Abstract
RhoA is overexpressed in human cancer and contributes to aberrant cell motility and metastatic progression; however, regulatory mechanisms controlling RhoA activity in cancer are poorly understood. Neuroepithelial transforming gene 1 (Net1) is a RhoA guanine nucleotide exchange factor that is overexpressed in human cancer. It encodes two isoforms, Net1 and Net1A, which cycle between the nucleus and plasma membrane. Net1 proteins must leave the nucleus to activate RhoA, but mechanisms controlling the extranuclear localization of Net1 isoforms have not been described. Here, we show that Rac1 activation causes relocalization of Net1 isoforms outside the nucleus and stimulates Net1A catalytic activity. These effects do not require Net1A catalytic activity, its pleckstrin homology domain, or its regulatory C terminus. We also show that Rac1 activation protects Net1A from proteasome-mediated degradation. Replating cells on collagen stimulates endogenous Rac1 to relocalize Net1A, and inhibition of proteasome activity extends the duration and magnitude of Net1A relocalization. Importantly, we demonstrate that Net1A, but not Net1, is required for cell spreading on collagen, myosin light chain phosphorylation, and focal adhesion maturation. These data identify the first physiological mechanism controlling the extranuclear localization of Net1 isoforms. They also demonstrate a previously unrecognized role for Net1A in regulating cell adhesion.
INTRODUCTION
Rho family small G proteins are critical regulators of actin cytoskeletal organization. In this role, they impact many aspects of cell function, including cell motility, extracellular matrix invasion, and oncogenic transformation (1–3). Rho GTPases fulfill this role by acting at the plasma membrane as molecular switches, cycling between their active, GTP-bound, and inactive, GDP-bound states. In their active forms, Rho proteins stimulate intracellular signaling by interacting with downstream effector proteins.
The best-characterized Rho proteins are Cdc42, Rac1, and RhoA, each of which makes important contributions to cell motility. For example, RhoA activation stimulates actomyosin contraction by promoting phosphorylation of the regulatory myosin light chain subunit (4, 5). This increased contractility drives F-actin stress fiber and focal adhesion formation (6). Within a migrating cell, RhoA is activated at both the leading and trailing edges (7, 8). At the leading edge, RhoA activation stimulates focal adhesion maturation, cortical actin polymerization, and retrograde actin flow (9, 10). At the trailing edge, RhoA activation promotes focal adhesion disassembly, thereby allowing trailing edge retraction (11).
Because of its crucial role in controlling cell motility and invasion, RhoA activation is thought to be a critical component of human cancer cell tumorigenic and invasive capacities. RhoA subfamily proteins are overexpressed in human cancers; however, unlike the related Ras GTPases, they are not activated by mutation (12–15). Thus, it is commonly believed that increased RhoA activation in human cancer occurs through alterations in the activity of up- and downstream regulatory proteins. The GTP activation cycle of Rho proteins is controlled by two large families of proteins known as GTPase-activating proteins (RhoGAPs) and guanine nucleotide exchange factors (RhoGEFs) (16, 17). RhoGAPs terminate downstream signaling by stimulating the intrinsic GTPase activity of Rho proteins, while RhoGEFs respond to extracellular stimuli to catalyze Rho protein exchange of GDP for GTP, thereby activating downstream signaling.
The neuroepithelial transforming gene 1 (Net1) is a RhoGEF specific for the RhoA subfamily of small G proteins. NET1 transcripts are overexpressed in a number of human cancers (18–20), and we have shown that coexpression of Net1 and β4 integrin is prognostic for decreased distant metastasis-free survival in estrogen receptor-positive breast cancer patients (21). Two isoforms of Net1 exist in most cells, known as Net1 and Net1A, which are expressed from different promoters and contain unique N-terminal regulatory domains (22, 23). By using small interfering RNAs (siRNAs) that target both Net1 isoforms, NET1 gene expression has recently been shown to be important for gastric and breast cancer cell motility and extracellular matrix invasion (18, 24, 25). NET1 expression is also necessary for cytoskeletal rearrangements associated with transforming growth factor β (TGF-β) signaling. Specifically, interfering with the function of both Net1 isoforms blocks TGF-β-stimulated RhoA activation and actin cytoskeletal reorganization, and siRNA-mediated knockdown of Net1A inhibits TGF-β-stimulated epithelial-mesenchymal transformation (EMT) (26–28). Thus, Net1 isoforms are emerging as important regulators of EMT and cell motility in both normal development and cancer progression.
A key aspect regulating the cellular activity of Net1 isoforms appears to be through control of their subcellular localization. Net1 proteins are unusual among RhoGEFs in that they localize to cell nuclei. For the Net1 isoform, this is at least partly due to the presence of two nuclear localization signal (NLS) sequences in its unique N-terminal regulatory domain (29). Mechanisms controlling the nuclear localization of Net1A are less well defined, but they clearly rely on the presence of an N-terminal region shared with Net1 (23). Importantly, Net1 isoforms must translocate to the plasma membrane to stimulate RhoA activation and actin cytoskeletal reorganization (23, 29). Thus, identification of regulatory mechanisms controlling the subcellular localization of Net1 proteins is crucial to understanding their function in normal and cancer cells.
In the present work, we demonstrate that activation of the small G protein Rac1 stimulates the accumulation of Net1 isoforms outside the nucleus. It also causes plasma membrane accumulation of Net1A, stimulates its activity toward RhoA, and protects Net1A from proteasome-mediated degradation. Furthermore, we demonstrate that the process of cell spreading results in a transient, Rac1-dependent relocalization of Net1A outside the nucleus and that inhibition of proteasome function extends the duration of Net1A extranuclear localization. Moreover, we demonstrate that Net1A expression is required for efficient cell spreading and is necessary for myosin light chain phosphorylation and focal adhesion maturation. These data provide for the first time an understanding of how Net1 isoform relocalization outside the nucleus is controlled and reveal a unique role for Net1A in regulating the process of cell spreading. In addition, this work demonstrates a clear functional distinction between Net1 isoforms, in both their regulation and downstream signaling activities within the cell.
MATERIALS AND METHODS
Cell culture and reagents.
MCF7 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with high glucose and glutamine (Sigma), supplemented with 10% fetal bovine serum (FBS; Invitrogen) and penicillin-streptomycin (HyClone) in 10% CO2. MDA-MB-231 cells were cultured in DMEM–F-12 plus 10% FBS in 5% CO2. Plasmids were transfected with Lipofectamine Plus reagent (Invitrogen) according to the manufacturer's instructions. siRNA transfections were performed with INTERFERin (PolyPlus). All siRNAs (Sigma) targeted human gene products and comprised the following sequences: Rac1-1, 5′-AAGGAGAUUGGUGCUGUAAAA-dTdT-3′; Rac1-2, 5′-AACCUUUGUACGCUUUGCUCA-dTdT-3′; Net1, 5′-GAAAACGCAGAGAGAAAGAUU-3′; Net1A #1, 5′-GGACCAUACGAGUCCUAGAUU-3′; Net1A #2, 5′-GCAUGGUGGCACAUGAUGAUU-3′; Net1/Net1A, 5′-GAGUGGACAUAAACUUUAC-dTdT-3′. The control siRNA sequence, 5′-GAUCAUACGUGCGAUCAGA-dTdT-3′, corresponds to a scrambled sequence targeting p21CIP1. Cells were assayed 96 h (Rac1-silenced cells) or 72 h (Net1-, Net1A-, Net1/Net1A-silenced cells) posttransfection. For experiments that included both siRNA and plasmid transfections, relevant plasmids were transfected 24 or 48 h after siRNA transfections.
Hemagglutinin (HA)-tagged mouse Net1 and Net1A were contained in pEFHA (23). Myc-tagged constitutively active forms of Pak1 (L107F), Rac1 (V12Rac1), Cdc42 (V12Cdc42), and RhoA (L63RhoA) were as described previously (30, 31). V12Rac1-C189S (-SAAX) was created by PCR using Pfu polymerase (Agilent), as were the Net1A point mutants W438L, S98A/S99A, and S98E/S99E and the deletion mutant Net1A 1–307. In all cases, the entire cDNA insert was sequenced to confirm correct amplification. The plasma membrane marker PM-mCherry was a generous gift from Guangwei Du (UT—Houston) (32).
Mouse collagen IV was obtained from BD Biosciences. Commercial antibodies used were anti-Rac1, anti-Cdc42, and antipaxillin from BD Biosciences, anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH), anti-glutathione S-transferase (anti-GST), anti-Net1, anti-HA, anti-superoxide dismutase 1 (anti-SOD1), anti-Na+/K+-ATPase (ATP1A1), and rabbit anti-Myc epitope from Santa Cruz Biotechnology, anti-RhoA from Cytoskeleton, anti-H3, mouse anti-pMLC2, and anti-phospho-FAK (Y397) from Cell Signaling, mouse anti-Myc epitope 9E10 (NCCC) and anti-α-tubulin from Sigma, and anti-THOC1 from GeneTex. Western blots were visualized with horseradish peroxidase-conjugated secondary antibodies (Kirkegaard & Perry Laboratories) via enhanced chemiluminescence (ECL). For immunofluorescence microscopy, Cy2- and Cy3-conjugated anti-mouse and anti-rabbit secondary antibodies were obtained from Jackson ImmunoResearch. Tetramethyl rhodamine isocyanate-phalloidin and 4′,6-diamidino-2-phenylindole (DAPI) were from Sigma. Alexa Fluor 647-phalloidin and Alexa Fluor 488-conjugated anti-mouse antibody were from Invitrogen.
Subcellular fractionation.
