Abstract
Numerous studies have reported the existence of sponge-specific 16S ribosomal RNA (rRNA) gene sequence clusters, representing bacteria found in sponges but not detected in other environments, such as seawater. The advent of deep-sequencing technologies allows us to examine the rare microbial biosphere in order to establish whether these bacteria are truly sponge specific, or are more widely distributed but only at abundances below the detection limit of conventional molecular approaches. We screened >12 million publicly available 16S rRNA gene pyrotags derived from 649 seawater, sediment, hydrothermal vent and coral samples from temperate, tropical and polar regions. We detected 77 of the 173 previously described sponge-specific clusters in seawater or other non-sponge samples, albeit generally at extremely low abundances. Sequences representing the candidate phylum ‘Poribacteria', previously thought to be largely restricted to sponges, were recovered from 46 (out of 411) seawater and 41 (out of 129) sediment samples. While the presence of an organism does not imply that it is active in situ, our results do suggest that many ‘sponge-specific' bacteria occur more widely outside of sponge hosts than previously thought.
Keywords: marine sponge, bacteria, specificity, 16S rRNA pyrosequencing
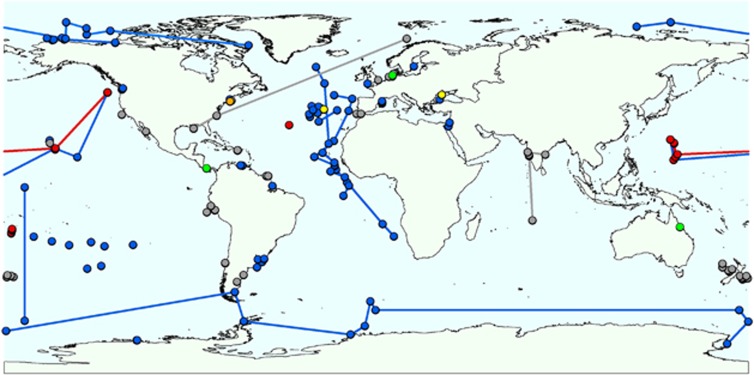
Marine sponges form relationships with a diverse range of microbes (Taylor et al., 2007; Webster and Taylor, 2012), and many of these associations are highly host specific (Schmitt et al., 2012). Numerous studies have reported the existence of monophyletic sponge-specific 16S ribosomal RNA (rRNA) gene sequence clusters, representing bacteria found in sponges but not detected in other environments, such as seawater (Hentschel et al., 2002; Taylor et al., 2007; Simister et al., 2012). In a recent 16S rRNA gene tag pyrosequencing study (Webster et al., 2010), we revealed the presence of sequences affiliated with ‘sponge-specific' clusters in seawater. While this finding suggests that ‘sponge-specific' bacteria can in fact reside outside of these hosts, the seawater was collected only 10 m away from the sampled sponges at the time of sponge spawning and thus a sponge origin for these bacteria could not be unequivocally ruled out. To clarify this fundamental issue in sponge symbiont biology, we examined >12 million 16S rRNA gene pyrotags (V6 region) generated under the auspices of the International Census of Marine Microbes (ICoMM; http://icomm.mbl.edu/). These sequences were derived from a range of seawater, sediment, hydrothermal vent and coral samples obtained from temperate, tropical and polar regions (Figure 1). The absence of ‘sponge-specific' bacteria from these samples would provide strong additional evidence for their host specificity, whereas their widespread occurrence among non-sponge samples would imply that they are capable of at least surviving in a free-living state.
Figure 1.
Geographic distribution of samples from the ICoMM, which were investigated in this study. Each line links together samples from a given ICoMM study. Colour codes represent sample types as shown in Supplementary Table 1: blue=seawater samples; grey=sediment; red=hydrothermal vent; green=host-associated; yellow=biofilm/microbial mat; brown=salt marsh.
