Abstract
Autophagy is a lysosomal degradation pathway important for cellular homeostasis, mammalian development, cancer and immunity. Many molecular components of autophagy have been identified, but little is known about regulatory mechanisms controlling their effector functions. Here, we show that, in contrast to other p38 MAP kinase activators, the growth arrest and DNA damage 45 beta (Gadd45β)–MAPK/ERK kinase kinase 4 (MEKK4) pathway specifically directs p38 to autophagosomes. This process results in an accumulation of autophagosomes through p38-mediated inhibition of lysosome fusion. Conversely, autophagic flux is increased in p38-deficient fibroblasts and Gadd45β-deficient cells. We further identified the underlying mechanism and demonstrate that phosphorylation of the autophagy regulator autophagy-related (Atg)5 at threonine 75 through p38 is responsible for inhibition of starvation-induced autophagy. Thus, we show for the first time that Atg5 activity is controlled by phosphorylation and, moreover, that the spatial regulation of p38 by Gadd45β/MEKK4 negatively regulates the autophagic process.
Keywords: Atg5, autophagy, Gadd45, p38, phosphorylation
Macroautophagy (hereafter referred to as autophagy) is a catabolic process, by which the cell degrades cytosolic content to supply metabolic processes with nutrients in order to maintain ATP production and macromolecular synthesis. Thus, autophagy acts as an efficient recycling mechanism in eukaryotic cells.1 Cellular stress, for example, nutrient deprivation, enhances autophagy as a survival mechanism during starvation. In addition, autophagy serves important functions in development, cancer, cell death and immunity in mammals.1, 2
Autophagy is controlled by conserved key regulators known as autophagy-related (Atg) proteins.3 At the onset of the autophagy cascade, Atg6/Beclin-1 forms a complex with the class III phosphatidylinoside kinase Vps34, which induces expansion of the precursor membrane vesicle, the phagophore, via recruitment of additional Atg proteins. During expansion, the double membrane vesicle surrounds cytosolic content, and the completed vesicle, called autophagosome, finally fuses with lysosomes to degrade the autophagosomal content.3 Maturation of autophagosomes is regulated by two ubiquitin-like conjugation systems, namely the Atg8-phosphatidylethanolamine (PE) and the Atg5-12/16L1 conjugation system. The Atg5–Atg12 conjugate interacts with Atg16L1, which tethers the complex to phagophores and autophagosomes. This complex then acts as an E3-like ubiquitin ligase for microtubule-associated protein 1 light chain 3 (LC3) lipidation. The conversion of LC3 to the PE-conjugated LC3-II form and its recruitment to the membrane serves as a well-accepted marker for autophagy.
Although Atg5-independent autophagy has been described,4 Atg5 is crucial for autophagy under most circumstances and Atg5-deficient mouse embryonic fibroblasts (MEFs) lack LC3 conversion and autophagy. Therefore, Atg5-deficient mice die postnatal owing to their inability to cope with starvation during the neonatal period.5 In addition, Atg5 seems to be directly involved in the induction of apoptosis, as a calpain-generated fragment of Atg5 associates with Bcl-xL at mitochondria, resulting in the activation of the intrinsic apoptosis pathway.6 Thus, Atg5 could have a key role in controlling cell fate.
Although the function of Atg5 in constituting a complex with Atg12 and Atg16L1 is well determined, much less is known about the regulation of Atg5 activity during the autophagic process. Given the importance of autophagy in various biological processes, it is not surprising that it is tightly regulated by post-translational modifications of Atg proteins. For instance, acetylation of Atg5, Atg7, Atg8 and Atg12 by p300 inhibits autophagy,7 while TIP60 acetylates ULK1 inducing autophagy.8 In addition, the ULK1–Atg13–FIP200 autophagy-initiating complex is activated by direct phosphorylation of ULK1 by AMPK and inhibited by mTORC1.9 Thus, acetylation and phosphorylation events might have opposing effects on the autophagic flux.
Growth arrest and DNA damage 45 beta (Gadd45β) is a member of a family comprising Gadd45α, Gadd45β/Myd118 and Gadd45γ/CR6.10 Gadd45 proteins are small acidic proteins without enzymatic activity that rather exert their function by protein–protein interactions. Best investigated is their regulatory role in mitogen-activated protein kinase (MAPK) signaling, and apoptosis signaling kinase 1 (ASK1), MAPK/ERK kinase kinase 4 (MEKK4) and MAP kinase kinase (MKK)7 have been identified as Gadd45 interaction partners.11, 12 Interestingly, while interaction of Gadd45β with MKK7 inhibits c-Jun N-terminal kinase (JNK) activation,11 binding of Gadd45β to MEKK4 activates the JNK and p38 pathway.12 T cells from Gadd45β-deficient mice show reduced activation of MAPKs, especially of p38, and impaired cytokine production upon T-cell receptor triggering.13 In myeloid cells and hepatocytes, Gadd45β has a survival function, which is thought to be mediated by inhibition of JNK.14, 15, 16
Here, we demonstrate that p38 MAPK is selectively targeted to autophagosomes when activated by the Gadd45β–MEKK4 signaling complex. At the autophagosomal membrane, active p38 phosphorylates Atg5 at threonine 75 leading to inhibition of autophagy. Thus, our data show for the first time that Atg5 activity is directly controlled by p38-mediated phosphorylation in a Gadd45β/MEKK4-dependent manner.
Results
Gadd45β links p38 MAPK to autophagy
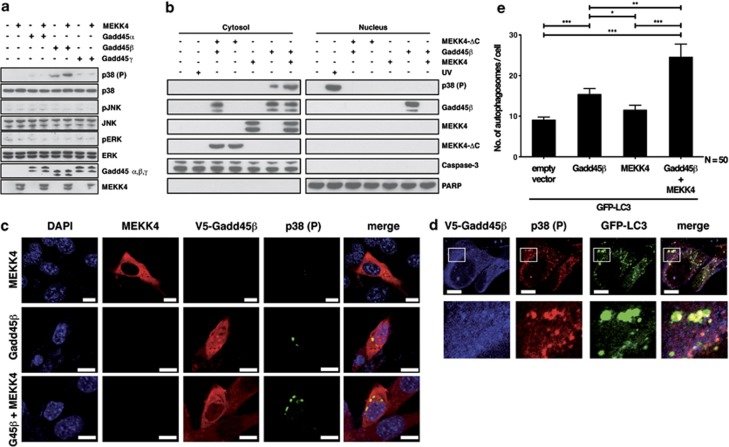
Gadd45 proteins have been described as regulators of MAPK signaling.11, 12 In NIH/3T3 fibroblasts coexpressing Gadd45β and MEKK4, we detected activation of p38 MAPK but not of JNK or ERK (Figure 1a and Supplementary Figure S1). Expression of Gadd45β alone resulted in detectable levels of phosphorylated p38, most likely owing to endogenous MEKK4 (Figure 1a and data not shown). Gadd45α- or Gadd45γ expression induced only minor p38 phosphorylation, which was not increased by MEKK4 coexpression (Figure 1a). As little is known about the spatial regulation of MAPK activation by Gadd45 proteins, we analyzed the subcellular localization of Gadd45-induced p38 activation. Thus, we prepared cytosolic and nuclear extracts of NIH/3T3 cells expressing Gadd45β, MEKK4 or both. Surprisingly, most of the phosphorylated p38 was found in the cytoplasmic fraction when activated by Gadd45β and MEKK4, in contrast to UV-irradiated cells, which showed nuclear localization of phosphorylated p38 (Figure 1b and Supplementary Figure S1e). Of note, coexpression of Gadd45β with a dominant-negative MEKK4-ΔC construct lacking the kinase domain did not result in p38 activation (Figure 1b).
Figure 1.
