Abstract
The ribonucleotide reductase (RNR) enzyme catalyzes an essential step in the production of deoxyribonucleotide triphosphates (dNTPs) in cells. Bulk biochemical measurements in synchronized Saccharomyces cerevisiae cells suggest that RNR mRNA production is maximal in late G1 and S phases; however, damaged DNA induces RNR transcription throughout the cell cycle. But such en masse measurements reveal neither cell-to-cell heterogeneity in responses nor direct correlations between transcript and protein expression or localization in single cells which may be central to function. We overcame these limitations by simultaneous detection of single RNR transcripts and also Rnr proteins in the same individual asynchronous S. cerevisiae cells, with and without DNA damage by methyl methanesulfonate (MMS). Surprisingly, RNR subunit mRNA levels were comparably low in both damaged and undamaged G1 cells and highly induced in damaged S/G2 cells. Transcript numbers became correlated with both protein levels and localization only upon DNA damage in a cell cycle-dependent manner. Further, we showed that the differential RNR response to DNA damage correlated with variable Mec1 kinase activity in the cell cycle in single cells. The transcription of RNR genes was found to be noisy and non-Poissonian in nature. Our results provide vital insight into cell cycle-dependent RNR regulation under conditions of genotoxic stress.
INTRODUCTION
Unrepaired DNA damage can result in cell growth arrest, apoptosis, premature aging, neurodegeneration, and cancer (1, 2). Because most DNA repair pathways require de novo synthesis of DNA, damaged DNA signals the increased production and activation of the ribonucleotide reductase (RNR) enzyme (3–5). In almost all eukaryotes, the functional RNR enzyme consists of a large and a small subunit (3). The Saccharomyces cerevisiae genes RNR1 and RNR3 code for the large-subunit proteins, while RNR2 and RNR4 code for the small-subunit proteins (Fig. 1). The active form of the small subunit is an Rnr2-Rnr4 heterodimer (6, 7), and it relocalizes to the cytoplasm from the nucleus upon DNA damage (4, 8) to make the functional holoenzyme with the large subunit. Additionally, upon DNA damage, the transcription of all RNR genes is induced by the Mec1-Rad53 pathway (9, 10), which also controls the subcellular localization of the Rnr2-Rnr4 heterodimer (11) and the activation of the RNR enzyme (12, 13). Much of our understanding of the response of RNR to DNA damage as a function of cell cycle stage comes from bulk biochemical studies involving the model eukaryote S. cerevisiae (Fig. 1) (14–16). However, the synchronization methods employed in these studies may alter normal cell behavior. Further, mean values probed in bulk population studies mask information on cell-to-cell variability in response, which is clearly resolvable with single-cell-level imaging (17–20). Moreover, mRNA and protein levels and localization are usually measured in separate experiments, and few studies have explored the measurement of both gene products in the same cells.
Fig 1.
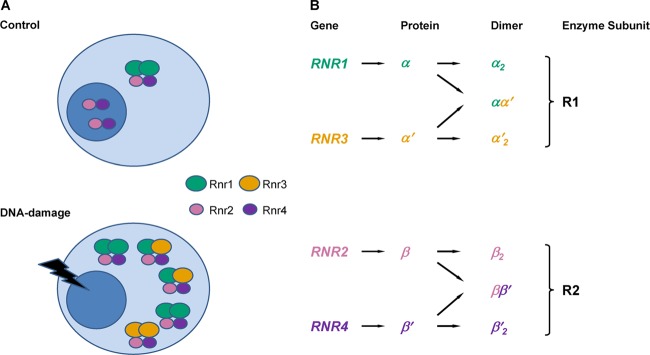
S. cerevisiae RNR enzyme response to damage. (A) The functional RNR holoenzyme consists of a large and a small subunit in almost all eukaryotes from yeast to humans. The form of the enzyme can be more complex than α2β2. Levels of all Rnr proteins go up, and Rnr2 and Rnr4 translocate to the cytoplasm upon DNA damage in S. cerevisiae. (B) The cytosolic Rnr1 and Rnr3 proteins constitute the large subunit (R1), and the Rnr2 and Rnr4 proteins constitute the small subunit (R2). The active form of the small subunit is an Rnr2-Rnr4 heterodimer (ββ′), which normally resides in the nucleus but relocalizes to the cytoplasm upon DNA damage. Rnr3 is not expressed in the absence of DNA damage.
As a consequence, it remains unclear whether RNR genes are induced uniformly across cells by DNA damage via a homogeneous amplification of the normal cell cycle transcript distributions or whether cell cycle stage-specific amplification of transcripts occurs. Additionally, correlated variation in protein and mRNA levels in individual cells in distinct stages of the cell cycle with and without genotoxic stress remains unexplored. For example, mRNA and protein levels were recently found to become correlated for a number of genes under conditions of osmotic stress using bulk mass spectrometry (21), whereas little to no correlation between mRNA and protein has been observed in several bulk and single-cell studies in unperturbed cells (20, 22, 23). This discrepancy is likely to be because of the longer half-lives of most proteins that result in slower fluctuations in their numbers with respect to mRNAs that typically degrade rapidly in a programmed manner (20, 24, 25).
To overcome these limitations and reveal the possible cell cycle dependence of Rnr mRNA and protein on DNA damage, we assayed the transcriptional response of the RNR subunit genes by imaging single transcripts with fluorescence in situ hybridization (FISH) (26–29) and subsequently combined this technique with immunofluorescence detection of Rnr proteins to simultaneously investigate their translational responses in the same individual cells as a function of the cell cycle.
MATERIALS AND METHODS
Cell growth and mRNA FISH.
