Abstract
Hedgehog (Hh) signaling is important for development and homeostasis in vertebrates and invertebrates. Ligand-independent, deregulated Hh signaling caused by loss of negative regulators such as Patched causes excessive cell proliferation, leading to overgrowth in Drosophila and tumors in humans, including basal-cell carcinoma and medulloblastoma. We show that in Drosophila deregulated Hh signaling also promotes cell survival by increasing the resistance to apoptosis. Surprisingly, cells with deregulated Hh activity do not protect themselves from apoptosis; instead, they promote cell survival of neighboring wild-type cells. This non-cell autonomous effect is mediated by Hh-induced Notch signaling, which elevates the protein levels of Drosophila inhibitor of apoptosis protein-1 (Diap-1), conferring resistance to apoptosis. In summary, we demonstrate that deregulated Hh signaling not only promotes proliferation but also cell survival of neighboring cells. This non-cell autonomous control of apoptosis highlights an underappreciated function of deregulated Hh signaling, which may help to generate a supportive micro-environment for tumor development.
Keywords: Hedgehog, Notch, non-cell autonomous effects, cell survival, Drosophila inhibitor of apoptosis
Programmed cell death or apoptosis is a normal feature of organ development that counterbalances growth and allows shaping of the organ by eliminating cells.1, 2 Control of apoptosis comes down to the control of specific cell death proteases, termed caspases.3 One class of caspase inhibitors are inhibitor of apoptosis proteins (IAPs). Drosophila IAP-1 (Diap-1) effectively inhibits the caspases Dronc (Caspase-9-like) and DrICE (Caspase-3-like).1, 2 The IAP antagonists Reaper, Hid and Grim stimulate ubiquitylation and degradation of Diap-1, releasing caspases from IAP inhibition.4 This mechanism is tightly coordinated with mechanisms that regulate proliferation and growth to maintain tissue homeostasis.5, 6 However, although much is known about the individual processes of proliferation, growth and apoptosis, how these mechanisms tie together is not well understood.
The development of the Drosophila eye depends on a changing balance of proliferative growth, differentiation and apoptosis, providing an excellent system to study how these processes interact.7, 8 During the first two stages of larval development, the eye-antennal imaginal disc proliferates extensively, forming a bi-lobed structure. The antennal lobe will make the adult antenna, while the eye lobe will form the head capsule and eye. In the third larval stage, a wave of differentiation begins at the most posterior part of the eye lobe and is marked by the formation of a groove called the morphogenetic furrow (MF) that moves anterior. Cells at the MF arrest proliferation and begin to differentiate in a well-defined pattern with the formation of photoreceptor neuron clusters followed by support cells that will separate each cluster. Cells that remain unspecified undergo apoptosis during pupal development.9
In the MF, signaling pathways coordinate the transition from proliferation to differentiation. In a simplified summary, cells in the MF arrest in G1 in response to Decapentaplegic (Dpp), which is induced by Hedgehog (Hh) signaling.10, 11, 12, 13 Hh and Dpp also induce the expression of the Notch (N) ligand Delta, which, in turn, induces a round of mitosis (second mitotic wave) in cells just posterior to the MF.11, 14 Thus, the Hh pathway is needed for MF progression (Figure 1h) and coordinates the transition from proliferation to differentiation, making it a critical target for homeostasis.
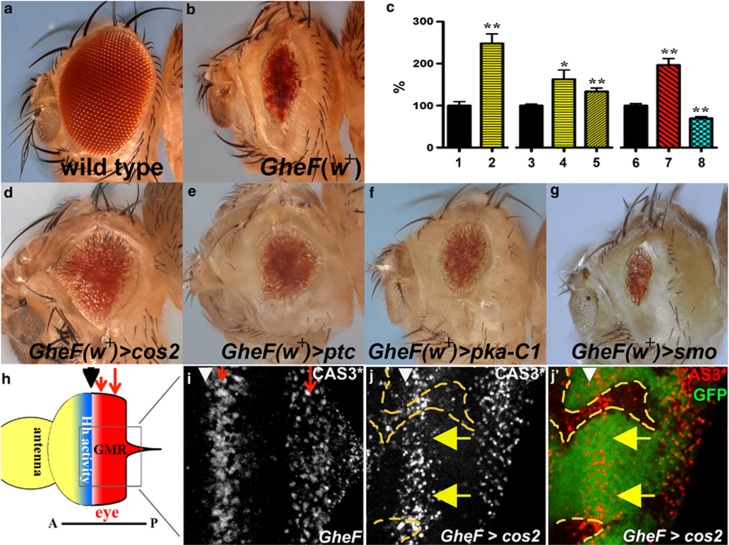
Figure 1.
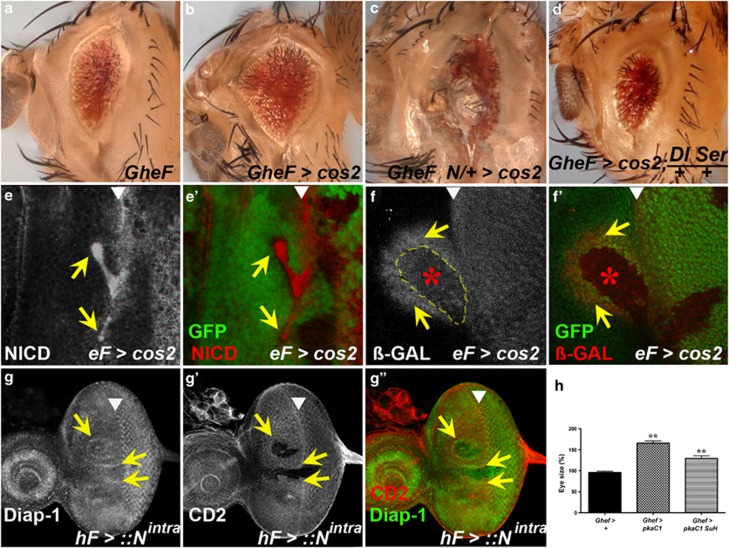
Mutants of negative regulators of Hh signaling suppress GMR-hid by non-cell autonomous inhibition of caspase activity. In this and the following figures, >denotes an FRT site, indicating mitotic or FLP-out clones. Adult eye images are of GMR-hid females unless otherwise specified. Anterior is to the left. The location of the MF is marked by arrowheads. (a) Wild-type eye. (b) The GMR-hid ey-FLP (GheF) eye ablation phenotype. Note, this transgene carries the P[w+] marker gene. (c) Histogram of relative eye size of different genotypes. Eyes from GMR-hid flies (black bars) are normalized to 100% 1 and 2 are male flies, 3–8 are females. cos2 (yellow bars) and pka-C1 (red bar) mosaics increase the average GMR-hid eye size, whereas smo mosaics (blue bar) decreases the average eye size. For each bar, 10 eyes were averaged, except 8 (5 eyes). *P-value ≤0.05 and **P-value ≤0.01. 1, GMR-hid ey-FLP/Y; FRT42D P[ubi-GFP]/CyO male. 2, GMR-hid ey-FLP/Y; FRT42D cos2H29/FRT42D P[ubi-GFP] male. 3, GMR-hid ey-FLP/y w; FRT42D P[ubi-GFP]/CyO female. 4, GMR-hid ey-FLP/y w; FRT42D cos2H29/FRT42D P[ubi-GFP] female. 5, GMR-hid ey-FLP/y w; FRT42D cos2L51/FRT42D P[ubi-GFP] female. 6, ey-FLP/y w; P(w+) FRT40A/ CyO; GMR-hid/+ female. 7, ey-FLP/y w; pka-c1K2 FRT40A/ P(w+) FRT40; GMR-hid/+ female. 8, ey-FLP/y w; smoD16 FRT40A/ P(w+) FRT40; GMR-hid/+ female. (d–f) The GheF phenotype is suppressed (eyes are larger) when flies are mosaic for either cos2 (d), ptc (e), or pka-C1 (f) mutations (quantified in (c)). (g) The GheF phenotype is enhanced when flies are mosaic for smo, a positive regulator of Hh signaling. (h) Schematic outline of an eye-antennal imaginal disc from a third instar larvae. The MF (arrowhead) separates anterior (A) and posterior (P) portions of the eye disc. Hh activity (blue) is required for anterior progression of the MF. GMR is expressed posterior to the MF (red). GMR-hid induces two apoptotic waves (red arrows). (i) In GMR-hid eye discs, cleaved Caspase-3 (CAS3*) antibody as apoptosis marker labels two distinct waves (red arrows) posterior to the MF.30(j and j′), A GMR-hid eye disc mosaic for cos2. cos2 clones are marked by the absence of GFP and outlined by yellow dashed lines. CAS3* labeling is high in cos2 clones but low in adjacent non-mutant tissue near the MF (yellow arrows). Genotypes: (b) y w GMR-hid ey-FLP; FRT42D P[ubi-GFP]/CyO. (d and e) y w GMR-hid ey-FLP; FRT42D cos2H29 (d) or ptcC (e)/FRT42D P[w+]. (f and g) y w GMR-hid ey-FLP; pka-c1B3 (f) or smoD16 (g) FRT40A/P[w+] FRT40. (i) y w GMR-hid ey-FLP; FRT42D P[ubi-GFP]/CyO. (j and j′) y w GMR-hid ey-FLP; FRT42 cos2H29/FRT42 P[ubi-GFP]
Hh signal transduction is highly conserved between flies and mammals.15, 16 In cells that are not exposed to the Hh ligand, the transmembrane protein Patched (Ptc; Ptch1 in mammals) blocks the availability of another transmembrane protein, Smoothened (Smo).17, 18 In the absence of Smo, Costal-2 (Cos2), which encodes a kinesin-like protein with similarity to mammalian Kif-7 (kinesin family member 7), associates in a complex with the transcription factor Cubitus interruptus (Ci) that promotes proteolytic processing of Ci to the truncated repressor, CiR.19, 20, 21, 22, 23, 24 When Hh binds Ptc, Smo becomes available, interacts with Cos2 and triggers release of full-length Ci, which can then function as a transcriptional activator (CiA).
