Abstract
Here, we present the first metagenomic study of viral communities from four perennial ponds (gueltas) located in the central Sahara (Mauritania). Three of the four gueltas (Ilij, Molomhar and Hamdoun) are located at the source of three different wadis belonging to the same hydrologic basin, whereas the fourth (El Berbera) belongs to a different basin. Overall, sequences belonging to tailed bacteriophages were the most abundant in all four metagenomes although electron microscopy and sequencing confirmed the presence of other viral groups, such as large DNA viruses. We observed a decrease in the local viral biodiversity in El Berbera, a guelta with sustained human activities, compared with the pristine Ilij and Molomhar, and sequences related to viruses infecting crop pests were also detected as a probable consequence of the agricultural use of the soil. However, the structure of the El Berbera viral community shared the common global characteristics of the pristine gueltas, that is, it was dominated by Myoviridae and, more particularly, by virulent phages infecting photosynthetic cyanobacteria, such as Prochlorococcus and Synechococcus spp. In contrast, the Hamdoun viral community was characterized by a larger proportion of phages with the potential for a temperate lifestyle and by dominant species related to phages infecting heterotrophic bacteria commonly found in terrestrial environments. We hypothesized that the differences observed in the structural and functional composition of the Hamdoun viral community resulted from the critically low water level experienced by the guelta.
Keywords: viral metagenomics, giant virus, Sahara desert, perennial water pond, Mauritania, Adrar plateau
Introduction
Viruses can colonize virtually all ecosystems on Earth and are found wherever cellular life exists (Le Romancer et al., 2007). In the ocean, viruses (the majority of which are bacteriophages) represent the most abundant biological component of the ecosystem and influence horizontal gene transfer, microbial diversity and biogeochemical cycling (Fuhrman, 1999; Suttle, 2005, 2007). Metagenomics (the sequence-based analysis of the collective genomes contained in an environmental sample) was first applied to environmental viral communities in marine waters 10 years ago (Breitbart et al., 2002). This study demonstrated that the viral fraction represents a vast reservoir of unexplored biodiversity. Since then, viral diversity has been investigated using metagenomics in a wide range of environments, including marine waters (Angly et al., 2006; Culley et al., 2006), freshwaters (Dinsdale et al., 2008a; Djikeng et al., 2009), hot springs (Schoenfeld et al., 2008), soils (Williamson et al., 2005; Fierer et al., 2007), stromatolites and thrombolites (Desnues et al., 2008), and animal-associated biomes (Breitbart et al., 2003; Zhang et al., 2006; Vega Thurber et al., 2008; Ng et al., 2011).
To date, studies on viral diversity have mainly focused on marine waters from temperate regions, and data exploring the extent of viral diversity and ubiquity in arid regions are largely scarce. Unfavorable conditions found in cold or hot deserts limit the development and the activity of eukaryotic life. In such environments, the study of viral assemblages is of particular interest because microbial communities are mainly regulated by viral lysis (Weinbauer, 2004; Laybourn-Parry, 2009). A recent study in the cold desert of Antarctica used a 0.45-μm size selective metagenomic analysis to show that the viral community of an ice-covered lake presented an unexpectedly high genetic richness, distinct from that of other aquatic viral metagenomes, and was dominated by small single-stranded DNA viruses infecting eukaryotes in the spring and by large double-stranded DNA (dsDNA) viruses (mostly Phycodnaviruses and Mimiviruses) and dsDNA bacteriophages in summer (Lopez-Bueno et al., 2009).
The Sahara is the largest non-polar desert on Earth. In the early Holocene period, the Sahara experienced humid episodes (Kuper and Kröpelin, 2006) that sustained the development of numerous lakes and wetlands; remnants of these aquatic environments still persist today. The Mauritanian Adrar is one of the mountainous massifs of the central Sahara and contains >20 perennial and semi-perennial freshwater bodies. Among them, rocky pools (gueltas) are found in the higher reaches of gorge-like watercourses and are alimented by subterranean seep and rainfall. Some of these gueltas are sites of permanent human settlements and are used for agricultural and animal-farming purposes.
Previous electron microscopy and pulse-field gel electrophoresis studies from bacteriophage particles induced from Namib and Sahara sands have revealed the presence of different morphotypes with genome sizes varying from 45 to 350 kb, suggesting the existence of large viral particles (Prigent et al., 2005; Prestel et al., 2008). The objective of this study was to fill the gaps in our knowledge of the ubiquity and diversity of viruses in hot desert environments. Using metagenomic approaches, we investigated the composition, taxonomy and functional diversity of the viral communities from four gueltas located in the Mauritanian Sahara.
Materials and methods
Geographic location of the sampling sites
During the dry season of 2009 (June), water samples were collected from four different gueltas: Ilij (20°38′046 N, 13°08′490 W), Molomhar (20°35′229 N, 13°08′794 W), Hamdoun (20°19′380 N, 13°08′550 W) and El Berbera (19°59′181 N, 12°49′3744 W) located in the Adrar plateau of Mauritania (Figure 1). Ilij, Molomhar and Hamdoun belong to the same hydrographic network (the Seguellîl wadi basin), whereas El Berbera belongs to a different network (the Timinit wadi basin). The local human population occasionally and permanently occupies the Hamdoun and El Berbera gueltas, respectively. Hamdoun is located close to the main Atar-Nouakchott road and may serve as a temporary open well, whereas El Berbera hosts a permanent human settlement and is a site of intensive date fruit production. During the driest periods of the year, the residual water volume of the Hamdoum guelta can drop to <2 m3, whereas the volumes of the other gueltas remain between 200 and 500 m3.
Figure 1.
Geographic localization and pictures of the samp ling sites. Central: A Google Earth map of the Adrar region showing mountains (gray) and sandstones (yellow). Il: Ilij guelta, Mo: Molomhar guelta, Ha: Hamdoun guelta, ElB: El Berbera guelta. The dashed line on the map indicates the separation between the two hydrologic systems: the Seguellîl wadi basin and the Timinit wadi basin. Images of the sampling sites are provided on the left and on the right of the map. Upper right: Localization of the 205 environmental metagenomic projects recorded in the Genome On Line Database (GOLD) as of 2012-01-10. Africa is outlined by a circle, and the Adrar plateau of Mauritania is indicated by a star.
