Abstract
The possibility of arsenic release and the potential role of Geobacter in arsenic biogeochemistry during in situ uranium bioremediation was investigated because increased availability of organic matter has been associated with substantial releases of arsenic in other subsurface environments. In a field experiment conducted at the Rifle, CO study site, groundwater arsenic concentrations increased when acetate was added. The number of transcripts from arrA, which codes for the α-subunit of dissimilatory As(V) reductase, and acr3, which codes for the arsenic pump protein Acr3, were determined with quantitative reverse transcription-PCR. Most of the arrA (>60%) and acr3-1 (>90%) sequences that were recovered were most similar to Geobacter species, while the majority of acr3-2 (>50%) sequences were most closely related to Rhodoferax ferrireducens. Analysis of transcript abundance demonstrated that transcription of acr3-1 by the subsurface Geobacter community was correlated with arsenic concentrations in the groundwater. In contrast, Geobacter arrA transcript numbers lagged behind the major arsenic release and remained high even after arsenic concentrations declined. This suggested that factors other than As(V) availability regulated the transcription of arrA in situ, even though the presence of As(V) increased the transcription of arrA in cultures of Geobacter lovleyi, which was capable of As(V) reduction. These results demonstrate that subsurface Geobacter species can tightly regulate their physiological response to changes in groundwater arsenic concentrations. The transcriptomic approach developed here should be useful for the study of a diversity of other environments in which Geobacter species are considered to have an important influence on arsenic biogeochemistry.
Keywords: Geobacter, arsenic respiration, arsenic resistance, gene transcription, uranium bioremediation
Introduction
The addition of organic electron donors to promote microbial reduction of soluble U(VI) to less soluble (IV) has shown promise as a strategy to prevent the spread of uranium-contaminated groundwater (Finneran et al., 2002; Holmes et al., 2002; Anderson et al., 2003; Istok et al., 2004; Vrionis et al., 2005; Wall and Krumholz, 2006; Wu et al., 2006, 2007; Luo et al., 2007; Shelobolina et al., 2008; Junier et al., 2010; Prakash et al., 2010; Williams et al., 2011). However, U(VI) reduction is accompanied by significant microbial reduction of Fe(III) oxides (Anderson et al., 2003; Vrionis et al., 2005), and possibly other electron acceptors, that might have undesirable consequences. For example, when cultured in sediments from a uranium-contaminated aquifer, the subsurface isolate G. uraniireducens increased the transcription of genes indicative of heavy metal stress, compared with growth in defined medium (Holmes et al., 2009). This might have been a response to the release of trace metals previously adsorbed onto Fe(III) oxides as the sediment Fe(III) oxides were reduced. G. uraniireducens also upregulated the transcription of a gene encoding an arsenite efflux pump, suggesting a need to respond to increased arsenic levels (Holmes et al., 2009).
Elevated concentrations of arsenic have been noted in some subsurface environments in which organic matter has been introduced (Dowdle et al., 1996; Ahmann et al., 1997; Rowland et al., 2004, 2007; Islam et al., 2004, 2005a; Lear et al., 2007; Héry et al., 2008, 2010). In some instances, these elevated arsenic concentrations could be the result of microbial reduction of less soluble As(V) to more soluble As(III) (Dowdle et al., 1996; Ahmann et al., 1997; Zobrist, 2000; Mukhopadhyay et al., 2002; Oremland and Stolz, 2003, 2005; Stolz et al., 2006). Alternatively, As(V) adsorbed to Fe(III) oxides can be released into solution when Fe(III) oxides are reduced (Devitre et al., 1991; Dixit and Hering, 2003; Campbell et al., 2005; Pedersen et al., 2006), or Fe(II) resulting from Fe(III) reduction can abiotically reduce As(V) (Islam et al., 2004; Tufano and Fendorf, 2008a, 2008b; Amstaetter et al., 2010).
A wide phylogenetic diversity of microorganisms can utilize As(V) as an electron acceptor to support anaerobic growth (Newman et al., 1997a; Oremland et al., 2000; Oremland and Stolz, 2005; Hollibaugh et al., 2006; Stolz et al., 2006). The gene arrA, which encodes the α-subunit of the dissimilatory arsenate reductase protein (ArrA), has been used to document the presence of dissimilatory As(V)-reducing bacteria in a diversity of subsurface environments (Kulp et al., 2004, 2006; Malasarn et al., 2004; Hollibaugh et al., 2006; Lear et al., 2007; Pederick et al., 2007). There are a number of previously described dissimilatory As(V)-reducing bacteria, such as Shewanella trabarsenatis ANA3 (Saltikov et al., 2003a; Saltikov and Newman, 2003b) and Sulfurospirillum barnesii, Sulfurospirillum arsenophilus (Stolz et al., 1999), Alkaliphilus oremlandii (Fisher et al., 2008), Bacillus arseniciselenatis (Blum et al., 1998), a Desulfosporosinus species (Perez–Jimenez et al., 2005) and Desulfotomaculum auripigmentum (Newman et al., 1997a, 1997b). The presence of genes for As(V) respiration in several Geobacter genomes suggests that certain Geobacter species might also have this ability (Lear et al., 2007; Duval et al., 2008), and preliminary studies have shown that G. uraniireducens, an organism isolated from the Rifle site, can grow with As(V) provided as an electron acceptor (Héry et al., 2008).
The cells of many bacteria and archaea have arsenic resistance systems that pump toxic arsenite (As(III)) out of the cell. The efflux pumps, ArsB or Acr3, are the most critical of these proteins, as the presence of these transporter proteins alone is sufficient to provide resistance to As(III) (Rosen, 1999, 2002; Oremland and Stolz, 2003). ArsB and Acr3 are unrelated families of arsenic transporters and homologs of each type are widespread throughout bacteria, archaea and fungi (Rosen, 1999; Mukhopadhyay et al., 2002). Although Acr3 is less well characterized, it is present in more phylogenetically distant species than ArsB (Rosen, 1999; Achour et al., 2007; Cai et al., 2009). On the basis of the phylogenetic dissimilarities, Acr3 members can be divided in two subfamilies, Acr3(1) and Acr3(2) (Rosen, 1999; Wysocki et al., 2003; Achour et al., 2007). The presence of arsenic resistance genes can serve as biomarkers of arsenic contamination in environmental samples (Anderson and Cook, 2004; Ford et al., 2005; Jackson et al., 2005; Cai et al., 2009).
More important than documenting the presence of a gene in the environment is quantifying the degree to which the gene is transcribed. Monitoring gene transcript abundance in subsurface microbial populations during in situ uranium bioremediation has been a useful strategy for documenting the physiological status of the microorganisms influencing groundwater quality (Holmes et al., 2004a, 2005, 2009; O'Neil et al., 2008; Mouser et al., 2009a, 2009b; N'Guessan et al., 2010). Here we report on the potential for release of arsenic during in situ uranium bioremediation and the transcription of genes related to groundwater arsenic biogeochemistry.
Materials and methods
Subsurface site and field experiment description
During August–October 2010, a study on bioremediation of uranium-contaminated groundwater was conducted at the Department of Energy's (DOEs) Integrated Field Research Challenge (IFRC) site near Rifle, CO, USA (Anderson et al., 2003; Vrionis et al., 2005; Wilkins et al., 2010; Williams et al., 2011). The aquifer is an ∼6.5-m thick heterogeneous alluvial deposit consisting of unconsolidated clay, silt, sand, gravel and cobbles lying on weathered claystone of the Wasatch formation. The groundwater table is ∼3.5 m below surface, and the flow is toward the Colorado river. The experimental plot comprised of a six injection wells gallery, nine downgradient monitoring wells and one background monitoring well (Supplementary Figure S1 in Supplementary material). As previously described (Williams et al., 2011), an acetate–bromide solution (50/20 mℳ) was prepared by mixing native groundwater pumped from an upgradient portion of the aquifer into a storage tank with sodium acetate (Sigma, St Louis, MO, USA) and sodium bromide (Sigma). This mixture was added to the subsurface via the injection wells to achieve target aquifer concentrations of 5 mℳ over the course of 30 days, as previously described (Anderson et al., 2003; Williams et al., 2011).
Groundwater sampling and geochemical analysis
Groundwater samples for chemical and molecular analyses were taken from wells CD01 and CD04 (Supplementary Figure S1). Samples for geochemical analyses were collected after purging 12 liters of groundwater from the wells using a peristaltic pump. Ferrous iron was measured spectrophotometrically, immediately after sampling using the phenanthroline method (AccuVac ampules, Hach Company, Loveland, CO, USA) for ferrous iron. After filtration through a 0.2-μm pore-size polytetrafluoroethylene ((Teflon)) filter (Alltech Associates Inc., Deerfield, IL, USA), acetate concentrations were measured using a Dionex ICS-1000 ion chromatograph equipped with a IonPac AS22 column, an ASRS 300 suppressor, and 4.5 mℳ carbonate–1.4 mℳ bicarbonate eluent (Dionex Corporation, Sunnyvale, CA, USA). Dissolved oxygen (DO) values were obtained using a luminescent oxygen sensor (YSI Inc., Yellow Springs, OH, USA).
Samples for arsenic analysis were filtered (polytetrafluoroethylene; 0.45 μm) and preserved with trace metal grade 12 𝒩 HNO3, and concentrations were determined using ion-coupled plasma mass spectrometry (Elan DRCII ICP-MS, PerkinElmer, CA, USA). In the 2010 field experiment, groundwater samples were not properly preserved for speciation analysis. However, samples from the same experimental plot taken in 2011 for arsenic speciation analysis showed that ∼95% of the total arsenic in the background wells (CU01) was arsenate (Stucker et al., submitted). It is likely that arsenate concentrations in the background well (CU01) in 2010 were similar to 2011, as other geochemical measurements were comparable.
Groundwater samples for molecular analyses were obtained after sampling for geochemical analyses by concentrating 40 liters of groundwater on a 0.2-μm pore size, 293-mm-diameter Supor-200 membrane filter (Pall Life Sciences, Ann Arbor, MI, USA). Filters were quickly sealed into a sterile whirl pack, flash frozen in an ethanol-dry ice bath, and stored at −80 °C until nucleic acid extraction.