Genetically, ptc and cos2 (and another component, protein kinase A (PKA)) are negative regulators of Hh signaling promoting the formation of CiR.15, 16 Thus, genetic inactivation of ptc, cos2 and pka triggers ligand-independent, deregulated Hh signaling due to accumulation of CiA. In humans, ligand-independent Hh signaling is associated with several tumors such as basal cell carcinoma, medulloblastoma, rhabdomyosarcoma and glioma.25 In most cases, either genetic inactivation of Ptch1 or activating missense mutations of smo are the underlying causes of these tumors.
Apoptosis can be induced in the larval eye disc to determine how the tissue responds when the balance between proliferation, differentiation and cell death is tilted by increased apoptosis. Using this system, we have identified several pathways that function in regulating tissue homeostasis.26, 27, 28, 29 Here, we show that in genetic mosaics, ligand-independent, deregulated Hh signaling due to loss of negative regulators suppresses excessive cell death. Interestingly, this control of apoptosis affects cells of the two genotypes differently. It is not the cells with increased Hh signaling that are resistant to apoptosis. Instead, these cells instruct neighboring wild-type cells to increase their apoptosis resistance. This non-cell autonomous effect is mediated through Hh-induced stimulation of the N pathway, which then transcriptionally increases the levels of Diap-1 in neighboring cells. This non-cell autonomous control of apoptosis highlights an underappreciated function of Hh signaling. The increased resistance to apoptosis in cells adjacent to cells with deregulated Hh signaling may change the micro-environment, providing an additional role for deregulated Hh signaling needed for efficient tumorigenesis in human cancer.
Results
Mutants of negative regulators of Hh signaling suppress apoptosis by non-cell autonomous inhibition of caspase activity
Expression of the pro-apoptotic gene hid posterior to the MF using the GMR promoter (GMR-hid) induces an eye-ablation phenotype due to massive apoptosis30 (Figures 1b, h and i). We are using the GMR-hid system to identify suppressor mutations that confer resistance to apoptosis. To identify suppressors, we performed a mutagenesis screen in a GMR-hid background by generating genetic mosaics using the ey-FLP/FRT system (GheF screen), as described.31, 32 Negative regulators of Hh signaling, including cos2, ptc and the catalytic subunit of PKA, pka-C1, were identified as moderate suppressors of GMR-hid (Figures 1d–f; quantified in Figure 1c; see Material and Methods section). Heterozygosity of these mutants does not dominantly suppress GMR-hid (see Supplementary Figure S1), indicating that they are recessive suppressors. By contrast, mosaic loss of the positive regulator smo enhances the GMR-hid eye phenotype (Figure 1g). Therefore, ligand-independent, deregulated Hh signaling by loss of negative regulators suppresses GMR-hid-induced apoptosis, whereas loss of positive regulators enhances it.
Components of the Hh pathway are known to regulate eye growth14, 33, 34, 35 but have not been described as regulating apoptosis previously. To further characterize the suppression of GMR-hid, we examined cos2 mosaic GMR-hid eye imaginal discs with cleaved Caspase-3 (CAS3*) antibody as apoptosis marker.30 GMR-hid induces two waves of apoptotic cells posterior to the MF (Figures 1h and i, red arrows).30 Surprisingly, in cos2 mosaic GMR-hid eye discs, CAS3* labeling is not reduced in cos2 clones overlapping with the apoptotic waves (Figures 1j and j′). By contrast, wild-type or heterozygous (referred to as non-mutant) cells immediately adjacent to cos2 clones show decreased CAS3* labeling (yellow arrows in Figures 1j and j′; see Supplementary Figure S2). Therefore, while cos2 clones themselves are unprotected from GMR-hid-induced apoptosis, they appear to increase the apoptosis resistance of neighboring non-mutant cells. The non-cell autonomous suppression of GMR-hid by cos2 clones occurs only in the first apoptotic wave located at the MF (Figures 1h i; see Supplementary Figure S2). cos2 clones do not affect the second apoptotic wave, explaining the moderate suppression of GMR-hid by cos2 (Figure 1d). Thus, mutants of negative regulators of the Hh pathway, which cause ligand-independent Hh activity, suppress GMR-hid through non-cell autonomous inhibition of caspase activity.
Non-cell autonomous suppression of GMR-hid by ligand-independent Hh signaling
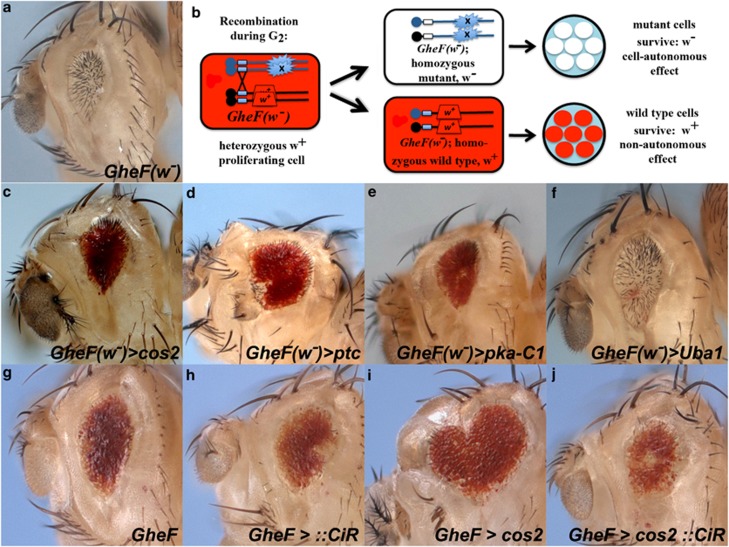
Because of the surprising observation that cos2 clones promote non-cell autonomous suppression of GMR-hid-induced apoptosis, we sought an unambiguous assay to identify the genetic identity of the rescued eye tissue in cos2, ptc and pka-C1 mosaic GMR-hid flies. The original GMR-hid transgene is marked with the white+ (w+) pigment marker, producing red eye pigment in mutant and wild-type cells (Figure 1b) and precluding an analysis of cell autonomy by eye pigmentation. Instead, we used a GMR-hid transgene without the w+ pigment gene causing a white eye of reduced size (referred to as GheF(w-)), Figure 2a). A w+ marker on the homologous chromosome arm allows determination of the genetic identity of the surviving tissue in genetic eye mosaics (Figure 2b). If the surviving tissue is white (w−) and thus mutant, the suppression is autonomous: a mosaic of Uba1, a known autonomous suppressor of GMR-hid,27 is shown in Figure 2f. However, if the surviving tissue is red (w+) and thus non-mutant, then the suppression is non-cell autonomous (Figure 2b). Mosaic cos2, ptc and pka-C1 eyes in a GheF(w-) background are composed of almost entirely red (w+) non-mutant tissue (Figures 2c–e), indicating non-cell autonomous suppression of GMR-hid, consistent with the CAS3* analysis in Figure 1j.
