Abstract
Short-term proteasome inhibition has been shown to prevent neuronal apoptosis. However, the key pro-survival proteins that must be degraded for triggering neuronal death are mostly unknown. Here, we show that Mcl-1, an anti-apoptotic Bcl-2 family member, is degraded by the proteasome during neuronal apoptosis. Using primary cultures of cerebellar granule neurons deprived of serum and KCl, we found that ubiquitination and proteasomal degradation of Mcl-1 depended on its prior phosphorylation by GSK3, providing the first insight into post-translational regulation of Mcl-1 in neurons. In a previous study, we have reported that the E3 ubiquitin-ligase Trim17 is both necessary and sufficient for neuronal apoptosis. Here, we identified Trim17 as a novel E3 ubiquitin-ligase for Mcl-1. Indeed, Trim17 co-immunoprecipitated with Mcl-1. Trim17 ubiquitinated Mcl-1 in vitro. Overexpression of Trim17 decreased the protein level of Mcl-1 in a phosphorylation- and proteasome-dependent manner. Finally, knock down of Trim17 expression reduced both ubiquitination and degradation of Mcl-1 in neurons. Moreover, impairment of Mcl-1 phosphorylation, by kinase inhibition or point mutations, not only decreased ubiquitination and degradation of Mcl-1, but also blocked the physical interaction between Trim17 and Mcl-1. As this stabilization of Mcl-1 increased its neuroprotective effect, our data strongly suggest that Trim17-mediated ubiquitination and degradation of Mcl-1 is necessary for initiating neuronal death.
Keywords: apoptosis, Trim17, E3 ubiquitin-ligase, Mcl-1, cerebellar granule neurons, proteasome
In the nervous system, apoptosis plays a critical role both during development and in neurodegenerative diseases.1 In many neuronal types, apoptosis is triggered through the intrinsic pathway of caspase activation that involves the release of cytochrome c from mitochondria. The proteins of the Bcl-2 family, that comprises both anti-apoptotic (Bcl-2, Bcl-xL, Mcl-1…) and pro-apoptotic members (Bax, Bak, Bim…), play an essential role in the regulation of apoptosis by controlling the integrity of the outer mitochondrial membrane and the release of apoptogenic factors such as cytochrome c.2 Amongst the anti-apoptotic proteins of the Bcl-2 family, Mcl-1 (Myeloid cell leukemia 1) is characterized by a short half-life.3 Its rapid degradation by the proteasome has been shown to be required for initiation of the apoptotic cascade in several cell lines.4, 5, 6, 7
For a long time, little attention was paid to a potential role for Mcl-1 in the nervous system because initial studies did not detect it in neurons. However, Mcl-1 is expressed in cerebellar granule neurons (CGNs) in vivo,8 and Mcl-1 expression has been associated with neuroprotection in the hippocampus.9 More recently, elegant studies using different conditional Mcl-1 mouse mutants, showed that Mcl-1 is required for neuronal development, that loss of Mcl-1 sensitizes neurons to DNA damage-induced apoptosis and that Mcl-1 negatively regulates autophagy in starved neurons.10, 11 Therefore, Mcl-1 appears to play a crucial role in neuronal survival. Nevertheless, post-translational regulation of Mcl-1 has never been studied in neurons.
In the present study, we report for the first time that Mcl-1 is degraded by the proteasome during neuronal apoptosis. We found that Mcl-1 degradation depended on its prior phosphorylation by glycogen synthase kinase 3 (GSK3), in primary cultures of CGNs deprived of serum and KCl. This phosphorylation of Mcl-1 favored its physical interaction with Trim17, a novel E3 ubiquitin-ligase involved in neuronal apoptosis. Moreover, our present data provide evidence that Trim17 contributes to the ubiquitination and the proteasomal degradation of Mcl-1 in neurons. As such, we define a novel molecular mechanism for regulation of Mcl-1 and initiation of apoptosis.
Results
Mcl-1 is degraded by the proteasome during CGN apoptosis
To study the regulation of Mcl-1 during neuronal death, we used primary cultures of cerebellar granule neurons (CGNs), one of the best characterized in vitro models of neuronal apoptosis. CGNs can survive and differentiate in culture in the presence of serum and depolarizing levels of extracellular KCl ([KCl]o=25 mM, K25) that mimic the excitatory activity required for CGN survival in vivo. Withdrawal of serum and lowering the KCl level to 5 mM (K5) triggers CGN apoptosis,12 recapitulating the programmed cell death that occurs in the cerebellum during postnatal development.13
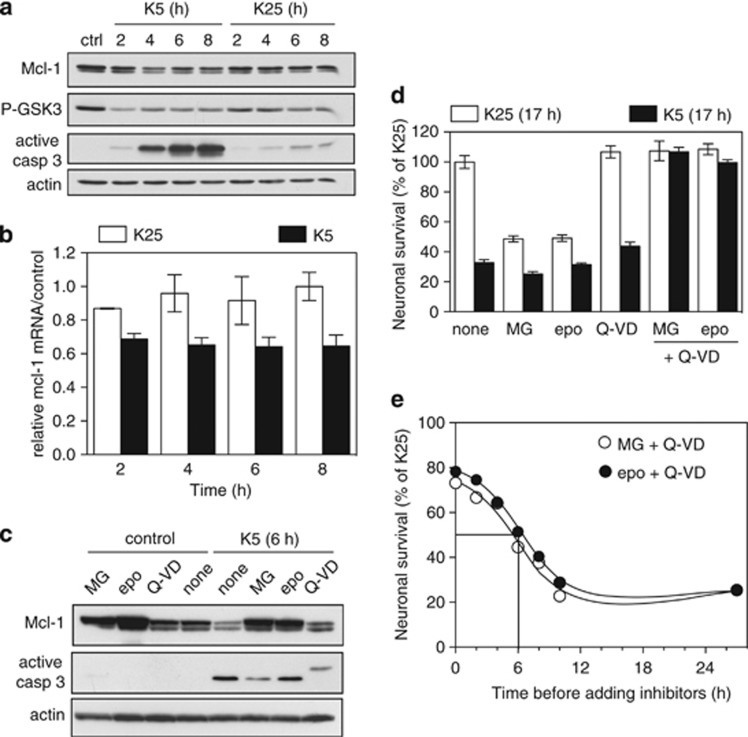
Using western blotting, we found that the protein level of Mcl-1 declined markedly during CGN apoptosis (Figure 1a). The minimum level of Mcl-1 (approximately 40% of the control) was reached 4–6 h after KCl and serum deprivation, depending on the experiments. This coincided with cytochrome c release from mitochondria,14 dephosphorylation (and thus activation) of GSK3 (Figure 1a) and caspase 3 activation (Figure 1a). The reduction of the Mcl-1 protein level was associated with a similar decrease in the mcl-1 mRNA level: about 35% reduction between K25 and K5 conditions after 4–8 h of deprivation (Figure 1b). Nevertheless, the decrease in Mcl-1 protein could be blocked by proteasome inhibition using two structurally unrelated molecules (MG-132 and epoxomicin) but not by the pancaspase inhibitor Q-VD-OPh (Figure 1c). Proteasome inhibitors also increased the level of Mcl-1 in survival conditions (Figure 1c), indicating that Mcl-1 is constitutively degraded by the proteasome. Taken together, our data thus suggest that Mcl-1 is mainly degraded by the proteasome in CGNs, and that its decline during apoptosis is due to the combined action of its proteasomal degradation and a reduction of its mRNA level.
Figure 1.