Preparation of G. lovleyi resting cell suspensions and measurement of arsenate
G. lovleyi SZ was cultured in an anaerobic basal NB medium (Bond and Lovley, 2003) containing 20 mℳ acetate as electron donor and 20 mℳ fumarate as electron acceptor. The previously published protocol (Shelobolina et al., 2007) was used for preparing resting cells and performing cell suspension experiments. Cells were harvested during late exponential phase, washed twice with NB medium and resuspended to an OD600 between 0.15 and 0.20 in NB medium containing acetate (20 mℳ), and As(V) (5 mℳ). A heat-killed control was prepared by autoclaving the cell suspension for 30 min before the addition of As(V). A control without acetate was also performed. The concentration of arsenate was monitored with a Dionex ICS-1000 ion chromatograph, fitted with a 4-mm ion exchange column (AS22-SC) and AG22-SC guard column (Dionex Corporation). Samples were eluted in 4 mℳ Na2CO3/1 mℳ NaHCO3 (flow rate 1.2 ml min−1; pressure 1.38610 Pa). Anions were detected by suppressed conductivity detection.
DNA, RNA extraction and reverse transcription
Genomic DNA and total RNA from G. lovleyi grown in NB medium containing acetate and fumarate, or acetate and fumarate+arsenate were extracted, as previously described (Miletto et al., 2011). RNA from environmental samples was extracted using a modified phenol–chloroform method, as previously described (Holmes et al., 2004a). RNA cleanup was performed using an RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany), and RNA was treated with DNase (DNA-free Kit, Ambion, Austin, TX, USA). Successful RNA isolation was checked by visualization on a 1% (w/v) agarose electrophoresis gel in 1 × TBE buffer. The absence of DNA contamination was confirmed by PCR amplification. RNA was quantified using a NanoDrop spectrophotometer (Thermo SCIENTIFIC, Wilmington, DE, USA) and stored at −80 °C until further analyses. An Enhanced Avian HS reverse transcription-PCR kit (Sigma) was used to generate cDNA as previously described (Miletto et al., 2011) cDNA was quantified using a NanoDrop spectrophotometer and stored at −80 °C until further analyses.
PCR regimens, clone library construction and phylogenetic analysis
PCRs were as follow: 1 × Q buffer (Qiagen), 0.4 mℳ of each dNTP, 1.5 mℳ MgCl2, 0.2 μℳ of each primer (Table1), 5 μg of bovine serum albumin, 2.5 U of Taq DNA polymerase (Qiagen) and 5 ng of cDNA matrix in a final volume of 50 μl. Environmental arrA genes were amplified using primer sets designed elsewhere (Kulp et al., 2006). Primers and conditions (Achour et al., 2007) to amplify environmental arsB/Acr3 genes were modified by Fahy et al. (submitted) (Table 1). All the PCR products were checked in 1% (w/v) agarose electrophoresis in 1 × TBE buffer and stained with ethidium bromide (0.2 μg ml−1). DNA bands were detected under ultraviolet light. For clone library construction, PCR products were purified with the Gel Extraction Kit (Qiagen), ligated into the pCRTOPO2.1 TA cloning vector (Invitrogen, Bleiswijk, The Netherlands) and transformed into competent Escherichia coli TOP 10 F' cells (Invitrogen), according to the manufacturer's instructions. Recombinant clones were analyzed by PCR using primers M13F (5′-GTAAAACGACGGCCAG-3′) and M13R (5′-CAGGAAACAGCTATGAC-3′). Inserts of clones from the different libraries were directly sequenced using the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems Inc., Foster City, CA, USA) with M13 primers, following the manufacturer's procedures. Sequences were assembled by using the software Sequencer v4.1.4 and were compared with sequences deposited in the GenBank DNA database by using the BLAST algorithm (Altschul et al., 1998). Alignments were achieved by using ClustalX v1.83 (Thompson et al., 1997) and corrected with ProSeq v2.9 (Filatov, 2002) before the construction of phylogenetic trees with Mega v4 (Tamura et al., 2007). The tree was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The evolutionary distances were computed using the Poisson correction method and all positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons. The confidence level of the phylogenetic tree topology was evaluated by performing 1000 bootstrap replications. Homologous coverage calculation and rarefaction analysis of each library was performed as previously described (Giloteaux et al., 2010).
Table 1. Primers used in this study for cDNA clone library construction and real-time PCR quantification of arsenic biotransformation-related genes (arrA and arsB/acr3) and the housekeeping gene recA.
| Target | Primer set | Forward primer (5'–3') | Reverse primer (5'–3') | Annealing temperature (°C) | Amplicon size (bp) | Reference or source | |
|---|---|---|---|---|---|---|---|
| arrA | Clone library | HAArrA-D1F/HAArr-G2R | CCGCTACTACACCGAGGGCWWYTGGGRNTA | CGTGCGGTCCTTGAGCTCNWDRTTCCACC | 53.5 | ∼500 | Kulp et al., 2006 |
| Real-time PCR | qArr1F/qARR1R | GATCCACGCTTCTCCACCTC | CCCCGGCTTTAAAGAGGTTC | 60 | ∼172 | This study | |
| arsB | Clone library | AarsB1F/AarsB1R | GAACATCGTCTGGAAYGCNAC | GTACACCACCAGRTACATNCC | 55 | ∼750 | Achour et al., 2007a |
| acr3-1 | Clone library | Aacr1F/Aacr2R | GGCCTGATCGTNATGATGTAYCC | GCGATGGCCAGCTCRAARTTRTT | 57b | ∼750 | Achour et al., 2007a |
| Real-time PCR | qA1-3F/qA1-2R | ATGGCCAGCTCGAAaTTGTT | 60 | ∼94 | This study | ||
| acr3-2 | Clone library | dacr5F/dacr4R | TGATCTGGGTCATGATCTTCCCVATGMTGVT | CGGCCACGGCCAGYTCRAARAARTT | 52 | ∼750 | Achour et al., 2007a |
| Real-time PCR | qA2-1F/qA2-1R | GGCCAGTTCGAAGAAGTTGG | GCCGATTTTGATCCAGGTGT | 60 | ∼127 | This study | |
| recA | Clone library | recA-48F/recA-583R | CCAGATHGARAARCAGTT | TTCATACGGATCTGGTTGAT | 55 | ∼540 | This study |
| Real-time PCR | qrecA-2F/qrecA-3R | TTGAGAAGCAGTTCGGCAAA | CGGTCCGAAGATCTCGATG | 60 | ∼156 | This study | |
Primers modified by Fahy et al., unpublished.
57°–0.5° for 10 cycles, 52° for 30 cycles.
Design of primers for real-time PCR
All quantitative PCR (qPCR) primers were designed according to the manufacturer's specifications (amplicon size 100–200 bp), and representative products from each of these primer sets were verified by sequencing clone libraries.
Primers Glov-arrA1637f (5′-ACCTTTCCATCCAGCAACAG-3′) and Glov-arrA1737r (5′-ACCCCTTGCCTTGAGTTTTT-3′) for quantitative reverse transcription PCR targeting the arrA gene from G. lovleyi were designed using Primer3 software (http://frodo.wi.mit.edu/). ArrA transcripts were normalized against the number of recA mRNA transcripts. The gene recA codes for DNA repair protein recombinase A, and transcription of this gene appears to be constitutive in pure cultures and in the environment (Holmes et al., 2005; O'Neil et al., 2008; Mouser et al., 2009b), and was selected as an external control for normalization of transcription levels. G. lovleyi recA gene was amplified with primers Glov-recA44f (5′-TTGAAAAGCAGTTTGGCAAA-3′) and Glov-recA199r (5′-AGGGCCATAGACCTCAATT-3′).
The predominant sequences detected in the arrA and acr3-1 cDNA clone libraries clustered with Geobacter and accounted for >65 and >85% of the sequences. qPCR primers were designed from these dominant sequences.
The housekeeping gene, recA, was used for normalization to correct for potential differences in mRNA extraction or other sampling discrepancies. Before in situ recA qPCR primers could be designed, it was necessary to identify the dominant recA sequences found in the groundwater during in situ bioremediation. Therefore, a degenerate PCR primer set (Table 1) that targeted an ∼550-bp region of recA was designed from nucleotide sequences extracted from the following genomes: G. sulfurreducens, G. metallireducens, Geobacter sp. FRC-32, G. uraniireducens Rf4, G. lovleyi SZ, Geobacter sp. M21, Geobacter sp. M18, G. bemidjiensis, Desulfuromonas acetoxidans, Pelobacter carbinolicus, Pelobacter propionicus, Dechloromonas aromatica and R. ferrireducens.
To determine which recA transcripts were most predominant in groundwater, cDNA made by reverse transcription of RNA extracted from the groundwater was used as the template for this degenerate primer set. A clone library was then constructed, and 200 clones were selected for analyses. Geobacter sequences detected in the recA clone library were most similar to the same Geobacter species observed in the arrA and acr3-1 libraries. qPCR primers were designed targeting these Geobacter species and sequences obtained from the arrA, acr3-1 and recA qPCR primer sets showed that they all targeted the same organisms.
The nucleotide sequences of arrA, acr3-1 and acr3-2 genes amplified from the uranium-contaminated aquifer have been deposited in the GenBank database under the accession numbers HE974855-HE974887.
Quantification of transcripts with real-time PCR
The 25 μl qPCR mixture contained 12.5 μl of Power SYBR green PCR Master Mix (Applied Biosystems), 1.5 μl of a 2.5 μℳ concentration of each primer, 5 ng of cDNA template in a final volume of 25 μl. PCR amplification was carried out with a 7500 real-time PCR System (Applied Biosystems). Cloned arrA, acr3-1, acr3-2 and recA genes from the cDNA libraries were chosen to create a standard curve (Smith et al., 2006). Standard curves covering eight orders of magnitude were constructed with serial dilutions of known amounts of purified DNA quantified with a NanoDrop ND-1000 spectrophotometer at an absorbance of 260 nm. Transcript abundances were calculated from appropriate standard curves. The qPCR efficiency (95–99%) was calculated on the basis of the slope of the standard curve. All qPCR assays were run in triplicate. Thermal cycling parameters consisted of an activation step at 50 °C for 2 min, a denaturation step at 95 °C for 10 min, and 50 cycles at 95 °C for 15 s and 60 °C for 1 min. This was followed by the construction of a dissociation curve by increasing the temperature from 60 to 95 °C at a ramp rate of 2%. A single predominant peak was observed in the dissociation curve of each gene, supporting the specificity of the PCR product.
