such as P1.
Clinically, only one of the sixteen known IFNGR2 heterozygous individuals tested displayed MSMD. The clinical penetrance of IFNGR2 haploinsufficiency is therefore low, at least for the clinical phenotype of MSMD. This is consistent with our previous demonstration of a correlation between the clinical severity of MSMD and the level of responsiveness to IFN-γ, as the cellular phenotype seen in IFNGR2 heterozygotes is milder than that of most other defects in the IFN-γ response pathway (11,16–20,22,25–27). It is similar to that of AR partial IFN-γR2 (R114C/R114C) or STAT1 deficiency (K201N/K201N). Moreover, the cells of P1 consistently presented one of the weakest responses of any of the cells heterozygous for IFNGR2 null alleles tested. The reasons for this remain unclear, as WES identified no other potential variant from the IFN-γ signaling pathway in P1. Moreover, our biochemical investigation of known factors in the IFN-γ signaling pathway revealed no abnormal expression (data not shown). Incomplete clinical penetrance has already been observed in the IFN-γ signaling pathway, particularly for AD STAT1 deficiency (for which the clinical penetrance of MSMD is low) (21,22,43,44). In addition, independently of the IFN-γ signaling pathway, other mutations may contribute to MSMD in P1. For example, we found a heterozygous ISG15 rare variant in P1 but not in her father. We recently showed that at least four individuals heterozygous for ISG15 loss-of-function alleles have no obvious clinical phenotype in terms of MSMD (10). However, heterozygous ISG15 mutation may play as a modifier in the context of another genetic lesion, such as AD IFN-γR2 deficiency, by reducing the amount of IFN-γ production. In any case, as previously suggested, MSMD is more likely to occur in individuals carrying dominant-negative IFNGR2 alleles, whose cellular response to IFN-γ is impaired (25). It is now tempting to speculate that IFNGR2 haploinsufficiency may also, and perhaps more commonly, underlie clinical tuberculosis in children living in areas of endemic disease, including those who did not develop clinical disease upon BCG vaccination (5,6,45,46).
MATERIALS AND METHODS
Ethics statement
All members of the families agreed to participate in this study, which was approved by the ethics committees of the hospitals concerned.
Case report
P1 is a Polish girl born to non-consanguineous parents in 1990. She was vaccinated by BCG injection into the left deltoid at birth and presented with enlarged and purulent homolateral axillary lymph nodes at the age of 2 months. She had no fever, no enlargement of the liver or spleen and no obvious abnormality on chest X-ray. Complete blood count and detailed T, B and NK immunophenotyping results were normal. P1 was then treated with lymphadectomy and local streptomycin. Histological examination of the excised lymph nodes showed cellular infiltration and granulomatosa mimicking tuberculosis lymphadenitis (type II granuloma). Acid-fast staining revealed the presence of positively stained bacilli. Cultures of pus from the axillary region yielded no pathogens. The infection resolved over a period of 4 months, with no oral antimycobacterial therapy. With the exception of this BCG disease, P1 had developed no other severe infections by her last follow-up visit at the age of 15 years (Fig. 1B). She had had mild respiratory tract infections about twice yearly and developed mumps at the age of 5 years and chicken pox at the age of 7 years, both of which ran a mild clinical course. Other vaccinations, including those with combined diphtheria, tetanus and poliovirus, attenuated rubella and mumps vaccine, hepatitis B virus (HBV) and flu, were uneventful. P1 was found to have protective antibodies against HBV. No specific family history for related MSMD was found. P1's maternal grandmother had hay fever. The parents of P1 remained healthy, although her father had heart arrhythmia.
We also retrospectively recruited 10 heterozygous relatives of previously reported and new MSMD patients homozygous for null IFNGR2 mutations (Fig. 1D; a detailed clinical report will be published elsewhere). One new patient (P2) with complete IFN-γR2 deficiency is homozygous for the 218delAA mutation (Fig. 1D). She is 6 years old, lives in France and was born to a consanguineous family from Pakistan. She suffered from BCG disease at 11 months of age and had a high serum IFN-γ (53 pg/ml) level. Two patients (P3 and P4) were born to a consanguineous family from Saudi Arabia. P3, a boy, suffered from severe BCG-osis shortly after birth and died. His sister (P4) was diagnosed with a recessive IFNGR2 Y235X mutation after birth. She was not vaccinated with BCG and has remained healthy. None of the 10 heterozygous relatives had ever displayed any detectable mycobacterial phenotype remotely reminiscent of MSMD, not even self-healing BCG-itis. Five of ten relatives had never been vaccinated with BCG and never had MSMD.