Gadd45β specifically activates p38 MAPK through MEKK4 and recruits p38 to autophagosomes. (a) NIH/3T3 cells were transfected with either V5-tagged Gadd45α or Gadd45β or Gadd45γ, HA-tagged MEKK4 or combinations thereof. As a negative control untransfected cells were included. At 24 h post transfection, cellular lysates were prepared and analyzed by immunoblotting using the indicated antibodies. (b) NIH/3T3 cells were transfected with either V5-tagged Gadd45β and HA-tagged MEKK4 or both. As negative and positive controls untransfected and UV-irradiated cells were included. As an additional control, a C-terminal deleted MEKK4 (MEKK4-ΔC) construct lacking the kinase domain was transfected together with Gadd45β. MEKK4-ΔC retains the ability to interact with Gadd45β but lacks enzymatic activity. At 24 h post transfection, cytosolic and nuclear extracts were prepared and analyzed by immunoblotting using the indicated antibodies. Caspase-3 and PARP were used as markers for cytosolic and nuclear compartments, respectively. (c) NIH/3T3 cells were transiently transfected with either V5-tagged Gadd45β and HA-tagged MEKK4 alone or with both constructs. At 24 h post transfection, the cells were fixed and stained with anti-HA (red), anti-V5 antibody (red), anti-phospho-p38 (green) and 4′,6′-diamidino-2-phenylindole dihydrochloride (DAPI) (blue). In the lower panel, MEKK4 is not visible as not more than three molecules could be stained owing to technical reasons. Nevertheless, cytoplasmic staining of Gadd45β in the absence of any nuclear signal indicates co-expression of MEKK4. Subsequently, samples were analyzed by confocal microscopy. Bars, 10 μm. (d) NIH/3T3 cells were transfected with V5-tagged Gadd45β, HA-tagged MEKK4 and GFP-LC3. The cells were stained with anti-V5 (blue), LC3-GFP (green) and anti-phospho-p38 (red), and analyzed as described in (c). Boxed areas are enlarged below. Bars, 10 μm. (e) NIH/3T3 cells stably expressing GFP-LC3 were transfected with the indicated constructs. After 24 h wide-field pictures were taken and the number of GFP-LC3-positive autophagosomes per cell was counted in 50 cells per condition. For statistics, two-tailed Student's t-test was performed (*P=0.0149, **P=0.001, ***P<0.0001)
In order to verify the subcellular localization of Gadd45β-mediated p38 activation, we expressed Gadd45β, MEKK4 or the combination of both in NIH/3T3 cells, stained for phosphorylated p38 and applied confocal microscopy. In agreement with previous results (Figure 1b and Takekawa and Saito12), MEKK4 localized cytosolically while Gadd45β showed a nuclear and cytosolic localization when expressed without MEKK4. In MEKK4–Gadd45β-coexpressing cells, Gadd45β showed a prominent cytosolic staining and colocalization with MEKK4, consistent with these two proteins being interaction partners (Figure 1c and Supplementary Figure S1).12 The expression of MEKK4 alone did not result in detectable p38 activation (Figure 1c, upper row), which is in agreement with the kinase being in a closed conformation when Gadd45 proteins are absent.17 Strikingly, phosphorylated p38 was detected in vesicular structures in the cytoplasm upon Gadd45β expression (Figure 1c, middle panel). This unexpected localization of phosphorylated p38 was enhanced upon coexpression of MEKK4 and Gadd45β (Figure 1c, lower panel), but was blocked by coexpression of Gadd45β with dominant-negative MEKK4-ΔC (Supplementary Figure S1f).
To identify the nature of these dot-like structures, we co-expressed Gadd45β and MEKK4 together with green fluorescent protein (GFP)-LC3, a widely accepted marker for autophagosomes.18 Indeed, phosphorylated p38 co-localized with GFP-LC3 at cytoplasmic vesicles upon expression of Gadd45β and MEKK4 (Figure 1d). In order to analyze whether activation of the Gadd45β–MEKK4 pathway affects the number of GFP-LC3-positive vesicles, we transfected Gadd45β, MEKK4 or both molecules into NIH/3T3 cells stably expressing GFP-LC3. At 24 h post transfection, the number of GFP-LC3-positive dots was counted in 50 cells per condition. Expression of Gadd45β alone showed a significantly higher number of GFP-LC3-positive puncta per cell compared with cells expressing GFP-LC3 alone (P=0.0002) or MEKK4 expression (P=0.0149, Figure 1e). Coexpression of Gadd45β and MEKK4 increased the number of GFP-LC3-positive vesicles per cell even more (P=0.001 compared with Gadd45β; P<0.0001 compared with MEKK4). Based on these results, we conclude that the Gadd45β–MEKK4-pathway directs activated p38 to autophagosomes, suggesting a role for p38 in autophagy regulation.
Gadd45β in conjunction with MEKK4 but not ASK1 directs p38 to autophagosomes
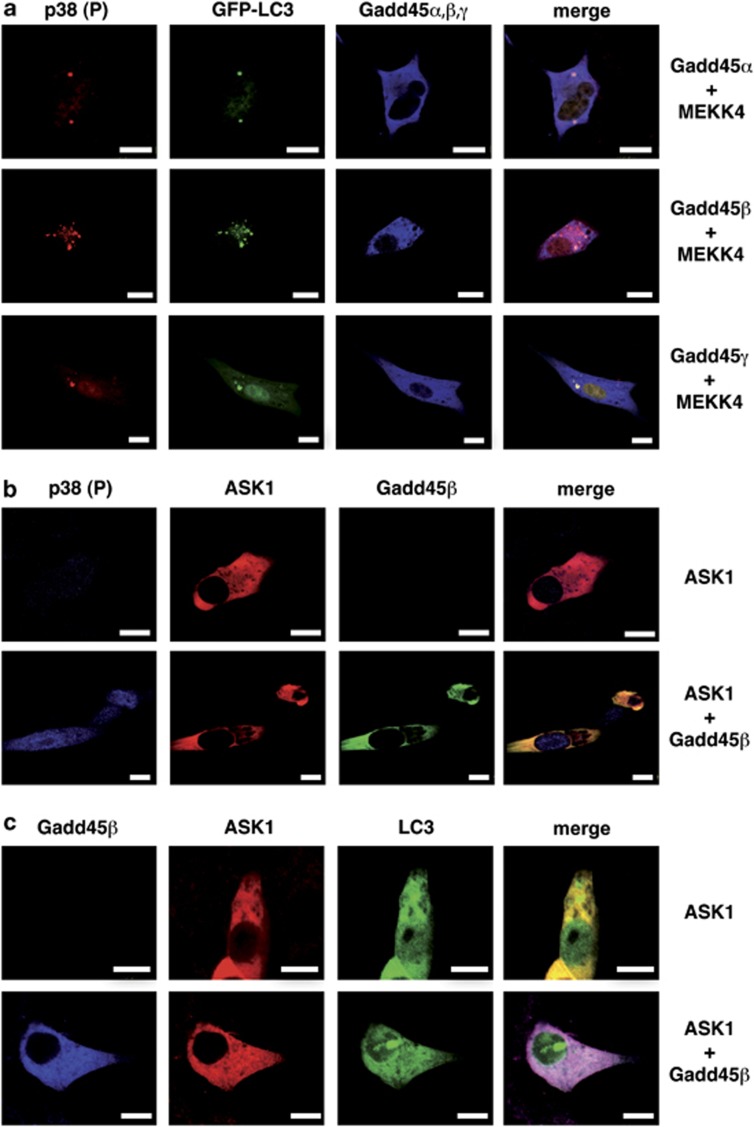
We asked whether autophagy is influenced specifically by Gadd45β and MEKK4. Thus, we investigated the potential role of other Gadd45 proteins and MAPK kinase kinases in p38 activation and autophagosome formation. Compared with other family members, Gadd45β induced a more pronounced punctuate localization of GFP-LC3, which co-localized with active p38, suggesting that Gadd45β but not other Gadd45 members are associated with autophagy (Figure 2a). Moreover, ASK1, another Gadd45-interacting kinase,11 did not result in punctual localization of phosphorylated p38 (Figure 2b) or GFP-LC3 (Figure 2c). Thus, we conclude that Gadd45β/MEKK4 but not Gadd45/ASK1 complexes direct p38 to autophagosomes.
Figure 2.
