Abstract
FGF receptor 3 (FGFR3) is activated by mutation or over-expression in many bladder cancers. Here, we identify an additional mechanism of activation via chromosomal re-arrangement to generate constitutively activated fusion genes. FGFR3–transforming acid coiled coil 3 (TACC3) fusions resulting from 4p16.3 re-arrangements and a t(4;7) that generates a FGFR3-BAI1-associated protein 2-like 1 (BAIAP2L1) fusion were identified in 4 of 43 bladder tumour cell lines and 2 of 32 selected tissue samples including the tumour from which one of the cell lines was derived. These are highly activated and transform NIH-3T3 cells. The FGFR3 component is identical in all cases and lacks the final exon that includes the phospholipase C gamma 1 (PLCγ1) binding site. Expression of the fusions in immortalized normal human urothelial cells (NHUC) induced activation of the mitogen-activated protein kinase pathway but not PLCγ1. A protein with loss of the terminal region alone was not as highly activated as the fusion proteins, indicating that the fusion partners are essential. The TACC3 fusions retain the TACC domain that mediates microtubule binding and the BAIAP2L1 fusion retains the IRSp53/MIM domain (IMD) that mediates actin binding and Rac interaction. As urothelial cell lines with FGFR3 fusions are extremely sensitive to FGFR-selective agents, the presence of a fusion gene may aid in selection of patients for FGFR-targeted therapy.
INTRODUCTION
Activating point mutations of FGF receptor 3 (FGFR3) are found in up to 80% of low-grade and -stage urothelial carcinoma (UC) of the bladder. The most common mutations favour ligand-independent receptor dimerization, leading to trans-phosphorylation and downstream signalling. Upregulated expression of FGFR3 protein is also found in a significant number of tumours, predominantly muscle invasive tumours, which lack point mutations (1). Thus, FGFR3 may be an important therapeutic target in both non-invasive and invasive UC. Several studies have shown that in preclinical models, downregulation of FGFR3 or inhibition using small molecules or antibodies has a profound inhibitory effect on some UC cells, e.g. (2–5) and clinical trials of such agents have been initiated.
In a recent study of the effect of the small molecule FGFR inhibitors PD173074, TKI-258 and SU5402, we showed that three UC cell lines, SW780, RT112 and RT4, that have upregulated expression of non-point-mutated FGFR3 and do not show high-level gene amplification, were much more sensitive to FGFR inhibition than cell lines with point mutations (2). High sensitivity of the same lines to NVP-BGJ398, a selective inhibitor of FGFRs 1, 2 and 3 has also been reported (6). RT112 cells are critically dependent on FGFR3 for both proliferation in vitro and as xenografts in vivo (2,3).
The high sensitivity of these ‘wild-type’ FGFR cell lines raises questions about the molecular mechanisms underlying their FGFR3 dependence compared with cell lines expressing point mutant FGFR3. Possibilities include autocrine signalling, effects of undefined interacting or co-operating proteins and the presence of hitherto undetected mutational events. Such molecular information may allow the prediction of sensitivity to FGFR inhibition in patients with FGFR-expressing but not point-mutated tumours. Here, we examined the FGFR3 protein, transcript and gene in non-point-mutated FGFR3-dependent UC cell lines and in primary tumours. This revealed the presence of genomic FGFR3 translocations involving two different fusion partners, which generate fusion proteins that are constitutively activated and oncogenic FGFR3 kinases.
RESULTS
High-molecular weight forms of FGFR3 protein in bladder cancer cell lines
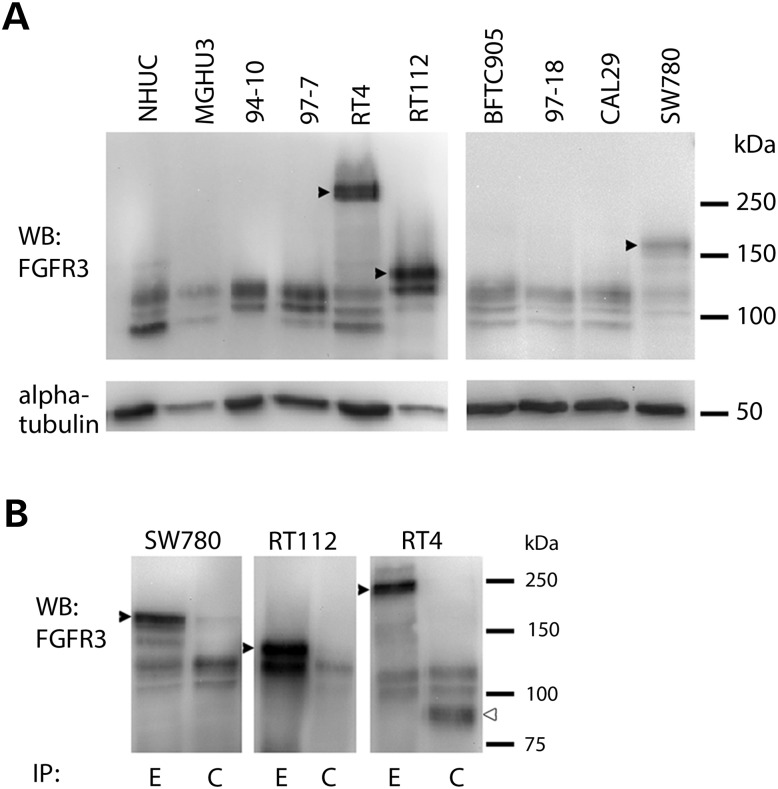
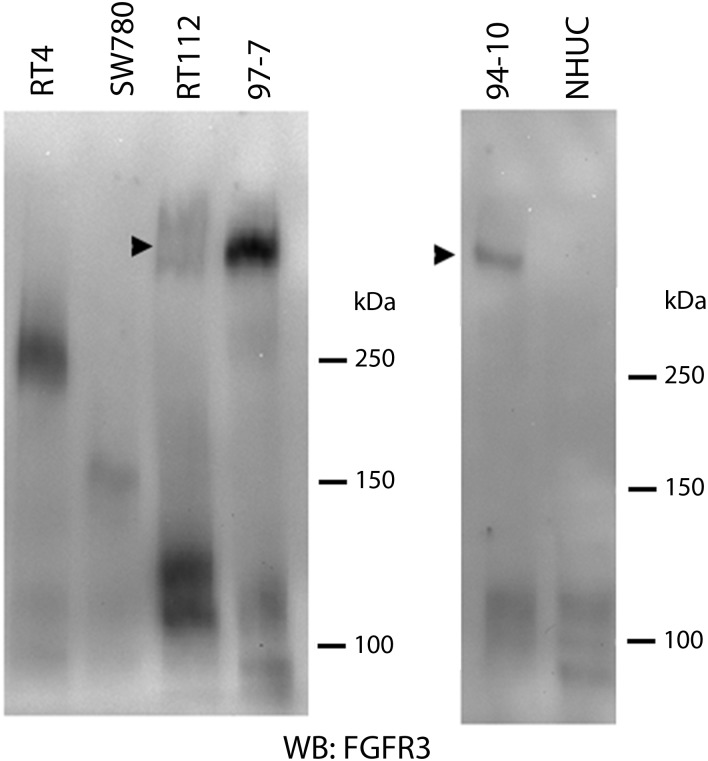
We screened 43 bladder tumour cell lines (Supplementary Material, Table S1) by western blotting and identified nine with detectable FGFR3 protein expression. Three (RT4, RT112, SW780) contained high-molecular weight forms of FGFR3 in addition to bands of the expected size (Fig. 1A).