All chemicals were from Sigma-Aldrich (St. Louis, MO), Invitrogen (Carlsbad, CA), or Ambion (Applied Biosystems, Austin, TX) unless otherwise noted. BY4741 cells were typically grown in yeast extract-peptone-dextrose (YPD) medium at 30°C with shaking. For experiments with RC634 cells, YPD medium with 0.003% adenine hemisulfate (YPDA) was used to avoid fluorescent purine precursors accumulating in the vacuoles. FISH was performed following earlier studies in yeast (26–29). Cells were diluted to an optical density at 600 nm (OD600) of 0.15 in the appropriate medium from an overnight saturated culture and allowed to grow to an OD600 of 0.5 in a 10-ml volume for each experiment. At this point, the culture was divided into two halves and cells were diluted in an equal volume of either control or methyl methanesulfonate (MMS)-containing medium and allowed to grow for another hour. At this time point, both broad cell cycle categories are still represented in the population. The final MMS concentration was 0.02%, as in previous works (4). For FISH experiments, cells were fixed for 45 min by direct addition of formaldehyde to a final concentration of 4%. Cells were then washed twice in buffer B (1.2 M sorbitol and 100 mM potassium phosphate in nuclease-free water), spheroplasted in buffer B with 100 mU/μl lyticase, 0.06 mg/ml phenylmethylsulfonyl fluoride (PMSF), 28 mM β-mercaptoethanol, and 10 mM vanadyl ribonucleoside complex (VRC; New England BioLabs, Ipswich, MA) at 30°C, and washed twice again in buffer B. The cells were then resuspended in 70% ethanol and left overnight at 4°C. The cells were then resuspended for 5 min in wash buffer (2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate] and 25% formamide in nuclease-free water) and resuspended in hybridization buffer (10 mM VRC, 1 mg/ml bovine serum albumin [BSA], 20× SSC, 0.5 mg/ml Escherichia coli tRNA, 0.5 mg/ml single-stranded DNA [ssDNA], 100 mg/ml dextran sulfate, 25% formamide, and 2× SSC in nuclease-free water) with Alexa Fluor 568-labeled probes against the target mRNA. mRNA probes were obtained from Biosearch Technologies (Novato, CA). Hybridization was allowed to proceed overnight at 30°C. The cells were then washed with wash buffer and stained for 30 min with 1 μg/ml DAPI (4′,6-diamidino-2-phenylindole) to stain the DNA. The cells were then washed and resuspended in 2× SSC and mounted in ProLong Gold antifade reagent on slides.
mRNA probe design.
Each RNR gene was targeted by 40 20-nucleotide-long DNA oligonucleotide probes, each with a 3′ Alexa Fluor 568 fluorophore. When designing probes, we used bioinformatics to eliminate any probe that can potentially cross-hybridize between genes like RNR1 and RNR3, which show large similarities (30) in nucleotide sequence (see Fig. S1 in the supplemental material). The efficacy of this approach is apparent in the fact that control untreated asynchronous cells expectedly do not show any RNR3 expression while a subpopulation of the same cells clearly stains for high numbers of RNR1 (Fig. 2B; see also Fig. S7 in the supplemental material). This indicates that RNR3 probes do not cross-hybridize with the ubiquitous RNR1 mRNA.
Fig 2.
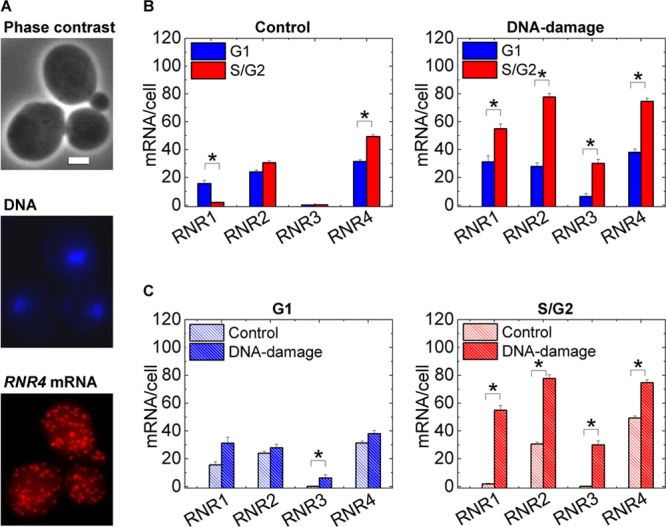
RNR mRNA induction depends on cell cycle stage. (A) A typical single-molecule mRNA FISH experiment is shown. RNR4 mRNA transcripts are targeted with Alexa Fluor 568-labeled DNA oligonucleotide probes. DAPI-stained DNA and phase-contrast images are also acquired to judge cell cycle stage. Scale bar, 2 μm. Z-projected images for the mRNA and DNA are shown. (B) Mean numbers computed from mRNA distribution histograms for approximately 90 to 120 such cells are plotted for RNR1, RNR2, RNR3, and RNR4 mRNA for control cells and under conditions of DNA damage by treatment with 0.02% MMS for 1 h. G1, unbudded G1 cells; S/G2, budded S/G2 cells. While absolute numbers of RNR1 mRNA are lower than absolute numbers of RNR2 and RNR4 mRNA in untreated cells, the relative fluctuation is greatest for RNR1 due to the near-complete absence in budded cells (see also Fig. 6 for RNR1 mRNA distributions). The relative distributions shift unexpectedly upon DNA damage. (C) The same data as in panel B, parsed according to the cell cycle stage to compare mRNA numbers in one cell cycle stage between control and damaged cells. In all cases, the error bars are standard errors. *, P < 10−3 in a Kolmogorov-Smirnov test (a nonparametric test is preferable given the nonnormal nature of some of the mRNA distributions).
