Abstract
mnd2 mice die prematurely as a result of neurodegeneration 30–40 days after birth due to loss of the enzymatic activity of the mitochondrial quality control protease HtrA2/Omi. Here, we show that transgenic expression of human HtrA2/Omi in the central nervous system of mnd2 mice rescues them from neurodegeneration and prevents their premature death. Interestingly, adult transgenic mnd2 mice develop accelerated aging phenotypes, such as premature weight loss, hair loss, reduced fertility, curvature of the spine, heart enlargement, increased autophagy, and death by 12–17 months of age. These mice also have elevated levels of clonally expanded mitochondrial DNA (mtDNA) deletions in their tissues. Our results provide direct genetic evidence linking mitochondrial protein quality control to mtDNA deletions and aging in mammals.
Keywords: HtrA2, mitochondria, quality control, aging, neurodegeneration, mitochondrial DNA
Mitochondria are dynamic organelles primarily involved in the production of adenosine triphosphate (ATP) through oxidative phosphorylation. They also play important roles in diverse cellular processes such as cell death, autophagy and innate immunity.1 As a consequence of oxidative phosphorylation, mitochondria produce reactive oxygen species (ROS), which damages mitochondrial proteins, lipids and nucleic acids, because of their proximity to the source of ROS production. Accumulation over time of mutations and deletions in mitochondrial DNA (mtDNA) together with increased protein misfolding, as a result of ROS damage, leads to age-associated decline in mitochondrial function, which is believed to be responsible for organismal aging and age-associated diseases.2, 3, 4
To maintain optimal mitochondrial function over time, a number of quality control mechanisms exist that monitor and regulate all aspects of mitochondrial physiology. Damaged and unfolded mitochondrial proteins are removed by mitochondrial quality control proteases, which recognize these proteins and degrade them.5, 6 The ATP-dependent AAA (ATPase-Associated with diverse cellular Activities) proteases, are among the best-characterized proteases implicated in mitochondrial protein quality control.7 The AAA proteases, ClpXP and Lon, located in the matrix are involved in quality control of matrix proteins. Although no specific mitochondrial substrate has been identified for the ClpXP protease, in vitro studies showed that Lon protease preferentially targets oxidatively damaged matrix aconitase.8 Two additional AAA proteases, Paraplegin (encoded by the SPG7 gene) and YME1L, are associated with the inner membrane with their catalytic sites facing the matrix and intermembrane space, respectively.7 These proteases are believed to be primarily involved in the degradation of damaged and unfolded membrane proteins of the electron transport chain. In addition, Paraplegin has been shown to process the mitochondrial ribosomal protein MrpL32,9 suggesting that it might also function in mitochondrial ribosome assembly. Loss-of-function mutations of Paraplegin cause hereditary spastic paraplegia in humans.10
Whole damaged mitochondria are specifically removed by a process called mitophagy, which is a specialized form of autophagy that targets damaged mitochondria for lysosomal degradation.6, 11 A number of mitochondrial proteases play important roles in mitochondrial quality control by cleaving proteins that regulate mitophagy. Optic atrophy 1 (OPA1) is a dynamin-related GTPases that regulates mitochondrial morphology and is also an important regulator of mitophagy.12 The activity of OPA1 is regulated by proteolytic cleavage. Paraplegin, YME1L and OMA1 proteases have all been shown to cleave OPA1,13, 14, 15, 16, 17 thus implicating them in the regulation of mitophagy. The mitochondrial rhomboid protease presenilin-associated rhomboid-like (PARL) is also implicated in regulating mitophagy by cleaving PINK1,18, 19 a kinase required for mitophagy.20 PARL has also been shown to regulate apoptosis by affecting the release of short OPA1 isoforms from mitochondria during apoptosis.21, 22 Loss-of-function of PARL in mice leads to increased apoptosis and premature death.22
HtrA2 (also known as Omi) is an ATP-independent serine protease structurally and functionally related to the bacterial quality control proteases DegP and DegS.23 HtrA2 resides in the intermembrane space of the mitochondrion, and its loss leads to accumulation of unfolded proteins in the mitochondria, oxidative stress, and defective mitochondrial respiration,24 suggesting that, like its bacterial counterparts, HtrA2 functions as a quality control protease. Mice lacking HtrA2 protease activity as a result of missense mutation (mnd2 mice) or targeted deletion (HtrA2 knockout mice) of the HtrA2 (Prss25) gene exhibit early onset neurodegeneration and motor abnormalities similar to Parkinson's disease, and die between postnatal day (dP) 30 and dP40.25, 26 mnd2 and HtrA2 knockout mice also fail to gain weight and show a dramatic decrease in organ weight (e.g., spleen and thymus) beginning at dP20 compared with wild-type (WT) mice.25, 26 To gain more insight into the function of HtrA2, we generated mice that are deficient in HtrA2 activity in non-neuronal tissues. We show here that HtrA2 deficiency in non-neuronal tissues does not cause early lethality, but leads to increased accumulation of mtDNA deletions and premature aging. Our results show for the first time that defective protein quality control in the intermembrane space of the mitochondria can lead to increased mtDNA deletions and aging in mammals.
Results
Neurodegeneration and early death in mnd2 mice is rescued by a neuron-targeted human HTRA2 transgene
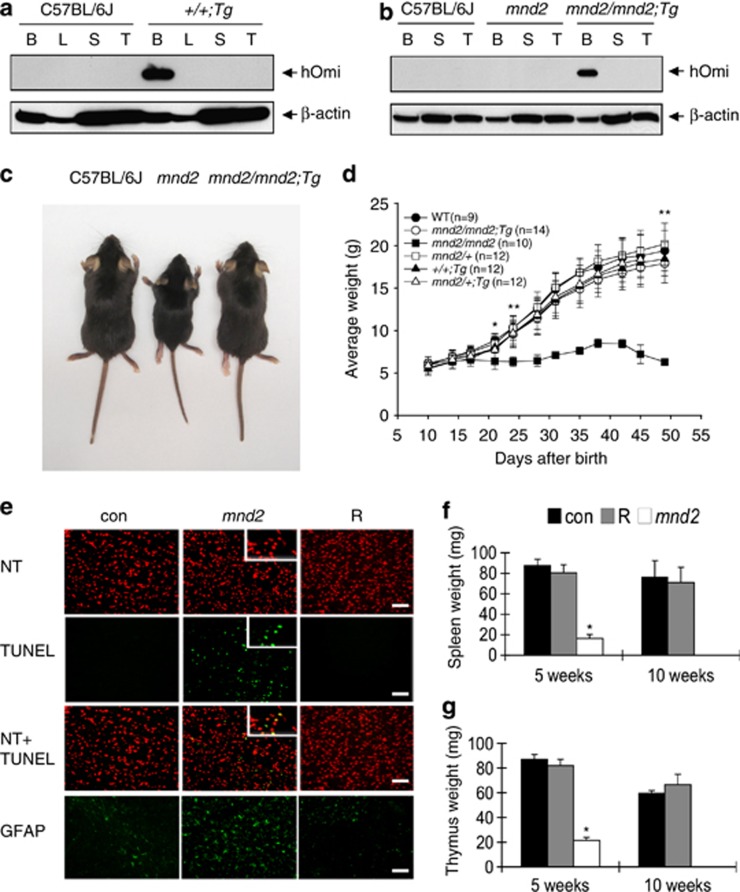
Previous studies in mnd2 and HtrA2 knockout mice provided evidence linking HtrA2 loss-of-function to early onset neurodegeneration in mice. However, it is not clear whether the loss of HtrA2 activity in neurons themselves, or other tissues, is the cause of neurodegeneration. Because of the early lethality of HtrA2 loss-of-function, it is also not fully clear what impact HtrA2 deficiency will have on non-neuronal tissues of adult mice. To address these important questions, we generated transgenic mice expressing a human WT HTRA2 (HTRA2) transgene (Tg) in the central nervous system (CNS) under the control of the neuron-specific enolase promoter. Four germline transmitting founder mice (+/+Tg) carrying the HTRA2 transgene were obtained. All four founder mice exhibited completely normal phenotype and behavior over their normal life span (>2-year period), indicating that transgenic expression of HTRA2 in neurons has no deleterious effects. We crossed the founder mice to heterozygous mnd2/+ mice to obtain F1 mice heterozygous for the mnd2 mutation and carrying the HTRA2 transgene (mnd2/+Tg). Next, the F1 mice were intercrossed to obtain mice homozygous for the mnd2 mutation and carry the HTRA2 transgene (mnd2/mnd2;Tg) hereafter denoted ‘rescued mnd2' mice. As expected, the product of the HTRA2 transgene was only expressed in the brain but was not detected in other tissues (e.g. liver, spleen, or thymus) of WT transgenic (+/+Tg) or rescued mnd2 mice (Figures 1a and b). Remarkably, although the rescued mnd2 mice carry the homozygous mnd2 mutation, the targeted expression of HTRA2 transgene in their neurons prevented the early lethality observed in the homozygous mnd2 mice. The external appearance, size and growth rate of these mice in the first 2 months after birth was comparable to that of their heterozygous mnd2/+Tg littermates or WT mice (Figures 1c and d). Analysis of cryostat sections of the Caudate-Putamen (CP) showed extensive cell death (as assayed by TUNEL) and astrocyte infiltration (increased glial fibrillary acidic protein-positive cells) in 4-week-old mnd2 but not in similar aged rescued mnd2 or WT mice (Figure 1e; Supplementary Figures 1a and b). These results demonstrate that targeted expression of HtrA2 in the CNS is sufficient to rescue the homozygous mnd2 mice from neurodegeneration, weight loss and early lethality. Surprisingly, targeted expression of HtrA2 in the CNS also rescued other associated abnormalities such as spleen and thymus atrophy (Figures 1f and g), which is caused by extensive apoptosis in these tissues (Supplementary Figures 1c and d). The ability of the neuron-targeted HTRA2 transgene to rescue all these phenotypic abnormalities suggests that the overt abnormalities observed in tissues of mnd2 mice (e.g., reduced body weight, muscle wasting, thymus, and spleen atrophy) are likely due to early onset neurodegeneration. Notably, the rescued mnd2 females were found to be infertile, whereas the males were fertile, indicating that HtrA2 plays an important role in the normal function or development of the female reproductive system.
