Abstract
In eukaryotic cells, sphingoid long chain bases (LCBs) such as sphingosine or phytosphingosine (PHS) behave as second messengers involved in various processes including programmed cell death (PCD). In plants, induction of PCD by LCBs has now been described, but the signalling pathway is still enigmatic. Using Arabidopsis, we identify new key steps in this pathway. We demonstrate that PHS induces activation of the calcium-dependent kinase CPK3, which phosphorylates its binding partners, the 14-3-3 proteins. This phosphorylation leads to the disruption of the complex and to CPK3 degradation. Using cpk3 knockout lines, we demonstrate that CPK3 is a positive regulator of LCB-mediated PCD. These findings establish 14-3-3-regulated CPK3 as a key component of the LCB pathway leading to PCD in plants.
Keywords: calcium signalling, programmed cell death, 14-3-3 proteins, CDPK, fumonisin B1, LCBs
In eukaryotes, programmed cell death (PCD) is an integral component of development and defence responses to various stresses.1, 2 PCD in animal cells is a complex mechanism induced by several death inducers including sphingolipids, a complex range of lipids in which fatty acids are linked by amide bonds to a sphingoid long chain base (LCB).3 Indeed, sphingosine, a sphingolipid metabolite and the most abundant free LCB in animals, induces apoptosis in many cell types through its accumulation in response to physiological activators.4 The molecular mechanisms involved in sphingosine-induced signalling leading to cell death remain unclear, however. Recently, direct binding of this LCB to 14-3-3 proteins was shown to control cell fate.5 The 14-3-3 proteins are a family of highly conserved proteins that have central regulatory roles in eukaryotes.6 14-3-3s Function as homo- and heterodimers that bind phosphopeptide motifs in diverse target proteins.7, 8 Through these interactions, 14-3-3s can modulate the subcellular localization, enzymatic activity, protein–protein interactions and proteolysis of their target proteins.9, 10 Binding of 14-3-3s to certain targets is associated with cell survival in mammalian cells, as 14-3-3s have been shown to inhibit multiple pro-apoptotic molecules such as BAD, BAX and Forkhead-box protein O class.11 Recently, it has been shown that sphingosine allows phosphorylation of 14-3-3ζ on a specific serine residue (Ser58).5 Sphingosine-dependent phosphorylation of 14-3-3ζ at Ser58 can be catalysed by two different protein kinases, namely sphingosine-dependent kinase 1 (SDK1), identified as the caspase-cleaved fragment of protein kinase C (PKC)δ,12, 13 and protein kinase A (PKA).14 Ser58 is located at the interface of the subunits in the 14-3-3 dimer, and its phosphorylation results in disruption of this dimeric structure.15 As the functions of 14-3-3s depend on their dimeric structure,16, 17 sphingosine might induce apoptosis through induction of 14-3-3 monomerization,5, 18 however, to the best of our knowledge, the involvement of this sphingosine-dependent mechanism in the dissociation of 14-3-3s from proapoptotic mediators has never been demonstrated.
Whereas the role of sphingolipids and their metabolites in induction of apoptosis in animal systems is well established,3 their involvement in plant PCD is emerging. Arabidopsis mutants impaired in LCB transport or sphingolipid metabolism exhibit spontaneous cell death phenotypes.19, 20 Fumonisin B1 (FB1), a toxin produced by the necrotrophic fungus Fusarium moniliforme, inhibits the enzyme ceramide synthase and induces plant PCD triggered by subsequent accumulation of dihydrosphingosine (d18:0, DHS) and phytosphingosine (PHS; t18:0, PHS).21, 22, 23 Moreover, a fast and sustained increase in PHS, due to de novo synthesis from DHS, was detected in Arabidopsis thaliana during the interaction with an avirulent strain of the bacterial pathogen Pseudomonas syringae pv. tomato, which typically leads to a localized PCD called the hypersensitive response.24 In a previous study, we showed that DHS-induced PCD in tobacco BY-2 cells is controlled by nuclear calcium,25 and recently the mitogen-activated protein kinase (MPK6) was described as a transducer in the LCB-mediated PCD in Arabidopsis.22
To get further insights into the still unknown signalling pathway of LCB-induced PCD in plants, we combined biochemistry and reverse genetics using the model plant Arabidopsis thaliana. We show that Arabidopsis CPK3, a member of the plant family of calcium-dependent Ser/Thr protein kinases (CDPKs or CPKs), has a critical role in LCB-mediated cell death. We found that CPK3 dissociates from 14-3-3s and is degraded during PHS-induced cell death. We also show that the equivalent of the Ser58 epitope in 14-3-3ζ is phosphorylated in Arabidopsis 14-3-3s in a PHS and calcium-dependent manner by CPK3, thus identifying the plant kinase able to phosphorylate 14-3-3s at this site in the dimer inferface. Moreover, gene knockout of CPK3 results in a FB1-resistant phenotype in Arabidopsis, revealing a new role for CPK3 as a positive regulator of plant PCD. On the basis of these findings, we provide a working model for how LCBs and calcium regulate CPK3 and 14-3-3 proteins in the context of plant cell death.
Results
PHS induces calcium-dependent PCD
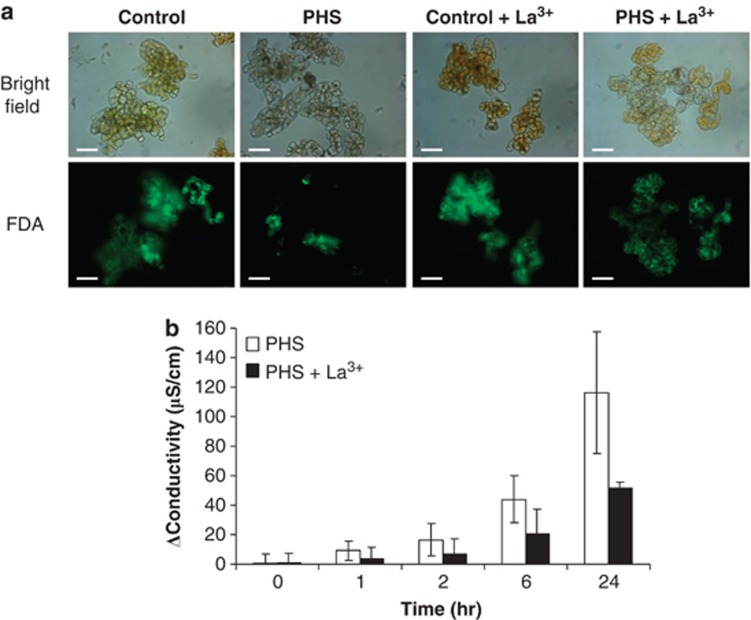
To investigate the effect of LCBs on plant cells, we assayed whether PHS was able to induce the death of Arabidopsis cells in culture qualitatively, by vital staining with fluorescein diacetate (FDA), and semi-quantitatively by using an electrolyte leakage assay (Figures 1a and b). Continuously applied PHS-induced cell death (Figure 1a) in a time-dependent manner (Figure 1b). As our previous work indicated that cell death induced by DHS is controlled by nuclear calcium in tobacco cells,25 we speculated that PHS-induced cell death might be Ca2+-dependent in Arabidopsis cells. To test this, we checked the effect of lanthanum ions (La3+), a general calcium channel blocker, on PHS-induced cell death. As shown in Figures 1a and b, La3+ efficiently blocked cell death, suggesting that a calcium influx is required for the response to PHS in Arabidopsis.
Figure 1.