Simultaneous detection of mRNA and protein.
mRNA FISH was performed as described before, followed by immunofluorescence for proteins. All reagents were specifically made from nuclease-free materials to avoid degradation of transcripts. We verified that largely the same mRNA numbers were obtained when FISH was performed alone and when FISH was performed with immunofluorescence (see Fig. S2 in the supplemental material). Following mRNA FISH, subsequent steps were performed in the blocking solution made from nuclease-free materials. Cells were blocked in 1% BSA in phosphate-buffered saline (PBS) for 1 h. Cells were stained with the primary antibodies at a 1:1,000 dilution for 3 h and then with the Alexa Fluor 647-tagged secondary antibodies at a 1:200 dilution for 1.5 h according to an earlier work (4). Cells were washed in 2× SSC and mounted in ProLong Gold antifade reagent on slides. The H2A-S129p antibody was obtained from Upstate (Millipore, Billerica, MA). All the Rnr antibodies used have been used in a previous study that demonstrated the translocation of Rnr2 and Rnr4 from the nucleus to the cytoplasm upon DNA damage (4). Rnr3 staining is not expected in wild-type (WT) cells in the absence of DNA damage. The weak basal staining we see in WT cells is comparable to that in a ΔRNR3 deletion strain (see Fig. S3 in the supplemental material). However, with DNA damage, there is a clear induction of Rnr3 expression in WT cells. The Rnr4 antibody worked well in assays in which the cells are processed for flow cytometry and showed proper nuclear localization in the absence of damage. A detergent permeabilization is used in this case. However, in FISH experiments, the permeabilization is in 70% ethanol, which can potentially affect the recognition of a protein by its antibody. In our experiments, the nuclear-to-cytoplasmic contrast of Rnr4 was poor when we attempted the simultaneous detection of RNR4 mRNA and Rnr4 protein. An induction of the signal was still able to be detected, but because of the lack of proper nuclear localization of Rnr4 in untreated cells, we have left this result out.
Antibody stains for flow cytometry.
Cells were grown and spheroplasted as described above (except without VRC), permeabilized in 0.2% Tween 20 in buffer B for 10 min, and blocked with 1% BSA in PBS for 1 h. Antibody stains were then performed as described above. Flow cytometry was performed on an Accuri C6 flow cytometer.
Image and statistical analyses.
Images were acquired on an Observer Z1 microscope (Carl Zeiss, Jena, Germany) with a Hamamatsu Orca-ER camera (Hamamatsu, Hamamatsu City, Japan). Z-stack images in all channels were obtained. For mRNA spot counting, we used an algorithm developed in a previous work (26). This algorithm was used previously to count mRNA numbers in yeast (28) and was verified in the present study to also be effective for the RNR genes (see Fig. S4 in the supplemental material). mRNA numbers were reproducible among different experiments, and the variation of the means did not reflect the large variation within the population (see Fig. S5 in the supplemental material). For evaluating total protein intensity, edge detection was performed on the phase image to extract cell contours, and subsequently the antibody stain intensity was evaluated within this mask. While cells have intrinsic autofluorescence, this is of weak intensity in the far-red wavelengths used. The mounting medium also introduces a weak level of background fluorescence. The effects of these two factors were eliminated by estimating the mean fluorescence levels in similarly mounted effectively unstained samples treated with only the secondary antibody and subtracting this mean intensity from the measured intensities. Representative images were processed with ImageJ, while all image analysis was performed in Matlab (MathWorks, Natick, MA). Statistical tests and graph plots were performed with Matlab and OriginPro 8.5 (OriginLab, Northampton, MA).
RESULTS
We first used single-mRNA-molecule FISH to measure RNR transcripts in a cell cycle-specific manner. Cell cycle stage was deduced from nucleus and cell images (Fig. 2; see also Fig. S6 in the supplemental material). In control, undamaged cells, we found a stark absence of RNR1 mRNA in nearly all budded cells, i.e., cells in S or G2, and only a subset of control, unbudded G1 cells had large amounts of RNR1 mRNA (Fig. 2; see also Fig. S7 in the supplemental material). These results are consistent with those of previous bulk Northern blot studies showing large fluctuations of RNR1 mRNA in the course of the normal cell cycle and transcript levels peaking in the late G1/early S phases (15), but the near-total absence of RNR1 mRNA in budded cells was surprising. This indicates that RNR1 mRNA numbers drop precipitously as cells initiate DNA synthesis. Also consistent with bulk studies (15, 16, 30), RNR3 mRNA was entirely absent throughout the cell cycle in undamaged log-phase cells and the cell cycle-dependent differences in RNR2 and RNR4 transcript numbers were relatively small, though significant, for RNR4 (Fig. 2B).
