Abstract
Protists (unicellular eukaryotes) arguably account for most eukaryotic diversity and are central players of the biosphere. Known protist diversity and biology is largely based on cultured strains. Yet, environmental molecular surveys have unveiled entirely novel lineages that, as their prokaryotic counterparts, are essentially uncultured. Culture bias is an important drawback for any microbe-related science and is particularly severe for heterotrophic protists, which depend on organic food sources for growth. Here, we show how ecologically significant bacterivorous protists have been brought into culture by mimicking in situ conditions. Single cells sorted by serial dilution or flow cytometry were inoculated into seawater amended with natural bacterial assemblage at nearly in situ abundances. Strains belonging to lineages only known so far from environmental sequencing were isolated. Among them, Minorisa minuta gen. nov. sp. nov. forms a novel branch within Rhizaria, holding a key evolutionary position, and with an average size of 1.4 μm represents one of the smallest bacterial grazers known to date. It has a worldwide planktonic distribution and can account for 5% of heterotrophic protists communities in coastal waters. Physiological features of this strain can partly explain its success in the environment. Culturing ecologically relevant but elusive protists provide invaluable material for ecophysiology, genomics, ecosystem modeling and evolutionary issues.
Keywords: culture, isolation, single cell, chlorarachnea, novelty, heterotrophic flagellates
Introduction
Unicellular organisms are major forces driving our planet ecosystems and are an outstanding reservoir of biological diversity (genes, molecules, metabolic pathways and cellular processes) yet to be discovered (Falkowski et al., 2008). They are also main actors in macro- and microevolutionary processes for life on Earth (Cavalier-Smith and Chao, 2006). Nowadays, culture independent approaches are regularly applied to investigate the diversity and function of microbes in the environment. Despite the valuable information provided by ‘omics' environmental studies, culture bias definitely remains as one of the most critical challenges faced by scientists aiming to achieve a full understanding of the ecological role of microbes (Giovannoni and Stingl, 2007) and is currently a bottleneck in ecosystem studies (Giovannoni et al., 2007; Raes and Bork, 2008). Environmental DNA surveys demonstrate the extent to which culturing efforts poorly capture in situ microbial diversity (Pedrós-Alió, 2006). Many lineages unveiled in the last few years and holding key phylogenetic positions to understand macroevolutionary patterns among eukaryotes are essentially composed of environmental sequences (Epstein and López-García, 2008). It is estimated that as little as 0.1% to 1% of bacterial and protist cells can be easily cultured (Caron et al., 1989; Amann et al., 1995). Unfortunately, some of the most highly represented taxa in the environment have not been cultured yet and some of those in culture do not appear to be among the most abundant (Massana et al., 2004; Sherr et al., 2007). Consequently, ultrastructural, physiological and genomic information for many ecologically relevant microorganisms is missing.
This culture bias can be overcome by using original culturing strategies (Button et al., 1993), as demonstrated for prokaryotes like Candidatus Pelagibacter ubique and Nitrosopumilus marinus (Rappé et al., 2002; Könneke et al., 2005), both initially detected through environmental molecular surveys and later identified as ecologically relevant taxa. Ca. P. ubique was brought into culture by mimicking oligotrophic conditions and Nitrosopumilus marinus were cultured in media amended with ammonia once molecular data revealed they were ammonia oxidizers. Similar culturing efforts have seldom been applied to marine protists, even though culture bias is perceived as a major limitation to investigate further the functional role and ecological significance of photosynthetic protists (Vaulot et al., 2008), being even more severe for the heterotrophic ones (Jürgens and Massana, 2008).
Marine heterotrophic flagellates perform key processes in microbial food webs as bacterial grazers, trophic linkers and nutrient remineralizers (Sherr and Sherr, 2002; Pernthaler, 2005). They exhibit a variety of trophic strategies and constitute a diverse assemblage of poorly identified species (Arndt et al., 2000; Vaulot et al., 2002). Up to now, bacterivorous flagellates have been invariably cultured using rich media composed of seawater supplemented with rice grains or yeast extract that promote the growth of very large bacteria at very-high densities. This strategy inexorably retrieves the same species complexes (that is, Cafeteria, Paraphysomonas, or Neobodo spp.) that are known to be rare in marine plankton (Jürgens and Massana, 2008). Abundant taxa identified by molecular surveys still remain uncultured, for example the bacterivorous MAST clades (Massana et al., 2006a).
In order to bring into culture ecologically relevant heterotrophic flagellates, we mimicked oligotrophic marine conditions by amending sterile seawater with a mix of natural bacteria collected from the same sampling site at abundances only slightly higher than in situ. To this media we added a single heterotrophic flagellate cell retrieved by dilution or by flow cytometry. This approach enabled us to obtain several interesting pre-cultures. Some of them were stabilized and maintained for months as stable cultures allowing its phylogenetic, morphological and ecological characterization.
Materials and methods
Isolation by single-cell dilution
Samples were collected at the Blanes Bay Microbial Observatory on the 25th of September 2007. Forty liters were filtered by gravity through 3 μm and incubated at 20 °C in the dark for 2 days. Then, this incubated sample was filtered by 0.45 μm and bacteria in the filtrate were concentrated by tangential flow filtration. Bacterial concentrates were quantified by epifluorescence microscopy using 4',6-diamidino-2-phenylindole (DAPI) staining (Porter et al., 1980) (typically ca 108 cells ml−1) and kept frozen. Multi-well plates of 0.1 or 1 ml were filled with aged seawater (3 μm filtered seawater kept in the dark for 2 months, 0.2 μm filtered and autoclaved before use), and an aliquot of the concentrate was added to attain a final concentration of 5 × 106 bacteria ml−1. Heterotrophic flagellates from the same 2-days incubated sample were counted by epifluorescence microscopy after DAPI staining and diluted to add one cell per well. Plates were incubated at 20 °C in the dark and inspected for flagellate growth by inverted microscopy every 3 days during a period of 3 weeks. Wells with observed growth were scaled to 30 ml to establish a pre-culture and later a stable culture.