Figure 1.
Targeted expression of HTRA2 in the CNS rescues mnd2 mice from neurodegeneration organ atrophy and early death. (a and b) Immunoblots showing the expression of the product of the HTRA2 transgene in the brains of transgenic WT (+/+;Tg) (a) and rescued mnd2 (mnd2/mnd2;Tg) (b) mice. B, brain; L, liver; S, spleen; T, thymus. (c) External appearance of 5-week-old rescued mnd2 (mnd2/mnd2;Tg, right), mnd2 (mnd2, middle) and C57BL/6J (left) mice. (d) Growth curves of WT (C57BL/6J), rescued mnd2 (mnd2/mnd2;Tg), homozygous mnd2 (mnd2/mnd2), heterozygous mnd2 (mnd2/+), HTRA2 transgenic WT (+/+Tg) and HTRA2 transgenic heterozygous mnd2 (mnd2/+Tg) mice over a 2-month period. Note the cessation of growth of the homozygous mnd2 mice at 15 days after birth. (e) Cryostat sections of the striatum (CP) of 4-week-old mnd2/+Tg control (con), mnd2/mnd2 (mnd2) and rescued mnd2 (R) mice stained successively with NeuroTrace (NT), TUNEL and glial fibrillary acidic protein (GFAP). Bar: 100 μm; insert: 60 μm. (f and g) Average weight of spleens (f) or thymi (g) from mnd2/+Tg control (con), rescued mnd2 (R) and mnd2/mnd2 (mnd2) mice at 5 and 10 weeks after birth. Homozygous mnd2/mnd2 mice did not survive beyond 6 weeks. Asterisk, P<0.01; double asterisk, P<0.001, Bonferroni test. All error bars indicate S.E.M.
Rescued mnd2 mice exhibit premature aging phenotype
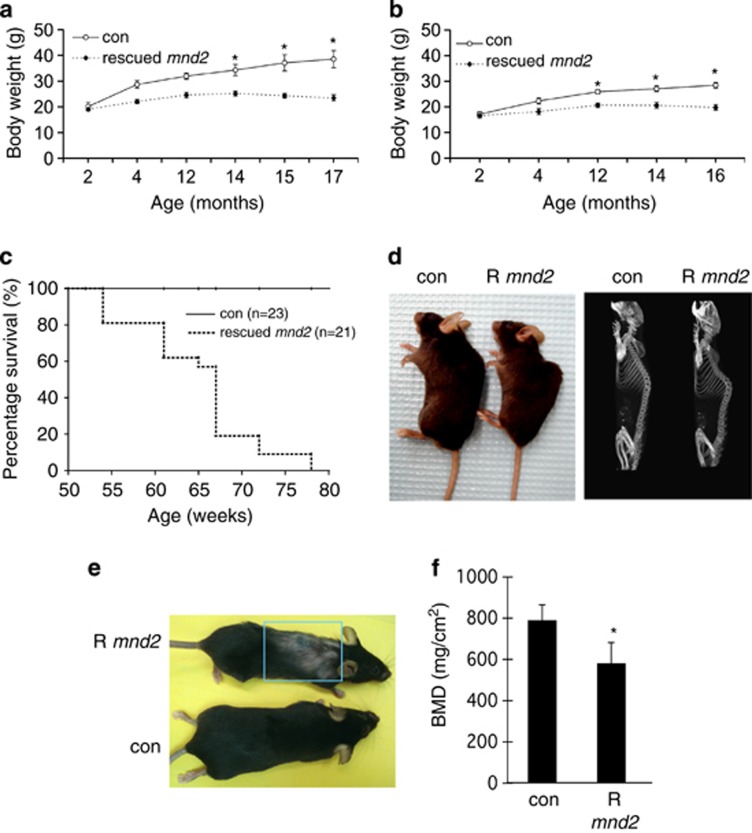
As the rescued mnd2 mice got older, they started showing a slower weight gain compared with control mice and began to lose weight at ∼14 months of age (Figures 2a and b). They had a mean lifespan of ∼67 weeks and the majority of them died before they reached 78 weeks of age (Figure 2c). The external appearance of rescued mnd2 mice was normal until the age of ∼21 weeks, when they started showing signs of slight curvature of the spine (lordokyphosis) (Figure 2d, left panel) and hair loss (alopecia) (Figure 2e), which became more pronounced as the mice got older. Consistent with the external appearance of these mice, radiographic imaging of bone tissues using microcomputer tomography showed clear lordokyphosis (Figure 2d, right panel). Quantitative analysis of bone mineral density (BMD) in dissected femurs of 15-month-old rescued mnd2 mice showed a clear reduction in BMD compared with control mice (Figure 2f). The reduced BMD is indicative of premature osteoporosis that is likely responsible for the notable lordokyphosis in these mice. We also observed a significant decrease in muscle fibers diameter in aged rescued mnd2 mice (12-month-old) compared with control mice indicating muscle atrophy (Supplementary Figure 2). However, there was no significant difference in muscle fibers diameter between young (3- and 5-month-old) rescued mnd2 mice and control mice (Supplementary Figure 2), indicating that the reduction in muscle fibers diameter is due to accelerated aging. Collectively, these abnormalities (weight loss, hair loss, osteoporosis, lordokyphosis, muscle atrophy, and reduced life span) are indicative of premature aging, and are common clinical features in human and rodent aging.2, 3, 27, 28, 29, 30, 31, 32
Figure 2.
Body weight, life span, and premature aging phenotypes of rescued mnd2 mice. (a) Body weight curves for rescued mnd2 male mice (n=5–7; dashed line) compared with control (con) mnd2/+Tg littermates (n=5–7; solid line). Asterisk, P<0.01. (b) Body weight curves for rescued mnd2 female mice (n=4–7; dashed line) compared with control (con) mnd2/+Tg littermates (n=4–7; solid line). Asterisk, P<0.01. (c) Survival plot for rescued mnd2 mice (dashed line) compared with control (con) mnd2/+Tg littermates (solid line) at different time points. (d–f) Curvature of the spine (d, left, external appearance; d, right, microcomputer tomography radiograph), hair loss (e), and BMD (f) in 15-month-old rescued mnd2 (R mnd2) mice compared with normal 15-month-old control (con) mnd2/+Tg littermates. P<0.05. All error bars indicate S.E.M.