Figure 1.
(A) High-molecular weight forms of FGF receptor 3 (FGFR3) (arrowheads) detected in three non-point mutated bladder tumour cell lines. MGHU3, 94–10 and 97–7 have FGFR3 point mutations. NHUC, normal human urothelial cells. (B) Immunoprecipitation with antibodies detecting epitopes in the extracellular (E) or cytoplasmic (C) regions of FGFR3 and detection with FGFR3 antibody B9, which recognizes amino acids 25–124. High-molecular weight forms (arrowheads) are precipitated by E but not C antibody. The FGFR3 Δ8–10 isoform which is expressed at highest level in RT4 is precipitated by C but not E antibody (open arrowhead in RT4-C).
When FGFR3 was immunoprecipitated with antibodies that recognize epitopes in the extracellular (E) (residues 359–372) or cytoplasmic (C) (residues 792–806) regions of the protein, these high-molecular weight forms could not be captured with the C epitope antibody (Fig. 1B). Deglycosylation reduces wild-type (WT) FGFR3 protein to 88 kDa (full-length IIIb isoform) and 76 kDa (Δ8–10 isoform) (7). Following deglycosylation, RT4, SW780 and RT112 retained higher molecular weight forms (Supplementary Material, Fig. S1A), raising the possibility that these represent fusion proteins.
Identification of a FGFR3-BAIAP2L1 fusion protein
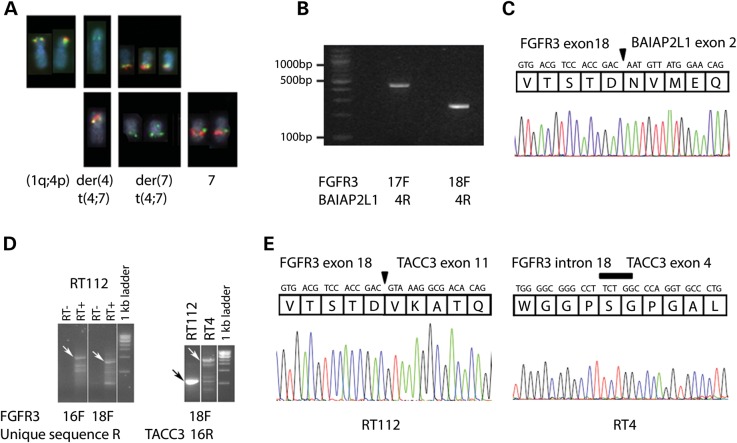
SNP array profiles were available for these cell lines and in each case we could detect apparent breakpoints and/or microamplification in 4p16.3 in the region of FGFR3 (Supplementary Material, Fig. S1B). We identified a reciprocal translocation t(4;7) in SW780 using fluorescence in situ hybridization (FISH), with breakpoints within bacterial artificial chromosome (BAC) probes containing FGFR3 on chromosome 4 and BAI1-associated protein 2-like 1 (BAIAP2L1), also known as IRTKS (insulin receptor kinase substrate) on chromosome 7 (Fig. 2A). The breakpoints were further defined using 3–5 kb PCR-generated FISH probes (Supplementary Material, Table S2and Fig. S1C).
Figure 2.
(A) FISH to SW780 to define t(4;7) breakpoints. Top row: FISH with bacterial artificial chromosome (BAC) clones from chromosome 4. SW780 has no normal chromosome 4; chromosome 4 which is not involved in t(4;7) is involved in an apparently balanced t(1;4)(q11;q1?) with an apparently intact 4p. RP11-138D6 (green, contains FGFR3) hybridizes to intact 4p and both t(4;7) translocation products, while the more distal RP11-572O17 (red) hybridizes to intact 4p and der(7) t(4;7) only. Bottom row: FISH with BACs from chromosome 7. RP11-307C18 (green) hybridizes to normal 7 and both t(4;7) translocation products, while the more distal RP11-473H4 (red) hybridizes to normal 7 and der(4) t(4;7) only. (B) RT-PCR products from SW780 cDNA with forward primers from FGFR3 exons 17 or 18 and a reverse primer from BAI1-associated protein 2-like 1 (BAIAP2L1) exon 4. (C) Sequence traces of these products showing an in-frame fusion between FGFR3 exon 18 and BAIAP2L1 exon 2. The junction is marked with an arrowhead. (D) RT112 cDNA made using a poly-dT primer with a unique 5′ sequence generated products following RT-PCR with forward primers from FGFR3 and a reverse primer from the unique sequence. Sequencing showed TACC3 involvement, and RT-PCR using a TACC3 reverse primer gave a unique band for RT112 and a series of bands for RT4. Arrows indicate FGFR3:TACC3 fusion transcripts that were confirmed by sequencing. RT− and RT+ are reverse transcriptase negative control and transcribed cDNA, respectively. (E) Sequence traces of FGFR3–TACC3 RT-PCR products showing the fusion gene junctions to be between FGFR3 exon 18 and TACC3 exon 11 in RT112 (arrowhead), and between FGFR3 within intron 18 and TACC3 within exon 4 in RT4 (bar).