Mcl-1 is degraded by the proteasome during KCl deprivation-induced apoptosis in CGNs. (a) CGN primary cultures were left untreated (ctrl) or washed and switched to serum free medium containing either 25 mM KCl (K25) or 5 mM KCl (K5) for increasing times. Total protein extracts were prepared and western blot analysis was performed with antibodies against Mcl-1, the phosphorylated form (Ser9) of GSK3, the active (cleaved) form of caspase 3 and actin (loading control). (b) CGNs were incubated in K25 or in K5 medium for the indicated times. Total RNA was extracted and mcl-1 mRNA levels were estimated by quantitative RT-PCR. Fold change was calculated by comparison with neurons maintained in the initial culture medium (ctrl). Data are means±S.D. of triplicates and are representative of five independent experiments. (c) CGNs were left untreated (control) or switched to K5 medium in the presence or absence of 20 μM MG-132 (MG), 10 μM epoxomicin (epo) or 100 μM Q-VD-OPh (Q-VD) for 6 h. Total protein extracts were analyzed by western blot using antibodies against Mcl-1, active caspase 3 and actin. (d) CGN primary cultures were switched to K25 or K5 medium in the presence or absence of 5 μM MG-132, 5 μM epoxomicin, 50 μM Q-VD-OPh or a combination of proteasome and caspase inhibitors for 17 h. Then, neuronal viability was determined by MTT assay. Data are the means±S.D. of triplicate determinations obtained in a typical experiment representative of five independent experiments. Results are expressed as the percentage of surviving neurons compared to neurons maintained in K25 medium. (e) To determine the proteasomal commitment point, CGNs were switched to K5 medium at time 0 to induce apoptosis. At increasing times after deprivation, proteasome inhibitors were added to the medium together with a caspase inhibitor: 5 μM MG-132+50 μM Q-VD-OPh (open circle) or 5 μM epoxomicin+50 μM Q-VD-OPh (closed circle), in order to rescue neurons. Neuronal viability for all conditions was estimated 27 h after KCl deprivation by MTT assay. Results are expressed as percentage of surviving neurons compared with cultures maintained in K25 throughout the experiment. Data are the means of two independent experiments performed with different neuronal cultures
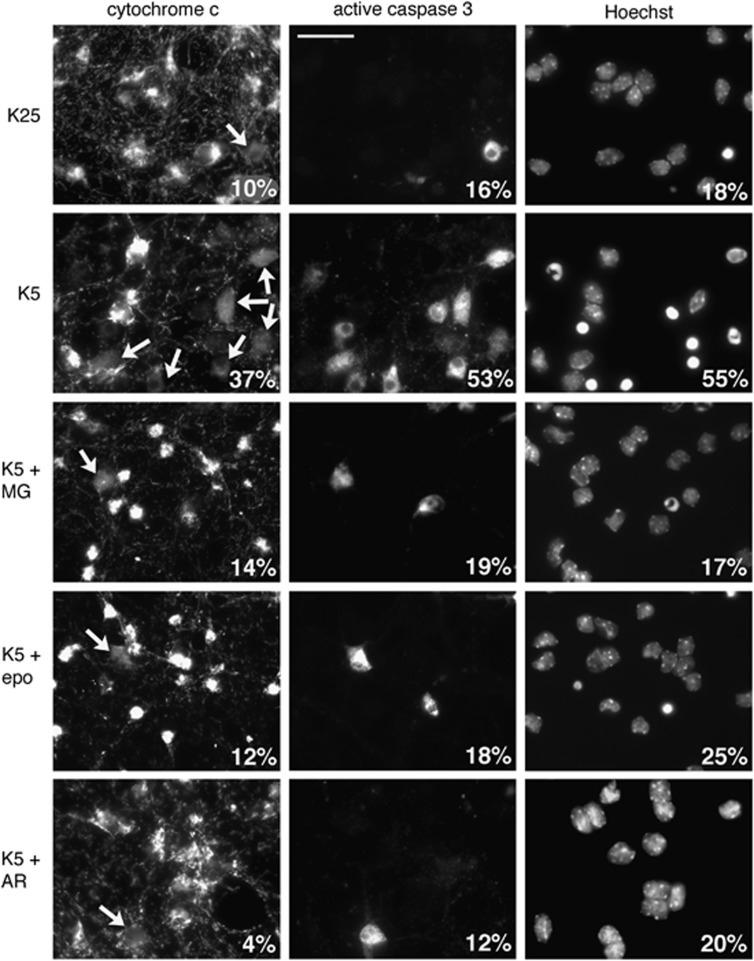
We next investigated whether Mcl-1 degradation correlates with the proteasomal commitment point in apoptotic CGNs. Indeed, we observed that inhibiting proteasome for 8 h prevented cytochrome c release, activation of caspase 3 and nuclear condensation in KCl-deprived CGNs (Figure 2), in agreement with previous studies.15, 16, 17 This suggests that key pro-survival proteins have to be degraded by the proteasome for apoptosis to be initiated in neurons. In contrast, incubation for 17 h with the same proteasome inhibitors was sufficient to induce 50% death in CGNs, even in the presence of 25 mM KCl (Figure 1d). This apparent discrepancy is due to the biphasic effect of proteasome inhibition on neuronal apoptosis (anti-apoptotic effect of short-term treatment versus pro-apoptotic effect of long term treatment) described by Butts et al.16 Interestingly, caspase inhibition completely prevented neuronal death triggered by a prolonged treatment with proteasome inhibitors (Figure 1d), although it could not significantly protect neurons from KCl deprivation (Figure 1d), as described previously.18, 19 Moreover, the combined action of proteasome and caspase inhibitors completely prevented neuronal death induced by 17 h KCl deprivation (Figure 1d). This may be explained by the ability of caspase inhibition to abrogate the adverse effects of prolonged proteasome inhibition, and thus to reveal the protective effect of proteasome inhibitors. We took advantage of this observation to assess the proteasomal commitment point of CGNs. For this purpose, neurons were deprived of KCl for increasing times before addition of proteasome and caspase inhibitors, and neuronal survival was assessed 27 h after the beginning of deprivation. Half of the neurons could no longer be rescued by the inhibitors 6 h after KCl deprivation (Figure 1e). This suggests that the proteins whose degradation is required for initiating neuronal apoptosis have already been degraded by the proteasome, at that time. Importantly, this proteasomal commitment point coincided with the maximal degradation of Mcl-1 (Figure 1a).
Figure 2.
Proteasome and GSK3 inhibitors protect neurons from apoptosis by preventing cytochrome c release and caspase activation. CGN primary cultures were washed and switched to serum-free medium containing either 25 mM KCl (K25) or 5 mM KCl (K5) in the presence or absence of 20 μM MG-132 (MG), 10 μM epoxomicin (epo) or 10 μM AR-A014418 for 8 h. After fixation, nuclear condensation was visualized by Hoechst 33258 staining. Cytochrome c subcellular localization and caspase 3 activation were detected by immunofluorescence. In healthy neurons, cytochrome c immunostaining is intense and punctate both in cell bodies and in neurites (axons and dendrites), indicating mitochondrial localization. In apoptotic neurons, the staining is faint and diffuse, indicating that cytochrome c has been released from mitochondria. At late stages of apoptosis, the staining disappears because cytochrome c is rapidly degraded after release. The percentages of neurons with a condensed nucleus, showing a diffuse staining for cytochrome c or positive for active caspase 3 are indicated for each condition (500 neurons scored in a blinded manner for each condition). Arrows indicate cell bodies with a diffuse cytochrome c staining. Scale bar is 20 μm. Data are representative of three independent experiments
Mcl-1 ubiquitination and degradation depend on its phosphorylation by GSK3 in CGNs
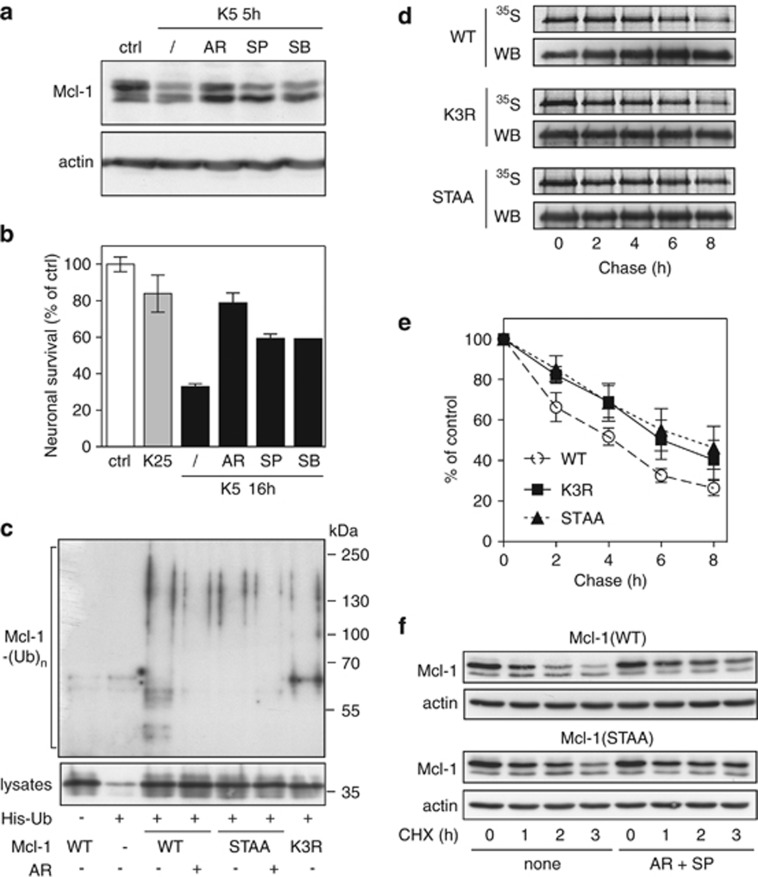
As prior phosphorylation of Mcl-1 by GSK3 has been shown to be required for its ubiquitination and degradation in different cell lines,5, 6, 20, 21 we addressed this question in CGNs. Indeed, the decrease in Mcl-1 following KCl deprivation was completely prevented by the specific GSK3 inhibitor AR-A014418 (Figure 3a). It has been demonstrated that c-Jun N-terminal protein kinase (JNK) is required for GSK3-mediated degradation of Mcl-1 in response to stress.22 Consistently, we found that the JNK inhibitor SP600125 partly prevented Mcl-1 elimination in CGNs, after KCl deprivation (Figure 3a). The p38 inhibitor SB 203580 had a similar effect (Figure 3a). However, only little in vitro phosphorylation of mouse Mcl-1 could be detected with p38 compared to JNK by Morel et al.22 Therefore, the stabilization of Mcl-1 by the p38 inhibitor in apoptotic CGNs (Figure 3a) might be explained by a possible indirect effect of p38 on JNK activity in neurons, as suggested by previous work.23 Interestingly, the ability of the different kinase inhibitors to sustain Mcl-1 level was directly correlated with their neuroprotective effect (Figures 2 and 3b).
Figure 3.