Loss of HtrA2 causes cardiac aging, increased extramedullary hematopoiesis and splenomegaly
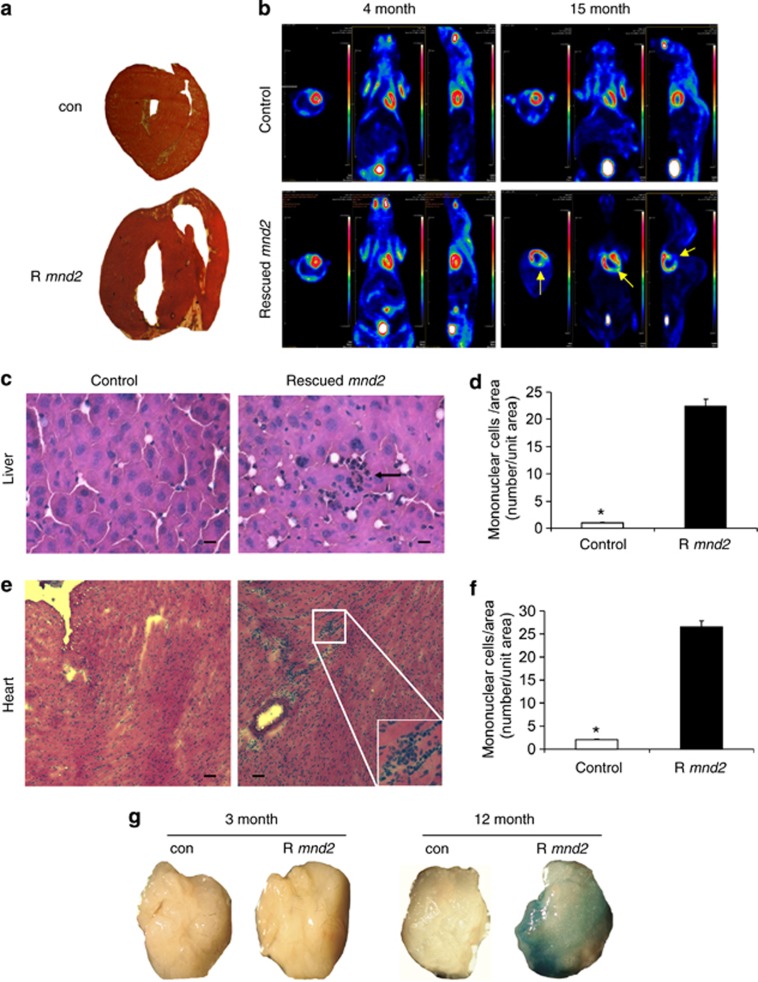
Gross anatomical examination of the heart from 15-month-old rescued mnd2 mice showed clear enlargement of the heart as compared with control animals (Figure 3a), with an increase in the left ventricular chamber size. Mosaic Positron Emission Tomography, which provides in vivo functional images of the myocardium using radio-labeled glucose as a metabolic probe, revealed a large decrease in glucose metabolism in large areas of the heart from 15-month-old rescued mnd2 mice (Figure 3b, right), but not in similar age control animals or young (4-month-old) rescued mnd2 mice (left). These metabolic abnormalities were most pronounced in the left ventricle of the heart, indicating severe cardiomyopathy. In addition, we found that aged rescued mnd2 mice have enlarged spleens with increased extramedullary hematopoiesis in the red pulp (Supplementary Figures 3a–c). In contrast, these mice had largely atrophied thymus (Supplementary Figure 3d). Foci of extramedullary hematopoiesis were also observed in the heart and liver of aged rescued mnd2 mice (Figures 3c–f). Cardiomyopathy, splenomegaly, thymus atrophy, and increased extramedullary hematopoiesis are commonly seen in aging humans and rodents.2, 32, 33
Figure 3.
Cardiomyopathy, increased extramedullary hematopoiesis, and senescence in aged rescued mnd2 mice. (a) Unstained thick sections of heart from 15-month-old rescued mnd2 mouse and control (con) mnd2/+Tg littermate. (b) In vivo mosaic positron emission tomography images of the myocardium using radio-labeled glucose as a metabolic probe of 4-month-old and 15-month-old rescued mnd2 (lower panels), and their control Tg;mnd2/+ littermates (upper panels), respectively. The metabolic activity in the myocardium is shown as intensely red areas. Arrows (yellow) show large regions of left ventricle in 15-month-old rescued mnd2 that are not visible because of lack of glucose metabolism. All panels, first images from left (top view); second images from left (front view); third images from left (left side view). (c and e) Cryostat sections of liver and heart from 12-month-old rescued mnd2 mice and control mnd2/+Tg littermates stained with HE. Extramedullary hematopoiesis is seen in liver (c, arrow) and heart (e, inset) of rescued mnd2 mice. Bar: 50 μm. (d and f) Graphs show an increase in the number of mononuclear cells in the liver and heart of 12-month-old rescued mnd2 mice (R mnd2) compared with control mnd2/+Tg littermates. Mononuclear cells were enumerated on at least five random sections of control and rescued mnd2 mice (n=6 mice). Asterisk, P<0.01. All error bars indicate s.e.m. (g) SA-β-Gal stained adipose tissues collected from 3- and 12-month-old control and rescued mnd2 mice (R mnd2) as indicated. Images are representative of three mice from each group
As cellular senescence has been implicated in generating age-related phenotypes,34 we also stained adipose tissues from control and rescued mnd2 mice with the senescence-associated-β-galactosidase (SA-β-Gal) biomarker. Adipose tissues from aged rescued mnd2 mice, but not from age-matched controls or young (3-month-old) rescued mnd2 mice, stained strongly for SA-β-Gal (Figure 3g). These results indicate that the age-related phenotypes in aged rescued mnd2 mice might be due to the induction of cellular senescence.
Loss of HtrA2 results in increased mtDNA deletions
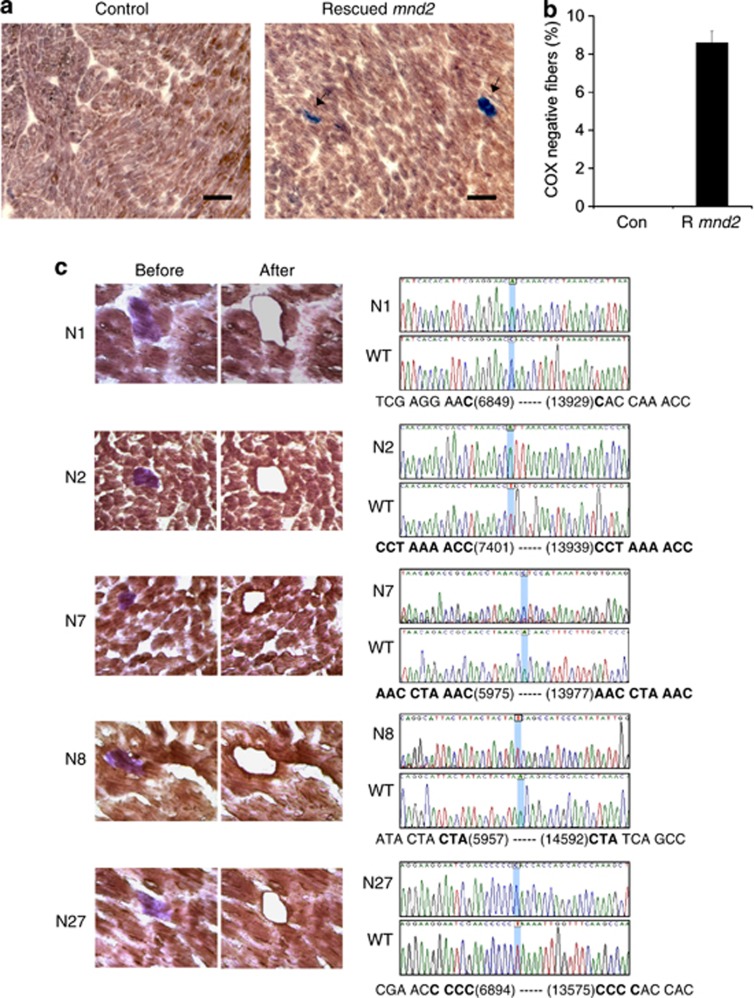
Cytochrome oxidase (COX) deficiency is a common finding in tissues from aging humans and rodents2, 35 and is attributed to clonally expanded mutations and deletions that affect mtDNA-encoded COX genes. Thus, we used histochemical staining to detect COX enzymatic activity in tissue sections from rescued mnd2 mice and their heterozygous mnd2/+Tg littermates. We found that heart, skeletal muscle, and duodenum tissues of 15-month-old rescued mnd2 mice contained many COX-negative cells (Figures 4a and b; Supplementary Figure 4). In contrast, we did not find COX-negative cells in the same tissues of mnd2/+Tg littermates (Figures 4a and b; Supplementary Figure 4), or in young (3- to 10-month-old) rescued mnd2 mice (not shown). COX-negative cells have also been described in brain tissues from premature aging mice.36 However, we did not detect COX-negative cells in brain tissue sections of rescued mnd2 mice or their heterozygous mnd2/+Tg littermates (Supplementary Figure 4), demonstrating that the lack of HtrA2 activity is responsible for the COX deficiency in non-neuronal tissues.