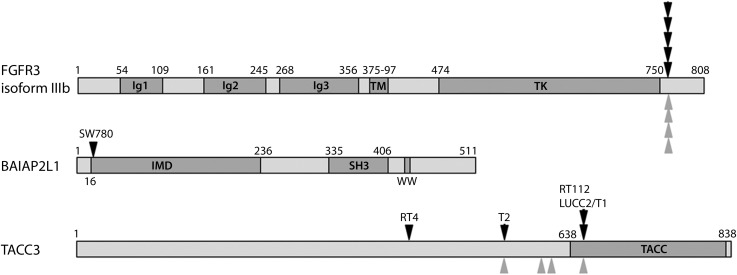
FGFR3 (ENSG00000068078) exons are numbered sequentially from 1 to 19, including the non-coding exon 1 and the alternate exons 8 (IIIb isoform, ENST00000340107) and 9 (IIIc isoform, ENST00000440486). RT-PCR with primers from FGFR3 and BAIAP2L1 generated products (Fig. 2B) that confirmed the fusion to be FGFR3 exon18::BAIAP2L1 exon 2, which comprised an in-frame fusion transcript (Fig. 2C) with a predicted translation product containing FGFR3 residues 1–760 (IIIb isoform) and BAIAP2L1 residues 18–522. Transcripts representing both the full-length IIIb and the Δ8–10 isoform (7) of FGFR3 were detected. BAIAP2L1 is a member of the IMD (IRSp53/MIM homology domain) family of proteins. In addition to the IMD domain (amino acids 1–250), it contains an SH3 domain (amino acids 375–438) and a putative WW domain interacting motif (PPPDY), all of which are retained in the FGFR3-BAIAP2L1 fusion protein (Fig. 3).
Figure 3.
Schematic of FGFR3, TACC3 and BAIAP2L1 proteins showing known domains and positions of the breakpoints (black arrowheads) identified in bladder tumours (T1 and T2) and cell lines and those reported in glioblastoma (grey arrowheads) (11).
FISH using a PCR-generated probe in BAIAP2L1 intron 1 (Supplementary Material, Table S2) showed that the genomic breakpoint is close to exon 1. PCR of genomic DNA with a forward primer from FGFR3 exon 18 and a reverse primer 270 bp into BAIAP2L1 intron 1 (Supplementary Material, Table S2) generated a product which included 145 nucleotides from FGFR3 intron 18, two nucleotides of unknown origin and BAIAP2L1 intron 1 excluding the first 41 nucleotides (Supplementary Material, Fig. S2). The t(4;7) was not precisely reciprocal as the expected reverse product was not found by PCR.
FGFR3–TACC3 fusions
We used a polyT primer with a unique 5′ sequence (Supplementary Material, Table S2) for first strand complementary DNA (cDNA) synthesis from RT112 RNA. PCR with an FGFR3 exon 18 forward primer and the unique sequence reverse primer generated a product (Fig. 2D and E) containing FGFR3 exon 18::TACC3 exon 11 which was predicted to generate an in-frame fusion protein containing FGFR3 residues 1–760 (IIIb isoform) and transforming acid coiled coil 3 (TACC3) residues 649–838. This contains virtually the entire TACC3 coiled-coil region implicated in the stabilization of kinetochore fibres during mitosis (8) (Fig. 3). PCR with primers on either side of the breakpoint (FGFR3 exon 18F and TACC3 exon 11R) gave a product that determined the genomic breakpoint (Supplementary Material, Fig. S2).
cDNA synthesis with polyT and FGFR3 exon 18 primers did not generate a product from RT4, but RT-PCR using FGFR3 and TACC3 primers generated a larger product than in RT112 (Fig. 2D). Sequencing of both the RT-PCR product and the genomic DNA showed fusion between breakpoints within FGFR3 intron 18 and within TACC3 exon 4 with a five-nucleotide sequence overlap at the breakpoint (Fig. 2E; Supplementary Material, Fig. S2). Splicing at the exon 18/intron 18 boundary was clearly disrupted by this translocation, and the protein product was an in-frame fusion predicted to encode FGFR3 residues 1–760 (IIIb isoform), 33 amino acids generated from FGFR3 intron 18 and TACC3 residues 433–838. RT4 expressed two transcripts from the fusion gene, one with this full-length product (termed RT4FUS) and one with 244 nucleotides spliced out from the 3′ end of FGFR3 exon 18, which is predicted to encode a truncated product (termed RT4del) including FGFR3 residues 1–760 and 3 codons from TACC3 exon 4 out of frame.
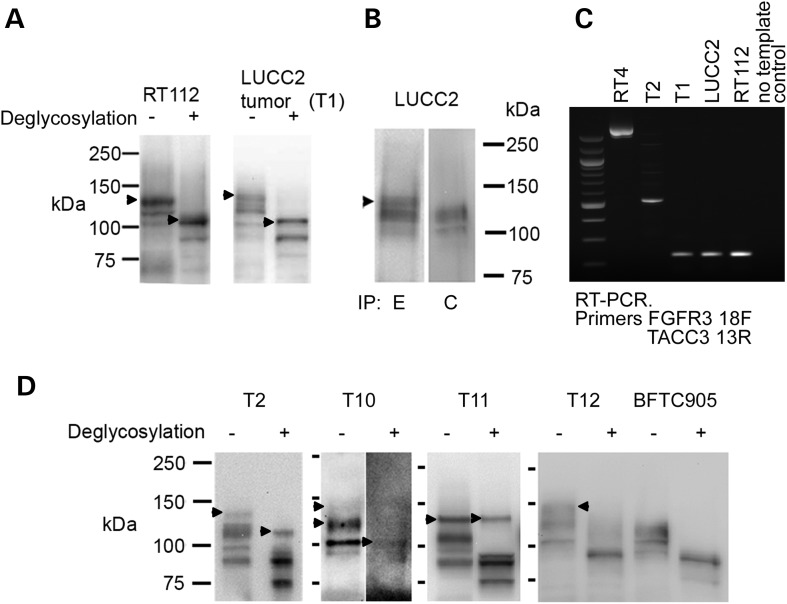
As loss of FGFR3 exon 19 was common to all three fusions, we used real-time RT-PCR to assess the loss of exon 18-19 product relative to the sequences retained in the fusion transcripts (exons 11-12) in 43 UC cell lines. We confirmed an over-representation of RNA from exons 11-12 compared with exons 18-19 in RT4, RT112 and SW780, and identified one other cell line, LUCC2, with a similar imbalance (Supplementary Material, Fig. S1D). LUCC2 is a cell line established in our laboratory from a muscle-invasive (stage T2 grade 3) bladder tumour, so we were able to examine tumour material as well as the cell line. LUCC2 cells express low levels of both normal FGFR3 and the fusion protein, but immunohistochemistry showed that the tumour contained high levels of FGFR3 proteins detected with an antibody that recognizes amino acids 25–124. Using fresh-frozen tumour tissue from this patient, we were able to confirm the presence of the fusion transcript, and show that it was the same size as the RT112 cell line transcript with and without deglycosylation (Fig. 4A). The LUCC2 fusion protein was immunoprecipitated by the E but not the C antibody (Fig. 4B).
Figure 4.