Prior phosphorylation of Mcl-1 is required for its ubiquitination and subsequent degradation. (a) CGNs were left untreated (ctrl) or washed and switched to K5 medium in the presence or absence of 10 μM AR-A014418, 10 μM SP600125 or 20 μM SB 203580 for 5 h. Total protein extracts were analyzed by immunoblotting using antibodies against Mcl-1 and actin. (b) CGN cultures were incubated in K25 or K5 medium in the presence or absence of 10 μM AR-A014418, 10 μM SP600125 or 20 μM SB 203580 for 16 h. Then, neuronal viability was determined by MTT assay. Data are the means±S.D. of triplicate determinations obtained in a typical experiment representative of three independent experiments. Results are expressed as the percentage of surviving neurons compared to neurons maintained in initial culture medium (ctrl). (c) CGNs were transfected with untagged Mcl-1(WT), Mcl-1(STAA) or Mcl-1(K3R), or with empty plasmid (−), together with His-tagged ubiquitin (+) or empty plasmid (−) for 18 h. Then, neurons were incubated for 6 h with 25 μM MG-132, in the presence or the absence of 10 μM AR-A011418 (AR). The ubiquitinated proteins were purified using nickel beads and analyzed by western blotting using anti-Mcl-1 antibody to detect ubiquitin-conjugated Mcl-1. In a separate SDS-PAGE, samples of the input lysates used for the purification were analyzed with anti-Mcl-1 antibody to estimate the expression level of the different forms of transfected Mcl-1. (d) CGNs were transfected with Mcl-1(WT)-GFP, Mcl-1(K3R)-GFP or Mcl-1(STAA)-GFP for 18 h. Then, neurons were metabolically labeled with [35S]-Met for 2 h (pulse) and harvested at different times after washing and incubation in K5 medium (chase). The different forms of Mcl-1-GFP were then immunoprecipitated using GFP-trap beads, separated by SDS-PAGE and visualized by both autoradiography (35S) and immunodetection of Mcl-1 (WB). Note that a higher amount of Mcl-1(WT) was immunoprecipitated compared to the mutants in the three last time points. This may give the feeling that it is more stable than it actually is. (e) The intensity of the bands on the autoradiograms and on the immunoblots of different experiments performed as in (d) was quantified. For each experiment, the radioactivity associated with the different forms of Mcl-1-GFP was normalized by the amount of protein immunoprecipitated in each condition and plotted against chase time. Data are the mean±S.E.M. of three independent experiments. (f) CGNs were transfected with untagged Mcl-1(WT) or Mcl-1(STAA). Eighteen hours after transfection, neurons were pre-incubated for 1 h in K5 medium in the presence or the absence of 10 μM AR-A011418 and 10 μM SP600125 (AR+SP). Then, 10 μg/ml cycloheximide (CHX) was added, and neurons were harvested after the indicated times after CHX addition. Proteins were analyzed by western blot using antibodies against Mcl-1 and actin
Several studies demonstrated that phosphorylation by GSK3 of Ser 159 (and possibly Thr 163) initiates proteasomal degradation of human MCL-1.5, 6, 20 In mouse Mcl-1, the corresponding Ser 140 and Thr 144 are also phosphorylated, and the phosphorylation of Thr 144 by JNK primes Mcl-1 for phosphorylation by GSK3.22 Therefore, we constructed the Mcl-1(STAA) mutant in which Ser 140 and Thr 144 were replaced by alanines, to generate a variant of mouse Mcl-1 that cannot be phosphorylated at these sites. In addition, human MCL-1 has been shown to be ubiquitinated on five lysine residues,20, 24 among which three are conserved in mouse Mcl-1. Therefore, we used the Mcl-1(K3R) mutant, in which these three conserved lysines were replaced with arginines, to investigate the influence of ubiquitination on Mcl-1 stability. We first assessed the ubiquitination level of the different forms of Mcl-1 when co-transfected with His-tagged ubiquitin in CGNs (Figure 3c). Ubiquitination was significantly reduced by mutation of Ser 140 and Thr 144 (STAA), indicating that phosphorylation of these sites plays a crucial role in mouse Mcl-1 ubiquitination. Consistently, GSK3 inhibition reduced the ubiquitination level of Mcl-1(WT) to a similar extent than these mutations, but did not significantly decrease the ubiquitination of Mcl-1(STAA). In addition, ubiquitination of the mutant lacking three lysines, Mcl-1(K3R), was also reduced, indicating that these residues are involved in ubiquitin conjugation to mouse Mcl-1 (Figure 3c). In order to test whether these changes in ubiquitination levels could have an impact on proteasomal degradation, we compared the protein half-lives of the different forms of Mcl-1 by both pulse-chase experiments and protein synthesis inhibition, in CGNs. Consistently Mcl-1(STAA) and Mcl-1(K3R) had a longer half-life than Mcl-1(WT) (Figure 3d and e). Moreover, kinase inhibition (AR+SP) induced a similar stabilization of Mcl-1 (WT) than mutation of the phosphorylation consensus site (Figure 3f). Taken together, our results suggest that phosphorylation is a prerequisite for Mcl-1 ubiquitination, and that ubiquitination is required for Mcl-1 degradation in neurons.
The neuroprotective effect of Mcl-1 increases with its stabilization
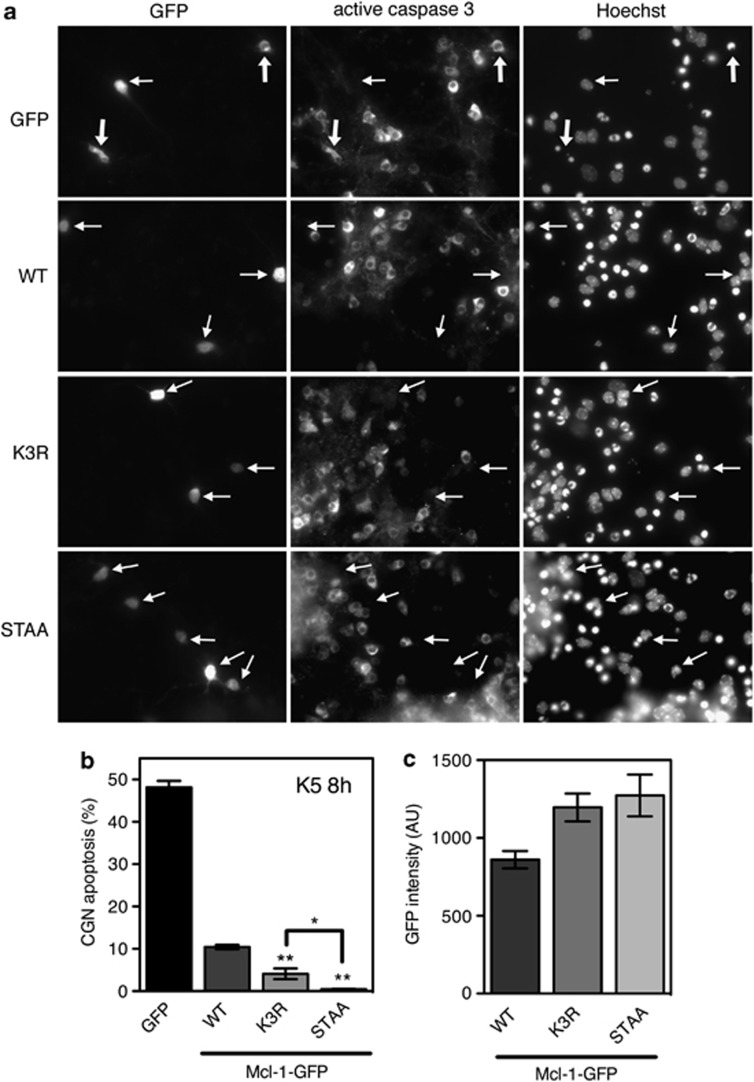
To determine whether Mcl-1 degradation plays an important role in the initiation of neuronal apoptosis, we examined whether overexpression of Mcl-1 fused to green fluorescent protein (GFP) could protect CGNs. Following 8 h KCl deprivation, about half of CGNs transfected with GFP showed apoptotic morphology, active caspase 3 immunostaining and chromatin condensation. In contrast, most neurons expressing Mcl-1(WT)-GFP were healthy although they were surrounded by many non-transfected apoptotic neurons (Figure 4a). While overexpression of Mcl-1 (WT) could reduce apoptosis from 50% to 10%, the two mutants of Mcl-1 provided a significantly higher protection (Figure 4b). Consistently, GFP intensity measured in transfected neurons was higher for Mcl-1(K3R)-GFP and Mcl-1(STAA)-GFP compared to Mcl-1(WT)-GFP (Figure 4c), confirming their increased stability (Figure 3d–f). Therefore, the ability of the different forms of Mcl-1 to protect CGNs from apoptosis was directly correlated with their stability. This suggests that proteasomal degradation of Mcl-1 is required for initiating neuronal apoptosis following survival factor withdrawal.
Figure 4.
Neuroprotective effect of the different forms of Mcl-1. (a) CGN primary cultures were transfected with GFP (used as a negative control) or the different forms of Mcl-1 fused to GFP, for 16 h. Then, neurons were incubated in K5 medium for 8 h. Caspase 3 activation was determined by immunofluorescence and nuclear condensation was visualized by Hoechst staining. Thick arrows indicate GFP-expressing neurons that undergo apoptosis, whereas thin arrows indicate healthy neurons expressing GFP or the different forms of Mcl-1-GFP. (b) The percentage of apoptosis among transfected neurons was assessed by examining cell morphology and nuclear condensation of GFP-positive neurons. Data are the mean±S.D. of three independent experiments. **P<0.001 significantly different from apoptosis observed in neurons expressing Mcl-1(WT)-GFP (ANOVA followed by Student-Newman–Keuls multiple comparison test). The difference observed between neurons expressing Mcl-1(K3R)-GFP and Mcl-1(STAA)-GFP is also significant (*P<0.01 using the same statistical tests). (c) GFP intensity (AU: Arbitrary Unit) in the cell body of transfected neurons was measured using MetaMorph. Data are the mean±S.E.M. of about 100 neurons for each condition
Trim17 mediates the degradation of Mcl-1 in CGNs
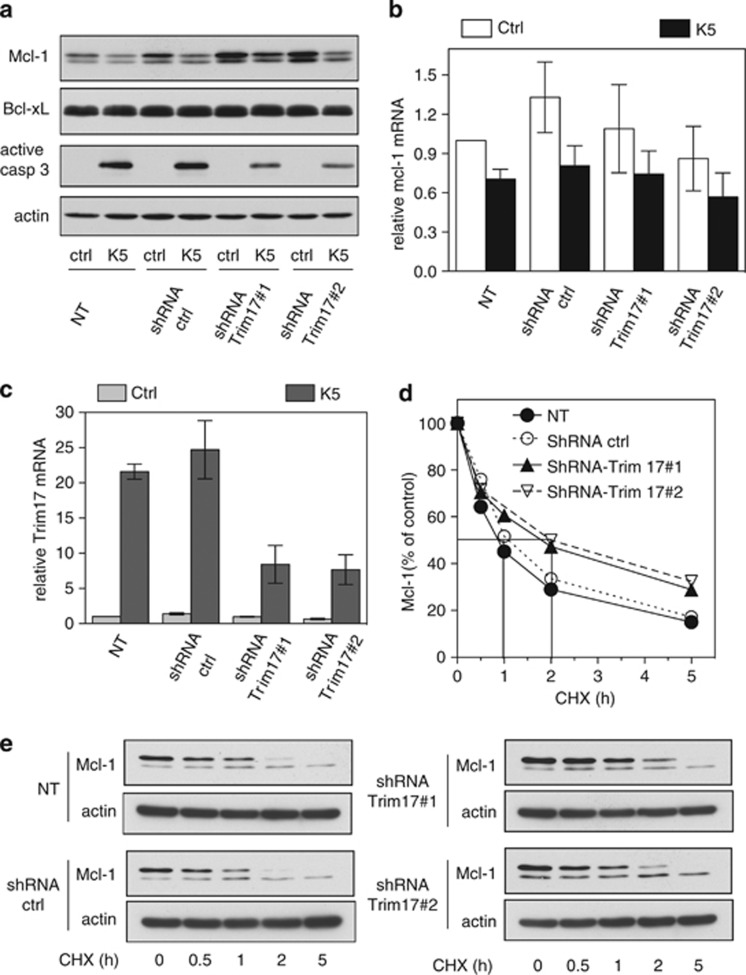
In a recent study, we have shown that Trim17 is an E3 ubiquitin-ligase whose expression is necessary for neuronal apoptosis.25 This prompted us to examine whether Trim17 could mediate the ubiquitination and subsequent proteasomal degradation of Mcl-1 in CGNs. For this purpose, we first estimated the protein level of endogenous Mcl-1 in neurons transduced with lentiviruses expressing two different short hairpin RNAs (shRNAs) that specifically target the Trim17 mRNA. In control condition, both shRNA-Trim17 sequences increased the amount of Mcl-1 more than two-fold compared to non transduced neurons or to neurons transduced with a non-targeting shRNA control (Figure 5a). However, after KCl deprivation, Mcl-1 protein level still decreased in neurons in which Trim17 expression was knocked down. To determine whether this decrease may be due to another E3 ubiquitin-ligase, we transduced neurons with shRNAs against two other E3 ubiquitin-ligases that have been shown to mediate the degradation of Mcl-1: Fbw720, 26 and Mule,24 either separately or together with a shRNA against Trim17 (Supplementary Figure). Silencing of Fbw7 had no effect on the protein level of Mcl-1 both in control and apoptotic conditions. Depletion of Mule alone slightly increased the level of endogenous Mcl-1, but only in control conditions and this effect was lower than that of Trim17 silencing. Moreover, depletion of Fbw7 or Mule, alone or in combination with Trim17, did not prevent the decrease in Mcl-1 protein level induced by KCl deprivation (Supplementary Figure). This decrease is certainly due to the reduction of the mcl-1 mRNA level that occurs during apoptosis (Figures 1b and 5b). Nevertheless, in Trim17-depleted neurons deprived of KCl, Mcl-1 level remained above that observed in non transduced neurons maintained in survival conditions (Figure 5a). The amount of Bcl-xL and actin did not change significantly in any condition. In contrast, activation of caspase 3 after KCl deprivation was reduced in CGNs depleted of Trim17 (Figure 5a), confirming the neuroprotective effect of Trim17 silencing.25
Figure 5.