PHS induces cell death in a time- and calcium-dependent manner. (a) Arabidopsis cells were treated ±25 μM PHS and ±1 mM LaCl3 for 6 h. FDA staining of living cells was monitored by fluorescence microscopy. Scale bars correspond to 50 μm. (b) Arabidopsis cells were treated ±25 μM PHS and ±1 mM LaCl3. The conductivity of the cell suspension was measured at the indicated times. For each time point, data are expressed as the differences between the PHS treatments and their corresponding controls. Data are shown as means±S.E.M. of four independent experiments. The data were analysed by a two-way analysis of covariance where time was the covariate. The increase in conductivity with time was highly significant (P<0.0001), and the effect of lanthanum ions was significant with P=0.012
PHS-induced phosphorylation of 14-3-3s is controlled by calcium
Given the importance of 14-3-3 phosphorylation at Ser58 for initiation of apoptosis by sphingosine in animal cells,5 we examined whether Arabidopsis 14-3-3s might also be phosphorylated in response to PHS. The Ser58 phosphorylation site in the human isoform 14-3-3ζ and the primary sequence surrounding it are highly conserved in yeast and plant 14-3-3 proteins (Supplementary Figure S1). Among the 13 Arabidopsis 14-3-3 isoforms,6 all except lambda and kappa possess a Ser at the site corresponding to Ser58 in 14-3-3ζ. We therefore tested whether Arabidopsis 14-3-3 isoforms were phosphorylated on this residue, and whether PHS was required for this phosphorylation event. The phospho-Ser58 (pSer58) antibody raised against a phosphopeptide corresponding to Gly53–Ser63 of 14-3-3ζ (Supplementary Figure S1) detected several 14-3-3 isoforms after PHS-dependent phosphorylation of a protein extract from Arabidopsis cells (Figure 2a). This suggests that an Arabidopsis protein kinase induces PHS-dependent phosphorylation of endogenous 14-3-3 proteins on an epitope equivalent to the Ser58 epitope in 14-3-3ζ. To determine whether this phosphorylation occurs in vivo in response to PHS, 14-3-3s were immmunoprecipitated from control or PHS-treated Arabidopsis cells using antibodies directed against plant 14-3-3s. Immunoprecipitates were analysed by immunoblotting with the antibody against pSer58. PHS treatment resulted in an increase in 14-3-3 phosphorylation, which was inhibited by La3+ (Figure 2b), indicating that calcium regulates PHS-dependent 14-3-3 phosphorylation.
Figure 2.
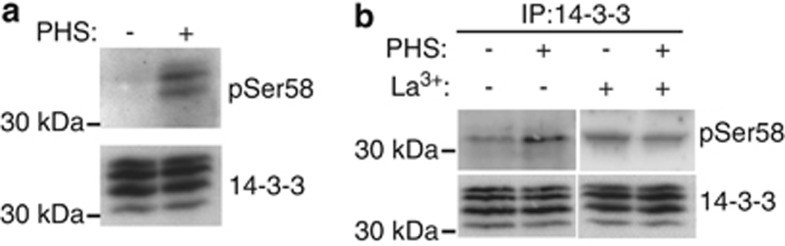
PHS induces calcium-dependent phosphorylation of 14-3-3 proteins on a conserved serine residue corresponding to Ser58 of human 14-3-3ζ. (a) In vitro phosphorylation assays were performed in the presence or absence (±) of 25 μM PHS on protein extracts of untreated Arabidopsis cells. Phosphorylated 14-3-3 proteins were detected by immunoblotting using antibodies to phosphorylated Ser58 (pSer58). Immunoblotting with an anti-14-3-3 protein antibody shows equal loading. The results presented are representative of three separate experiments. (b) Arabidopsis cells were treated ±25 μM PHS and ±1 mM LaCl3 for 2 h. 14-3-3 Immunoprecipitates from cell extracts were immunoblotted with the indicated antibodies. The results presented are representative of three separate experiments
14-3-3-Binding and proteolysis of CPK3 are regulated by PHS
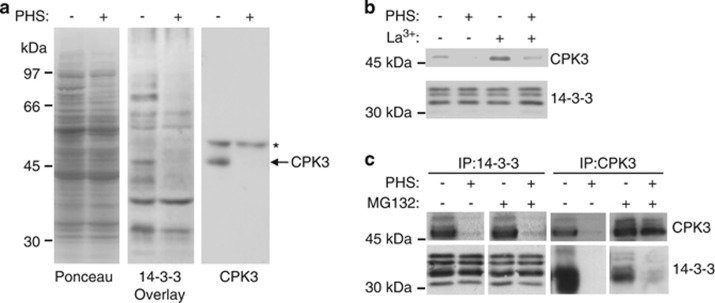
Phosphorylation of the 14-3-3ζ dimer at Ser58 induces its monomerization,15 which is expected to disrupt 14-3-3 interactions with ‘client' proteins, as many targets bind to 14-3-3 dimers.17, 26 Moreover, loss of binding to 14-3-3 can induce proteolysis and/or dephosphorylation of target proteins,9, 10 making them unable to subsequently bind in vitro to 14-3-3s. To examine the effect of PHS-induced 14-3-3 phosphorylation on the global 14-3-3-binding status of target proteins, we used the digoxigenin (DIG)-14-3-3 overlay assay.7 Treatment of Arabidopsis cells with PHS resulted in a decrease in the intensities of several DIG-14-3-3-binding proteins (Figure 3a, middle panel). One of these had an apparent molecular mass of around 50 kDa, consistent with the size of a calcium-dependent protein kinase (CDPK or CPK) previously identified as a 14-3-3-binding protein.7, 9 To determine whether this 50 kDa protein was a CPK, we immunoblotted the samples analysed in 14-3-3 overlay assays, probing the blots with an antibody raised against 6-His-tagged CPK3 (also known as CDPK6). This antibody was specific for CPK3 as indicated by the unique band detected in immunoblots of protein extracts from leaves of wild-type Arabidopsis plants and the absence of a signal in cpk3 knockout mutant plants (Supplementary Figure S2B). By using this CPK3-specific antibody to immunoblot Arabidopsis cell extracts and comparing the electrophoretic mobility of the recognized band to those detected in 14-3-3 overlay assays, we assigned the 50-kDa protein to CPK3 (Figure 3a). Moreover, western blotting revealed that the loss of CPK3 binding to 14-3-3s in the overlay assay correlated with depletion of the CPK3 protein in extracts of PHS-treated cells, suggesting that PHS might induce proteolysis of CPK3 (Figure 3b). Interestingly, the CPK3 band was not depleted when cells were treated with La3+, consistent with Ca2+-dependent degradation of the protein.
Figure 3.