Figure 2.
Negative regulators of Hh signaling are non-cell autonomous suppressors of GMR-hid. In this and the following figures, :: denotes expression from a UAS-based transgene. (a) The GMR-hid(w-) ey-FLP (GheF(w-)) eye ablation phenotype. (b) Schematic for determination of the autonomy/non-autonomy of surviving tissue in mutant mosaics in GheF(w-) background. (c–e) The suppressed GheF(w-) eye by cos2 (c), ptc (d) or pka-C1 (e) mosaics is nearly all red (w+), thus comprised of non-mutant tissue and indicating non-cell autonomous suppression. (f) Uba1, a known autonomous suppressor,27 serves as a positive control for autonomous suppression of GheF(w-) as revealed by a white (w-) suppressed eye. (g–j) Ectopic expression of CiR using the MARCM system (Material and Methods section) abrogates cos2 suppression of GMR-hid. Suppression of GMR-hid by cos2 mosaics (i) is reversed by co-expression of CiR (j). Expression of CiR alone (h) has no effects on GMR-hid (g). Genotypes: (c and d) y w ey-FLP; FRT42D cos2H29(c) or ptcC(d)/FRT42D P[w+] GMR-hid(w-). (e) y w ey-FLP; pka-c1K2 FRT40/P[w+] FRT40; GMR-hid(w-). (f) y w ey-FLP; FRT42D Uba1H42/FRT42D P[w+] GMR-hid(w-). (g) y w GMR-hid ey-FLP; FRT42 tubP-GAL80/CyO; tubP-GAL4/UAS-CiCE. (h) y w GMR-hid ey-FLP; FRT42 tubP-GAL80/FRT42; tubP-GAL4/UAS-CiCE. (i) y w GMR-hid ey-FLP; FRT42 cos2H29/FRT42 tubP-Gal80; tubP-GAL4/TM6B. (j) y w GMR-hid ey-FLP; FRT42 cos2H29/FRT42 tubP-Gal80; tubP-GAL4/UAS-CiCE
This result is remarkable. Normally, wild-type and heterozygous cos2, ptc and pka-C1 cells are susceptible to GMR-hid-induced apoptosis (see Supplementary Figure S1). However, while cos2, ptc and pka-C1 mutant clones are unprotected from GMR-hid-induced apoptosis and do not contribute to the rescued eye tissue, they appear to increase the apoptosis resistance of neighboring non-mutant tissue, leading to suppression of the strong apoptotic phenotype of GMR-hid. These observations imply that cells with deregulated Hh activity produce a signal that increases the apoptosis resistance of neighboring cells.
Hh activity is mediated by the transcription factor Ci.15, 16 In the absence of Hh, Ci is proteolytically processed to the repressor form CiR.15, 36 Hh signaling or loss of negative Hh pathway regulators maintain full-length Ci (CiA).15 To test whether activation of Ci accounts for the suppression of GMR-hid (Figures 2g and i), we blocked Ci activity in cos2 clones by overexpression of CiR. Consistently, expression of CiR in cos2 clones reverts the suppression of GMR-hid by cos2 (Figure 2j) and restores the normal CAS3* pattern (Supplementary Figure S3). Expression of CiR alone does not grossly modify GMR-hid (Figure 2h). These data suggest that the non-cell autonomous resistance to apoptosis in cos2 mosaics is mediated by inappropriate Ci activation.
Non-cell autonomous upregulation of Diap1 by ligand-independent Hh signaling
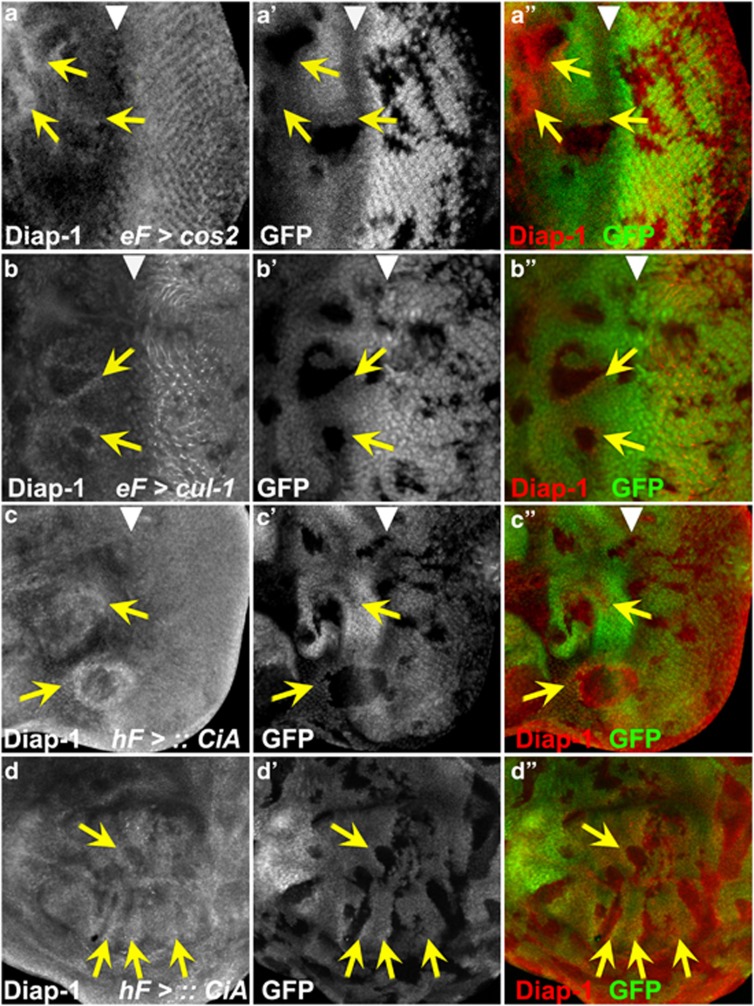
To determine the mechanism of increased non-cell autonomous resistance to apoptosis, we examined the protein levels of Diap-1, an inhibitor of apoptosis,37, 38, 39, 40, 41 in cos2 mosaic eye discs (without GMR-hid). Diap-1 is the rate-limiting component in the apoptosis pathway, and Diap-1 protein levels determine the apoptosis threshold. Significantly, Diap-1 protein accumulates just outside of cos2 clones (Figures 3a, a′ and a′′, arrows), consistent with the non-cell autonomous suppression of apoptosis. Furthermore, heterozygosity of diap-1 reverts the suppression of GMR-hid in cos2 mosaics (see Supplementary Figure S4), suggesting that Diap-1 is genetically required for suppression of GMR-hid in cos2 mosaics. We did not detect any significant changes in the protein levels of other cell death pathway components, including the caspases Dronc and DrICE. Therefore, cells with deregulated Hh signaling promote upregulation of Diap-1 in neighboring cells, which increases their apoptosis resistance and protects them from GMR-hid-induced apoptosis. The non-cell autonomous upregulation of Diap-1 is best detectable in or anterior to the MF (Figures 3a, a′ and a′′). It is difficult to judge whether cos2 clones posterior to the MF also increase Diap-1 levels non-cell autonomously because endogenous Diap-1 levels are high (Figures 3a, a′ and a′′). However, as shown below, Hh activity is also increased immediately posterior to the MF.
Figure 3.