Silencing of Trim17 favours Mcl-1 stabilization. (a) CGNs were left untreated (non-transduced: NT) or were transduced with lentiviral particles expressing shRNA sequences (one control and two against Trim17) one day after plating. At DIV 6, neurons were incubated for 6 h in K5 medium or maintained in the initial culture medium (ctrl). Then, proteins were analyzed by western blot using antibodies against Mcl-1, Bcl-x, active caspase 3 and actin. (b and c) CGNs were transduced or not as in (a). Then, neurons were incubated for 4 h in K5 medium or maintained in the initial culture medium (ctrl). Total RNA was extracted and the mRNA levels of mcl-1 (b) and Trim17 (c) were estimated under each condition by quantitative RT-PCR. Fold change was calculated by comparison with non-transduced neurons maintained in the initial culture medium. Data are means±s.d. of triplicate measurements of one experiment that is representative of three independent experiments. (d and e) CGNs were transduced or not as in (a) and incubated for increasing times with 10 μg/ml cycloheximide (CHX). Proteins were analyzed by western blot using antibodies against Mcl-1 and actin. (d) The intensity of the Mcl-1 bands presented in a representative experiment performed as in (e) was estimated and expressed as a percentage of the corresponding value for time zero
Whereas both shRNAs against Trim17 efficiently reduced the level of Trim17 mRNA (Figure 5c), they did not significantly affect the mRNA level of mcl-1 both in control and apoptotic conditions (Figure 5b). This suggests that the accumulation of Mcl-1 induced by Trim17 depletion (Figure 5a) is not due to a transcriptional effect. Therefore, to test whether Trim17 may affect the stability of Mcl-1, we estimated the half-life of endogenous Mcl-1 in transduced CGNs treated with the protein synthesis inhibitor cycloheximide. Strikingly, the half-life of Mcl-1 was increased from 1 to 2 h in neurons transduced with the two shRNA-Trim17 compared to non transduced neurons or to neurons transduced with the control shRNA (Figures 5d and e). Taken together, these data thus indicate that Trim17 is involved in the degradation of Mcl-1 in CGNs.
The interaction of Mcl-1 with Trim17 depends on its phosphorylation
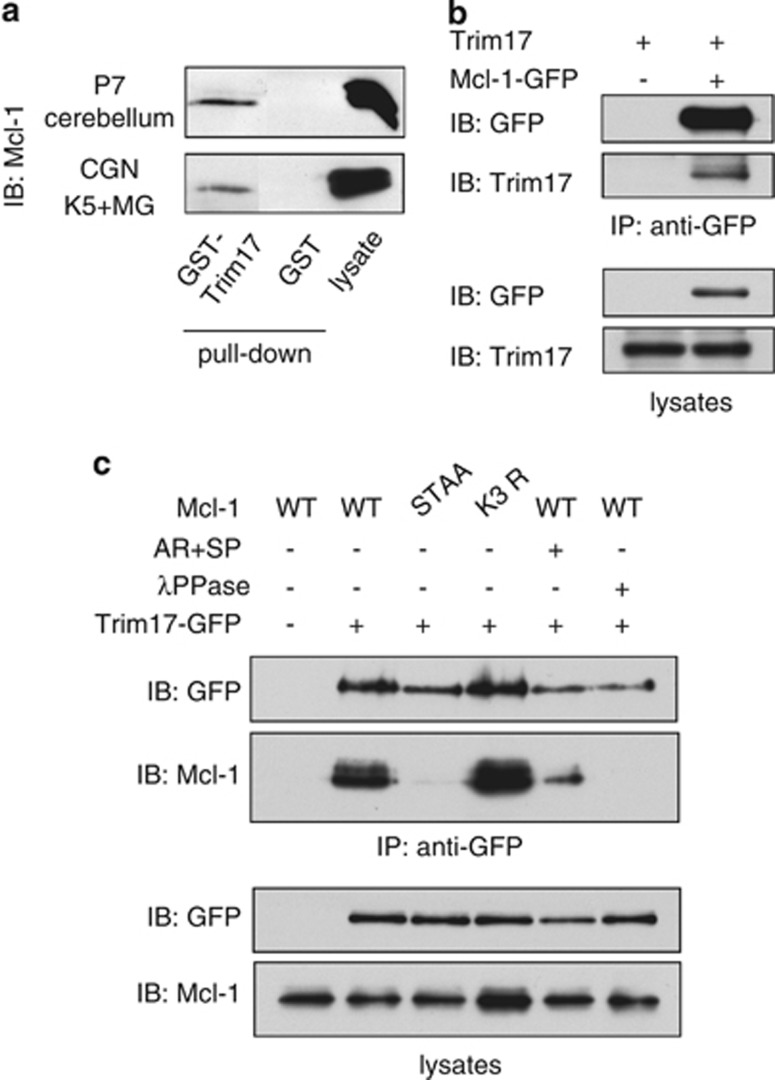
To test whether Trim17 can directly interact with Mcl-1, we first used recombinant GST-Trim17 to pull down proteins from cell extracts derived from mouse cerebellum, or from KCl-deprived CGN. GST-Trim17 pulled down endogenous Mcl-1 from both lysates while no interaction between GST and Mcl-1 was detected (Figure 6a). Then, we performed co-immunoprecipitation in the presence of phosphatase inhibitors and MG-132 in Neuro2A cells. When Mcl-1-GFP and untagged Trim17 were co-transfected, GFP-trap beads precipitated Trim17 together with Mcl-1-GFP (Figure 6b). Inversely, untagged Mcl-1 was immunoprecipitated together with Trim17-GFP (Figure 6c). Interestingly, mutation of the phosphorylation site of Mcl-1 (STAA) abolished its interaction with Trim17, whereas mutation of three lysine residues (K3R) did not modify it (Figure 6c). Pharmacological inhibition of GSK3 and JNK before cell lysis also strongly decreased the amount of Mcl-1 co-precipitated with Trim17 (Figure 6c). Moreover, λ-phosphatase treatment of the beads used for co-immunoprecipitation totally disrupted the interaction between Mcl-1 and Trim17 (Figure 6c). Taken together, these data indicate that phosphorylation of Mcl-1 is required for its physical interaction with Trim17.
Figure 6.
Trim17 interacts with phosphorylated Mcl-1. (a) GST pull-down assays were performed with lysates from P7 mice cerebella, or from CGNs incubated for 6 h in K5 medium with 20 μM MG-132, using recombinant GST-Trim17 or GST (as a negative control) purified on glutathione magnetic beads. Materials bound to recombinant proteins (pull-down) and input lysates were analyzed by western blot using anti-Mcl-1 antibody. (b) Neuro2A cells were transfected with Trim17 together with Mcl-1-GFP or GFP alone (as a negative control) for 24 h. Then, cells were treated for 8 h with 20 μM MG-132 before cell harvesting. Cell lysates were subjected to immunoprecipitation in the presence of phosphatase inhibitors using GFP-trap beads. Immunoprecipitates and lysates were analyzed by western blot using anti-GFP and anti-Trim17 antibodies. (c) Neuro2A cells were transfected with the different untagged variants of Mcl-1 together with Trim17-GFP or GFP alone for 24 h. Then, cells were treated for 8 h with 20 μM MG-132, in the absence or the presence of 10 μM AR-A014418 and 10 μM SP600125 (AR+SP) before cell harvesting. Cell lysates were subjected to immunoprecipitation as in (b). When indicated, beads were treated with λ-phosphatase (λPPase) after immunoprecipitation, and were further washed before elution. Immunoprecipitates and lysates were analyzed by western blot using anti-GFP and anti-Mcl-1 antibodies
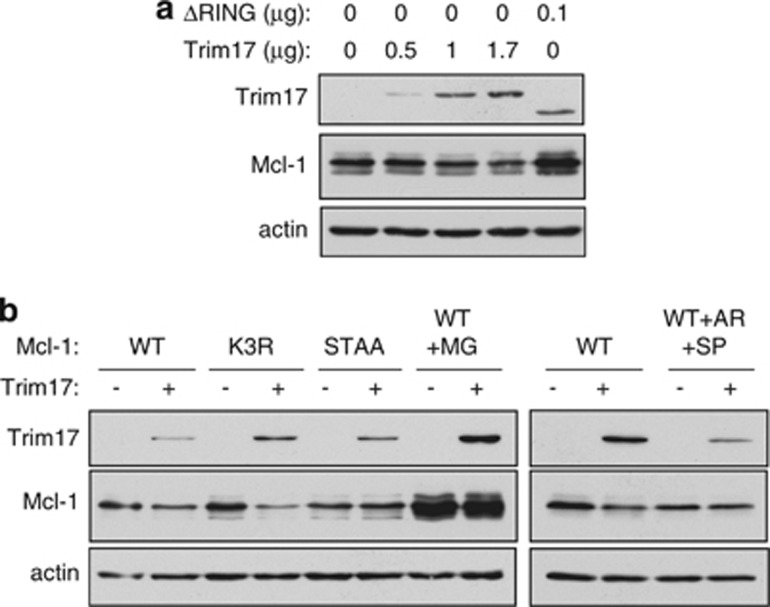
Consistently with a role of Trim17 in the degradation of Mcl-1, Trim17 overexpression decreased the protein level of Mcl-1 in a dose-dependent manner, whereas the dominant-negative mutant Trim17(ΔRING)25 slightly increased Mcl-1 level (Figure 7a). In addition, the decrease of Mcl-1 induced by Trim17 was prevented by MG-132, by mutation of the phosphorylation motif of Mcl-1(STAA) and by pharmacological inhibition of GSK3 and JNK (Figure 7b). Taken together, these results strongly suggest that the phosphorylation-dependent interaction of Mcl-1 with Trim17 is necessary for the proteasomal degradation of Mcl-1 triggered by Trim17.