Figure 4.
COX deficiency and mtDNA deletions in the heart of rescued mnd2 mice. (a) Cryostat sections of heart from 15-month-old rescued mnd2 and control mnd2/+Tg mice double stained for COX and SDH activities. Cardiomyocytes that contain both COX and SDH activities are stained brown, while cardiomyocytes that lack COX activity (arrows) are stained purple-blue. Bar: 60 μm. (b) Graph shows an increase in the number of COX-negative fibers (expressed as percentage of total fibers) in the heart of 15-month-old rescued mnd2 (R mnd2) mice. COX-negative fibers were enumerated on at least five random sections of the heart from control and aged rescued mnd2 mice (n=6). No COX-negative fibers could be seen in control mice. (c) (Left) Micrographs of five representative cryostat sections from 15-month-old rescued mnd2 mice cardiac muscles stained sequentially for COX (brown) and SDH-activity (purple) (micrographs labeled ‘before'). Using laser microdissection, COX-negative fibers were excised (micrographs labeled ‘after') and their mtDNA amplified by PCR. The sequences of the PCR products of these COX-negative fibers are shown to the right of the micrographs. Bold letters indicate direct repeat sequences. Accession number for mouse mtDNA sequence used is NC_005089
To confirm that the COX deficiency is due to mtDNA deletions of COX genes, we microdissected COX-negative cardiac muscle fibers from rescued mnd2 mice and polymerase chain reaction (PCR) amplified their mtDNA, using PCR primers flanking a 10.5-Kb region containing the COX genes. These COX-negative fibers generated PCR products ranging in size from 2 to 4 Kb. DNA sequencing of these products (Figure 4c) revealed 6.5–8.5 Kb deletions that contain the COX genes. All of these deletions occurred in regions that contain 1–9 bp direct DNA repeats. These results indicate that HtrA2 deficiency in non-neuronal tissues causes increased clonally expanded mtDNA deletions, which are believed to be a major contributing factor leading to premature aging and decreased life span.2, 3, 36
Loss of HtrA2 causes increased autophagosome activity
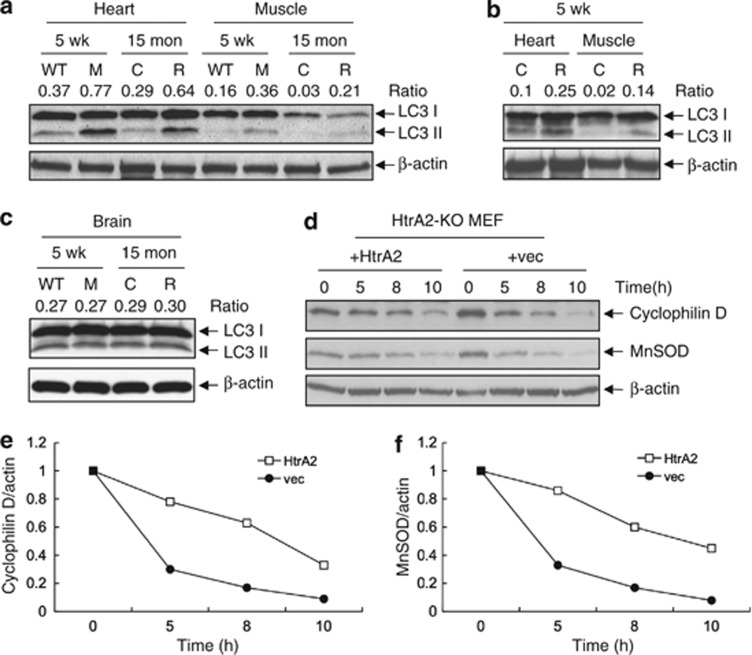
Increased autophagy has been observed in premature aging mice.31, 37 To investigate whether rescued mnd2 mice exhibit increased autophagosome activity, we examined the ratio of autophagosome marker LC3-II/LC3-I in cardiac and skeletal muscles of rescued mnd2 and control animals. Indeed, the ratio of LC3-II/LC3-I was significantly higher in cardiac and skeletal muscles of aged rescued mnd2 mice than in corresponding tissues of control littermates (Figure 5a). Interestingly, autophagosome activity was also higher in the same tissues of 5-week-old mnd2 and rescued mnd2 mice compared with WT control mice (Figures 5a and b), suggesting that the increased autophagosome activity is trigged early in life by mitochondrial dysfunction in order to eliminate defective mitochondria. The ratio of LC3-II/LC3-I was not significantly altered in brain tissues from mnd2 or rescued mnd2 mice (Figure 5c). These results indicate that HtrA2 deficiency in the mitochondria of non-neuronal tissues leads to increased autophagosome activity perhaps as a compensatory quality control mechanism to remove dysfunctional mitochondria by mitophagy. Consistent with this, the mitochondrial proteins, cyclophilin D and manganese superoxide dismutase (MnSOD), were degraded at a faster rate in HtrA2-deficient MEFs than in HtrA2-containing MEFs after induction of autophagy by starvation (Figures 5d–f). No differences were observed in the rate of degradation of the cytosolic protein β-actin. These results indicate that HtrA2-deficiancy increases mitophagy. Similar results were previously reported in MEFs deficient in the mitochondrial proteins OPA1 or mitofusins (MFN) 1 and 2,38 suggesting that HtrA2-deficiancy might impact the function of these proteins, resulting in impaired mitochondrial remodeling, which triggers mitochondrial degradation by mitophagy. Supporting this, a previous study showed that HtrA2-deficiency modulate the level of soluble OPA1 and affects mitochondria morphology.39
Figure 5.
HtrA2 deficiency causes increased autophagy/mitophagy. (a–c) Western blotting for LC3-I and LC3-II in the heart, muscle, and brain of 5-week-old WT and mnd2 (M), or 15-month-old rescued mnd2 (R) and control Tg;mnd2/+ littermates (C) (a and c), or in the heart and muscle of 5-week-old rescued mnd2 (R) and control Tg;mnd2/+littermates (C) (b). (d) Western blotting for cyclophilin D, MnSOD and β-actin in HtrA2-deficient (HtrA2-KO) MEF stably reconstituted with HtrA2 (+HtrA2) or empty vector (+vec) and starved for the indicated periods of time (hours). (e and f) Ratio between the densitometric levels of cyclophilin D (e) or MnSOD (f) and those of β-actin in the western blots shown in (d)
Discussion
We have shown here that a neuron-targeted human HTRA2 transgene can rescue mnd2 mice from rapid onset neurodegeneration, multiple tissue atrophy and early lethality, indicating that the specific loss of HtrA2 activity in the CNS, but not in other tissues of these mice, is directly responsible for these phenotypic abnormalities. This also indicates that the human and the mouse HtrA2 genes are functional orthologs. We have also identified a previously unrecognized role for HtrA2 in aging.
Protein quality in the mitochondria is monitored by nuclear-encoded proteases that survey the mitochondrial compartments for damaged and unfolded proteins.5 Although defects in mitochondrial protein quality control have been cited as the cause of many neurodegenerative and age-related diseases, there is no direct genetic evidence to date linking any of the mitochondrial quality control proteases to aging in mammals. This could be attributed to the fact that deficiencies or mutations in mitochondrial quality control proteases (e.g., HtrA2, PARL) usually lead to neurodegeneration and early lethality,22, 25, 26 which hampers and complicates long-term studies of their loss-of-function in non-neuronal tissues. Thus, by preventing neurodegeneration in mnd2 mice using a neuron-targeted human HTRA2 transgene, we were able to show that loss of HtrA2 activity in non-neuronal tissues of rescued mnd2 mice leads to premature aging as evidenced by specific age-related phenotypic abnormalities.