CiA-dependent non-cell autonomous accumulation of Diap-1 anterior to the MF. (a, a′ and a′′) cos2 and (b, b′ and b′′) cul-1 mosaic eye discs were labeled with anti-Diap-1 antibody. Mutant clones are marked by the absence of GFP. Clones located in or anterior to the MF (arrowhead) promote non-cell autonomous increase of Diap-1 levels (arrows). In contrast to cos2, mutant clones of cul-1 posterior to the MF contain reduced levels of Diap-1, indicating a requirement of cul-1 for Diap-1 regulation in addition to its role in Hh signaling. eF=ey-FLP. (c and d) Ectopic expression of CiA induces non-cell autonomous increase of Diap-1 levels in (c, c′ and c′′) eye and (d, d′ and d′′) wing discs (arrows). CiA-expressing clones are marked by the absence of GFP. hF=hs-FLP. Panels (a–d) show the Diap-1 labelings only; panels (a′–d′) show the GFP channel only; panels (a′′–d′′) show the merge of Diap-1 and GFP. Genotypes: (a and b) y w ey-FLP; FRT42D cos2H29(a) or cul-1EX (b)/FRT42D P[ubi-GFP]. (c and d) y w hs-FLP; P[tubP>GFP>GAL4]; UAS-CiA. (> denotes FRT)
To address whether Ci mediates the non-cell autonomous increase of Diap-1, we examined Diap-1 levels in mosaic discs of regulators of Ci processing. Anterior to the MF, processing of CiA to CiR requires a Cullin-1 (Cul-1) and Nedd8-dependent ubiquitylation event, mediated by the ubiquitin ligase Slimb (Slmb).36, 42, 43, 44 Loss of these genes causes accumulation of active CiA,36 similar to cos2 mutants. Consistently, non-mutant cells immediately adjacent to cul-1, nedd8 and slmb clones have increased levels of Diap-1 in or anterior to the MF similar to cos2 (Figures 3b and b′ and see Supplementary Figures S4d and e), suggesting that lack of CiA processing promotes non-cell autonomous increase of Diap-1. Furthermore, clonal overexpression of CiA induces a non-cell autonomous increase of Diap-1 in eye discs (Figure 3c). This effect is best visible in or anterior to the MF, but can also be detected immediately posterior to the MF (Figure 3c). In addition, CiA-expressing clones in wing imaginal discs also increased Diap-1 levels non-cell autonomously in a position-dependent manner in the wing pouch (Figure 3d).
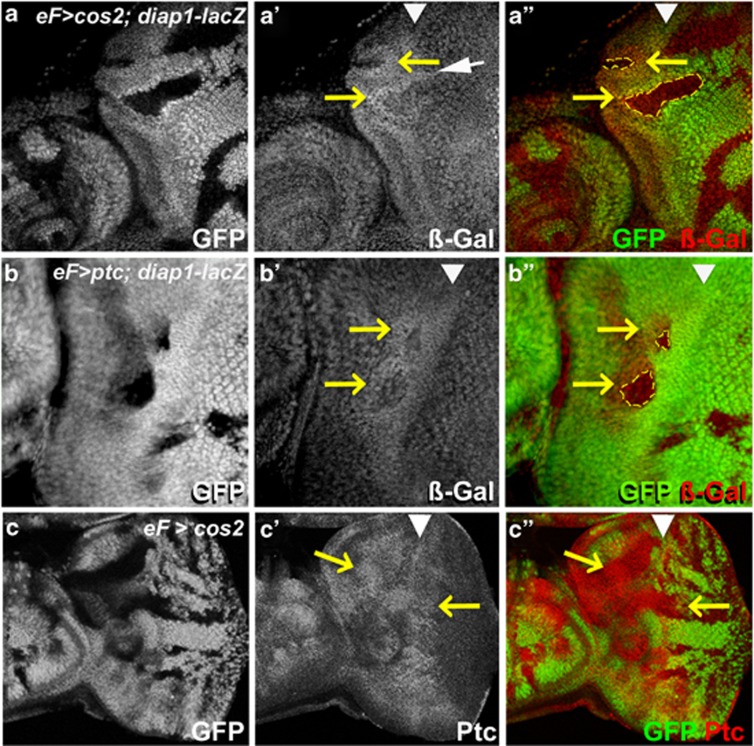
The non-cell autonomous accumulation of Diap-1 occurs transcriptionally, as indicated by the non-cell autonomous induction of the transcriptional reporter diap1-lacZ in cos2 and ptc mosaics (Figures 4a and b). The upregulation of the diap1-lacZ reporter is best detectable in or anterior to the MF, but some clones also show an effect immediately posterior to the MF (white arrow in Figure 4a′). To further clarify the position-dependence in cos2 mosaics, we used a different marker of Hh signaling. Ptc is a transcriptional target of CiA45 and can be used as a Hh marker. Consistent with the Diap1 and diap1-lacZ labelings, Ptc protein is upregulated in cos2 mutant clones anterior to the MF and in the antennal dics (Figure 4c). Interestingly, while cos2 clones located in the far posterior section of the eye disc do not change Ptc protein levels, cos2 clones located immediately posterior to the MF also accumulate Ptc protein (Figure 4c). Therefore, while clones with deregulated Hh activity show the strongest phenotypes anterior to the MF, they also affect the area immediately posterior to the MF. This area overlaps with the first apoptotic wave (Figure 1i) explaining why only the first apoptotic wave of GMR-hid is suppressed in cos2 mosaics (Figure 1j; Supplementary Figure S2).
Figure 4.
Non-cell autonomous upregulation of diap1 transcription by deregulated Hh signaling. (a and b) An enhancer trap insertion in the diap1 locus, diap1-lacZ (thj5c8), was used as a reporter for diap1 transcription. In both cos2 (a) and ptc (b) mosaics, ß-GAL levels are increased (yellow arrows in a′, a′′, b′ and b′′) anterior to the MF (white arrowheads) outside of the mutant clones (yellow outline in a′′ and b′′). However, there is also non-cell autonomous induction of diap1-lacZ immediately posterior to the MF (white arrow in a′). (c, c′, c′′) A cos2 mosaic eye disc labeled with anti-Ptc antibody. Genotypes: (a) ey-FLP; FRT42 cos2H29/FRT42 P[ubi-GFP]; thj5c8/+. (b) ey-FLP; FRT42 ptcX115/FRT42 P[ubi-GFP]; thj5c8/+. (c) ey-FLP; FRT42 cos2H29/FRT42 P[ubi-GFP]
We considered several signaling pathways that are activated in signal-receiving cells by deregulated Hh signaling in cos2 clones, including the Hippo/Warts/Yorkie pathway, a growth control pathway known to control diap1 transcription.46 To monitor Hippo/Warts/Yorkie signaling, we used an enhancer trap insertion in the expanded (ex) gene (ex-lacZ), a target of Hippo/Warts/Yorkie signaling. However, ex-lacZ labeling is not significantly induced in cos2 and ptc mosaic eye imaginal discs (Supplementary Figure S5), excluding the Hippo/Warts/Yorkie pathway as the target of deregulated Hh signaling in eye discs.
Notch is required for cos2 suppression of GMR-hid and promotes Diap-1 accumulation non-cell autonomously
Hh activity produces several signaling molecules in the MF that activate Dpp, EGFR and N pathways for regulation of proliferation and differentiation in the eye disc.8, 11, 13 Although we cannot exclude a role of EGFR and Dpp signaling, five lines of evidence implicate the N pathway for control of non-cell autonomous survival. First, the suppression of GMR-hid in cos2 mosaics is abrogated by reduced N pathway activity (e.g., heterozygosity for N or the ligands Dl and Serrate (Ser)) (Figures 5a–d). Furthermore, heterozygosity of Dl and Ser suppresses the non-autonomous accumulation of Diap1 in cos2 mosaics (Supplementary Figure S6). Therefore, the suppression of GMR-hid and non-autonomous accumulation of Diap1 in cos2 mosaics depends on N activity. Second, protein levels of Dl are increased in cos2 mutant clones (see Supplementary Figure S7a). The accumulation of Dl is dependent on Ci activity, because overexpression of CiR in cos2 clones suppresses the upregulation of Dl (Supplementary Figure S7b). Surprisingly, protein levels of N itself also accumulate in cos2 clones in and anterior to the MF (Figures 5e and e′). It is unknown how N levels accumulate in cells with deregulated Hh activity, but it suggests that Hh not only controls Dl but also N.
Figure 5.