MCF7 cells were transfected with HA-Net1A, with or without Myc-V12Rac1. MDA-MB-231 cells were transfected with control or Rac1-specific siRNAs. For fractionation, cells were washed in phosphate-buffered saline (PBS) and scraped into cold hypotonic lysis buffer (50 mM Tris-HCl [pH 7.4], 1 mM EDTA, 1 mM dithiothreitol [DTT], 50 mM NaF, 80 mM β-glycerophosphate, 1 mM sodium orthovanadate, 10 μg/ml pepstatin A, 10 μg/ml leupeptin, 2 μg/ml aprotinin, 1 mM phenylmethylsulfonyl fluoride [PMSF]). The cells were allowed to swell on ice for 10 min and then lysed with 20 strokes in a Dounce homogenizer. Nuclei and unbroken cells were pelleted by centrifugation (1,500 × g, 10 min, 4°C). The pellet was washed once with hypotonic lysis buffer and solubilized with 2% SDS buffer (2% SDS, 20 mM Tris-HCl [pH 8.0], 50 mM NaF, 80 mM β-glycerophosphate, 1 mM sodium orthovanadate, 10 μg/ml pepstatin A, 10 μg/ml leupeptin, 2 μg/ml aprotinin, 1 mM PMSF). DNA within the pellet was fragmented by sonication. The supernatant from the initial centrifugation step was separated into membrane and cytosol fractions by high-speed centrifugation (100,000 × g, 30 min, 4°C). The membrane pellet was resolubilized with 2% SDS buffer. Protein concentrations were determined by a bicinchoninic acid (BCA) assay (Pierce), and equal amounts of protein were resolved by SDS-PAGE. After transfer to a polyvinylidene difluoride (PVDF) membrane, the presence of the indicated proteins was assessed by Western blotting.
Half-life determinations.
MCF7 cells were transfected with HA-Net1A with or without Myc-V12Rac1 and then treated with 10 μg/ml cycloheximide (Fisher) for the indicated times. The cells were washed with PBS and lysed in 2% SDS buffer, and protein concentrations were determined via a BCA assay. Equal amounts of protein were resolved by SDS-PAGE and transferred to a PVDF membrane. Levels of HA-Net1A and other proteins were determined by Western blotting and normalized to α-tubulin. Data were plotted using Prism software and fitted using a single-phase exponential decay curve or linear regression, where indicated.
Net1 activity assays.
G17A-RhoA in pGEX-KG was created by PCR and verified by DNA sequencing. Expression and purification of GST or GST-A17RhoA protein were as previously described (33). Briefly, cultures of Escherichia coli BL21(DE3) bearing the relevant plasmids were cultured to an optical density at 600 nm (OD600) of 0.8, and protein expression was induced for 12 to 16 h at room temperature following the addition of 50 μM isopropyl-β-d-thiogalactopyranoside (IPTG). Bacterial cells were lysed in buffer containing 20 mM HEPES (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 1% Triton X-100, 1 mM DTT, 1 mM PMSF, 1 mg/ml lysozyme (Fisher), and 10 μg/ml each of aprotinin, leupeptin, and pepstatin A, and insoluble material was pelleted by centrifugation (20,000 × g, 30 min, 4°C). GST or GST-A17RhoA in the soluble fraction was purified by incubation with glutathione-agarose beads (Sigma) for 1 h at 4°C, followed by two washes with lysis buffer and two washes with lysis buffer lacking Triton X-100 and protease inhibitors. Protein purity was assessed by Coomassie staining, and concentrations were determined by BCA assay. Proteins were left attached to the beads and snap-frozen in aliquots.
Active Net1 pulldown assays were performed essentially as described previously (33). Briefly, cells were washed with PBS and lysed in lysis buffer (20 mM HEPES [pH 7.5], 150 mM NaCl, 5 mM MgCl2, 1% Triton X-100, 1 mM DTT, 1 mM PMSF, and 10 μg/ml each of aprotinin, leupeptin, and pepstatin A), sonicated for 30 s, and clarified by centrifugation (16,100 × g, 10 min, 4°C). Lysate concentrations were determined by BCA assay, and equal amounts of lysate were mixed for 1 h at 4°C with 20 μg of GST or GST-A17RhoA beads. Beads were pelleted by centrifugation and washed 3 times in lysis buffer, resuspended in 25 μl Laemmli sample buffer, boiled for 5 min, separated by SDS-PAGE, and transferred to a PVDF membrane for Western blot analysis.
Rac1 activity assays.
Rac1 activity was measured by GST pulldown using the p21-binding domain (PBD) of Pak1 cloned into pGEX-2T (Catherine Denicourt, UT—Houston). Prokaryotic expression of the pGEX-2T/PBD construct was performed as described previously (34). Briefly, E. coli BL21(DE3) cells containing pGEX-KG or pGEX-2T/PBD constructs were grown to an OD600 of 0.8, and expression of the GST or GST-PBD proteins was induced with 400 μM IPTG for 3 h at 30°C. Bacterial cells were pelleted by centrifugation and resuspended in buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 1 mg/ml lysozyme, and 10 μg/ml each of aprotinin, leupeptin, and pepstatin A. After sonication, insoluble material was pelleted by centrifugation (12,000 × g, 10 min, 4°C), and soluble proteins were incubated with glutathione-agarose beads for 1 h at 4°C. Beads were then pelleted by centrifugation and washed using buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, 1 mM PMSF, and 10 μg/ml each of aprotinin, leupeptin, and pepstatin A. Protein purity was assessed by Coomassie staining, and concentrations were determined by BCA assay. Proteins were left attached to the beads and snap-frozen in aliquots.
Active Rac1 pulldown experiments were performed as described previously (34). Briefly, suspended and adherent cells were washed with PBS and lysed in lysis buffer (50 mM Tris-HCl [pH 7.5], 200 mM NaCl, 10 mM MgCl2, 1% NP-40, 5% glycerol, 1 mM PMSF, and 10 μg/ml each of aprotinin, leupeptin, and pepstatin A) and incubated on ice for 5 min. Insoluble material was pelleted by centrifugation (13,000 × g, 10 min, 4°C). Lysate concentrations were determined by BCA assay, and equal amounts were incubated with GST or GST-PBD beads for 1 h at 4°C. The beads were pelleted and washed 3 times in wash buffer (25 mM Tris-HCl [pH 7.5], 40 mM NaCl, 30 mM MgCl2, 1% NP-40, 1 mM DTT, 1 mM PMSF, and 10 μg/ml each of aprotinin, leupeptin, and pepstatin A), followed by two washes with wash buffer lacking NP-40. Lysates were subsequently prepared for SDS-PAGE as described above.
Cell spreading assays.
MCF7 cells were serum starved for 16 h in DMEM supplemented with antibiotics and 0.5% FBS, then trypsinized, washed three times with PBS, and resuspended in DMEM plus 0.5% delipidated bovine serum albumin (BSA; Sigma) for 1 h at 37°C (35). Cells were then replated on tissue culture dishes or glass coverslips previously coated with collagen (10 μg/ml). Cells were harvested at the indicated times for biochemical or microscopic analysis. Where noted, cells were resuspended in the presence 10 μM MG132 (Calbiochem) or an equivalent volume of dimethyl sulfoxide (DMSO).
Immunofluorescence microscopy.
Cells grown or plated on collagen-coated coverslips were washed in PBS, fixed in 4.0% paraformaldehyde (Electron Microscopy) in PBS for 5 min at 37°C, and permeabilized with 0.2% Triton X-100 in PBS for 5 min at room temperature. Coverslips were then washed in PBS plus 0.1% Tween 20 (PBST), followed by blocking with 1% BSA in PBST for 30 min. Cells were incubated with primary antibody diluted to 1 to 3 μg/ml in PBST plus 1% BSA for 1 h at 37°C. Coverslips were washed 3 times in PBST for 5 min and incubated with secondary antibodies diluted to 0.5 to 2 μg/ml in PBST plus 1% BSA for 1 h at 37°C. For pMLC staining, cells were fixed in 2% paraformaldehyde in PBS for 15 min at room temperature and permeabilized in 0.2% Triton X-100 in PBS for 5 min at room temperature. Coverslips were then blocked in 3% BSA in PBS for 30 min at room temperature, followed by incubation for 1 h at 37°C with mouse anti-pMLC2 diluted in PBS plus 3% BSA. Coverslips were incubated with secondary antibodies diluted in 3% BSA in PBS for 1 h at room temperature and then washed in PBS and rinsed with deionized H2O. All other coverslips were washed with PBST and rinsed in deionized H2O. All coverslips were mounted on slides with FluorSave reagent (Calbiochem). Epifluorescence images were captured with a Zeiss Axioskop 40 microscope equipped with a Zeiss AxioCam MRm MC100 Spot digital camera and AxioVision software. Confocal images were taken using the Nikon A1R confocal system. For quantitative analysis, images were serially acquired with the same illumination and exposure parameters, and the average fluorescence intensity in regions of interest for each transfected cell was determined using ImageJ software. The ratio of cytosolic to nuclear HA-Net1A was determined as the total fluorescence intensity of staining for HA minus the intensity of HA staining in the nuclear region as defined by DAPI staining, divided by the intensity in the nucleus. The total cell area of MCF7 cells was determined by costaining with phalloidin and was quantified using ImageJ software. Focal adhesions were quantified with ImageJ, using dual staining for paxillin and phospho-Y397 FAK. Cell shape was defined as the length of the cell between the two most distant points divided by the width of the cell perpendicular to the axis defined by those points.