Sampling procedure, virus purification, transmission electron microscopy, nucleic acid extraction and sequencing
During a 2-day mission, one liter of water was collected from each guelta and filtered through a 0.45-μm pore filter. Virus-size particles contained in the filtrate were precipitated on site using PEG (10%) and NaCl (1 ℳ final) in bottles maintained at 4 °C in a cool-box. Samples (kept at 4 °C) were brought to the laboratory immediately (within the 48 h) for further processing. Precipitated viral particles were purified using CsCl density gradient ultracentrifugation and DNAse treated as previously described (Thurber et al., 2009). Purified viral particles were stained with 3.5% uranyl acetate and lead citrate and then examined by transmission electron microscopy (TEM) (Philips Morgagni 268D, FEI Co., Eindhoven, The Netherlands). Nucleic acids were extracted using the formamide procedure (Thurber et al., 2009) and amplified using the illustra GenomiPhi V2 DNA Amplification Kit (GE Healthcare Life Sciences, Freiburg, Germany). Because phi29 DNA polymerase has been shown to preferentially amplify circular DNA and genomes from single-stranded DNA viruses (Kim et al., 2008; Kim and Bae, 2011), duplicate reactions were performed to minimize this bias, as previously suggested (Thurber et al., 2009). Amplification products were then pooled, ethanol purified and pyrosequenced on a Roche Applied Sciences (454 Life Sciences, Basel, Switzerland) GS20 platform. Metagenomes are freely accessible on the MG-RAST annotation server with the following accession numbers: El Berbera 2 (4446033.3), Molomhar Guelta (4445718.3), Ilij Guelta (4445716.3) and Hamdoun Guelta (4445715.3).
Taxonomic and functional annotations
Metagenomes were annotated using MG-RAST version 2 (Meyer et al., 2008) with an E-value cutoff of 10–5. The MG-RAST server produces automated taxonomic assignments using Blastx searches against the SEED non-redundant database and other accessory databases (rRNA, chloroplast and mitochondrial databases) and also produces metabolic profiles of metagenomes by Blastx comparisons using the SEED-Subsystem data set. Pairwise comparisons of the metabolic profiles were performed using XIPE-TOTEC (Rodriguez-Brito et al., 2006), a non-parametric pairwise bootstrap statistical test that was specifically developed for metagenomic functional comparisons and is based on median difference analysis. This test locates statistically significant differences and identifies subsystems that are overrepresented in each comparison. The confidence level chosen for the test was 98%.
Sequence analysis
The GC content of the four metagenomes was analyzed using the geecee function of EMBOSS. The average GC fraction was computed for each metagenome as a whole or separately for subsets of bacterial- and viral-annotated reads.
Assembly and phylogeny
The assembly of each metagenome was performed using the Genome Sequencer (GS) De Novo Assembler version 2.0.01 (Roche Diagnostics, Meylan, France), an application especially suited to the analysis of GS-FLX data. We chose a minimum overlap length of 20 bp and a minimum overlap identity of 95%. We only kept contigs longer than 300 bp for subsequent analyses because the average read length was 251–258 bp. Open Reading Frames (ORFs) were searched on large contigs (>1500 bp) by Prodigal (Hyatt et al., 2010) and MetaGeneMark (Zhu et al., 2010). Phylogenetic trees were constructed for ORFs with at least 10 homologs, according to a Blastx search against the NCBI non-redundant database (E-value<1e−10). An ORF and its homologs were aligned using MUSCLE (Edgar, 2004), and the alignment was curated using Gblocks (Castresana, 2000). Phylogenetic trees were constructed using PhyML (Guindon et al., 2010), with 100 bootstrap replicates, and visualized using MEGA v5 software (Tamura et al., 2011). A specific research of phages and prophages has also been performed on assembled contigs by a Blastn search (E-value<1e−05) against the Aclame database (Leplae et al., 2004).
Mapping
For each of the most abundant organisms found in the MG-RAST analysis, metagenomic reads were mapped (that is, aligned against a reference sequence) on the genome of that organism. Mapping was performed using the GS Reference Mapper version 2.0.01 (Roche).
Population modeling
Information about community structure and diversity was obtained for each metagenome using the following workflow: (i) computation of the community contig spectrum using the free Circonspect software, (ii) evaluation of average genome sizes using GAAS free software (Angly et al., 2009) with an E-value cutoff of 10–3 and (iii) mathematical modeling of the community structure and diversity by PHACCS (Angly et al., 2005).
Phylogenetic tests
Several phylogenetic tests were performed using the FastUniFrac tool (Hamady et al., 2009) to find statistically significant differences among the four metagenomes. The analyses were performed on the subset of viral sequences of each metagenome. For each metagenome, FastUniFrac uses phylogenetic information to assemble metagenomic sequences into a tree. P-test and UniFrac metric capture significant diversity between the trees associated with the different metagenomes, and they account only for tree topology or for both tree topology and branch length, respectively. The two tests can be used for multiple or pairwise comparisons or to compare one particular tree with all others. Principal Component Analysis (PCA) and hierarchical clustering were also performed using FastUniFrac. The robustness of the clustering results to the sampling effort and evenness was determined using the Jackknife Environment Clusters analysis option.
Comparative metagenomics of viral communities from freshwater environments
A multiple comparison of the phylogenetic profiles of different natural and non-natural freshwater viral communities was performed using the MG-RAST server (Meyer et al., 2008). Ten viral metagenomes were compared; the four analyzed in this work, two from two different temperate freshwater lakes (Roux et al., 2012), one from an Antarctic lake sampled in the spring and summer seasons (Lopez-Bueno et al. 2009), and two from an aquaculture system (Dinsdale et al., 2008a). The phylogenetic profile was based on the sequence taxonomic assignment according to a Blastx search (E-value<1e−05) against the NCBI GenBank non-redundant database. Multiple comparisons were performed by PCA on the MG-RAST server using normalized values and a Bray-Curtis distance matrix. P-values were computed on the MG-RAST server (Meyer et al., 2008).