PHS induces CPK3 release from 14-3-3 proteins and CPK3 proteolysis. (a) Protein extracts from Arabidopsis cells treated ±25 μM PHS for 2 h were assayed by DIG-14-3-3 overlay and immunoblotting with an anti-CPK3 antibody. Left panel shows Ponceau protein stain of the same samples. Asterisk indicates an uncharacterised band recognized by the CPK3 antibody only in Arabidopsis cells and arrow indicates CPK3. The results presented are representative of three separate experiments. (b) Protein extracts from Arabidopsis cells treated ±25 μM PHS and ±1 mM LaCl3 for 2 h were immunoblotted for CPK3. Immunoblotting with an anti-14-3-3 protein antibody shows equal loading. The results presented are representative of three separate experiments. (c) Arabidopsis cells were pretreated ±100 μM MG132 for 15 min before treatment with ±25 μM PHS for 2 h. 14-3-3 Protein and CPK3 immunoprecipitates were analysed by immunoblotting. The results presented are representative of three separate experiments
Given the effect of PHS on the binding of CPK3 to 14-3-3s in the overlay assay, we next examined whether treating Arabidopsis cells with PHS would disrupt any endogenous interaction between 14-3-3s and CPK3. In Arabidopsis cell extracts, CPK3 was co-immunoprecipitated with an anti-14-3-3 antibody and conversely 14-3-3s were precipitated with an anti-CPK3 antibody, confirming that CPK3 bind to endogenous 14-3-3s (Figure 3c). In accordance, CPK3 was found associated with 14-3-3s in untreated leaves of Arabidopsis wild-type plants whereas CPK3 was not co-immunoprecipitated with an anti-14-3-3 antibody from leaves of cpk3 knockout plants (Supplementary Figure S3). Consistent with the DIG-14-3-3 overlay assay (Figure 3a), association of CPK3 with 14-3-3s was much lower in PHS-treated cells than in control cells (Figure 3c). Moreover, very little CPK3 was immunoprecipitated with the anti-CPK3 antibody from PHS-treated cells (Figure 3c), consistent with the depletion of CPK3 from extracts of PHS-treated cells seen above (Figures 3a and b). Previously, we have shown that CPK3, like many other targets of 14-3-3s, is cleaved in Arabidopsis cells starved of sugars, and that this starvation-induced degradation is blocked by the cysteine protease inhibitor MG132.9 We asked, therefore, whether MG132 would stabilize CPK3 in PHS-treated cells. Indeed, MG132 did protect CPK3 against PHS-induced degradation, as indicated by immunoprecipitation and immunoblotting (Figure 3c). Interestingly, however, 14-3-3s were not associated with this CPK3, confirming that treating Arabidopsis cells with PHS disrupts the interaction between CPK3 and 14-3-3s.
PHS mediates 14-3-3 phosphorylation on Ser58 site by Ca2+-activated CPK3
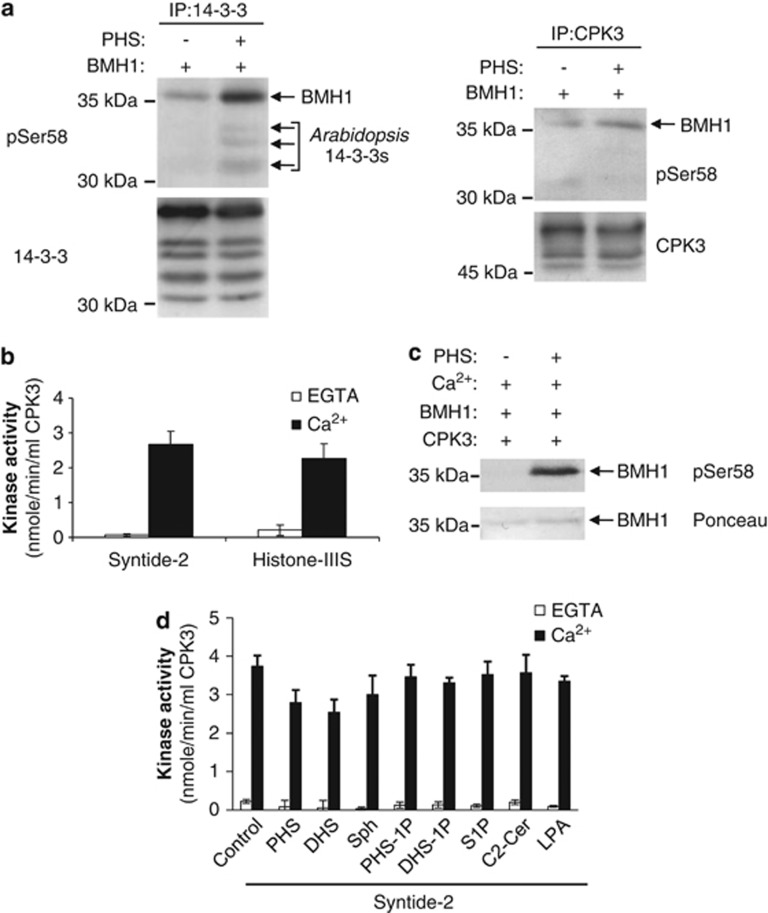
Sphingosine-dependent protein kinase (SDK1) activity co-purifies with 14-3-3s in extracts of mouse Balb/c 3T3 cells, suggesting that this kinase is physically associated with 14-3-3s, either directly or indirectly.13 To investigate whether a similar kinase exists in plants, we attempted to phosphorylate the yeast 14-3-3 BMH1 with immunoprecipitates of 14-3-3s from Arabidopsis cells as the source of kinase activity, in the presence or absence of PHS. We detected little phosphorylation of BMH1 by the immunoprecipitates in the absence of PHS, whereas phosphorylation was markedly enhanced in the presence of PHS (as detected with the pSer58 antibody; Figure 4a and Supplementary Figure S4). Not only was the yeast 14-3-3 phosphorylated upon incubation with PHS but also the endogenous immunoprecipitated Arabidopsis 14-3-3s were phosphorylated on a site recognized by the pSer58 antibody (Figure 4a). These data indicate that a PHS-dependent kinase is associated with endogenous 14-3-3s in Arabidopsis. Given that CPK3 binds to 14-3-3s (Figure 3c), we speculated that this PHS-dependent kinase might be CPK3. To test this, we examined whether CPK3 immunoprecipitated from Arabidopsis cells could phosphorylate BMH1 in vitro. As shown in Figure 4a, PHS-dependent phosphorylation of BMH1 by immunoprecipitates of CPK3 was observed with the anti-pSer58 antibody, suggesting that CPK3 is a LCB-dependent plant kinase that phosphorylates the Ser58 epitope in 14-3-3s.
Figure 4.
CPK3 phosphorylates 14-3-3 Ser58 site in the presence of PHS. (a) Proteins from untreated Arabidopsis cells were immunoprecipitated with the indicated antibodies. Immunoprecipitates were used as kinase sources in nonradioactive in vitro phosphorylation assays performed with recombinant BMH1 and ±25 μM PHS. Proteins from these assays were blotted with the indicated antibodies. The results presented are representative of three separate experiments. See also Supplementary Figure S4. (b) The activity of recombinant Arabidopsis CPK3 was assayed in the presence (Ca2+) or absence (EGTA) of Ca2+ using a synthetic peptide (syntide-2) or histone-IIIS substrates. Data are shown as means±S.E.M. (n=3). (c) Recombinant BMH1 was used as the substrate in kinase assays with recombinant Arabidopsis CPK3 assayed in the presence of Ca2+ and ±25 μM PHS. Proteins were analysed by immunoblotting with anti-pSer58 antibodies and Ponceau staining. The results presented are representative of three separate experiments. See also Supplementary Figure S5. (d) The activity of recombinant Arabidopsis CPK3 was assayed using syntide-2 in the presence (Ca2+) or absence (EGTA) of Ca2+ and ±25 μM of the indicated lipid: PHS (t18:0); DHS (d18:0); sphingosine, Sph (d18:1); PHS-1-phosphate, PHS-1P; DHS-1-phosphate, DHS-1P; sphingosine-1-phosphate, S1P; C2-ceramide, C2-Cer; and oleoyl-ℒ-α-lysophosphatidic acid, LPA. Data are means±S.E.M. (n=3)
To confirm direct PHS-dependent phosphorylation of 14-3-3s by CPK3, we performed in vitro kinase assays using recombinant His-tagged CPK3. Recombinant Arabidopsis CPK3 had CDPK activity when tested with a peptide substrate, syntide-2, and a protein substrate, histone-IIIS, with similar activities toward both substrates (Figure 4b). To test whether CPK3 functions as a 14-3-3 Ser58 kinase, we assessed the ability of recombinant Ca2+-activated CPK3 to phosphorylate recombinant BMH1 in vitro. Thereby, in the presence of Ca2+ and PHS, CPK3 was able to phosphorylate the Ser58 site of the yeast 14-3-3 protein (Figure 4c and Supplementary Figure S5). To determine whether the PHS dependency of this phosphorylation is due to substrate modulation or kinase activation, a panel of sphingolipids and related molecules were examined for their effect on phosphorylation of syntide-2 by CPK3 in the presence or absence of Ca2+. Neither PHS nor any of the other lipids we tested activated CPK3's kinase activity toward the peptide substrate (Figure 4d), indicating that PHS is not acting on the kinase. Collectively, these data demonstrate that Ca2+-activated CPK3 is not a LCB-dependent protein kinase, but is able to phosphorylate 14-3-3 Ser58 in the presence of PHS, suggesting that the LCB modulates 14-3-3 proteins for CPK3 phosphorylation at this site.