For Net1A colocalization studies with PM-Cherry, MCF7 cells grown on glass coverslips were cotransfected with expression plasmids for HA-Net1A and PM-Cherry (plasma membrane marker), with or without Myc-V12Rac1. After 48 h, the coverslips were fixed in 4% paraformaldehyde in PBS for 20 min at room temperature. Cells were permeabilized with 0.5% Triton X-100 in PBS for 10 min. After washing with PBST, the cells were incubated in primary and secondary antibodies as described above and mounted on slides with FluorSave reagent. Confocal images were taken using the Nikon A1R confocal system in Galvano mode. z-stack images were acquired in 0.4-μm steps by using the 60× Apo TIRF/1.49-numerical-aperture oil objective lens and visualized with NIS Elements software (Nikon).
RESULTS
Active Rac1 coexpression preferentially stimulates Net1A relocalization to the plasma membrane and Net1A activation.
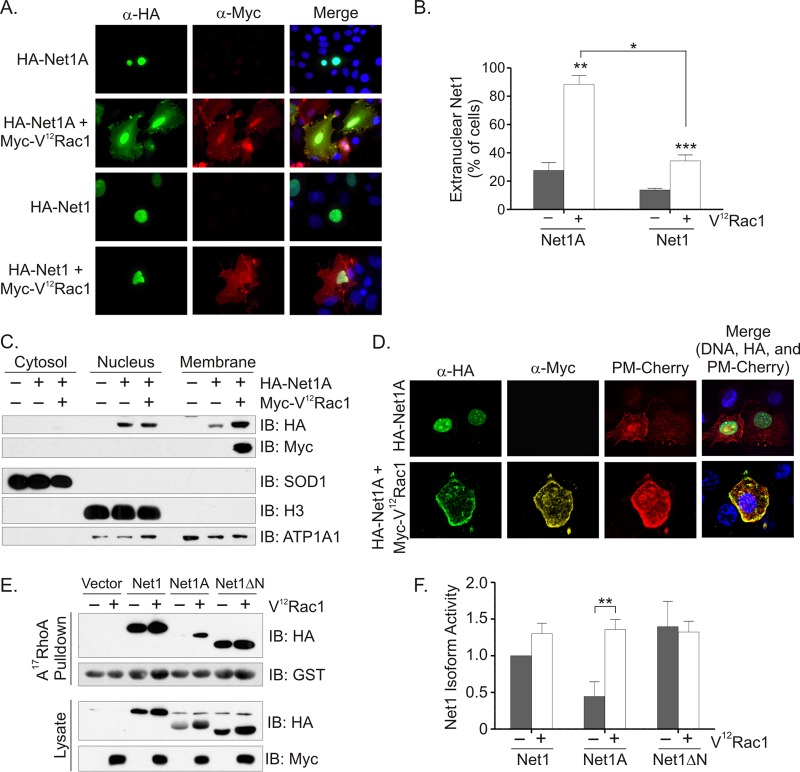
We and others have shown that Net1 isoforms must be localized outside the nucleus to stimulate RhoA activation and actin cytoskeletal rearrangement (23, 29). In order to find candidate proteins that control Net1 isoform relocalization, we cotransfected MCF7 breast cancer cells with various regulators of Rho GTPase signaling pathways plus Net1 or Net1A. The localization of Net1 proteins was then assessed by immunofluorescence microscopy. In these assays, we transfected HA epitope-tagged Net1 isoforms, because antibodies suitable for detection of endogenous Net1 isoforms by immunofluorescence were not available. We found that coexpression of constitutively active Rac1 (V12Rac1) caused a dramatic relocalization of Net1A outside the nucleus. Although significant, this effect was less apparent for the Net1 isoform (Fig. 1A). Quantification of these results demonstrated that coexpression of V12Rac1 caused extranuclear localization of Net1A and Net1 in nearly 90% and 40% of the cells, respectively (Fig. 1B). Because the effect on Net1A localization was greater, we focused on understanding how Net1A function was impacted by Rac1.
Fig 1.
Coexpression of V12Rac1 relocalizes Net1A to the plasma membrane. (A) MCF7 cells were transfected with expression plasmids for HA-Net1 or HA-Net1A, with or without Myc-V12Rac1. Two days later the cells were fixed and stained for HA epitope (green), Myc epitope (red), and DNA (blue). Results of a representative experiment are shown. (B) Quantification of subcellular localization of Net1 proteins. Shown are the averages of at least three independent experiments. Error bars show standard errors of the means. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) Subcellular fractionation of MCF7 cells transfected with expression plasmids for HA-Net1A alone or with Myc-V12Rac1. Proteins were analyzed by Western blotting with the indicated antibodies. SOD-1, histone H3 (H3), and Na+/K+-ATPase 1 (ATP1A1) were monitored as controls. Shown are results of a representative experiment. (D) MCF7 cells were transfected with expression plasmids for HA-Net1A and the plasma membrane marker PM-Cherry, without or with Myc-V12Rac1. The cells were fixed and stained for HA-Net1A (green), Myc-V12Rac1 (yellow), and DNA (blue) and analyzed by confocal microscopy. Shown are z-plane images from a representative experiment. (E) MCF7 cells were transfected with expression plasmids for HA-Net1, HA-Net1A, or HA-Net1ΔN, with or without Myc-V12Rac1. The activity of Net1 protein was assessed in a GST-A17RhoA pulldown assay. (F) Quantification of Net1 protein activity from GST-A17RhoA pulldown assays. Averages are from three independent experiments. Error bars are standard errors of the means. **, P < 0.01.
Relocalized Net1A appeared to accumulate in the cell periphery (Fig. 1A). To ascertain whether this reflected an increase in membrane localization, we used subcellular fractionation to biochemically assess its localization in cells coexpressing active Rac1. In these assays, we observed a significant increase in the amount of HA-Net1A in the membrane fraction in V12Rac1-coexpressing cells (Fig. 1C). Because this membrane fraction also contains cytoskeletal elements, we further assessed Net1A localization by testing for colocalization with the plasma membrane marker PM-Cherry. PM-Cherry contains the N-terminal myristoylation and palmitoylation domain of the Lyn tyrosine kinase, which targets it to the plasma membrane, fused to the fluorescent protein mCherry (32). MCF7 cells were cotransfected with PM-Cherry plus Net1A, with or without V12Rac1, and Net1A localization was assessed by confocal microscopy. As shown in Fig. 1D, coexpression of V12Rac1 caused clear colocalization of Net1A with the plasma membrane marker PM-Cherry. Taken together, these data indicate that active Rac1 stimulates the relocalization of the Net1A isoform to the plasma membrane.
Because relocalized Net1A would be expected to be active toward RhoA, we tested whether interaction with A17RhoA, which is a nucleotide-free version of RhoA that binds tightly only to active RhoGEFs (33). MCF7 cells were transfected with Net1, Net1A, or the constitutively active deletion mutant Net1ΔN (36), with or without V12Rac1. Two days later, the cells were lysed and incubated with GST-A17RhoA bound to glutathione-agarose. Net1 proteins bound to GST-A17RhoA, or in the cell lysates, were detected by Western blotting. Net1 isoform activity was quantified by dividing the amount of Net1 protein in the pulldown fraction versus that in the lysate. Although V12Rac1 coexpression tended to increase expression of all Net1 proteins, quantification of these protein levels clearly demonstrated that active Rac1 coexpression significantly increased Net1A activity (Fig. 1E and F). On the other hand, both Net1 and Net1ΔN displayed a relatively high degree of activity that was not significantly enhanced by coexpression of V12Rac1. Therefore, these data indicate that Rac1-stimulated relocalization of Net1A is accompanied by a robust increase in Net1A activity.
Rac1 and Cdc42 can both stimulate Net1A relocalization.
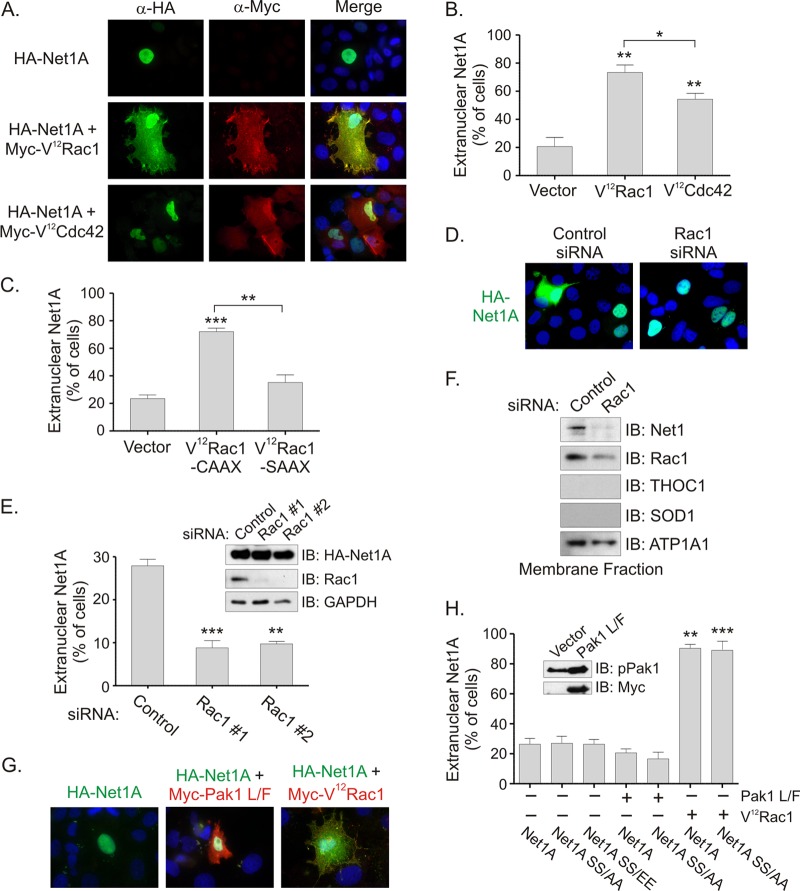
Because Rac1 shares many effector proteins with the related GTPase Cdc42 (37), we tested whether coexpression of constitutively active Cdc42 stimulated Net1A relocalization. MCF7 cells were transfected with Net1A alone or with constitutively active Rac1 or constitutively active Cdc42 (V12Cdc42). The cells were fixed only 1 day after transfection, as expression of V12Cdc42 for longer periods of time was toxic to MCF7 cells (not shown). In these experiments, we observed that coexpression of either V12Rac1 or V12Cdc42 caused Net1A relocalization, although V12Rac1 was more efficient in this regard (Fig. 2A and B). Coexpression of constitutively active RhoA (L63RhoA) did not affect Net1A localization (data not shown). We then examined whether membrane targeting of Rac1 was required for effects on Net1A localization by coexpressing a V12Rac1 mutant in which the geranylgeranylated cysteine is mutated to serine (V12Rac1-SAAX) (38–40). As shown in Fig. 2C, mutation of this residue this completely inhibited the ability of V12Rac1 to stimulate Net1A relocalization. Thus, these data indicate that both Rac1 and Cdc42 are capable of stimulating Net1A plasma membrane localization and that membrane localization of Rac1 is required for this effect. Because Rac1 was somewhat more effective than Cdc42 at stimulating Net1A relocalization, these results may also indicate that Rac1 couples to the relevant effector more efficiently than Cdc42.