Cell culture and stimulation, DNA extraction, PCR, microarray transcriptome analysis and sequencing
Genomic DNA was extracted from fresh blood cells, as previously described (20). Primers and PCR conditions are available upon request. Sequencing was carried out on an ABI 3130× (Applied Biosystems) sequencer. Peripheral blood mononuclear cells (PBMCs) were isolated with Ficoll as previously described (8). EBV-transformed B lymphocytes (EBV-B cells) were cultured as previously described (47). EBV-B cells, monocytes, MDMs, naive T cells and Th2 cells were stimulated with the indicated doses of IFN-γ (Imukin, Boehringer Ingelheim), IFN-α 2b (IntronA, Schering Plough) and IL-27 (R&D). Electroporation and Lipofectamin methods were used to transfect EBV-B cells and fibroblasts, respectively, with a WT construct, with or without mutant IFNGR2 plasmids (pcDNA3.1-IFNGR2), as previously reported (27). We incubated five million EBV-B cells with or without 104IU/ml IFN-γ for 2 h and then subjected them to microarray analysis as previously described (48). The procedure of WES and analysis were carried out as previously described (49,50). Briefly, variant calls were made with GATK UnifiedGenotyper. All calls with a read coverage ≤10× and a Phred-scaled SNP quality of ≤30 were removed from consideration.
Electrophoretic mobility shift assay (EMSA) and western blotting
EMSA was carried out as previously described (17–19). Briefly, cells were stimulated by incubation for 20 min with IFN-γ or IFN-α, at the indicated doses. We incubated 10 µg of nuclear extract with a 32P-labeled (α-dATP) GAS (from the FCGR1 promoter) probe and subjected the mixture to electrophoresis in a polyacrylamide gel. The GAF DNA-binding activity was assessed with ImageQuant (Amersham Bioscience). We used antibodies recognizing STAT1 and tyrosine 701-phosphorylated STAT1 for western blot analysis, as previously described (30).
Flow cytometry analysis
We used fluorescence-activated cell sorting (FACS) to assess IFN-γR2 expression on the surface of EBV-B cells, PBMCs and monocytes. Briefly, 0.5 to 1 million PBMCs or EBV-B cells were first blocked by incubation with 10 μl of FcR blocking reagent in 1% SVF + 1× PBS for 10 min. We then added 125 ng of anti-IFN-γR2-APC (FAB773A, R&D systems) and incubated the cells for 30 min on ice before washing them twice with 1% SVF + 1× PBS. Goat IgG control APC (IC108A, R&D systems) served as the isotype control. Tyr-701-phosphorylated STAT1 levels were assessed as previously described. Antibodies directed against CD3 (560366, BD), CD14 (557742, BD), CD19 (555413, BD) and CD56 (557699, BD) conjugated with different fluorochromes were used to distinguish between the various cell populations in PBMCs, and dead cells were excluded by Aqua Live/Dead staining (Invitrogen). Tyr-701-phosphorylated STAT1 levels were assessed by activating cells by incubation with IFN-γ or IFN-α for 30 min and incubating with an antibody specific for Tyr 701-phosphorylated STAT1. Flurokon signals were measured with a BD LSR II and analyzed with Flowjo (Tree Star, Inc.).
Differentiation of macrophage and CD4+ T cells and statistics
We isolated CD14+ monocytes from PBMCs using a positive selection kit from Miltenyi Biotec (8). Briefly, 0.5 million monocytes were incubated in 24-well plates with 0.5 ng/ml GM-CSF (215-MC-050, R&D systems) and 0.5 ng/ml TNF-α (210-TA-050), or with 50 ng/ml M-CSF (216-MC-025, R&D systems) and 50 ng/ml IL-4 (204-IL-050, R&D systems), to induce macrophage differentiation, as previously described (8,51). CD4+ naive T cells were obtained using Miltenyi MACS naive CD4+ T cell isolation kit II (No.130-094-131). In total, 0.5 million naive T cells in 1 ml RPMI-1640 supplemented with 10% human plasma were allowed to differentiate in each of the wells of 24-well plates, as previously described (52), with 5 μg/ml anti-CD3 antibody, 750 ng/ml anti-CD28 antibody and 20 IU/ml rIL-2, with either 1 ng/ml IL-12 or 50 ng/ml IL-4, for Th1 and Th2 cell differentiation, respectively. The cells were stimulated with IFN-γ or IL-27 on the fourth day after differentiation. ANOVA was used to assess the difference between multiple groups, and Student's t-tests were then carried out to assess the significance of differences in quantitative variables between two groups.