Isolation by single-cell sorting
Seawater from a second sampling (30th of September 2008) was filtered through 3 μm, and sent to Bigelow Laboratory for Ocean Sciences (Boothbay Harbor, ME, USA) for cell sorting in a MoFlo Flow Cytometer (Dako-Cytomation, Glostrup, Denmark). Digestive vacuoles of heterotrophic protists were stained using the vital stain LysoTracker (Life Technologies, New York, NY, USA) and cells were sorted based on their green fluorescence (LysoTracker fluorescence) and the absence of chlorophyll fluorescence. Details of the staining protocol and flow cytometer setup are described elsewhere (Rose et al., 2004; Heywood et al., 2011). Side scatter was used to select only the smallest protists, approximately <10 μm in diameter. Individual target cells were deposited into 24-well plates, in which some wells were dedicated for positive controls (10 cells per well) and negative controls (0 cells per well). All wells on the microplates contained 1-ml aged seawater, together with natural bacteria as before. Multi-well plates were sent back to the Institut de Ciències del Mar (Barcelona, Catalonia, Spain) by plane at room temperature (12 h) and treated as above.
Culture maintenance
Cultures were maintained in 50 ml flasks and transferred every 2 to 4 weeks to fresh media (aged seawater with added bacteria as described above) at 1/10 dilution. The full culturing protocol is outlined in Supplementary Figure 1. Physiological parameters were inferred from a batch culture where the abundance of flagellates and bacteria were followed twice a day by epifluorescence microscopy after DAPI staining. Growth rate was calculated from the exponential increase of flagellate cells. Grazing rate was calculated using the exponential decrease of bacterial cells and the average concentration of flagellates and bacteria during the incubation (calculated according to Frost, 1972). Growth efficiency was calculated from the ratio of protist biomass produced as compared with bacterial biomass consumed.
Sequencing and phylogenetic analysis
For molecular analysis, the whole culture was filtered on 0.6 μm polycarbonate filters, DNA was extracted by standard procedures, and 18S rDNA genes were PCR amplified with eukaryote-specific primers (Díez et al., 2001). Complete sequences of 18S rDNA were obtained with five internal primers by the MACROGEN Genomics Sequencing Services (accession numbers JX272635-JX272636). Sequences were identified and inspected for chimeras by BLAST and KeyDNATools (http://www.keydnatools.com) and were aligned with a subset of relative sequences using MAFFT (Katoh et al., 2002). Alignments were checked with Seaview 3.2 (Galtier et al., 1996) and highly variable regions of the alignment were removed using Gblocks (Castresana, 2000). Maximum likelihood (ML) phylogenetic trees were constructed with the fast ML method RAxML (Stamatakis, 2006) using the evolutionary model GTRMIXI (which conducted inference under GRTCAT and calculated best tree under GTRGAMMA, but with estimate of proportion of invariable sites). Phylogenetic analyses were done in the freely available University of Oslo Bioportal (www.bioportal.uio.no). Repeated runs on distinct starting trees were carried out to select the tree with the best topology (the one having the best Likelihood of 1000 alternative trees). Bootstrap ML analysis was done with 1000 pseudo-replicates and the consensus tree was computed with MrBayes (Huelsenbeck and Ronquist, 2001).
Scanning electron microscopy (SEM)
SEM was performed as in Garcés et al. (2006). Samples were fixed for 2 h in 2% OsO4 diluted in seawater, or with 2% glutaraldehyde. Cells were subsequently washed with distilled water (2 h) and filtered onto a 0.6 μm pore size Nuclepore filter (Whatman, Maidstone, UK). Samples were dehydrated in an ethanol series (30%, 50%, 70%, 96 and 100%) for 15 min each, followed by an acetone series (25%, 50%, 75% and 100%) for 15 min each. Samples were critical point-dried in liquid CO2 using a BAL-TEC CPD 030 critical point drying apparatus (Balzers Union, Balzers, Germany). Filters were subsequently glued to SEM-stubs with colloidal silver, sputter coated with gold palladium, and examined with a Hitachi S-3500 N (Nissei Sangyo Co. Ltd., Tokyo, Japan) SEM operating at 5 kV.