Results
Taxonomic composition of the viral metagenomes
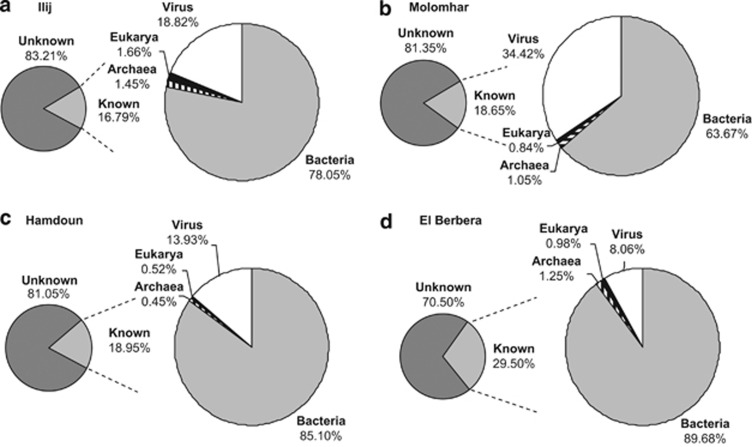
A total of 82 814 818 bp of sequence was generated from the four samples (Ilij ∼17 Mbp, Molomhar ∼25 Mbp, Hamdoun ∼15 Mbp, El Berbera ∼24 Mbp), corresponding to 324 603 sequences with an average length of 250 bp. Annotation of the sequence fragments by MG-RAST using an E-value cutoff of 1e−05 indicated that 70.50–83.21% of these fragments had no significant hits to known sequences stored in the SEED non-redundant database or other accessory databases (Figure 2). According to the MG-RAST annotation, 8.06–34.42% of the known reads were classified as viruses. The majority of viral reads belonged to dsDNA viruses (Table 1) and, among these, >92% matched with Caudovirales. Sequences belonging to the Myoviridae were the most abundant in all metagenomes followed by Podoviridae and Siphoviridae. The presence of tailed phages was confirmed by TEM (Figures 3a and b). Viral morphotypes were usually between 50 and 200 nm in diameter, but some viral particles with diameters <50 nm (Figures 3d–f, arrows) and >200 nm (Figure 3c) were also observed. Among the dsDNA viruses, sequences belonging to eukaryotic viruses were also found (Tables 1 and 2). Four out of the seven families of nucleocytoplasmic large DNA viruses group were represented (Table 1, in bold). Viruses from the Phycodnaviridae (infecting algae) and Mimiviridae (infecting amoebas and algae) were more abundant in Hamdoun (4.25%) and El Berbera (3.18%). Poxviridae sequences were also more frequently found in the El Berbera metagenome. Only a few reads were associated with single-stranded DNA viruses. The majority of these reads were found in the Molomhar metagenome and were related to Microviridae (2.92%). To analyze the taxonomic composition more accurately, we used GAAS that normalizes the number of hits for the genome size and then provides a more realistic description of species abundances. According to the GAAS analysis, the most represented viral genotype was that of Prochlorococcus phage, found in three out of the four gueltas (Ilij, Molomhar and El Berbera) with relative abundances ranging between 31.54 and 55.24% (Table 2). In contrast, Hamdoun was dominated by viruses that infect members of the genus Microbacterium (Microbacterium phage Min1), which represented >44% of the total viral genotype abundance (Table 2).
Figure 2.
Reads classification according to their Best Blast Hit (E-value<10–5) in the MG-RAST analysis. (a) Ilij guelta, (b) Molomhar guelta, (c) Hamdoun guelta and (d) El Berbera guelta.
Table 1. Classification of reads hitting viral sequences.
| Group | Order | Family | Ilij (%) | Molomhar (%) | Hamdoun (%) | El Berbera (%) |
|---|---|---|---|---|---|---|
| Unclassified | 5.78 | 2.86 | 5.16 | 5.92 | ||
| Caudovirales | Myoviridae | 63.33 | 73.04 | 48.03 | 56.43 | |
| Podoviridae | 11.75 | 8.45 | 22.61 | 18.92 | ||
| Siphoviridae | 12.50 | 8.20 | 16.39 | 11.93 | ||
| Herpesvirales | Herpesviridae | 0.05 | 0.02 | 0.00 | 0.00 | |
| dsDNA | – | Tectiviridae | 0.05 | 0.06 | 0.00 | 0.00 |
| – | Iridoviridae | 0.67 | 1.11 | 0.83 | 0.54 | |
| – | Phycodnaviridae | 2.08 | 1.20 | 4.25 | 2.25 | |
| – | Poxviridae | 0.28 | 0.02 | 0.38 | 0.54 | |
| – | Mimiviridae | 1.90 | 0.84 | 0.99 | 3.18 | |
| – | Baculoviridae | 0.00 | 0.00 | 0.00 | 0.15 | |
| ssDNA | – | Circoviridae | 0.00 | 0.14 | 0.00 | 0.05 |
| – | Microviridae | 0.00 | 2.92 | 0.15 | 0.09 | |
| – | Geminiviridae | 0.00 | 0.02 | 0.08 | 0.00 | |
| – | Nanoviridae | 0.05 | 0.14 | 0.08 | 0.00 | |
| – | Parvoviridae | 0.00 | 0.00 | 0.08 | 0.00 | |
| Retroviridae | – | 0.00 | 0.00 | 0.08 | 0.00 | |
| Unclassified phages/viruses | – | 1.56 | 0.98 | 0.91 | 0.00 |
Abbreviations: dsDNA, double-stranded DNA; ssDNA, single-stranded DNA.
Assignment was made according to the best Blastx hit (E-value<10−5) in the MG-RAST analysis.