SUPPLEMENTARY MATERIAL
FUNDING
X.F.K. was supported by the Stony Wold-Herbert Fund, Choh-Hao Li Memorial Fund Scholar award and the Shanghai Educational Development Foundation, Y.I. is supported by the AXA Research Fund, V.L.B. is supported by the Stony Wold-Herbert Fund and A.K. is supported by the Fondation Médicale Medische Stichting Mathilde E. Horlait-Dapens. The Laboratory of Human Genetics of Infectious Diseases is supported in part by grants from the St Giles Foundation, the Jeffrey Modell Foundation, The Rockefeller University Center for Clinical and Translational Science grant number 8UL1TR000043 from the National Center for Research Resources and the National Center for Advancing Sciences (NCATS), National Institutes of Health, the National Institute of Allergy and Infectious Diseases grant number 5R01AI089970-02, The Rockefeller University and the European Research Council (ERC). J.L.C. received a research grant from the Jeffrey Modell Foundation in collaboration with Talecris BioTherapeutics. The funder was not involved in the study design; collection, analysis and interpretation of data; writing of the paper or decision to submit for publication.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Bertrand Boisson and Minji Byun for helpful discussions and critical reading. We thank Yelena Nemirovskaya, Eric Anderson, Tiffany Nivare, Tatiana Kochetkov, Lucile Jannière, Martine Courat, Devon Davenport, Jacqueline Rose and Jacqueline Feinberg for technical and secretarial assistance and all members of the Laboratory of Human Genetics of Infectious Diseases for helpful discussions.
Conflict of Interest statement: None declared.
REFERENCES
- 1.Alcais A., Abel L., Casanova J.L. Human genetics of infectious diseases: between proof of principle and paradigm. J. Clin. Invest. 2009;119:2506–2514. doi: 10.1172/JCI38111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casanova J.L., Abel L. Genetic dissection of immunity to mycobacteria: the human model. Annu. Rev. Immunol. 2002;20:581–620. doi: 10.1146/annurev.immunol.20.081501.125851. [DOI] [PubMed] [Google Scholar]
- 3.Casanova J.L., Abel L. The human model: a genetic dissection of immunity to infection in natural conditions. Nat. Rev. Immunol. 2004;4:55–66. doi: 10.1038/nri1264. [DOI] [PubMed] [Google Scholar]
- 4.Fortin A., Abel L., Casanova J.L., Gros P. Host genetics of mycobacterial diseases in mice and men: forward genetic studies of BCG-osis and tuberculosis. Annu. Rev. Genomics Hum. Genet. 2007;8:163–192. doi: 10.1146/annurev.genom.8.080706.092315. [DOI] [PubMed] [Google Scholar]
- 5.Alcais A., Fieschi C., Abel L., Casanova J.L. Tuberculosis in children and adults: two distinct genetic diseases. J. Exp. Med. 2005;202:1617–1621. doi: 10.1084/jem.20052302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boisson-Dupuis S., El Baghdadi J., Parvaneh N., Bousfiha A., Bustamante J., Feinberg J., Samarina A., Grant A.V., Janniere L., El Hafidi N., et al. IL-12Rbeta1 deficiency in two of fifty children with severe tuberculosis from Iran, Morocco, and Turkey. PLoS One. 2011;6:e18524. doi: 10.1371/journal.pone.0018524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Filipe-Santos O., Bustamante J., Chapgier A., Vogt G., de Beaucoudrey L., Feinberg J., Jouanguy E., Boisson-Dupuis S., Fieschi C., Picard C., et al. Inborn errors of IL-12/23- and IFN-gamma-mediated immunity: molecular, cellular, and clinical features. Semin. Immunol. 2006;18:347–361. doi: 10.1016/j.smim.