Results and discussion
Biogeochemistry and arsenic release during acetate amendments
Pre-injection levels of DO in the Rifle aquifer were well below 0.5 mg l−1, with most values <0.1 mg l−1. Low DO levels are a characteristic feature of the Rifle groundwater and inferred to result from a combination of organic (for example, lignitic carbon) and inorganic (for example, Fe(II) or Fe(II)-bearing minerals) reductants present within the aquifer and associated with aquifer sediments. Following acetate amendment, DO values fell to levels at or below the detection limit (<0.01 mg l−1) of the luminescent oxygen sensor used to quantify DO, and remained at these low levels for the duration of the experiment. DO values for locations not impacted by acetate varied from 0.2 to 0.5 mg l−1 over the experimental period. pH values were largely stable during the experiment, ranging from 7.1 to 7.2, owing to the well-buffered nature of the groundwater and sediments.
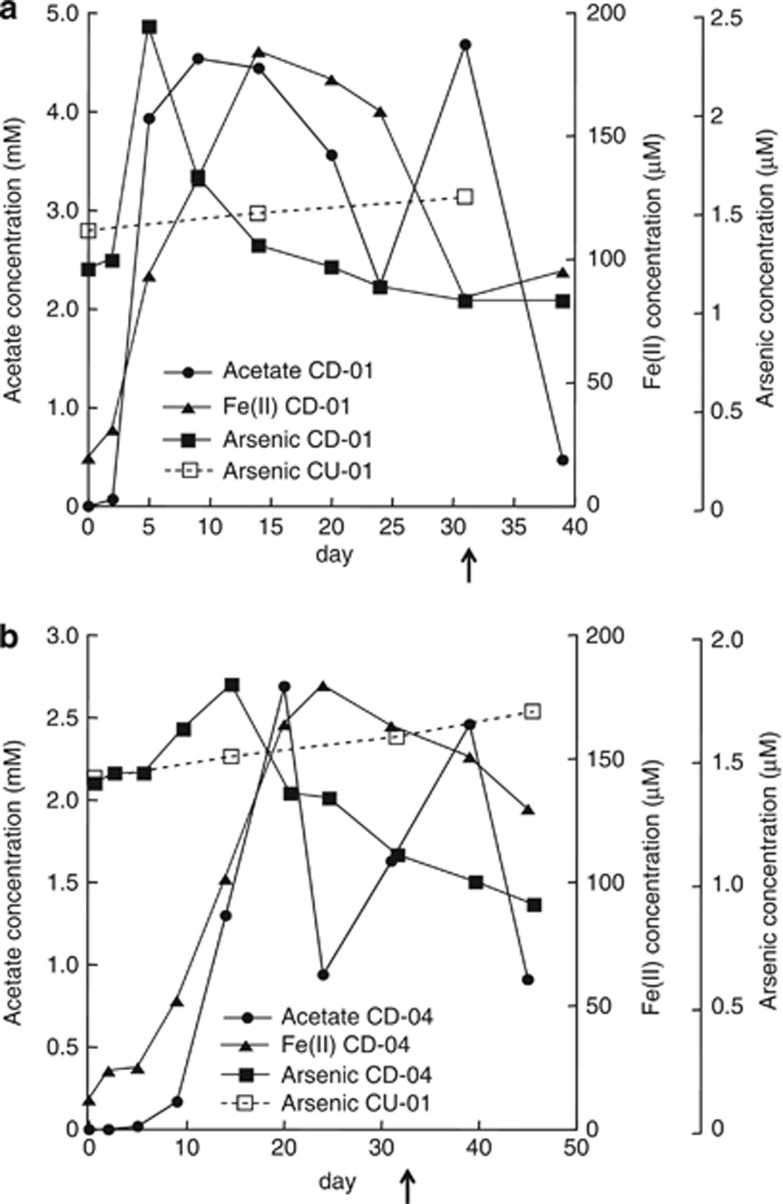
Before acetate amendments, groundwater arsenic concentrations were ca. 1.5 μℳ and arsenic remained at these levels in background wells not amended with acetate throughout the field experiment (Figure 1). With the addition of acetate, there was a rapid increase in arsenic in well CD01, the well closest to the injection gallery (Figure 1a). The increase in arsenic was associated with an accumulation of Fe(II), which continued accumulating even as arsenic declined. Previous studies have demonstrated a similar coincident release of arsenic and Fe(II) under anaerobic conditions (Rochette et al., 1997, 2000). Arsenic adsorbed to the surface of Fe(III) oxides or incorporated within the Fe(III)-containing minerals may be released when Fe(III) is reduced (Cummings et al., 1999; Nickson et al., 2000; McArthur et al., 2001; Bose and Sharma, 2002; Harvey et al., 2002; Nicholas et al., 2003; Horneman et al., 2004; Van Geen et al., 2004), and microbial reduction of As(V) to As(III) can enhance mobilization (Nicholas et al., 2003; Oremland and Stolz, 2003; Lloyd and Oremland, 2006).
Figure 1.
Acetate, Fe(II) and arsenic concentrations in well CD01 (a) and CD04 (b) and arsenic concentrations in the background well CU-01 (a) during acetate amendment at the Rifle site. The arrow indicates the end of acetate injection at day 31.
After the initial arsenic release, arsenic concentrations slowly started to decline after day 5 in CD01 and day 14 in CD04, even though acetate continued to be injected until day 31 (Figure 1). One potential explanation for this decline in arsenic is the fact that the divalent cations Ca2+, Mg2+ and Fe2+ increased significantly in the groundwater (Supplementary Figure S2), and studies have shown that these ions promote As precipitation (Appelo et al., 2002; Smedley and Kinniburgh, 2002; Smith et al., 2002). Precipitation with sulfide (Newman et al., 1997b) and a change in pH (Smedley and Kinniburgh, 2002) were unlikely to have affected arsenic release as sulfide was not detected, and the pH was neutral throughout this field experiment.
Arsenic concentrations in the downgradient well, CD04, were lower than those observed in CD01 and increased before significant acetate concentration reached the site. The complex dynamics of dissolved arsenic noted above make it difficult to determine which of the arsenic release and precipitation possibilities were responsible for this pattern, but the initial increase in dissolved arsenic in CD04 may have resulted from dissolved arsenic moving downgradient from CD01.
Although the arsenic in the groundwater was not speciated, the following year, acetate addition and the onset of reducing conditions resulted in increased concentrations of As(III) in groundwater collected from the same well (45–65% of the arsenic was As(III)) (Stucker et al., submitted). Although we cannot rule out the formation of other arsenic species during the 2010 amendment experiment, that is, thioarsenate (Stucker et al., submitted), it is likely that arsenic speciation patterns in 2010 were similar.
Phylogenetic analysis of transcribed arsenic functional genes detected from Rifle groundwater
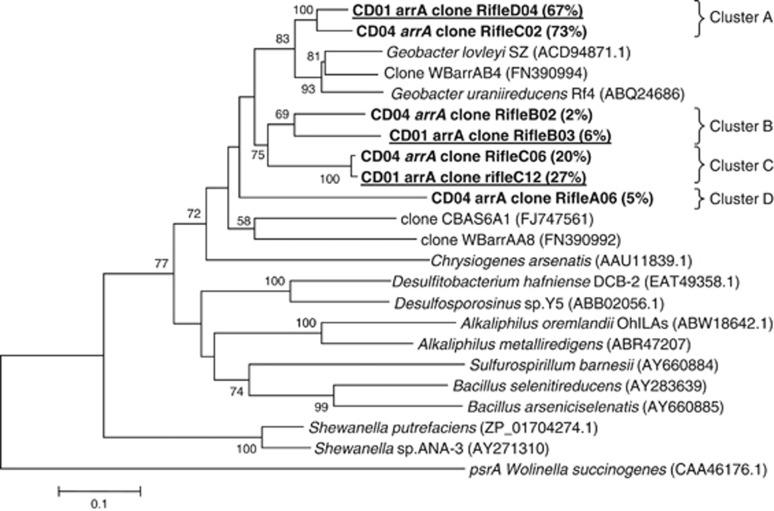
To determine which arrA, arsB and acr3 sequences were being transcribed during the in situ bioremediation experiment, mRNA was extracted from the groundwater and cDNA libraries were made from arrA, arsB and acr3 transcripts. The predicted amino-acid sequences for the arsenic reductase subunit ArrA in the transcribed genes recovered from CD01 and CD04 formed four distinct phylogenetic clusters, sharing 60–98% similarity with each other (139 amino-acid sequences considered) (Figure 2). Sequences in cluster A accounted for 67 and 73% of the ArrA sequences recovered from CD01 and CD04, respectively, and shared 64–78% and 64–82% amino-acid sequence similarity with G. lovleyi and G. uraniireducens ArrA proteins, respectively. Clusters B and C branched separately in the phylogenetic tree and represented 22% and 33% of the clones from CD04 and CD01, respectively. These sequences could not be affiliated with confidence to any known sequences of previously described As(V)-respiring bacteria. Sequences from cluster D represented 5% of the clones from CD04 samples, branched separately in the phylogenetic tree, and thus could not be affiliated with any known As(V) respirers.
Figure 2.
Phylogenetic comparison of ArrA sequences found in the groundwater to ArrA from other known arsenate respiring bacteria (139 amino acids considered). Clones obtained from both CD01 and CD04 wells in this study are in bold, and clones from CD01 are underlined. The percentage of clones is indicated in brackets. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown above or near the relevant nodes.
Overall, the majority (67% and 73% for CD01 and CD04, respectively) of arrA sequences were most closely related to sequences previously described in Geobacter species, with little nucleotide diversity among the sequences (Figure 2). Rarefaction analysis revealed high diversity coverage values (0.96 and 0.93 for CD01 and CD04, respectively), and the rarefaction curves reached a plateau (see Supplementary data; Figure S3). The most predominant species were most similar to G. uraniireducens Rf4, which was isolated from the Rifle site (Shelobolina et al., 2008) and G. lovleyi. The high similarity of the arrA clones to putative arrA genes from Geobacter spp. retrieved in this study is of particular interest as Geobacter spp. are the dominant Fe(III)-reducing bacteria in Rifle (Holmes et al., 2002, 2007; Anderson et al., 2003), and their presence at relatively high abundance in bacterial communities has previously been correlated with As release in Asian aquifers (Islam et al., 2004; Lear et al., 2007; Rowland et al., 2007; Héry et al., 2008).