Figure 7.
Overexpressed Trim17 decreases the protein level of Mcl-1. (a) Neuro2A cells were transfected with 0.3 μg pCI-Mcl-1(WT) together with the indicated amounts of pCI-Trim17 or pCI-Trim17(ΔRING). Twenty four hours after transfection, cell lysates were analyzed by western blot using antibodies against Trim17, Mcl-1 and actin. (b) Neuro2A cells were transfected for 24 h with the different forms of Mcl-1 (0.5 μg of each plasmid), together with 1 μg pCI-Trim17 when indicated. Then, cells were treated for 5 h with 25 μM MG-132 (MG), or 10 μM AR-A014418 and 10 μM SP600125 (AR+SP), before cell harvesting, when indicated. Cell lysates were analyzed by western blot using antibodies against Trim17, Mcl-1 and actin
Trim17 directly promotes Mcl-1 ubiquitination
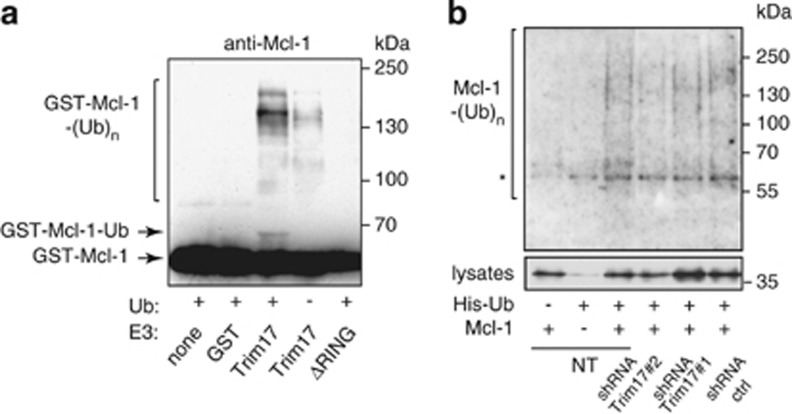
To test whether Trim17 can be an E3 ubiquitin-ligase for Mcl-1, we carried out an in vitro ubiquitination assay using only purified recombinant proteins. Following in vitro phosphorylation of Mcl-1 by recombinant GSK3 and JNK, addition of GST-Trim17 but not GST nor the inactive mutant Trim17(ΔRING) stimulated Mcl-1 ubiquitination in the presence of ubiquitin, E1 and E2 enzymes (Figure 8a). These data indicate that Mcl-1 is a direct substrate for the E3 ubiquitin-ligase activity of Trim17. To investigate whether Trim17 can also ubiquitinate Mcl-1 in neurons, we assessed the ubiquitination level of Mcl-1 co-transfected with His-ubiquitin in CGNs transduced with different shRNAs. Ubiquitination of Mcl-1 was significantly reduced by both shRNA specifically targeting Trim17 compared to control shRNA (Figure 8b), suggesting that Trim17 actually contributes to the ubiquitination of Mcl-1 in neurons.
Figure 8.
Trim17 promotes Mcl-1 ubiquitination both in vitro and in vivo. (a) Purified recombinant mouse Mcl-1 was first phosphorylated by recombinant JNK and GSK3 in vitro. Then, it was incubated for 2 h in the in vitro ubiquitination reaction mix with different recombinant proteins: GST, GST-Trim17(WT) or GST-Trim17(ΔRING), in the presence or the absence of ubiquitin, as indicated. Mcl-1 ubiquitination was examined by anti-Mcl-1 immunoblotting. The arrows indicate unmodified Mcl-1 (GST-Mcl-1) and mono- or di-ubiquitinated form of Mcl-1 (GST-Mcl-1-Ub). Higher bands are poly- or multi-ubiquitinated forms of Mcl-1 (GST-Mcl-1-(Ub)n). (b) CGNs were left untreated (non transduced: NT) or were transduced with lentiviral particles expressing shRNA sequences (one control and two against Trim17), one day after plating. At DIV 5, neurons were transfected with Mcl-1(WT), His-tagged ubiquitin or both for 18 h. The ubiquitinated proteins were purified using nickel beads, separated by SDS-PAGE and visualized by immunoblotting with anti-Mcl-1 antibody to detect ubiquitin-conjugated Mcl-1. In a separate SDS-PAGE, samples of the input lysates were analyzed with anti-Mcl-1 antibody to estimate the expression level of transfected Mcl-1 in the different conditions. Normalization of the ubiquitination signal by the expression level of Mcl-1 in lysates shows that ubiquitination is approximately reduced by half after transduction with both shRNA-Trim17#1 and #2 compared to shRNA ctrl. *Indicates a non-specific band
Discussion
Growing evidence indicate that the ubiquitin-proteasome system regulates apoptosis, by controlling the level or the function of key factors.27, 28 Notably, acute treatment with proteasome inhibitors has been shown to block apoptosis in several types of neurons.17, 29, 30 However, the proteins that must be eliminated by the proteasome to initiate neuronal death are mostly unknown. In the present study, we report for the first time that the anti-apoptotic protein Mcl-1 is degraded by the proteasome during neuronal apoptosis. In addition, our data indicate that prior phosphorylation by GSK3 is required for ubiquitination and subsequent proteasomal degradation of Mcl-1, in KCl-deprived CGNs. Indeed, the decrease in Mcl-1 level was abrogated by a specific GSK3 inhibitor and was partly reduced by inhibiting protein kinases that prime Mcl-1 for GSK3 phosphorylation. Moreover, mutation of residues phosphorylated by GSK3 and priming kinases resulted in both a decreased level of ubiquitination and an increased half-life of Mcl-1. More importantly, we provide experimental evidence that the E3 ubiquitin-ligase Trim17 mediates Mcl-1 ubiquitination and directs its degradation in CGNs, in a GSK3-dependent manner. As such, our results provide the first insight into Mcl-1 post-translational regulation in neurons. In addition, stabilization of Mcl-1, by either pharmacological inhibition or point mutations, resulted in improved neuroprotection. Our study thus strongly suggests that proteasomal degradation of Mcl-1 is required for the initiation of apoptosis in neurons.
The involvement of ubiquitination in Mcl-1 proteasomal degradation has recently been questioned by a study using a non-ubiquitinable Mcl-1 mutant in which all 14 lysine residues were replaced by arginines.31 Despite failing to be ubiquitinated, this mutant protein was eliminated at a rate similar to Mcl-1(WT).31 This work suggests that Mcl-1 degradation can occur independently of ubiquitination. However, it does not exclude that ubiquitination may accelerate Mcl-1 elimination. Indeed, overexpression of Mule enhanced degradation of Mcl-1(WT) whereas it did not influence the turnover of the non-ubiquitinable mutant.31 Mutagenesis of five critical lysine residues has been found to both abolish ubiquitination and extend the half-life of human MCL-1 in HeLa and 293T cells.20, 24 In the present study, we show that mutation of three of these five lysines in mouse Mcl-1 increases its half-life in CGNs. Therefore, our results support the notion that ubiquitination of Mcl-1 is required for its proteasomal degradation, at least in some conditions.
Trim17 (also known as terf) is a member of the TRIM protein family that is defined by the tripartite motif, comprising a RING finger, one or two B-Box domains and a coiled-coil.32 Accumulating data indicate that the TRIM family represents one of the largest classes of single-protein, RING-containing, E3 ubiquitin-ligases.33, 34 We and others have demonstrated that Trim17 is a bona fide E3 ubiquitin-ligase.25, 35 In addition, we have shown that the E3 activity of Trim17 is both necessary and sufficient for the initiation of neuronal apoptosis.25 Recently, human TRIM17 has been found to stimulate the proteasome-dependent degradation of the kinetochore protein ZWINT.36 Nevertheless, whether ZWINT is a genuine substrate of TRIM17 has not been demonstrated.
Here, we present several lines of evidence indicating that Mcl-1 is a substrate for Trim17. First, Trim17 physically interacted with both overexpressed and endogenous Mcl-1. Second, Trim17 ubiquitinated Mcl-1 in vitro. Third, overexpression of Trim17 decreased the protein level of Mcl-1 whereas knock down of Trim17 expression both reduced the ubiquitination level of Mcl-1 and increased its half-life. Finally, preventing Mcl-1 phosphorylation by kinase inhibition or point mutations not only resulted in decreased ubiquitination and degradation of Mcl-1, but also blocked the physical interaction between Trim17 and Mcl-1. Taken together, these data support the hypothesis that Trim17 is a physiological E3 ubiquitin-ligase for Mcl-1. The ubiquitin-mediated degradation of Mcl-1 may therefore underlie, at least partially, the pro-apoptotic effect of Trim17 in neurons. Moreover, by identifying Mcl-1 as the first genuine substrate of Trim17, we define a novel molecular mechanism for regulation of neuronal apoptosis by the ubiquitin-proteasome system.