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 8.Bustamante J., Arias A.A., Vogt G., Picard C., Galicia L.B., Prando C., Grant A.V., Marchal C.C., Hubeau M., Chapgier A., et al. Germline CYBB mutations that selectively affect macrophages in kindreds with X-linked predisposition to tuberculous mycobacterial disease. Nat. Immunol. 2011;12:213–221. doi: 10.1038/ni.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hambleton S., Salem S., Bustamante J., Bigley V., Boisson-Dupuis S., Azevedo J., Fortin A., Haniffa M., Ceron-Gutierrez L., Bacon C.M., et al. IRF8 mutations and human dendritic-cell immunodeficiency. N. Engl. J. Med. 2011;365:127–138. doi: 10.1056/NEJMoa1100066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bogunovic D., Byun M., Durfee L.A., Abhyankar A., Sanal O., Mansouri D., Salem S., Radovanovic I., Grant A.V., Adimi P., et al. Mycobacterial disease and impaired IFN-gamma immunity in humans with inherited ISG15 deficiency. Science. 2012;337:1684–1688. doi: 10.1126/science.1224026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupuis S., Doffinger R., Picard C., Fieschi C., Altare F., Jouanguy E., Abel L., Casanova J.L. Human interferon-gamma-mediated immunity is a genetically controlled continuous trait that determines the outcome of mycobacterial invasion. Immunol. Rev. 2000;178:129–137. doi: 10.1034/j.1600-065x.2000.17810.x. [DOI] [PubMed] [Google Scholar]
- 12.Altare F., Durandy A., Lammas D., Emile J.F., Lamhamedi S., Le Deist F., Drysdale P., Jouanguy E., Doffinger R., Bernaudin F., et al. Impairment of mycobacterial immunity in human interleukin-12 receptor deficiency. Science. 1998;280:1432–1435. doi: 10.1126/science.280.5368.1432. [DOI] [PubMed] [Google Scholar]
- 13.de Jong R., Altare F., Haagen I.A., Elferink D.G., Boer T., van Breda Vriesman P.J., Kabel P.J., Draaisma J.M., van Dissel J.T., Kroon F.P., et al. Severe mycobacterial and Salmonella infections in interleukin-12 receptor-deficient patients. Science. 1998;280:1435–1438. doi: 10.1126/science.280.5368.1435. [DOI] [PubMed] [Google Scholar]
- 14.Picard C., Fieschi C., Altare F., Al-Jumaah S., Al-Hajjar S., Feinberg J., Dupuis S., Soudais C., Al-Mohsen I.Z., Genin E., et al. Inherited interleukin-12 deficiency: IL12B genotype and clinical phenotype of 13 patients from six kindreds. Am. J. Hum. Genet. 2002;70:336–348. doi: 10.1086/338625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filipe-Santos O., Bustamante J., Haverkamp M.H., Vinolo E., Ku C.L., Puel A., Frucht D.M., Christel K., von Bernuth H., Jouanguy E., et al. X-linked susceptibility to mycobacteria is caused by mutations in NEMO impairing CD40-dependent IL-12 production. J. Exp. Med. 2006;203:1745–1759. doi: 10.1084/jem.20060085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jouanguy E., Altare F., Lamhamedi S., Revy P., Emile J.F., Newport M., Levin M., Blanche S., Seboun E., Fischer A., et al. Interferon-gamma-receptor deficiency in an infant with fatal bacille Calmette-Guerin infection. N. Engl. J. Med. 1996;335:1956–1961. doi: 10.1056/NEJM199612263352604. [DOI] [PubMed] [Google Scholar]
- 17.Jouanguy E., Dupuis S., Pallier A., Doffinger R., Fondaneche M.C., Fieschi C., Lamhamedi-Cherradi S., Altare F., Emile J.F., Lutz P., et al. In a novel form of IFN-gamma receptor 1 deficiency, cell surface receptors fail to bind IFN-gamma. J. Clin. Invest. 2000;105:1429–1436. doi: 10.1172/JCI9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jouanguy E., Lamhamedi-Cherradi S., Altare F., Fondaneche M.C., Tuerlinckx D., Blanche S., Emile J.F., Gaillard J.L., Schreiber R., Levin M., et al. Partial interferon-gamma receptor 1 deficiency in a child with tuberculoid bacillus Calmette-Guerin infection and a sibling with clinical tuberculosis. J. Clin. Invest. 1997;100:2658–2664. doi: 10.1172/JCI119810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jouanguy E., Lamhamedi-Cherradi S., Lammas D., Dorman S.E., Fondaneche M.C., Dupuis S., Doffinger R., Altare F., Girdlestone J., Emile J.F., et al. A human IFNGR1 small deletion hotspot associated with dominant susceptibility to mycobacterial infection. Nat. Genet. 1999;21:370–378. doi: 10.1038/7701. [DOI] [PubMed] [Google Scholar]