Fig 2.
Net1A relocalization in MCF7 cells is regulated by Rac1 and is not controlled by Pak1. (A) MCF7 cells were transfected with expression plasmids for HA-Net1A (green), with or without Myc-V12Rac1 or Myc-V12Cdc42 (both red). Results from a representative experiment are shown. (B) Quantification of Net1A subcellular localization in V12Rac1- or V12Cdc42-transfected cells. Shown are the averages from three independent experiments. *, P < 0.05; **, P < 0.01. (C) MCF7 cells were transfected with HA-Net1A plus vector, V12Rac1-CAAX, or V12Rac1-SAAX and then fixed and stained for HA-Net1A localization. Shown is the quantification from three independent experiments. **, P < 0.01; ***, P < 0.001. (D) MCF7 cells were transfected with control or Rac1-specific siRNAs. One day later, they were retransfected with an expression plasmid for HA-Net1A. Two days later, the cells were fixed and stained for HA-Net1A (green) and DNA (blue). Shown are the results of a representative experiment. (E) Quantification of effects of Rac1 knockdown on HA-Net1A localization. Shown are the averages of three independent experiments. Insets are Western blots for HA-Net1A and Rac1 expression. **, P < 0.01; ***, P < 0.001. (F) MDA-MB-231 cells were transfected with control or Rac1-specific siRNAs. Three days later, the membrane fraction was isolated by subcellular fractionation. The presence of Net1 proteins and controls were detected by Western blotting. THOC1 is a nuclear matrix protein. (G) MCF7 cells were transfected with expression plasmids for HA-Net1A (green), alone or together with constitutively active Myc-Pak1-L107F or Myc-V12Rac1 (red). Shown are results from a representative experiment. (H) Quantification of localization of wild-type and mutant Net1A proteins, without or with Pak1 L/F or V12Rac1. Shown are the averages of three independent experiments. Error bars are standard errors of the means. **, P < 0.01; ***, P < 0.001. The inset shows Western blots for activated Pak1 and the Myc epitope tag in control or Myc-Pak1 L107F-transfected cells.
Endogenous Rac1 controls Net1A localization.
Net1A localizes outside the nucleus in 20% to 30% of MCF7 cells growing in FBS (Fig. 1B and 2B). To determine whether this was due to endogenous Rac1 activity, MCF7 cells were transfected with control or Rac1-specific siRNAs. One day later, the cells were transfected with a Net1A expression plasmid, and 2 days after that the cells were fixed and tested for Net1A localization by immunofluorescence microscopy. In these experiments, we observed that transfection of two different siRNAs targeting Rac1 reduced the number of cells exhibiting extranuclear Net1A localization from 28% to 9% (Fig. 2D and E). Both siRNAs were equally effective at inhibiting Rac1 expressio and did not affect HA-Net1A expression (Fig. 2E, inset). In separate experiments, transfection of siRNAs targeting Cdc42 did not affect Net1A localization, and cotransfection of Rac1 and Cdc42 siRNAs did not result in a greater reduction of extranuclear Net1A (data not shown). Thus, these data indicate that endogenous Rac1 expression is required for extranuclear localization of Net1A in actively growing MCF7 cells. They also indicate that the subcellular localization of Net1A is largely controlled by Rac1 rather than Cdc42 in these cells.
We then tested whether the plasma membrane localization of endogenous Net1A was controlled by endogenous Rac1. For these experiments, we used MDA-MB-231 cells, which express higher levels of plasma membrane-localized Net1A than MCF7 cells. Cells were transfected with control or Rac1-specific siRNAs, and the membrane fraction was isolated by subcellular fractionation. We observed that inhibition of Rac1 expression significantly reduced the localization of endogenous Net1 and Net1A in membranes (Fig. 2F). Thus, these data demonstrate that Rac1 controls the subcellular localization of endogenous Net1 isoforms, and they also show that the effects of Rac1 on Net1 isoform localization are not restricted to MCF7 cells.
The Rac1 effector Pak1 does not control Net1A localization.
We showed previously that the Rac1 effector Pak1 phosphorylates two residues within the N terminus of Net1, serines 152 and 153, which inhibits the catalytic activity of Net1 toward RhoA (31). These sites are conserved in Net1A, but effects of Pak1 on Net1A activity or localization have not been tested. To determine whether Pak1 mediates the effects of Rac1 on Net1A localization, MCF7 cells were transfected with Net1A, with or without constitutively active PAK1 L107F (Pak1 L/F) (41). Two days later, the cells were fixed and tested for Net1A localization by immunofluorescence. In these experiments we observed that coexpression of active Pak1 did not stimulate Net1A relocalization (Fig. 2G). In related experiments, we examined whether alanine or glutamate substitutions of Net1A at residues corresponding to the Pak1 phosphorylation sites in Net1 affected Net1A localization. In these experiments, we observed that replacement of serines 98 and 99 with alanines did not prevent relocalization of Net1A by V12Rac1. Similarly, replacement of these serines with glutamates, which mimics the effect of Pak1 phosphorylation on the activity of Net1 (31), did not stimulate Net1A relocalization in the absence of V12Rac1 expression (Fig. 2H). Taken together, these data indicate that Pak1 cannot substitute for Rac1 in stimulating Net1A relocalization and that the putative Pak1 phosphorylation sites in Net1A are not required for Rac1-stimulated Net1A relocalization.
Net1A catalytic activity and pleckstrin homology (PH) domain function are not required for Rac1-stimulated relocalization.
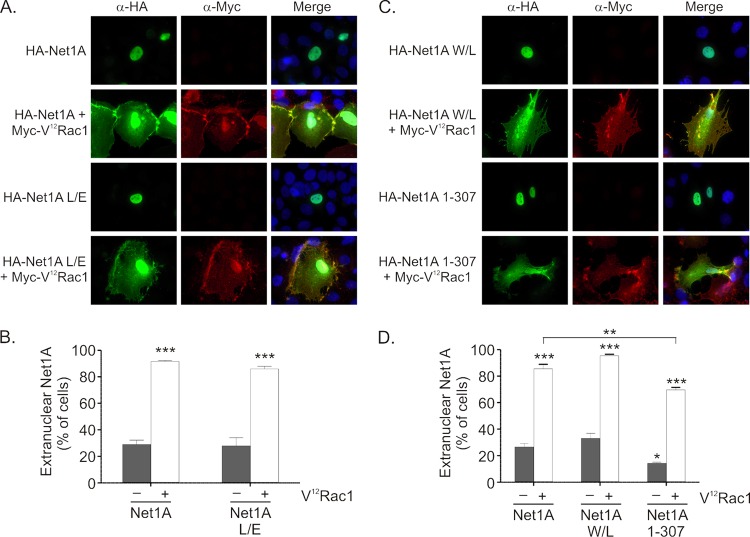
We next examined whether Net1A activity toward RhoA was required for V12Rac1-stimulated relocalization. MCF7 cells were transfected with wild-type or catalytically inactive Net1A L267E (Net1A L/E) (23), with or without V12Rac1, and the localization of Net1A was assessed by immunofluorescence microscopy. We observed that Net1A catalytic activity was not required for Rac1-stimulated relocalization (Fig. 3A and B). These data are consistent with the lack of effect of constitutively active RhoA coexpression on Net1A localization.
Fig 3.
Rac1-stimulated relocalization of Net1A does not require Net1A enzymatic activity or the Net1A PH domain. (A) MCF7 cells were transfected with expression plasmids for wild-type or catalytically inactive HA-Net1A (Net1A L267E), without or with Myc-V12Rac1. The cells were then fixed and stained for HA-Net1A proteins (green), Myc-V12Rac1 (red), and DNA (blue). Results of a representative experiment are shown. (B) Quantification of the localization of wild-type Net1A and Net1A L/E. Shown are the averages of three independent experiments. Error bars are standard errors of the means. ***, P < 0.001. (C) Cells were transfected with expression plasmids for HA-Net1A containing an inactivating mutation in its PH domain (W438L) or the truncation mutant lacking the PH domain and the C terminus (Net1A 1–307), without or with Myc-V12Rac1. Cells were then fixed and stained for HA-Net1 proteins (green), Myc-V12Rac1 (red), and DNA (blue). Results of a representative experiment are shown. (D) Quantification of the localization of wild-type Net1A, Net1A W/L, and Net1A 1–307. Averages are from three independent experiments. Error bars are standard errors of the means. *, P < 0.05; **, P < 0.01; ***, P < 0.0001.