Localization of p38 in dot-like structures is specific for the interaction between Gadd45 proteins and MEKK4. (a) NIH/3T3 cells were transiently transfected with either V5-tagged Gadd45α, V5-tagged Gadd45β or V5-tagged Gadd45γ together with GFP-tagged LC3 and HA-tagged MEKK4. At 24 h post transfection, the cells were stained with anti-V5 (blue), LC3-GFP (green) and anti-phospho-p38 (red). Subsequently, samples were analyzed by confocal microscopy. Bars, 10 μm. (b) NIH/3T3 cells were transiently transfected with V5-tagged Gadd45β and HA-tagged ASK1. At 24 h after transfection, the staining was performed with anti-V5 (green), anti-HA (red) and endogenous phosphorylated p38 (blue). Subsequently, samples were analyzed by confocal microscopy. Bars, 10 μm. (c) NIH/3T3 cells were transfected with V5-tagged Gadd45β HA-tagged ASK1 and GFP-LC3. After 18 h, cells were fixed and stained with anti-HA (red) and anti-V5 antibody (blue). Bars, 10 μm
Gadd45β expression inhibits autophagic flux
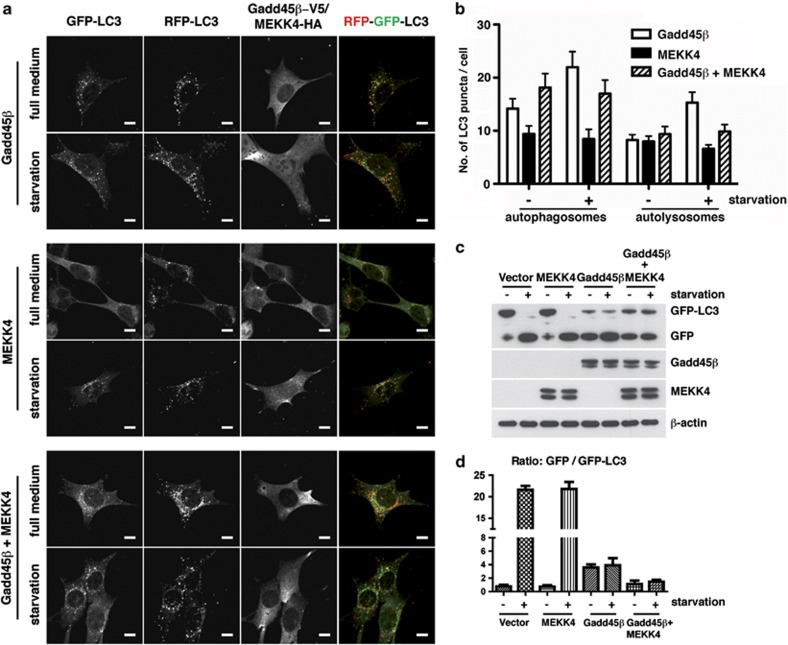
As autophagy is a dynamic process, the appearance of GFP-LC3-positive autophagosomes might be caused either by enhanced autophagy owing to accelerated autophagosome formation or by the accumulation of autophagosomes resulting from reduced fusion with lysosomes.19 To analyze the autophagic flux, we transfected Gadd45β, MEKK4 or both together with a tandem reporter construct. This allows identification of autophagosomes (red fluorescent protein (RFP)+GFP+) and autolysosomes (RFP+GFP−), as GFP fluorescence is lost upon lysosomal acidification while RFP fluorescence remains stable.20 While LC3 puncta formation was increased upon starvation in MEKK4 single-transfected cells, starvation had no effect on puncta formation in Gadd45β only and Gadd45β/MEKK4 transfected cells (Figure 3a). Importantly, double-transfected cells showed increased autophagosome numbers but no change in autolysosomes upon starvation, suggesting inhibition of autophagic flux upon Gadd45β and MEKK4 expression (Figure 3b). As an independent measure of autophagic flux we analyzed degradation of GFP-LC3 by immunoblotting, as LC3 is rapidly degraded but GFP is quite resistant to lysosomal hydrolysis.19 In line with the microscopy data, cleavage of GFP-LC3 was impaired in GFP-LC3-expressing NIH/3T3 cells when Gadd45β was expressed with or without MEKK4, while MEKK4 alone had no effect compared with the control cells (Figures 3c and d).
Figure 3.
Gadd45β inhibits starvation-induced autophagic flux. (a) NIH/3T3 cells stably expressing RFP-GFP-LC3 were transiently transfected with V5-tagged Gadd45β and HA-tagged MEKK4. At 24 h post transfection, the cells were fixed and stained with anti-HA or anti-V5 (AF633). Subsequently, samples were analyzed by confocal microscopy. Bars, 10 μm. (b) Quantification of the experiments shown in (a). The number of autophagosomes (RFP+GFP+) and autolysosomes (RFP+GFP−) per cell was counted in 50 cells per condition. (c) NIH/3T3 cells stably expressing GFP-LC3 were transiently transfected as in (a). At 24 h post transfection, the cells were either starved in HBSS for 4 h or kept in full medium. Subsequently, processing of GFP-LC3 was analyzed by immunoblotting. (d) Densitometric quantification of the GFP to GFP-LC3 ratio of results shown in (b)
Active p38 inhibits autophagy
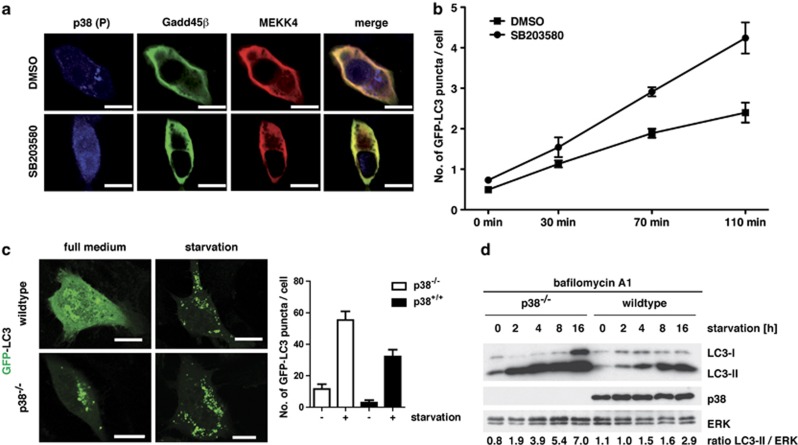
Next, we asked whether or not the kinase activity of p38 was instrumental to the inhibitory effect on autolysosome formation. To address this question, we expressed Gadd45β and MEKK4 in NIH/3T3 cells in the absence or presence of the p38 inhibitor SB203580 and analyzed the localization of active p38. In control samples the expected vesicular localization of active p38 was detected, whereas inhibition of p38 activity by SB203580 resulted in a homogenous distribution of phosphorylated p38 in the cytoplasm as well as in the nucleus (Figure 4a). Therefore, kinase activity is required to target p38 to autophagosomes.
Figure 4.
Lack of active p38 accelerates autophagy. (a) NIH/3T3 cells were transiently transfected with V5-tagged Gadd45β and HA-tagged MEKK4. At 24 h post transfection, the cells were fixed and stained for endogenous phosphorylated p38 (blue), HA-tagged MEKK4 (red) and the V5-tagged Gadd45β (green). In addition, the cells were treated with 500 nM of the p38 inhibitor SB203580 or DMSO as solvent control. Bars, 10 μm. (b) GFP-LC3 expressing NIH/3T3 cells were cultured in the presence or absence of 500 nM SB203580 in HBSS and followed by life cell imaging for the indicated time points. Autophagosomes were counted in 40 cells and the mean number of autophagosomes per cell was calculated from two independent visual fields. Error bars represent S.E.M. (c) WT and p38−/− MEFs were transfected with GFP-LC3 as a marker for autophagy. At 24 h post transfection, cells were cultured for 4 h in HBSS medium to induce autophagy. Subsequently, samples were fixed and analyzed by confocal microscopy (left panel). Bars, 10 μm. The number of autophagosomes was counted in 40 cells per condition (right panel). (d) WT and p38−/− MEFs were cultured for up to 16 h in HBSS medium to induce autophagy. In order to prevent LC3-II degradation, cells were pretreated with 100 nM bafilomycin A1 for 1 h. Subsequently, cellular lysates were prepared and analyzed by immunoblotting using the indicated antibodies. Densitometric quantification of the LC3-II to ERK ratio is depicted at the bottom
To investigate the impact of p38 kinase activity on autophagy in more detail, we used the GFP-LC3-expressing NIH/3T3 cells and cultured them in Hank's buffered salt solution (HBSS) to induce autophagy. Cells were grown in the absence or presence of SB203580 to inhibit p38, and autophagosome formation was monitored by life cell imaging. Quantification of GFP-LC3 puncta per cell at different time points revealed that the number of autophagosomes was about doubled when p38 kinase activity was inhibited (Figure 4b).