We downloaded Bacteria-derived 16S rRNA gene pyrotag sequences that were publicly accessible via the ICoMM website in May 2011. All sequence reads had previously been de-replicated into 3% operational taxonomic units by ICoMM, and all singleton operational taxonomic units (that is, those operational taxonomic units represented in the entire ICoMM data set by only a single sequence read) were removed before analysis. This yielded a total of 12 312 433 sequences obtained from 649 samples in 42 different studies (see Supplementary Table S1 for detailed sample information). These sequences were screened against a manually curated SILVA 16S rRNA database (Pruesse et al., 2007) containing 366 026 bacterial sequences, using a phylogenetic assignment procedure based on those described previously (Webster et al., 2010; Schmitt et al., 2012). One-hundred and seventy-three sponge-specific clusters (SC1–SC173) and 32 sponge-coral-specific clusters (SCC 1–SCC 32) were identified in an earlier study (Simister et al., 2012) and were represented in the SILVA database. In brief, each ICoMM-derived pyrotag was subjected to a BLAST (Altschul et al., 1990) search against the curated SILVA database, and the 50 best hits were aligned in order to determine sequence similarities. The most similar pyrotag sequence to the respective reference sequence was then assigned (or not) to an SC or SCC cluster based on application of a 75% sequence similarity threshold (that is, a sequence read was only assigned to a cluster if it was more similar to the members of that cluster than to other sequences outside the cluster and its similarity to the most similar sequence within that cluster was above 75%). In cases where the assignment of the most similar sequences was inconsistent, a majority rule was applied, and the pyrotag was only assigned to an SC or SCC if at least 60% of the reference sequences were affiliated with this cluster.
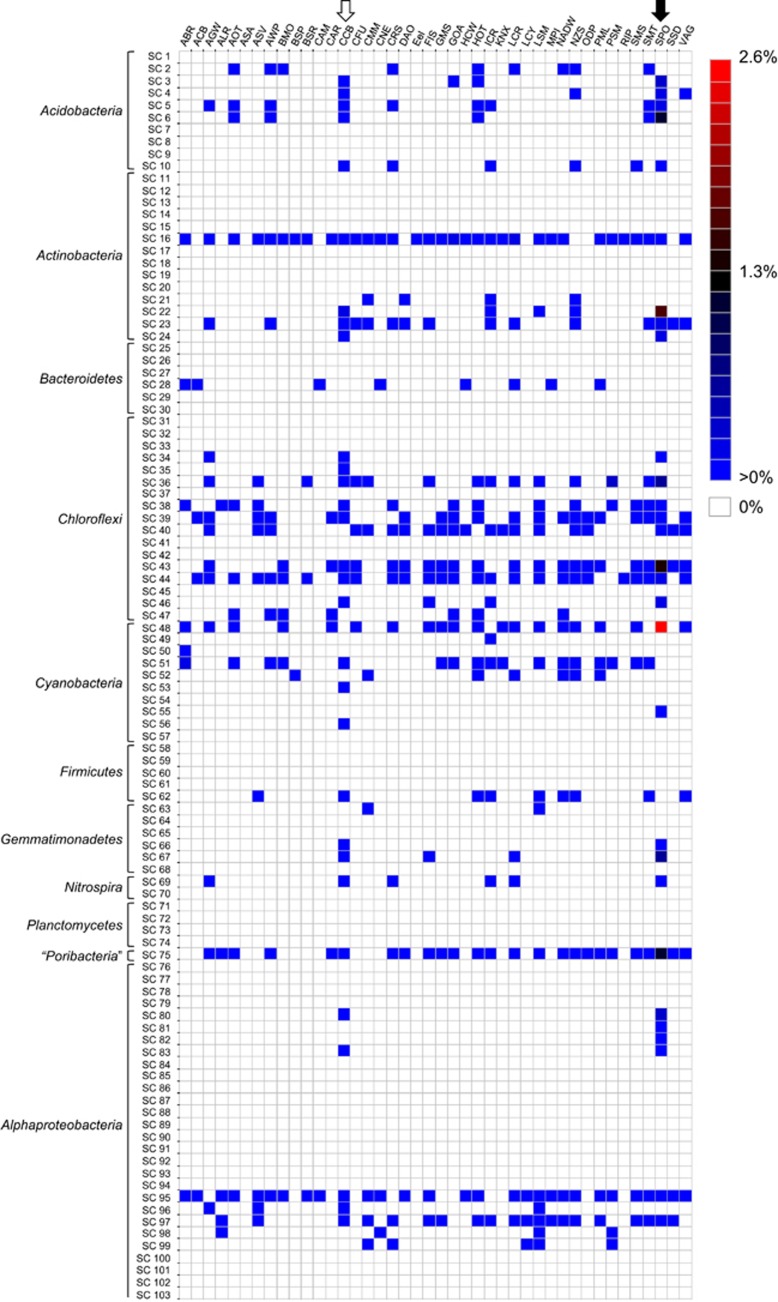
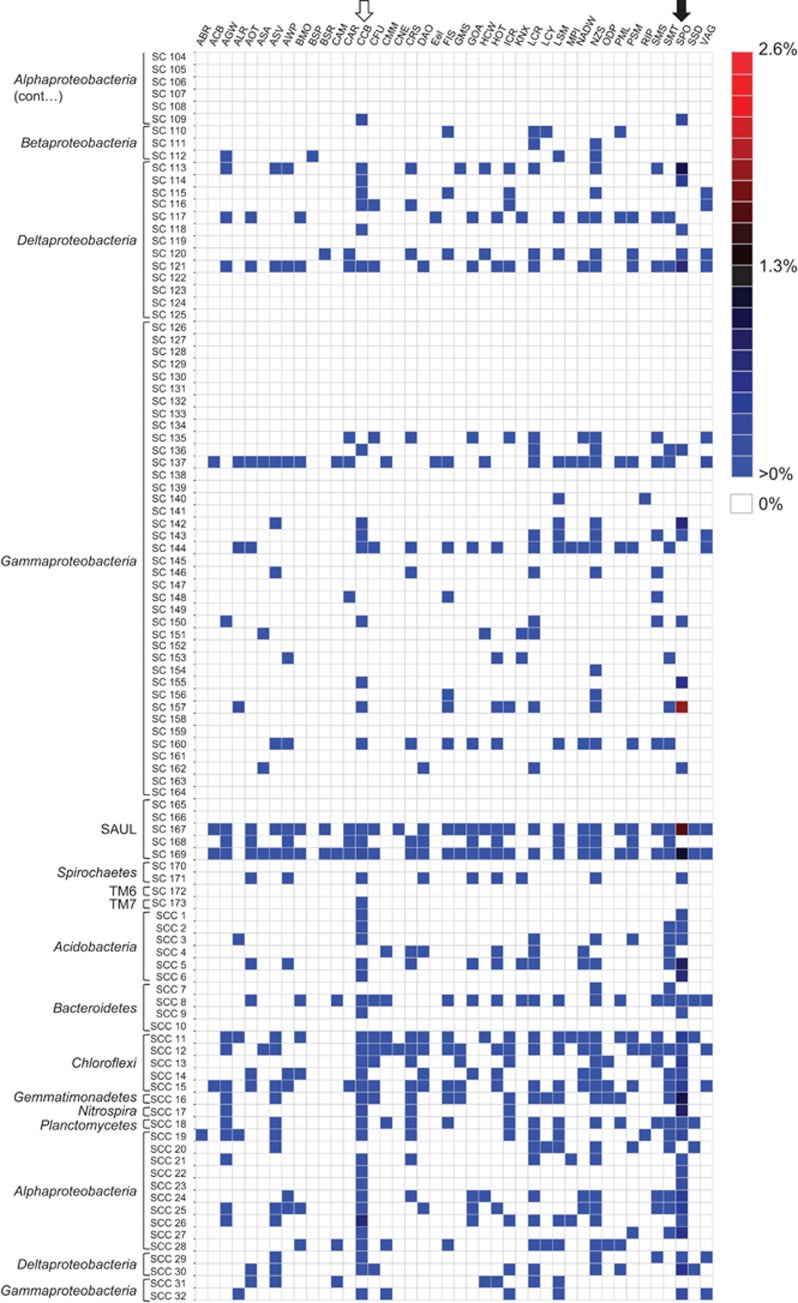