Transcripts of the genes arsB and acr3, which encode membrane subunits from an arsenic oxyanion translocation pump (Rosen, 1999, 2002), were evaluated in order gain insight into arsenic resistance. The ArsB sequences recovered were highly diverse, with clones mainly related to the Gammaproteobacteria and Firmicutes classes (data not shown). Gammaproteobacteria and Firmicutes only accounted for 0–5% and 2–9% of the 16S rRNA sequences detected during active uranium bioremediation in this field experiment (Holmes et al., submitted), and thus the AarsB genes were not studied further.
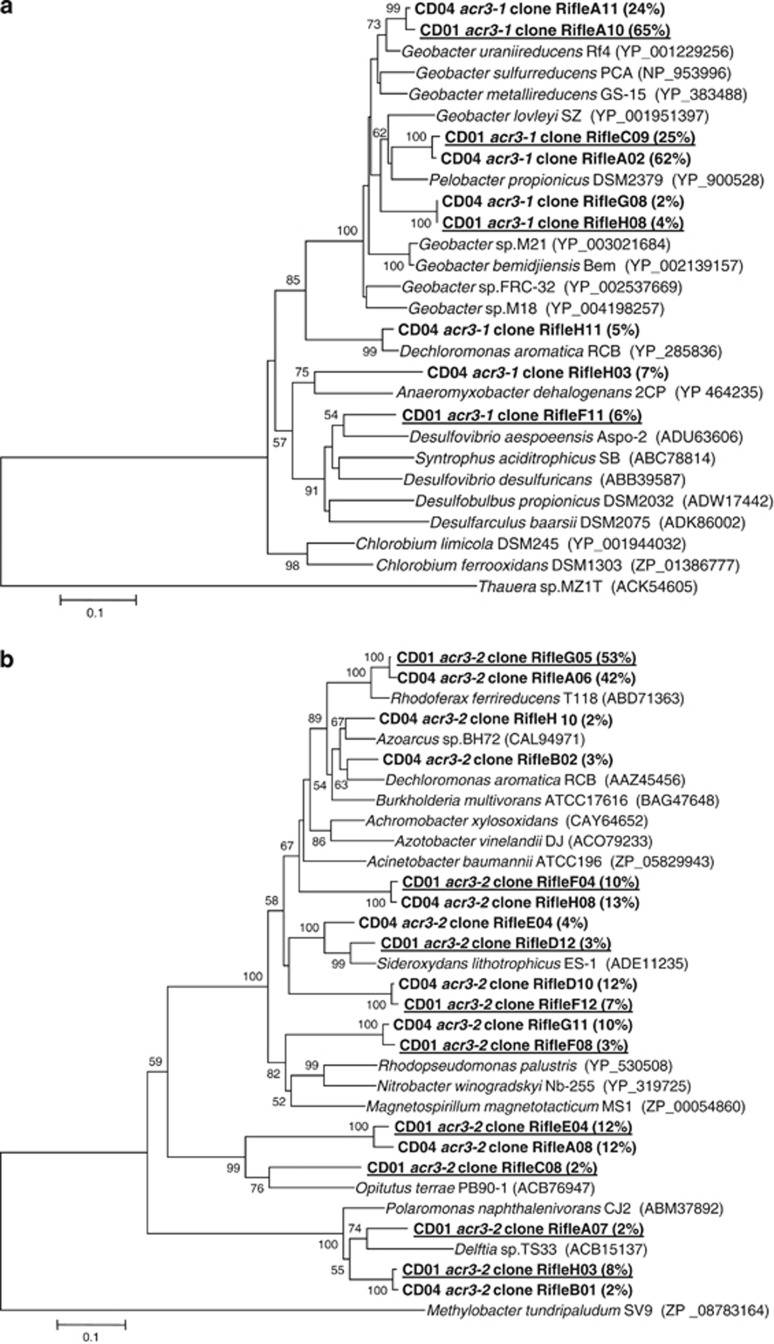
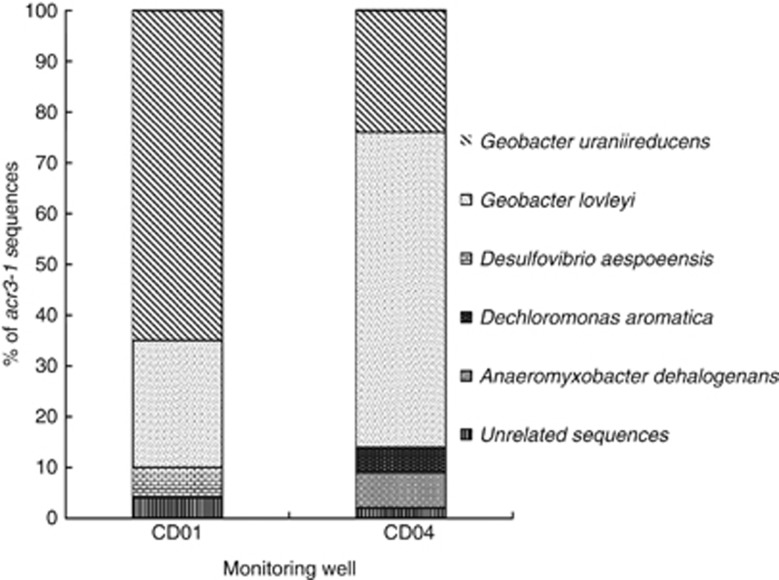
Phylogenetic studies have shown that the other arsenic resistance gene, acr3, is very diverse and two acr3 subfamilies, acr3-1 and acr3-2, are seen among bacteria (Wysocki et al., 2003; Achour et al., 2007). Sequences from both subfamilies were recovered in groundwater collected from this site (Figure 3). The majority, that is, 90% and 86% for CD01 and CD04, respectively, of acr3-1 sequences (Figures 3a and 4) were most similar to Geobacter species (Figures 3a and 4) and shared 86–96% and 83–91% amino-acid similarity to G. uraniireducens, an isolate recovered from the Rifle site (Shelobolina et al., 2008) and G. lovleyi ACR3-1 proteins (Figure 3a). These numbers are consistent with results from 16S rRNA clone libraries and FISH analysis showing that up to 89% of the 16S rRNA sequences from well CD04 were most similar to Geobacter species during active Fe(III) reduction (Holmes et al., submitted).
Figure 3.
(a) Phylogenetic comparison of Acr3-1 sequences found in the groundwater to Acr3-1 from other known arsenic-resistant bacteria (257 amino acids considered) and (b) phylogenetic comparison of Acr3-2 sequences found in the groundwater to Acr3-2 from other known arsenic-resistant bacteria (259 amino acids considered). Clones obtained from both CD01 and CD04 wells in this study are in bold, and clones from CD01 are underlined. The percentage of clones is indicated in brackets. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown above or near the relevant nodes.
Figure 4.
Relative abundance of acr3-1 sequences from two downgradient monitoring wells CD01 and CD04.
Less abundant were other sequences in the Deltaproteobacteria, most closely related to Desulfovibrio or Anaeromyxobacteria sequences. Sequences which shared 97% similarity with the Betaproteobacteria member D. aromatica were also detected (Figures 3 and 4). The high coverage values calculated for acr3-1 clone libraries (0.83 and 0.8 for CD01 and CD04, respectively) together with rarefaction analysis suggested that the almost entire diversity of acr3-1 genes was covered with these clone libraries (Supplementary Figure S3).
Sequences that fell within the ACR3-2 subfamily of arsenic transporters were related to the Proteobacteria and Verrucomicrobia phyla (Figure 3b). More than 65% of the acr3-2 sequences were affiliated with members of the Betaproteobacteria class, with the majority of clones (>50%) being most closely related to acr3-2 sequences from the Fe(III)-reducing bacterium, R. ferrireducens (Figure 3b). The diversity of acr3-2 sequences was significantly higher than what was seen in acr3-1 clone libraries in contrast with previous studies (Achour et al., 2007; Cai et al., 2009) and clone library coverage values from acr3-2 libraries were 0.61 and 0.65 for CD01 and CD04 samples respectively.
Some bacteria have genes coding for proteins involved in both arsenic reduction and arsenic resistance (that is, arr and ars operons). This has been observed in Shewanella strain ANA-3 (Saltikov and Olson, 2002; Saltikov et al., 2003a, 2005; Saltikov and Newman, 2003b), G. uraniireducens and G. lovleyi. In many of these organisms, all of these genes are located in an arsenic island within the genome. For example, the locus tag for the arr operon in G. uraniireducens is Gura_0469-Gura_0471 and the locus tag for the arsenic resistance genes is Gura_0468 to Gura_0467.
Analysis of As(V) respiration in Geobacter
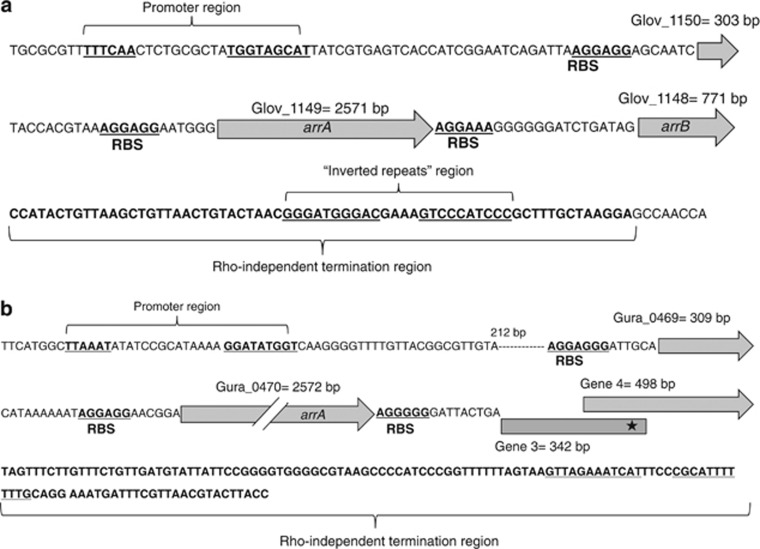
A putative arr operon is present in the genomes of both G. lovleyi and G. uraniireducens. Preliminary analysis of this operon shows that it consists of three functional genes: a gene coding for a hypothetical protein (Glov_1150 and Gura_0469); a gene coding for a protein that has a molybdopterin cofactor binding site and is homologous to the α-subunit of the arsenate respiratory reductase protein ArrA (Glov_1149 and Gura_0470); and a gene coding for a protein that is homologous to the beta subunit of arsenate respiratory reductase ArrB (Glov_1148 and genes 3 and 4 in G. uraniireducens). The G. uraniireducens arr operon actually has four putative genes; however, only three of them appear to be functional. There are two ferredoxin genes that could potentially encode ArrB; however, one of them (gene 3) contains a stop codon in the 342 basepair (Figure 5b). Further analysis of the arr operon in both G. uraniireducens and G. lovleyi showed that it contains Shine-Delgarno ribosome binding sites upstream from all three start codons, a promoter region, and a rho-independent terminator region (Figure 5).