Next, we used p38α knockout MEFs as a model system lacking p38 kinase activity.21 p38α-deficient MEFs and their wild-type (WT) counterparts were transfected with GFP-LC3. At 24 h post transfection, cells were starved in HBSS and GFP-LC3 puncta were analyzed by confocal microscopy. Upon starvation, p38α-deficient MEFs showed a higher number of GFP-LC3 puncta than control WT MEFs (Figure 4c). Thus, the number of GFP-LC3-positive puncta increased with both activation and inhibition of p38 (compare Figures 1c and e versus 4c). This at first glance contradictory result prompted us to investigate the autophagic flux in p38α-deficient cells. To this end, we starved WT and p38α-deficient MEFs for up to 16 h and analyzed the conversion of endogenous LC3-I to the PE-conjugated LC3-II form by immunoblotting. Cells were cultured in the presence of bafilomycin A1 for the last 2 h in order to prevent lysosomal degradation of LC3-II. In the absence of bafilomycin A1, LC3-I and LC3-II were hardly detectable in these MEFs (data not shown). p38α-deficient MEFs showed an earlier accumulation of LC3-II and in higher numbers compared with WT MEFs (Figure 4d). Thus, autophagic flux was strongly promoted in the absence of p38.
p38 phosphorylates Atg5
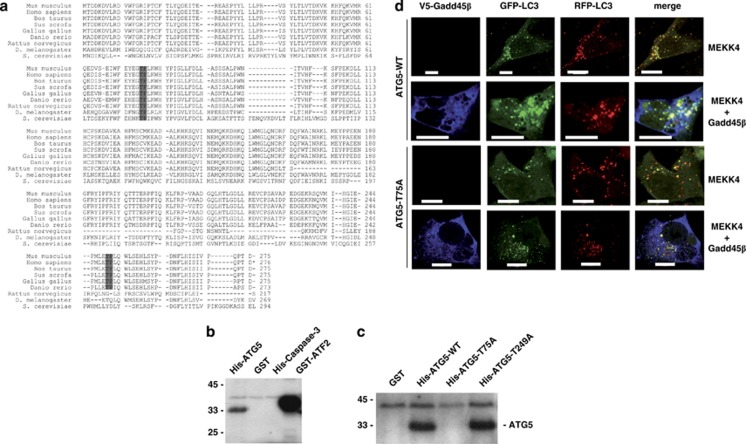
So far, we established that active p38 was recruited to autophagosomes and its kinase activity was necessary for the inhibition of autophagosome–lysosome fusion. Therefore, we reasoned that p38 phosphorylates a target protein on autophagosomal vesicles and that this phosphorylation event influenced autophagic activity. In search for a potential kinase substrate, we analyzed the sequences of those Atg proteins that are essential components of autophagosomes. The objective was to find conserved S/T-P motifs recognized by MAPKs.22 We identified two potential phosphorylation sites in Atg5 at T75 and T249 (Figure 5a). The TP motif at position 75/76 is conserved in all species analyzed ranging from yeast to men, while the TP motif at position 249/250 is not present in Saccharomyces cerevisiae, Drosophila melanogaster and Rattus norvegicus. To test whether p38 directly phosphorylates Atg5, we purified recombinant Atg5 and incubated it with active p38 and radioactive ATP. Caspase-3 and GST were used as negative controls, while ATF2, a known p38 target, served as positive control. Although ATF2 phosphorylation was stronger, phosphorylation of Atg5 by p38 could readily be detected (Figure 5b). To determine whether p38 phosphorylates Atg5 at T75 or T249, recombinant Atg5 proteins with alanine substitutions were analyzed by in vitro kinase assays. The p38 kinase phosphorylated the T249A mutant as efficiently as WT Atg5, whereas the T75A mutant was not phosphorylated by p38, indicating that p38 phosphorylates Atg5 at T75 (Figure 5c).
Figure 5.
Atg5 is a substrate for p38. (a) The protein sequences of Atg5 proteins from different species were analyzed for potential phosphorylation sites of p38 (XS/TPX consensus). Two putative phosphorylation sites could be identified at threonine 75 and threonine 249 of murine Atg5. (b) The indicated proteins were tested for phosphorylation by p38 in an in vitro kinase assay. His-Caspase-3 and GST served as a negative control, whereas GST-ATF2 was used as a positive control. (c) In vitro kinase assay with WT His-Atg5 as well as with His-Atg5, in which T75 and T249 had been mutated to alanine, respectively. GST served as a negative control. (d) Atg5−/− MEF cells stably reconstituted with WT or a T75A mutant of Atg5 were transfected with MEKK4 and an RFP-GFP-LC3 reporter either with or without Gadd45β co-expression. At 24 h post transfection, cells were starved for 4 h in HBSS. Subsequently, cells were fixed, stained for Gadd45β expression and analyzed by confocal microscopy for RFP-GFP-LC3 localization. Bars, 10 μm
To show that p38-mediated phosphorylation of Atg5 is important for autophagy regulation in living cells, we took advantage of Atg5-deficient MEFs,5 which were reconstituted either with WT or the phosphorylation-defective T75A mutant of Atg5. These MEFs were transfected with the RFP-GFP-LC3 tandem reporter and with MEKK4 together with either Gadd45β or an empty vector control. At 24 h post transfection, cells were starved for 4 h and analyzed by confocal microscopy. While WT Atg5-expressing cells showed an accumulation of large RFP+GFP+ autophagosomes and decreased RFP+GFP− autolysosomes upon Gadd45β expression, the appearance of autophagosomes and autolysosomes was not affected by Gadd45β expression in Atg5 T75A-expressing MEFs (Figure 5d). Thus, we conclude that the effect of the Gadd45β–MEKK4-p38 pathway on autophagy is mediated by phosphorylation of Atg5 at T75.