We detected 77 of 173 previously described SC in seawater or other non-sponge samples, albeit generally at extremely low abundances (for example, 2 sequence reads out of 20 000 for a given sample) (Figure 2; Supplementary Table S1). Twelve of these 77 clusters were identified only in coral samples (Caribbean Coral Bacteria, CCB study). Of 32 previously described SCC, 27 were recorded outside of either sponges or corals (Supplementary Table S1). Those that were present in higher numbers included clusters within the Gammaproteobacteria (SC157, comprising 1.96% of sequence reads in the hydrothermal vent sample ALR_0008_2005_05_04; Supplementary Table S1), Actinobacteria (SC16, comprising 1.28% of sequences in the coastal seawater sample LCR_0002_2009_11_13), and a lineage of uncertain affiliation (sponge-associated unidentified lineage (Schmitt et al., 2012)) related to the Planctomycetes–Verrucomicrobia–Chlamydiae superphylum (SC169, comprising 1.07% of sequences in the coral sample CCB_0001_2008_03_26; 0.36% in the seawater samples HOT177_5 and HOT186_175). One important caveat to keep in mind when considering these data is that, while 16S rRNA gene pyrotags may be represented in many samples, this does not necessarily mean that the exact same microorganisms are so widespread. This is, of course, an acknowledged limitation of the 16S rRNA gene as a phylogenetic marker and is not unique to this study. It is also worth bearing in mind that the original definition of SC is based on phylogenetic reconstructions, whereas our assignment of ICoMM pyrotags to these clusters is based on sequence similarity. Although we do not believe that this has a substantial effect on our data or its interpretation, we do acknowledge that some sequence tags that are assigned to a given cluster could actually fall adjacent to (rather than within) that cluster.
Figure 2.