Figure 5.
The arr operon of G. lovleyi (a) and G. uraniireducens (b), including the promoter region and termination site. RBS, ribosome-binding site
Further evidence of functionality of the Arr protein found in G. lovleyi comes from the fact that ArrA and ArrB from G. lovleyi are 60.3% and 53% similar to ArrA and ArrB found in Shewanella sp. ANA-3, a known arsenate respiring bacterium (Saltikov and Newman, 2003b). As shown in Supplementary Figure S4, the arrA gene from both organisms codes for a molybdopterin oxidoreductase protein, and contains: (i) a cysteine-rich motif (C-X2-C-X3-C-X27-C) that is predicted to coordinate an iron–sulfur cluster; (ii) a conserved cysteine residue of the molybdenum-binding domain, which is likely to represent the amino acid that coordinates to the molybdenum; (iii) a twin arginine translocation (TAT) signal (RRDFLK); and (iv) a potential cleavage site at Ala-31, which is indicative of localization to the cytoplasmic membrane. Comparative analysis of ArrB from G. lovleyi showed that this gene codes for a 4Fe-4 S ferredoxin iron-sulfur binding domain protein similar to ArrB found in the Shewanella sp. ANA-3 (Supplementary figure S4).
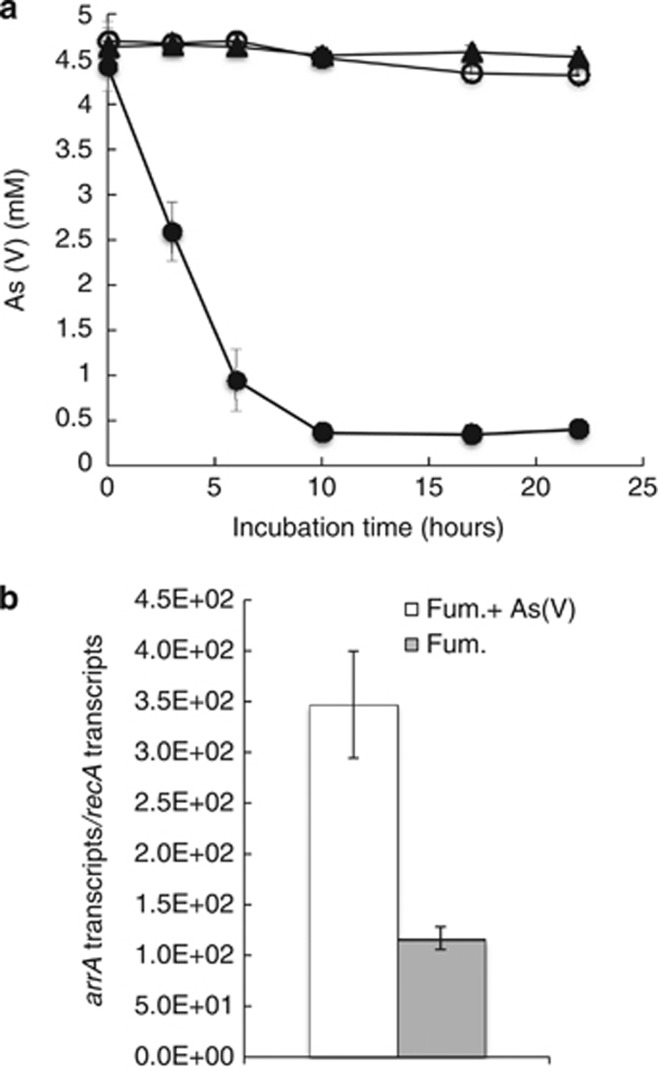
It has also been reported that G. uraniireducens can grow with As(V) provided as an electron acceptor (Héry et al., 2008), and cell suspensions of G. lovleyi reduced As(V) with acetate as the electron donor (Figure 6a). There was no As(V) reduction in acetate-free controls. Attempts to grow G. lovleyi with As(V) as the sole electron acceptor were not successful. However, when G. lovleyi was grown with fumarate as an electron acceptor in addition to As(V), arrA transcripts were 3 fold more abundant than in cultures grown with fumarate as the electron acceptor in the absence of As(V) (Figure 6b), demonstrating a transcriptional response to the presence of As(V).
Figure 6.
(a) Measurement of As(V) concentrations in G. lovleyi cell suspensions provided 5 mℳ As(V). (•) Resting cells with acetate (20 mℳ) added as an electron donor; (▪) Heat-killed control cells; and (○) Resting cells without acetate. (b) Transcription levels of G. lovleyi arrA gene, normalized against G. lovleyi recA, in cells grown with either fumarate or fumarate and As(V) as potential electron acceptors.
Transcription and abundance of arsenic functional genes over time
It was not feasible to track gene transcription patterns for the full diversity of arrA, acr3-1 and acr3-2 genes over time. Therefore, this study focused on transcription of the dominant sequences from the arrA and acr3-1 clone libraries from CD01 and CD04; which were most similar to Geobacter species, the predominant members of the microbial community.
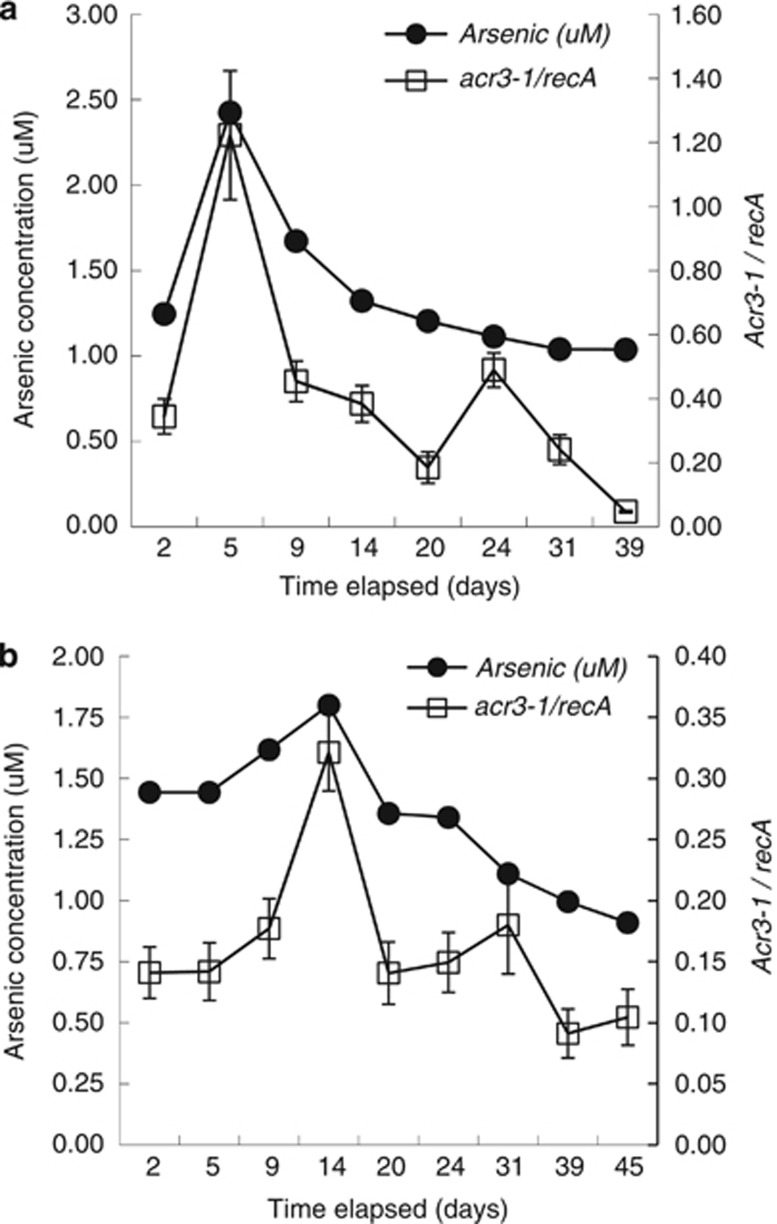
The abundance of transcripts for the arsenic resistance acr3-1 genes associated with Geobacteraceae species closely tracked the abundance of arsenic in the groundwater (Figure 7). There was a significant positive correlation between the number of Geobacter acr3-1 transcripts and arsenic concentrations (Pearson's correlation r=0.91 and 0.84 (P<0.05) and Spearman's correlation R=0.67 and 0.81 (P<0.05) for CD01 and CD04, respectively). Transcript abundance did not correspond well with acetate (r=0.31 and −0.11 (P>0.05) and R=0.21 and −0.33 (P>0.05)) or Fe(II) concentrations (r=−0.08 and −0.16 (P>0.05) and R=0.04 and −0.24 (P>0.05)).
Figure 7.
Arsenic concentrations and transcripts levels of acr3-1 normalized to recA in well CD01 (a) and CD04 (b) over the course of the biostimulation experiment at Rifle, CO, USA. Error bars represent propagation of errors of triplicate real-time PCR reactions. Open squares, acr3-1 genes; closed circles, arsenic.