Notch is required for cos2 suppression of GMR-hid and promotes Diap-1 accumulation non-cell autonomously. (a–d) Heterozygosity of N (c) or Dl Ser (d) reverts the suppression of GMR-hid in cos2 mosaics (b), suggesting that Notch signaling is required for suppression of GMR-hid in cos2 mosaics. (e and e′) N protein strongly accumulates in cos2 clones (arrows) in or anterior to the MF (arrowhead). The N antibody (clone No. C17-9C6) was raised against the intracellular domain of N (NICD). (f and f′) cos2 induces N activity autonomously (red star) and non-cell autonomously (yellow arrows) anterior to the MF (arrowhead) as shown by an E(spl)m8-2.61-lacZ reporter expressing ß-GAL. (g, g′ and g′′) Ectopic expression of the active form of N, Nintra, induces Diap-1 levels non-cell autonomously immediately adjacent to Nintra-expressing clones (arrows), located in or anterior to the MF (arrowhead). Nintra-expressing clones are marked by the absence of the CD2 marker. (h) Statistical analysis of the suppression of GMR-hid by pka-C1 mosaics and pka-C1 Su(H) double mosaics. Eyes from GMR-hid flies are normalized to 100%. The suppression is partially reversed in double mosaics. For each bar, 10 eyes were averaged. **P-value ≤0.01. Genotypes: (a) y w GMR-hid ey-FLP/+; FRT42D P[w+]/CyO. (b) y w GMR-hid ey-FLP/+; FRT42D cos2H29/FRT42D P[w+]. (c) y w GMR-hid ey-FLP/N8; FRT42D cos2H29/FRT42D P[w+]. (d) y w GMR-hid ey-FLP/+; FRT42D cos2H29/FRT42D P[w+]; DlRevF10 SerRX82/+. (e and e′) y w ey-FLP; FRT42D cos2H29/FRT42D P[ubi-GFP]. (f and f′) y w ey-FLP; FRT42D cos2H29P[E(spl)m8-2.61-lacZ]/FRT42D P[ubi-GFP] P[E(spl)m8-2.61-lacZ]. (g, g′ and g′′) y w hs-FLP/+; P[Act5c>CD2>GAL4]; UAS-Nintra/+. (h) GMR-hid ey-FLP/+; P[ubi-GFP] FRT40/FRT40; GMR-hid ey-FLP/+; P[ubi-GFP] FRT40/pka-C1B3 FRT40; GMR-hid ey-FLP/+; P[ubi-GFP] FRT40/pka-C1B3 Su(H)Δ47 FRT40
Third, cos2 clones induce N activity autonomously and non-cell autonomously. Eye discs with cos2 clones in or anterior to the MF show an autonomous increase of the N activity marker E(spl)m8-2.61-lacZ (Figures 5f and f′, red star). Importantly, there is also a strong non-cell autonomous increase of E(spl)m8-2.61-lacZ expression in non-mutant cells adjacent to anteriorly localized clones (Figures 5f and f′, yellow arrows), indicating increased non-cell autonomous N activity. We characterized the autonomous and non-cell autonomous N activity further. Both components of N activity are dependent on Ci signaling, because overexpression of CiR in cos2 clones blocks E(spl)m8-2.61-lacZ expression anterior to the MF (Supplementary Figure S7c). As expected, the non-cell autonomous component of N activity is dependent on Dl (Supplementary Figure S7d and e), consistent with the observations that removing Dl reverts the suppression of GMR-hid (Figure 5d) and suppresses the non-autonomous accumulation of Diap1 in cos2 mosaics (Supplementary Figure S6). Fourth, clones of a different mutant, vps25, which is characterized by strong N activity, cause non-autonomous accumulation of Diap-1 protein.26
Fifth, clones of cells expressing the active form of N (Nintra) cause non-cell autonomous accumulation of Diap-1 in neighboring cells (Figures 5g, g′ and g′′). This observation demonstrates that N does not directly induce diap1 transcription, but generates another signal that promotes diap1 expression in neighboring cells (see Discussion section). Therefore, N relays the signal from cells with deregulated Hh signaling for Diap-1 induction in neighboring cells.
Because of the induction of both autonomous and non-cell autonomous N signaling in clones with deregulated Hh signaling, we also examined whether both the components of N signaling contribute to the suppression of GMR-hid by deregulated Hh signaling. For that purpose, we blocked autonomous N activity in pka-C1 mutant clones by removing Su(H) function, which is required for transcriptional activity of N.47 Interestingly, the suppression of GMR-hid by pka-C1 mosaics is partially reverted by loss of Su(H) function and thus loss of autonomous N signaling (Figure 5h). These observations suggest that both autonomous and non-cell autonomous N activation contribute to the suppression of GMR-hid.
Discussion
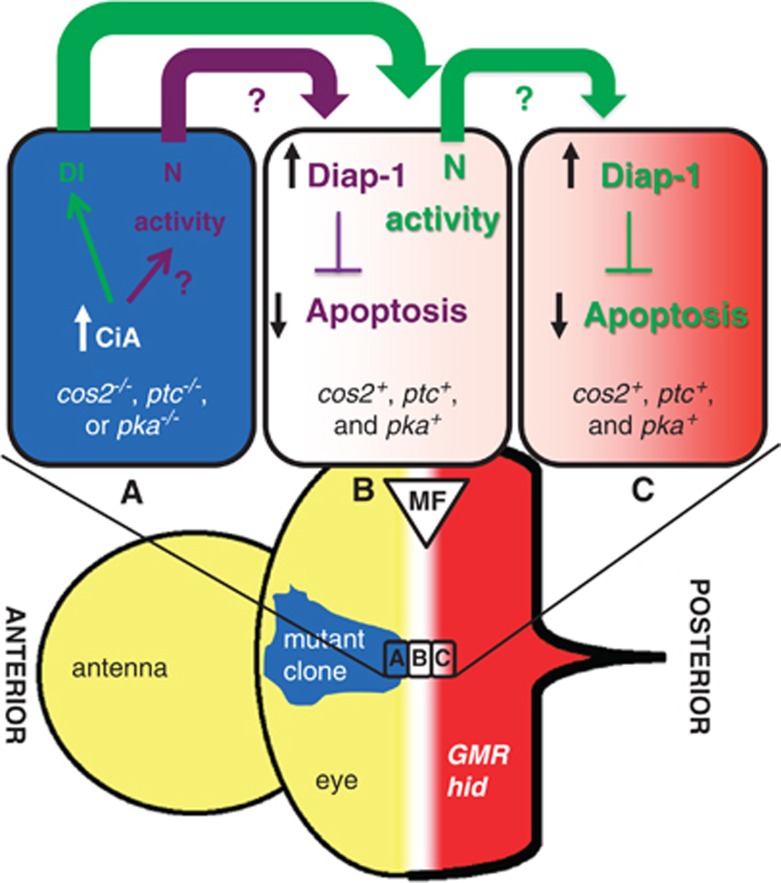
We showed that deregulated Hh signaling in the eye disc through loss of the negative regulators cos2, ptc or pka triggers both autonomous and non-cell autonomous N activation in a CiA-dependent manner (see model in Figure 6). Although it is unknown how deregulated Hh signaling promotes autonomous N activity, the non-cell autonomous component of N signaling is mediated by Hh-dependent Dl expression (Supplementary Figure S7a).11, 14 Once N has been activated, it promotes non-cell autonomous expression of diap1 in neighboring cells (Figure 5g). Therefore, at a clonal boundary of deregulated Hh signaling, there are two parallel events leading to N-dependent induction of diap1 transcription in neighboring wild-type cells (Figure 6). In the first event, autonomous N signaling in cos2 mutant cells directly generates an extracellular signal that induces diap1 transcription in neighboring non-mutant cells (illustrated in purple in Figure 6). The molecular identity of this signal is unknown. In the second event, Dl triggers the non-cell autonomous component of N activation in neighboring non-mutant cells (illustrated in green in Figure 6). Non-cell autonomous N likely produces the same extracellular signal as autonomous N and triggers upregulation of diap1 one cell further, thus acting as a relay of the Hh-derived signal for Diap-1 upregulation in neighboring cells. In this manner, upregulation of Diap-1 occurs in a stripe of at least two cells wide surrounding clones of deregulated Hh signaling. Depending on the range of the signal released by N, Diap-1 upregulation may extend even further. In addition, we observe non-cell autonomous N activity further away from the clone than one cell diameter (Figure 5f). This expansion may be due to cytoplasmic extensions such as cytonemes or additional relay mechanisms, and may increase Diap-1 levels further away from the clone. Therefore, when the MF in GMR-hid discs approaches a clone of deregulated Hh activity, it encounters higher levels of Diap-1, which protect neighboring cells from hid-induced apoptosis and causes the suppression of the GMR-hid eye phenotype.