Occurrence of ‘sponge-specific' 16S rRNA gene sequence clusters in ICoMM samples. Heatmap showing the distribution of representatives of previously described ‘sponge-specific' 16S rRNA gene sequence clusters (Simister et al., 2012) among the V6 pyrotag sequences downloaded from the ICoMM website. Clusters with an SC prefix contain sequences previously reported only from sponges; SCC prefix signifies clusters containing only sponge- and coral-derived sequences. Displayed data for a given project represent all samples from that project pooled together; data for individual samples is provided in Supplementary Table S1. Black arrow at the top denotes the ‘SPO' study which included 12 marine sponge samples from the Great Barrier Reef (Webster et al., 2010). White arrow denotes the ‘CCB' study of Caribbean corals (Sunagawa et al., 2010). The colours indicate relative abundance of sequence reads, from blue (low abundance) via black to red (high abundance); white indicates that no sequences were assigned to the respective cluster. ICoMM project names: ABR, Active But Rare; ACB, Arctic Chuki Beaufort; AGW, Amazon-Guianas Water; ALR, Lau Hydrothermal Vent; AOT, Atlantic Ocean Transect; ASA, Amundsen Sea Antarctica; ASV, Azorean Shallow Vents; AWP, Azores Waters Project; BMO, Blanes Bay Microbial Observatory; BSP, Baltic Sea Proper; BSR, Black Sea Redox; CAM, Census Antarctic Marine; CAR, Cariaco Basin; CCB, Caribbean Coral Bacteria; CFU, Deep Subseafloor Sediment; CMM, Coastal Microbial Mats; CNE, Coastal New England; CRS, Coral Reef Sediment; DAO, Deep Arctic Ocean; Eel, Eel Pond Winter Pilot Study; FIS, Frisian Island Sylt; GMS, Guaymas Methane Seeps; GOA, Gulf of Aqaba; HCW, Hood Canal Washington; HOT, Hawaiian Ocean Time Series; ICR, IOMM Cooperative Run; KNX, Station KNOX South Pacific Gyre Ocean Drilling Project; LCR, LaCAR Cooperative Run; LCY, Lost City; LSM, Salt Marsh study; MPI, Helgoland; NADW, North Atlantic Deep Water Flow; NZS, New Zealand Sediment; ODP, Ocean Drilling Project; PML, English Channel; PSM, Pilot Project—Seamounts; RIP, Dead Sea Project; SMS, Station M Sediments; SMT, Hydrothermal seamounts; SPO, sponges; SSD, Spatial Scaling Diversity; VAG, Humboldt Marine Ecosystem.