(A) Western blot shows the presence of a high-molecular weight form of FGFR3 (arrowheads) in the tumour from which LUCC2 was derived (T1), and that it is the same size as that seen in RT112 (arrowheads), with and without deglycosylation. (B) Immunoprecipitation of LUCC2 protein with antibodies detecting epitopes in the extracellular (E) or cytoplasmic (C) regions of FGFR3 and detection with FGFR3 antibody B9, which recognizes amino acids 25–124. A high-molecular weight form (arrowhead) is precipitated by E but not C antibody. Because of the lower FGFR3 expression, 20 times more total protein was used for the LUCC2 IP than for the cell lines shown in Fig. 1. (C) RT-PCR using primers from FGFR3 exon 18 and TACC3 exon 13 shows fusion transcripts in tumour samples T1 and T2 and cell lines RT4, RT112 and LUCC2. (D) Tumours T2, T10 and T11 retain high-molecular weight forms of FGFR3 after deglycosylation (arrowheads). Tumour T12 shows a high-molecular weight form of FGFR3 (arrowhead), but this is reduced to the normal molecular weight by deglycosylation. Cell line BFTC905 shows the expected result for untranslocated FGFR3.
RT-PCR using FGFR3 exon 18 and TACC3 primers amplified an FGFR3–TACC3 fusion transcript identical to that in RT112 in both the tumour and LUCC2 cell line (Figs 3 and 4C) but with different genomic breakpoints (Supplementary Material, Fig. S2).
We hypothesized that FGFR3 fusions may be more common in tumours in which high levels of FGFR3 proteins are expressed but no point mutations can be detected. We identified 46 invasive tumours with no detectable point mutations, 32 of which showed upregulated FGFR3 expression by immunohistochemistry. As two distinct fusion partners had been identified, we used western blotting as an unbiased means to search for FGFR3 proteins of abnormal size. Nine samples showed bands of higher molecular weight than normal (Fig. 4D). Five of these were found to represent highly glycoslylated versions of the normal protein form (e.g. T12 in Fig. 4D). One sample was too small to analyse further. RT-PCR with FGFR3 forward and TACC3 or BAIAP2L1 reverse primers on cDNA from the remaining 3 tumours showed that one of these contained a fusion between FGFR3 exon 18 and TACC3 exon 8 (T2 in Figs 3 and 4C). The other two high-molecular weight forms have yet to be characterized (T10 and T11 in Fig. 4D). We used RT-PCR with primers from FGFR3 exon 18 and TACC3 exon 16 or BAIAP2L1 3′UTR to screen cDNA from FGFR3 WT tumours that were stage T2+ or stage T1 grade 3 without selection for FGFR3 expression levels. We did not find any fusion genes (0/46 for TACC3, 0/27 for BAIAP2L1).
Activation status and oncogenic potential of FGFR3 fusion genes
Some activating mutations of FGFR3, including the common mutations found in bladder cancer, act by constitutive stabilization of receptor dimers. Examination of FGFR3 proteins in serum-starved cells under non-denaturing conditions revealed constitutive dimers in the S249C mutant cell lines 94–10 and 97–7, but not in normal human urothelial cells (NHUC), SW780 or RT4. RT112 contained a low level of dimers, but the majority of the fusion protein was present as a monomer, suggesting that constitutive dimerization is not the principle method of activation (Fig. 5).
Figure 5.
Non-denaturing gel showing the presence of constitutively dimerized S249C mutant protein in 94-10 and 97-7 cells which contain point mutant (S249C) FGFR3 but not in NHUC, RT4 or SW780. A small amount of the dimer is present in RT112 (arrow).
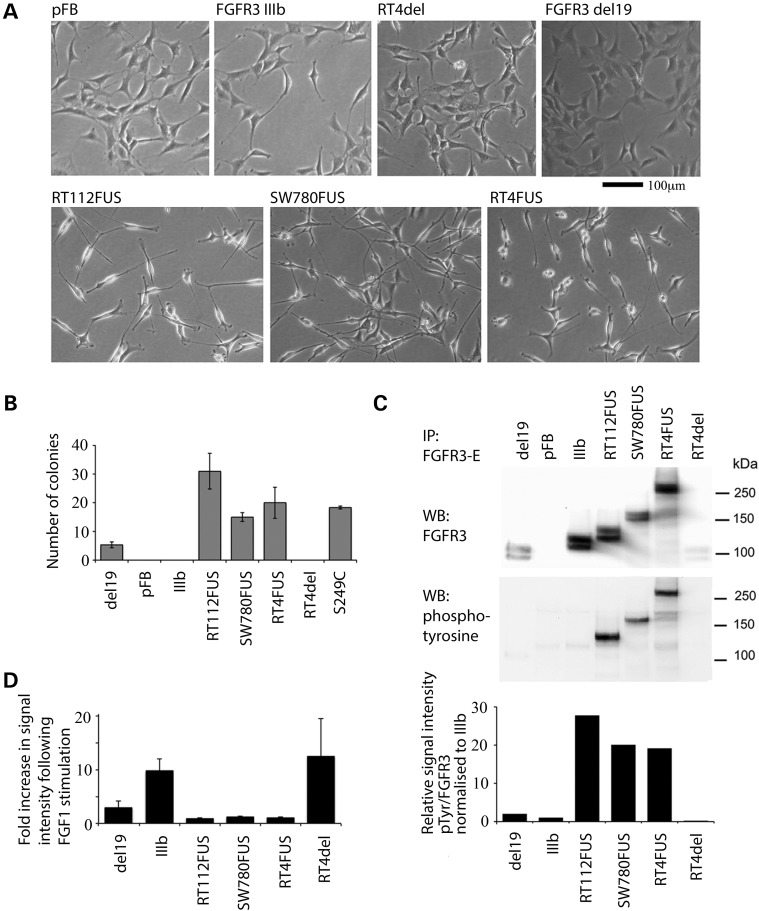
Retroviral vectors were constructed to express the fusion transcripts from SW780, RT112 and RT4 (RT4FUS and RT4del). To determine whether loss of the last exon of FGFR3 is functionally relevant, we also made a construct in which a stop codon was introduced at the end of wild-type FGFR3 exon 18 (del19). All full-length fusions induced change to a more spindle-type cellular morphology in NIH-3T3 cells. Wild-type FGFR3 and RT4del and del19 did not alter the cell morphology (Fig. 6A). All fusions and an S249C mutant FGFR3 positive control induced anchorage independent growth with potency in the order: RT112FUS > RT4FUS > S249C > SW780FUS. RT4del behaved like wild-type FGFR3. del19 produced an intermediate phenotype, suggesting that the loss of the terminal exon per se is not potently oncogenic (Fig. 6B). All full-length fusion proteins were constitutively phosphorylated and showed no increase in phosphorylation following FGF1 stimulation (Fig. 6C and D). RT4del was not constitutively phosphorylated, and del19 showed some phosphorylation which was stimulated by FGF1. All full-length fusion proteins were more highly phosphorylated than the fully stimulated FGFR3 IIIb wild-type form.
Figure 6.