Figure 3.
Viral morphotypes observed under TEM in the gueltas of the Adrar plateau. Example of tailed phages belonging to the Myoviridae family (a, b). Viral morphotypes were usually between 50 and 200 nm in diameter (d–f), but some small viral particles with diameters <50 nm (d–f, arrows) and large Mimivirus-like particles (c) were also observed. Images (a) and (d) are from Ilij, (b) and (f) are from Hamdoun, and (c) and (e) are from Molomhar.
Table 2. Most represented viral genotypes among the viral hits according to GAAS analysis.
| Metagenome | Viral species | Relative abundance (%) | Host |
|---|---|---|---|
| Ilij | Prochlorococcus phages | 55.2449 | B |
| Burkholderia phages | 18.9451 | B | |
| Synechococcusphages | 15.9518 | B | |
| Roseobacter phage SIO1 | 5.8638 | B | |
| Acanthocystis turfacea Chlorella virus 1 | 2.4366 | E | |
| Aeromonas phages | 1.3597 | B | |
| Molomhar | Prochlorococcus phages | 46.0733 | B |
| Synechococcus phages | 35.6391 | B | |
| Mycobacterium phages | 10.2428 | B | |
| Bordetella phages | 2.0430 | B | |
| Acyrthosiphon pisum secondary endosymbiont phage | 1.1061 | E | |
| Acanthocystis turfacea Chlorella virus 1 | 1.0322 | E | |
| Hamdoun | Microbacterium phage Min1 | 44.3371 | B |
| Synechococcus phages | 29.9623 | B | |
| Prochlorococcus phages | 11.5155 | B | |
| Acanthocystis turfacea Chlorella virus 1 | 10.7050 | E | |
| Acanthamoeba polyphaga mimivirus | 3.4801 | E | |
| El Berbera | Prochlorococcus phages | 31.5358 | B |
| Synechococcus phages | 26.8707 | B | |
| Mycobacterium phages | 14.3964 | B | |
| Lactobacillus phage phiJL-1 | 9.7423 | B | |
| Burkholderia phage phi644-2 | 7.3403 | B | |
| Spodoptera litura NPV | 2.7741 | E | |
| Musca domestica salivary gland hypertrophy virus | 2.6775 | E | |
| Acanthocystis turfacea Chlorella virus 1 | 2.4808 | E | |
| Ostreococcus virus OsV5 | 1.7077 | E |
Abbreviations: B, Bacteria; E, Eukaryotes.
Only viral genotypes with a relative abundance superior to 1% are indicated. Viral species infecting cyanobacteria are shown in bold.
Although no bacterial cells could be detected via electron microscopy, 63.67–89.68% of the reads were classified as bacterial in the viral metagenomes (Figure 2; Supplementary Table 1). Using GAAS, all bacteria-annotated reads were dominated by sequences related to the Acinetobacter genus (Supplementary Table 2). Between 3 and 20 sequences matching bacterial 16S rRNA genes were also found for each metagenome (Supplementary Table 3).
Functional annotation and metabolic analysis
The metabolic profile of the four metagenomes was explored using MG-RAST, which assigns sequences to metabolic categories based on their Best Blastx Hit against the SEED database (E-value<1e−05). Only 6.22–17.43% of the sequences could be functionally classified in this way. The most represented categories were related to the metabolism of carbohydrates, amino acids, proteins, cofactors, vitamins, DNA, and nucleosides/nucleotides (Figure 4). We compared these data with the metabolic profile derived from the combined analysis of 42 viral metagenomes (subterranean, hypersaline, marine, aquaculture freshwater, coral, microbialites, fish, terrestrial animals and mosquito) described in a previous study (Dinsdale et al., 2008a). The guelta metagenomes were depleted in virulence subsystems compared with the average value found for the other 42 viral metagenomes. Metabolic profile comparisons using XIPE-TOTEC showed that respiration, regulation and cell signaling, and motility and chemotaxis subsystems were overrepresented in the Hamdoun metagenome compared with the other gueltas (P-value<0.02). Deeper in the respiration subsystem hierarchical levels, the electron donating reaction of the Hamdoun metagenome was dominated by the respiratory dehydrogenase I subsystem, which was mainly represented by the proline dehydrogenase. In contrast, the other three gueltas were dominated by the NAD(P)H dehydrogenase complex, which is classified as a respiratory complex I subsystem. An overrepresentation of RNA metabolism was also evidenced by the XIPE-TOTEC analyses of the El Berbera metagenome, whereas the Molomhar metagenome displayed a statistically significantly higher number of sequences related to photosynthesis and nucleoside/nucleotide metabolism (P-value<0.02).
Figure 4.
Relative abundances of sequences assigned to each metabolic subsystem by MG-RAST. The metabolic categorization is based on the sequences best Blast hits in the SEED database curated subsystems (E-value <1e−05). Asterisks: metabolic subsystems for which pairwise comparisons were performed by XIPE-TOTEC to identify statistically significant differences (P<0.05) between the four guelta metagenomes.
Assembly, contig analysis and mapping
Contigs were assembled using the GS De Novo Assembler, and only contigs longer than 300 bp were kept (Supplementary Table 4). The average contig length was from 742 to 990 bp, and large contigs were also obtained (for example, 55 kbp in El Berbera and 52 kbp in Hamdoun). Overall, 29.90–37.06% of the contigs had similarities to phage and prophage sequences in the Aclame database (Supplementary Table 5). Viral assemblies were dominated by phage genomes. However, mapping to fully sequenced genomes of the most abundant phages in the metagenomes resulted in low coverages (<5% Supplementary Figure 1). Low coverage was also found for plasmids; the maximum plasmid coverage was 14.24% for the Acinetobacter venetianus pAV2 genome.