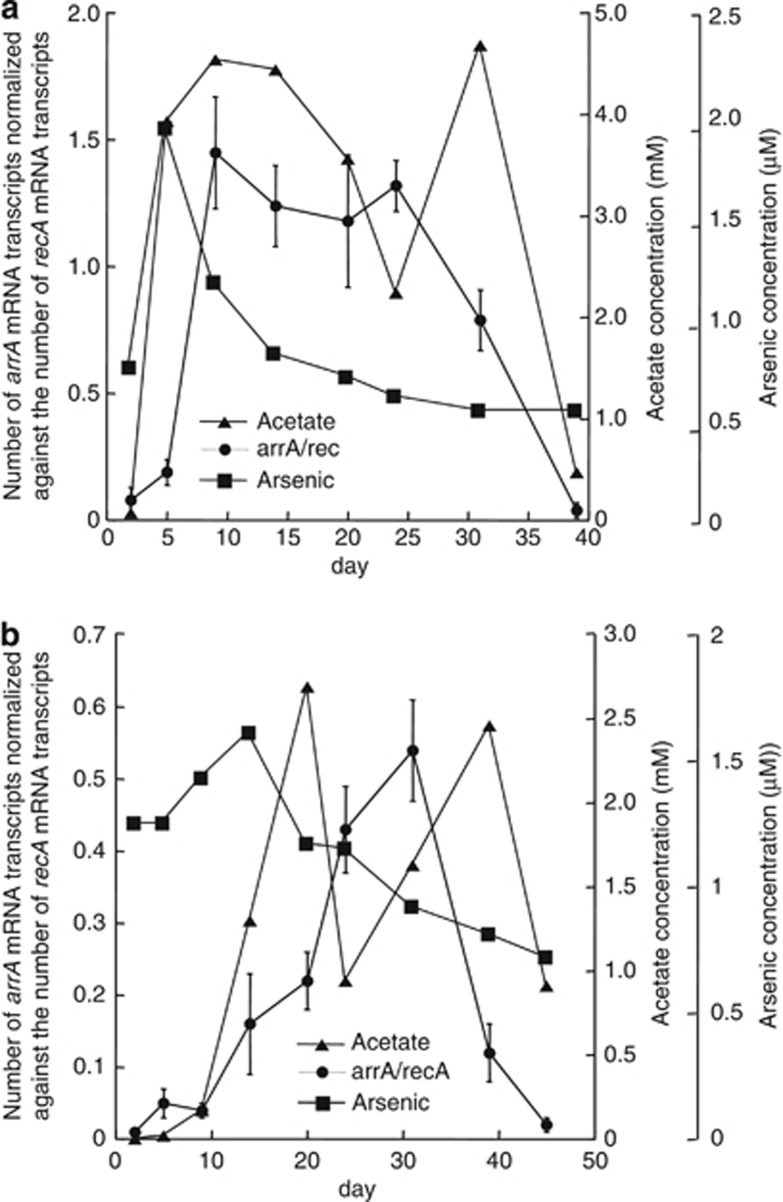
The abundance of Geobacter arrA transcripts in the subsurface community lagged behind the increases in dissolved arsenic and remained high even after groundwater arsenic concentrations declined (Figure 8). There was some correspondence between the number of arrA transcripts and acetate (r=0.63 and 0.52 (P<0.05) and R=0.52 and 0.74 (P<0.05) for CD01 and CD04) and Fe(II) concentrations (r=0.8 and 0.64 (P<0.05) and R=0.62 and 0.79 (P<0.05) for CD01 and CD04).
Figure 8.
Arsenic and acetate concentrations and transcripts levels of arrA normalized to recA in well CD01 (a) and CD04 (b) over the course of the biostimulation experiment at Rifle, CO, USA. Error bars represent propagation of errors of triplicate real-time PCR reactions. Closed circles, arrA genes; closed triangles, acetate; closed squares, arsenic.
When acetate concentrations declined after the acetate amendments were stopped on day 31, there was a dramatic decline in the relative transcription of arrA (Figure 8). This gene transcription pattern is similar to that previously noted for central metabolic genes (Holmes et al., 2004a, 2005, 2007), such as citrate synthase (Holmes et al., 2005), and suggests that increased transcription of this respiratory gene could be part of an overall increase in genes involved in energy generation, rather than a specific response to the availability of As(V) as an electron acceptor.
Implications
The results demonstrate that the subsurface Geobacter population rapidly responded to changes in arsenic concentrations in the groundwater by modulating the transcription of a gene for arsenic resistance. This result further emphasizes that transcriptome analysis is a powerful method for diagnosing the physiological status of subsurface microbial communities.
The factors controlling the transcription of arrA, presumed to encode a subunit of a dissimilatory As(V) reductase, is less clear. G. lovleyi, which was shown to be capable of acetate-dependent As(V) reduction, increased transcription of arrA in the presence of As(V). However, during uranium bioremediation arrA transcription patterns appeared to be correlated with the availability of acetate rather than arsenic concentrations in the subsurface. This enhanced transcription of arrA when acetate levels were high might be associated with a generalized increase in anaerobic respiratory genes under these conditions. More information on the availability of As(V) as an electron acceptor throughout the bioremediation process will be required to better interpret this data.
Quantifying the abundance of 16S rRNA gene sequences or genes more directly related to arsenic have demonstrated an association of Geobacter species with arsenic release in aquifer sediments and arsenic-rich lake sediments (Islam et al., 2004; Héry et al., 2008, 2010). The transcriptomic approach reported here has the potential to provide more in-depth analysis of the metabolic activity associated with arsenic biogeochemistry in such environments because gene transcription is more directly related to physiological activity than gene presence. A similar transcriptomic approach should be applicable to the study of subsurface arsenic biogeochemistry in environments in which organisms other than Geobacter species predominate.
Acknowledgments
Research at the University of Massachusetts was funded by the Office of Science (BER), US Department of Energy, Awards no. DE-SC0004080 and DE-SC0004814 and Cooperative Agreement no. DE-FC02-02ER63446. Additional support for field research was equally supported through the Integrated Field Research Challenge Site (IFRC) at Rifle, CO, USA and the Lawrence Berkeley National Laboratory's Sustainable Systems Scientific Focus Area. The US Department of Energy (DOE), Office of Science, Office of Biological and Environmental Research funded the work under contract DE-AC02-05CH11231 (Lawrence Berkeley National Laboratory; operated by the University of California).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Achour AR, Bauda P, Billard P. Diversity of arsenite transporter genes from arsenic-resistant soil bacteria. Res Microbiol. 2007;158:128–137. doi: 10.1016/j.resmic.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Ahmann D, Krumholz LR, Hemond HF, Lovley DR, Morel FMM. Microbial mobilization of arsenic from sediments of the Aberjona watershed. Environ Sci Technol. 1997;31:2923–2930. [Google Scholar]
- Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, et al. Gapped blast and psi-blast: a new generation of protein database search programs. Nucleic Acids Res. 1998;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amstaetter K, Borch T, Larese-Casanova P, Kappler A. Redox transformation of arsenic by Fe(II)-activated goethite (alpha-FeOOH) Environ Sci Technol. 2010;44:102–108. doi: 10.1021/es901274s. [DOI] [PubMed] [Google Scholar]
- Anderson CR, Cook GM. Isolation and characterization of arsenate-reducing bacteria from arsenic-contaminated sites in New Zealand. Curr Microbiol. 2004;48:341–347. doi: 10.1007/s00284-003-4205-3. [DOI] [PubMed] [Google Scholar]
- Anderson RT, Vrionis HA, Ortiz-Bernad I, Resch CT, Long PE, Dayvault R, et al. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl Environ Microbiol. 2003;69:5884–5891. doi: 10.1128/AEM.69.10.5884-5891.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelo CAJ, Van der Weiden MJJ, Tournassat C, Charlet L. Surface complexation of ferrous iron and carbonate on ferrihydrite and the mobilization of arsenic. Environ Sci Technol. 2002;36:3096–3103. doi: 10.1021/es010130n. [DOI] [PubMed] [Google Scholar]
- Blum JS, Bindi AB, Buzzelli J, Stolz JF, Oremland RS. Bacillus arsenicoselenatis, sp nov, and Bacillus selenitireducens, sp nov: two haloalkaliphiles from Mono Lake, California that respire oxyanions of selenium and arsenic. Arch Microbiol. 1998;171:19–30. doi: 10.1007/s002030050673. [DOI] [PubMed] [Google Scholar]
- Bond DR, Lovley DR. Electricity production by Geobacter sulfurreducens attached to electrodes. Appl Environ Microbiol. 2003;69:1548–1555. doi: 10.1128/AEM.69.3.1548-1555.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose P, Sharma A. Role of iron in controlling speciation and mobilization of arsenic in subsurface environment. Water Res. 2002;36:4916–4926. doi: 10.1016/s0043-1354(02)00203-8. [DOI] [PubMed] [Google Scholar]
- Cai L, Liu GH, Rensing C, Wang GJ. Genes involved in arsenic transformation and resistance associated with different levels of arsenic-contaminated soils. BMC Microbiol. 2009;9:4. doi: 10.1186/1471-2180-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell KM, Malasarn D, Saltikov CW, Newman DK, Hering JG. Effect of sorbed arsenic species on bacterial reduction of HFO. Geochim Cosmochim Acta. 2005;69:A357. [Google Scholar]
- Cummings DE, Caccavo F, Fendorf S, Rosenzweig RF. Arsenic mobilization by the dissimilatory Fe(III)-reducing bacterium Shewanella alga BrY. Environ Sci Technol. 1999;33:723–729. [Google Scholar]
- Devitre R, Belzile N, Tessier A. Speciation and adsoprtion of arsenic on diagenetic iron oxyhydroxides. Limnol Oceanogr. 1991;36:1480–1485. [Google Scholar]
- Dixit S, Hering JG. Comparison of arsenic(V) and arsenic(III) sorption onto iron oxide minerals: implications for arsenic mobility. Environ Sci Technol. 2003;37:4182–4189. doi: 10.1021/es030309t. [DOI] [PubMed] [Google Scholar]
- Dowdle PR, Laverman AM, Oremland RS. Bacterial dissimilatory reduction of arsenic(V) to arsenic(III) in anoxic sediments. Appl Environ Microbiol. 1996;62:1664–1669. doi: 10.1128/aem.62.5.1664-1669.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval S, Ducluzeau A-L, Nitschke W, Schoepp-Cothenet B. Enzyme phylogenies as markers for the oxidation state of the environment: the case of respiratory arsenate reductase and related enzymes. BMC Evol Biol. 2008;8:206. doi: 10.1186/1471-2148-8-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy A, Giloteaux L, Bertin P, Le Paslier D, Médigue C, Weissenbach J, et al. 201216S rRNA and functional diversity: contrasting fingerprints in arsenic-rich sediments from an acid mine drainage Mol Ecol(submitted). [DOI] [PubMed]
- Filatov DA. PROSEQ: A software for preparation and evolutionary analysis of DNA sequence data sets. Mol Ecol Notes. 2002;2:621–624. [Google Scholar]
- Finneran KT, Anderson RT, Nevin KP, Lovley DR. Potential for Bioremediation of uranium-contaminated aquifers with microbial U(VI) reduction. Soil Sediment Contam. 2002;11:339–357. [Google Scholar]
- Fisher E, Dawson AM, Polshyna G, Lisak J, Crable B, Perera E, et al. Transformation of inorganic and organic arsenic by Alkaliphilus oremlandii sp nov strain OhILAs. Am NY Acad Sci. 2008;1125:230–241. doi: 10.1196/annals.1419.006. [DOI] [PubMed] [Google Scholar]