The candidate phylum ‘Poribacteria' occurs in many marine sponges, often at very high abundances (Fieseler et al., 2004; Lafi et al., 2009). Having never been detected outside of sponges using conventional molecular approaches such as 16S rRNA gene library construction or fluorescence in situ hybridisation, ‘Poribacteria' were generally considered to be restricted to sponge hosts. However, their recent detection in seawater by pyrotag (Webster et al., 2010) and fosmid (Pham et al., 2008) sequencing suggested their wider occurrence in non-sponge habitats. In this study, we detected sequences affiliated with ‘Poribacteria' (cluster SC75) in 46 seawater, 41 sediment, 14 hydrothermal vent, 14 salt marsh and 3 coral samples collected from around the world (Figure 2, Supplementary Table S1). Invariably, numbers were low, never comprising >0.19% of sequences in seawater or sediment samples. Interestingly, the highest non-sponge occurrence of ‘Poribacteria' was in a coral, where they comprised 0.25% of sequence reads in the sample CCB_0001_2008_03_26 (Supplementary Table S1). SC vary greatly in size, with clusters such as the ‘Poribacteria' (SC75) containing 136 sequences whereas other, smaller clusters such as the Planctomycetes cluster SC74 contain as few as three sequences. To determine whether the size of a sponge-specific cluster increases the likelihood of a non-sponge-derived sequence being assigned to it, we performed a simple analysis to compare SC/SCC size with the number of matching ICoMM pyrotags (data not shown). Cluster size was a very poor predictor of sequence tag assignment (R2=0.0124), indicating that this factor has only a negligible effect.
This study provides compelling evidence for the occurrence of many so-called sponge-specific bacteria in other marine environments. However, even upon examination of more than 12 million sequences, 96 of the 173 previously defined SC were undetected outside of sponges and, therefore, remain putatively sponge specific. These include clusters within the Acidobacteria, Actinobacteria, Chloroflexi, Cyanobacteria, Gemmatimonadetes, Alphaproteobacteria and Gammaproteobacteria (Figure 2). If these bacteria are indeed absent outside of sponges, then the symbiotic association must be maintained solely via vertical transmission. The transmission of diverse microbial communities between sponge generations is well documented (Sharp et al., 2007; Schmitt et al., 2008; Webster et al., 2010). It is worth noting that the absence of these clusters from the analysed ICoMM data set does not confirm their complete absence from the environment and further sequencing could ultimately reveal their presence outside of sponge hosts.
The detection of an organism's DNA in a sample does not necessarily mean that this organism is active in situ. Hence, the finding of ‘sponge-specific' bacteria in diverse habitats outside of sponges could theoretically reflect the presence of metabolically inactive dispersal stages for these bacteria. This type of rare biosphere ‘seed bank' for colonisation of sponges has been suggested before (Webster et al., 2010) and could explain the apparent lack of co-speciation in sponge–microbe associations. It is often suggested that cellular rRNA concentrations are correlated with growth rate and activity, hence rRNA may reflect which members of the community are active (Kamke et al., 2010). To explore whether these ‘sponge-specific' bacteria were metabolically active outside of their host we examined the only ICoMM study to have included both RNA- and DNA-derived samples (Coral Reef Sediment, (CRS); Gaidos et al., 2011). Of the sponge-specific and sponge-coral-specific clusters identified in the CRS study, 25 contained CRS sequences found only in DNA-derived samples, 9 contained sequences in both DNA- and RNA-derived samples and only 1 contained sequences exclusively from RNA-derived samples. While the majority of sponge-specific sequences were recovered only from the reef sediment DNA fraction, the finding of any sponge-specific bacteria within the RNA fraction suggests that at least some of these microorganisms may be active outside the confines of the host. Sponge-specific bacteria in the environment could also originate from adult sponge tissue via damage and/or release of reproductive stages, although this would seem unlikely for many of the environments included in this study such as deep-sea hydrothermal vents and open ocean waters.
The occurrence of putatively sponge-specific bacteria in so many non-sponge samples strongly suggests that they are capable of surviving outside of sponge hosts. The analysed data demonstrate the power of next-generation sequencing technologies for improving our understanding of marine symbioses and should enable further identification of hitherto unknown reservoirs for these bacteria.