Figure 6.
Model of non-cell autonomous induction of Diap-1 by cells with ligand-independent deregulated Hh activity. Drawn are three cells at a clonal boundary in the MF, one mutant cell (cell A) with deregulated Hh activity (blue) and two non-mutant cells (cells B and C; white to light red). The mutant cell accumulates CiA, which promotes autonomous N activity (purple arrow) and stimulates Dl expression (green arrow). Autonomous N signaling releases an unknown extracellular factor that promotes transcription of diap1 in the first signal-receiving non-mutant cell, cell B. In the same non-mutant cell, Dl induces non-cell autonomous N activity, which in turn promotes transcription of diap1 in the second non-mutant cell, cell C. In this manner, cells with deregulated Hh signaling transmit increased apoptosis resistance to neighboring cells by upregulation of Diap-1. When the MF moves into clones of deregulated Hh activity, GMR-driven hid expression (red) will be inhibited by increased Diap-1 levels, resulting in suppression of the GMR-hid eye phenotype. The white-to-red color gradient in cells B and C indicates the approaching hid-expressing wave driven by GMR
The identity of the extracellular signal generated by N that triggers expression of diap1 in neighboring wild-type cells is unknown. We tested the Hippo/Warts/Yorkie growth control pathway, which is known to regulate diap1 transcription. However, our data did not support an involvement of this pathway for non-autonomous induction of diap1 in mosaic cos2 and ptc eye imaginal discs (Supplementary Figure S5).
Interestingly, in eye imaginal discs, this non-autonomous activity is position-dependent and occurs only anterior and immediately posterior to the MF. We also observed a position-dependence in the wing imaginal disc where the non-cell autonomous upregulation is restricted to the wing pouch (Figure 3d). The reason for the position-dependence is unknown, but it shows that not every tissue with deregulated Hh signaling responds to it. Some tissues appear to be inert to it. This position-dependent effect of deregulated Hh signaling has also been observed in human cancer55 and may explain why oncogenic Hh signaling causes only certain types of cancers but not others. It is also noteworthy that even when the tissue responds to deregulated Hh signaling, the outcome can be different, too. For example, mosaic wing discs, which contain clones of activated N or are doubly mutant for ptc and ark (Apaf-1-related killer), stimulate Hippo/Warts/Yorkie signaling.35, 56 These responses appear to be a wing-specific effect, as the eye does not respond in this way (Supplementary Figure S5).
It is estimated that deregulated Hh signaling is associated with up to 25% of human tumors.48 Ligand-independent Hh signaling is associated with several tumors in humans such as basal cell carcinoma, medulloblastoma, rhabdomyosarcoma and glioma.25 In most cases, genetic inactivation of Ptch1 or activating missense mutations of Smo are the underlying cause of these tumors. Gene amplification of Gli1 and Gli2 also promotes ligand-independent Hh signaling and causes glioma and medulloblastoma.25 Recent work in a mouse model for breast cancer that induced ligand-independent Hh signaling by expression of a constitutively active smo allele demonstrated a non-cell autonomous effect on proliferation that may support tumor growth.49 Furthermore, in samples from human breast cancer patients, cells with deregulated Hh activity did not label for the proliferation marker Ki67, implying a non-cell autonomous effect of deregulated Hh signaling.50 Although these studies focused on proliferation, we demonstrate here that deregulated Hh signaling also increases resistance to apoptosis non-cell autonomously.
The control of N by Hh signaling is conserved in mammals and occurs during normal development and in tumors, including medulloblastomas and breast cancer.49, 51, 52 IAPs are upregulated in many human cancers and contribute to increased tumor cell survival.53, 54 The non-cell autonomous control of IAP levels by deregulated Hh and N signaling reveals a novel mechanism by which tumor cells and the tumor micro-environment increase cell survival.
In summary, we demonstrated that aberrant Hh signaling not only affects proliferation but also cell survival of neighboring cells. This non-cell autonomous control of apoptosis highlights an underappreciated function of Hh signaling. Potentially, the combined effects of non-cell autonomous proliferation and apoptosis resistance on the tumor micro-environment may be needed for efficient tumorigenesis.
Materials and Methods
Mutagenesis and fly stocks
The GheF screens are described elsewhere.31, 32 Two complementation groups were isolated in the GheF mutagenesis screen for 2R (right arm of chromosome 2). The first group of three alleles is mutant for cos2, because they fail to complement two known alleles of cos2, cos25 and cos2k16101. Two new alleles, cos2H29 and cos2P50, were chosen for sequencing. Both carry premature termination codons: cos2H29 at position 15 (Arg15) and cos2P50 at position 580 (Gln580). Because cos2H29 has an early premature termination codon and is likely a null allele, most of the data presented in this paper were obtained for cos2H29. The second group was identified as ptc because they fail to complement the ptcS2 allele, and point mutations were identified for two alleles. ptcC carries a premature termination codon at position 361 (Gln361) and ptcL49 contains a missense mutation changing Trp775 to Arg. Mutants of pka-C1, located on 2L, were tested separately and also found to suppress GMR-hid in mosaics.
The following mutants and transgenic lines were used: cos2H29, cos2P50, cosL51; ptcC, ptcL49 (this study); cos25, cos2k16101 (Bloomington); ptcS2 (kind gift of Phil Ingham); pka-C1K2, pka-C1B3, smoD16, Su(H)Δ47, UAS-CiA (kind gift of Dan Kalderon); UAS-CiR≡UAS-ciCE (kind gift of Konrad Basler); cul-1EX, nedd8AN015, slmb1 (kind gift of Cheng-Ting Chien); diap1-lacZ is thj5c8 (Bloomington); N8, DlRevF10 SerRX82, UAS-Nintra (kind gift of Hugo Bellen); Uba1H42; tubulin-1>y+, GFP>GAL4 (kind gift of Hyung Don Ryoo); GMR-hid; GMR-hid ey-FLP; GMR-hid[w-] E(spl)m8-2.61-lacZ and ex-lacZ (ex697) (kind gift of Georg Halder); UAS-Dl RNAi37287 (obtained from the Vienna DRC). Other stocks were obtained from the Bloomington stock center. E(spl)m8-2.61-lacZ is inserted on 2R. For analysis in cos2 and pka-C1 background, E(spl)m8-2.61-lacZ was recombined onto the cos2, the homologous FRT42D P[ubi-GFP] and the P[ubi-GFP] FRT40 chromosomes.
Mosaic analysis
Mosaics were induced using several techniques. Generally, we used the FLP/FRT system,57 with ey-FLP58 as the enzymatic source and marking the non-mutant tissue using either P[ubi-GFP] to express GFP in the larval tissue or P[w+] to generate red eye pigment in adults in a w- background. The autonomy of GMR-hid suppression was examined with GMR-hid[w-], an insertion on the third chromosome.26 hs-FLP was used to induce mosaic wing discs. Mosaics were also generated using the MARCM (mosaic analysis with a repressible cell marker) technique, which allows expression of transgenes such as UAS-CiR in mutant clones.59
hs-FLP was also used as the source of enzymatic activity to induce recombination using the FLP-out technique60 using FLP-out cassettes that contain 2 FRT (>) sites flanking the marker genes; FLP-induced excision of the marker gene allows the promoter (tub or Act5C) to drive expression of GAL4. The following FLP-out cassettes were used: hs-FLP; P[tubP>GFP>GAL4] (Figures 3c and d) and hs-FLP/+ P[Act5c>CD2>GAL4] (Figure 4g). First instar larvae were heat-shocked for 1 h at 37 °C to induce clones.
Immunohistochemistry
Imaginal discs were dissected from third instar larvae and stained using the standard protocols. Antibodies to the following primary antigens were used: cleaved caspase 3 (CAS3* Cell Signaling Technology, Danvers, MA, USA);30 Diap-1 (gifts from Pascal Meier and Hyung Don Ryoo); ß-GAL (Promega, Madison, WI, USA); CD2 (Santa Cruz Biotechnology, Santa Cruz, CA, USA); Notch (clones C458.2H and C17.9C6; DSHB, Iowa City, IA, USA) and Delta (C594.9B; DSHB); Ptc (clone Apa 1; DSHB). Cy3-conjugated anti-guinea pig, anti-rat and anti-rabbit (Jackson ImmunoResearch, West Grove, PA, USA) and AlexaFluor 546-conjugated anti-mouse and anti-rabbit (Invitrogen, Grand Island, NY, USA) were used as secondary antibodies.