Phosphorylation of Atg5 at Thr75 inhibits autophagy
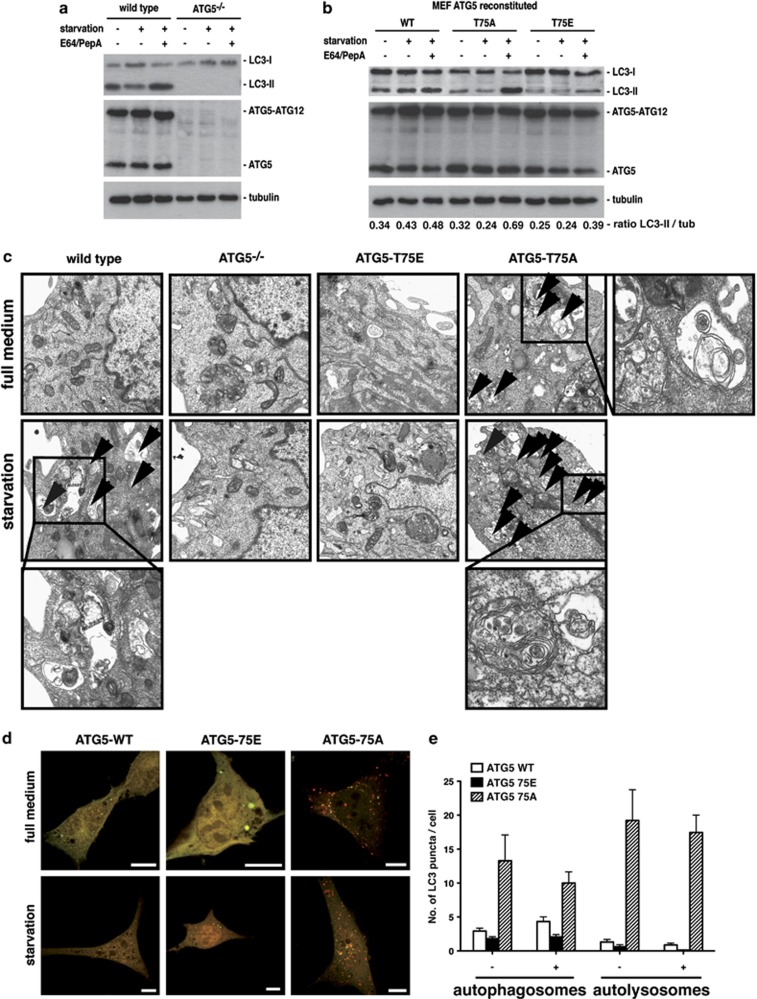
In order to investigate the functional consequences of Atg5 phosphorylation, we used reconstituted Atg5-deficient MEFs as above and additionally introduced a T75E mutant, which mimics constitutive phosphorylation. First, we analyzed autophagic flux by immunoblotting for endogenous LC3. Cells were either starved in HBSS or cultured in complete medium. Moreover, starvation was performed in the absence or presence of E64 and pepstatin A to inhibit lysosomal proteases and prevent degradation of LC3-II. In agreement with previous studies,5 Atg5-deficient MEFs showed no conversion of LC3-I to the lipidated LC3-II form (Figure 6a). As expected, WT MEFs accumulated LC3-II upon starvation when lysosomal proteases were inhibited by E64 and pepstatin A, consistent with enhanced autophagic flux. WT Atg5-reconstituted MEFs showed a similar LC3 pattern as MEFs from WT mice (Figure 6b). Of note, the levels of the Atg5–Atg12 complex were similar in the reconstituted MEFs when compared with WT MEFs (Figures 6a and b). Importantly, Atg5 T75A mutant cells showed higher amounts of LC3-II when starved in the presence of protease inhibitors, indicating a higher autophagic activity (Figure 6b). In contrast, less LC3-II was detectable in MEFs reconstituted with the T75E mutant. Similar results were obtained when autophagy was monitored by acridine orange, a lysosomotropic fluorescent dye, and flow cytometry (Supplementary Figure S2). Taken together, these data strongly suggest that p38-mediated phosphorylation of Atg5 at T75 inhibits autophagy.
Figure 6.
Atg5 phosphorylation by p38 inhibits the process of autophagy. (a) WT and Atg5−/− MEF were cultured for 4 h in HBSS medium to induce autophagy or left in full culture medium. E64 and pepstatin A were used to prevent lysosomal degradation of LC3-II. Subsequently, cellular lysates were prepared and analyzed by immunoblotting using anti-LC3 and anti-Atg5 antibodies. Tubulin served as a loading control. (b) MEFs reconstituted with WT, T75A or T75E-mutated Atg5 were treated and analyzed as described in (a). Densitometric quantification of the LC3-II to tubulin ratio is depicted below the loading control. (c) WT, Atg5-deficient (Atg5−/−) and Atg5−/− MEF cells stably reconstituted with T75A or T75E mutants of Atg5 were cultured for 4 h in HBSS to induce autophagy. Control cells were left in full culture medium. Subsequently, samples were fixed and analyzed by transmission electron microscopy. (d) Atg5-deficient MEF cells reconstituted with WT Atg5, Atg5-T75A or Atg5-T75E were transfected with RFP-GFP-LC3. At 24 h post transfection, cells were either starved in HBSS for 4 h or left in full culture medium. Subsequently, cells were fixed and analyzed by confocal microscopy for RFP-GFP-LC3 localization. Bars, 10 μm. (e) Quantification of the results shown in (d). Numbers of autophagosomes and autolysosomes were counted in 50 cells per condition
Next, we analyzed authophagosome formation by electron microscopy. WT but not Atg5-deficient cells showed typical double-membrane structures, that is, autophagosomes, upon starvation (Figure 6c). MEFs expressing the T75A mutants contained autophagosomes even when cultured in complete medium, suggesting enhanced basal autophagy (Figure 6c). In contrast, T75E-expressing MEFs showed no autophagosome formation, even under starvation conditions (Figure 6c). Furthermore, reconstituted MEFs were transfected with a RFP-GFP-LC3 reporter and starved for 4 h or left in complete culture medium. RFP-GFP-LC3 was mostly diffusely distributed in cells expressing WT or the T75E mutant Atg5, which were cultured in complete medium, with only a few RFP+GFP+ vesicles (Figure 6d). As expected, RFP+GFP+ LC3 puncta were increased upon starvation in cells expressing WT Atg5, but not in T75E mutant cells (Figures 6d and e). In both genotypes, we detected only few RFP+GFP- dots, that is, autolysosomes. Importantly, T75A-expressing MEFs had highly increased numbers of LC3 puncta, especially of RFP+GFP− autolysosomes, suggesting that preventing phosphorylation of Atg5 strongly promotes autophagic flux (Figures 6d and e).
Increased autophagy in Gadd45β-deficient cells upon lipopolysaccharide (LPS) stimulation
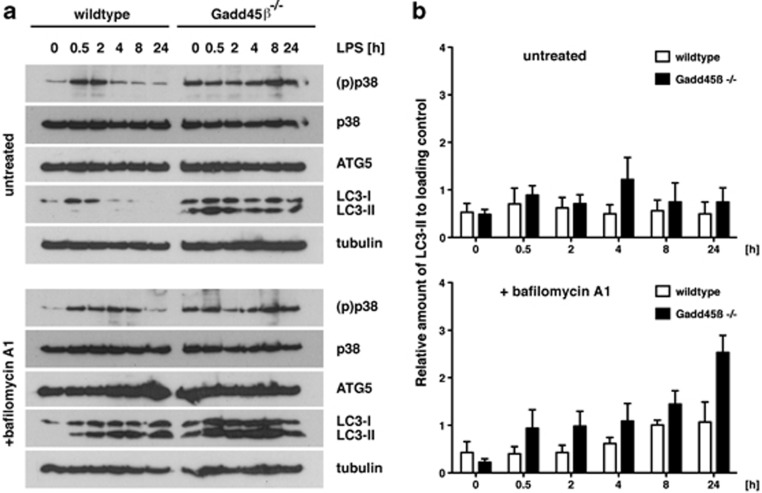
Finally, we asked whether Gadd45β-mediated regulation of autophagy has a physiological role. As Gadd45β expression is induced by LPS23 and autophagy is initiated upon Toll-like receptor (TLR) activation,24 we investigated whether autophagy was altered in Gadd45β-deficient cells upon LPS treatment. Therefore, we activated WT and Gadd45β-deficient MEFs with LPS in the absence and presence of bafilomycin A1. Without inhibition of lysosome acidification, the ratio of LC3-I to LC3-II was comparable between Gadd45β-deficient and WT MEFs, although total LC3 expression was increased in the knockout cells (Figure 7a). This higher LC3 expression in Gadd45β-deficient MEFs could indicate a general increase in autophagy in these cells, as expression of autophagy genes is induced during ongoing autophagy.19 Indeed, blocking lysosomal degradation by bafilomycin A1 showed a LPS- and time-dependent accumulation of LC3-II, which was much more pronounced in Gadd45β-deficient compared with WT MEFs (Figure 7a). Quantification of LC3 abundance over three independent experiments substantiated the observed increase in autophagic flux upon LPS stimulation in the absence of Gadd45β (Figure 7b).
Figure 7.
Gadd45β suppresses autophagy upon LPS stimulation in MEFs. (a) MEFs from WT and Gadd45β-deficient mice were stimulated in vitro with 100 ng ml−1 LPS for the indicated time periods. In order to prevent LC3-II degradation, half of the cells were treated with 100 nM bafilomycin A1 for the last 2 h of culturing. Subsequently, cellular lysates were prepared and analyzed by immunoblotting for expression of phospho-p38, total p38, Atg5 and LC3. Tubulin served as a loading control. Results shown are representative for three independent experiments. (b) Quantification of the results shown in (a). The intensities of LC3-II bands were normalized to the tubulin loading control. Data are represented as mean of three independent experiments±S.E.M.