Some of the more prominent pathological findings in rescued mnd2 mice included weight loss, osteoporosis, curvature of the spine, muscle atrophy, and heart enlargement, are all commonly seen in aging humans and rodents.2, 3, 27, 28, 29, 30, 31, 32 Pathological heart enlargement, particularly left ventricular hypertrophy, is a physiological change associated with cardiac aging and a major risk factor affecting life span. Adult rescued mnd2 mice exhibit clear features of cardiac aging including left ventricular hypertrophy, decreased glucose metabolism, increased mtDNA deletions and increased autophagosome activity. Similar age-related changes have been described in mitochondrial mutator mice that carry a homozygous mutation in the catalytic subunit of mtDNA polymerase γ (PolgA).2, 3 Interestingly, these changes were partially attenuated in mitochondrial mutator mice overexpressing catalase targeted to mitochondria (mCAT),40 suggesting that mitochondrial ROS might play a role in these age-related changes.
Another important pathological finding in tissues of rescued mnd2 mice is the presence of clonally expanded large mtDNA deletions that span most of the respiratory chain enzymes. Although the mechanism by which these mtDNA deletions or mutations arise and their exact contribution to aging is not clear at present, our results suggest that decreased protein quality control in the intermembrane space of the mitochondria is a likely mechanism, as this could lead to increased ROS production. Indeed, increased ROS production was previously reported in thymocytes and MEFs from HtrA2-deficient mice.24 ROS could damage the respiratory complexes and other mitochondrial proteins including PolgA, resulting in decreased respiratory chain activity, and increased mutation and deletions of mtDNA. Eventually, these deleterious effects can cause mitochondrial dysfunction, which triggers mitophagy to clear the defective mitochondria. In support of this, our results showed an increase in autophagosome activity in rescued mnd2 mice perhaps as a result of increased mitophagy. Furthermore, there was an increase in the degradation rate of two mitochondrial proteins, cyclophilin D and MnSOD, in response to starvation in HtrA2-deficient MEFs compared with HtrA2-expressing MEFs, indicating increased mitophagy. This compensatory quality control mechanism, however, did not prevent the decline in cellular function that caused premature aging in these mice. On the contrary, we speculate that this increased autophagy/mitophagy might have inadvertently accelerated aging in this case, as increased mitophagy is usually compensated by increased mitochondrial biogenesis to meet energetic demand in somatic cells. This can likely lead to clonal expansion of mitochondria that carry deleterious mtDNA deletions or mutations, which once they reach to a certain phenotypic threshold, may further reduce the energy output of the mitochondria, hence exacerbating the decline in cellular functions leading to cell death.
Based on our data, a possible unifying mechanism that could explain the similar age-related changes observed in our rescued mnd2 mice and mitochondrial mutator mice is a decline in the catalytic activities of the mitochondrial respiratory chain complexes. Several subunits of the respiratory complexes I, III, and IV are encoded by mtDNA and the remainder by nuclear DNA.41 Accumulation of deletion and mutations in mtDNA of mitochondrial mutator mice will likely affect the proper assembly of not only mtDNA-encoded, but also nuclear DNA-encoded subunits of the respiratory complexes. Similarly, the loss of HtrA2 activity in rescued mnd2 mice could impact the folding and assembly of both mitochondrially and nuclearly encoded subunits of the respiratory chain complexes, as accumulation of oxidatively damaged subunits perhaps as a result of ROS damage will likely lead to aggregation of these complexes followed by loss of their catalytic activities (Figure 6). Indeed, studies with isolated mitochondria from HtrA2 knockout mice demonstrated a generalized respiratory chain dysfunction and presence of unfolded subunits of respiratory complexes I–IV.24 Therefore, defects in the catalytic activities of the respiratory chain complexes as a result of mtDNA deletions or defective protein quality control can put severe stress on the mitochondria to meet the energetic demand of cells of high energy-requiring tissues such as the heart, muscle, and brain. This in turn might cause increased ROS production, which further damages respiratory chain complexes and mtDNA, leading to mitochondrial dysfunction. Ultimately, these changes can lead to cellular senescence, and tissue atrophy and loss as seen in the mitochondrial mutator and rescued mnd2 mice.
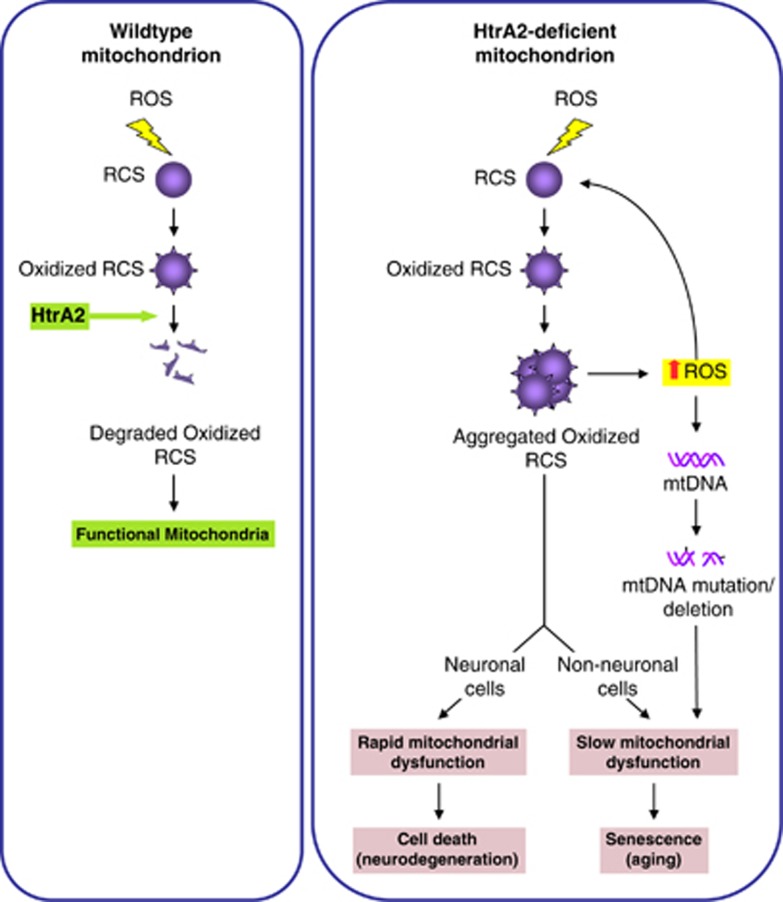
Figure 6.
A putative model depicting how HtrA2 deficiency could lead to neurodegeneration and aging. ROS production during normal mitochondrial respiration can cause oxidative damage to the respiratory complexes subunits (RCS). In WT mitochondria, these oxidized RCS are degraded by HtrA2. However, in HtrA2-deficient mitochondria, the oxidized RCS aggregate causing a decline in the activity of the respiratory complexes, which results in more ROS production that causes further damage to the RCS. As a result, this causes rapid mitochondrial dysfunction in neuronal cells, leading to early onset neurodegenration. In contrast, in non-neuronal cells, the presence of aggregated RCS causes slow mitochondrial dysfunction that eventually leads to cellular senescence as seen during aging. In addition, the increased ROS production in these cells might be responsible for induction of mtDNA mutations and deletions that further contribute to the decline in mitochondrial function
Although the loss of either HtrA2 or PolgA activity causes premature aging in mice, it is not clear why the loss of HtrA2 activity in neuronal tissues leads to early cell death and neurodegeneration (in mnd2 mice), whereas the loss of PolgA activity does not causes an overt neurodegenerative phenotype.2, 3 A likely explanation is that HtrA2 deficiency leads to rapid accumulation of unfolded respiratory complexes24 and possibly other protein aggregates in the mitochondria of neuronal cells, which causes early mitochondrial dysfunction and death of these cells (Figure 6). In contrast, PolgA deficiency is not likely to cause such a rapid accumulation of unfolded respiratory complexes and mitochondrial dysfunction. This is because mitochondria contain a functional complementation system that facilitates the exchange of genetic material between mitochondria through fusion and fission.42, 43 Therefore, even when the cell harbors large number of mitochondria with deleterious mtDNA mutations or deletions, the cell may remain healthy if it still contains >20% mitochondria with WT mtDNA. Hence, it is possible that the presence of WT copies of the respiratory complexes may phenotypically mask the mtDNA mutational burden in neuronal cells of the mitochondrial mutator mice for a long time. However, the continuous accumulation of mtDNA mutations and clonal expansion of these mutations can reach to a level that exceeds the ability of the complementation system to alleviate it, eventually leading to mitochondrial dysfunction.