In neurons transduced with specific shRNAs against Trim17, the ubiquitination level of Mcl-1 was reduced and its half-life was doubled, indicating that Trim17 is involved in the turnover of Mcl-1. However, Mcl-1 ubiquitination and degradation were not completely abolished. This may be due to the incomplete knock down of Trim17 expression, as revealed by our quantitative PCR measurements. It is also possible that other E3 ubiquitin-ligases contribute to the ubiquitination of Mcl-1 in neurons in addition to Trim17. Three different E3 ubiquitin-ligases have previously been implicated in mediating MCL-1 ubiquitination. The first one, Mule/ARF-BP1, is a HECT-domain family E3 ubiquitin-ligase.24 The second one, SCFβ-TrCP, is a SKP1-cullin-1-F-box protein (SCF) family member. It requires prior MCL-1 phosphorylation by GSK3 to mediate ubiquitination.5 Finally, two recent studies identified another SCF complex as a third E3 ubiquitin-ligase for MCL-1 in cancer cells.20, 26 The tumor suppressor FBW7, the F-box protein of this complex, binds MCL-1 and targets it for ubiquitination and proteasomal degradation also in a GSK3-dependent manner.20 Moreover, the deubiquitinase USP9X has been shown to bind MCL-1 and to remove poly-ubiquitinated chains, resulting in MCL-1 stabilization.37 In CGNs, silencing of Fbw7 had no effect, and silencing of Mule had a smaller effect on the protein level of Mcl-1 than Trim17 knock down. This suggests that Trim17 is the major E3 ubiquitin ligase regulating the stability of Mcl-1, at least in CGNs. Determining the relative contribution of Trim17, other E3 ubiquitin-ligases and USP9X in the regulation of Mcl-1 level in other neuronal types requires further investigation. This is of crucial importance, as targeting the specific interaction between Mcl-1 and the enzymes regulating its stability might be used to improve neuronal survival in pathological conditions.
High levels of Trim17 mRNA have been detected not only in apoptotic neurons25 but also in organs with a high rate of apoptosis such as testis, spleen and thymus.35 Future work will determine whether Trim17 can mediate the degradation of Mcl-1 and control the initiation of apoptosis in non-neuronal cells. As Mcl-1 is a major anti-apoptotic protein, the activities that regulate its protein level are potentially important in many physiological and pathological situations. Notably, Mcl-1 overexpression has been found in many human malignancies and often correlates with poor prognosis and drug resistance.3 By stimulating Mcl-1 degradation, Trim17 might thus contribute to tumor suppression.
Materials and Methods
Unless otherwise stated, data are representative of at least three independent experiments.
Materials
Rabbit polyclonal antibodies against mouse Mcl-1 and against the cleaved form (Asp175) of caspase 3 were from Rockland (600-401-394) and Cell Signaling Technology (#9661), respectively. Rabbit monoclonal antibody against phospho-GSK-3β (Ser9) (5B3) was from Cell Signaling Technology (#9323). Mouse monoclonal antibodies against actin and against the native form of cytochrome c were from Chemicon International (MAB1501R) and from BD Pharmingen (#556432), respectively. Fluorescent and horseradish peroxidase-conjugated goat anti-rabbit and anti-mouse secondary antibodies were from Molecular Probes and Jackson ImmunoResearch Laboratories Inc., respectively. MG-132, epoxomicin and AR-A014418 (GSK-3β inhibitor VIII) were from Calbiochem, Q-VD-OPh was from Enzyme Systems Products, SB 203580 was from LC Laboratories (Woburn, MA, USA). Protein A-agarose beads and GFP-Trap-A beads were respectively from Roche and Chromotek. Culture media were from GIBCO Invitrogen. Poly-𝒟-lysine was from Becton Dickinson Biosciences. The HIV-derived lentiviral vectors pLKO.1 containing a non target shRNA (shRNA control) and the shRNAs TRCN0000037334 (shRNA Trim17#1) and TRCN0000037337 (shRNA Trim17#2) were obtained from Sigma and Open Biosystems. Recombinant E1 enzyme, UbcH5b and UbcH5c E2 enzymes, and ubiquitin were from BostonBiochem. Fetal calf serum, other culture reagents, protease inhibitor cocktail, N-ethylmaleimide, methionine and cysteine-free Dulbecco's modified Eagle's medium (DMEM), Hoechst 33258, MTT, SP600125 and all other chemicals were from Sigma-Aldrich.
Cell cultures
Primary cultures of CGNs were prepared from 7-day-old murine pups (C57Bl/6 J mice) as described previously.25 Briefly, freshly dissected cerebella were dissociated by trypsinization and mechanical disruption, and plated in Basal Medium Eagle (BME) medium supplemented with 10% fetal bovine serum, 2 mM ℒ-Gln, 10 mM HEPES, penicillin-streptomycin 100 IU/ml-100 μg/ml and 20 mM KCl.
Lenti-X 293 T cells (Clontech) and the murine neuroblastoma cells Neuro2A were cultured in Glutamax-containing DMEM supplemented with 10% FCS and penicillin-streptomycin 100 IU/ml–100 μg/ml.
KCl deprivation, survival assay and immunofluorescence
After 6 days in vitro (DIV), CGNs were washed once and incubated for indicated times in serum-free BME supplemented with ℒ-Gln, HEPES, antibiotics and 1 μM (+)-MK-801, and containing either 25 mM KCl (K25 medium) or 5 mM KCl (K5 medium). Neuronal viability was assessed in triplicate by MTT assay as described previously.25 For immunofluorescence analysis, CGNs were seeded onto glass coverslips coated with poly-𝒟-lysine and laminin. After induction of apoptosis in the presence or absence of inhibitors, neurons were fixed with paraformaldehyde and detection of nuclear condensation, caspase 3 activation and cytochrome c release was performed as described previously.25
Cycloheximide treatment and western blot analysis
For measurement of the half-life of Mcl-1, 10 μg/ml cycloheximide was added to medium for increasing times before cell lysis and protein extraction. CGNs were harvested and lysed by thorough vortexing in lysis buffer A (50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 10 mM NaF, 5 mM sodium pyrophosphate, 25 mM β-glycerophosphate, 5 mM EDTA, 25 μM MG-132, and protease inhibitor cocktail). Cell debris were removed by centrifugation at 1000 × g for 5 min at 4°C. Protein concentration of supernatants was estimated with Biorad protein assay with bovine serum albumin as the standard. Proteins (40–60 μg) were separated by 12% SDS-PAGE and transferred to Hybond-P PVDF membrane (GE Healthcare). Blocking, probing with antibodies and visualization of immunoreactive proteins were performed as previously described.25 ImageJ software was used for optical density quantitation of western blots.
RNA preparation and real time quantitative RT-PCR
Total RNA was extracted using the RNAqueous kit (Ambion) and treated with the DNase I from the DNA-free kit (Ambion) according to manufacturer's instructions. RNAs were used to perform a two-step reverse-transcription polymerase chain reaction (RT-PCR) to amplify mouse mcl-1 (forward primer: 5′-AGATGGCGTAACAAACTGGG-3′ reverse primer: 5′-TGGAAGAACTCCACAAACCC-3′) and mouse Trim17 (forward primer: 5′-AGGGAGTATAAGCTCAAGTTGGA-3′ reverse primer: 5′-CCTGCCACTCAGTTAAGGTCT-3′), as described previously.25 Data were analyzed and relative amounts of specifically amplified cDNA were calculated with MxPro software (Agilent) using the β-2 microglobulin amplicon (forward primer: 5′-TATGCTATCCAGAAAACCCCTCAA-3′ reverse primer: 5′-GTATGTTCGGCTTCCCATTCTC-3′) as a reference.
Site-directed mutagenesis of Mcl-1
The cDNA of mouse mcl-1 in pBluescript SK plasmid was obtained from Dr. Joseph Opferman (St. Jude Children's Research Hospital, Memphis, TN, USA). It was subcloned between the EcoRI and XbaI sites of pCI (Promega) to create pCI-Mcl-1. The pCI-Mcl-1-GFP fusion was obtained by overlap extension PCR. Two overlapping fragments were amplified in two separate PCR reactions. The first one used pCI-Mcl-1 as template, with the following primers: 5′-AGGGCATGCTCCGGAAAC-3′ (forward) and 5′-GCCCTTGCTCACGGATCCTCTTATTAGATATGCCAGACCAGCC-3′ (reverse). The second PCR reaction used pCI-Trim17-GFP25 as a template with the following primers: 5′-GGTCTGGCATATCTAATAAGAGGATCCGTGAGCAAGGGC-3′ (forward) and 5′-AAATCTAGACTATTACTTGTACAGCTCGTCCATG-3′ (reverse). The two amplicons were purified on a 1% agarose gel. Ten ng of each were mixed and used as template for a third PCR amplification using primers 5′-ATAGAATTCATGTTTGGCCTGCGGAG-3′ (forward) and 5′-AAATCTAGACTATTACTTGTACAGCTCGTCCATG-3′ (reverse). The recombinant PCR product was used to replace an Sph1-Xba1 fragment deleted from the coding region of Mcl-1 in the pCI-Mcl-1 plasmid. The pCI-Mcl-1(STAA) mutant, in which Ser 140 and Thr 144 (corresponding to Ser 159 and Thr 163 in human MCL-1) were replaced by alanines, was obtained by site-directed mutagenesis of pCI-Mcl-1, using the QuikChange II XL kit (Agilent Technologies). Primers 5′-GGGCCGACGGAGCTCTGCCCTCCACGC-3′ and 5′-GCGTGGAGGGCAGAGCTCCGTCGGCCC-3′ were first used to create pCI-Mcl-1(S140A), that was then used as a template to produce pCI-Mcl-1(STAA) using primers 5′-GCTCTCTGCCCTCGGCGCCGCCGCC-3′ and 5′-GGCGGCGGCGCCGAGGGCAGAGAGC-3′. The pCI-Mcl-1(K3R) mutant, in which lysines 117, 175 and 178 (corresponding to lysines 136, 194 and 197 in human MCL-1) were replaced by arginines, was obtained in a similar manner. Primers 5′-GCCATCGGCAGGCGCCCGGCCG-3′ and 5′-CGGCCGGGCGCCTGCCGATGGC-3′ were first used to create pCI-Mcl-1(K117R), that was then used as a template to produce pCI-Mcl-1(K117R/K175R/K178R) using primers 5′-CGACCGGCTCCAGGGACTCGAGGCCTCTGGGC-3′ and 5′-GCCCAGAGGCCTCGAGTCCCTGGAGCCGGTCG-3′. The sequences of all the constructs were confirmed by automatic sequencing.