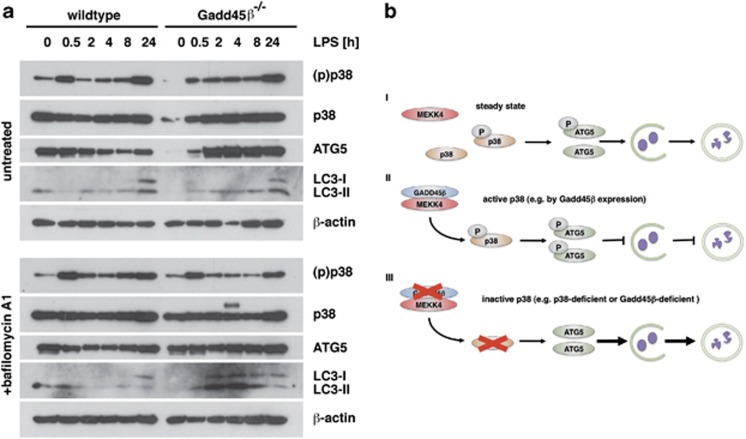
In order to substantiate our findings, we analyzed the role of Gadd45β on autophagy in primary cells. To this end, bone marrow-derived macrophages (BMDMs) were generated from WT and Gadd45β-deficient mice, stimulated with LPS and analyzed by immunoblotting. Similar to MEFs, a LPS- and time-dependent accumulation of LC3-II was observed in Gadd45β-deficient BMDMs in the presence of bafilomycin A1 (Figure 8a). In contrast, WT BMDMs did not show increased LC3-II. Therefore, we conclude that Gadd45β controls the amplitude of TLR-mediated autophagy.
Figure 8.
Gadd45β deficiency enhances LPS-induced autophagy in BMDMs. (a) BMDMs from WT and Gadd45β-deficient mice were stimulated in vitro with 100 ng ml−1 LPS for the indicated time periods. In order to prevent LC3-II degradation, half of the cells were treated with 100 nM bafilomycin A1 for the last 2 h of culturing. Subsequently, cellular lysates were prepared and analyzed by immunoblotting for expression of phospho-p38, total p38, Atg5 and LC3. β-actin served as a loading control. Results shown are representative for two independent experiments. (b) Schematic representation of the role of the Gadd45β-MEKK4-p38 pathway on Atg5 and autophagic flux. See discussion for details
Discussion
Although the pathways mediating autophagy have been elucidated in recent years, regulatory mechanisms modulating this process are poorly characterized. Most previous work focused on two kinase complexes, the class III PI3K complex and mTORC1, which are positive and negative regulators of autophagy, respectively.3 Both kinase complexes act in the very early phase of autophagy induction, but very little is known about how autophagy is regulated at later stages. Here, we show that phosphorylation of Atg5 blocks autophagosome maturation downstream of initiation processes. We furthermore identified p38 as the regulator that mediates phosphorylation of Atg5 and identified threonine-75 as its substrate amino acid. Importantly, threonine-75 is evolutionary conserved from yeast to men indicating the relevance of this post-translational modification. Interestingly, activation of the Gadd45β–MEKK4 pathway specifically and not other p38 stimuli such as ASK1 (another Gadd45β-interacting kinase) or UV irradiation induced p38 translocation to autophagosomes. In summary, we provide a detailed molecular mechanism implicating p38 in autophagy regulation downstream of mTOR and class III PI3K complexes by phosphorylation of Atg5.
At first glance, it is surprising that autophagosome numbers increase upon p38 activation, for example, by expression of Gadd45β and MEKK4, as well as upon p38 inactivation, that is, by pharmacological inhibition or gene knockout. How can these results be explained? An increase in autophagosome numbers can be detected either because fusion of autophagosomes with lysosomes is blocked, or because of a general increase in autophagy, which can lead to increased expression of Atg genes.19 Thus, the mere number of autophagosomes does not allow conclusions about enhanced or repressed autophagy. In addition, one has to keep in mind that next to stimulation-induced (e.g., starvation-induced) autophagy every cell has a certain level of basal autophagy.25 We show here that the expression of Gadd45β leads to p38-mediated phosphorylation of Atg5, which blocks autolysosome formation. In this situation, more autophagosomes can be detected as basal autophagy is inhibited. In contrast, in the p38-deficient situation, we detected more autophagosomes upon starvation, that is, stimulation-induced autophagy. In this setting, autophagic flux is enhanced as Atg5 cannot be phosphorylated by p38 and, thus, fusion of autophagosomes with lysosomes proceeds.
What could be the physiological role for autophagy regulation by Gadd45β and MEKK4? Gadd45β is induced upon different cellular stresses, inflammatory cytokines and LPS.10, 23 As LPS signals via TLR4, which was shown to induce autophagy,24 we investigated whether Gadd45β affects LPS-induced autophagy. Indeed, we show that autophagy was increased in the absence of Gadd45β suggesting that induction of Gadd45β by the LPS-TLR4 pathway acts as a negative feedback loop to dampen TLR-induced autophagy.
MAPKs, especially JNK1, have been implicated in autophagy regulation before.3 JNK1 phosphorylates Bcl-2 at serine and threonine residues in a N-terminal regulatory loop, releasing Beclin1 from Bcl-2 and initiating autophagy.26, 27 Thus, JNK1 affects an early step of autophagy while the Gadd45β–MEKK4-p38 pathway described here acts further downstream. Furthermore, the role of p38 in the regulation of autophagy is controversial, as both promotion and inhibition of autophagy by p38 have been reported.28, 29, 30, 31 However, none of these studies provided insights into the underlying molecular mechanisms.
Recently, p38-interacting protein (p38IP) was reported to interact with Atg9 and to be required for starvation-induced trafficking of Atg9.32 Active p38 sequesters p38IP away from Atg9 and, thus, indirectly impairs Atg9 trafficking and autophagosome formation.32 Unfortunately, the enzymatic activity of p38 was not addressed, suggesting that not kinase activity but competition between p38 and Atg9 for p38IP is crucial in this context. We show here an additional and direct role for p38 in autophagy regulation, involving its enzymatic activity. p38-mediated phosphorylation of Atg5 inhibits autophagy upstream of the fusion of autophagosomes with lysosomes. Importantly, the Atg5 conjugation system, which is also an essential component of the classical autophagy machinery, operates independently from Atg9.33 Another distinction between our study and the one performed by Webber and Tooze is the p38-activating stimulus. We show that p38 is specifically directed to autophagosomes upon activation of the Gadd45β–MEKK4 pathway. The Atg9–p38IP interaction was inhibited by UV or anisomycin treatment, both of which activate p38 and JNK via MLK7, a MAPK kinase kinase different from MEKK4,34 suggesting that these two regulatory pathways are activated by different stimuli. In conclusion, our work provides first insights into a novel role of Gadd45β in autophagy, showing for the first time a selective and spatial regulation of Atg5 activity through Gadd45β–MEKK4-mediated p38 activation, which exerts a negative regulatory role for autophagosome maturation.
Materials and Methods
Antibodies and reagents
The antibodies used for immunoblotting and immunoprecipitation were: Atg5 (AP1812a, Abgent, San Diego, CA, USA), Atg5 (D1G9), caspase-3, ERK, LC3 (G40), phospho-ERK, JNK, phospho-JNK, p38, phospho-p38, PARP (all from Cell Signaling, Danvers, MA, USA), LC3 (2G6; Nanotools, Teningen, Germany), MEKK4 (Sigma, Deisenhofen, Germany), Flag (M2; Sigma), HA (12CA5, Roche, Mannheim, Germany), c-MYC (9E10; Santa Cruz Biotechnology, Santa Cruz, CA, USA), V5 (Invitrogen, Carlsbad, CA, USA) and tubulin (Sigma). HRP-conjugated goat anti-rabbit IgG was obtained from Santa Cruz Biotechnology. HRP-conjugated goat anti-rat IgG, donkey anti-goat IgG, goat anti-mouse IgG1, IgG2a and IgG2b were acquired from SouthernBiotech (Birmingham, AL, USA). Bafilomycin A1, E64, LPS and pepstatin A were purchased from Sigma. Lysotracker was obtained from Invitrogen.