Another important question is why loss of HtrA2 activity in neuronal tissues of mnd2 mice leads to cell death, whereas in non-neuronal tissues it leads to cellular aging. This could be explained by differences in the response between neuronal cells and other cell types to the presence of unfolded and aggregated proteins in the mitochondria. Non-neuronal cells may respond by undergoing senescence, rather than cell death, which could underline the premature aging phenotype. Indeed, our results show that the SA-β-Gal biomarker is highly elevated in adipose tissues from aged rescued mnd2 mice, indicating induction of cellular senescence. In contrast, neuronal cells might undergo a rapid neuron-specific cell death that leads to neurodegeneration. Recent studies in HtrA2 knockout mice have shown that loss of HtrA2 leads to induction of the proapoptotic stress-inducible transcription factor CHOP exclusively in neuronal tissues.24 CHOP plays important roles in neuronal cell death and its deletion delays neurodegeneration in HtrA2 knockout mice, as well as other neurodegeneration mouse models.24, 44, 45 The upregulation of CHOP, and possibly other stress genes, in neuronal tissues of HtrA2 knockout and mnd2 mice might therefore contribute to the early neurodegenerative phenotype in these mice.
Collectively, our results provide the first genetic evidence linking protein quality control in the intermembrane space of the mitochondria to mtDNA deletions and mammalian aging. We suggest that loss of HtrA2 activity impacts the folding and assembly of the respiratory chain complexes leading to their aggregation, mitochondrial dysfunction, and cell death. As mitochondrial quality control proteases such as HtrA2, might become less efficient with age, therapeutic strategies aimed at enhancing their activities might prove effective in treating neurodegenerative and other age-related diseases.
Materials and Methods
Generation of transgenic mice
The full-length Flag C-tagged human HTRA2 cDNA was cloned into the pNSE-Ex4 vector (kindly provided by Dr. Cravatt, Scripps Research Institute, La Jolla, CA, USA) by blunt end cloning for expression under the neural-specific enolase promoter. The resulting HTRA2/Flag C-pNSE construct was digested with SalI to remove the prokaryote sequences. Transgenic animals carrying the HTRA2 transgene (+/+Tg) were produced on a C57BL/6J background by the Kimmel Cancer Center Transgenic and Gene Targeting Core and were identified by PCR using a primer set corresponding to bp3986–4009 (5′-CTGGCCTCAGGCTCCACCCTTCTA′) of pNSE-Ex4 vector and bp810–837 of HTRA2 cDNA (5′-CTGAGCAGAGGCAACAATGCCGGATGTG). The transgenic founder mice (+/+Tg) were crossed with mnd2/+ mice to obtain F1 mice heterozygous for the mnd2 mutation and carry the HTRA2 transgene (mnd2/+Tg). Heterozygous mnd2/+Tg mice derived from two of the four founders were intercrossed for the generation of rescued mnd2 mice (mnd2/mnd2;Tg). mnd2 phenotyping was done as we described previously.25 All mice were used in experiments following protocols approved by the Institutional Animal Care and Use Committee, Thomas Jefferson University.
Antibodies and reagents
Human HtrA2-specific monoclonal antibody was developed in the laboratory. Rabbit anti-LC3 (NB100-2220) was obtained from NOVUS Biologicals (Littleton, CO, USA). Anti-cyclophilin D monoclonal antibody and anti-MnSOD polyclonal antibody were from Stressgen (Vancouver, Canada). For immunohistochemistry, several primary antibodies were used: mouse anti-GFAP (anti-glial fibrillary acidic protein) (IgG2b; 1 : 100) (BD Pharmingen, Franklin Lakes, NJ, USA), rabbit anti-active form of caspase-3 (IgG; 1 : 100) (Santa Cruz, Santa Cruz, CA, USA), rat antibodies to CD3 (IgG2a; 1 : 100) and to CD45 (IgG2b; 1 : 100) (Santa Cruz Biotech). Fluorescein IsoThioCyanate- and Tetramethyl Rhodamine IsoThioCyanate-conjugated secondary antibodies (IgG; 1 : 100) were obtained from Sigma (St Louis, MO, USA) or Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA, USA). NeuroTrace green and red fluorescent Nissl stains were obtained from Life technologies (Grand Island, NY, USA). Sodium dibasic phosphate, sodium monobasic phosphate, sucrose, catalase, cytochrome c, nitroblue tetrazolium, sodium succinate, and β-actin monoclonal antibody were from Sigma. Diaminobenzidine was from Santa Cruz. SA-β-Gal staining kit was from Cell Signaling Technology (Danvers, MA, USA).
Western blotting of tissue lysates
For determining transgenic HtrA2 expression in transgenic mice, the brain was removed from mice and homogenized in lysis buffer (50 mM Tris-HCl, 4 mM EDTA, 250 mM Sucrose, 1 mM DTT, and 1 mM PMSF). Liver, spleen, and thymus were also harvested and homogenized in Triton lysis buffer (1% Triton X-100, 50 mM Tris-HCl, 150 mM NaCl, 1 mM PMSF and complete protease inhibitor cocktail). In all, 30 μg of each protein was resolved on SDS-PAGE and transferred onto a PVDF membrane (Life technologies). Blots were probed with anti-human HtrA2 monoclonal antibody and HRP-conjugated anti-mouse IgG antibody and developed by using the ECL kit (Amersham, Arlington Heights, IL, USA). For measurements of LC3 in mouse tissues, the brain, heart, and muscle were harvested and homogenized in lysis buffer (150 mM NaCl, 10 mM Tris-HCl, 5 mM EDTA, 1% Triton X-100, 0.1% SDS, 1% Deoxycholate, 1mM PMSF and complete protease inhibitor cocktail). In all, 30 μg of each protein was resolved on 15% SDS-PAGE and transferred onto a PVDF membrane (Invitrogen), immunoblotted with rabbit anti-LC3 and developed by ECL.
Assay of mitophagy
Immortalized HtrA2-knockout (HtrA2-KO) MEFs stably transfected with HtrA2 (HtrA2-KO+HtrA2) or vector (HtrA2+vec) were used. Cells were starved by washing four times with phosphate buffer saline (PBS) followed by incubation in Hanks balanced salt solution supplemented with 10 mM Hepes pH 7.4, at 37°C for the indicated time periods. In all, 2.7 × 105 cells were lysed and 30 μg of each lysate was resolved on SDS-PAGE and immunoblotted with antibodies against cyclophilin D, MnSOD, and β-actin. The ratio between the levels of cyclophilin D or MnSOD and those of β-actin were calculated by densitometric scanning of the immunoblots.
Tissue processing for morphology analysis
Animals were euthanized using CO2. The skeletal muscles, hearts, livers, spleens, thymi, and duodenums were frozen in methyl butane cooled in liquid nitrogen. Rat brains were dissected out, placed in 4% paraformaldehyde for 24 h, followed by incubation in 30% sucrose solution for 24 h, then frozen in methyl butane cooled in liquid nitrogen. The samples were cut on a cryostat (10 μm sections).
Immunohistochemistry
Coronal cryostat sections (10 μm thick) were processed for indirect immunofluorescence. Blocking was performed by incubating 60 min with 10% goat, or 10% donkey, serum in PBS (pH 7.4). Then, sections were incubated with antibodies diluted according to the manufacturer's recommendations: 1 h with primary antibody, then 1 h with secondary antibody diluted 1 : 100, all at room temperature. Double immunofluorescence was performed as previously described.46 All incubations were followed by extensive washing with PBS. To stain nuclei, we used mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA, USA). Specimens were finally examined under a Leica DMRBE microscope (Leica Microsystems, Buffalo Grove, IL, USA) under × 4, × 10, and × 20 × objectives with appropriate filters. Images were captured using Spot Advance software (Diagnostic Instruments Inc., Sterling Heights, MI, USA). As positive controls for antibodies anti-caspase-3 and GFAP, we used coronal sections of brains injected or not with HIV-1 gp120, known to present apoptosis and gliosis.47 Data presented are representative of at least three independent experiments.
Staining of neurons
NeuroTrace (NT) staining was performed as previously described.48 After rehydration in 0.1 M PBS, pH 7.4, sections were treated with PBS plus 0.1% Triton X-100 10 min, washed twice for 5 min in PBS then stained with NT (1 : 100), for 20 min at room temperature. Sections were washed in PBS plus 0.1% Triton X-100 then × 2 with PBS, then left to stand for 2 h at room temperature in PBS before being counterstained with DAPI. Combination of NT and antibody staining was performed with the primary and secondary antibodies staining first, followed by staining with the NT fluorescent Nissl stain. All experiments were repeated three times and test and control slides were stained the same day.