Tyramide signal amplification-fluorescent in situ hybridization (TSA-FISH) analysis)
Samples were fixed with formaldehyde (2% final concentration), filtered on 0.6 μm pore size Nucleopore filters and kept frozen. The probe CRN02 (5′-TACTTAGCTCTCAGAACC-3′) was designed with the ARB package (Ludwig et al., 2004) to have no mismatches with the target sequences (coverage in Figure 1a) and at least two mismatches with nontarget sequences. The probe was purchased with a 5′-aminolink (C6) from Thermo Fisher Scientific (Waltham, MA, USA) and was labeled with the enzyme horseradish peroxidase, HRP (Roche Diagnostic Boehringer, Basel, Switzerland) as described elsewhere (Urdea et al., 1988; Amann et al., 1992). The hybridization was carried out on filter pieces using the temperatures and conditions described in Not et al. (2002). The signal amplification was done using a tyramide (as the substrate of HRP) labeled with Alexa 488 as in Pernthaler et al. (2004). The probe was tested against the positive culture in a gradient of formamide (10% to 70%) in the hybridization buffer. Optimized conditions were 35 °C and 30% formamide in the buffer. The probe gave a negative signal with several nontarget cultures. Filters were counterstained with DAPI, mounted in a slide with an oil mix containing Vecta Shield, and observed by epifluorescence microscopy at 1000 × under ultraviolet (DAPI signal) and blue light excitation (TSA-FISH signal). Two transects (of around 15 mm) were inspected per filter piece. Counts for Blanes samples were done triplicated (s.e. were typically 20% of the mean), whereas no replicates were done for the worldwide and culture samples. The minimal abundance quantified by this counting strategy was around one cell ml−1.
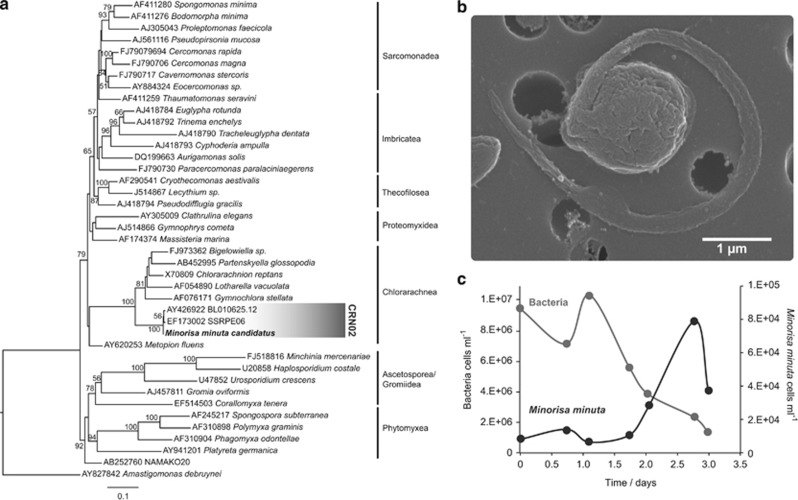
Figure 1.
(a) ML phylogenetic tree with complete 18S rDNA sequences showing the position of Minorisa minuta within the Cercozoa. The scale bar indicates 0.1 substitutions per position. The coverage of the specific probe CRN02 is shown in grey. (b) SEM image of a Minorisa minuta cell of 1.6 μm in size and possessing a single flagellum. (c) Growth of Minorisa minuta with natural bacteria as prey in a batch culture.
Results
Pre-cultures based on serial dilution yielded a 5.2% success rate (growth observed in 25 of the 480 inoculated wells). Out of the 25 pre-cultures, 12 were stable and were scaled to 30 ml volumes. Based on their 18 S rDNA, four pre-cultures were identified as Paraphysomonas spp. and the others represented taxa closer to environmental sequences than to any known culture, including a cercozoan, a rhizarian (two pre-cultures), a choanoflagellate, a chrysophyte, two unclassified stramenopiles, and a MAST (Table 1). After a second step of single cell inoculation from the latter eight flasks, some pre-cultures were lost and others were substituted by a different cell type. The two resulting unieukaryote clonal and stable cultures included a rhizaria related to chlorarachniophytes (Figure 1a) and a stramenopile distantly related to Developayella elegans (Supplementary Figure 2). Single-cell sorting by flow cytometry was carried out to avoid time-consuming serial dilution steps. From the 400 wells inoculated using single-cell sorting, growth was detected in five wells (1.25% success rate). All of them were identified as the same rhizarian lineage obtained by the dilution method. SEM performed on the two stable cultures revealed extremely small cells with little morphological features (Figure 1b and Supplementary Figure 3). Based on these SEM images we observed that rhizarian cells (40 cells measured) had one flagellum and measured 0.8–2 μm in width (mean of 1.3 μm, s.d. of 0.4) and 1.0–2.1 μm in length (mean of 1.5 μm, s.d.=0.4), whereas Developayella-like cells (13 cells measured) had two flagella (one with hairs) and measured 1.2–3 μm in width (mean of 2.1 μm, s.d.=0.6) and 1.5–3.0 μm in length (mean of 2.3 μm, s.d.=0.5).
Table 1. Taxonomic affiliation of the obtained pre-cultures and stable pure cultures based on blast similarity (March 2012).