FB1 disrupts the interaction between CPK3 and 14-3-3s and induces CPK3 cleavage
In Arabidopsis plants, LCBs mediate PCD induced by the fungus toxin FB1.22, 23 To examine the effect of FB1 on the interaction between 14-3-3s and CPK3 in plants, we immunoprecipitated 14-3-3s from Arabidopsis leaves after treatment with FB1 or with the control using antibodies directed against plant 14-3-3s. Immunoblotting revealed that the same amount of 14-3-3s were immunoprecipitated from FB1-treated plants and from the control plants (Figure 5a). By contrast, less CPK3 was detected in the immunoprecipitates from FB1-treated leaves than from control leaves (Figure 5a), indicating that FB1 disrupts CPK3 interaction with 14-3-3s in Arabidopsis plants. To determine whether this process was accompanied by CPK3 proteolysis, we looked at protein levels in the same extracts by immunoblotting. In extracts of FB1-treated leaves, intact CPK3 was depleted and faster-migrating immunoreactive bands were seen (Figure 5b). By contrast, FB1 did not alter the abundance of total 14-3-3s or of glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a protein known to bind 14-3-3s9 (Figure 5b). These findings suggest that CPK3 is selectively cleaved following FB1 treatment of Arabidopsis leaves.
Figure 5.
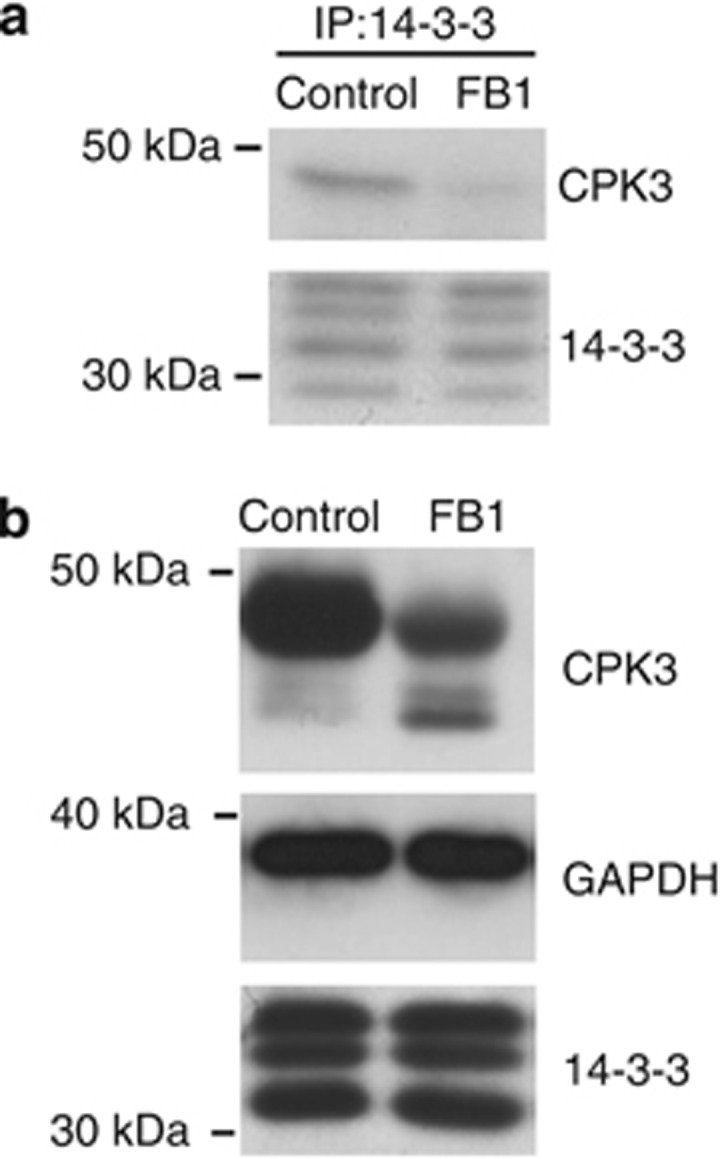
FB1 promotes CPK3 release from 14-3-3 proteins and CPK3 proteolysis. (a) Arabidopsis leaves were treated ±10 μM FB1 for 48 h. 14-3-3 immunoprecipitates were immunoblotted with the indicated antibodies. The results presented are representative of three separate experiments. (b) Protein extracts from Arabidopsis leaves treated as in (a) were blotted for the indicated proteins. The results presented are representative of three separate experiments
CPK3 knockout plants display a FB1-resistant phenotype
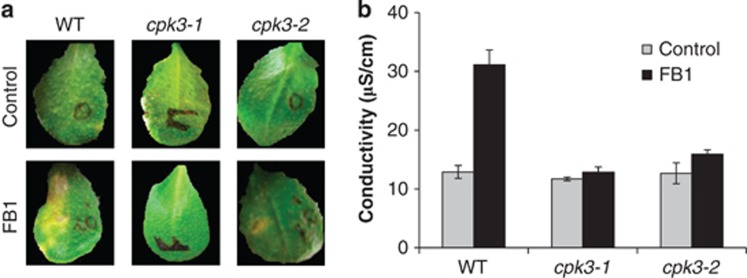
To address whether CPK3 has a role in LCB-mediated PCD in plants, we analysed CPK3 knockout plant lines for their response to FB1 treatment. We used two independent T-DNA insertion lines, cpk3-1 (SALK_107620) and cpk3-2 (SALK_022862), which have a single T-DNA inserted into the first exon and into the first intron, respectively27 (Supplementary Figure S2A). The cpk3-1 and cpk3-2 lines were verified as null mutants by immunoblotting (Supplementary Figure S2B). We compared the responses of these cpk3 mutants with that of the corresponding wild-type when FB1 was infiltrated into leaves left attached to growing plants. Four days after infiltration, severe lesions had appeared in wild-type plants, whereas the mutant plants exhibited no obvious symptoms (Figure 6a). To quantify cell death at this stage, ion leakage from leaf discs was measured. Compared with wild-type plants, the extent of FB1-induced cell death was significantly diminished in cpk3-1 and cpk3-2 (Figure 6b), indicating that they were resistant to FB1. Taken together, these results demonstrate that CPK3 is a positive regulator of FB1-induced PCD in Arabidopsis.
Figure 6.