In contrast, cells damaged with the alkylating agent MMS for 1 h exhibited clear induction of all four RNR mRNAs. RNR1 mRNA was highly induced from near absence in S/G2 cells, and for all RNR genes, cell cycle-dependent differences in mRNA numbers that were negligible in control cells became pronounced upon damage (Fig. 2B). Thus, overall RNR transcriptional inductions observed upon DNA damage in bulk studies are not mere amplifications of relative distributions of mRNA numbers across the cell cycle in control, untreated cells. Remarkably, G1 mRNA numbers were largely comparable between control and damaged cells for all three normal cell cycle RNR genes (RNR1, RNR2, and RNR4) whereas S/G2 numbers were significantly different (Fig. 2C). This was unexpected, as in previous work, under conditions of DNA damage, cells exhibited induction of RNR1, RNR2, and RNR3 mRNA in α-factor-arrested G1 cells with Northern blot measurements (14, 15), leading to the conclusion that RNR gene induction is independent of the cell cycle. And while a clear induction was seen, it should be noted that even in these studies, the induction of RNR2 and RNR3 was lower in α-factor-arrested cells than in asynchronous cells. We investigated this discrepancy by using the same S. cerevisiae strain and conditions used in the previous studies and found that the perceived induction was likely due to a small subpopulation of budded S/G2 cells that escape arrest; this subpopulation had an overwhelming RNR response to DNA damage, greatly biasing the mean (see Fig. S8 in the supplemental material). Importantly, “shmooed” G1 cells showed no significant RNR2 induction. Also, it is possible that α-factor-arrested cells activate DNA damage checkpoints differently from G1 cells in asynchronous cultures. This underscores the importance of studying cells in a normal asynchronous cycling population versus under α-factor arrest and also the importance of single-cell response studies as opposed to bulk-cell responses. Cell cycle-dependent responses in the previous studies were performed with alkylation damage by MMS, though other forms of genotoxic stress were also shown to induce RNR expression. It can be expected that the RNR response in the cell cycle would be different for other forms of lesions, like double-strand breaks (DSBs) or those caused by UV radiation. We tested this possibility for damage by the UV-mimetic agent 4-NQO and the radio-mimetic DSB-causing agent bleomycin in terms of the transcriptional responses of the large-subunit (R1) gene RNR1 and the small-subunit (R2) gene RNR2. For both these agents, we found that the transcriptional induction response was much larger in S/G2 cells than G1 cells. The induction of RNR2 in G1 was significant but still much smaller than that in S/G2 cells (see Fig. S9 in the supplemental material). Thus, the cell cycle-dependent induction of RNR genes seems to be a general feature of at least three different forms of genotoxic stress. RNR induction, when present, is severely abrogated in G1 cells in asynchronous cultures.
Next, we determined whether the protein induction correlates with transcript induction and how transcript induction relates to protein localization. We detected endogenous RNR mRNA and Rnr protein in the same cells by FISH and antibody staining, respectively. Rnr protein levels showed significant induction in S/G2 cells upon damage (Fig. 3A). By staining mRNA in the same cells, we were able to correlate RNR1, RNR2, and RNR3 gene products on a cell-by-cell basis (Fig. 3B). Fluctuations in mRNA in the normal cell cycle may not reflect in protein levels. But under conditions of stress, cell cycle-dependent induction of both transcript and protein were observed. Whereas levels were heterogeneous across individual cells, clear induction of mean levels over cells was seen for both mRNA and protein. Unfortunately, the Rnr4 antibody did not work in the assay for simultaneous detection of mRNA and protein, and this is discussed in Materials and Methods.
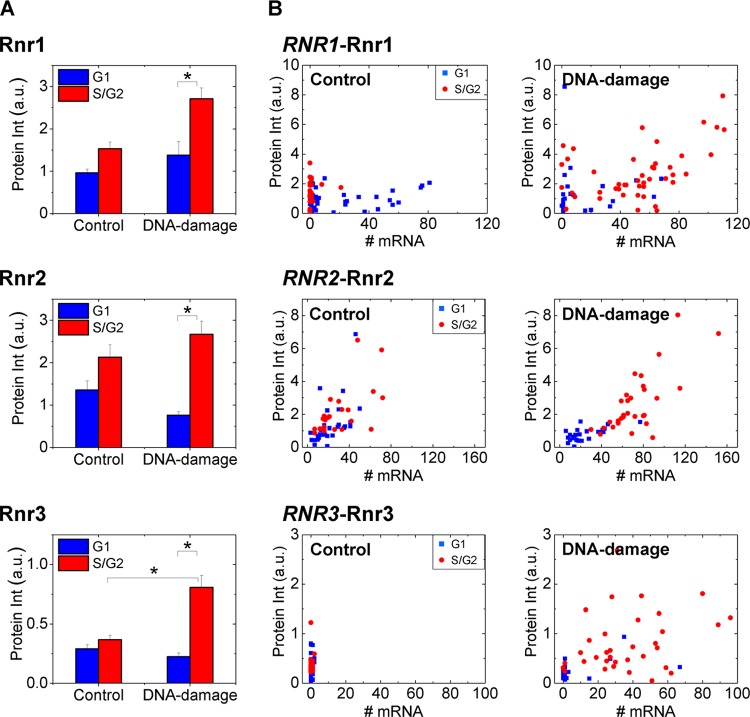
Fig 3.
RNR transcript numbers show a cell cycle-dependent relation to protein levels and localization upon DNA damage. Mean Rnr protein intensities with standard errors (A) and mRNA numbers and protein intensities on a cell-by-cell basis (B) are plotted for Rnr1 (n = 71 cells), Rnr2 (n = 57 cells), and Rnr3 (n = 64 cells). Equal numbers of cells were considered for the control and DNA damage (1-h) samples. The staining for Rnr3 in the absence of damage was nonspecific. Note that while S/G2 cells have little or no RNR1 mRNA (like Fig. 2) compared to G1 cells, the protein levels are similar in untreated control cells. A clear separation of G1 and S/G2 cells in MMS-treated samples was observed. *, P < 10−3 in a Kolmogorov-Smirnov test.