| Closest cultured match | % | Lineage | Closest match | % | |
|---|---|---|---|---|---|
| Pre-cultures | |||||
| Rhizaria | |||||
| IE1.2.D2 | Chlorarachnion reptans | 90.5 | Chlorarachniophyte | SSRPE06 | 99.8 |
| IE1.4.B4 | Chlorarachnion reptans | 90.9 | Chlorarachniophyte | SSRPE06 | 99.9 |
| IE4.4.A1a | Cercozoa sp. CC-2009d | 94.4 | Cercozoan | SA2.4G8 | 96.8 |
| Opisthokonta | |||||
| IE1.4.D1 | Stephanoeca cauliculata | 91.6 | Choanoflagellate | Band 4DB38 | 97.7 |
| Stramenopiles | |||||
| IE1.1.A5 | Paraphysomonas foraminifera | 99.4 | Chrysophyte | ||
| IE1.3.B3 | Paraphysomonas imperforate | 99.4 | Chrysophyte | ||
| IE1.3.B6 | Developayella elegans | 94.4 | Stramenopile | RM2-SGM49 | 96.6 |
| IE1.4.D5b | Pseudobodo tremulans | 95.7 | Bicosoecida | Biosope_T123.046 | 95.7 |
| IE2.4.A6a | Chrysophyceae sp. CCMP2296 | 91.5 | Chrysophyte | CD8.S17 | 98.8 |
| IE3.4.B1 | Bolidomonas pacifica | 88.3 | MAST-3 | NIF.1E11 | 97.0 |
| IE4.6.B5 | Paraphysomonas imperforate | 98.7 | Chrysophyte | ||
| IE4.8.C6 | Paraphysomonas imperforate | 99.9 | Chrysophyte | ||
| Stable cultures | |||||
| Rhizaria | |||||
| Minorisa minutac | Chlorarachnion reptans | 99.6 | Chlorarachniophyte | SSRPE06 | 99.6 |
| Stramenopiles | |||||
| Developayella-like | Developayella elegans | 94.4 | Stramenopile | RM2-SGM49 | 96.6 |
Sequences with ambiguities.
Lost after 1 year of being stable.
Culture isolated by Flow Cytometry.
The rhizarian isolate was distant to any described organism, its 18 S rDNA sequence being only 90.6% similar to Chlorarachnion reptans. The 18 S rDNA sequence of our isolate was highly similar to environmental sequences retrieved from the Mediterranean Sea (BL000921.31 and BL010625.12; Massana et al., 2004), the Sargasso Sea (SSRPE06; Not et al., 2007), and the English Channel (RA070625T.047; Marie et al., 2010). Therefore, we decided to focus in the rhizarian isolate for the rest of our study, given its taxonomic novelty and its similarity with environmental marine sequences. This small heterotrophic flagellate has been named Minorisa minuta gen. nov. sp. nov. (see formal description at the end of this section).
Following Minorisa minuta abundance and that of the bacterial prey for several days in a batch culture has assessed its physiological properties. This flagellate grew relatively fast on natural bacteria (doubling time of 10.6 h) and reduced bacterial abundances from 107 cells ml−1 to around 106 cells ml−1 (Figure 1c). The estimated growth efficiency (bacterial biomass converted to protist biomass) of this protist in this batch culture was 30%, and its grazing rate was seven bacteria flagellate−1 h−1.
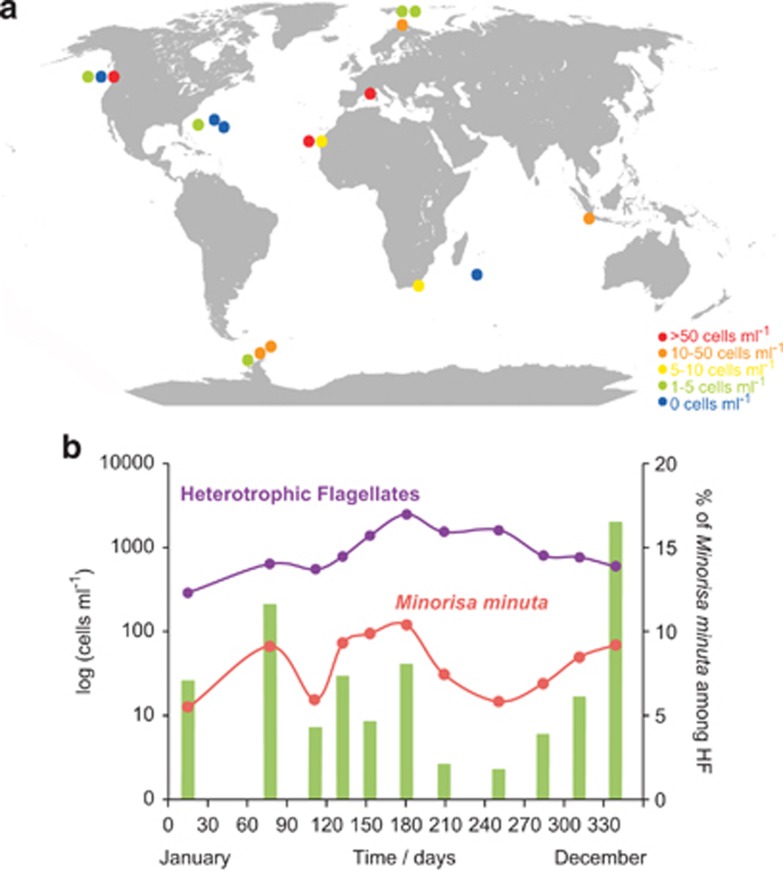
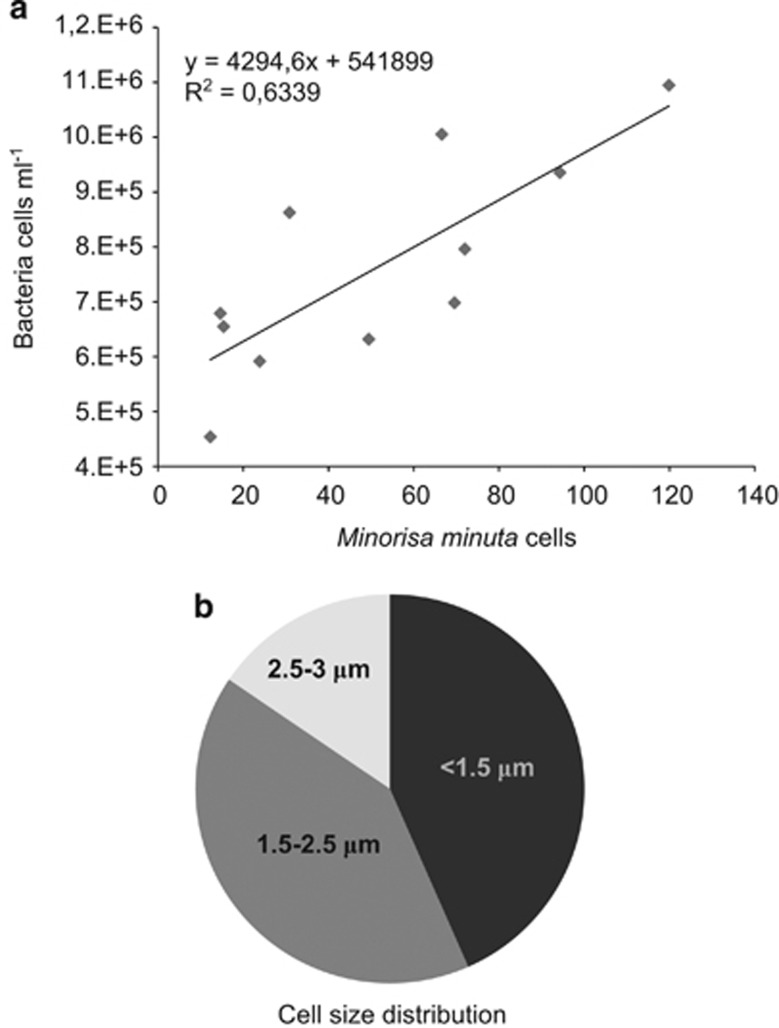