- 20.Kong X.F., Vogt G., Chapgier A., Lamaze C., Bustamante J., Prando C., Fortin A., Puel A., Feinberg J., Zhang X.X., et al. A novel form of cell type-specific partial IFN-gammaR1 deficiency caused by a germ line mutation of the IFNGR1 initiation codon. Hum. Mol. Genet. 2010;19:434–444. doi: 10.1093/hmg/ddp507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapgier A., Boisson-Dupuis S., Jouanguy E., Vogt G., Feinberg J., Prochnicka-Chalufour A., Casrouge A., Yang K., Soudais C., Fieschi C., et al. Novel STAT1 alleles in otherwise healthy patients with mycobacterial disease. PLoS Genet. 2006;2:e131. doi: 10.1371/journal.pgen.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dupuis S., Dargemont C., Fieschi C., Thomassin N., Rosenzweig S., Harris J., Holland S.M., Schreiber R.D., Casanova J.L. Impairment of mycobacterial but not viral immunity by a germline human STAT1 mutation. Science. 2001;293:300–303. doi: 10.1126/science.1061154. [DOI] [PubMed] [Google Scholar]
- 23.Doffinger R., Jouanguy E., Dupuis S., Fondaneche M.C., Stephan J.L., Emile J.F., Lamhamedi-Cherradi S., Altare F., Pallier A., Barcenas-Morales G., et al. Partial interferon-gamma receptor signaling chain deficiency in a patient with bacille Calmette-Guerin and Mycobacterium abscessus infection. J. Infect. Dis. 2000;181:379–384. doi: 10.1086/315197. [DOI] [PubMed] [Google Scholar]
- 24.Dorman S.E., Holland S.M. Mutation in the signal-transducing chain of the interferon-gamma receptor and susceptibility to mycobacterial infection. J. Clin. Invest. 1998;101:2364–2369. doi: 10.1172/JCI2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenzweig S.D., Dorman S.E., Uzel G., Shaw S., Scurlock A., Brown M.R., Buckley R.H., Holland S.M. A novel mutation in IFN-gamma receptor 2 with dominant negative activity: biological consequences of homozygous and heterozygous states. J. Immunol. 2004;173:4000–4008. doi: 10.4049/jimmunol.173.6.4000. [DOI] [PubMed] [Google Scholar]
- 26.Vogt G., Bustamante J., Chapgier A., Feinberg J., Boisson Dupuis S., Picard C., Mahlaoui N., Gineau L., Alcais A., Lamaze C., et al. Complementation of a pathogenic IFNGR2 misfolding mutation with modifiers of N-glycosylation. J. Exp. Med. 2008;205:1729–1737. doi: 10.1084/jem.20071987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vogt G., Chapgier A., Yang K., Chuzhanova N., Feinberg J., Fieschi C., Boisson-Dupuis S., Alcais A., Filipe-Santos O., Bustamante J., et al. Gains of glycosylation comprise an unexpectedly large group of pathogenic mutations. Nat. Genet. 2005;37:692–700. doi: 10.1038/ng1581. [DOI] [PubMed] [Google Scholar]
- 28.Toyoda H., Ido M., Nakanishi K., Nakano T., Kamiya H., Matsumine A., Uchida A., Mizutani H., de Beaucoudrey L., Vogt G., et al. Multiple cutaneous squamous cell carcinomas in a patient with interferon gamma receptor 2 (IFN gamma R2) deficiency. J. Med. Genet. 2010;47:631–634. doi: 10.1136/jmg.2009.072108. [DOI] [PubMed] [Google Scholar]