Acknowledgments
We gratefully acknowledge funding support from University of Auckland Faculty Research Development Fund grants (3609286 and 3622989) to MWT, a German Research Foundation (DFG) grant (SCHM 2559/1-1) to SS, and a Feodor Lynen Research Fellowship from the Alexander von Humboldt Foundation to PD. L Taylor is thanked for insightful comments on the manuscript. We also thank ICoMM for the public availability of 16S rRNA gene sequence data.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Fieseler L, Horn M, Wagner M, Hentschel U. Discovery of the novel candidate phylum "Poribacteria" in marine sponges. Appl Environ Microbiol. 2004;70:3724–3732. doi: 10.1128/AEM.70.6.3724-3732.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaidos E, Rusch A, Ilardo M. Ribosomal tag pyrosequencing of DNA and RNA from benthic coral reef microbiota: community spatial structure, rare members and nitrogen-cycling guilds. Environ Microbiol. 2011;13:1138–1152. doi: 10.1111/j.1462-2920.2010.02392.x. [DOI] [PubMed] [Google Scholar]
- Hentschel U, Hopke J, Horn M, Friedrich AB, Wagner M, Hacker J, et al. Molecular evidence for a uniform microbial community in sponges from different oceans. Appl Environ Microbiol. 2002;68:4431–4440. doi: 10.1128/AEM.68.9.4431-4440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamke J, Taylor MW, Schmitt S. Activity profiles for marine sponge-associated bacteria obtained by 16S rRNA vs 16S rRNA gene comparisons. ISME J. 2010;4:498–508. doi: 10.1038/ismej.2009.143. [DOI] [PubMed] [Google Scholar]
- Lafi FF, Fuerst JA, Fieseler L, Engels C, Goh WW, Hentschel U. Widespread distribution of Poribacteria in Demospongiae. Appl Environ Microbiol. 2009;75:5695–5699. doi: 10.1128/AEM.00035-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham VD, Konstantinidis KT, Palden T, DeLong EF. Phylogenetic analyses of ribosomal DNA containing bacterioplankton genome fragments from a 4000 m vertical profile in the North Pacific Subtropical Gyre. Environ Microbiol. 2008;10:2313–2330. doi: 10.1111/j.1462-2920.2008.01657.x. [DOI] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S, Angermeier H, Schiller R, Lindquist N, Hentschel U. Molecular microbial diversity survey of sponge reproductive stages and mechanistic insights into vertical transmission of microbial symbionts. Appl Environ Microbiol. 2008;74:7694–7708. doi: 10.1128/AEM.00878-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S, Tsai P, Bell J, Fromont J, Ilan M, Lindquist N, et al. Assessing the complex sponge microbiota: core, variable and species-specific bacterial communities in marine sponges. ISME J. 2012;6:564–576. doi: 10.1038/ismej.2011.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp KH, Eam B, Faulkner DJ, Haygood MG. Vertical transmission of diverse microbes in the tropical sponge Corticium sp. Appl Environ Microbiol. 2007;73:622–629. doi: 10.1128/AEM.01493-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simister RL, Deines P, Botte ES, Webster NS, Taylor MW. Sponge-specific clusters revisited: a comprehensive phylogeny of sponge-associated microorganisms. Environ Microbiol. 2012;14:517–524. doi: 10.1111/j.1462-2920.2011.02664.x. [DOI] [PubMed] [Google Scholar]
- Sunagawa S, Woodley CM, Medina M. Threatened corals provide underexplored microbial habitats. PloS One. 2010;5:e9554. doi: 10.1371/journal.pone.0009554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MW, Radax R, Steger D, Wagner M. Sponge-associated microorganisms: evolution, ecology, and biotechnological potential. Microbiol Mol Biol Rev. 2007;71:295–347. doi: 10.1128/MMBR.00040-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster NS, Taylor MW. Marine sponges and their microbial symbionts: love and other relationships. Environ Microbiol. 2012;14:335–346. doi: 10.1111/j.1462-2920.2011.02460.x. [DOI] [PubMed] [Google Scholar]
- Webster NS, Taylor MW, Behnam F, Lücker S, Rattei T, Whalan S, et al. Deep sequencing reveals exceptional diversity and modes of transmission for bacterial sponge symbionts. Environ Microbiol. 2010;12:2070–2082. doi: 10.1111/j.1462-2920.2009.02065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.