Transfection of CGNs and protection against apoptosis
Primary CGNs grown on glass coverslips in 24-well plates were transfected at DIV 5 with 1 μg of pCI expression vectors encoding GFP (as a control) or different forms of Mcl-1 fused to GFP, together with 1 μg of empty pCI. For this purpose, we used a calcium phosphate protocol optimized for neuronal cultures as previously described.25 Eighteen hours after transfection, the medium was changed to K5 medium and the neurons were further incubated for 8 h. Then, CGNs were fixed, stained with Hoechst and mounted on glass slides in Mowiol. Among GFP-positive neurons, apoptosis was assessed by detection of nuclear condensation or fragmentation by Hoechst staining. For each experiment and each condition, at least 200 GFP-positive neurons were scored in a blinded manner. Statistical analyses were performed using GraphPad InStat version 3.0 for Mac (GraphPad Software, San Diego, CA, USA, www.graphpad.com). GFP intensity was estimated using MetaMorph software.
In vivo ubiquitination of Mcl-1
CGNs cultured in 60 mm dishes were transfected at DIV 5, using calcium phosphate precipitation, with 6 μg of plasmids expressing the different forms of untagged Mcl-1 (or empty pCI), together with 14 μg of a plasmid that expresses eight His6-tagged ubiquitin (His-Ub), or empty pCI. Eighteen hours after transfection, the medium was supplemented with 25 μM MG-132, and neurons were further cultured for 6 h. Then, neurons were washed once in PBS and homogenized in 700 μl lysis buffer B (6M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, 10 mM Tris-HCl (pH 8.0) supplemented with 0.3M NaCl. The lysates were sonicated and cleared by centrifugation at 1500 × g for 5 min at room temperature. Aliquots (50 μl) of resulting supernatants were precipitated with TCA for transfection efficiency control. In parallel, the remaining 650 μl of each extract were supplemented with 10 mM imidazole (pH 8.0), added to 20 μl magnetic nickel beads (MagneHis Ni-Particles, Promega) and rotated for 2 h at room temperature to purify ubiquitinated proteins. Beads were washed once with lysis buffer B supplemented with 10 mM imidazole, once with 8 M urea, 0.1 M Na2HPO4/NaH2PO4, 10 mM Tris-HCl (pH 8.0) 10 mM imidazole, once with 8 M urea, 0.1 M Na2HPO4/NaH2PO4, 10 mM Tris-HCl (pH 6.3) 10 mM imidazole, 0.2% Triton X-100, 0.5 M NaCl and three times with 8 M urea, 0.1 M Na2HPO4/NaH2PO4, 10 mM Tris-HCl (pH 6.3) 10 mM imidazole, 0.2% Triton X-100. Materials bound to the beads were eluted by the addition of 3 × Laemmli sample buffer and boiling for 10 min. These purification products were resolved by SDS-PAGE and blotted for ubiquitinated Mcl-1 products using anti-Mcl-1 antibody. TCA precipitates of the input lysates were analyzed with anti-Mcl-1 antibody in a separate SDS-PAGE.
Half-life measurement by pulse-chase experiments
Primary CGNs cultured in 6-well plates were transfected at DIV 5 with 10 μg of plasmids expressing the different forms of Mcl-1 fused to GFP, using calcium phosphate precipitation as described above. Eighteen hours after transfection, neurons were washed twice and incubated in labeling medium (methionine and cysteine-free DMEM supplemented with 50 μM ℒ-cysteine-HCl and GlutaMAX-I) for 1 h to deplete intracellular stores of methionine. Labeling medium containing 25 μCi of Easy Tag Methionine ℒ-[35S]-(1175 Ci/mmol; Perkin Elmer) in 1 ml was then added for 2 h for metabolic labeling. Then, neurons were washed three times in PBS and either harvested immediately (time 0) or washed twice and incubated for 2, 4, 6 or 8 h in normal K5 medium, before harvesting in lysis buffer A. Cell lysates were diluted seven times in dilution buffer (10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.5 mM EDTA, 25 μM MG-132, and protease inhibitor cocktail) and cell debris were removed by centrifugation at 3000 × g for 5 min at 4°C. Resulting supernatants were precleared by rotation for 1 h at 4°C with 10 μl protein A-agarose beads and then rotated for 2 h at 4°C with 12 μl GFP-Trap-A beads to immunoprecipitate Mcl-1 and its mutants fused to GFP. Beads were recovered by centrifugation and washed four times with dilution buffer. Materials bound to the beads were eluted by the addition of 3 × Laemmli sample buffer and boiling for 5 min. Precipitated proteins were separated by SDS–PAGE, transferred to PVDF membranes and analyzed by autoradiography. Then, membranes were probed with anti-Mcl-1 antibody. The radioactivity associated to each band was quantified and normalized by the amount of Mcl-1 precipitated in each condition.
Lentivirus preparation and lentiviral transduction of CGN
Lentiviruses were produced as described.38 CGNs were transduced one day after plating. Approximately 500 ng p24 of each lentiviral preparation were added, per million neurons, directly to the culture medium for 8 h. Neurons were then replaced in fresh medium and culture was continued until 6 DIV.
GST pull down
Recombinant GST and GST-Trim17 fusion proteins were produced in Escherichia coli and purified on glutathione magnetic beads as described previously.25 For pull down assays, cerebella from P7 mice, or CGN incubated in K5 medium supplemented with 20 μM MG-132 for 6 h, were lysed in lysis buffer C (50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 20 μM MG-132 and protease inhibitor cocktail (Sigma)). The lysates were first precleared on free glutathione magnetic beads on a rotating wheel for 1 h at 4°C. Precleared lysates were then added to GST or GST-Trim17 recombinant proteins bound to glutathione magnetic beads and rotated overnight at 4°C. Beads were washed three times with lysis buffer C and once with the same buffer supplemented with 0.3 M NaCl. Materials bound to the beads were eluted by the addition of 3 × Laemmli sample buffer and boiling for 10 min. The samples were analyzed by western blot.
Co-transfection and co-immunoprecipitation in Neuro2A cells
Neuro2A cells, plated on six well plates, were transfected with indicated plasmids, using the GenJet transfection reagent for Neuro2A cells, according to the instructions of the manufacturer (SignaGen Laboratories). Twenty-four hours after transfection, cells were harvested in lysis buffer B and protein extracts were analyzed by western blot. For co-immunoprecipitation, Neuro2A cells were incubated for 8 h with 20 μM MG-132, in the absence or the presence of 10 μM AR-A014418 and 10 μM SP600125. Cells were lysed and GFP or Trim17-GFP were immunoprecipitated as described above, except that all buffers used from cell lysis to washing of the beads contained phosphatase inhibitors (10 mM NaF, 5 mM sodium pyrophosphate, 25 mM disodium β-glycerophosphate). In one condition, following immunoprecipitation and washes, the beads were incubated with 800 U λ-phosphatase (New England Biolabs), in 20 μl of the reaction buffer supplied by the manufacturer, for 30 min at 30°C. After phosphatase treatment, the beads were washed three times before elution. Precipitated proteins were analyzed by western blot.
Production of recombinant GST-Mcl-1 and in vitro ubiquitination assay
In order to produce recombinant GST-Mcl-1 in Escherichia coli, an N-terminal GST tagged Mcl-1 was produced by PCR amplification of the mcl-1 coding region using pCI-Mcl-1 as a template and primers 5′-ATACCCGGGGCTCTTATTAGATATGCCAGACCAGC-3′ and 5′-ATAGAATTCTTTGGCCTGCGGAGAAAC-3′. The PCR product was cloned between the EcoRI and SmaI sites of the pGEX-5X-1 vector (Amersham). The resulting pGEX-Mcl-1 was transformed in BL21 Escherichia coli. Protein expression was induced by the addition of 200 μM IPTG and was carried out at 20°C for 4 h. Bacteria were lysed by sonication at 4°C and GST fusion proteins were isolated by binding to glutathione beads (Glutathione Sepharose 4 fast flow, GE Healthcare): the soluble protein fraction was incubated with the beads for 2 h at 4°C on a rotating wheel and washed three times with PBS supplemented with 0.1% NP-40. Bound protein was eluted with 50 mM reduced glutathione in 50 mM Tris-HCl (pH 8). Purest fractions were pooled and protein concentration was determined using Biorad protein assay.
Recombinant Mcl-1 was first submitted to in vitro phosphorylation: 2.45 μg of purified GST-Mcl-1 was incubated in the presence of 1000 U recombinant GSK3 (New England BioLabs), 0.2 μg recombinant active JNK1 (BioVision), and 1 mM ATP in 50 μl JNK reaction buffer (25 mM MOPS (pH 7.2), 12.5 mM β-glycerophosphate, 25 mM MgCl2, 5 mM EGTA, 2 mM EDTA, 0.25 mM DTT) for 1 h at 30°C. For each ubiquitination reaction, 5 μl of the phophorylation reaction were incubated in 20 μl of ubiquitination reaction buffer (25 mM Tris-HCl (pH 7.5), 50 mM NaCl, 4 mM ATP, 4 mM MgCl2, 2 mM DTT, 10 mM phosphocreatine, 0.5 U creatine kinase, 20 μM ZnCl2), in the presence of 50 ng recombinant E1 enzyme, 250 ng recombinant UbcH5b and 250 ng recombinant UbcH5c E2 enzymes, 10 μg ubiquitin, and ≈1 μg of purified recombinant GST, GST-Trim17(WT) or GST-Trim17(ΔRING). Reactions were incubated at 37°C for 2 h, and stopped by adding 10 μl of 3 × Laemmli sample buffer and heating at 95°C for 5 min. The samples were analyzed by SDS-PAGE and immunoblotting with anti-Mcl-1 antibody.