Cell culture
NIH/3T3 murine fibroblasts were cultured as described previously.35 NIH/3T3 cells stably expressing GFP-LC3 or RFP-GFP-LC3 were generated by retroviral transduction. MEFs deficient of p38α and their WT counterparts were kindly provided by Dr Angel Nebreda.36 Atg5-deficient and respective control MEFs were a kind gift of Dr Noboru Mizushima.5 Gadd45β-deficient MEFs and their WT counterparts were generated according to standard protocols and immortalized by transfection of a plasmid encoding the SV40 large T antigen. Gadd45β-deficient mice have been described before.14 All MEFs were cultured in Dulbecco's Modified Eagle Medium (DMEM high glucose, Invitrogen) supplemented with 10% fetal calf serum (PAA, Coelbe, Germany) and 50 μg ml−1 of each penicillin and streptomycin (Invitrogen). Cells were transfected with Lipofectamine 2000 (Invitrogen), Dreamfect (OZ Biogene, Marseille, France), JetPei (Peqlab, Erlangen, Germany) or Nanofectin (PAA) according to manufacturers' instructions. MEFs stably expressing Atg5 WT and mutants were selected using 1 mg ml−1 G418. BMDMs were differentiated out of bone marrow from femurs and tibias of either WT or Gadd45β-deficient mice, 8 to 10 weeks of age, according to published protocols.37 Briefly, bone marrow cells were cultured for 7 days in DMEM containing 10% fetal bovine serum (Sigma), 2 mM glutamine, 50 μg ml−1 of each penicillin and streptomycin and 5% macrophage colony-stimulating factor (M-CSF)-containing supernatant from 3T3 cells, with feeding on day 1, 3 and 5. On day 7, cells were detached from the surface with trypsin, put into new M-CSF-free medium and subsequently used for stimulation experiments. Successful differentiation was validated by F4/80 staining and flow cytometry.
Plasmids, recombinant protein expression and site-directed mutagenesis
Expression constructs for Gadd45 proteins,38 MEKK4,39 Atg5,6 Ask1,40 GFP-LC318 and RFP-GFP-LC320 have been described previously. A dominant-negative mutant of MEKK4 lacking the C-terminal kinase domain (MEKK4-ΔC) was cloned into pEF4-myc/his (Invitrogen) using the following primers: forward 5′-GGGGTACCATGAGAGACGCCATCGCC-3′ reverse 5′-GCTCTAGACTCCACTGGACGGTCGTCC-3′. Expression and purification of recombinant Atg5 was done as reported before.6 Atg5 mutants were generated by using the QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) and verified by DNA sequencing. Primer sequences are available upon request. Atg5 WT and mutants were cloned into either pET15b (Novagen, Darmstadt, Germany) for recombinant protein expression or into pEF-myc-cyto (Invitrogen) for eukaryotic cell transfection.
Fluorescence microscopy
4 × 104 NIH/3T3 cells grown on a coverslip in a 12-well dish were transiently transfected with 4 μg plasmid DNA using Optifect (Invitrogen) or JetPei (Peqlab) transfection reagent. After 24 h, cells were washed with phosphate-buffered saline (PBS) and fixed with 3.7% paraformaldehyde in PBS for 15 min. The cells were washed again and permeabilized with 0.05% saponin and 4% BSA in PBS for 30 min. Thereafter, cells were incubated with the appropriate antibody overnight in blocking solution (4% BSA and 0.05% saponin in PBS) at 4 °C. Coverslips were washed several times with PBS and incubated with Alexa-Fluor-conjugated secondary antibodies (Invitrogen) for 1 h at room temperature. For nuclear staining, the cells were washed several times and incubated with 4′,6′-diamidino-2-phenylindole dihydrochloride (100 ng ml−1) for 10 min. Coverslips were then mounted in fluorescence mounting medium (DakoCytomation, Hamburg, Germany) and analyzed in confocal laser scanning microscopes (LS2 and LS5, Leica Microsystems (Wetzlar, Germany), and LSM780, Zeiss (Jena, Germany)) or on a spinning disc microscope (Perkin Elmer, Waltham, MA, USA).
Transmission electron microscopy
For electron microscopy, MEFs were plated at a density of 2 × 106 cells in 175 cm2 cell culture flasks. After 24 h, the cells were starved in HBSS minimal medium or cultivated in supplemented DMEM (Invitrogen) and harvested 24 h later. Subsequently, cultured cells were harvested, washed in PBS, centrifuged for 5 min at 200 g and fixed in 2% glutaraldehyde/0.1 M sodium cacodylate buffer (pH 7.4) at room temperature for 1 h. Cells were washed twice in PBS and postfixed in 2% OsO4/PBS at room temperature for 1 h, dehydrated in ethanol and embedded in Epon (Serva, Heidelberg, Germany). Thin sections of 50 nm were contrasted with uranylacetate and lead citrate, and examined on a transmission electron microscope (Zeiss EM 902A, Zeiss).
Immunoblotting and immunoprecipitations
Immunoblotting was performed as described previously.35 For co-immunoprecipitation analysis, 2 × 106 NIH/3T3 cells were transiently transfected as described above. Cells were lysed in lysis buffer (20 mM Tris/HCl, pH 7.4, 1% Triton X-100, 10% glycerol, 150 mM NaCl, 1 mM PMSF and 1 μg ml−1 each of leupeptin, aprotinin, chymostatin and pepstatin A) for 15 min on ice and centrifuged (15 min, 20 000 g). Subsequently, the immunoprecipitation was performed with 2 μg antibody coupled to protein G beads (Sigma) for 4 h at 4 °C. Finally, beads were washed three times with 750-μl ice-cold lysis buffer and analyzed by immunoblotting.
In vitro kinase assay
The radioactive kinase assay cocktail contained 250 μCi ml−1 [γ-32P]ATP, 10 mM MgCl2, 5 mM MnCl2, 25 mM β-glycerophosphate, 25 mM HEPES pH 7.5, 5 mM benzamidine, 0.5 mM dithiothreitol and 1 mM Na2VO3. Two hundred nanograms of recombinant active p38 (ProQinase/Biomol) was incubated with 1 μg of recombinant substrate proteins for 1 h at 37 °C while shaking. The reactions were stopped by adding 5 × SDS-sample buffer (155 mM Tris-HCl pH 6.8, 5% SDS, 50% glycerol and 25% β-mercaptoethanol) and denaturing at 96 °C for 5 min. The samples were analyzed by 12% SDS-PAGE. The gel was transferred onto PVDF membrane, followed by autoradiography.
Acknowledgments
We thank Stephanie Grosch, Birgit Lamik-Wolters, Moritz Lemke, Daniel Scholtyssik and Sabrina Schumann for their expert technical assistance. We wish to thank Dr Melanie Brinkmann, Dr Maximiliano Gutierrez and Alisha Walker for critically reading the manuscript as well as Marion Nissen and Dr Heinz Mehlhorn for help with electron microscopy. We are grateful to Drs Pär Gerwins, Antje Gohla, Dietmar Kültz, Stephan Ludwig, Hans-Uwe Simon, Björn Stork, Osamu Takeuchi, Sebastian Wesselborg and Tamotsu Yoshimori for various plasmids and to Dr Ute Fischer for recombinant caspase-3. We thank Dr Angel Nebreda for p38-deficient MEFs and Dr Noboru Mizushima for Atg5-deficient MEFs. This work was supported by grants GK1033, GK1302, SFB685 and SFB773 (to KS-O), Leibniz (to KP) and SCHM1586/3-1 (to IS) of the Deutsche Forschungsgemeinschaft and by the Fritz-Thyssen-Stiftung (to IS).
