Abstract
Photochemical reaction centers and rhodopsins are the only phototrophic mechanisms known to have evolved on Earth. The minimal cost of bearing a rhodopsin-based phototrophic mechanism in comparison to maintaining a photochemical reaction center suggests that rhodopsin is the more abundant of the two. We tested this hypothesis by conducting a global abundance calculation of phototrophic mechanisms from 116 marine and terrestrial microbial metagenomes. On average, 48% of the cells from which these metagenomes were generated harbored a rhodopsin gene, exceeding the reaction center abundance by threefold. Evidence from metatranscriptomic data suggests that this genomic potential is realized to a substantial extent, at least for the small-sized (>0.8 μm) of microbial fractions.
Keywords: metagenomics, phototrophy, rhodopsin
The two light-harvesting mechanisms known to have independently evolved on Earth, photochemical reaction centers and retinal-activated proton pumps (Bryant and Frigaard, 2006), have evolved in dramatically different directions. Photochemical reaction centers have radiated and increased in complexity throughout their evolution, forming subcellular mechanisms composed of dozens of proteins and pigments capable of not only harvesting solar energy but also of using it to fix carbon by generating a reductive force. Retinal-activated proton pumps, on the other hand, have retained a simple mechanism throughout their evolutionary course, using a single membrane protein—rhodopsin—to form a proton gradient employed to activate ATPase (Béjà et al., 2000; Spudich and Jung, 2005; Frigaard et al., 2006).
These two parallel mechanisms represent opposing evolutionary strategies: the machinery comprising photochemical reaction centers allows the utilization of light at a high quantum yield (Wraight and Clayton, 1974) and at an efficient coverage of the solar spectrum (Hohmann-Marriott and Blankenship, 2011). More importantly, photochemical reaction centers can generate the reducing power used to fix carbon in addition to a proton motive force. However, the cost of occupying extensive membrane real estate (Molenaar et al., 2009), as well as that of high repair and maintenance due to photodamage (Blokhina et al., 2003), greatly exceeds that of a monomeric proton pump. Furthermore, the complexity of the photochemical reaction center is likely to render its lateral transfer a relatively rare event. Rhodopsins, on the other hand, do not provide sufficient energy for cellular growth and are not known to support carbon fixation, but they require the expression of only one membrane protein and are simple enough to be expected to proliferate by lateral gene transfer (Frigaard et al., 2006).
One of the first discoveries made possible by metagenomics was the apparent abundance and diversity of rhodopsins in marine environments (Béjà et al., 2000; Rusch et al., 2007; Fuhrman et al., 2008); these proteins have been found in diverse taxa, including SAR11 (Giovannoni et al., 2005), a contender to the title of ‘the most abundant organism on the Earth'. However, despite the plethora of increasingly available metagenomic data, the abundance of rhodopsins was not systematically compared to that of photochemical reaction centers.