- Ford T, Jay J, Patel A, Kile M, Prommasith P, Galloway T, et al. Use of ecotoxicological tools to evaluate the health of New Bedford Harbor sediments: a microbial biomarker approach. Environ Health Perspect. 2005;113:186–191. doi: 10.1289/ehp.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giloteaux L, Goni-Urriza M, Duran R. Nested PCR and new primers for analysis of sulfate-reducing bacteria in low-cell-biomass environments. Appl Environ Microbiol. 2010;76:2856–2865. doi: 10.1128/AEM.02023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey CF, Swartz CH, Badruzzaman AB, Keon-Blute N, Yu W, Ali MA, et al. Arsenic mobility and groundwater extraction in Bangladesh. Science. 2002;298:1602–1606. doi: 10.1126/science.1076978. [DOI] [PubMed] [Google Scholar]
- Héry M, Gault AG, Rowland HAL, Lear G, Polya DA, Lloyd JR. Molecular and cultivation-dependent analysis of metal-reducing bacteria implicated in arsenic mobilisation in south-east asian aquifers. Appl Geochem. 2008;23:3215–3223. [Google Scholar]
- Héry M, van Dongen BE, Gill F, Mondal D, Vaughan DJ, Pancost RD, et al. Arsenic release and attenuation in low organic carbon aquifer sediments from West Bengal. Geobiology. 2010;8:155–168. doi: 10.1111/j.1472-4669.2010.00233.x. [DOI] [PubMed] [Google Scholar]
- Hollibaugh JT, Budinoff C, Hollibaugh RA, Ransom B, Bano N. Sulfide oxidation coupled to arsenate reduction by a diverse microbial community in a Soda Lake. Appl Environ Microbiol. 2006;72:2043–2049. doi: 10.1128/AEM.72.3.2043-2049.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes DE, Nevin KP, Lovley DR. In situ expression of nifD in Geobacteraceae in subsurface sediments. Appl Environ Microbiol. 2004a;70:7251–7259. doi: 10.1128/AEM.70.12.7251-7259.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes DE, Finneran KT, O'Neil RA, Lovley DR. Enrichment of members of the family Geobacteraceae associated with stimulation of dissimilatory metal reduction in uranium-contaminated aquifer sediments. Appl Environ Microbiol. 2002;68:2300–2306. doi: 10.1128/AEM.68.5.2300-2306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes DE, Nevin KP, O'Neil RA, Ward JE, Adams LA, Woodard TL, et al. Potential for quantifying expression of the Geobacteraceae citrate synthase gene to assess the activity of Geobacteraceae in the subsurface and on current-harvesting electrodes. Appl Environ Microbiol. 2005;71:6870–6877. doi: 10.1128/AEM.71.11.6870-6877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes DE, O'Neil RA, Chavan MA, N'Guessan LA, Vrionis HA, Perpetua LA, et al. Transcriptome of Geobacter uraniireducens growing in uranium-contaminated subsurface sediments. ISME J. 2009;3:216–230. doi: 10.1038/ismej.2008.89. [DOI] [PubMed] [Google Scholar]
- Holmes DE, Giloteaux L, Barlett M, Chavan MA, Smith JA, Risso C, et al. 2012. Molecular analysis of the growth rate of subsurface geobacter species during in situ uranium bioremediation (submitted). [DOI] [PMC free article] [PubMed]
- Holmes DE, O'Neil RA, Vrionis HA, N'Guessan LA, Ortiz-Bernad I, Larrahondo MJ, et al. Subsurface clade of Geobacteraceae that predominates in a diversity of Fe(III)-reducing subsurface environments. ISME J. 2007;1:663–677. doi: 10.1038/ismej.2007.85. [DOI] [PubMed] [Google Scholar]
- Horneman A, Van Geen A, Kent DV, Mathe PE, Zheng Y, Dhar RK, et al. Decoupling of As and Fe release to Bangladesh groundwater under reducing conditions. Part 1: evidence from sediment profiles. Geochim Cosmochim Acta. 2004;68:3459–3473. [Google Scholar]
- Islam FS, Gault AG, Boothman C, Polya DA, Charnock JM, Chatterjee D, et al. Role of metal-reducing bacteria in arsenic release from Bengal delta sediments. Nature. 2004;430:68–71. doi: 10.1038/nature02638. [DOI] [PubMed] [Google Scholar]
- Islam FS, Pederick RL, Gault AG, Adams LK, Polya DA, Charnock JM, et al. Interactions between the Fe(III)-reducing bacterium Geobacter sulfurreducens and arsenate, and capture of the metalloid by biogenic Fe(II) Appl Environ Microbiol. 2005a;71:8642–8648. doi: 10.1128/AEM.71.12.8642-8648.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam FS, Pederick RL, Polya DA, et al. Reduction of Fe(III) by Geobacter sulfurreducens and the capture of arsenic by biogenic Fe(II) minerals. Geochim Cosmochim Acta. 2004b;68:A518–A518. [Google Scholar]
- Istok JD, Senko JM, Krumholz LR, Bogle MA, Peacock A, Chang YJ, et al. In situ bioreduction of technetium and uranium in a nitrate-contaminated aquifer. Environ Sci Technol. 2004;38:468–475. doi: 10.1021/es034639p. [DOI] [PubMed] [Google Scholar]
- Jackson CR, Harrison KG, Dugas SL. Enumeration and characterization of culturable arsenate resistant bacteria in a large estuary. Syst Appl Microbiol. 2005;28:727–734. doi: 10.1016/j.syapm.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Junier P, Suvorova EI, Bernier-Latmani R. Effect of competing electron acceptors on the reduction of U(VI) by Desulfotomaculum reducens. Geomicrobiol J. 2010;27:435–443. [Google Scholar]
- Kulp TR, Hoeft SE, Oremland RS. Redox transformations of arsenic oxyanions in periphyton communities. Appl Environ Microbiol. 2004;70:6428–6434. doi: 10.1128/AEM.70.11.6428-6434.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp TR, Hoeft SE, Miller LG, Saltikov C, Murphy JN, Han S, et al. Dissimilatory arsenate and sulfate reduction in sediments of two hypersaline, arsenic-rich soda lakes: Mono and Searles lakes, California. Appl Environ Microbiol. 2006;72:6514–6526. doi: 10.1128/AEM.01066-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lear G, Song B, Gault AG, Polya DA, Lloyd JR. Molecular analysis of arsenate-reducing bacteria within Cambodian sediments following amendment with acetate. Appl Environ Microbiol. 2007;73:1041–1048. doi: 10.1128/AEM.01654-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd JR, Oremland RS. Microbial transformations of arsenic in the environment: from soda lakes to aquifers. Elements. 2006;2:85–90. [Google Scholar]
- Luo W, Wu W-M, Yan T, Criddle CS, Jardine PM, Zhou J, et al. Influence of bicarbonate, sulfate, and electron donors on biological reduction of uranium and microbial community composition. Appl Microbiol Biotechnol. 2007;77:713–721. doi: 10.1007/s00253-007-1183-6. [DOI] [PubMed] [Google Scholar]
- Malasarn D, Saltikov W, Campbell KM, Santini JM, Hering JG, Newman DK. arrA is a reliable marker for As(V) respiration. Science. 2004;306:455–455. doi: 10.1126/science.1102374. [DOI] [PubMed] [Google Scholar]
- McArthur JM, Ravenscroft P, Safiulla S, Thirlwall MF. Arsenic in groundwater: testing pollution mechanisms for sedimentary aquifers in Bangladesh. Water Resour Res. 2001;37:109–117. [Google Scholar]
- Miletto M, Williams KH, N'Guessan AL, Lovley DR. Molecular analysis of the metabolic rates of discrete subsurface populations of sulfate reducers. Appl Environ Microbiol. 2011;77:6502–6509. doi: 10.1128/AEM.00576-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouser PJ, Holmes DE, Perpetua LA, DiDonato R, Postier B, Liu A, et al. Quantifying expression of Geobacter spp. oxidative stress genes in pure culture and during in situ uranium bioremediation. ISME J. 2009a;3:454–465. doi: 10.1038/ismej.2008.126. [DOI] [PubMed] [Google Scholar]
- Mouser PJ, N'Guessan AL, Elifantz H, Holmes DE, Williams KH, Wilkins MJ, et al. Influence of heterogeneous ammonium availability on bacterial community structure and the expression of nitrogen fixation and ammonium transporter genes during in situ bioremediation of uranium-contaminated groundwater. Environ Sci Technol. 2009b;43:4386–4392. doi: 10.1021/es8031055. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay R, Rosen BP, Pung LT, Silver S. Microbial arsenic: from geocycles to genes and enzymes. FEMS Microbiol Rev. 2002;26:311–325. doi: 10.1111/j.1574-6976.2002.tb00617.x. [DOI] [PubMed] [Google Scholar]
- N'Guessan AL, Elifantz H, Nevin KP, Mouser PJ, Methé B, Woodard TL, et al. Molecular analysis of phosphate limitation in Geobacteraceae during the bioremediation of a uranium-contaminated aquifer. ISME J. 2010;4:253–266. doi: 10.1038/ismej.2009.115. [DOI] [PubMed] [Google Scholar]
- Newman DK, Beveridge TJ, Morel FMM. Precipitation of arsenic trisulfide by Desulfotomaculum auripigmentum. Appl Environ Microbiol. 1997b;63:2022–2028. doi: 10.1128/aem.63.5.2022-2028.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman DK, Kennedy EK, Coates JD, Ahmann D, Ellis DJ, Lovley DR, et al. Dissimilatory arsenate and sulfate reduction in Desulfotomaculum auripigmentum sp. nov. Arch Microbiol. 1997a;168:380–388. doi: 10.1007/s002030050512. [DOI] [PubMed] [Google Scholar]
- Nicholas DR, Ramamoorthy S, Palace V, Spring S, Moore JN, Rosenzweig RF. Biogeochemical transformations of arsenic in circumneutral freshwater sediments. Biodegradation. 2003;14:123–137. doi: 10.1023/a:1024031700533. [DOI] [PubMed] [Google Scholar]
- Nickson RT, McArthur JM, Ravenscroft P, Burgess WG, Ahmed KM. Mechanism of arsenic release to groundwater, Bangladesh and West Bengal. Appl Geochem. 2000;15:403–413. [Google Scholar]