Glossary
- ASK1
apoptosis signaling kinase 1
- Atg
autophagy-related
- BMDM
bone marrow-derived macrophage
- Gadd45β
growth arrest and DNA damage 45 beta
- GFP
green fluorescent protein
- JNK
c-Jun N-terminal kinase
- LC3
microtubule-associated protein 1 light chain 3
- LPS
lipopolysaccharide
- MAPK
mitogen-activated protein kinase
- M-CSF
macrophage colony-stimulating factor
- MEF
mouse embryonic fibroblast
- MEKK4
MAPK/ERK kinase kinase 4
- MKK
MAP kinase kinase
- PE
phosphatidylethanolamine
- RFP
red fluorescent protein
- TLR
Toll-like receptor
- WT
wild-type
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by H-U Simon
Supplementary Material
References
- Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Yousefi S, Perozzo R, Schmid I, Ziemiecki A, Schaffner T, Scapozza L, et al. Calpain-mediated cleavage of Atg5 switches autophagy to apoptosis. Nat Cell Biol. 2006;8:1124–1132. doi: 10.1038/ncb1482. [DOI] [PubMed] [Google Scholar]
- Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem. 2009;284:6322–6328. doi: 10.1074/jbc.M807135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Li TY, Liu Q, Zhang C, Li X, Chen Y, et al. GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science. 2012;336:477–481. doi: 10.1126/science.1217032. [DOI] [PubMed] [Google Scholar]
- Alers S, Loffler AS, Wesselborg S, Stork B. Role of AMPK-mTOR-Ulk1/2 in the regulation of autophagy: cross talk, shortcuts, and feedbacks. Mol Cell Biol. 2012;32:2–11. doi: 10.1128/MCB.06159-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebermann DA, Hoffman B. Myeloid differentiation (MyD) primary response genes in hematopoiesis. Oncogene. 2002;21:3391–3402. doi: 10.1038/sj.onc.1205312. [DOI] [PubMed] [Google Scholar]
- Papa S, Zazzeroni F, Bubici C, Jayawardena S, Alvarez K, Matsuda S, et al. Gadd45 beta mediates the NF-kappa B suppression of JNK signalling by targeting MKK7/JNKK2. Nat Cell Biol. 2004;6:146–153. doi: 10.1038/ncb1093. [DOI] [PubMed] [Google Scholar]
- Takekawa M, Saito H. A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell. 1998;95:521–530. doi: 10.1016/s0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]
- Lu B, Ferrandino AF, Flavell RA. Gadd45beta is important for perpetuating cognate and inflammatory signals in T cells. Nat Immunol. 2004;5:38–44. doi: 10.1038/ni1020. [DOI] [PubMed] [Google Scholar]
- Gupta M, Gupta SK, Balliet AG, Hollander MC, Fornace AJ, Hoffman B, et al. Hematopoietic cells from Gadd45a- and Gadd45b-deficient mice are sensitized to genotoxic-stress-induced apoptosis. Oncogene. 2005;24:7170–7179. doi: 10.1038/sj.onc.1208847. [DOI] [PubMed] [Google Scholar]
- Gupta SK, Gupta M, Hoffman B, Liebermann DA. Hematopoietic cells from gadd45a-deficient and gadd45b-deficient mice exhibit impaired stress responses to acute stimulation with cytokines, myeloablation and inflammation. Oncogene. 2006;25:5537–5546. doi: 10.1038/sj.onc.1209555. [DOI] [PubMed] [Google Scholar]
- Papa S, Zazzeroni F, Fu YX, Bubici C, Alvarez K, Dean K, et al. Gadd45beta promotes hepatocyte survival during liver regeneration in mice by modulating JNK signaling. J Clin Invest. 2008;118:1911–1923. doi: 10.1172/JCI33913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita H, Tsutsui J, Takekawa M, Witten EA, Saito H. Regulation of MTK1/MEKK4 kinase activity by its N-terminal autoinhibitory domain and GADD45 binding. Mol Cell Biol. 2002;22:4544–4555. doi: 10.1128/MCB.22.13.4544-4555.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Adams RH, Porras A, Alonso G, Jones M, Vintersten K, Panelli S, et al. Essential role of p38alpha MAP kinase in placental but not embryonic cardiovascular development. Mol Cell. 2000;6:109–116. [PubMed] [Google Scholar]
- Shaul YD, Seger R. The MEK/ERK cascade: from signaling specificity to diverse functions. Biochim Biophys Acta. 2007;1773:1213–1226. doi: 10.1016/j.bbamcr.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Zhang N, Ahsan MH, Zhu L, Sambucetti LC, Purchio AF, West DB. NF-kappaB and not the MAPK signaling pathway regulates GADD45beta expression during acute inflammation. J Biol Chem. 2005;280:21400–21408. doi: 10.1074/jbc.M411952200. [DOI] [PubMed] [Google Scholar]
- Xu Y, Jagannath C, Liu XD, Sharafkhaneh A, Kolodziejska KE, Eissa NT. Toll-like receptor 4 is a sensor for autophagy associated with innate immunity. Immunity. 2007;27:135–144. doi: 10.1016/j.immuni.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattingre S, Bauvy C, Carpentier S, Levade T, Levine B, Codogno P. Role of JNK1-dependent Bcl-2 phosphorylation in ceramide-induced macroautophagy. J Biol Chem. 2009;284:2719–2728. doi: 10.1074/jbc.M805920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell. 2008;30:678–688. doi: 10.1016/j.molcel.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comes F, Matrone A, Lastella P, Nico B, Susca FC, Bagnulo R, et al. A novel cell type-specific role of p38alpha in the control of autophagy and cell death in colorectal cancer cells. Cell Death Differ. 2007;14:693–702. doi: 10.1038/sj.cdd.4402076. [DOI] [PubMed] [Google Scholar]
- Haussinger D, Schliess F, Dombrowski F, Vom Dahl S. Involvement of p38MAPK in the regulation of proteolysis by liver cell hydration. Gastroenterology. 1999;116:921–935. doi: 10.1016/s0016-5085(99)70076-4. [DOI] [PubMed] [Google Scholar]
- Prick T, Thumm M, Kohrer K, Haussinger D, Vom Dahl S. In yeast, loss of Hog1 leads to osmosensitivity of autophagy. Biochem J. 2006;394:153–161. doi: 10.1042/BJ20051243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang G, Yue Z, Talloczy Z, Hagemann T, Cho W, Messing A, et al. Autophagy induced by Alexander disease-mutant GFAP accumulation is regulated by p38/MAPK and mTOR signaling pathways. Hum Mol Genet. 2008;17:1540–1555. doi: 10.1093/hmg/ddn042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber JL, Tooze SA. Coordinated regulation of autophagy by p38alpha MAPK through mAtg9 and p38IP. EMBO J. 2010;29:27–40. doi: 10.1038/emboj.2009.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci USA. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Mader MM, Toth JE, Yu X, Jin N, Campbell RM, et al. Complete inhibition of anisomycin and UV radiation but not cytokine induced JNK and p38 activation by an aryl-substituted dihydropyrrolopyrazole quinoline and mixed lineage kinase 7 small interfering RNA. J Biol Chem. 2005;280:19298–19305. doi: 10.1074/jbc.M413059200. [DOI] [PubMed] [Google Scholar]
- Ueffing N, Keil E, Freund C, Kuhne R, Schulze-Osthoff K, Schmitz I. Mutational analyses of c-FLIPR, the only murine short FLIP isoform, reveal requirements for DISC recruitment. Cell Death Differ. 2008;15:773–782. doi: 10.1038/sj.cdd.4402314. [DOI] [PubMed] [Google Scholar]
- Porras A, Zuluaga S, Black E, Valladares A, Alvarez AM, Ambrosino C, et al. P38 alpha mitogen-activated protein kinase sensitizes cells to apoptosis induced by different stimuli. Mol Biol Cell. 2004;15:922–933. doi: 10.1091/mbc.E03-08-0592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhirter SM, Barbalat R, Monroe KM, Fontana MF, Hyodo M, Joncker NT, et al. A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J Exp Med. 2009;206:1899–1911. doi: 10.1084/jem.20082874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak SK, Kultz D. Gadd45 proteins induce G2/M arrest and modulate apoptosis in kidney cells exposed to hyperosmotic stress. J Biol Chem. 2004;279:39075–39084. doi: 10.1074/jbc.M406643200. [DOI] [PubMed] [Google Scholar]
- Gerwins P, Blank JL, Johnson GL. Cloning of a novel mitogen-activated protein kinase kinase kinase, MEKK4, that selectively regulates the c-Jun amino terminal kinase pathway. J Biol Chem. 1997;272:8288–8295. doi: 10.1074/jbc.272.13.8288. [DOI] [PubMed] [Google Scholar]
- Ichijo H, Nishida E, Irie K, ten Dijke P, Saitoh M, Moriguchi T, et al. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science. 1997;275:90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.