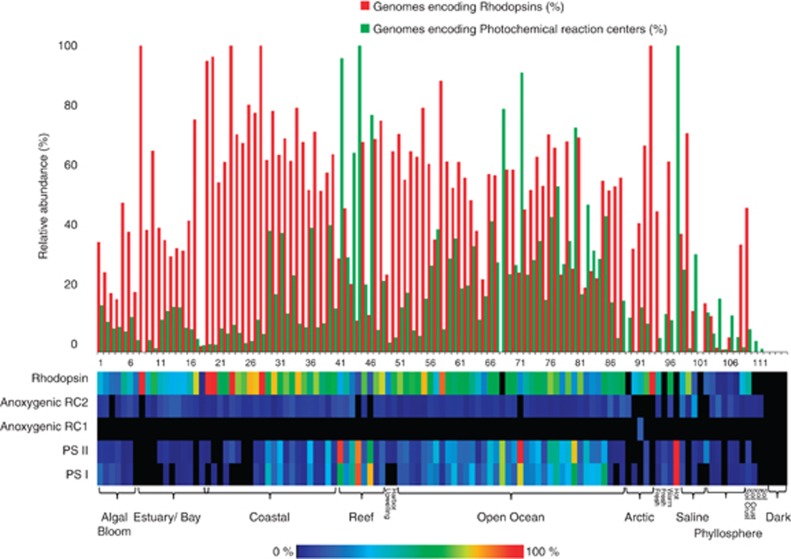
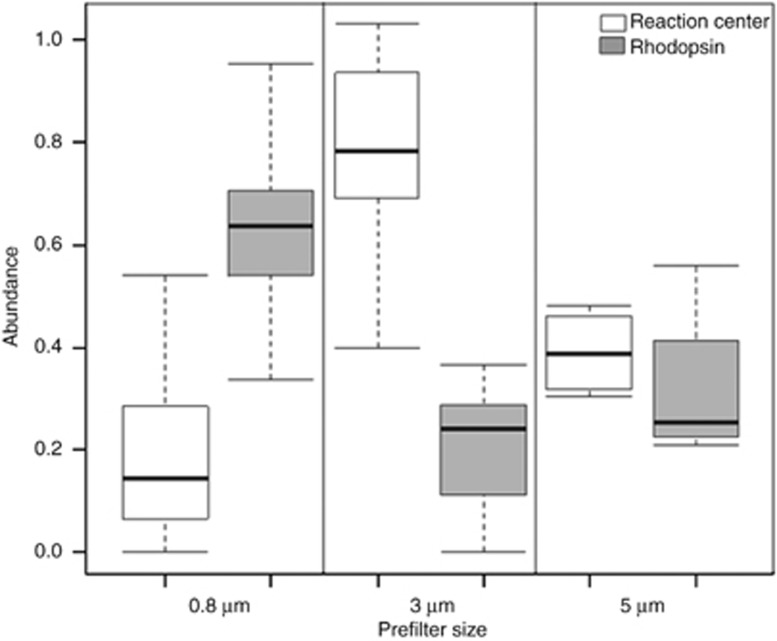
Here we present a systematic abundance profile of genes encoding for photochemical reaction centers and for rhodopsins in publically available metagenomes. Using the MG-RAST metagenomic analysis server (Meyer et al., 2008), we compiled and normalized the number of hits to oxygenic photosystem I and II genes, to anoxygenic RC1 and RC2 photosystem genes, and to rhodopsin homologs from 115 marine and terrestrial metagenomes (Supplementary Table 1). Genes representing the different groups were chosen according to their occurrence profiles in sequenced genomes (Supplementary Table 2), selecting genes that were (a) nearly single-copy (Supplementary Table 2) and (b) ubiquitous within their category. For both criteria, the maximal deviation allowed was 20%. For example, psaB was not used as it appeared 232 times in 75 genomes and over 3 times per genome, whereas psaH was not used as it was found in only 14 out of the 75 PS I-bearing genomes. Abundance profiles were generated by normalizing the hit number to gene size and to an average abundance of 35 independent single copy genes (Supplementary Table 3). As a measure of quality control, the calculated abundances of PS I and PS II were plotted against each other and were found to be at a nearly 1:1 ratio, as expected (Supplementary Figure S1). Rhodopsin genes, found in nearly all photic environments, were both more abundant and more ubiquitous than all four photochemical reaction centers combined. On average, 48% of the cells from which these metagenomes were generated harbored a rhodopsin gene, in comparison with 18% harboring a photochemical reaction center (Figure 1). This trend appears to apply only for the fraction of particles smaller than 0.8 μm. Samples that were prefiltered with a larger pore size displayed an opposite trend, dominated by photochemical reaction centers (Figure 2), indicating that rhodopsin-based phototrophy is a prominently prokaryotic process. Most terrestrial environments (soil and phyllosphere) had a relatively high proportion of photochemical reaction centers as well, presumably due to a large proportion of eukaryotic microorganisms in these samples. Interestingly, although oxygenic photosystems were found to be the most abundant photochemical reaction centers, anoxygenic photosystems were found to be more ubiquitous, as they were present in nearly all metagenomes. In 13 of the data sets, the summed abundance of rhodopsins and of reaction centers exceeds 100%. Furthermore, in six of these cases, the abundance of PS genes or rhodopsin genes alone exceeds 100%. One possible explanation for this apparent anomaly is that some sequenced cyanobacterial as well as eukaryotic genomes harbor both PS and rhodopsins (for example, Nostoc sp. PCC 7120 (de la Torre et al., 2003) and uncultivated oceanic diatoms (Marchetti et al., 2012)). This co-occurence of both light-harvesting genes may be more common than currently thought. Furthermore, the occurrence profile of PS genes in the sequenced genomes may not properly represent their profile in nature. Finally, it is also possible that PS genes from phage genomes (Mann et al., 2003; Lindell et al., 2004; Millard et al., 2004; Sullivan et al., 2005, 2006; Zeidner et al., 2005; Sharon et al., 2007, 2009; Alperovitch et al., 2011; Béjà et al., 2012) may have been included in the samples.