We then tested whether the function or presence of the PH domain within Net1A was necessary for Rac1-stimulated relocalization. Although the PH domain of Net1A has not been shown to bind phosphoinositides or other proteins, in some RhoGEFs this domain may contribute to plasma membrane localization (16). To test this possibility, we assessed Rac1-stimulated relocalization of Net1A containing a W438L mutation within its PH domain (Net1A W/L), which is predicted to preclude PH domain function and has been shown to inhibit catalytic activity of the Net1 isoform (36). We also tested whether the localization of a Net1A deletion mutant lacking the PH domain and the entire C terminus (Net1A 1–307) was regulated by Rac1. In these experiments, we observed that Rac1 stimulated the relocalization of Net1A W438L as efficiently as wild-type Net1A (Fig. 3C and D). Net1A lacking its C terminus and PH domain was also relocalized outside the nucleus by V12Rac1 coexpression. Although relocalization of the Net1A 1–307 mutant was slightly less efficient than for wild-type Net1A, the percentage of cells expressing extranuclear Net1A 1–307 in the absence of active Rac1 was also reduced, indicating that the magnitude of the effect of active Rac1 on this mutant was maintained. Taken together, these data indicate that neither the catalytic activity of Net1A nor the presence of its PH domain or C terminus is required for V12Rac1-stimulated relocalization.
Rac1 activation enhances Net1A stability.
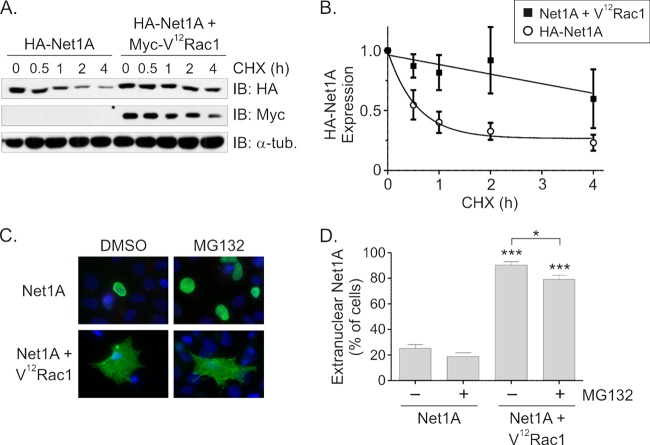
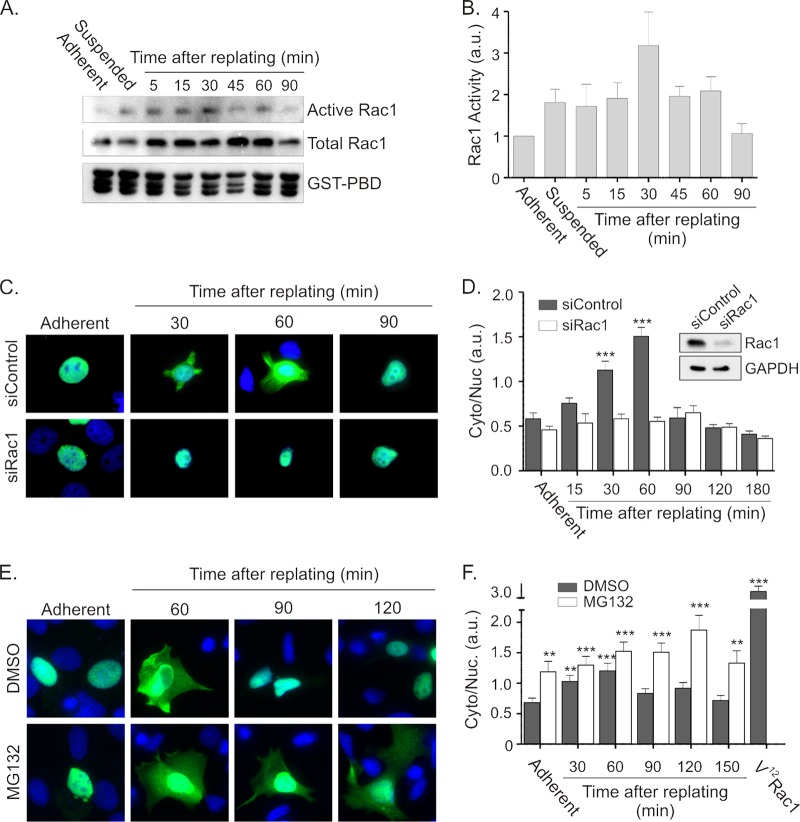
We have shown that Net1A is subject to proteasome-mediated degradation (42). Thus, a possible mechanism accounting for Rac1-stimulated Net1A accumulation outside the nucleus might be through enhancement of Net1A stability. To test this, we measured the half-life of Net1A in cells coexpressing V12Rac1. MCF7 cells were transfected with HA-Net1A, alone or together with V12Rac1. After 2 days, the protein translation inhibitor cycloheximide was added to the medium for different periods of time, and the expression of Net1A was then tested by Western blotting. In these experiments, Net1A expressed alone had a half-life of 40 min, consistent with our previous observations (42). Importantly, coexpression of V12Rac1 significantly stabilized Net1A, increasing its half-life to 5.8 h (Fig. 4A and B). Thus, these data indicate that V12Rac1 coexpression strongly enhances Net1A stability. Because Net1A is subject to degradation by the proteasome, we then examined whether treatment with the proteasome inhibitor MG132 is sufficient to cause Net1A relocalization on its own or to enhance V12Rac1-stimulated relocalization. We observed that in the absence of V12Rac1 coexpression, MG132 treatment increased the number of cells staining positive for Net1A within the nucleus, indicating that it protected Net1A from degradation (Fig. 4C). However, it did not enhance Net1A localization outside the nucleus in these cells, nor did it increase the already high percentage of cells with relocalized Net1A caused by cotransfection of V12Rac1 (Fig. 4C and D). Thus, although active Rac1 stabilizes Net1A, protection from proteasome-mediated degradation is not sufficient to promote Net1A relocalization outside the nucleus.
Fig 4.
Active Rac1 coexpression enhances Net1A stability. (A) MCF7 cells were transfected with HA-Net1A alone or with Myc-V12Rac1. Two days later the cells were incubated with the protein synthesis inhibitor cycloheximide (CHX) for different lengths of time. After lysis, the amount of HA-Net1A was determined by Western blotting. Shown are results of a representative experiment. (B) Quantification of Net1A stability. Shown are the averages of three independent experiments. Error bars are standard errors of the means. (C) Addition of the proteasome inhibitor MG132 is not sufficient to alter Net1A localization. MCF7 cells were transfected with expression plasmids for HA-Net1A, alone or with Myc-V12Rac1. Cells were incubated with MG132 overnight prior to fixation and stained for HA-Net1A (green) and DNA (blue). (D) Quantification of the effects of MG132 on Net1A localization. Shown are the averages of three independent experiments. Error bars are standard errors of the means. *, P < 0.05; ***, P < 0.001.
Cell spreading stimulates Rac1-dependent relocalization of Net1A.
Replating fibroblasts on fibronectin or collagen has been reported to result in a transient activation of Rac1 (43, 44). To assess whether stimuli that activate endogenous Rac1 also control Net1A localization, we tested whether replating cells on the extracellular matrix collagen affected the Net1A distribution. We first examined the kinetics of endogenous Rac1 activation after replating MCF7 cells on collagen, as this has not been reported previously. MCF7 cells were serum starved, trypsinized, held in suspension for an hour, and then replated on collagen-coated dishes. After different periods of time, the cells were lysed, and Rac1 activity was assessed in a GST-PBD pulldown assay (34). In these assays, we observed that endogenous Rac1 was maximally active within 30 min of replating (Fig. 5A and B), consistent with previously published data in other cell types (43, 44).
Fig 5.
Cell spreading stimulates Net1A relocalization in a Rac1-dependent and proteasome-regulated manner. (A) MCF7 cells were replated on collagen-coated dishes for different lengths of time and then lysed and tested for endogenous Rac1 activation in GST-PBD assays. Shown are results of a representative experiment. (B) Quantification of Rac1 activation after replating MCF7 cells on collagen. Results shown are the average of three independent experiments. Error bars are standard errors of the means. (C) MCF7 cells were transfected with control of Rac1-specific siRNAs. Two days later, the cells were transfected with an HA-Net1A expression plasmid. Cells were starved overnight and replated on collagen for different lengths of time. Cells were fixed and stained for HA-Net1A (green) and DNA (blue). Results of a representative experiment are shown. (D) Quantification of Net1A subcellular localization. Localization is represented as the ratio of HA-Net1A in the extranuclear space (Cyto) divided by that in the nucleus (Nuc). Error bars are standard errors of the means. Shown in the inset is a representative Western blot, demonstrating Rac1 knockdown. ***, P < 0.001. (E) Proteasome inhibition extends the time of Net1A localization outside the nucleus during cell spreading. MCF7 cells were transfected with an HA-Net1A expression plasmid and then starved and replated on collagen-coated coverslips for different lengths of time in the presence of DMSO or MG132. Cells were then fixed and stained for HA-Net1A (green) and DNA (blue). Shown are results of a representative experiment. (F) Quantification of HA-Net1A localization. Shown are the averages of three independent experiments. Error bars are standard errors of the means. **, P < 0.01; ***, P < 0.001.