SA-β-galactosidase staining
Adipose tissues were harvested from 3- and 12-month-old control and rescued mnd2 mice. Tissues were fixed and then stained for SA-β-Gal activity for 12 h at 37°C in the dark according to the manufacturer's protocol (Cell Signaling Technology, Danvers, MA, USA). After staining, tissues were washed with PBS and viewed by bright field microscopy.
TUNEL assay
TUNEL assay was performed according to the manufacturer's recommendations (Roche Diagnostics, Indianapolis, IN, USA).
Histochemistry for COX and succinate dehydrogenase
Succinate dehydrogenase (SDH) staining solution contained 10 ml 0.2 phosphate buffer, pH 7.6, 270 mg sodium succinate, nitroblue tetrazolium 10 mg. For COX staining solution, 750 mg sucrose was first dissolved in 7.5 ml deionized H2O, then 2.5 ml 0.2 phosphate buffer, pH 7.6 was added, as well as 5 mg diaminobenzidine, 10 mg cytochrome C and 20 mg catalase. Frozen cryosections were incubated with COX solution at room temperature first, then with SDH staining solution at 37°C. Incubation times for COX staining solution were 60 min (brain), 20 min (heart), and 40 min (duodenum). Sections were washed in PBS (1 × ) and incubated with SDH staining solution for 45 min (brain), 10 min (heart), and 15 min (duodenum). Sections were washed in 1 × PBS then either washed in 70% ethanol and left at room temperature for laser microdissection or coverslipped mounting.
HE staining and morphometry
Briefly, cryostat sections were stained with hematoxylin for 3 min, rinsed in deionized and tap water, dipped in acid ethanol, rinsed in tap and deionized water and finally stained with eosin for 30 sec. Sections were dehydrated in 95 then 100% ethanol before being cleared in xylene and mounted in Permount (Fischer Scientific, Pittsburgh, PA, USA).47 Mononuclear cells (expressed as the number per unit area) were enumerated in HE-stained sections of liver and heart in at least five consecutive sections using a computerized imaging system (Image-Pro Plus, MediaCybernetics, Rockville, MD,USA) as previously described.49, 50 CD3- and CD45-positive cells were enumerated on sections of the spleen immunostained with the corresponding antibodies. COX-negative fibers (% of the total number of heart fibers) were counted in heart sections sequentially stained by COX then SDH. In all cases, the final number was an average of results measured in different sections. Skeletal muscle fibers diameters were measured in HE-stained sections of the Quadriceps muscle as previously described.51
Microdissection and isolation of DNA
Laser microdissected was performed by using Positioning Ablation Laser Micro beam (PALM) (Carl Zeiss, Thornwood, NY, USA). Microdisected heart fibers were placed in a sterile microfuge tube containing 30 μl of lysis buffer and genomic DNA was prepared by following QIAamp DNA micro kit protocol (QIAGEN, Valencia, CA, USA).
Detection of mtDNA deletions
First PCR reactions were performed using the forward 5297 and reverse 129 primers and second PCR reactions were done with internal forward 5781 and internal reverse 48 primers: (forward 5297, 5′-TGTAAAACGACGGCCAGTCAGTCTAATGCTTACTCAGCC; reverse 129, 5′-CAGGAAACAGCTATGACCATGGAGGTTTGCATGTGTAA; internal forward 5781, 5′-TGTAAAACGACGGCCAGTTAGCTGGAGTGTCATCTATTT; internal reverse 48, 5′-CAGGAAACAGCTATGACCCCATCTAAGCATTTTCAGTGC). All primers are tagged with M13 sequence to facilitate the direct sequencing of PCR-amplified products. The reactions were performed with the Expand Long Template PCR System (Roche Applied Science), using buffer 2, 500 μmol/l dNTPs, 0.33 μmol/l of each primer, and 100 ng of genomic DNA. Hot-start PCR was performed as indicated by the manufacturer (Roche Applied Science). The total PCR reaction volume was 50 μl. The PCR cycling profile was as follows: denaturation at 93°C for 3 min; 10 cycles at 93°C for 30 s, 49°C for 30 s, 68°C for 12 min; 25 cycles at 93°C for 30 s, 49°C for 30 s, 68°C for12 min, plus a 20-s increment for each cycle; and a final extension at 68°C for 10 min.
Acknowledgments
We thank Julian Downward and L Miguel Martins for the immortalized HtrA2-KO MEFs. This work was supported by NIH Grants GM076176 and AG14357 to ESA.
Glossary
- mtDNA
mitochondrial DNA
- ROS
reactive oxygen species
- AAA
ATPase-Associated with diverse cellular Activities
- CP
Caudate-Putamen
- BMD
bone mineral density
- COX
cytochrome oxidase
- CNS
central nervous system
- GFAP
glial fibrillary acidic protein
- EDTA
ethylenediaminetetraacetic acid
- PMSF
phenylmethylsulfonyl fluoride
- DTT
dithiothreitol
- DAPI
diamidino-2-phenylindole
- NT
NeuroTrace
- PBS
phosphate buffer saline
- SDH
succinate dehydrogenase
- HE
hematoxylin–eosin
- MEF
mouse embryonic fibroblast
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by G Melino
Supplementary Material
References
- Green DR, Galluzzi L, Kroemer G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science. 2011;333:1109–1112. doi: 10.1126/science.1201940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Kujoth GC, Hiona A, Pugh TD, Someya S, Panzer K, Wohlgemuth SE, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Baker BM, Haynes CM. Mitochondrial protein quality control during biogenesis and aging. Trends Biochem Sci. 2011;36:254–261. doi: 10.1016/j.tibs.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Rugarli EI, Langer T. Mitochondrial quality control: a matter of life and death for neurons. EMBO J. 2012;31:1336–1349. doi: 10.1038/emboj.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes F, Tatsuta T, Langer T. Mitochondrial AAA proteases—towards a molecular understanding of membrane-bound proteolytic machines. Biochim Biophys Acta. 2012;1823:49–55. doi: 10.1016/j.bbamcr.2011.09.015. [DOI] [PubMed] [Google Scholar]
- Bota DA, Davies KJ. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat Cell Biol. 2002;4:674–680. doi: 10.1038/ncb836. [DOI] [PubMed] [Google Scholar]
- Nolden M, Ehses S, Koppen M, Bernacchia A, Rugarli EI, Langer T. The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell. 2005;123:277–289. doi: 10.1016/j.cell.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Casari G, De Fusco M, Ciarmatori S, Zeviani M, Mora M, Fernandez P, et al. Spastic paraplegia and OXPHOS impairment caused by mutations in paraplegin, a nuclear-encoded mitochondrial metalloprotease. Cell. 1998;93:973–983. doi: 10.1016/s0092-8674(00)81203-9. [DOI] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Fujita Y, Oka T, Mihara K. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 2006;25:2966–2977. doi: 10.1038/sj.emboj.7601184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehses S, Raschke I, Mancuso G, Bernacchia A, Geimer S, Tondera D, et al. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J Cell Biol. 2009;187:1023–1036. doi: 10.1083/jcb.200906084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Chen H, Fiket M, Alexander C, Chan DC. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J Cell Biol. 2007;178:749–755. doi: 10.1083/jcb.200704110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head B, Griparic L, Amiri M, Gandre-Babbe S, van der Bliek AM. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J Cell Biol. 2009;187:959–966. doi: 10.1083/jcb.200906083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvezin-Caubet S, Jagasia R, Wagener J, Hofmann S, Trifunovic A, Hansson A, et al. Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology. J Biol Chem. 2006;281:37972–37979. doi: 10.1074/jbc.M606059200. [DOI] [PubMed] [Google Scholar]