We were able to assemble 30.33–68.72% of all reads (Supplementary Table 4). Interestingly, between 34.98 and 93.03% of the ‘unknown' reads identified by the MG-RAST annotation system were assembled into contigs (Supplementary Table 4). The assembly of these unknown reads into contigs could facilitate their taxonomic assignment. Indeed, short sequences are less likely than long sequences to retrieve statistically significant similarities in Blast searches and sequence assembly into longer contigs is helpful to overcome this difficulty (Wommack et al., 2008). Moreover, long contigs can contain unknown reads and reads with far homologies to known sequences, which are suggestive of the putative phylogenetic origin of the whole contig. The largest contigs (>1500 bp) were annotated by ORF prediction and Blast search (Supplementary Table 6). When possible, phylogenetic analysis was performed to confirm the origin of the predicted ORFs (Supplementary Figure 2). A few large contigs contained a relevant proportion of predicted ORFs with similarities to phage sequences and coding for some specific conserved phage proteins, that is, terminases, structural proteins (mainly related to Caudovirales tail structures) and phage DNA polymerases (Supplementary Table 6). It has been previously shown that viral genomes contain more ORFans (that is, ORFs without homologs in the databases) than do bacteria (∼30 and ∼10% for viral and bacterial genomes, respectively) (Yin and Fischer, 2006; Boyer et al., 2010) and that viral (meta)genomes tend to be more AT rich than those of their hosts (Rocha and Danchin, 2002; Willner et al., 2009). Thus, contigs with >50% ORFans, low GC%, and for which conserved viral protein-encoding genes have been identified, can confidently be considered as of viral origin (for example, contigs ElBerbera_882 or Hamdoun_439 in Supplementary Table 6).
Community phylogenetic structure and diversity across sampling sites
Viral community structure and diversity estimations were performed using the PHACCS analysis system on each metagenome (Supplementary Figure 3). Briefly, contig spectra were generated using the Circonspect tool and the average genome size was estimated by GAAS. These parameters were passed to PHACCS for alpha-diversity analysis. Computed community structures (defined by richness (R), evenness (E) and diversity (H')) are graphically represented as rank-abundance curves in Supplementary Figure 3. Based on the obtained results, the samples with the highest viral diversity index were the pristine Ilij and Molomhar gueltas (H'=4.83 and H'=4.33, respectively), followed by El Berbera (H'=4.19) and Hamdoun (H'=2.21). The phylogenetic composition of the viral communities of the four metagenomes was then considered, and comparisons were computed using the FastUniFrac tool on the subset of viral annotated metagenomic sequences. Statistically significant differences were measured between two samples (P-value<0.05) using the ‘UniFrac significance analysis' test and further confirmed using the P-test. PCA, which was used to visualize multiple comparisons between samples (Supplementary Figure 4), showed that Hamdoun is isolated from the other samples. To evaluate the robustness of this clustering pattern, we performed a Jackknife environment cluster analysis. The results of this bootstrap procedure confirmed the confidence in the Hamdoun cluster node.
Comparisons with viral communities from other freshwater environments
The phylogenetic profile of the four gueltas was compared with that of other natural or non-natural freshwater environments, two temperate freshwater lakes (Roux et al., 2012), an Antarctic lake sampled in the spring and summer seasons (Lopez-Bueno et al., 2009) and two freshwater samples from a human-controlled aquaculture system (Dinsdale et al., 2008a; Figure 5). As a phylogenetic profile representation, we used the metagenomic sequences classification according to their best Blast hit in a Blastx search against the NCBI GenBank non-redundant database (E-value<1e−05). Multiple comparisons were performed and visualized by PCA on the MG-RAST server. The results showed a geographic clustering pattern with the Mauritanian gueltas clustering together, separate from the others (and Hamdoun again separated from the other three Mauritanian gueltas) (Figure 5). Similarly, metagenomes from temperate natural and artificial freshwaters group together and are separate from the metagenomes of other environments. The two metagenomes from the Antarctic lake did not group together, which is consistent with the phylogenetic profile differences observed between the spring and summer communities from this lake (Lopez-Bueno et al., 2009). The metagenome-clustering pattern showed statistically significant differences in the viral domain (P-value=0.006).
Figure 5.
First two principal coordinates from the principal coordinate analysis of the viral communities in freshwater samples from different environments. The PCA was run in MG-RAST to visualize the overall patterns of variation between the samples.
Discussion
With the advent of metagenomics, an increasing number of studies describing viral and bacterial diversity have been conducted. Currently, only a few investigations have focused on viral assemblages in freshwaters, and most of them concern freshwaters from non-natural or polluted ecosystems. For example, viral communities have been described from aquaculture ponds (Dinsdale et al., 2008a; Rodriguez-Brito et al., 2010), a cattle farm pond (Rooks et al.), reclaimed and potable waters (Rosario et al., 2009), hydrocarbon-polluted groundwater (Abbai et al., 2012), and a man-made recreational lake in MD, USA (Bench et al., 2007). Viral communities in natural freshwater systems have only been described in an ice-covered lake in Antarctica (Lopez-Bueno et al., 2009) and, more recently from two temperate freshwater lakes in France (Roux et al., 2012). By combining electron microscopy and metagenomics, we provide here the first comprehensive analysis of viral communities from freshwater ponds in the Sahara desert of Mauritania.
Most studies focusing on viral diversity in the environment use a 0.2-μm filtration step to separate viruses and bacteria on size criteria. One drawback of this method is that it may fail to recover large viral particles, which are supposed to be common in aquatic ecosystems (Claverie, 2005). However, using a 0.45-micron pore size filter, Lopez-Bueno et al. (2009) were able to identify sequences associated with large dsDNA viruses (mainly from the Phycodnaviridae and Mimiviridae families) from an Antarctic freshwater lake in summer. In this study, up to 6.51% of the sequences matched large dsDNA viruses, and the presence of large viral particles (>200 nm) with Mimivirus-like morphologies was confirmed by electron microscopy (Figure 3c). These results further support that large DNA viruses are common in the environment (Ghedin and Claverie, 2005; Monier et al., 2008a, 2008b) and that the 0.2-μm filtration step currently used to prepare environmental viral metagenomes most likely leads to an underestimation of their genetic diversity.