Figure 1.
Relative abundance of four types of photochemical reaction centers and of rhodopsins in 115 metagenomes. Reaction center abundances were calculated using averages of single-copy components of the different photosystems: anoxygenic RC2—pufM, pufL, H subunit, Cyt. C Subunit; anoxygenic RC1—pscA, pscB, pscC, pscD; PSII—psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbO, psbW, psbY, psbZ, psb27; PSI—psaA, psaC, pdsD, psaE, psaF, psaJ, psaL. Rhodopsin abundances were calculated using abundances of genes annotated as proteorhodopsin, xanthorhodopsin and bacteriorhodopsin. Metagenomes used are listed in Supplementary Table 1.
Figure 2.
Relative abundance of photochemical reaction centers and of rhodopsins as a function of prefilter size. Left panel: particles of the size range 0.1−0.8 μm (n=63); middle panel: particles 0.1−3 μm (n=8); right panel: particles 0.22−5 μm (n=4).
One important caveat of the abundance profile presented above is the fact that the data only refer to genomic abundance and not to the expression or function of these genes. However, transcriptomic and proteomic data, rapidly increasing in scale and depth, suggest that rhodopsin is abundantly expressed in a variety of ocean sites (Béjà et al., 2001; Frias-Lopez et al., 2008; Poretsky et al., 2009; Shi et al., 2010; Gifford et al., 2011). In fact, four of the samples used in our analysis (samples 5, 6, 75 and 76) have also been subjected to metatranscriptomic analysis (Gilbert et al., 2008). A positive correlation was found between the abundance of functional groups in the respective DNA and RNA samples (Supplementary Figure S2). No expression of PS I or PS II was detected in the mRNA samples, while the expression of rhodopsin and RC2 genes was on average 74% and 10% of the expression of the 35 aforementioned marker genes, respectively. This indicates that at least for these cases, high abundance of genomic sequences accurately predicted high expression levels.
Although this line of evidence suggests that the majority of prokaryotic cells in the photic biosphere bear phototrophic potential, and that many of them contain rhodopsin genes, a precise assessment of their actual expression and activity needs to be carried out at the protein and functional levels. We hope that the intriguing rhodopsin abundance profiles suggested by our analysis will trigger more accurate measurements of rhodopsin activity in nature.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alperovitch A, Sharon I, Rohwer F, Aro E-M, Glaser F, Milo R, et al. Reconstructing a puzzle: existence of cyanophages containing both photosystem-I & photosystem-II gene suites inferred from oceanic metagenomic datasets. Environ Microbiol. 2011;13:24–32. doi: 10.1111/j.1462-2920.2010.02304.x. [DOI] [PubMed] [Google Scholar]
- Béjà O, Aravind L, Koonin EV, Suzuki MT, Haad A, Nguyen LP, et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289:1902. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- Béjà O, Fridman S, Glaser F. Viral clones from the GOS expedition with an unusual photosystem-I gene cassette organization. ISME J. 2012;6:1617–1620. doi: 10.1038/ismej.2012.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411:786–789. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- Blokhina O, Virolainen E, Fagerstedt KV. Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann Bot. 2003;91:179. doi: 10.1093/aob/mcf118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant DA, Frigaard N-U. Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol. 2006;14:488. doi: 10.1016/j.tim.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. Science. 1998. p. 237. [DOI] [PubMed]