- Jin SM, Lazarou M, Wang C, Kane LA, Narendra DP, Youle RJ. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191:933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deas E, Plun-Favreau H, Gandhi S, Desmond H, Kjaer S, Loh SH, et al. PINK1 cleavage at position A103 by the mitochondrial protease PARL. Hum Mol Genet. 2011;20:867–879. doi: 10.1093/hmg/ddq526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SM, Youle RJ. PINK1- and Parkin-mediated mitophagy at a glance. J Cell Sci. 2012;125:795–799. doi: 10.1242/jcs.093849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell. 2006;126:177–189. doi: 10.1016/j.cell.2006.06.025. [DOI] [PubMed] [Google Scholar]
- Cipolat S, Rudka T, Hartmann D, Costa V, Serneels L, Craessaerts K, et al. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell. 2006;126:163–175. doi: 10.1016/j.cell.2006.06.021. [DOI] [PubMed] [Google Scholar]
- Clausen T, Kaiser M, Huber R, Ehrmann M. HTRA proteases: regulated proteolysis in protein quality control. Nat Rev Mol Cell Biol. 2011;12:152–162. doi: 10.1038/nrm3065. [DOI] [PubMed] [Google Scholar]
- Moisoi N, Klupsch K, Fedele V, East P, Sharma S, Renton A, et al. Mitochondrial dysfunction triggered by loss of HtrA2 results in the activation of a brain-specific transcriptional stress response. Cell Death Differ. 2009;16:449–464. doi: 10.1038/cdd.2008.166. [DOI] [PubMed] [Google Scholar]
- Jones JM, Datta P, Srinivasula SM, Ji W, Gupta S, Zhang Z, et al. Loss of Omi mitochondrial protease activity causes the neuromuscular disorder of mnd2 mutant mice. Nature. 2003;425:721–727. doi: 10.1038/nature02052. [DOI] [PubMed] [Google Scholar]
- Martins LM, Morrison A, Klupsch K, Fedele V, Moisoi N, Teismann P, et al. Neuroprotective role of the Reaper-related serine protease HtrA2/Omi revealed by targeted deletion in mice. Mol Cell Biol. 2004;24:9848–9862. doi: 10.1128/MCB.24.22.9848-9862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arking R.Biology of Aging Sunderlanded.Sinauer Associates, Inc.: Massachusetts; 1998 [Google Scholar]
- de Boer J, Andressoo JO, de Wit J, Huijmans J, Beems RB, van Steeg H, et al. Premature aging in mice deficient in DNA repair and transcription. Science. 2002;296:1276–1279. doi: 10.1126/science.1070174. [DOI] [PubMed] [Google Scholar]
- Sun LQ, Lee DW, Zhang Q, Xiao W, Raabe EH, Meeker A, et al. Growth retardation and premature aging phenotypes in mice with disruption of the SNF2-like gene, PASG. Genes Dev. 2004;18:1035–1046. doi: 10.1101/gad.1176104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George SK, Jiao Y, Bishop CE, Lu B. Mitochondrial peptidase IMMP2L mutation causes early onset of age-associated disorders and impairs adult stem cell self-renewal. Aging Cell. 2011;10:584–594. doi: 10.1111/j.1474-9726.2011.00686.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Kao CH, Chen YT, Wang CH, Wu CY, Tsai CY, et al. Cisd2 deficiency drives premature aging and causes mitochondria-mediated defects in mice. Genes Dev. 2009;23:1183–1194. doi: 10.1101/gad.1779509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines DC, Chattopadhyay S, Ward JM. Pathology of aging B6;129 mice. Toxicol Pathol. 2001;29:653–661. doi: 10.1080/019262301753385988. [DOI] [PubMed] [Google Scholar]
- Dai DF, Chen T, Johnson S, Szeto HH, Rabinovitch PS. Cardiac aging: from molecular mechanisms to significance in human health and disease. Antioxid Redox Signal. 2012;16:1492–1526. doi: 10.1089/ars.2011.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Wijshake T, Tchkonia T, LeBrasseur NK, Childs BG, van de Sluis B, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Hocker J. Cytochrome-c-oxidase deficient cardiomyocytes in the human heart—an age-related phenomenon. A histochemical ultracytochemical study. Am J Pathol. 1989;134:1167–1173. [PMC free article] [PubMed] [Google Scholar]
- Vermulst M, Wanagat J, Kujoth GC, Bielas JH, Rabinovitch PS, Prolla TA, et al. DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nat Genet. 2008;40:392–394. doi: 10.1038/ng.95. [DOI] [PubMed] [Google Scholar]
- Marino G, Ugalde AP, Salvador-Montoliu N, Varela I, Quiros PM, Cadinanos J, et al. Premature aging in mice activates a systemic metabolic response involving autophagy induction. Hum Mol Genet. 2008;17:2196–2211. doi: 10.1093/hmg/ddn120. [DOI] [PubMed] [Google Scholar]
- Gomes LC, Di Benedetto G, Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieper N, Holmstrom KM, Ciceri D, Fiesel FC, Wolburg H, Ziviani E, et al. Modulation of mitochondrial function and morphology by interaction of Omi/HtrA2 with the mitochondrial fusion factor OPA1. Exp Cell Res. 2010;316:1213–1224. doi: 10.1016/j.yexcr.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai DF, Santana LF, Vermulst M, Tomazela DM, Emond MJ, MacCoss MJ, et al. Overexpression of catalase targeted to mitochondria attenuates murine cardiac aging. Circulation. 2009;119:2789–2797. doi: 10.1161/CIRCULATIONAHA.108.822403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabi B, Mousson de Camaret B, Chevrollier A, Boisgard S, Stepien G. Random mtDNA deletions and functional consequence in aged human skeletal muscle. Biochem Biophys Res Commun. 2005;332:542–549. doi: 10.1016/j.bbrc.2005.04.153. [DOI] [PubMed] [Google Scholar]
- Attardi G, Yoneda M, Chomyn A. Complementation and segregation behavior of disease-causing mitochondrial DNA mutations in cellular model systems. Biochim Biophys Acta. 1995;1271:241–248. doi: 10.1016/0925-4439(95)00034-2. [DOI] [PubMed] [Google Scholar]
- Schon EA, Gilkerson RW. Functional complementation of mitochondrial DNAs: mobilizing mitochondrial genetics against dysfunction. Biochim Biophys Acta. 2010;1800:245–249. doi: 10.1016/j.bbagen.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Tajiri S, Oyadomari S, Yano S, Morioka M, Gotoh T, Hamada JI, et al. Ischemia-induced neuronal cell death is mediated by the endoplasmic reticulum stress pathway involving CHOP. Cell Death Differ. 2004;11:403–415. doi: 10.1038/sj.cdd.4401365. [DOI] [PubMed] [Google Scholar]
- Pennuto M, Tinelli E, Malaguti M, Del Carro U, D'Antonio M, Ron D, et al. Ablation of the UPR-mediator CHOP restores motor function and reduces demyelination in Charcot-Marie-Tooth 1B mice. Neuron. 2008;57:393–405. doi: 10.1016/j.neuron.2007.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouger K, Louboutin JP, Villanova M, Cherel Y, Fardeau M. X-linked vacuolated myopathy: TNF-alpha and IFN-gamma expression in muscle fibers with MHC class I on sarcolemma. Am J Pathol. 2001;158:355–359. doi: 10.1016/s0002-9440(10)63976-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louboutin JP, Agrawal L, Reyes BA, Van Bockstaele EJ, Strayer DS. HIV-1 gp120-induced injury to the blood-brain barrier: role of metalloproteinases 2 and 9 and relationship to oxidative stress. J Neuropathol Exp Neurol. 2010;69:801–816. doi: 10.1097/NEN.0b013e3181e8c96f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louboutin JP, Liu B, Reyes BA, Van Bockstaele EJ, Strayer DS. Rat bone marrow progenitor cells transduced in situ by rSV40 vectors differentiate into multiple central nervous system cell lineages. Stem Cells. 2006;24:2801–2809. doi: 10.1634/stemcells.2006-0124. [DOI] [PubMed] [Google Scholar]
- Louboutin JP, Chekmasova AA, Marusich E, Chowdhury JR, Strayer DS. Efficient CNS gene delivery by intravenous injection. Nat Methods. 2010;7:905–907. doi: 10.1038/nmeth.1518. [DOI] [PubMed] [Google Scholar]
- Louboutin JP, Chekmasova A, Marusich E, Agrawal L, Strayer DS. Role of CCR5 and its ligands in the control of vascular inflammation and leukocyte recruitment required for acute excitotoxic seizure induction and neural damage. FASEB J. 2011;25:737–753. doi: 10.1096/fj.10-161851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louboutin JP, Fichter-Gagnepain V, Pastoret C, Thaon E, Noireaud J, Sebille A, et al. Morphological and functional study of extensor digitorum longus muscle regeneration after iterative crush lesions in mdx mouse. Neuromuscul Disord. 1995;5:489–500. doi: 10.1016/0960-8966(95)00006-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.