Minorisa minuta was a significant component of marine heterotrophic flagellates on a global scale, being both widely distributed and abundant (Figure 2a). TSA-FISH counts using a newly designed specific probe revealed abundances up to 60 cells ml−1 (17 cells ml−1 on average) in the Atlantic, Pacific, Indian and Southern Oceans and the Mediterranean Sea. It accounted for 1.8% of heterotrophic flagellates in these samples, a value that increased to 5% when considering coastal sites only (Table 2). Minorisa minuta was detected all year long in a coastal oligotrophic station in the NW Mediterranean Sea (Figure 2b), ranging from 12 to 120 cells ml−1 (52 cells ml−1 on average) and accounting for 1–12% of heterotrophic flagellates (5% on average). In this site, its abundance was well correlated with bacterial cells numbers (Figure 3a). Sizing Minorisa minuta cells in natural marine assemblages using TSA-FISH observations confirmed its picoeukaryotic character with cell size varying from 1 to 3 μm (Figure 3b).
Figure 2.
(a) Abundances of Minorisa minuta cells at various sampling sites as estimated by TSA-FISH counts. (b) Abundances of Minorisa minuta cells during a temporal study at Blanes Bay (year 2007), together with the abundance of heterotrophic flagellates, estimated by epifluorescence (log scale). Bars represent the contribution (in %) of Minorisa minuta cells to heterotrophic flagellates.
Table 2. Counts of Minorisa minuta (cells ml−1) in a set of samples from diverse oceans and comparison with the counts of heterotrophic flagellates (cells ml−1). Data of heterotrophic flagellates and codes of samples are from Massana et al. (2006a).
| System | Date | Minorisa minuta | Heterotrophic flagellates | %-Minorisa minuta |
|---|---|---|---|---|
| Atlantic ocean | ||||
| ATL1 | 24-Aug-02 | 4 | 652 | 0.7 |
| ATL2 | 27-Aug-02 | 9 | 1003 | 0.9 |
| ATL3 | 29-Aug-02 | 9 | 584 | 1.6 |
| ATL4 | 23-Sep-02 | 54 | 1302 | 4.1 |
| ATL5 | 30-Sep-02 | 13 | 814 | 1.6 |
| ATL6 | 12-Jul-04 | 9 | 214 | 4.1 |
| ATL7 | 15-Jul-04 | 0 | 443 | 0.0 |
| ATL8 | 30-Jul-04 | 0 | 593 | 0.0 |
| Pacific ocean | ||||
| PAC1 | 09-May-02 | 12 | – | – |
| PAC2 | 15-May-02 | 0 | – | – |
| PAC3 | 18-May-02 | 60 | – | – |
| Indian ocean | ||||
| INO1 | 16-May-03 | 33 | 687 | 4.9 |
| INO2 | 23-May-03 | 0 | 626 | 0.0 |
| INO3 | 06-Jun-03 | 5 | 719 | 0.7 |
| Southern ocean | ||||
| ANT1 | 03-Dec-02 | 7 | 1562 | 0.4 |
| ANT2 | 05-Dec-02 | 4 | 1668 | 0.3 |
| ANT3 | 11-Dec-02 | 2 | 2367 | 0.1 |
| Mediterranean sea | ||||
| BL07 | 2007a | 52 | 1039 | 5.0 |
Average of a whole-year sampling.
Figure 3.
(a) Correlation between the abundances of Minorisa minuta and natural bacteria during the seasonal study (year 2007) at Blanes Bay. (b) Cell size distribution of Minorisa minuta in these samples (1148 cells measured by TSA-FISH).
Minorisa minuta J. del Campo gen. nov. sp. nov.