CPK3 knockout plants are resistant to FB1. (a) Leaves of Arabidopsis wild-type (WT) or mutant plants (cpk3-1 and cpk3-2) were infiltrated in their left halves ±10 μM FB1 and photographed after 4 days. (b) Quantification of cell death was performed by electrolyte leakage assay. Plants of the indicated genotypes were treated as in (a) for 4 days and leaf discs were excised to measure conductivity. Data are shown are means±S.E.M. (n=4). See also Supplementary Figure S2
Discussion
Despite evidence that LCBs mediate PCD in plants, the underlying mechanisms are essentially unknown. We report here that the 14-3-3 target CPK3 is a critical component of the LCB-induced plant PCD, and we provide a model for the regulation of CPK3 and 14-3-3s by PHS in the context of plant cell death.
PHS regulates 14-3-3 phosphorylation by CPK3
In animals, sphingosine-dependent phosphorylation of 14-3-3ζ on Ser58 is achieved by both PKA14 and a ‘sphingosine-dependent kinase', SDK1, now known to be the kinase domain of PKCδ produced by the action of the apoptotic protease caspase-3.12 In plants, although several 14-3-3 isoforms are phosphorylated in vivo,28, 29, 30 phosphorylation at the equivalent of the 14-3-3ζ Ser58 site has not been reported previously. We show that this conserved site is phosphorylated in plants in response to PHS by the calcium-dependent kinase CPK3. Consistent with our findings, three Arabidopsis 14-3-3 isoforms (ψ, υ and ω) have been identified in a recent proteomic study as potential CPK3 substrates.31 CPK3 is a member of the large CDPK family comprising 34 members in Arabidopsis.32 CDPKs are Ca2+-binding Ser/Thr protein kinases that are not found in animals or fungi.33 These kinases are defined by a unique structure in which the catalytic kinase domain in the N-terminal half of the protein is directly tethered via an autoinhibitory junction domain to a regulatory calmodulin-like domain that contains four functional Ca2+-binding EF-hands. In conventional CDPKs, binding of Ca2+ to the C-terminal EF-hands induces a conformational change, resulting in relief of inhibition of the kinase activity.33 CPK3 is a CDPK whose activity is induced by Ca2+.34 In our study, calcium activation of CPK3 is not sufficient to promote 14-3-3 phosphorylation but PHS is also required to allow this phosphorylation at the Ser58 site. As PHS does not increase the Ca2+-induced activity of recombinant CPK3 (at least when assayed against an artificial peptide substrate), our findings suggest that the LCB modulates the structure of 14-3-3s, making the Ser58 site accessible for phosphorylation by CPK3. How might PHS mediate this effect? The conserved Ser58 site is masked in the 14-3-3 dimer interface and therefore not readily accessible to kinases.35 A recent report shows that the PHS analogue sphingosine functions as a substrate regulator for 14-3-3s, binding to them and making the protein available for Ser58 phosphorylation by PKA or the caspase-cleaved form of PKCδ.5 In the same study, the substrate modulation of mammalian 14-3-3s for phosphorylation was found to be specific to non-acylated sphingolipids that carry a net positive charge, such as PHS. As members of the eukaryotic 14-3-3 family are highly conserved, it is conceivable that PHS might also bind to plant 14-3-3s to open up the Ser58 site sufficiently to allow phosphorylation by CPK3.
14-3-3 Phosphorylation promotes CPK3 release leading to its proteolysis
In plants, the biochemical and functional effects of 14-3-3 phosphorylation in vivo have not been characterized, whereas in mammals, phosphorylation at specific sites is known to affect the interaction with targets.36 We demonstrate here that PHS induces 14-3-3 phosphorylation on Ser58 site. In animal systems, sphingosine-dependent phosphorylation of Ser58 induces 14-3-3 monomerization,15 which may mediate the release of pro-apoptotic proteins bound to 14-3-3s leading to subsequent activation of the apoptotic pathway.5, 18 Consistent with such a LCB-dependent mechanism in plants, we found that CPK3 is released from 14-3-3s in response to PHS. Moreover, this loss of 14-3-3 binding coincides with CPK3 proteolysis. As 14-3-3 binding to target proteins is known to block their proteolysis,9 we presume that 14-3-3s dissociate from CPK3 before proteolysis occurs. Consistent with this, we observed that the protease inhibitor MG132 blocked PHS-induced proteolysis of CPK3 but this did not permit its binding to 14-3-3s in vivo. PHS-induced proteolysis of CPK3 was also partially blocked by the general calcium channel blocker lanthanum chloride, suggesting that this process is Ca2+-dependent. However, PHS might also induce in parallel a Ca2+-independent degradation of CPK3, as CPK3 was less abundant in cells treated with PHS and La3+ than in cells treated with La3+ alone. Dissociation of the 14-3-3-CPK3 complex might be triggered by modification of CPK3 or by phosphorylation of 14-3-3s by the associated Ca2+-activated CPK3. In the latter case, a PHS-induced increase in Ca2+ concentration might activate 14-3-3-bound CPK3, which then phosphorylates 14-3-3 leading to dissociation of the 14-3-3-CPK3 complex and CPK3 proteolysis. Consistent with this hypothesis, we demonstrate in this study that all these events are inhibited by the broad calcium channel inhibitor lanthanum chloride. Moreover, we show that CPK3 dissociates from 14-3-3s and is less abundant in response to FB1 in Arabidopsis, suggesting that the PHS-dependent signalling complex consisting of calcium, 14-3-3 proteins and CPK3 is required for induction of plant PCD.
CPK3 is a positive regulator of LCB-mediated PCD
Little is known about the signalling proteins that control LCB-mediated PCD in plants. Only the MPK6 has been implicated so far as an essential transducer in the pathway leading to LCB-induced PCD in Arabidopsis.22 Here, by using T-DNA insertional mutants from the SALK collection in combination with FB1 treatment, we reveal a role for CPK3 in the regulation of cell death in plants. In our study, we used two independent allelic mutants and both gave the same resistant phenotype to FB1, confirming that CPK3 has a pro-cell death function in Arabidopsis. Plant Ca2+-dependent kinases regulate diverse processes in plant growth and development, and signalling in biotic and abiotic stress responses.32, 33 Analysis of knockout mutants in Arabidopsis has revealed some of the biological functions of some CPK isoforms: CPK3 and CPK6 regulate Ca2+-permeable channels and S-type anion channel currents in guard cells and are involved in abscisic acid signalling leading to stomatal closure.27 Moreover, CPK3 is required for plants to acclimatize to salt stress31 and together with CPK13, it is part of the plant signalling pathway involved in the response to a herbivore attack.37 As CPK3 induces the flagellin-responsive gene NHL10 in Arabidopsis, it may also be involved in plant innate immune signalling.38
Despite data indicating a role for CPK3 in plant defence, no pro-cell death function of this kinase has been reported to our knowledge. Another CDPK from tobacco, Nicotiana tabacum, CDPK2 (NtCDPK2), is essential for the hypersensitive response-like PCD involved in gene-for-gene (Avr9–Cf9) fungal resistance: virus-induced gene silencing of NtCDPK2 and its closely related homologues in Nicotiana benthamiana resulted in plants that were compromised in the induction of the Avr9–Cf9-dependent hypersensitive cell death response.39 In complementary gain-of-function experiments, ectopic expression of a truncated form of NtCDPK2 resulted in enhanced plant defence responses, including induction of symptoms typical of hypersensitive response-like cell death.40 This deregulated version of NtCDPK2 lacked its C-terminal regulatory autoinhibitory and Ca2+-binding domains and was therefore expected to act as a dominant, constitutively active kinase. Interestingly, we found that CPK3 is selectively cleaved in response to FB1 treatment in Arabidopsis. Further elucidation of the biochemical properties of cleaved CPK3 and identification of its substrates or partners will likely provide new insights into the mechanisms that underlie LCB-mediated PCD in plants.