No bacterial cells were observed under electron microscopy, and the number of reads annotated as bacteria (Figure 2) was similar to those reported for other environmental viral metagenomes (Edwards and Rohwer, 2005), indicating a low bacterial contamination of the metagenomes. In addition, the high proportion of unassigned sequences and relatively low number of 16S rRNA matching sequences (Supplementary Table 3) supported a viral origin for the bacterially annotated reads. Because bacterial genes can be packaged into generalized transducing phage particles (Beumer and Robinson, 2005; Ghosh et al., 2008; Del Casale et al., 2011), the bacterial-like sequences in the guelta metagenomes might come from excised prophages mistakenly annotated as bacterial and/or from genes of bacterial origins that were transferred to their phages.
Blast searches performed on the viral metagenomes showed that >70% of the sequences before assembly did not have homologs in current sequence databases (Figure 2). This result is consistent with results of previously published viral metagenomic projects (Breitbart et al., 2002; Desnues et al., 2008; Lopez-Bueno et al., 2009) and again emphasizes that most of the biological diversity in the viral world is still unknown. In this case, sequence assembly using low stringency parameters (20 bp coverage and 95% identity) was of particular interest in classifying the unknown sequences. For example, the Hamdoun metagenome contained >90% unknown reads that could be assembled into contigs (Supplementary Table 4). The downstream identification of structurally conserved viral genes in these contigs (Supplementary Table 6) has provided information about the putative viral origin of these ORFans.
The guelta viral metagenomes were largely dominated by Caudovirales reads, and TEM confirmed the presence of tailed phages (Figure 3) along with other viral morphotypes. Caudovirales are common in the environment and are the dominant viral type recovered from metagenomic analyses in marine environments (Breitbart et al., 2002; Suttle, 2005). Myoviruses, Siphoviruses and Podoviruses were also the most frequently observed viral particles in samples of Namib and Sahara desert sands after the mitomycin C induction of prophages and sonication to release pseudo-lysogens (Prigent et al., 2005; Prestel et al., 2008). The Molomhar metagenome presented the largest number of reads (73% of all reads) that were related to Myoviruses and only 8.2% of reads represented Siphoviruses. At a deeper taxonomic level, viruses infecting photosynthetic bacteria (for example, Prochlorococcus and Synechococcus phages; in bold, Table 2) were the most abundant in both absolute percentage and rank in the El Berbera, Ilij and Molomhar metagenomes.
Despite being a site for a permanent human settlement, the El Berbera viral community presented a viral community structure similar to those of the Ilij and Molomhar pristine gueltas. It has been previously shown that human activities can affect the diversity and the composition of microbial communities and, thus, their viral predators (reviewed in Horner-Devine et al., 2004). For instance, microbial and viral communities from four coral atolls in the Pacific Ocean dramatically changed along a gradient of human disturbance; the most human-impacted atoll was dominated by heterotrophic microbes, including a large percentage of potential human pathogens (Dinsdale et al., 2008b). In this study, the index of viral biodiversity was inversely correlated with human presence (Supplementary Figure 3), with El Berbera displaying a lower diversity index than the pristine Ilij and Molomhar gueltas. In addition, reads matching Spodoptera litura NPV, a baculovirus infecting S. litura (Lepidoptera: Noctuidae) a crop pest in tropical regions (Rao et al., 1993), were found in the El Berbera metagenome; its presence is most likely linked to the agricultural activity (date production) that developed around the guelta. However, no significant change in the taxonomic structure of the viral communities was observed between the El Berbera human-populated and the Ilij and Molomhar non-populated gueltas. This stability reflects either that the magnitude of human disturbance is weak enough to be absorbed by the system or that the sequencing depth is not sufficient to statistically support finer differences in the phylogenetic profiles between the El Berbera, Ilij and Molomhar metagenomes.
In comparison with the Ilij, Molomhar and El Berbera metagenomes, the Hamdoun metagenome contains dramatically fewer reads matching Myoviruses but more reads related to Siphoviruses (Table 1). This result shows that as it is the case for terrestrial and sediment phage communities (Breitbart et al., 2004; Williamson et al., 2007), viruses with the potential for temperate lifestyles were common in this environment. This is also confirmed by the viral community taxonomic profile, which is dominated by the Microbacterium phage Min1 (Akimkina et al., 2007) a potentially temperate phage that infects Microbacterium sp., a versatile heterotrophic bacteria that is frequently isolated from the rhizosphere and soils (Takeuchi and Hatano, 1998). The presence of viruses with the ability for being temperate in the free-viral fraction may be related to high nutrient availability, high bacterial density or to environmental stress that leads to virus induction (McDaniel et al., 2002; Breitbart et al., 2004). For decades, the Sahara has experienced a dramatic rainfall deficit, and a recent study has stressed that, with only 1.7 m3 of remaining water in July 2007, Hamdoun is one of the most endangered gueltas of the Adrar plateau (Trape, 2009). We hypothesize that the high proportion of induced lysogens in the Hamdoun viral community compared with the other gueltas reflects stress associated with the unprecedentedly low water content of the pond. Further studies will be required (for example, after the rainy season) to confirm this hypothesis and to determine whether the current structure of the viral community is maintained over time.
Acknowledgments
We thank Florent Angly for his help with GAAS and Jean-François Trape for the valuable assistance during the field surveys. This work was funded by the Centre National de la Recherche Scientifique (crédits récurrents).