We then examined whether replating cells on collagen stimulated Net1A relocalization outside the nucleus and if this required endogenous Rac1 activity. Because Net1A relocalization stimulated by transient Rac1 activation was expected to be less robust than that caused by V12Rac1 expression, we more precisely quantified Net1A relocalization by measuring the immunofluorescent signal of Net1A in the nuclear and cytoplasmic compartments. The subcellular localization of Net1A was then represented as the ratio of cytoplasmic to nuclear staining. To monitor Rac1-dependent relocalization of Net1A during cell spreading, MCF7 cells were transfected with control or Rac1-specific siRNAs. Two days later the cells were retransfected with an HA-Net1A expression plasmid. Two days after that, the cells were serum starved overnight, trypsinized, and replated on collagen-coated coverslips for different periods of time. The cells were then fixed, and the localization of Net1A was assessed. In these assays we observed that Net1A was relocalized outside the nucleus immediately as cells began to attach to the collagen. This reached a peak by 1 h after replating, and by 90 min the Net1A was relocalized to the nucleus. Furthermore, Net1A relocalization required Rac1 expression, since there was no extranuclear localization of Net1A in the Rac1 siRNA-transfected cells (Fig. 5C and D). These results clearly demonstrated that Net1A is transiently exported from the nucleus during cell spreading and that this requires endogenous Rac1 expression.
Proteasome-mediated degradation terminates extranuclear localization of Net1A during cell spreading.
Because V12Rac1 expression enhanced Net1A stability (Fig. 4), we tested whether the reduction in Net1A localization outside the nucleus 90 min after replating required proteasome activity. Thus, MCF7 cells expressing Net1A were replated on collagen for different periods of time in the presence of vehicle (DMSO) or the proteasome inhibitor MG132. Consistent with previous experiments, the DMSO-treated cells reached peak relocalization at 60 min, and by 90 min the Net1A was largely relocalized within the nucleus. Strikingly, the cells replated in the presence of MG132 maintained Net1A in the cytoplasm for the duration of the experiment (Fig. 5E and F). Thus, these data indicate that proteasome activity is required to remove Net1A from the cytoplasm once cells have completed the spreading process. Since Net1A is subject to proteasome-mediated degradation (42), this suggests that extranuclear Net1A is cleared from the cell by degradation.
Net1A is required for myosin light chain phosphorylation and focal adhesion maturation during cell spreading.
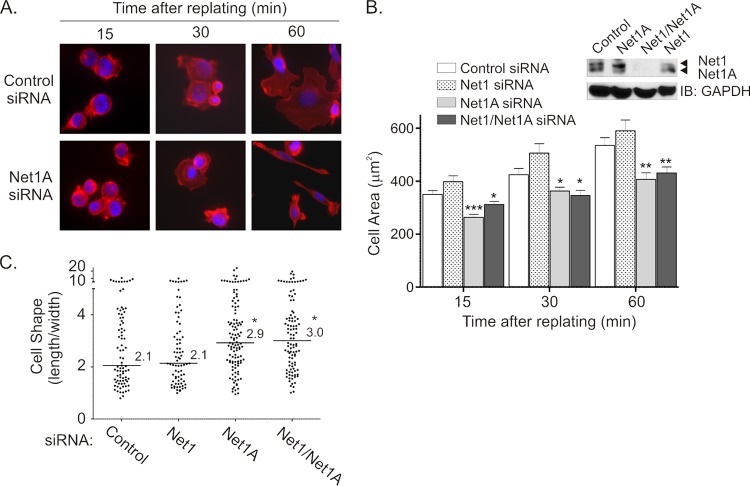
Cell spreading on an extracellular matrix is generally controlled by an early burst of Rac1 activation, which drives lamellar spreading and initial focal contact formation. At later times, RhoA is activated to stimulate cortical actin polymerization and F-actin stress fiber formation. This, in turn, promotes actomyosin contraction and focal adhesion maturation (45). Since Net1A relocalization was stimulated by cell spreading, we examined whether it contributed to this process. MCF7 cells were transfected with control, Net1, Net1A, or dual isoform-specific siRNAs and then replated on collagen-coated coverslips. At different times, the cells were fixed and stained for F-actin. In these experiments, depletion of either Net1A alone or both Net1 isoforms together equally lowered the rate of cell spreading, such that the total cell area was smaller at all times measured (Fig. 6A and B). Importantly, depletion of the Net1 isoform was without effect (Fig. 6B). Knockdown of Net1A also significantly altered cell morphology at later times, so that by 60 min many of the cells had assumed an elongated morphology (Fig. 6A and C). This type of morphology is consistent with reduced actomyosin contractility during cell spreading (46).
Fig 6.
Net1A expression is required for efficient cell spreading. (A) MCF7 cells were transfected with control or Net1A-specific siRNAs. Three days later the cells were serum starved, trypsinized, and replated on collagen-coated coverslips. At different times, the cells were fixed and stained for F-actin (red) and DNA (blue). Shown are results of a representative experiment. (B) Quantification of cell area in siRNA-transfected cells. MCF7 cells were transfected with the control of siRNAs specific for Net1, Net1A, or the dual isoform (Net1/Net1A). The cell area after replating was measured using ImageJ software. Shown are the averages of three independent experiments. Error bars are standard errors of the means. *, P < 0.05; **, P < 0.01; ***, P < 0.001. The inset shows a representative Western blot for Net1 isoform-specific knockdown. (C) Quantification of cell shape. Cells were transfected with the siRNAs shown and replated on collagen-coated coverslips for 60 min. Cell shape (length/width) was measured. Shown are the averages of three independent experiments. Bars are median values. *, P < 0.05.
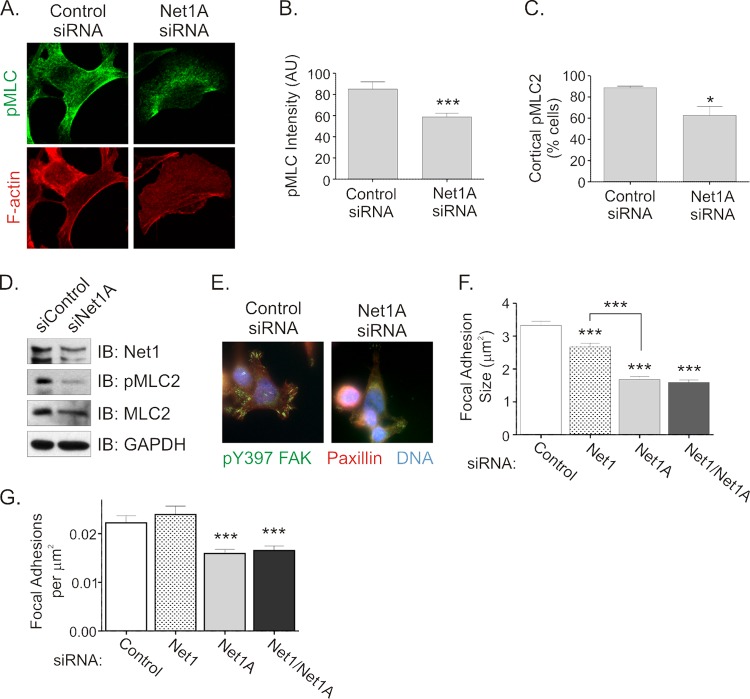
Because RhoA controls actomyosin contraction in part by stimulating phosphorylation of the regulatory myosin light chain (MLC) subunit (4, 5, 45), we examined whether Net1A was required for MLC phosphorylation during cell spreading. In these experiments, we observed that Net1A knockdown reduced overall MLC phosphorylation in cells 60 min after replating and also resulted in a loss of the cortical pMLC signal (Fig. 7A to D). This was accompanied by a decrease in F-actin staining (Fig. 7A). These data indicate that Net1A is required for efficient MLC phosphorylation and F-actin polymerization during cell spreading, which is consistent with a reduction in actomyosin contractility.
Fig 7.
Net1A controls MLC phosphorylation and focal adhesion formation during cell spreading. (A) MCF7 cells were transfected with control or Net1A-specific siRNAs and then replated on collagen-coated coverslips for 1 h. Cells were then fixed and stained for pMLC and F-actin. Shown are representative confocal images. (B) Quantification of pMLC fluorescence intensity. Shown are the averages of three independent experiments. Error bars are standard errors of the means. ***, P < 0.001. (C) Quantification of cortical pMLC staining. Shown are the averages of three independent experiments. Error bars are standard errors of the means. *, P < 0.05. (D) Western blot for pMLC in control or Net1A siRNA-transfected cells. Shown are results of a representative experiment. (E) Net1A knockdown inhibits focal adhesion maturation. MCF7 cells were transfected with control or Net1A-specific siRNAs. Three days later, the cells were replated on collagen-coated coverslips for 1 h. The cells were then fixed and stained for pY397-FAK (green), paxillin (red), and DNA (blue). Shown are representative images. (F) Quantification of focal adhesion size. MCF7 cells were transfected with the siRNAs shown and replated on collagen-coated coverslips for 1 h. Focal adhesions containing pY397-FAK and paxillin were quantified. Error bars are standard errors of the means. ***, P < 0.001. (G) Quantification of focal adhesions per cell area (in μm2). Cells were transfected and processed as for panel F. Phospho-Y397-FAK- and paxillin-positive focal adhesions were quantified, and values were then divided by the areas of the cells. Error bars are standard errors of the means. ***, P < 0.001.
Because Net1A knockdown affected the rate at which cells spread, we also measured effects on focal adhesion formation. As shown in Fig. 7E, control siRNA-transfected cells exhibited numerous, large, phospho-FAK-positive focal adhesions that were distributed throughout the cell 60 min after replating. However, Net1A siRNA-transfected cells had fewer focal adhesions that were generally smaller and restricted to the cell periphery. Quantification of focal adhesion size and number indicated that siRNA-mediated knockdown of Net1A alone, or of both Net1 isoforms together, reduced focal adhesion maturation equally. However, knockdown of the Net1 isoform minimally affected focal adhesion size and did not affect focal adhesion number (Fig. 7F and G). Taken together, these data indicate that Net1A expression, but not Net1, is required for focal adhesion maturation during cell spreading.