- O'Neil RA, Holmes DE, Coppi MV, Adams LA, Larrahondo MJ, Ward JE, et al. Gene transcript analysis of assimilatory iron limitation in Geobacteraceae during groundwater bioremediation. Environ Microbiol. 2008;10:1218–1230. doi: 10.1111/j.1462-2920.2007.01537.x. [DOI] [PubMed] [Google Scholar]
- Oremland RS, Stolz JF. The ecology of arsenic. Science. 2003;300:939–944. doi: 10.1126/science.1081903. [DOI] [PubMed] [Google Scholar]
- Oremland RS, Stolz JF. Arsenic, microbes and contaminated aquifers. Trends Microbiol. 2005;13:45–49. doi: 10.1016/j.tim.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Oremland RS, Dowdle PR, Hoeft S, Hoeft S, Sharp JO, Schaefer JK, et al. Bacterial dissimilatory reduction of arsenate and sulfate in meromictic Mono Lake, California. Geochim Cosmochim Acta. 2000;64:3073–3084. [Google Scholar]
- Pederick RL, Gault AG, Charnock JM, Polya DA, Lloyd JR. Probing the biogeochemistry of arsenic: response of two contrasting aquifer sediments from Cambodia to stimulation by arsenate and ferric iron. J Environ Sci Health A Tox Hazard Subst Environ Eng. 2007;42:1763–1774. doi: 10.1080/10934520701564269. [DOI] [PubMed] [Google Scholar]
- Pedersen HD, Postma D, Jakobsen R. Release of arsenic associated with the reduction and transformation of iron oxides. Geochim Cosmochim Acta. 2006;70:4116–4129. [Google Scholar]
- Perez-Jimenez JR, DeFraia C, Young LY. Arsenate respiratory reductase gene (arrA) for Desulfosporosinus sp strain Y5. Biochem Biophys Res Commun. 2005;338:825–829. doi: 10.1016/j.bbrc.2005.10.011. [DOI] [PubMed] [Google Scholar]
- Prakash O, Gihring TM, Dalton DD, Chin KJ, Green SJ, Akob DM, et al. Geobacter daltonii sp. nov., an Fe(III)- and uranium(VI)-reducing bacterium isolated from a shallow subsurface exposed to mixed heavy metal and hydrocarbon contamination. Int J Syst Evol Microbiol. 2010;60:546–553. doi: 10.1099/ijs.0.010843-0. [DOI] [PubMed] [Google Scholar]
- Rochette EA, Fendorf SE, Reynolds JG.1997Arsenic speciation of biotically-reduced systems by XANES spectroscopy Abstracts Papers Am Chem Soc 21460-GEOC. [Google Scholar]
- Rochette EA, Bostick BC, Li GC, Fendorf S. Kinetics of arsenate reduction by dissolved sulfide. Environ Sci Technol. 2000;34:4714–4720. [Google Scholar]
- Rosen BP. Families of arsenic transporters. Trends Microbiol. 1999;7:207–212. doi: 10.1016/s0966-842x(99)01494-8. [DOI] [PubMed] [Google Scholar]
- Rosen BP. Biochemistry of arsenic detoxification. FEBS Lett. 2002;529:86–92. doi: 10.1016/s0014-5793(02)03186-1. [DOI] [PubMed] [Google Scholar]
- Rowland HAL, Polya DA, Gault AG, Charnock JM, Pederick RL, Lloyd JR. Microcosm studies of microbially mediated arsenic release from contrasting Cambodian sediments. Geochim Cosmochim Acta. 2004;68:A390–A390. [Google Scholar]
- Rowland HAL, Pederick RL, Polya DA, Pancost RD, Van Dongen BE, Gault AG, et al. The control of organic matter on microbially mediated iron reduction and arsenic release in shallow alluvial aquifers, Cambodia. Geobiology. 2007;5:281–292. [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Saltikov CW, Olson BH. Homology of Escherichia coli R773 arsA, arsB, and arsC genes in arsenic-resistant bacteria isolated from raw sewage and arsenic-enriched creek waters. Appl Environ Microbiol. 2002;68:280–288. doi: 10.1128/AEM.68.1.280-288.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltikov CW, Newman DK. Genetic identification of a respiratory arsenate reductase. Proc Natl Acad Sci USA. 2003b;100:10983–10988. doi: 10.1073/pnas.1834303100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltikov CW, Wildman RA, Newman DK. Expression dynamics of arsenic respiration and detoxification in Shewanella sp strain ANA-3. J Bacteriol. 2005;187:7390–7396. doi: 10.1128/JB.187.21.7390-7396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltikov CW, Cifuentes A, Venkateswaran K, Newman DK. The ars detoxification system is advantageous but not required for As(V) respiration by the genetically tractable Shewanella species strain ANA-3. Appl Environ Microbiol. 2003a;69:2800–2809. doi: 10.1128/AEM.69.5.2800-2809.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelobolina ES, Vrionis HA, Findlay RH, Lovley DR. Geobacter uraniireducens sp nov., isolated from subsurface sediment undergoing uranium bioremediation. Int J Syst Evol Microbiol. 2008;58:1075–1078. doi: 10.1099/ijs.0.65377-0. [DOI] [PubMed] [Google Scholar]
- Shelobolina ES, Coppi MV, Korenevsky AA, DiDonato LN, Sullivan SA, Konishi H, et al. Importance of c-type cytochromes for U(VI) reduction by Geobacter sulfurreducens. BMC Microbiol. 2007;7:16. doi: 10.1186/1471-2180-7-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smedley PL, Kinniburgh DG. A review of the source, behaviour and distribution of arsenic in natural waters. Appl Geochem. 2002;17:517–568. [Google Scholar]
- Smith CJ, Nedwell DB, Dong LF, Osborn AM. Evaluation of quantitative polymerase chain reaction-based approaches for determining gene copy and gene transcript numbers in environmental samples. Environ Microbiol. 2006;8:804–815. doi: 10.1111/j.1462-2920.2005.00963.x. [DOI] [PubMed] [Google Scholar]
- Smith E, Naidu R, Alston AM. Chemistry of inorganic arsenic in soils: II. Effect of phosphorus, sodium, and calcium on arsenic sorption. J Environ Qual. 2002;31:557–563. [PubMed] [Google Scholar]
- Stolz JF, Basu P, Santini JM, Oremland RS. Arsenic and selenium in microbial metabolism. Annu Rev Microbiol. 2006. pp. 107–130. [DOI] [PubMed]
- Stolz JF, Ellis DJ, Blum JS, Ahmann D, Lovley DR, Oremland RS. Sulfurospirillum barnesii sp nov and Sulfurospirillum arsenophilum sp nov., new members of the Sulfurospirillum clade of the epsilon Proteobacteria. Int J Syst Bacteriol. 1999;49:1177–1180. doi: 10.1099/00207713-49-3-1177. [DOI] [PubMed] [Google Scholar]
- Stucker VT, Williams KH, Robbins MJ, Ranville JF.2012Arsenic geochemistry in a biostimulated aquifer: an aqueous speciation study Environ Toxicol Chem(submitted). [DOI] [PubMed]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tufano KJ, Fendorf S. Confounding impacts of iron reduction on arsenic retention. Environ Sci Technol. 2008a;42:4777–4783. doi: 10.1021/es702625e. [DOI] [PubMed] [Google Scholar]
- Tufano KJ, Reyes C, Saltikov CW, Fendorf S. Reductive processes controlling arsenic retention: revealing the relative importance of iron and arsenic reduction. Environ Sci Technol. 2008b;42:8283–8289. doi: 10.1021/es801059s. [DOI] [PubMed] [Google Scholar]
- Van Geen A, Rose J, Thoral S, Garnier JM, Zheng Y, Bottero JY. Decoupling of As and Fe release to Bangladesh groundwater under reducing conditions. Part II: evidence from sediment incubations. Geochim Cosmochim Acta. 2004;68:3475–3486. [Google Scholar]
- Vrionis HA, Anderson RT, Ortiz-Bernad I, O'Neill KR, Resch CT, Peacock AD, et al. Microbiological and geochemical heterogeneity in an in situ uranium bioremediation field site. Appl Environ Microbiol. 2005;71:6308–6318. doi: 10.1128/AEM.71.10.6308-6318.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD, Krumholz LR. Uranium reduction. Annu Rev Microbiol. 2006;60:149–166. doi: 10.1146/annurev.micro.59.030804.121357. [DOI] [PubMed] [Google Scholar]
- Wilkins MJ, Callister SJ, Miletto M, Williams KH, Nicora CD, Lovley D, et al. Development of a biomarker for Geobacter activity and strain composition; Proteogenomic analysis of the citrate synthase protein during bioremediation of U(VI) Microb Biotechnol. 2010;4:55–63. doi: 10.1111/j.1751-7915.2010.00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KH, Long PE, Davis JA, Wilkins MJ, N'Guessan AL, Steefel CI, et al. Acetate availability and its influence on sustainable bioremediation of uranium-contaminated groundwater. Geomicrobiol J. 2011;28:519–539. [Google Scholar]
- Wu W-M, Carley J, Gentry T, Ginder-Vogel MA, Fienen M, Mehlhorn T, et al. Pilot-scale in situ bioremedation of uranium in a highly contaminated aquifer. 2. Reduction of U(VI) and geochemical control of U(VI) bioavailability. Environ Sci Technol. 2006;40:3986–3995. doi: 10.1021/es051960u. [DOI] [PubMed] [Google Scholar]
- Wu W-M, Carley J, Luo J, Ginder-Vogel MA, Cardenas E, Leigh MB, et al. In situ bioreduction of uranium (VI) to submicromolar levels and reoxidation by dissolved oxygen. Environ Sci Technol. 2007;41:5716–5723. doi: 10.1021/es062657b. [DOI] [PubMed] [Google Scholar]
- Wysocki R, Clemens S, Augustyniak D, Golik P, Maciaszczyk E, Tamas MJ, et al. Metalloid tolerance based on phytochelatins is not functionally equivalent to the arsenite transporter Acr3p. Biochem Biophys Res Commun. 2003;304:293–300. doi: 10.1016/s0006-291x(03)00584-9. [DOI] [PubMed] [Google Scholar]
- Zobrist J, Dowdle PR, Davis JA, Oremland RS. Mobilization of arsenite by dissimilatory reduction of adsorbed arsenate. Environ Sci Technol. 2000;34:4747–4753. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.