Naked and spherical or ovoid cells: 0.8–2 μm (mean=1.3) in width and 1.0–2.1 μm (mean=1.5) in length. They possess a single flagella that could be as long as four times the cell length. They are swimmers and active bacterial grazers.
Holotype: Figure 1b from culture BMO.6.C1, isolated from Blanes Bay Microbial Observatory (NW Mediterranean Sea) and maintained at the Institut de Ciències del Mar (Barcelona). Scanning microscopy samples and FISH samples are also preserved there. 18S rDNA sequence deposited in GenBank under accession number: JX272635.
Etymology: Latin Minorisa, Manresa, author′s born town. Latin minuta, tiny, refers to the small size of the organisms.
Type locality: Blanes Bay (Catalonia, Spain).
Habitat and ecology: In Blanes Bay, the organism was detected in coastal plankton all year long. It has been also detected in different world oceans, but the maximum abundances have always been observed in coastal areas.
Discussion
Using the culturing approach developed here, we isolated several small heterotrophic flagellates belonging to previously uncultured taxa and from distant lineages within the eukaryotic tree of life (Table 1). When applied at different temporal and spatial scales, this strategy will potentially give access to a wealth of novel heterotrophic protists in culture. This is important because the presence of heterotrophic flagellates in culture collections is scarce (Gachon et al., 2007), and among the ones present in culture collections there are few with a significant representation in the environment (Lim et al., 1999).
The tiny uniflagellated Minorisa minuta stands up as one of the smallest bacterivore known so far, together with the minuscule phagotrophic bicosoecid Symbiomonas scintillans isolated from open oligotrophic waters (Guillou et al., 1999). Moreover, it represents the only heterotrophic representative within the chlorarachniophytes, the single photosynthetic lineage within rhizaria. Chlorarachnyophytes are marine amoeboflagellated cercozoans (Cavalier-Smith and Chao, 2003) that together with the Euglenophytes represent the two unrelated eukaryotic groups that acquired a chloroplast by secondary endosymbiosis with a green alga (Delwiche, 1999). The discovery of this unpigmented protist could be another evidence indicating that the acquisition of the green chloroplast took place independently in both lineages (Rogers et al., 2007). Nevertheless, this is not a definitive clue as the absence of chloroplast could also derive from a posterior loss of it. Therefore, the genome analysis of Minorisa minuta is of primary interest to study the transition to secondary plastid endosymbiosis and, as for its photosynthetic counterparts in the oceans, is expected to reveal unprecedented cellular, biochemical and evolutionary pathways (Worden et al., 2009).
We have shown here that Minorisa minuta has a worldwide marine distribution and is a significant member of heterotrophic flagellate assemblages, particularly in coastal waters. In the same set of samples considered for the present study, Minorisa minuta followed in abundance the uncultured MAST-4, MAST-1C and MAST-1B groups (Massana et al., 2006a), which is remarkable given that its probe is clearly more specific than MAST probes (roughly species versus family level). Our data suggest that the importance of this organism in coastal waters is comparable to the importance of MAST in open waters, considered as abundant bacterivores in the sea. Minorisa minuta represents one of the main players in the eukaryotic picoplankton of coastal ecosystems, possibly having a relevant role in the carbon fluxes and in controlling bacterial populations. Nevertheless, many more samples need to be processed in order to verify the patterns of its abundance, distribution and coastal preference. Its growth rate is double of that observed in MAST in unamended incubations (Massana et al., 2006b), and similar to or lower than maximal growth rates observed for other cultured heterotrophic flagellates (Eccleston-Parry and Leadbeater, 1994, Boenigk et al., 2007). The estimated growth efficiency and its grazing rate are again within the range of known cultured strains. The functional response of Minuta minuta exhibits a half-saturation constant much lower than that of other cultured flagellates, below 106 cells ml−1 (Rodríguez-Martínez in preparation). This suggests that this grazer is adapted to live at the usual bacterioplankton concentrations in marine waters. The physiological properties of Minorisa minuta can explain its ecological success and set this species as a good example of dominant marine heterotrophic flagellates (Montagnes et al., 2012), whose parameters could be used to improve ecological models (Caron et al., 2009). New experiments must be performed in order to determine its functional behavior and the degree of its influence over the coastal microbial community.
This is the first time that a widely distributed and abundant heterotrophic flagellate has been obtained in culture. Getting the environmentally relevant bacteria Ca. P. ubique in culture led to a leap forward toward a better understating of microbes′ function in the oceans and opened up several research directions (Tripp et al., 2008). Taming small marine predators with ecological relevance holds promise for similar future discoveries.