In addition to R1 (Rnr1 and Rnr3) levels, active RNR enzyme numbers are regulated by the nuclear-to-cytoplasmic translocation of the R2 proteins (Rnr2 and Rnr4) upon DNA damage (4, 16) (Fig. 1) and Sml1-mediated inhibition of the RNR enzyme (12, 13). There was no obvious relation between the nuclear-to-cytoplasmic ratio (NCR) of Rnr2 and the number of RNR2 transcripts in control cells. But after 1 h of DNA damage, we observed that the cells that still had nuclear Rnr2 were typically in G1 and that these cells had low RNR2 transcripts. In contrast, S/G2 cells exhibited clearly homogeneous or cytosolic Rnr2 and high numbers of RNR2 transcripts (Fig. 4). While it is known that the Mec1-Rad53 pathway controls both the transcriptional induction of the RNR genes (9, 10) and the subcellular relocalization of Rnr2-Rnr4 (11), we show here that both of these responses are cell cycle dependent in asynchronous cell populations. In previous studies, no nuclear-to-cytoplasmic translocation of Rnr2 or Rnr4 was observed in α-factor-arrested G1 cells, and this was attributed to a possible lower activation of the Mec1-Rad53 pathway in these cells (4). However, recent research has demonstrated that the Mec1 kinase can be activated throughout the cell cycle by two independent mechanisms dependent on the 9-1-1 complex and DNA polymerase (Pol) ε (31). This study used the DNA damage-dependent phosphorylation of the yeast histone H2A at serine 129 (H2A-S129p) as a direct readout of Mec1 kinase activity (31). Hence, we next adapted our approach of simultaneous detection of protein and mRNA to determine whether Mec1 kinase activity varies in the cell cycle in a manner similar to that of the RNR transcriptional response.
Fig 4.
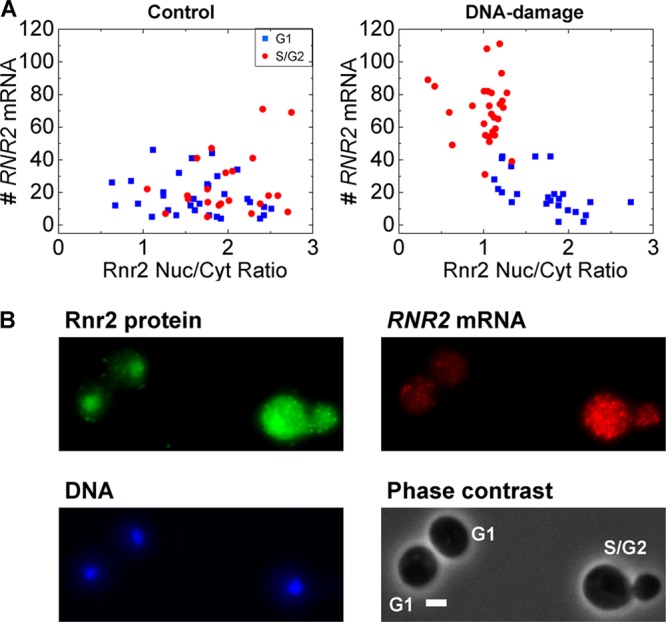
RNR2 transcript numbers show a cell cycle-dependent relation to Rnr2 protein localization upon DNA damage. (A) In the control population, RNR2 mRNA numbers in cells are uncorrelated with the nuclear-to-cytoplasmic (Nuc/Cyt) ratio of Rnr2 and there is no obvious segregation in the cell cycle. However, upon DNA damage, the S/G2 cells show a higher accumulation of Rnr2 in the cytoplasm and higher induction of RNR2 mRNA (n = 53 cells each). (B) A typical image is shown for the small-subunit Rnr2 upon DNA damage. Rnr2 is normally nucleus localized in control cells. At the 1-h time point, there are still cells with nuclear Rnr2. The G1 cells with nuclear Rnr2 have fewer RNR2 transcripts, while the S/G2 cell shows visibly greater RNR2 expression and a homogenous distribution of the Rnr2 protein. Scale bar, 2 μm. Z-projected images for the DNA, mRNA, and protein are shown. Cell cycle stages are indicated.
Asynchronous and α-factor-arrested cells showed similar relative inductions of H2A-S129p upon DNA damage in terms of the mean response (see Fig. S10 in the supplemental material). When we performed simultaneous detection of RNR2 mRNA and H2A-S129p in the same cells in an asynchronous population, we found an expected correlation between Mec1 kinase activity and RNR2 induction upon DNA damage (Fig. 5). However, both responses were cell cycle dependent and S/G2 cells clearly separated from G1 cells upon damage. The means show similar trends for RNR2 and H2A-S129p induction, and the few G1 cells that showed high H2A-S129p staining also generally had higher RNR2 mRNA levels. Thus, in response to MMS damage, G1 cells display much lower Mec1 kinase activity than S/G2 cells. While lower RNR2 expression in G1 cells was expected, the corresponding lower Mec1 kinase activity was somewhat surprising because a previous study has shown that Mec1 can be activated throughout the cell cycle (31), and we, too, detected Mec1 activity in α-factor-arrested cells (see Fig. S10 in the supplemental material). Future work will explore if this is a peculiarity of the damage caused by MMS or if the 9-1-1-dependent pathway operating in G1 is less efficient at activating Mec1 than the Pol ε-dependent pathway, which operates in the S phase in conjunction with 9-1-1 (31).
Fig 5.