Imaging and analysis
Bright field imaging was performed on a Zeiss AxioImager using ApoTome technology and CZ Projection software (Zeiss, Jena, Germany). Statistical analysis was performed on bright field images using Adobe Photoshop CS4 (San Jose, CA, USA) software to quantify the size of the eye using at least 10 images per genotype; GraphPad Prism 5 (La Jolla, CA, USA) was used to generate graphs. Images of eye imaginal discs were taken using either an Olympus Fluoview 500 or Fluoview 1000 Laser Confocal Microscope (Center Valley, PA, USA) and digital images were processed using the associated software.
Acknowledgments
We are grateful to our colleagues who have shared their knowledge and resources, especially Konrad Basler, Hugo Bellen, Cheng-Ting Chien, Steve Cohen, Phil Ingham, Georg Halder, Dan Kalderon, Graeme Mardon, Pascal Meier and Hyung Don Ryoo, the Bloomington Stock Center in Indiana and the Developmental Studies Hybridoma Bank in Iowa. Heather Scherr performed the GheF screen for 2R. Aimée Anderson and J Henri Bayle improved the quality of the manuscript. Funding: This research was supported in part by the Cancer Center Support Grant CA No. 16672 to the DNA Analysis Facility. AB is grateful for the support by the NIH (GM068016).
Glossary
- 2R
right arm of chromosome 2
- CAS3*
cleaved Caspase-3
- Ci
Cubitus interruptus
- CiA
Cubitus interruptus, activator
- CiR
Cubitus interruptus, repressor
- Cos2
Costal-2
- Cul-1
Cullin-1
- Diap-1
Drosophila inhibitor of apoptosis protein-1
- Dl
Delta
- Dpp
Decapentaplegic
- DrICE
Drosophila interleukin-1β-converting enzyme
- Dronc
Drosophila Nedd2-like caspase
- E(spl)
enhancer of split
- ex
expanded
- ey
eyeless
- FLP
flippase
- FRT
flippase recombination target
- GFP
green fluorescent protein
- GheF
GMR-hid ey-FLP
- GMR
glass multimer reporter
- Hh
Hedgehog
- hid
head involution defective
- hs
heat shock
- IAP
inhibitor of apoptosis protein
- MF
morphogenetic furrow
- N
Notch
- PKA
protein kinase A
- PKA-C1
protein kinase A, catalytic subunit 1
- Ptc
Patched
- Ptch-1
Patched-1
- Ser
Serrate
- Slmb
Slimb
- Smo
Smoothened
- Su(H)
suppressor of hairless
- ubi-GFP
ubiquitously expressed GFP
- Uba1
ubiquitin-activating enzyme
- UAS
upstream activation sequence
- w
white
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by JA Cidlowski
Supplementary Material
References
- Xu D, Woodfield SE, Lee TV, Fan Y, Antonio C, Bergmann A. Genetic control of programmed cell death (apoptosis) in Drosophila. Fly (Austin Tex) 2009;3:78–90. doi: 10.4161/fly.3.1.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs Y, Steller H. Programmed cell death in animal development and disease. Cell. 2011;147:742–758. doi: 10.1016/j.cell.2011.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. Caspase function in programmed cell death. Cell Death Differ. 2007;14:32–43. doi: 10.1038/sj.cdd.4402060. [DOI] [PubMed] [Google Scholar]
- Bergmann A. The role of ubiquitylation for the control of cell death in Drosophila. Cell Death Differ. 2010;17:61–67. doi: 10.1038/cdd.2009.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann A, Steller H. Apoptosis, stem cells, and tissue regeneration. Sci Signal. 2010;3:re8. doi: 10.1126/scisignal.3145re8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Bergmann A. Apoptosis-induced compensatory proliferation. The Cell is dead. Long live the Cell! Trends Cell Biol. 2008;18:467–473. doi: 10.1016/j.tcb.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NE. Patterning signals and proliferation in Drosophila imaginal discs. Curr Opin Genet Dev. 2007;17:287–293. doi: 10.1016/j.gde.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Roignant JY, Treisman JE. Pattern formation in the Drosophila eye disc. Int J Dev Biol. 2009;53:795–804. doi: 10.1387/ijdb.072483jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Cagan RL. Patterning the fly eye: the role of apoptosis. Trends Genet. 2003;19:91–96. doi: 10.1016/S0168-9525(02)00041-0. [DOI] [PubMed] [Google Scholar]
- Horsfield J, Penton A, Secombe J, Hoffman FM, Richardson H. Decapentaplegic is required for arrest in G1 phase during Drosophila eye development. Development. 1998;125:5069–5078. doi: 10.1242/dev.125.24.5069. [DOI] [PubMed] [Google Scholar]
- Firth LC, Baker NE. Extracellular signals responsible for spatially regulated proliferation in the differentiating Drosophila eye. Dev Cell. 2005;8:541–551. doi: 10.1016/j.devcel.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Dominguez M, Hafen E. Hedgehog directly controls initiation and propagation of retinal differentiation in the Drosophila eye. Genes Dev. 1997;11:3254–3264. doi: 10.1101/gad.11.23.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberlein U, Wolff T, Rubin GM. The TGF beta homolog dpp and the segment polarity gene hedgehog are required for propagation of a morphogenetic wave in the Drosophila retina. Cell. 1993;75:913–926. doi: 10.1016/0092-8674(93)90535-x. [DOI] [PubMed] [Google Scholar]
- Baonza A, Freeman M. Control of cell proliferation in the Drosophila eye by Notch signaling. Dev Cell. 2005;8:529–539. doi: 10.1016/j.devcel.2005.01.019. [DOI] [PubMed] [Google Scholar]
- Huangfu D, Anderson KV. Signaling from Smo to Ci/Gli: conservation and divergence of Hedgehog pathways from Drosophila to vertebrates. Development. 2006;133:3–14. doi: 10.1242/dev.02169. [DOI] [PubMed] [Google Scholar]
- Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15:801–812. doi: 10.1016/j.devcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW, Nystedt S, Nakano Y, Brown W, Stark D, van den Heuvel M, et al. Patched represses the Hedgehog signalling pathway by promoting modification of the Smoothened protein. Curr Biol. 2000;10:1315–1318. doi: 10.1016/s0960-9822(00)00755-7. [DOI] [PubMed] [Google Scholar]
- Denef N, Neubuser D, Perez L, Cohen SM. Hedgehog induces opposite changes in turnover and subcellular localization of Patched and Smoothened. Cell. 2000;102:521–531. doi: 10.1016/s0092-8674(00)00056-8. [DOI] [PubMed] [Google Scholar]
- Robbins DJ, Nybakken KE, Kobayashi R, Sisson JC, Bishop JM, Therond PP. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell. 1997;90:225–234. doi: 10.1016/s0092-8674(00)80331-1. [DOI] [PubMed] [Google Scholar]
- Cheung HO, Zhang X, Ribeiro A, Mo R, Makino S, Puviindran V, et al. The kinesin protein Kif7 is a critical regulator of Gli transcription factors in mammalian hedgehog signaling. Sci Signal. 2009;2:ra29. doi: 10.1126/scisignal.2000405. [DOI] [PubMed] [Google Scholar]
- Endoh-Yamagami S, Evangelista M, Wilson D, Wen X, Theunissen JW, Phamluong K, et al. The mammalian Cos2 homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr Biol. 2009;19:1320–1326. doi: 10.1016/j.cub.2009.06.046. [DOI] [PubMed] [Google Scholar]
- Farzan SF, Ascano M, Ogden SK, Sanial M, Brigui A, Plessis A, et al. Costal2 functions as a kinesin-like protein in the hedgehog signal transduction pathway. Curr Biol. 2008;18:1215–1220. doi: 10.1016/j.cub.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisson JC, Ho KS, Suyama K, Scott MP. Costal2, a novel kinesin-related protein in the Hedgehog signaling pathway. Cell. 1997;90:235–245. doi: 10.1016/s0092-8674(00)80332-3. [DOI] [PubMed] [Google Scholar]