(A) Morphological changes in NIH-3T3 cells induced by ectopic expression of SW780FUS, RT112FUS and RT4 full-length (RT4FUS) fusion genes, but not with RT4-truncated form (RT4del), FGFR3IIIb, FGFR3 with exon 19 deleted (del19) or vector control. (B) Number of anchorage-independent colonies > 100 µm diameter in 15 × 1 mm2 areas formed by NIH-3T3 cells expressing FGFR3 fusion proteins and controls. (C) NIH-3T3 cells expressing fusion genes and control constructs were serum starved for 1 h, immunoprecipitated with FGFR3 extracellular antibody E and detected using FGFR3 antibody, which recognizes amino acids 25–124. The presence of FGFR3 of the appropriate size is detected in each case; anti-phosphotyrosine antibody (middle panel) shows constitutive phosphorylation of RT112FUS, SW780FUS and RT4FUS fusion genes. Chart (lower panel) shows the relative intensities of signal from the two antibodies. (D) Fold increase in phospho-FGFR3 following stimulation with FGF1, presented as phospho-FGFR3:total FGFR3 following 10 min of FGF1 stimulation. Error bars are ± 1 SE.
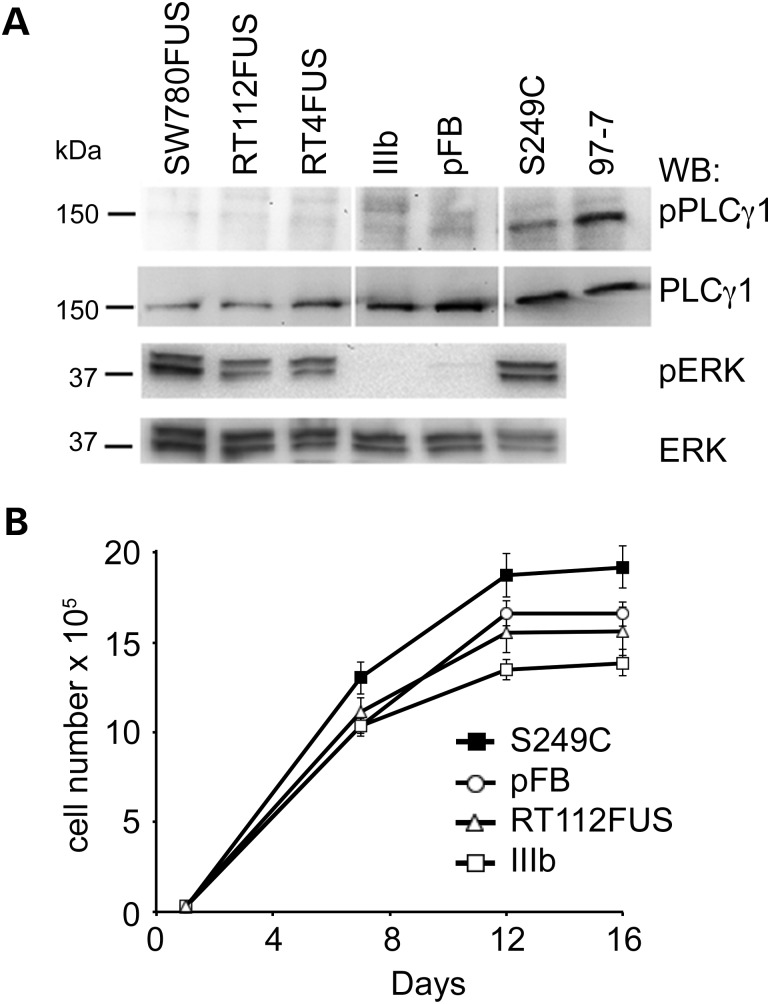
As we have shown previously that FGFR3 induces cell context-dependent signalling and phenotypic outputs (9), we also expressed the fusion genes in immortalized NHUC (TERT-NHUC). In these cells, S249C-FGFR3 activates the mitogen activated protein kinase (MAPK) pathway and phospholipase c gamma 1 (PLCγ1) and induces PLCγ1-dependent overgrowth at confluence (9). As the tyrosine residue responsible for interaction with PLCγ1 is reported to be in exon 19 (10) and all of the fusion genes have lost this, we predicted that their expression would not lead to PLCγ1 phosphorylation or overgrowth at confluence and this was confirmed (Fig. 7A and B). The ability to activate extracellular signal-regulated kinase (ERK) was retained by all active forms of FGFR3 (Fig. 7A). Fluorescence-activated cell sorting (FACS) analysis of TERT-NHUC expressing IIIb or RT112FUS fusion FGFR3 showed that both the fusion protein and IIIb forms were present at the cell surface (Supplementary Material, Fig. S3).
Figure 7.
(A) Western blots showing levels of phospho-phospholipase C gamma 1 (PLCγ1) and phospho extracellular signal-regulated kinase (ERK) in TERT-NHUC expressing FGFR3 fusion proteins and controls. pFB, vector control; TERT-NHUC S249C and 97-7 cell line containing point-mutated FGFR3 (S249C) are positive controls. The PLCγ1 images are obtained from a single blot with some lanes removed. (B) Growth curves of TERT-NHUC expressing FGFR3 fusion proteins and controls. Cells expressing S249C-FGFR3 reach a higher confluent density than those expressing FGFR3–TACC3 fusion (RT112FUS) and controls. Error bars are ± 1 SE.
DISCUSSION
FGFR3 is implicated as an oncogene in the majority (∼80%) of low-grade non-invasive (stage Ta) bladder tumours and its upregulated expression in ∼40% of invasive bladder tumours implies that it also plays an oncogenic role in these (1). Our finding of FGFR3 fusion proteins identifies an additional class of mutational events in a subset of bladder cancers with upregulated FGFR3 expression.
The cell lines RT112, RT4 and SW780 that contain fusion proteins all show low IC50 values (15, 5 and 50 nm, respectively) for the FGFR-selective inhibitor PD173074, indicating high FGFR3 dependence (2). These cell lines are widely used in preclinical assays as examples of ‘wild-type’ bladder tumour cell lines. We hypothesize that their high FGFR3 dependence and sensitivity to FGFR-targeted agents are likely to be related to the specific function(s) of the fusion genes, rather than to the overall level of FGFR3 expression. It will now be critical to examine a large series of tumours with upregulated expression of non-point-mutated FGFR3 to determine the frequencies of truly wild-type and translocation-containing cases and to test FGFR3 dependence in cell lines of each type.