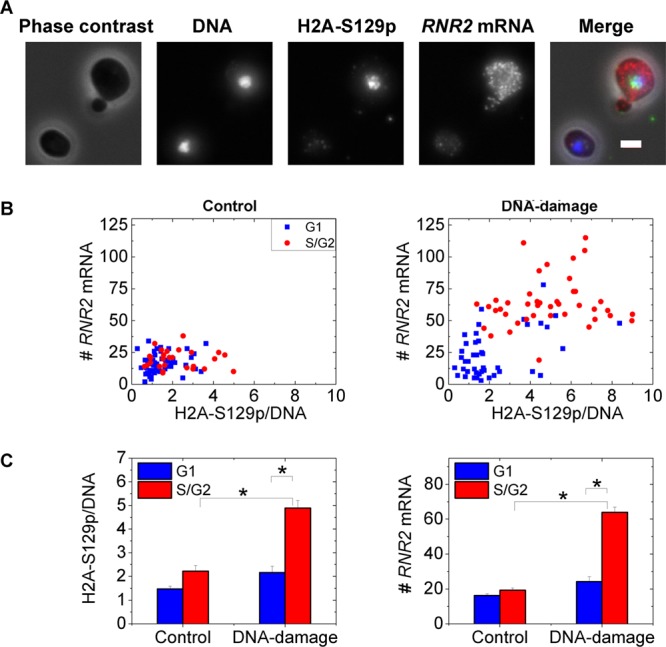
RNR2 induction correlates with variable Mec1 kinase activity in the cell cycle. (A) Two typical cells from an MMS-treated sample are shown. Note that the higher H2A-S129p staining, indicative of Mec1 kinase activity, in the budded cell correlates with higher RNR2 mRNA numbers. Scale bar, 2 μm. In the merged image, DNA is blue, H2A-S129p is green, RNR2 mRNA is red, and the phase image is gray. (B) RNR2 mRNA numbers are plotted against H2A-S129p stain intensity in control untreated cells and MMS-treated cells (n = 85 cells each). The Pearson's r value for the untreated sample is 0.16, while it is 0.6 with DNA damage. The H2A-S129p stain intensity is normalized by the DNA intensity evaluated in the same nuclear mask to ensure that the differential response between G1 and S/G2 cells is not merely a function of DNA synthesis. (C) Mean values for the H2A-S129p stain intensity and RNR2 mRNA from the graphs in panel B. *, P < 10−3 in a Kolmogorov-Smirnov test.
Finally, a core strength of investigating single-cell responses is that forms of the underlying distributions across cell populations can be assessed in addition to the means. The RNR2 mRNA distributions appeared bimodal when cell cycle stage was ignored, but the two peaks resolved into two overlapping unimodal distributions when cells were classified according to cell cycle. The two peaks were not as well resolved in the RNR4 data. Single-cell-level variability, or “noise,” in RNR mRNA expression generally increased upon DNA damage, with the large subunits exhibiting greater variability than the small subunits (Fig. 6) when resolved according to the cell cycle stage. Fano factors (σ2/μ; variance by mean of the distributions) quantify this noise, and a Poissonian distribution has a Fano factor of 1, as expected for mRNA production with constant probability in time (19, 20). “Transcriptional bursting” can, however, result in larger variability within the population and, consequently, higher Fano factors (19). Control, untreated mRNA distributions for all RNRs exhibited Fano factors greater than 1, indicative of noisy, non-Poissonian transcriptional processes (19, 20). While expression noise generally increased upon induction by DNA damage for most of the RNRs when parsed according to the cell cycle, the assumption of a steady state that is required to mechanistically interpret these distributions is not satisfied due to the transient nature of the DNA damage response. Similar Fano factors cannot be calculated for the protein distributions, as absolute numbers are not measured (19), but these exhibit forms different from those of the mRNA distributions (see Fig. S11 in the supplemental material).
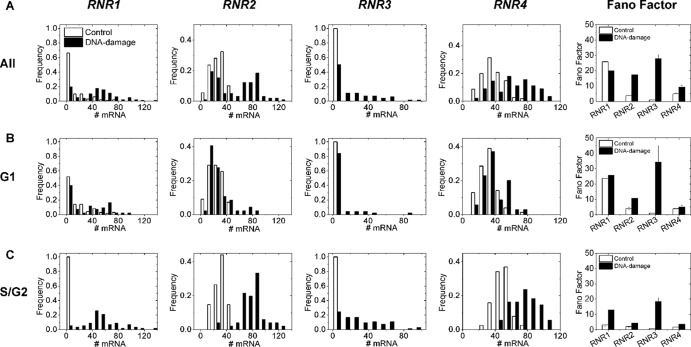
Fig 6.
mRNA histograms capture heterogeneity within the cell population. mRNA histograms and corresponding Fano factors for the studied RNR genes for all cells (A), G1 cells (B), and S/G2 cells (C). When expressed, all RNR genes have Fano factors greater than 1, indicating non-Poissonian transcription processes. Note the generally higher Fano factors for damaged cells when parsed according to the cell cycle, though this is within error bars for RNR1 in G1. Also, when they are expressed, R1 genes have higher Fano factors than R2 genes. The error bars of the Fano factors are standard deviations obtained by bootstrapping from the distributions on the left.
DISCUSSION
The principal conclusion of this work is that the RNR response to DNA damage does not operate similarly across the cell cycle at either the transcript or the protein level. We also show that these responses correlate even at the single-cell level with each other and with Mec1 kinase activity across the cell cycle. Control of Rnr protein levels and localization in turn regulates RNR enzyme numbers and implies that the deoxynucleoside triphosphate (dNTP) synthesis potential of cell subpopulations varies according to cell cycle stage under conditions of genotoxic stress. Such fine-tuning of dNTP levels possibly minimize spontaneous mutations within the population. Our results concur with those of a previous study showing that dNTP levels are low in G1 and high in S phase and that constitutively high dNTP levels transiently arrest cells in late G1 and inhibit the DNA damage checkpoint (32). It is not well understood why dNTP levels should necessarily be low in G1. Lesion bypass by DNA polymerases has been shown to be dependent on dNTP concentrations (33). In an in vitro assay, the replicative DNA polymerase ε was not able to bypass 4-NQO-induced 8-oxoG lesions at normal S phase concentrations of dNTP but was able to bypass it when the concentrations were comparable to those in the DNA damage-induced state (33). Another independent line of evidence has demonstrated abundant incorporation of ribonucleotides into DNA by yeast replicative polymerases that, if left unrepaired, can block Pol ε (34). This, in turn, may activate the Mec1-Rad53 pathway (31) and the downstream RNR transcriptional response (9, 10). Given the large molar excess of ribonucleoside triphosphates (rNTPs) over dNTPs in cells (34), upregulating dNTP production may reduce rNTP misincorporation into DNA. However, it is well known that while dNTPs are essential for responding to genotoxic stress, high dNTP levels are mutagenic and the RNR enzyme is subject to dATP feedback inhibition (35). The Mec1-Rad53-Dun1 target Sml1 also regulates the activity of the RNR enzyme (12, 13). Thus, cells have evolved a number of mechanisms for regulating dNTP concentrations by controlling the levels, localization, and activation state of the RNR enzyme components. Our work shows that the observed low dNTP levels in G1 can, at least in part, be due to low absolute numbers of the active enzyme in this cell cycle stage.