DISCUSSION
RhoA is a critical regulator of actin cytoskeletal organization and cell motility (2). Moreover, wild-type forms of RhoA are commonly overexpressed in human cancer, including breast cancer (12–15). However, the regulatory mechanisms governing RhoA activation in human cancers have not been fully elucidated. It has been shown that the ability of Net1 proteins to activate RhoA and thereby stimulate cytoskeletal reorganization and cell motility is inhibited by their nuclear localization. However, mechanisms controlling their extranuclear localization have not been identified. In the present work, we showed that Rac1 activation potently stimulates Net1 isoform relocalization outside the nucleus, and in the case of Net1A it causes a marked accumulation at the plasma membrane. We also showed that cell spreading on the extracellular matrix collagen activates endogenous Rac1 to cause a transient relocalization of Net1A and that removal of Net1A from the extranuclear space requires proteasome activity. Moreover, we demonstrated that the Net1A isoform is specifically required for proper cell spreading. In particular, loss of Net1A expression inhibits myosin light chain phosphorylation, actin stress fiber formation, and focal adhesion maturation. These data represent the first identified mechanism controlling Net1 isoform relocalization outside the nucleus, and they also demonstrate an isoform-specific function for Net1A in controlling actin cytoskeletal organization.
We surmise that Rac1 controls the subcellular localization of Net1 isoforms by affecting their rates of nuclear import and/or export, as well as their stability. This conclusion is based on a number of observations. For example, protection from degradation was not sufficient for plasma membrane accumulation of Net1A, as treatment with the proteasome inhibitor MG132 significantly increased the half-life of Net1A and augmented its nuclear expression without affecting its extranuclear localization (Fig. 4). However, stabilization of Net1A outside the nucleus was also clearly important for its localization, as proteasome inhibition caused a significant extension in the duration of Net1A localization outside the nucleus during cell spreading (Fig. 5). Thus, we favor a model in which Rac1 activation alters the nuclear import/export dynamics of Net1A, thereby promoting its accumulation at the plasma membrane and access to RhoA. Once localized at the membrane, Rac1 signaling protects Net1A from proteasome-mediated degradation. Cessation of Rac1 signaling would promote the termination of Net1A activity by allowing proteasome-dependent degradation to occur. In support of a role for the proteasome in controlling Net1A activity, we showed previously that Net1A is subject to ubiquitylation and degradation by the proteasome in MCF7 cells (42), and Papadimitriou et al. similarly observed that Net1A is degraded by the proteasome in TGF-β-treated HaCaT keratinocytes (27).
A question that arises from this work is precisely how Rac1 activation in MCF7 cells stimulates Net1A relocalization. This is unlikely to occur through direct interaction, as Net1A and V12Rac1 did not interact in coimmunoprecipitation assays (data not shown). A more likely explanation is that Rac1 initiates a signaling cascade to stimulate Net1A relocalization. Rac1 controls cell signaling by activating downstream effectors, including such well-studied proteins as Paks 1 to 3, phosphatidylinositol 3-kinase (PI3K), PI4-P5K, Wave/Scar, and IQGAP1 to -3 (37). Because we observed that expression of constitutively active Cdc42 can also stimulate Net1A relocalization (Fig. 2), we surmise that an effector shared by Rac1 and Cdc42 may mediate this effect. Of these, it is unlikely that class I Paks (Paks 1 to 3) control Net1A localization, since we showed that expression of constitutively active Pak1 did not cause Net1A relocalization (Fig. 2), and these kinases have very similar substrate specificities (47, 48). Similarly, Net1A relocalization is not likely to be mediated by PI3K, as it was not inhibited by addition of the PI3K inhibitor LY29004, by coexpression of a dominant-negative p85 PI3K subunit, or by overexpression of PTEN (data not shown). We also observed that coexpression of constitutively active PI4-5PK or PLD2 did not stimulate Net1A relocalization (data not shown). Thus, many of the commonly studied Rac1 and Cdc42 effectors do not mediate Net1A relocalization.
Rac1 caused accumulation of Net1A outside the nucleus more frequently than that of Net1. An explanation for this may be that mechanisms controlling the nuclear localization of Net1A are less robust than for Net1. For example, Schmidt and Hall showed that Net1 contains two NLS sequences within its unique N terminus (29), and we have found that Net1 contains two additional NLS sequences in a region common to both Net1 isoforms (H. S. Carr and J. A. Frost, unpublished data). Thus, Net1A may be poised to respond to plasma membrane accumulation stimuli, such as that generated by Rac1. This fits with our prior observation that transfected fibroblasts are more sensitive to actin cytoskeletal reorganization following Net1A overexpression than with Net1 (23). A more stringent nuclear localization of Net1 is also consistent with the prior studies of Schmidt and Hall, in which they were unable to find stimuli that caused relocalization of the Net1 isoform in transfected fibroblasts (29). Importantly, some of the stimuli that were listed by Schmidt et al. would be expected to have activated Rac1, further supporting the divergent regulation of Net1 isoforms that we have observed.
An important observation of this work is that Net1A, but not Net1, is required for efficient cell spreading and focal adhesion maturation. To date, only two studies have touched on differing roles for Net1 isoforms in cell function. Duterte et al. showed that estrogen treatment preferentially stimulated the expression of Net1 over Net1A in MCF7 cells. They also showed data indicating that siRNA-mediated knockdown of Net1 reduced the proliferation of these cells more than Net1A, while Net1A knockdown caused cells to detach from the culture dish (22). Alternatively, Papadimitriou et al. showed that Net1A expression was preferentially induced by TGF-β treatment and that its expression was necessary for early actin cytoskeletal reorganization events associated with EMT (27). Our work clearly showed that Net1A knockdown reduced the rate of cell spreading on collagen (Fig. 6). It also significantly changed cell morphology during this process, such that the cells assumed an elongated, spindly shape. We also observed that Net1A knockdown blocked myosin light chain phosphorylation and inhibited focal adhesion maturation, leaving cells with fewer focal adhesions that were smaller and confined to the cell periphery. All of these effects are consistent with a reduction in actomyosin contractility, which is known to be required for maintenance of cell shape during spreading, as well as for focal adhesion maturation (45, 49).
Since the mechanics of cell spreading are similar to those required for cell motility, it is likely that Net1A mediates many of the effects on cancer cell motility and invasion that have hitherto been ascribed to both Net1 isoforms (18, 24, 25). As cells move, they generally require Rac1 activation at the leading edge to promote lamellar extension and to initiate the formation of focal contacts. On the other hand, RhoA activation at the leading edge contributes to cortical actin polymerization and the maturation of focal contacts to larger focal adhesions. RhoA activation is also necessary at the trailing edge, where it generates actomyosin contraction to promote focal adhesion disassembly and trailing edge retraction (10, 11, 45). In future studies, it will be important to determine what aspects of leading and trailing edge dynamics are regulated by Net1A to control planar cell movement and extracellular matrix invasion, and also to understand how these mechanisms may be coopted in metastatic cancer cells.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant CA116356 to J.A.F. from the National Cancer Institute.
We thank other members in the lab for helpful advice and discussions and Rebecca Berdeaux for critical reading of the manuscript.