Acknowledgments
This study was supported by projects MICROVIS (CTM2007–62140/MAR, MEC), FLAME (CGL2010–16304, MICINN) and BioMarKs (2008–6530, ERA-net Biodiversa, EU). JdC was funded by the I3P program (I3PPRE-06–00676, CSIC) and FN by the Marie-Curie fellowship ESUMAST (MEIF-CT-2005–025000). We thank Marco Álvarez for help in TSA-FISH, Vanessa Balagué for help in molecular analysis, José Manuel Fortuño for help in SEM and Nicole Poulton for help in flow cytometry.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Amann RI, Zarda B, Stahl DA, Schleifer KH. Identification of individual prokaryotic cells by using enzyme-labeled, rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1992;58:3007–3011. doi: 10.1128/aem.58.9.3007-3011.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt H, Dietrich D, Auer B, Cleven EJ, Gräfenhan T, Weitere M, et al. 2000Functional diversity of heterotrophic flagellates in aquatic ecosystemsIn: Leadbeater BSC, Green JC, (eds).The Flagellates: Unity, Diversity and Evolution Taylor & Francis Group: London, UK; 240–268. [Google Scholar]
- Boenigk J, Jost S, Stoeck T, Garstecki T. Differential thermal adaptation of clonal strains of a protist morphospecies originating from different climatic zones. Environ Microbiol. 2007;9:593–602. doi: 10.1111/j.1462-2920.2006.01175.x. [DOI] [PubMed] [Google Scholar]
- Button DK, Schut F, Quang P, Martin R, Robertson BR. Viability and isolation of marine bacteria by dilution culture: theory, procedures and initial results. Appl Environ Microbiol. 1993;59:881–891. doi: 10.1128/aem.59.3.881-891.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron DA, Davis PG, Sieburth JMcN. Factors responsible for the differences in cultural estimates and direct microscopical counts of populations of bacterivorous microflagellates. Microb Ecol. 1989;18:89–104. doi: 10.1007/BF02030118. [DOI] [PubMed] [Google Scholar]
- Caron DA, Worden AZ, Countway PD, Demir E, Heidelberg KB. Protists are microbes too: a perspective. ISME J. 2009;3:4–12. doi: 10.1038/ismej.2008.101. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EEY. Phylogeny and classification of phylum cercozoa (Protozoa) Protist. 2003;154:341–358. doi: 10.1078/143446103322454112. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T, Chao EEY. Phylogeny and megasystematics of phagotrophic heterokonts (Kingdom Chromista) J Mol Evol. 2006;62:388–420. doi: 10.1007/s00239-004-0353-8. [DOI] [PubMed] [Google Scholar]
- del Campo J, Massana R. Emerging diversity within chrysophytes, choanoflagellates and bicosoecids based on molecular surveys. Protist. 2011;162:435–448. doi: 10.1016/j.protis.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Delwiche C. Tracing the thread of plastid diversity through the tapestry of life. Am Nat. 1999;154:S164–S177. doi: 10.1086/303291. [DOI] [PubMed] [Google Scholar]
- Díez B, Pedrós-Alió C, Massana R. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl Environ Microbiol. 2001;67:2932–2941. doi: 10.1128/AEM.67.7.2932-2941.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccleston-Parry JD, Leadbeater BSC. A comparison of the growth kinetics of six marine heterotrophic nanoflagellates fed with one bacterial species. Mar Ecol Prog Ser. 1994;105:167–177. [Google Scholar]
- Epstein S, López-García P. “Missing” protists: a molecular prospective. Biodivers Conserv. 2008;8:27–42. [Google Scholar]
- Falkowski PG, Fenchel T, Delong EF. The microbial engines that drive Earth's biogeochemical cycles. Science. 2008;320:1034–1039. doi: 10.1126/science.1153213. [DOI] [PubMed] [Google Scholar]
- Frost BW. Effects of size and concentration of food particles on the feeding behavior of the marine planktonic copepod Calanus pacificus. Limnol Oceanogr. 1972;17:805–815. [Google Scholar]
- Gachon CMM, Day JG, Campbell CN, Pröschold T, Saxon RJ, Küpper FC. The Culture Collection of Algae and Protozoa (CCAP): a biological resource for protistan genomics. Gene. 2007;406:51–57. doi: 10.1016/j.gene.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Garcés E, Fernandez M, Penna A, Van Lenning K, Gutierrez A, Camp J. Characterization of NW Mediterranean Karlodinium spp. (Dinophyceae) strains using morphological, molecular, chemical, and physiological methodologies. J Phycol. 2006;42:1096–1112. [Google Scholar]
- Giovannoni SJ, Foster RA, Rappé MS, Epstein S. New cultivation strategies bring more microbial plankton species into the laboratory. Oceanography. 2007;20:62–69. [Google Scholar]
- Giovannoni SJ, Stingl U. The importance of culturing bacterioplankton in the ‘omics' age. Nature Rev Microbiol. 2007;5:820–826. doi: 10.1038/nrmicro1752. [DOI] [PubMed] [Google Scholar]
- Guillou L, Chrétiennot-Dinet MJ, Boulben S, Moon-van der Staay SY, Vaulot D. Symbiomonas scintillans gen. et sp. nov. and Picophagus flagellatus gen. et sp. nov. (Heterokonta): two new heterotrophic flagellates of picoplanktonic size. Protist. 1999;150:383–398. doi: 10.1016/S1434-4610(99)70040-4. [DOI] [PubMed] [Google Scholar]