- 29.Chapgier A., Kong X.F., Boisson-Dupuis S., Jouanguy E., Averbuch D., Feinberg J., Zhang S.Y., Bustamante J., Vogt G., Lejeune J., et al. A partial form of recessive STAT1 deficiency in humans. J. Clin. Invest. 2009;119:1502–1514. doi: 10.1172/JCI37083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kong X.F., Ciancanelli M., Al-Hajjar S., Alsina L., Zumwalt T., Bustamante J., Feinberg J., Audry M., Prando C., Bryant V., et al. A novel form of human STAT1 deficiency impairing early but not late responses to interferons. Blood. 2010;116:5895–5906. doi: 10.1182/blood-2010-04-280586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D'Haene B., Attanasio C., Beysen D., Dostie J., Lemire E., Bouchard P., Field M., Jones K., Lorenz B., Menten B., et al. Disease-causing 7.4 kb cis-regulatory deletion disrupting conserved non-coding sequences and their interaction with the FOXL2 promotor: implications for mutation screening. PLoS Genet. 2009;5:e1000522. doi: 10.1371/journal.pgen.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dorman S.E., Picard C., Lammas D., Heyne K., van Dissel J.T., Baretto R., Rosenzweig S.D., Newport M., Levin M., Roesler J., et al. Clinical features of dominant and recessive interferon gamma receptor 1 deficiencies. Lancet. 2004;364:2113–2121. doi: 10.1016/S0140-6736(04)17552-1. [DOI] [PubMed] [Google Scholar]
- 33.Bach E.A., Aguet M., Schreiber R.D. The IFN gamma receptor: a paradigm for cytokine receptor signaling. Annu. Rev. Immunol. 1997;15:563–591. doi: 10.1146/annurev.immunol.15.1.563. [DOI] [PubMed] [Google Scholar]
- 34.Bernabei P., Coccia E.M., Rigamonti L., Bosticardo M., Forni G., Pestka S., Krause C.D., Battistini A., Novelli F. Interferon-gamma receptor 2 expression as the deciding factor in human T, B, and myeloid cell proliferation or death. J. Leukoc. Biol. 2001;70:950–960. [PubMed] [Google Scholar]
- 35.van Boxel-Dezaire A.H., Stark G.R. Cell type-specific signaling in response to interferon-gamma. Curr. Top. Microbiol. Immunol. 2007;316:119–154. doi: 10.1007/978-3-540-71329-6_7. [DOI] [PubMed] [Google Scholar]
- 36.Bach E.A., Szabo S.J., Dighe A.S., Ashkenazi A., Aguet M., Murphy K.M., Schreiber R.D. Ligand-induced autoregulation of IFN-gamma receptor beta chain expression in T helper cell subsets. Science. 1995;270:1215–1218. doi: 10.1126/science.270.5239.1215. [DOI] [PubMed] [Google Scholar]
- 37.Bach E.A., Tanner J.W., Marsters S., Ashkenazi A., Aguet M., Shaw A.S., Schreiber R.D. Ligand-induced assembly and activation of the gamma interferon receptor in intact cells. Mol. Cell. Biol. 1996;16:3214–3221. doi: 10.1128/mcb.16.6.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kotenko S.V., Izotova L.S., Pollack B.P., Mariano T.M., Donnelly R.J., Muthukumaran G., Cook J.R., Garotta G., Silvennoinen O., Ihle J.N., et al. Interaction between the components of the interferon gamma receptor complex. J. Biol. Chem. 1995;270:20915–20921. doi: 10.1074/jbc.270.36.20915. [DOI] [PubMed] [Google Scholar]
- 39.Pernis A., Gupta S., Gollob K.J., Garfein E., Coffman R.L., Schindler C., Rothman P. Lack of interferon gamma receptor beta chain and the prevention of interferon gamma signaling in TH1 cells. Science. 1995;269:245–247. doi: 10.1126/science.7618088. [DOI] [PubMed] [Google Scholar]
- 40.Gajewski T.F., Goldwasser E., Fitch F.W. Anti-proliferative effect of IFN-gamma in immune regulation. II. IFN-gamma inhibits the proliferation of murine bone marrow cells stimulated with IL-3, IL-4, or granulocyte-macrophage colony-stimulating factor. J. Immunol. 1988;141:2635–2642. [PubMed] [Google Scholar]