Expressions of RNR2, RNR3, and RNR4 genes are controlled by the transcriptional repressor Crt1, while RNR1 is under the regulation of the activator Ixr1 through a Dun1-independent branch of the Mec1-Rad53 pathway (9, 10). The resultant highly heterogeneous mRNA distributions are consistent with models of transcriptional bursting of the RNR genes (19). Unlike mammalian cells, only a small subset of yeast genes are thought to undergo bursting, and promoter regions in these genes are enriched in TATA elements (29). Only 20% of yeast genes have TATA boxes in their promoters, and these are also enriched in stress-related genes (36, 37), which have been shown to exhibit particularly noisy expression (36). The RNR genes also have TATA regulatory elements in their promoters (37, 38), supporting the observed non-Poissonian nature of RNR transcription under control conditions. Functional consequences of this variability in expression may be important to ensure survival of subpopulations of cells under challenging environmental conditions (17, 19). Future work will explore how the heterogeneity in RNR expression promotes cell survival.
In the broader context of gene expression, a previous study that explored simultaneous detection of yellow fluorescent protein (YFP)-tagged E. coli proteins and the transcripts that encoded them found little correlation between the levels of these two gene products (20). However, fluorescent protein signals are severely attenuated in most fixation procedures, and both mRNA numbers and protein levels can be affected by the addition of tags. Further, mRNA-protein correlations under conditions of stress have not been explored at the single-cell level, as reported here in the model eukaryote S. cerevisiae. The methods developed here for monitoring endogenous mRNA and protein levels simultaneously offer important insight into RNR enzyme regulation in eukaryotes, showing clear cell cycle-dependent partitioning of the RNR response in terms of both the mRNA and protein induction and the subcellular trafficking of Rnr subunits. RNR genes are overexpressed in many cancers (39–41). This work establishes an experimental platform for subsequent studies on the effects of DNA damage in metazoan cells that may serve to investigate the development and progression of cancer, which requires understanding the misregulation of expression patterns at the single-cell level that result in disease phenotype.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by MIT Faculty Start-up Funds, the Samuel A. Goldblith Professorship, and a CEHS Pilot Project grant (deriving from NIH P30-ES002109) (to M.B., K.T., and A.M.), NIH R01-CA055042 and DP1-ES022576 (to L.D.S.), and the CSBi Merck-MIT fellowship (to A.M.).
We thank JoAnne Stubbe for the gift of Rnr antibodies and for insightful discussions. We thank Narendra Maheshri for insightful discussions, Arjun Raj for initial guidance with the FISH experiments, Katharina Ribbeck for access to the Zeiss Observer Z1 microscope, and Gerald Fink for the gift of the RC634 strain. The fluorophore-labeled DNA oligonucleotide probes were purified at the MIT Biopolymers facility.
Footnotes
Published ahead of print 26 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01020-12.
REFERENCES
- 1. Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. 2006. DNA repair and mutagenesis. ASM Press, Washington, DC [Google Scholar]
- 2. Hoeijmakers JH. 2007. Genome maintenance mechanisms are critical for preventing cancer as well as other aging-associated diseases. Mech. Ageing Dev. 128: 460– 462 [DOI] [PubMed] [Google Scholar]
- 3. Nordlund P, Reichard P. 2006. Ribonucleotide reductases. Annu. Rev. Biochem. 75: 681– 706 [DOI] [PubMed] [Google Scholar]
- 4. Yao R, Zhang Z, An X, Bucci B, Perlstein DL, Stubbe J, Huang M. 2003. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc. Natl. Acad. Sci. U. S. A. 100: 6628– 6633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhao X, Muller EG, Rothstein R. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2: 329– 340 [DOI] [PubMed] [Google Scholar]
- 6. Chabes A, Domkin V, Larsson G, Liu A, Graslund A, Wijmenga S, Thelander L. 2000. Yeast ribonucleotide reductase has a heterodimeric iron-radical-containing subunit. Proc. Natl. Acad. Sci. U. S. A. 97: 2474– 2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Perlstein DL, Ge J, Ortigosa AD, Robblee JH, Zhang Z, Huang M, Stubbe J. 2005. The active form of the Saccharomyces cerevisiae ribonucleotide reductase small subunit is a heterodimer in vitro and in vivo. Biochemistry 44: 15366– 15377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. An X, Zhang Z, Yang K, Huang M. 2006. Cotransport of the heterodimeric small subunit of the Saccharomyces cerevisiae ribonucleotide reductase between the nucleus and the cytoplasm. Genetics 173: 63– 73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang M, Zhou Z, Elledge SJ. 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94: 595– 605 [DOI] [PubMed] [Google Scholar]
- 10. Tsaponina O, Barsoum E, Astrom SU, Chabes A. 2011. Ixr1 is required for the expression of the ribonucleotide reductase Rnr1 and maintenance of dNTP pools. PLoS Genet. 7: e1002061 doi:10.1371/journal.pgen.1002061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee YD, Wang J, Stubbe J, Elledge SJ. 2008. Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol. Cell 32:70– 80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R. 2001. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 20: 3544– 3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao X, Rothstein R. 2002. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc. Natl. Acad. Sci. U. S. A. 99: 3746– 3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Elledge SJ, Davis RW. 1989. DNA damage induction of ribonucleotide reductase. Mol. Cell. Biol. 9: 4932– 4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Elledge SJ, Davis RW. 1990. Two genes differentially regulated in the cell cycle and by DNA-damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev. 4: 740– 751 [DOI] [PubMed] [Google Scholar]