A model for LCB-mediated PCD
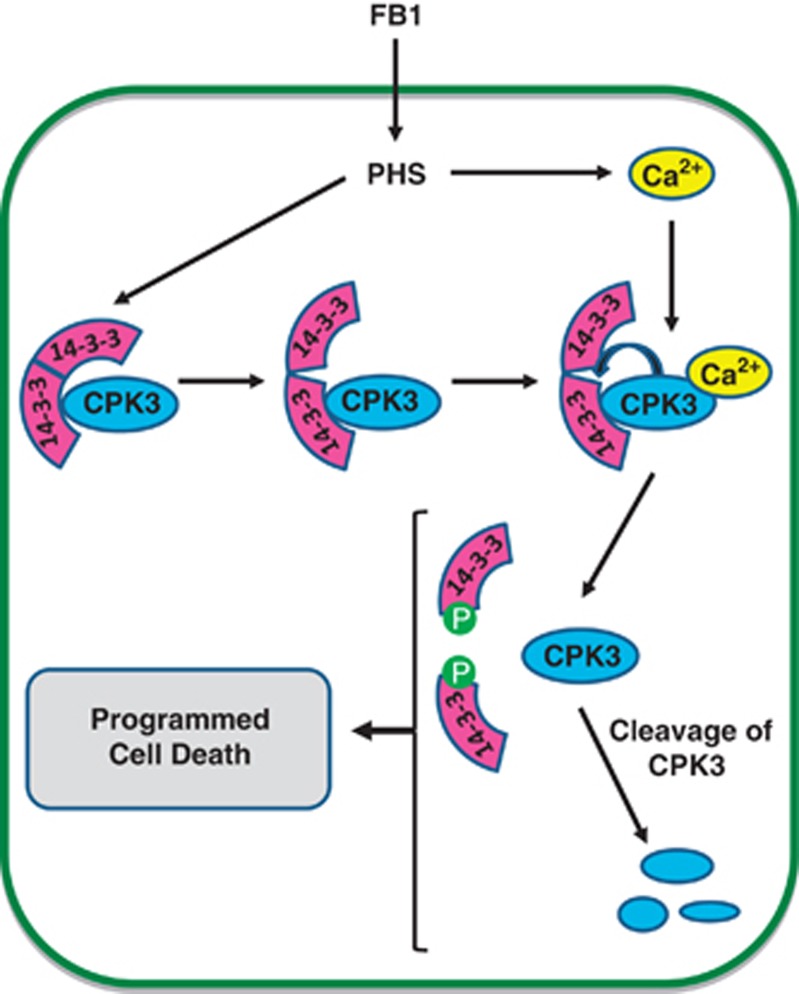
We suggest that the following events control FB1-induced PCD (Figure 7): in the absence of a cell death signal, CPK3 is bound to 14-3-3s. Upon exposure to FB1, ceramide synthase is inhibited and PHS accumulates22, 23 leading to elevation of the intracellular calcium ion concentration.25 PHS also modulates 14-3-3s to allow phosphorylation of the Ser58 site by the associated Ca2+-activated CPK3. We speculate, based on previous published work, that phosphorylation at this site disrupts the 14-3-3 dimer15 promoting dissociation of the 14-3-3-CPK3 complex, thus releasing CPK3 and subsequently allowing its proteolysis.
Figure 7.
A model for LCB-mediated PCD in Arabidopsis. In the absence of FB1, dimeric 14-3-3 proteins bind to CPK3. In response to FB1, the amount of cellular PHS increases leading to elevation of the intracellular calcium concentration. Concomitantly, the accumulation of PHS modulates 14-3-3 proteins allowing their phosphorylation by the associated Ca2+-activated CPK3. Phosphorylation of 14-3-3 proteins at the dimer interface disrupts their dimeric structure, promoting dissociation of the 14-3-3–CPK3 complex, thus releasing CPK3 and subsequently allowing its proteolysis. Collectively, these events promote FB1-induced programmed cell death
Materials and Methods
Cell culture, plant material and growth conditions
Arabidopsis cells were cultured in suspension as previously described.9 Arabidopsis thaliana plants (Columbia ecotype) were grown in soil in a growth room at 50% humidity with daily cycles of 9 h of light and 15 h of dark at a photon fluence rate of 250 μmol/m2/s and a temperature of 20 °C. CPK3 T-DNA insertion lines cpk3-1 (Salk_107620) and cpk3-2 (Salk_022862) were obtained from Julian Schroeder27 and Bernhard Wurzinger31, respectively.
Chemicals
Sphingoid bases and C2-ceramide (d18:1/2:0) were purchased from Avanti Polar Lipids (Coger, Paris, France). FB1, oleoyl-ℒ-α-lysophosphatidic acid, LaCl3 and FDA were from Sigma (Saint-Quentin Fallavier, France). MG132 was provided by Calbiochem (Merck-Millipore, Molsheim, France). The following solvents were used for the preparation of stock solutions: methanol (FB1), water (LaCl3), acetone (FDA), dimethyl sulfoxide (MG132) and ethanol (all others).
Cell and plant treatments
One-week-old cells were treated with 25 μM PHS as previously described.25 LaCl3 (1 mM) was added simultaneously with PHS to the cell incubation medium. MG132 (100 μM) was applied to cells 15 min before treatment with LCBs. Solvents were used as controls for chemical treatments. Treatments of 4-week-old plants were performed by infiltration of FB1 aqueous solution (10 μM) or methanol 0.5% (v/v) as a control into the mesophyll of half leaves by means of a syringe. After treatment, cell and leaf samples were collected and frozen in liquid nitrogen for protein extraction.
Cell viability assessment and electrolyte leakage assay
Cell viability was assessed by using the vital stain FDA as a marker of viability: 0.001% (w/v) FDA was added to a cell suspension and the fluorescence of living cells was visualized with an epifluorescence microscope (DMIRBE, Leica, Nanterre, France) at the excitation wavelength of 490 nm and emission wavelength of 528 nm. Quantification of cell death by lesion formation in infiltrated leaves was assayed by measuring ion leakage from leaf discs 96 h after FB1 treatment. For each measurement, four discs of 4 mm diameter were cut from the leaves and floated on 1.5 ml of distilled water for 1 h at room temperature. After incubation, the conductivity of the bathing solution was measured with a conductivity meter (model C532, Consort, Turnhout, Belgium).
Antibodies
Antibodies that recognize several, or possibly all, plant 14-3-3 isoforms were raised against 14-3-3s purified from spinach leaves.7 Anti-phophoSer58 14-3-3ζ antibodies were purchased from Abcam (Paris, France). Sheep polyclonal antibodies against CPK3 were raised against purified 6His-Arabidopsis CPK3 at the MRC Protein Phosphorylation Unit (Dundee, UK). The antibodies were purified from sheep serum by affinity chromatography on CH-Sepharose (GE Healthcare, Velizy-Villacoublay, France) coupled to 6His-Arabidopsis CPK3.