- Heywood JL, Sieracki ME, Bellows W, Poulton NJ, Stepanauskas R. Capturing diversity of marine heterotrophic protists: one cell at a time. ISME J. 2011;5:674–684. doi: 10.1038/ismej.2010.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinfomatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Jürgens K, Massana R.2008Protistan grazing on marine bacterioplanktonIn: Kirchman DL, (ed.)Microbial Ecology of the Oceans2nd EditionJohn Wiley & Sons, Inc.: New York, USA; 383–441. [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Könneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- Lim EL, Dennet MR, Caron DA. The ecology of Paraphysomonas imperforata based on studies employing oligonucleotide probe identification in coastal water samples ans enrichment cultures. Limnol Oceanogr. 1999;44:37–51. [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1671. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie D, Shi XL, Rigaut-Jalabert F, Vaulot D. Use of flow cytometric sorting to better assess the diversity of small photosynthetic eukaryotes in the English Channel. FEMS Microbiol Ecol. 2010;72:165–178. doi: 10.1111/j.1574-6941.2010.00842.x. [DOI] [PubMed] [Google Scholar]
- Massana R, Balagué V, Guillou L, Pedrós-Alió C. Picoeukaryotic diversity in an oligotrophic coastal site studied by molecular and culturing approaches. FEMS Microbiol Ecol. 2004;50:231–243. doi: 10.1016/j.femsec.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Massana R, Terrado R, Forn I, Lovejoy C, Pedrós-Alió C. Distribution and abundance of uncultured heterotrophic flagellates in the world oceans. Environ Microbiol. 2006a;8:1515–1522. doi: 10.1111/j.1462-2920.2006.01042.x. [DOI] [PubMed] [Google Scholar]
- Massana R, Guillou L, Terrado R, Forn I, Pedrós-Alió C. Growth of uncultured heterotrophic flagellates in unamended seawater incubations. Aquat Microb Ecol. 2006b;45:171–180. [Google Scholar]
- Montagnes D, Roberts E, Lukeš J, Lowe C. The rise of model protozoa. Trends Microbiol. 2012;20:184–191. doi: 10.1016/j.tim.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Not F, Simon N, Biegala IC, Vaulot D. Application of fluorescent in situ hybridization coupled with tyramide signal amplification (FISH-TSA) to assess eukaryotic picoplankton composition. Aquat Microb Ecol. 2002;28:157–166. [Google Scholar]
- Not F, Gausling R, Azam F, Heidelberg JF, Worden AZ. Vertical distribution of picoeukaryotic diversity in the Sargasso Sea. Environ Microbiol. 2007;9:1233–1252. doi: 10.1111/j.1462-2920.2007.01247.x. [DOI] [PubMed] [Google Scholar]
- Pedrós-Alió C. Marine microbial diversity: can it be determined. Trends Microbiol. 2006;14:257–263. doi: 10.1016/j.tim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Pernthaler A, Pernthaler J, Amann R.2004Sensitive multicolor fluorescence in situ hybridization for the identification of environmental microorganismsIn: Akkermans ADL, de Bruijn FJ, van Elsas JD, (eds).Molecular Microbial Ecology Manual2nd EditionKluwer Academic Publishers: Dordrecht, Netherlands; 711–726. [Google Scholar]
- Pernthaler J. Predation on prokaryotes in the water column and its ecological implications. Nature Rev Microbiol. 2005;3:537–546. doi: 10.1038/nrmicro1180. [DOI] [PubMed] [Google Scholar]
- Porter KG, Feig YS. The use of DAPI for identifying and counting aquatic microflora. Limnol Oceanogr. 1980;25:943–948. [Google Scholar]
- Raes J, Bork P. Molecular eco-systems biology: towards an understanding of community function. Nature Rev Microbiol. 2008;6:683–699. doi: 10.1038/nrmicro1935. [DOI] [PubMed] [Google Scholar]
- Rappé MS, Connon SA, Vergin KL, Giovannoni SJ. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature. 2002;418:630–633. doi: 10.1038/nature00917. [DOI] [PubMed] [Google Scholar]
- Rogers MB, Gilson PR, Su V, McFadden GI, Keeling PJ. The complete chloroplast genome of the chlorarachniophyte Bigelowiella natans: evidence for independent origins of chlorarachniophyte and euglenid secondary endosymbionts. Mol Biol Evol. 2007;24:54–62. doi: 10.1093/molbev/msl129. [DOI] [PubMed] [Google Scholar]
- Rose JM, Caron DA, Sieracki ME, Poulton NJ. Counting heterotrophic nanoplanktonic protists in cultures and aquatic communities by flow cytometry. Aquat Microb Ecol. 2004;34:263–277. [Google Scholar]
- Sherr EB, Sherr BF. Significance of predation by protists in aquatic microbial food webs. Anton van Leeuwen. 2002;81:293–308. doi: 10.1023/a:1020591307260. [DOI] [PubMed] [Google Scholar]
- Sherr BF, Sherr EB, Caron DA, Vaulot D. Oceanic protists. Oceanography. 2007;20:130–134. [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Tripp HJ, Kitner JB, Schwalbach MS, Dacey JWH, Wilhelm LJ, Giovannoni SJ. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature. 2008;452:741–744. doi: 10.1038/nature06776. [DOI] [PubMed] [Google Scholar]
- Urdea MS, Warner BD, Running JA, Stempien M, Clyne J, Horn T. A comparison of non-radioisotopic hybridization assay methods using fluorescent, chemiluminescent, and enzyme labeled oligodeoxyribonucleotide probes. Nucleic Acids Res. 1988;16:4937–4956. doi: 10.1093/nar/16.11.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaulot D, Romari K, Not F. Are autotrophs less diverse than heterotrophs in marine picoplankton. Trends Microbiol. 2002;10:266–267. doi: 10.1016/s0966-842x(02)02366-1. [DOI] [PubMed] [Google Scholar]
- Vaulot D, Eikrem W, Viprey M, Moreau H. The diversity of small eukaryotic phytoplankton (⩽ 3 μm) in marine ecosystems. FEMS Microbiol Ecol. 2008;32:795–820. doi: 10.1111/j.1574-6976.2008.00121.x. [DOI] [PubMed] [Google Scholar]
- Worden AZ, Lee JH, Mock T, Rouzé P, Simmons MP, Aerts AL, et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes. Micromonas. Science. 2009;324:268–272. doi: 10.1126/science.1167222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.