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abbai N, Govender A, Shaik R, Pillay B. Pyrosequence analysis of unamplified and whole genome amplified DNA from hydrocarbon-contaminated groundwater. Mol Biotechnol. 2012;50:39–48. doi: 10.1007/s12033-011-9412-8. [DOI] [PubMed] [Google Scholar]
- Akimkina T, Venien-Bryan C, Hodgkin J. Isolation, characterization and complete nucleotide sequence of a novel temperate bacteriophage Min1, isolated from the nematode pathogen Microbacterium nematophilum. Res Microbiol. 2007;158:582–590. doi: 10.1016/j.resmic.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Angly F, Rodriguez-Brito B, Bangor D, McNairnie P, Breitbart M, Salamon P, et al. PHACCS, an online tool for estimating the structure and diversity of uncultured viral communities using metagenomic information. BMC Bioinformatics. 2005;6:41. doi: 10.1186/1471-2105-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angly FE, Felts B, Breitbart M, Salamon P, Edwards RA, Carlson C, et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4:e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angly FE, Willner D, Prieto-Davo A, Edwards RA, Schmieder R, Vega-Thurber R, et al. The GAAS metagenomic tool and its estimations of viral and microbial average genome size in four major biomes. PLoS Comput Biol. 2009;5:e1000593. doi: 10.1371/journal.pcbi.1000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bench SR, Hanson TE, Williamson KE, Ghosh D, Radosovich M, Wang K, et al. Metagenomic Characterization of Chesapeake Bay Virioplankton. Appl Environ Microbiol. 2007;73:7629–7641. doi: 10.1128/AEM.00938-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beumer A, Robinson JB. A broad-host-range, generalized transducing phage (SN-T) acquires 16S rRNA genes from different genera of bacteria. Appl Environ Microbiol. 2005;71:8301–8304. doi: 10.1128/AEM.71.12.8301-8304.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer M, Gimenez G, Suzan-Monti M, Raoult D. Classification and determination of possible origins of ORFans through analysis of nucleocytoplasmic large DNA viruses. Intervirology. 2010;53:310–320. doi: 10.1159/000312916. [DOI] [PubMed] [Google Scholar]
- Breitbart M, Felts B, Kelley S, Mahaffy JM, Nulton J, Salamon P, et al. Diversity and population structure of a near-shore marine-sediment viral community. Proc Biol Sci. 2004;271:565–574. doi: 10.1098/rspb.2003.2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, et al. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol. 2003;185:6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitbart M, Salamon P, Andresen B, Mahaffy JM, Segall AM, Mead D, et al. Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci USA. 2002;99:14250–14255. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Claverie J-M. Giant viruses in the oceans: the 4th Algal Virus Workshop. Virol J. 2005;2:52. doi: 10.1186/1743-422X-2-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culley AI, Lang AS, Suttle CA. Metagenomic analysis of coastal RNA virus communities. Science. 2006;312:1795–1798. doi: 10.1126/science.1127404. [DOI] [PubMed] [Google Scholar]
- Del Casale A, Flanagan PV, Larkin MJ, Allen CCR, Kulakov LA. Extent and variation of phage-borne bacterial 16S rRNA gene sequences in wastewater environments. Appl Environ Microbiol. 2011;77:5529–5532. doi: 10.1128/AEM.00457-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnues C, Rodriguez-Brito B, Rayhawk S, Kelley S, Tran T, Haynes M, et al. Biodiversity and biogeography of phages in modern stromatolites and thrombolites. Nature. 2008;452:340–343. doi: 10.1038/nature06735. [DOI] [PubMed] [Google Scholar]
- Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM, et al. Functional metagenomic profiling of nine biomes. Nature. 2008a;452:629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- Dinsdale EA, Pantos O, Smriga S, Edwards RA, Angly F, Wegley L, et al. Microbial ecology of four coral atolls in the Northern Line Islands. PLoS One. 2008b;3:e1584. doi: 10.1371/journal.pone.0001584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng A, Kuzmickas R, Anderson NG, Spiro DJ. Metagenomic analysis of RNA viruses in a fresh water lake. PLoS One. 2009;4:e7264. doi: 10.1371/journal.pone.0007264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RA, Rohwer F. Viral metagenomics. Nat Rev Microbiol. 2005;3:504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- Fierer N, Breitbart M, Nulton J, Salamon P, Lozupone C, Jones R, et al. Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl Environ Microbiol. 2007;73:7059–7066. doi: 10.1128/AEM.00358-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–548. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- Ghedin E, Claverie JM. Mimivirus relatives in the Sargasso sea. Virol J. 2005;2:62. doi: 10.1186/1743-422X-2-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh D, Roy K, Williamson KE, White DC, Wommack KE, Sublette KL, et al. Prevalence of lysogeny among soil bacteria and presence of 16S rRNA and trzN genes in viral-community DNA. Appl Environ Microbiol. 2008;74:495–502. doi: 10.1128/AEM.01435-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2009;4:17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner-Devine MC, Carney KM, Bohannan BJ. An ecological perspective on bacterial biodiversity. Proc Biol Sci. 2004;271:113–122. doi: 10.1098/rspb.2003.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K-H, Bae J-W. Amplification methods bias metagenomic libraries of uncultured single-stranded and double-stranded DNA viruses. Appl Environ Microbiol. 2011;77:7663–7668. doi: 10.1128/AEM.00289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Chang HW, Nam YD, Roh SW, Kim MS, Sung Y, et al. Amplification of uncultured single-stranded DNA viruses from rice paddy soil. Appl Environ Microbiol. 2008;74:5975–5985. doi: 10.1128/AEM.01275-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuper R, Kröpelin S. Climate-Controlled Holocene Occupation in the Sahara: Motor of Africa's Evolution. Science. 2006;313:803–807. doi: 10.1126/science.1130989. [DOI] [PubMed] [Google Scholar]
- Laybourn-Parry J. Microbiology. No place too cold. Science. 2009;324:1521–1522. doi: 10.1126/science.1173645. [DOI] [PubMed] [Google Scholar]