14-3-3 Overlays, immunoblotting and immunoprecipitation
To prepare protein extracts, Arabidopsis cells or leaves were ground in liquid nitrogen and extracted in buffer containing 50 mM HEPES-NaOH pH 7.5, 10 mM MgCl2, 1 mM DTT, 50 mM K4P2O7, 10 mM Na3VO4, 5 mM NaF, 100 μM PMSF, 2 μM leupeptin, 50 μM MG132 and the protease inhibitor cocktail for plant extracts (Sigma). After removal of insoluble materials by centrifugation at 6000 g for 30 min at 4 °C, the protein concentrations of the extracts were determined and normalized by using the Bradford assay (Uptima, Interchim, Montluçon, France). Production of recombinant 14-3-3 proteins, DIG-14-3-3 overlays, immunoprecipitation and western blotting were performed as described.7
Production of 6His-Arabidopsis CPK3
The Arabidopsis CPK3 cDNA was cloned into the expression vector pDEST17 (Invitrogen, Life Technologies, Saint Aubin, France) using Gateway technology (Invitrogen) and expressed in Escherichia coli BL21 cells. Expression was induced with isopropyl-β-𝒟-thiogalactopyranoside in E. coli BL21 cells (Invitrogen). The 6-His-tagged CPK3 was purified by Ni-NTA agarose chromatography (Qiagen, Courtaboeuf, France).
In vitro 14-3-3 phosphorylation
For in vitro phosphorylation of BMH1, either extracts (0.5 μg) or immunoprecipitates (10 μl) from Arabidopsis cells, or purified recombinant CPK3 (1 μg) were used as a kinase source. Purified recombinant BMH1 (1 μg) was incubated with the kinase source in phosphorylation buffer containing 50 mM Tris-HCl pH 7.5, 3 mM DTT, 15 mM magnesium acetate and 25 μM ATP in the presence or absence of 25 μM PHS in a total reaction volume of 30 μl at 30 °C for 15 min. When phosphorylation assays were performed with CPK3, the phosphorylation buffer was supplemented with 150 μM EGTA and 450 μM CaCl2. In this buffer, the free calcium concentration is estimated to be 300 μM. For in vitro phosphorylation of Arabidopsis 14-3-3s, reactions were carried out as for BMH1 phosphorylation reactions but using 5 μg of Arabidopsis cell extracts in place of the kinase source and BMH1. Following incubation, reactions were stopped by addition of SDS loading buffer and heating to 100 °C. The reaction products were separated by SDS-PAGE, and 14-3-3 phosphorylation was analysed by immunoblotting with pSer58 antibody.
CPK3 kinase assays
The reaction mix (45 μl) contained purified CPK3 (1 μg) in 50 mM HEPES-NaOH pH 7.5, 10 mM MgCl2, 1 mM DTT, with 25 μg of histone III-S (Sigma) or 40 μM syntide-2 (Santa Cruz Biotechnology, Heidelberg, Germany) as a substrate. Control levels of phosphorylation were determined by chelating free Ca2+ with 150 μM EGTA. To test the effect of Ca2+, the reaction mix with EGTA was supplemented with 450 μM CaCl2. To examine the effect of sphingolipids and related molecules, the enzymatic activity was measured in the presence of Ca2+ and 25 μM of the tested lipid. Reactions were initiated by adding 5 μl of 100 mM magnesium acetate and 1 mM γ-33P-ATP (500 c.p.m./pmol). After 20 min at 30 °C, 40 μl aliquots were applied to phosphocellulose papers (2 × 2 cm), which were immersed in 75 mM orthophosphoric acid, washed four times (5 min per wash) in orthophosphoric acid to remove unreacted γ-33P-ATP and subjected to scintillation counting. In control incubations the substrate was replaced by buffer.
Acknowledgments
We thank Julian I Schroeder (University of California San Diego, CA, USA) for providing cpk3-1 seeds, Bernhard Wurzinger (University of Vienna, Austria) for providing cpk3-2 seeds, Ming-Che Shih (Academia Sinica, Taipei, Taiwan) for providing anti-GAPDH antibodies and Alexandre Perochon for providing CPK3 entry clone. We are grateful to Carol MacKintosh (MRC, Dundee, UK) for anti-14-3-3 antibodies and for other helpful tools. We thank Mercedes Pozuelo Rubio, Carole Pichereaux, Gisèle Borderies and Michel Rossignol for scientific discussions. We also thank Pauline Blanquet for help in experiments performed on plants. This work was supported by the Centre National de la Recherche Scientifique and the Université de Toulouse and was carried out in the Laboratoire de Recherche en Sciences Végétales, part of the Laboratoire d'Excellence entitled TULIP (ANR-10-LABX-41).
Glossary
- CDPK or CPK
calcium-dependent protein kinase
- DHS
dihydrosphingosine
- DIG
digoxigenin
- FB1
fumonisin B1
- FDA
fluorescein diacetate
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- LCB
sphingoid long chain base
- MPK
mitogen-activated protein kinase
- PCD
programmed cell death
- PHS
phytosphingosine
- PKA
protein kinase A
- PKC
protein kinase C
- SDK
sphingosine-dependent protein kinase
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by P Bozhkov
Supplementary Material
References
- Lam E. Controlled cell death, plant survival and development. Nat Rev Mol Cell Biol. 2004;5:305–315. doi: 10.1038/nrm1358. [DOI] [PubMed] [Google Scholar]
- Vaux DL, Korsmeyer SJ. Cell death in development. Cell. 1999;96:245–254. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- Cuvillier O. Sphingosine in apoptosis signaling. Biochim Biophys Acta. 2002;1585:153–162. doi: 10.1016/s1388-1981(02)00336-0. [DOI] [PubMed] [Google Scholar]
- Woodcock JM, Ma Y, Coolen C, Pham D, Jones C, Lopez AF, et al. Sphingosine and FTY720 directly bind pro-survival 14-3-3 proteins to regulate their function. Cell Signal. 2010;22:1291–1299. doi: 10.1016/j.cellsig.2010.04.004. [DOI] [PubMed] [Google Scholar]
- MacKintosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem J. 2004;381:329–342. doi: 10.1042/BJ20031332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorhead G, Douglas P, Cotelle V, Harthill J, Morrice N, Meek S, et al. Phosphorylation-dependent interactions between enzymes of plant metabolism and 14-3-3 proteins. Plant J. 1999;18:1–12. doi: 10.1046/j.1365-313x.1999.00417.x. [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, et al. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- Cotelle V, Meek SEM, Provan F, Milne FC, Morrice N, MacKintosh C. 14-3-3s regulate global cleavage of their diverse binding partners in sugar-starved Arabidopsis cells. Embo J. 2000;19:2869–2876. doi: 10.1093/emboj/19.12.2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzivion G, Avruch J. 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J Biol Chem. 2002;277:3061–3064. doi: 10.1074/jbc.R100059200. [DOI] [PubMed] [Google Scholar]
- Porter GW, Khuri FR, Fu H. Dynamic 14-3-3/client protein interactions integrate survival and apoptotic pathways. Semin Cancer Biol. 2006;16:193–202. doi: 10.1016/j.semcancer.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Hamaguchi A, Suzuki E, Murayama K, Fujimura T, Hikita T, Iwabuchi K, et al. Sphingosine-dependent protein kinase-1, directed to 14-3-3, is identified as the kinase domain of protein kinase C delta. J Biol Chem. 2003;278:41557–41565. doi: 10.1074/jbc.M305294200. [DOI] [PubMed] [Google Scholar]
- Megidish T, Cooper J, Zhang L, Fu H, Hakomori S. A novel sphingosine-dependent protein kinase (SDK1) specifically phosphorylates certain isoforms of 14-3-3 protein. J Biol Chem. 1998;273:21834–21845. doi: 10.1074/jbc.273.34.21834. [DOI] [PubMed] [Google Scholar]