- 41.Levin D., Harari D., Schreiber G. Stochastic receptor expression determines cell fate upon interferon treatment. Mol. Cell. Biol. 2011;31:3252–3266. doi: 10.1128/MCB.05251-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas C., Moraga I., Levin D., Krutzik P.O., Podoplelova Y., Trejo A., Lee C., Yarden G., Vleck S.E., Glenn J.S., et al. Structural linkage between ligand discrimination and receptor activation by type I interferons. Cell. 2011;146:621–632. doi: 10.1016/j.cell.2011.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sampaio E.P., Bax H.I., Hsu A.P., Kristosturyan E., Pechacek J., Chandrasekaran P., Paulson M.L., Dias D.L., Spalding C., Uzel G., et al. A novel STAT1 mutation associated with disseminated mycobacterial disease. J. Clin. Immunol. 2012;32:681–689. doi: 10.1007/s10875-012-9659-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsumura M., Okada S., Sakai H., Yasunaga S., Ohtsubo M., Murata T., Obata H., Yasumi T., Kong X.F., Abhyankar A., et al. Dominant-negative STAT1 SH2 domain mutations in unrelated patients with mendelian susceptibility to mycobacterial disease. Hum. Mutat. 2012;33:1377–1387. doi: 10.1002/humu.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alcais A., Quintana-Murci L., Thaler D.S., Schurr E., Abel L., Casanova J.L. Life-threatening infectious diseases of childhood: single-gene inborn errors of immunity? Ann. N. Y. Acad. Sci. 2010;1214:18–33. doi: 10.1111/j.1749-6632.2010.05834.x. [DOI] [PubMed] [Google Scholar]
- 46.Tabarsi P., Marjani M., Mansouri N., Farnia P., Boisson-Dupuis S., Bustamante J., Abel L., Adimi P., Casanova J.L., Mansouri D. Lethal tuberculosis in a previously healthy adult with IL-12 receptor deficiency. J. Clin. Immunol. 2011;31:537–539. doi: 10.1007/s10875-011-9523-9. [DOI] [PubMed] [Google Scholar]
- 47.Kong X.F., Zhang X.X., Gong Q.M., Gao J., Zhang S.Y., Wang L., Xu J., Han Y., Jin G.D., Jiang J.H., et al. MxA induction may predict sustained virologic responses of chronic hepatitis B patients with IFN-alpha treatment. J. Interferon Cytokine Res. 2007;27:809–818. doi: 10.1089/jir.2006.0163. [DOI] [PubMed] [Google Scholar]
- 48.Sancho-Shimizu V., Perez de Diego R., Lorenzo L., Halwani R., Alangari A., Israelsson E., Fabrega S., Cardon A., Maluenda J., Tatematsu M., et al. Herpes simplex encephalitis in children with autosomal recessive and dominant TRIF deficiency. J. Clin. Invest. 2011;121:4889–4902. doi: 10.1172/JCI59259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolze A., Byun M., McDonald D., Morgan N.V., Abhyankar A., Premkumar L., Puel A., Bacon C.M., Rieux-Laucat F., Pang K., et al. Whole-exome-sequencing-based discovery of human FADD deficiency. Am. J. Hum. Genet. 2010;87:873–881. doi: 10.1016/j.ajhg.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Byun M., Abhyankar A., Lelarge V., Plancoulaine S., Palanduz A., Telhan L., Boisson B., Picard C., Dewell S., Zhao C., et al. Whole-exome sequencing-based discovery of STIM1 deficiency in a child with fatal classic Kaposi sarcoma. J. Exp. Med. 2010;207:2307–2312. doi: 10.1084/jem.20101597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vogt G., Nathan C. In vitro differentiation of human macrophages with enhanced antimycobacterial activity. J. Clin. Invest. 2011;121:3889–3901. doi: 10.1172/JCI57235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma C.S., Hare N.J., Nichols K.E., Dupre L., Andolfi G., Roncarolo M.G., Adelstein S., Hodgkin P.D., Tangye S.G. Impaired humoral immunity in X-linked lymphoproliferative disease is associated with defective IL-10 production by CD4+ T cells. J. Clin. Invest. 2005;115:1049–1059. doi: 10.1172/JCI23139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.