- Le Romancer M, Gaillard M, Geslin C, Prieur D. Viruses in extreme environments. Rev Environ Sci Biotechnol. 2007;6:17–31. [Google Scholar]
- Leplae R, Hebrant A, Wodak SJ, Toussaint A. ACLAME: a CLAssification of Mobile genetic Elements. Nucleic Acids Res. 2004;32:D45–D49. doi: 10.1093/nar/gkh084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bueno A, Tamames J, Velazquez D, Moya A, Quesada A, Alcami A. High diversity of the viral community from an Antarctic lake. Science. 2009;326:858–861. doi: 10.1126/science.1179287. [DOI] [PubMed] [Google Scholar]
- McDaniel L, Houchin LA, Williamson SJ, Paul JH. Lysogeny in marine Synechococcus. Nature. 2002;415:496. doi: 10.1038/415496a. [DOI] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier A, Claverie JM, Ogata H. Taxonomic distribution of large DNA viruses in the sea. Genome Biol. 2008a;9:R106. doi: 10.1186/gb-2008-9-7-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier A, Larsen JB, Sandaa RA, Bratbak G, Claverie JM, Ogata H. Marine mimivirus relatives are probably large algal viruses. Virol J. 2008b;5:12. doi: 10.1186/1743-422X-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TFF, Willner DL, Lim YW, Schmieder R, Chau B, Nilsson C, et al. Broad surveys of DNA Viral diversity obtained through viral metagenomics of mosquitoes. PLoS One. 2011;6:e20579. doi: 10.1371/journal.pone.0020579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestel E, Salamitou S, DuBow MS. An examination of the bacteriophages and bacteria of the Namib desert. J Microbiol. 2008;46:364–372. doi: 10.1007/s12275-008-0007-4. [DOI] [PubMed] [Google Scholar]
- Prigent M, Leroy M, Confalonieri F, Dutertre M, DuBow MS. A diversity of bacteriophage forms and genomes can be isolated from the surface sands of the Sahara Desert. Extremophiles. 2005;9:289–296. doi: 10.1007/s00792-005-0444-5. [DOI] [PubMed] [Google Scholar]
- Rao GVR, Wightman JA, Rao DVR. World review of the natural enemies and diseases of Spodoptera litura (F.) (Lepidoptera: Noctuiidae) Insect Sci Appl. 1993;14:273–284. [Google Scholar]
- Rocha EP, Danchin A. Base composition bias might result from competition for metabolic resources. Trends Genet. 2002;18:291–294. doi: 10.1016/S0168-9525(02)02690-2. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Brito B, Li L, Wegley L, Furlan M, Angly F, Breitbart M, et al. Viral and microbial community dynamics in four aquatic environments. ISME J. 2010;4:739–751. doi: 10.1038/ismej.2010.1. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Brito B, Rohwer F, Edwards RA. An application of statistics to comparative metagenomics. BMC Bioinformatics. 2006;7:162. doi: 10.1186/1471-2105-7-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooks DJ, Smith DL, McDonald JE, Woodward MJ, McCarthy AJ, Allison HE. 454-Pyrosequencing: a molecular Battiscope for freshwater viral ecology. Genes. 1:210–226. doi: 10.3390/genes1020210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario K, Nilsson C, Lim YW, Ruan Y, Breitbart M. Metagenomic analysis of viruses in reclaimed water. Environ Microbiol. 2009;11:2806–2820. doi: 10.1111/j.1462-2920.2009.01964.x. [DOI] [PubMed] [Google Scholar]
- Roux S, Enault F, Robin As, Ravet V, Personnic Sb, Theil Sb, et al. Assessing the diversity and specificity of two freshwater viral communities through metagenomics. PLoS One. 2012;7:e33641. doi: 10.1371/journal.pone.0033641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfeld T, Patterson M, Richardson PM, Wommack KE, Young M, Mead D. Assembly of Viral Metagenomes from Yellowstone Hot Springs. Appl Environ Microbiol. 2008;74:4164–4174. doi: 10.1128/AEM.02598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle CA. Viruses in the sea. Nature. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- Suttle CA. Marine viruses—major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- Takeuchi M, Hatano K. Union of the genera Microbacterium Orla-Jensen and Aureobacterium Collins et al. in a redefined genus Microbacterium. Int J Syst Bacteriol. 1998;48:739–747. doi: 10.1099/00207713-48-3-739. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F. Laboratory procedures to generate viral metagenomes. Nat Protoc. 2009;4:470–483. doi: 10.1038/nprot.2009.10. [DOI] [PubMed] [Google Scholar]
- Trape S. Impact of climate change on the relict tropical fish fauna of central Sahara: threat for the survival of Adrar mountains fishes, Mauritania. PLoS One. 2009;4:e4400. doi: 10.1371/journal.pone.0004400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega Thurber RL, Barott KL, Hall D, Liu H, Rodriguez-Mueller B, Desnues C, et al. Metagenomic analysis indicates that stressors induce production of herpes-like viruses in the coral Porites compressa. Proc Natl Acad Sci USA. 2008;105:18413–18418. doi: 10.1073/pnas.0808985105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbauer MG. Ecology of prokaryotic viruses. FEMS Microbiol Rev. 2004;28:127–181. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Williamson KE, Radosevich M, Smith DW, Wommack KE. Incidence of lysogeny within temperate and extreme soil environments. Environ Microbiol. 2007;9:2563–2574. doi: 10.1111/j.1462-2920.2007.01374.x. [DOI] [PubMed] [Google Scholar]
- Williamson KE, Radosevich M, Wommack KE. Abundance and diversity of viruses in six Delaware soils. Appl Environ Microbiol. 2005;71:3119–3125. doi: 10.1128/AEM.71.6.3119-3125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willner D, Thurber RV, Rohwer F. Metagenomic signatures of 86 microbial and viral metagenomes. Environ Microbiol. 2009;11:1752–1766. doi: 10.1111/j.1462-2920.2009.01901.x. [DOI] [PubMed] [Google Scholar]
- Wommack KE, Bhavsar J, Ravel J. Metagenomics: read length matters. Appl Environ Microbiol. 2008;74:1453–1463. doi: 10.1128/AEM.02181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Fischer D. On the origin of microbial ORFans: quantifying the strength of the evidence for viral lateral transfer. BMC Evol Biol. 2006;6:63. doi: 10.1186/1471-2148-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Breitbart M, Lee WH, Run JQ, Wei CL, Soh SW, et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006;4:e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38:e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.