The two fusion partners we have identified show no apparent homology or overlap in their reported functions. Initially, we considered that the loss of the C-terminus of FGFR3 may be sufficient to cause activation of the kinase, as it has been reported that tyrosine 770 in the final exon is inhibitory to the transforming activities of FGFR3 (10). Although truncation had a small effect on NIH-3T3 cell anchorage independence, it did not induce the high level of activation and transforming ability of the fusion proteins. Thus, it is predicted that the presence of the fusion partner is important. All the fusions contain the same FGFR3 coding sequence including the entire kinase domain, indicating that retention of kinase activity rather than the loss of the final exon is critical. It is notable that we did not identify any microhomology at the genomic breakpoints, which were different in each case.
The fusion proteins are highly activated and induce signalling via the MAPK pathway in urothelial cells. All lack the ability to activate PLCγ1, a function previously found to be important for phenotypic effect in normal urothelial cells. To date, we have not identified any phenotypic consequences induced by the fusion proteins in normal urothelial cells. We predict that they have novel kinase-related functions that are dependent on the fusion partners and these remain to be elucidated.
The findings of several FGFR3–TACC3 fusions indicate that TACC3 is a common partner. The gene is located within 48 kb of FGFR3 on 4p16.3, and this spatial proximity may favour recombination. A similar relationship exists for FGFR1 and TACC1 on 8p and indeed, an FGFR1–TACC1 fusion was identified in glioblastoma (11). TACC3 is a member of the TACC family of proteins that share a conserved C-terminal TACC domain that is involved in microtubule binding. TACC3 plays an important role in stabilization and organization of the mitotic spindle to allow proper chromosome segregation (12). Depletion results in aberrant mitotic events and cell cycle arrest (13,14) and disruption in tumours in vivo leads to regression (15), suggesting that it may represent a useful therapeutic target. Upregulated expression has been reported in several cancer types, e.g. non-small-cell lung carcinoma (16) and glioblastoma (17), and recently four FGFR3–TACC3 fusions were identified in glioblastoma (Fig. 3). Ectopic expression of one of these in Ink4A;Arf-/- mouse astrocytes revealed localization of the fusion protein at the metaphase spindle poles and its continued expression led to aneuploidy (11).
Three of the fusion transcripts identified here are identical to two reported in glioblastoma. The two UC cell lines in which FGFR3–TACC3 fusions were highly expressed have the least diversity of chromosome counts of all 43 cell lines that we have characterized. In addition to the well-studied role of TACC3 in mitotic regulation, several other potential functions have been reported. Binding to a range of other proteins including Notch proteins (18), ARNT (19), MBD2 (20) and FOG-1 (21) has been identified, suggesting that its functions are not confined to mitosis.
The second fusion partner BAIAP2L1 is known to be expressed in mouse bladder, liver, testes, heart and lung. This protein is a substrate for insulin receptor tyrosine kinase and has been shown to bind Rac and to cause actin filament bundling and plasma membrane deformation (22). It is also a phosphorylation target of Src that enhances cell migration in a Src-dependent manner (23) and also binds p53 and MDM2, modulating MDM2-mediated p53 low-level ubiquitination in unstressed cells (24). To date, we have not found any other fusions involving this gene. However, the finding of a second fusion partner apparently unrelated to TACC3 and the identification of some aberrant FGFR3 proteins in tumour samples that we have been unable to confirm as either TACC3 or BAIAP2L1 fusions suggest that further partners remain to be identified and that the function of these fusions may be heterogeneous.
Our initial impetus to study FGFR3 in so-called ‘wild-type’ bladder tumour cell lines came from their high sensitivity to FGFR-targeted agents. Although it is not clear how the function of the fusion proteins differs from that of point mutant FGFR3, the lack of activation of PLCγ1 may affect their signalling output, and the binding properties of the fusion partners may lead to some altered localization, allowing signalling in a different sub-cellular context. However, as shown here, much of the FGFR3–TACC3 fusion protein is retained at the cell surface like FGFR3 IIIb. Based on the in vitro studies that show greater FGFR-dependence of the tumour cell lines with FGFR fusions than those with point mutations (2,3), we suggest that the presence of a fusion protein may be a useful predictive biomarker in the selection of patients for FGFR-targeted therapy.
MATERIALS AND METHODS
Tumour samples
Tumour tissues were obtained with approval of the Leeds-East Research Ethics Committee and informed consent was obtained from all patients. Cold cup biopsies were snap-frozen and stored in liquid nitrogen and the remainder of the sample embedded in paraffin for diagnostic assessment.
Cell culture, constructs and infections
NHUC and telomerase immortalized NHUC (TERT-NHUC) were as described (25). The conditions for these and NIH-3T3 cells are described in the Supplementary Material. A panel of 43 UC cell lines was used (Supplementary Material, Table S1). Cell line identity was verified by short tandem repeat DNA typing using a Powerplex 16 kit (Promega). The profiles were compared with publically available data (ATCC, DSMZ) or when no reference profiles were available, these were confirmed as unique.
Wild-type and S249C mutant FGFR3 in the retroviral expression vector pFB (Stratagene) were as described (9). Further constructs were made for the SW780 fusion gene (SW780FUS), RT112 fusion gene (RT112FUS), RT4 fusion gene (RT4FUS) and RT4 deletion isoform (RT4del) (Supplementary Material). A truncated version of wild-type FGFR3 (del19) was made using a reverse primer to introduce a stop codon at the end of exon 18 (Supplementary Material, Table S2). Vectors were transfected into Phoenix A cells using TransIT-293 transfection reagent (Mirus). Cells were incubated with a retroviral supernatant containing 8 μg/ml polybrene and selected with the appropriate antibiotic after 48–72 h.
Protein extraction, western blotting and immunoprecipitation
Cell line protein extraction was as described (7). For tumour protein extraction, 10–20 × 10 μm sections were lysed in 40 μl buffer (25 mM Tris–HCl, pH7.0, 2 mM EDTA, 2% SDS) with three freeze–thaw cycles, boiled for 5 min and cleared by centrifugation at 12 700g at 4°C. Thirty micrograms of the cell line protein or 5 μl of the tumour protein was run under denaturing or non-denaturing conditions (Supplementary Material). Immunoprecipitation was as described (9). Deglycosylation used a glycoprotein deglycosylation kit (Calbiochem) according to the manufacturer's instructions.
Primary antibodies used were 4G10 anti-phosphotyrosine (Millipore); B9 anti-FGFR3 (Santa Cruz); anti-TACC3 (R&D Systems); anti-BAIAP2L1 (Abcam); anti-phosphoPLCgamma-1(Tyr783) (Cell Signaling Technology); anti-PLC gamma-1 (Cell Signalling Technology); E4 anti-phospho-ERK and K23 anti-ERK(Santa-Cruz) and anti-tubulin alpha (AbD Serotec). For conditions, see Supplementary Material.