Footnotes
Published ahead of print 26 November 2012
REFERENCES
- 1. Coleman ML, Marshall CJ, Olson MF. 2004. RAS and RHO GTPases in G1-phase cell-cycle regulation. Nat. Rev. Mol. Cell Biol. 5: 355– 366 [DOI] [PubMed] [Google Scholar]
- 2. Jaffe AB, Hall A. 2005. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21: 247– 269 [DOI] [PubMed] [Google Scholar]
- 3. Ridley AJ. 2004. Rho proteins and cancer. Breast Cancer Res. Treat. 84: 13– 19 [DOI] [PubMed] [Google Scholar]
- 4. Amano M, Ito M, Kimura K, Fukata Y, Chihara K, Nakano T, Matsuura Y, Kaibuchi K. 1996. Phosphorylation and activation of myosin by Rho-associated kinase (Rho-kinase). J. Biol. Chem. 271: 20246– 20249 [DOI] [PubMed] [Google Scholar]
- 5. Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. 1996. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase). Science 273: 245– 248 [DOI] [PubMed] [Google Scholar]
- 6. Chrzanowska-Wodnicka M, Burridge K. 1996. Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J. Cell Biol. 133: 1403– 1415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. 2009. Coordination of Rho GTPase activities during cell protrusion. Nature 461: 99– 103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pertz O, Hodgson L, Klemke RL, Hahn KM. 2006. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature 440: 1069– 1072 [DOI] [PubMed] [Google Scholar]
- 9. Ridley AJ. 2011. Life at the leading edge. Cell 145: 1012– 1022 [DOI] [PubMed] [Google Scholar]
- 10. Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. 2003. Cell migration: integrating signals from front to back. Science 302: 1704– 1709 [DOI] [PubMed] [Google Scholar]
- 11. Broussard JA, Webb DJ, Kaverina I. 2008. Asymmetric focal adhesion disassembly in motile cells. Curr. Opin. Cell Biol. 20: 85– 90 [DOI] [PubMed] [Google Scholar]
- 12. Fritz G, Brachetti C, Bahlmann F, Schmidt M, Kaina B. 2002. Rho GTPases in human breast tumours: expression and mutation analyses and correlation with clinical parameters. Br. J. Cancer 87: 635– 644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fritz G, Just I, Kaina B. 1999. Rho GTPases are over-expressed in human tumors. Int. J. Cancer 81: 682– 687 [DOI] [PubMed] [Google Scholar]
- 14. Horiuchi A, Imai T, Wang C, Ohira S, Feng Y, Nikaido T, Konishi I. 2003. Up-regulation of small GTPases, RhoA and RhoC, is associated with tumor progression in ovarian carcinoma. Lab. Invest. 83: 861– 870 [DOI] [PubMed] [Google Scholar]
- 15. Moscow JA, He R, Gnarra JR, Knutsen T, Weng Y, Zhao WP, Whang-Peng J, Linehan WM, Cowan KH. 1994. Examination of human tumors for rhoA mutations. Oncogene 9: 189– 194 [PubMed] [Google Scholar]
- 16. Rossman KL, Der CJ, Sondek J. 2005. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 6: 167– 180 [DOI] [PubMed] [Google Scholar]
- 17. Tcherkezian J, Lamarche-Vane N. 2007. Current knowledge of the large RhoGAP family of proteins. Biol. Cell 99: 67– 86 [DOI] [PubMed] [Google Scholar]
- 18. Leyden J, Murray D, Moss A, Arumuguma M, Doyle E, McEntee G, O'Keane C, Doran P, MacMathuna P. 2006. Net1 and Myeov: computationally identified mediators of gastric cancer. Br. J. Cancer 94: 1204– 1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shen SQ, Li K, Zhu N, Nakao A. 2008. Expression and clinical significance of NET-1 and PCNA in hepatocellular carcinoma. Med. Oncol. 25: 341– 345 [DOI] [PubMed] [Google Scholar]
- 20. Tu Y, Lu J, Fu J, Cao Y, Fu G, Kang R, Tian X, Wang B. 2010. Over-expression of neuroepithelial-transforming protein 1 confers poor prognosis of patients with gliomas. Jpn. J. Clin. Oncol. 40: 388– 394 [DOI] [PubMed] [Google Scholar]
- 21. Gilcrease MZ, Kilpatrick SK, Woodward WA, Zhou X, Nicolas MM, Corley LJ, Fuller GN, Tucker SL, Diaz LK, Buchholz TA, Frost JA. 2009. Coexpression of α6β4 integrin and guanine nucleotide exchange factor Net1 identifies node-positive breast cancer patients at high risk for distant metastasis. Cancer Epidemiol. Biomarkers Prev. 18: 80– 86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dutertre M, Gratadou L, Dardenne E, Germann S, Samaan S, Lidereau R, Driouch K, de la Grange P, Auboeuf D. 2010. Estrogen regulation and physiopathologic significance of alternative promoters in breast cancer. Cancer Res. 70: 3760– 3770 [DOI] [PubMed] [Google Scholar]
- 23. Qin H, Carr HS, Wu X, Muallem D, Tran NH, Frost JA. 2005. Characterization of the biochemical and transforming properties of the neuroepithelial transforming protein 1. J. Biol. Chem. 280: 7603– 7613 [DOI] [PubMed] [Google Scholar]
- 24. Ecimovic P, Murray D, Doran P, McDonald J, Lambert DG, Buggy DJ. 2011. Direct effect of morphine on breast cancer cell function in vitro: role of the NET1 gene. Br. J. Anaesth. 107: 916– 923 [DOI] [PubMed] [Google Scholar]
- 25. Murray D, Horgan G, MacMathuna P, Doran P. 2008. NET1-mediated RhoA activation facilitates lysophosphatidic acid-induced cell migration and invasion in gastric cancer. Br. J. Cancer 99: 1322– 1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee J, Moon HJ, Lee JM, Joo CK. 2010. Smad3 regulates Rho signaling via NET1 in the transforming growth factor-beta-induced epithelial-mesenchymal transition of human retinal pigment epithelial cells. J. Biol. Chem. 285: 26618– 26627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Papadimitriou E, Vasilaki E, Vorvis C, Iliopoulos D, Moustakas A, Kardassis D, Stournaras C. 2012. Differential regulation of the two RhoA-specific GEF isoforms Net1/Net1A by TGF-beta and miR-24: role in epithelial-to-mesenchymal transition. Oncogene 31: 2862– 2875 [DOI] [PubMed] [Google Scholar]
- 28. Shen X, Li J, Hu PP, Waddell D, Zhang J, Wang XF. 2001. The activity of guanine exchange factor NET1 is essential for transforming growth factor-beta-mediated stress fiber formation. J. Biol. Chem. 276: 15362– 15368 [DOI] [PubMed] [Google Scholar]
- 29. Schmidt A, Hall A. 2002. The Rho exchange factor Net1 is regulated by nuclear sequestration. J. Biol. Chem. 277: 14581– 14588 [DOI] [PubMed] [Google Scholar]
- 30. Ahn SJ, Chung KW, Lee RA, Park IA, Lee SH, Park DE, Noh DY. 2003. Overexpression of βPix-a in human breast cancer tissues. Cancer Lett. 193: 99– 107 [DOI] [PubMed] [Google Scholar]
- 31. Alberts AS, Qin H, Carr HS, Frost JA. 2005. PAK1 negatively regulates the activity of the Rho exchange factor NET1. J. Biol. Chem. 280: 12152– 12161 [DOI] [PubMed] [Google Scholar]
- 32. Yeung T, Terebiznik M, Yu L, Silvius J, Abidi WM, Philips M, Levine T, Kapus A, Grinstein S. 2006. Receptor activation alters inner surface potential during phagocytosis. Science 313: 347– 351 [DOI] [PubMed] [Google Scholar]
- 33. Garcia-Mata R, Wennerberg K, Arthur WT, Noren NK, Ellerbroek SM, Burridge K. 2006. Analysis of activated GAPs and GEFs in cell lysates. Methods Enzymol. 406: 425– 437 [DOI] [PubMed] [Google Scholar]
- 34. Sander EE, ten Klooster JP, van Delft S, van der Kammen RA, Collard JG. 1999. Rac downregulates Rho activity: reciprocal balance between both GTPases determines cellular morphology and migratory behavior. J. Cell Biol. 147: 1009– 1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dubash AD, Wennerberg K, Garcia-Mata R, Menold MM, Arthur WT, Burridge K. 2007. A novel role for Lsc/p115 RhoGEF and LARG in regulating RhoA activity downstream of adhesion to fibronectin. J. Cell Sci. 120: 3989– 3998 [DOI] [PubMed] [Google Scholar]
- 36. Alberts AS, Treisman R. 1998. Activation of RhoA and SAPK/JNK signalling pathways by the RhoA-specific exchange factor mNET1. EMBO J. 17: 4075– 4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bishop AL, Hall A. 2000. Rho GTPases and their effector proteins. Biochem. J. 348: 241– 255 [PMC free article] [PubMed] [Google Scholar]
- 38. Gorzalczany Y, Sigal N, Itan M, Lotan O, Pick E. 2000. Targeting of Rac1 to the phagocyte membrane is sufficient for the induction of NADPH oxidase assembly. J. Biol. Chem. 275: 40073– 40081 [DOI] [PubMed] [Google Scholar]
- 39. Kinsella BT, Erdman RA, Maltese WA. 1991. Carboxyl-terminal isoprenylation of ras-related GTP-binding proteins encoded by rac1, rac2, and ralA. J. Biol. Chem. 266: 9786– 9794 [PubMed] [Google Scholar]
- 40. Mizuno T, Kaibuchi K, Ando S, Musha T, Hiraoka K, Takaishi K, Asada M, Nunoi H, Matsuda I, Takai Y. 1992. Regulation of the superoxide-generating NADPH oxidase by a small GTP-binding protein and its stimulatory and inhibitory GDP/GTP exchange proteins. J. Biol. Chem. 267: 10215– 10218 [PubMed] [Google Scholar]
- 41. Frost JA, Khokhlatchev A, Stippec S, White MA, Cobb MH. 1998. Differential effects of PAK1-activating mutations reveal activity-dependent and -independent effects on cytoskeletal regulation. J. Biol. Chem. 273: 28191– 28198 [DOI] [PubMed] [Google Scholar]
- 42. Carr HS, Cai C, Keinanen K, Frost JA. 2009. Interaction of the RhoA exchange factor Net1 with discs large homolog 1 protects it from proteasome mediated degradation and potentiates Net1 activity. J. Biol. Chem. 284: 24269– 24280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Berrier AL, Martinez R, Bokoch GM, LaFlamme SE. 2002. The integrin beta tail is required and sufficient to regulate adhesion signaling to Rac1. J. Cell Sci. 115: 4285– 4291 [DOI] [PubMed] [Google Scholar]
- 44. Price LS, Leng J, Schwartz MA, Bokoch GM. 1998. Activation of Rac and Cdc42 by integrins mediates cell spreading. Mol. Biol. Cell 9: 1863– 1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Burridge K, Wennerberg K. 2004. Rho and Rac take center stage. Cell 116: 167– 179 [DOI] [PubMed] [Google Scholar]
- 46. Sastry SK, Rajfur Z, Liu BP, Cote JF, Tremblay ML, Burridge K. 2006. PTP-PEST couples membrane protrusion and tail retraction via VAV2 and p190RhoGAP. J. Biol. Chem. 281: 11627– 11636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Arias-Romero LE, Chernoff J. 2008. A tale of two Paks. Biol. Cell 100: 97– 108 [DOI] [PubMed] [Google Scholar]
- 48. Bokoch GM. 2003. Biology of the p21-activated kinases. Annu. Rev. Biochem. 72: 743– 781 [DOI] [PubMed] [Google Scholar]
- 49. Olson MF, Sahai E. 2009. The actin cytoskeleton in cancer cell motility. Clin. Exp. Metastasis 26: 273– 287 [DOI] [PubMed] [Google Scholar]