- Ma Y, Pitson S, Hercus T, Murphy J, Lopez A, Woodcock J. Sphingosine activates protein kinase A type II by a novel cAMP-independent mechanism. J Biol Chem. 2005;280:26011–26017. doi: 10.1074/jbc.M409081200. [DOI] [PubMed] [Google Scholar]
- Woodcock JM, Murphy J, Stomski FC, Berndt MC, Lopez AF. The dimeric versus monomeric status of 14-3-3zeta is controlled by phosphorylation of Ser58 at the dimer interface. J Biol Chem. 2003;278:36323–36327. doi: 10.1074/jbc.M304689200. [DOI] [PubMed] [Google Scholar]
- Messaritou G, Grammenoudi S, Skoulakis EMC. Dimerization is essential for 14-3-3zeta stability and function in vivo. J Biol Chem. 2010;285:1692–1700. doi: 10.1074/jbc.M109.045989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzivion G, Luo Z, Avruch J. A dimeric 14-3-3 protein is an essential cofactor for Raf kinase activity. Nature. 1998;394:88–92. doi: 10.1038/27938. [DOI] [PubMed] [Google Scholar]
- Kanno T, Nishizaki T. Sphingosine induces apoptosis in hippocampal neurons and astrocytes by activating caspase-3/-9 via a mitochondrial pathway linked to SDK/14-3-3 Protein/Bax/Cytochrome c. J Cell Physiol. 2011;226:2329–2337. doi: 10.1002/jcp.22571. [DOI] [PubMed] [Google Scholar]
- Brodersen P, Petersen M, Pike HM, Olszak B, Skov S, Ødum N, et al. Knockout of Arabidopsis accelerated-cell-death11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 2002;16:490–502. doi: 10.1101/gad.218202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg JT, Silverman FP, Liang H. Uncoupling salicylic acid-dependent cell death and defense-related responses from disease resistance in the Arabidopsis mutant acd5. Genetics. 2000;156:341–350. doi: 10.1093/genetics/156.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas HK, Tanaka T, Duke SO, Porter JK, Wray EM, Hodges L, et al. Fumonisin- and AAL-toxin-induced disruption of sphingolipid metabolism with accumulation of free sphingoid bases. Plant Physiol. 1994;106:1085–1093. doi: 10.1104/pp.106.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo-García M, Guevara-García A, González-Solís A, Cruz-García F, Vázquez-Santana S, Markham JE, et al. MPK6, sphinganine and the LCB2a gene from serine palmitoyltransferase are required in the signaling pathway that mediates cell death induced by long chain bases in Arabidopsis. New Phytol. 2011;191:943–957. doi: 10.1111/j.1469-8137.2011.03727.x. [DOI] [PubMed] [Google Scholar]
- Shi L, Bielawski J, Mu J, Dong H, Teng C, Zhang J, et al. Involvement of sphingoid bases in mediating reactive oxygen intermediate production and programmed cell death in Arabidopsis. Cell Res. 2007;17:1030–1040. doi: 10.1038/cr.2007.100. [DOI] [PubMed] [Google Scholar]
- Peer M, Stegmann M, Mueller MJ, Waller F. Pseudomonas syringae infection triggers de novo synthesis of PHS from sphinganine in Arabidopsis thaliana. FEBS Lett. 2010;584:4053–4056. doi: 10.1016/j.febslet.2010.08.027. [DOI] [PubMed] [Google Scholar]
- Lachaud C, Da Silva D, Cotelle V, Thuleau P, Xiong TC, Jauneau A, et al. Nuclear calcium controls the apoptotic-like cell death induced by D-erythro-sphinganine in tobacco cells. Cell Calcium. 2010;47:92–100. doi: 10.1016/j.ceca.2009.11.011. [DOI] [PubMed] [Google Scholar]
- Shen YH, Godlewski J, Bronisz A, Zhu J, Comb MJ, Avruch J, et al. Significance of 14-3-3 self-dimerization for phosphorylation-dependent target binding. Mol Biol Cell. 2003;14:4721–4733. doi: 10.1091/mbc.E02-12-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori IC, Murata Y, Yang Y, Munemasa S, Wang YF, Andreoli S, et al. CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+-permeable channels and stomatal closure. PLoS Biol. 2006;4:327. doi: 10.1371/journal.pbio.0040327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrawal GK, Thelen JJ. Large scale identification and quantitative profiling of phosphoproteins expressed during seed filling in oilseed rape. Mol Cell Proteomics. 2006;5:2044–2059. doi: 10.1074/mcp.M600084-MCP200. [DOI] [PubMed] [Google Scholar]
- Barjaktarovíc Z, Schütz W, Madlung J, Fladerer C, Nordheim A, Hampp R. Changes in the effective gravitational field strength affect the state of phosphorylation of stress-related proteins in callus cultures of Arabidopsis thaliana. J Exp Bot. 2009;60:779–789. doi: 10.1093/jxb/ern324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Alvarez S, Burch AY, Jez JM, Schachtman DP. Phosphoproteomic identification of targets of the Arabidopsis sucrose nonfermenting-like kinase SnRK2.8 reveals a connection to metabolic processes. Proc Natl Acad Sci USA. 2007;104:6460–6465. doi: 10.1073/pnas.0610208104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlmer N, Wurzinger B, Stael S, Hofmann-Rodrigues D, Csaszar E, Pfister B, et al. The Ca2+-dependent protein kinase CPK3 is required for MAPK-independent salt-stress acclimation in Arabidopsis. Plant J. 2010;63:484–498. doi: 10.1111/j.1365-313X.2010.04257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SH, Willmann MR, Chen HC, Sheen J. Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 2002;129:469–485. doi: 10.1104/pp.005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JF, Harmon A. Plants, symbiosis and parasites: a calcium signalling connection. Nat Rev Mol Cell Biol. 2005;6:555–566. doi: 10.1038/nrm1679. [DOI] [PubMed] [Google Scholar]
- Hong Y, Takano M, Liu CM, Gasch A, Chye ML, Chua NH. Expression of three members of the calcium-dependent protein kinase gene family in Arabidopsis thaliana. Plant Mol Biol. 1996;30:1259–1275. doi: 10.1007/BF00019557. [DOI] [PubMed] [Google Scholar]
- Dubois T, Howell S, Amess B, Kerai P, Learmonth M, Madrazo J, et al. Structure and sites of phosphorylation of 14-3-3 protein: role in coordinating signal transduction pathways. J Protein Chem. 1997;16:513–522. doi: 10.1023/a:1026321813463. [DOI] [PubMed] [Google Scholar]
- Aitken A. Post-translational modification of 14-3-3 isoforms and regulation of cellular function. Semin Cell Dev Biol. 2011;22:673–680. doi: 10.1016/j.semcdb.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Kanchiswamy CN, Takahashi H, Quadro S, Maffei ME, Bossi S, Bertea C, et al. Regulation of Arabidopsis defense responses against Spodoptera littoralis by CPK-mediated calcium signaling. BMC Plant Biol. 2010;10:97. doi: 10.1186/1471-2229-10-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M, Willmann MR, McCormack M, Lee H, Shan L, He P, et al. Differential innate immune signalling via Ca2+ sensor protein kinases. Nature. 2010;464:418–422. doi: 10.1038/nature08794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeis T, Ludwig AA, Martin R, Jones JDG. Calcium-dependent protein kinases play an essential role in a plant defence response. Embo J. 2001;20:5556–5567. doi: 10.1093/emboj/20.20.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig AA, Saitoh H, Felix G, Freymark G, Miersch O, Wasternack C, et al. Ethylene-mediated cross-talk between calcium-dependent protein kinase and MAPK signalling controls stress responses in plants. Proc Natl Acad Sci USA. 2005;102:10736–10741. doi: 10.1073/pnas.0502954102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.