Acknowledgments
We would like to thank the staff of the Animal facilities of the IGMM for the breeding of mice and the RIO imaging platform for technical assistance. We also thank Dr. Joseph Opferman for providing mouse mcl-1 cDNA. We are grateful to Dr. Jonathan Ham and Pr. Jean-Claude Martinou for critical reading of the manuscript. This work was supported by the Centre National de la Recherche Scientifique (CNRS), the Institut National de la Santé et de la Recherche Médicale (INSERM) and Université Montpellier 2.
Glossary
- CGN
cerebellar granule neurons
- GSK3
Glycogen Synthase Kinase 3
- [KCl]o
extracellular concentration of KCl
- K25
serum-free medium containing 25 mM KCl
- K5
serum-free medium containing 5 mM KCl
- JNK
c-Jun N-terminal protein kinase
- WT
wild type
- GFP
green-fluorescent protein
- shRNA
short hairpin RNA
- DIV
days in vitro
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by C Borner
Supplementary Material
References
- Yuan J, Yankner BA. Apoptosis in the nervous system. Nature. 2000;407:802–809. doi: 10.1038/35037739. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Moldoveanu T, Llambi F, Parsons MJ, Green DR. The BCL-2 family reunion. Mol Cell. 2010;37:299–310. doi: 10.1016/j.molcel.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warr MR, Shore GC. Unique biology of Mcl-1: therapeutic opportunities in cancer. Curr Mol Med. 2008;8:138–147. doi: 10.2174/156652408783769580. [DOI] [PubMed] [Google Scholar]
- Nijhawan D, Fang M, Traer E, Zhong Q, Gao W, Du F, et al. Elimination of Mcl-1 is required for the initiation of apoptosis following ultraviolet irradiation. Genes Dev. 2003;17:1475–1486. doi: 10.1101/gad.1093903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, He X, Hsu JM, Xia W, Chen CT, Li LY, et al. Degradation of Mcl-1 by beta-TrCP mediates glycogen synthase kinase 3-induced tumor suppression and chemosensitization. Mol Cell Biol. 2007;27:4006–4017. doi: 10.1128/MCB.00620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol Cell. 2006;21:749–760. doi: 10.1016/j.molcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- Sitailo LA, Tibudan SS, Denning MF. The protein kinase C delta catalytic fragment targets Mcl-1 for degradation to trigger apoptosis. J Biol Chem. 2006;281:29703–29710. doi: 10.1074/jbc.M607351200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, D'Ercole AJ. Expression of Mcl-1 in cerebellar granule neurons is regulated by IGF-I in a developmentally specific fashion. Brain Res Dev Brain Res. 2004;152:255–263. doi: 10.1016/j.devbrainres.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Mori M, Burgess DL, Gefrides LA, Foreman PJ, Opferman JT, Korsmeyer SJ, et al. Expression of apoptosis inhibitor protein Mcl1 linked to neuroprotection in CNS neurons. Cell Death Differ. 2004;11:1223–1233. doi: 10.1038/sj.cdd.4401483. [DOI] [PubMed] [Google Scholar]
- Arbour N, Vanderluit JL, Le Grand JN, Jahani-Asl A, Ruzhynsky VA, Cheung EC, et al. Mcl-1 is a key regulator of apoptosis during CNS development and after DNA damage. J Neurosci. 2008;28:6068–6078. doi: 10.1523/JNEUROSCI.4940-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain M, Nguyen AP, Le Grand JN, Arbour N, Vanderluit JL, Park DS, et al. MCL-1 is a stress sensor that regulates autophagy in a developmentally regulated manner. EMBO J. 2011;30:395–407. doi: 10.1038/emboj.2010.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Mello SR, Galli C, Ciotti T, Calissano P. Induction of apoptosis in cerebellar granule neurons by low potassium: inhibition of death by insulin-like growth factor I and cAMP. Proc Natl Acad Sci USA. 1993;90:10989–10993. doi: 10.1073/pnas.90.23.10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood KA, Dipasquale B, Youle RJ. In situ labelling of granule cells for apoptosis-associated DNA fragmentation reveals different mechanisms of cell loss in developing cerebellum. Neuron. 1993;11:621–632. doi: 10.1016/0896-6273(93)90074-2. [DOI] [PubMed] [Google Scholar]
- Desagher S, Severac D, Lipkin A, Bernis C, Ritchie W, Le Digarcher A, et al. Genes regulated in neurons undergoing transcription-dependent apoptosis belong to signaling pathways rather than the apoptotic machinery. J Biol Chem. 2005;280:5693–5702. doi: 10.1074/jbc.M408971200. [DOI] [PubMed] [Google Scholar]
- Bobba A, Canu N, Atlante A, Petragallo V, Calissano P, Marra E. Proteasome inhibitors prevent cytochrome c release during apoptosis but not in excitotoxic death of cerebellar granule neurons. FEBS Lett. 2002;515:8–12. doi: 10.1016/s0014-5793(02)02231-7. [DOI] [PubMed] [Google Scholar]
- Butts BD, Hudson HR, Linseman DA, Le SS, Ryan KR, Bouchard RJ, et al. Proteasome inhibition elicits a biphasic effect on neuronal apoptosis via differential regulation of pro-survival and pro-apoptotic transcription factors. Mol Cell Neurosci. 2005;30:279–289. doi: 10.1016/j.mcn.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Canu N, Barbato C, Ciotti MT, Serafino A, Dus L, Calissano P. Proteasome involvement and accumulation of ubiquitinated proteins in cerebellar granule neurons undergoing apoptosis. J Neurosci. 2000;20:589–599. doi: 10.1523/JNEUROSCI.20-02-00589.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Mello SR, Kuan CY, Flavell RA, Rakic P. Caspase-3 is required for apoptosis-associated DNA fragmentation but not for cell death in neurons deprived of potassium. J Neurosci Res. 2000;59:24–31. [PubMed] [Google Scholar]
- Porcile C, Piccioli P, Stanzione S, Bajetto A, Bonavia R, Barbero S, et al. Proteasome inhibitors induce cerebellar granule cell death: inhibition of nuclear factor-kB activation. Ann NY Acad Sci. 2002;973:402–413. doi: 10.1111/j.1749-6632.2002.tb04673.x. [DOI] [PubMed] [Google Scholar]
- Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, et al. SCFFBW7 regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Altman BJ, Coloff JL, Herman CE, Jacobs SR, Wieman HL, et al. Glycogen synthase kinase 3{alpha} and 3{beta} mediate a glucose-sensitive antiapoptotic signaling pathway to stabilize Mcl-1. Mol Cell Biol. 2007;27:4328–4339. doi: 10.1128/MCB.00153-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel C, Carlson SM, White FM, Davis RJ. Mcl-1 integrates the opposing actions of signaling pathways that mediate survival and apoptosis. Mol Cell Biol. 2009;29:3845–3852. doi: 10.1128/MCB.00279-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada J, Sugimoto M. An inhibitor of p38 and JNK MAP kinases prevents activation of caspase and apoptosis of cultured cerebellar granule neurons. Jpn J Pharmacol. 1999;79:369–378. doi: 10.1254/jjp.79.369. [DOI] [PubMed] [Google Scholar]
- Zhong Q, Gao W, Du F, Wang X. Mule/ARF-BP1, a BH3-only E3 ubiquitin ligase, catalyzes the polyubiquitination of Mcl-1 and regulates apoptosis. Cell. 2005;121:1085–1095. doi: 10.1016/j.cell.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Lassot I, Robbins I, Kristiansen M, Rahmeh R, Jaudon F, Magiera MM, et al. Trim17, a novel E3 ubiquitin-ligase, initiates neuronal apoptosis. Cell Death Differ. 2010;17:1928–1941. doi: 10.1038/cdd.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–114. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- Broemer M, Meier P. Ubiquitin-mediated regulation of apoptosis. Trends Cell Biol. 2009;19:130–140. doi: 10.1016/j.tcb.2009.01.004. [DOI] [PubMed] [Google Scholar]
- Vucic D, Dixit VM, Wertz IE. Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol. 2011;12:439–452. doi: 10.1038/nrm3143. [DOI] [PubMed] [Google Scholar]
- Sadoul R, Fernandez P-A, Quiquerez A-L, Martinou I, Maki M, Schröter M, et al. Involvement of the proteasome in the programmed cell death of NGF-deprived sympathetic neurons. EMBO J. 1996;15:3845–3852. [PMC free article] [PubMed] [Google Scholar]
- Suh J, Lee YA, Gwag BJ. Induction and attenuation of neuronal apoptosis by proteasome inhibitors in murine cortical cell cultures. J Neurochem. 2005;95:684–694. doi: 10.1111/j.1471-4159.2005.03393.x. [DOI] [PubMed] [Google Scholar]
- Stewart DP, Koss B, Bathina M, Perciavalle RM, Bisanz K, Opferman JT. Ubiquitin-independent degradation of antiapoptotic MCL-1. Mol Cell Biol. 2010;30:3099–3110. doi: 10.1128/MCB.01266-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger' E3 ubiquitin ligases. Bioessays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- Napolitano LM, Meroni G. TRIM family: Pleiotropy and diversification through homomultimer and heteromultimer formation. IUBMB Life. 2012;64:64–71. doi: 10.1002/iub.580. [DOI] [PubMed] [Google Scholar]
- Urano T, Usui T, Takeda S, Ikeda K, Okada A, Ishida Y, et al. TRIM44 interacts with and stabilizes terf, a TRIM ubiquitin E3 ligase. Biochem Biophys Res Commun. 2009;383:263–268. doi: 10.1016/j.bbrc.2009.04.010. [DOI] [PubMed] [Google Scholar]
- Endo H, Ikeda K, Urano T, Horie-Inoue K, Inoue S. Terf/TRIM17 stimulates degradation of kinetochore protein ZWINT and regulates cell proliferation. J Biochem (Tokyo) 2012;151:139–144. doi: 10.1093/jb/mvr128. [DOI] [PubMed] [Google Scholar]
- Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, et al. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- Swainson L, Mongellaz C, Adjali O, Vicente R, Taylor N. Lentiviral transduction of immune cells. Methods Mol Biol. 2008;415:301–320. doi: 10.1007/978-1-59745-570-1_18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.