FGF1 stimulation and measurement of phosphorylation
The cells were starved in supplement-free medium for 1 h, then stimulated with 20 ng/ml FGF1 (R&D Systems) and 1 IU/ml of heparin (Leo Labs Ltd), or heparin alone for 10 min. Protein was immunoprecipitated, run in SDS polyacrylamide gels and blotted as above. The intensity of the signal with anti-phosphotyrosine primary antibody was determined relative to B9 anti-FGFR3 primary antibody, using Quantity One software (BioRad) and compared between FGF1-stimulated and heparin-only-stimulated cells.
Fluorescence in situ hybridization
The cells were harvested and the slides prepared using standard cytogenetic techniques. BAC clones from the RP11 BAC library were obtained from the Wellcome Trust Sanger Centre (http://www.sanger.ac.uk). PCR-generated probes used BAC DNA as a template, with primers and PCR conditions as described in Supplementary Material, Table S2. BAC and PCR probes were labelled with biotin-14-dATP or digoxygenin-11-dUTP using the BioPrime DNA labelling system (Invitrogen). Chromosome-specific centromere probes (Appligene Oncor) were hybridized according to the manufacturer's instructions.
DNA and RNA extraction and cDNA synthesis
DNA was extracted either by standard phenol/chloroform extraction or using a QIAamp® DNA mini kit (Qiagen). RNA was extracted using an RNeasy kit (Qiagen). First strand cDNA was synthesized using Superscript™ II RT and random primers (Invitrogen) or the Transcriptor High Fidelity cDNA synthesis kit (Roche) according to the manufacturer's instructions. The cDNA for fusion gene partner detection was made using a tagged polyT primer with a unique 5′ sequence (Supplementary Material, Table S2) in place of random primers. Primers for RT-PCR are listed in Supplementary Material, Table S2.
Real-time reverse transcriptase PCR
Gene expression was measured in triplicate using TaqMan assays (Applied Biosystems). The results were normalized to a pool of cultured NHUC. The results from assays for different regions of FGFR3 were compared to assess whether or not there was a loss of exon 19 (Hs00997393_g1) relative to the exons 11 and 12 (Hs00997400_g1) of FGFR3. Comparison with SDHA expression (Hs00417200_m1) was used to assess the overall levels of FGFR3 expression. Analysis was carried out using 7500 analysis software (Applied Biosystems).
Anchorage independent growth
Transduced NIH-3T3 cells were cultured in medium containing 0.3% agarose with weekly feeding, after seeding in triplicate into six-well plates at 1.06 × 104 cells/cm2 on bases containing 0.6% agarose. Colonies >100 μm diameter were counted on days 21 or 22, after staining with 8 μm p-iodonitrotetrazolium violet (Sigma-Aldrich). The results were obtained from three independent assays.
Measurement of saturation density
TERT-NHUC were seeded in triplicate into six-well plates at 3.2 × 103 cells/cm2 and the medium was changed every 2 to 3 days. Triplicate wells for each were counted after 1, 7, 12 and 16 days using a Z2 Coulter analyser (Beckman Coulter). The results were obtained from three independent assays.
FACS analysis
Sub-confluent TERT-NHUC were removed from flasks by treatment with accutase (Sigma) and allowed to rest in full medium for at least 20 min before staining with monoclonal anti-human FGFR3-phycoerythrin antibody (R&D Systems) according to the manufacturer's instructions. The cells were analysed on a FacsCalibur machine (BD Biosciences) using acquisition software Cell QuestPro (BD Biosciences). Data were analysed using FlowJo (Treestar). The results were gated using forward scatter/side scatter to select intact cellular bodies.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by Yorkshire Cancer Research (grant number L367) (S.V.W.) and Cancer Research UK (grant number C6228/A5437) (C.D.H., M.A.K.). Funding to pay the Open Access publication charges for this article was provided by Yorkshire Cancer Research.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dr Darren Tomlinson for help in cloning of the fusion transcripts and to Dr Erica di Martino for providing TERT-NHUC and NIH-3T3 expressing FGFR3-S249C and advice on the assessment of downstream signalling in these cells.
Conflict of Interest statement. None declared.
REFERENCES
- 1.di Martino E., Tomlinson D.C., Knowles M.A. A decade of FGF receptor research in bladder cancer: past, present, and future challenges. Adv. Urol. 2012;2012:429213. doi: 10.1155/2012/429213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamont F.R., Tomlinson D.C., Cooper P.A., Shnyder S.D., Chester J.D., Knowles M.A. Small molecule FGF receptor inhibitors block FGFR-dependent urothelial carcinoma growth in vitro and in vivo. Br. J. Cancer. 2011;104:75–82. doi: 10.1038/sj.bjc.6606016. doi:10.1038/sj.bjc.6606016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qing J., Du X., Chen Y., Chan P., Li H., Wu P., Marsters S., Stawicki S., Tien J., Totpal K., et al. Antibody-based targeting of FGFR3 in bladder carcinoma and t(4;14)-positive multiple myeloma in mice. J. Clin. Invest. 2009;119:1216–1229. doi: 10.1172/JCI38017. doi:10.1172/JCI38017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tomlinson D.C., Hurst C.D., Knowles M.A. Knockdown by shRNA identifies S249C mutant FGFR3 as a potential therapeutic target in bladder cancer. Oncogene. 2007;26:5889–5899. doi: 10.1038/sj.onc.1210399. doi:10.1038/sj.onc.1210399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernard-Pierrot I., Brams A., Dunois-Lardé C., Caillault A., de Medina S.G.D., Cappellen D., Graff G., Thiery J.P., Chopin D., Ricol D., et al. Oncogenic properties of the mutated forms of fibroblast growth factor receptor 3b. Carcinogenesis. 2006;27:740–747. doi: 10.1093/carcin/bgi290. doi:10.1093/carcin/bgi290. [DOI] [PubMed] [Google Scholar]
- 6.Guagnano V., Furet P., Spanka C., Bordas V., Le Douget M., Stamm C., Brueggen J., Jensen M.R., Schnell C., Schmid H., et al. Discovery of 3-(2,6-dichloro-3,5-dimethoxy-phenyl)-1-{6-[4-(4-ethyl-piperazin- 1-yl)-phenylamin o]-pyrimidin-4-yl}-1-methyl-urea (NVP-BGJ398), a potent and selective inhibitor of the fibroblast growth factor receptor family of receptor tyrosine kinase. J. Med. Chem. 2011;54:7066–7083. doi: 10.1021/jm2006222. doi:10.1021/jm2006222. [DOI] [PubMed] [Google Scholar]