- 16. Huang M, Elledge SJ. 1997. Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 6105– 6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Altschuler SJ, Wu LF. 2010. Cellular heterogeneity: do differences make a difference? Cell 141: 559– 563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cai L, Friedman N, Xie XS. 2006. Stochastic protein expression in individual cells at the single molecule level. Nature 440: 358– 362 [DOI] [PubMed] [Google Scholar]
- 19. Raj A, van Oudenaarden A. 2009. Single-molecule approaches to stochastic gene expression. Annu. Rev. Biophys. 38: 255– 270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. 2010. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329: 533– 538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee MV, Topper SE, Hubler SL, Hose J, Wenger CD, Coon JJ, Gasch AP. 2011. A dynamic model of proteome changes reveals new roles for transcript alteration in yeast. Mol. Syst. Biol. 7: 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. de Godoy LM, Olsen JV, Cox J, Nielsen ML, Hubner NC, Frohlich F, Walther TC, Mann M. 2008. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455:1251– 1254 [DOI] [PubMed] [Google Scholar]
- 23. Gygi SP, Rochon Y, Franza BR, Aebersold R. 1999. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19: 1720– 1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bregman A, Avraham-Kelbert M, Barkai O, Duek L, Guterman A, Choder M. 2011. Promoter elements regulate cytoplasmic mRNA decay. Cell 147: 1473– 1483 [DOI] [PubMed] [Google Scholar]
- 25. Trcek T, Larson DR, Moldon A, Query CC, Singer RH. 2011. Single-molecule mRNA decay measurements reveal promoter-regulated mRNA stability in yeast. Cell 147: 1484– 1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. 2008. Imaging individual mRNA molecules using multiple singly labeled probes. Nat. Methods 5: 877– 879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tan RZ, van Oudenaarden A. 2010. Transcript counting in single cells reveals dynamics of rDNA transcription. Mol. Syst. Biol. 6: 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Youk H, Raj A, van Oudenaarden A. 2010. Imaging single mRNA molecules in yeast. Methods Enzymol. 470:429– 446 [DOI] [PubMed] [Google Scholar]
- 29. Zenklusen D, Larson DR, Singer RH. 2008. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat. Struct. Mol. Biol. 15: 1263– 1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Domkin V, Thelander L, Chabes A. 2002. Yeast DNA damage-inducible Rnr3 has a very low catalytic activity strongly stimulated after the formation of a cross-talking Rnr1/Rnr3 complex. J. Biol. Chem. 277: 18574– 18578 [DOI] [PubMed] [Google Scholar]
- 31. Puddu F, Piergiovanni G, Plevani P, Muzi-Falconi M. 2011. Sensing of replication stress and Mec1 activation act through two independent pathways involving the 9-1-1 complex and DNA polymerase epsilon. PLoS Genet. 7: e1002022 doi:10.1371/journal.pgen.1002022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chabes A, Stillman B. 2007. Constitutively high dNTP concentration inhibits cell cycle progression and the DNA damage checkpoint in yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 104:1183– 1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sabouri N, Viberg J, Goyal DK, Johansson E, Chabes A. 2008. Evidence for lesion bypass by yeast replicative DNA polymerases during DNA damage. Nucleic Acids Res. 36: 5660– 5667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nick McElhinny SA, Watts BE, Kumar D, Watt DL, Lundstrom EB, Burgers PM, Johansson E, Chabes A, Kunkel TA. 2010. Abundant ribonucleotide incorporation into DNA by yeast replicative polymerases. Proc. Natl. Acad. Sci. U. S. A. 107: 4949– 4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L. 2003. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 112: 391– 401 [DOI] [PubMed] [Google Scholar]
- 36. Bar-Even A, Paulsson J, Maheshri N, Carmi M, O'Shea E, Pilpel Y, Barkai N. 2006. Noise in protein expression scales with natural protein abundance. Nat. Genet. 38: 636– 643 [DOI] [PubMed] [Google Scholar]
- 37. Basehoar AD, Zanton SJ, Pugh BF. 2004. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116: 699– 709 [DOI] [PubMed] [Google Scholar]
- 38. Tomar RS, Zheng S, Brunke-Reese D, Wolcott HN, Reese JC. 2008. Yeast Rap1 contributes to genomic integrity by activating DNA damage repair genes. EMBO J. 27: 1575– 1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cerqueira NM, Fernandes PA, Ramos MJ. 2007. Ribonucleotide reductase: a critical enzyme for cancer chemotherapy and antiviral agents. Recent Pat. Anticancer Drug Discov. 2: 11– 29 [DOI] [PubMed] [Google Scholar]
- 40. Cerqueira NM, Fernandes PA, Ramos MJ. 2007. Understanding ribonucleotide reductase inactivation by gemcitabine. Chemistry 13: 8507– 8515 [DOI] [PubMed] [Google Scholar]
- 41. Jordheim LP, Seve P, Tredan O, Dumontet C. 2011. The ribonucleotide reductase large subunit (RRM1) as a predictive factor in patients with cancer. Lancet Oncol. 12: 693– 702 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.