- Marks SA, Kalderon D. Regulation of mammalian Gli proteins by Costal 2 and PKA in Drosophila reveals Hedgehog pathway conservation. Development. 2011;138:2533–2542. doi: 10.1242/dev.063479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teglund S, Toftgard R. Hedgehog beyond medulloblastoma and basal cell carcinoma. Biochim Biophys Acta. 2010;1805:181–208. doi: 10.1016/j.bbcan.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Herz HM, Chen Z, Scherr H, Lackey M, Bolduc C, Bergmann A. vps25 mosaics display non-autonomous cell survival and overgrowth, and autonomous apoptosis. Development. 2006;133:1871–1880. doi: 10.1242/dev.02356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TV, Ding T, Chen Z, Rajendran V, Scherr H, Lackey M, et al. The E1 ubiquitin-activating enzyme Uba1 in Drosophila controls apoptosis autonomously and tissue growth non-autonomously. Development. 2008;135:43–52. doi: 10.1242/dev.011288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chen Z, Bergmann A. Regulation of EGFR and Notch signaling by distinct isoforms of D-cbl during Drosophila development. Dev Biol. 2010;342:1–10. doi: 10.1016/j.ydbio.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Werz C, Xu D, Chen Z, Li Y, Hafen E, et al. Drosophila cbl is essential for control of cell death and cell differentiation during eye development. PloS ONE. 2008;3:e1447. doi: 10.1371/journal.pone.0001447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Bergmann A. The cleaved-Caspase-3 antibody is a marker of Caspase-9-like DRONC activity in Drosophila. Cell Death Differ. 2010;17:534–539. doi: 10.1038/cdd.2009.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Li Y, Arcaro M, Lackey M, Bergmann A. The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila. Development. 2005;132:2125–2134. doi: 10.1242/dev.01790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Scherr H, Lackey M, Xu D, Chen Z, Lu J, et al. ARK, the Apaf-1 related killer in Drosophila, requires diverse domains for its apoptotic activity. Cell Death Differ. 2007;14 (1:92–102. doi: 10.1038/sj.cdd.4401931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman-Scheel M, Weng L, Xin S, Du W. Hedgehog regulates cell growth and proliferation by inducing Cyclin D and Cyclin E. Nature. 2002;417:299–304. doi: 10.1038/417299a. [DOI] [PubMed] [Google Scholar]
- Christiansen AE, Ding T, Bergmann A. Ligand-independent activation of the Hedgehog pathway displays non-cell autonomous proliferation during eye development in Drosophila. Mech Dev. 2012;129:98–108. doi: 10.1016/j.mod.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey JD, Brown JA, Moberg KH.Regulation of Yorkie activity in Drosophila imaginal discs by the Hedgehog receptor gene patched Mech Dev 2012. e-pub ahead of print 15 June 2012; doi: 10.1016/j.mod.2012.05.007 [DOI] [PMC free article] [PubMed]
- Ou CY, Lin YF, Chen YJ, Chien CT. Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev. 2002;16:2403–2414. doi: 10.1101/gad.1011402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay BA, Wassarman DA, Rubin GM. Drosophila homologs of baculovirus inhibitor of apoptosis proteins function to block cell death. Cell. 1995;83:1253–1262. doi: 10.1016/0092-8674(95)90150-7. [DOI] [PubMed] [Google Scholar]
- Wang SL, Hawkins CJ, Yoo SJ, Muller HA, Hay BA. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- Lisi S, Mazzon I, White K. Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics. 2000;154:669–678. doi: 10.1093/genetics/154.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier P, Silke J, Leevers SJ, Evan GI. The Drosophila caspase DRONC is regulated by DIAP1. EMBO J. 2000;19:598–611. doi: 10.1093/emboj/19.4.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal L, McCall K, Agapite J, Hartwieg E, Steller H. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 2000;19:589–597. doi: 10.1093/emboj/19.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aza-Blanc P, Ramirez-Weber FA, Laget MP, Schwartz C, Kornberg TB. Proteolysis that is inhibited by hedgehog targets Cubitus interruptus protein to the nucleus and converts it to a repressor. Cell. 1997;89:1043–1053. doi: 10.1016/s0092-8674(00)80292-5. [DOI] [PubMed] [Google Scholar]
- Smelkinson MG, Kalderon D. Processing of the Drosophila hedgehog signaling effector Ci-155 to the repressor Ci-75 is mediated by direct binding to the SCF component Slimb. Curr Biol. 2006;16:110–116. doi: 10.1016/j.cub.2005.12.012. [DOI] [PubMed] [Google Scholar]
- Ou CY, Wang CH, Jiang J, Chien CT. Suppression of Hedgehog signaling by Cul3 ligases in proliferation control of retinal precursors. Dev Biol. 2007;308:106–119. doi: 10.1016/j.ydbio.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Hepker J, Wang QT, Motzny CK, Holmgren R, Orenic TV. Drosophila cubitus interruptus forms a negative feedback loop with patched and regulates expression of Hedgehog target genes. Development. 1997;124:549–558. doi: 10.1242/dev.124.2.549. [DOI] [PubMed] [Google Scholar]
- Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey AM, Posakony JW. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Therond P. Hedgehog signaling: from the Drosophila cuticle to anti-cancer drugs. Dev Cell. 2005;8:143–151. doi: 10.1016/j.devcel.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Visbal AP, LaMarca HL, Villanueva H, Toneff MJ, Li Y, Rosen JM, et al. Altered differentiation and paracrine stimulation of mammary epithelial cell proliferation by conditionally activated Smoothened. Dev Biol. 2011;352:116–127. doi: 10.1016/j.ydbio.2011.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes RC, Zhang X, Harrington N, Fung JY, Wu MF, Hilsenbeck SG, et al. Constitutive activation of Smoothened (SMO) in mammary glands of transgenic mice leads to increased proliferation, altered differentiation and ductal dysplasia. Development. 2007;134:1231–1242. doi: 10.1242/dev.02797. [DOI] [PubMed] [Google Scholar]
- Hallahan AR, Pritchard JI, Hansen S, Benson M, Stoeck J, Hatton BA, et al. The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 2004;64:7794–7800. doi: 10.1158/0008-5472.CAN-04-1813. [DOI] [PubMed] [Google Scholar]
- Sengupta A, Banerjee D, Chandra S, Banerji SK, Ghosh R, Roy R, et al. Deregulation and cross talk among Sonic hedgehog, Wnt, Hox and Notch signaling in chronic myeloid leukemia progression. Leukemia. 2007;21:949–955. doi: 10.1038/sj.leu.2404657. [DOI] [PubMed] [Google Scholar]
- LaCasse EC, Mahoney DJ, Cheung HH, Plenchette S, Baird S, Korneluk RG. IAP-targeted therapies for cancer. Oncogene. 2008;27:6252–6275. doi: 10.1038/onc.2008.302. [DOI] [PubMed] [Google Scholar]
- Gyrd-Hansen M, Meier P. IAPs: from caspase inhibitors to modulators of NF-kappaB, inflammation and cancer. Nat Rev Cancer. 2010;10:561–574. doi: 10.1038/nrc2889. [DOI] [PubMed] [Google Scholar]
- Stecca B, Ruiz IAA. Context-dependent regulation of the GLI code in cancer by HEDGEHOG and non-HEDGEHOG signals. J Mol Cell Biol. 2010;2:84–95. doi: 10.1093/jmcb/mjp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves HK, Woodfield SE, Yang CC, Halder G, Bergmann A. Notch signaling activates Yorkie non-cell autonomously in Drosophila. PloS One. 2012;7:e37615. doi: 10.1371/journal.pone.0037615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Rubin GM. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 1993;117:1223–1237. doi: 10.1242/dev.117.4.1223. [DOI] [PubMed] [Google Scholar]
- Newsome TP, Asling B, Dickson BJ. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development. 2000;127:851–860. doi: 10.1242/dev.127.4.851. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Struhl G, Basler K. Organizing activity of wingless protein in Drosophila. Cell. 1993;72:527–540. doi: 10.1016/0092-8674(93)90072-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.