- 7.Tomlinson D.C., L'Hôte C.G., Kennedy W., Pitt E., Knowles M.A. Alternative splicing of fibroblast growth factor receptor 3 produces a secreted isoform that inhibits fibroblast growth factor-induced proliferation and is repressed in urothelial carcinoma cell lines. Cancer Res. 2005;65:10441–10449. doi: 10.1158/0008-5472.CAN-05-1718. doi:10.1158/0008-5472.CAN-05-1718. [DOI] [PubMed] [Google Scholar]
- 8.Booth D.G., Hood F.E., Prior I.A., Royle S.J. A TACC3/ch-TOG/clathrin complex stabilises kinetochore fibres by inter-microtubule bridging. EMBO J. 2011;30:906–919. doi: 10.1038/emboj.2011.15. doi:10.1038/emboj.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.di Martino E., L'Hôte C.G., Kennedy W., Tomlinson D.C., Knowles M.A. Mutant fibroblast growth factor receptor 3 induces intracellular signaling and cellular transformation in a cell type- and mutation-specific manner. Oncogene. 2009;28:4306–4316. doi: 10.1038/onc.2009.280. doi:10.1038/onc.2009.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hart K.C., Robertson S.C., Donoghue D.J. Identification of tyrosine residues in constitutively activated fibroblast growth factor receptor 3 involved in mitogenesis, Stat activation, and phosphatidylinositol 3-kinase activation. Mol. Biol. Cell. 2001;12:931–942. doi: 10.1091/mbc.12.4.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh D., Chan J.M., Zoppoli P., Niola F., Sullivan R., Castano A., Liu E.M., Reichel J., Porrati P., Pellegatta S., et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science. 2012;337:1231–1235. doi: 10.1126/science.1220834. doi:10.1126/science.1220834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hood F.E., Royle S.J. Pulling it together: The mitotic function of TACC3. Bioarchitecture. 2011;1:105–109. doi: 10.4161/bioa.1.3.16518. doi:10.4161/bioa.1.3.16518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schneider L., Essmann F., Kletke A., Rio P., Hanenberg H., Wetzel W., Schulze-Osthoff K., Nurnberg B., Piekorz R.P. The transforming acidic coiled coil 3 protein is essential for spindle-dependent chromosome alignment and mitotic survival. J. Biol. Chem. 2007;282:29273–29283. doi: 10.1074/jbc.M704151200. doi:10.1074/jbc.M704151200. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt S., Schneider L., Essmann F., Cirstea I.C., Kuck F., Kletke A., Janicke R.U., Wiek C., Hanenberg H., Ahmadian M.R., et al. The centrosomal protein TACC3 controls paclitaxel sensitivity by modulating a premature senescence program. Oncogene. 2010;29:6184–6192. doi: 10.1038/onc.2010.354. doi:10.1038/onc.2010.354. [DOI] [PubMed] [Google Scholar]
- 15.Yao R., Natsume Y., Saiki Y., Shioya H., Takeuchi K., Yamori T., Toki H., Aoki I., Saga T., Noda T. Disruption of Tacc3 function leads to in vivo tumor regression. Oncogene. 2012;31:135–148. doi: 10.1038/onc.2011.235. doi:10.1038/onc.2011.235. [DOI] [PubMed] [Google Scholar]
- 16.Jung C.K., Jung J.H., Park G.S., Lee A., Kang C.S., Lee K.Y. Expression of transforming acidic coiled-coil containing protein 3 is a novel independent prognostic marker in non-small cell lung cancer. Pathol. Int. 2006;56:503–509. doi: 10.1111/j.1440-1827.2006.01998.x. doi:10.1111/j.1440-1827.2006.01998.x. [DOI] [PubMed] [Google Scholar]
- 17.Duncan C.G., Killela P.J., Payne C.A., Lampson B., Chen W.C., Liu J., Solomon D., Waldman T., Towers A.J., Gregory S.G., et al. Integrated genomic analyses identify ERRFI1 and TACC3 as glioblastoma-targeted genes. Oncotarget. 2010;1:265–277. doi: 10.18632/oncotarget.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bargo S., Raafat A., McCurdy D., Amirjazil I., Shu Y., Traicoff J., Plant J., Vonderhaar B.K., Callahan R. Transforming acidic coiled-coil protein-3 (Tacc3) acts as a negative regulator of Notch signaling through binding to CDC10/Ankyrin repeats. Biochem. Biophys. Res. Commun. 2010;400:606–612. doi: 10.1016/j.bbrc.2010.08.111. doi:10.1016/j.bbrc.2010.08.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Partch C.L., Gardner K.H. Coactivators necessary for transcriptional output of the hypoxia inducible factor, HIF, are directly recruited by ARNT PAS-B. Proc. Natl Acad. Sci. USA. 2011;108:7739–7744. doi: 10.1073/pnas.1101357108. doi:10.1073/pnas.1101357108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Angrisano T., Lembo F., Pero R., Natale F., Fusco A., Avvedimento V.E., Bruni C.B., Chiariotti L. TACC3 mediates the association of MBD2 with histone acetyltransferases and relieves transcriptional repression of methylated promoters. Nucleic Acids Res. 2006;34:364–372. doi: 10.1093/nar/gkj400. doi:10.1093/nar/gkj400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garriga-Canut M., Orkin S.H. Transforming acidic coiled-coil protein 3 (TACC3) controls friend of GATA-1 (FOG-1) subcellular localization and regulates the association between GATA-1 and FOG-1 during hematopoiesis. J. Biol. Chem. 2004;279:23597–23605. doi: 10.1074/jbc.M313987200. doi:10.1074/jbc.M313987200. [DOI] [PubMed] [Google Scholar]
- 22.Millard T.H., Dawson J., Machesky L.M. Characterisation of IRTKS, a novel IRSp53/MIM family actin regulator with distinct filament bundling properties. J. Cell Sci. 2007;120:1663–1672. doi: 10.1242/jcs.001776. doi:10.1242/jcs.001776. [DOI] [PubMed] [Google Scholar]
- 23.Chen G., Li T., Zhang L., Yi M., Chen F., Wang Z., Zhang X. Src-stimulated IRTKS phosphorylation enhances cell migration. FEBS Lett. 2011;585:2972–2978. doi: 10.1016/j.febslet.2011.08.005. doi:10.1016/j.febslet.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Wang K.S., Chen G., Shen H.L., Li T.T., Chen F., Wang Q.W., Wang Z.Q., Han Z.G., Zhang X. Insulin receptor tyrosine kinase substrate enhances low levels of MDM2-mediated p53 ubiquitination. PLoS One. 2011;6:e23571. doi: 10.1371/journal.pone.0023571. doi:10.1371/journal.pone.0023571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chapman E.J., Hurst C.D., Pitt E., Chambers P., Aveyard J.S., Knowles M.A. Expression of hTERT immortalises normal human urothelial cells without inactivation of the p16/Rb pathway. Oncogene. 2006;25:5037–5045. doi: 10.1038/sj.onc.1209513. doi:10.1038/sj.onc.1209513. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.