- de la Torre JR, Christianson LM, Béjà O, Suzuki MT, Karl DM, Heidelberg J, et al. Proteorhodopsin genes are distributed among divergent marine bacterial taxa. Proc Natl Acad Sci USA. 2003;100:12830–12835. doi: 10.1073/pnas.2133554100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, et al. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigaard N-U, Martinez A, Mincer TJ, DeLong EF. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature. 2006;439:847. doi: 10.1038/nature04435. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Schwalbach MS, Sting U. Proteorhodopsins: an array of physiological roles. Nat Rev Microbiol. 2008;6:488. doi: 10.1038/nrmicro1893. [DOI] [PubMed] [Google Scholar]
- Gifford SM, Sharma S, Rinta-Kanto JM, Moran MA. Quantitative analysis of a deeply sequenced marine microbial metatranscriptome. ISME J. 2011;5:461–472. doi: 10.1038/ismej.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Dawn F, Huang Y, Edwards. Li W, Gilna P, Joint I. Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PloS One. 2008;3:e3024. doi: 10.1371/journal.pone.0003042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Bibbs L, Cho JC, Stapels MD, Desiderio R, Vergin KL, et al. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature. 2005;438:82. doi: 10.1038/nature04032. [DOI] [PubMed] [Google Scholar]
- Hohmann-Marriott MF, Blankenship RE. Evolution of photosynthesis. Annu Rev Plant Biol. 2011;62:515. doi: 10.1146/annurev-arplant-042110-103811. [DOI] [PubMed] [Google Scholar]
- Lindell D, Sullivan MB, Johnson ZI, Tolonen AC, Rohwer F, Chisholm SW. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc Natl Acad Sci USA. 2004;101:11013–11018. doi: 10.1073/pnas.0401526101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann NH, Cook A, Millard A, Bailey S, Clokie M. Bacterial photosynthesis genes in a virus. Nature. 2003;424:741. doi: 10.1038/424741a. [DOI] [PubMed] [Google Scholar]
- Marchetti A, Schruth DM, Durkin CA, Parker MS, Kodner RB, Berthiaume CT, et al. Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. Proc Natl Acad Sci USA. 2012;109:E317–E325. doi: 10.1073/pnas.1118408109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, et al. The metagenomics RAST server–a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard A, Clokie MRJ, Shub DA, Mann NH. Genetic organization of the psbAD region in phages infecting marine Synechococcus strains. Proc Natl Acad Sci USA. 2004;101:11007–11012. doi: 10.1073/pnas.0401478101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar D, van Berlo R, de Ridder D, Teusink B. Shifts in growth strategies reflect tradeoffs in cellular economics. Mol Syst Biol. 2009;5:323. doi: 10.1038/msb.2009.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poretsky RS, Hewson I, Sun S, Allen AE, Zehr JP, Moran MA. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ Microbiol. 2009;11:1358–1375. doi: 10.1111/j.1462-2920.2008.01863.x. [DOI] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The sorcerer ii global ocean sampling expedition: northwest atlantic through eastern tropical pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon I, Alperovitch A, Rohwer F, Haynes M, Glaser F, Atamna-Ismaeel N, et al. Photosystem-I gene cassettes are present in marine virus genomes. Nature. 2009;461:258–262. doi: 10.1038/nature08284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon I, Tzahor S, Williamson S, Shmoish M, Man-Aharonovich D, Rusch DB, et al. Viral photosynthetic reaction centre genes and transcripts in the marine environment. ISME J. 2007;1:492–501. doi: 10.1038/ismej.2007.67. [DOI] [PubMed] [Google Scholar]
- Shi Y, Tyson GW, Eppley JM, Delong EF. Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J. 2010;5:999–1013. doi: 10.1038/ismej.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JL, Jung KH.2005Microbial rhodopsins: phylogenetic and functional diversityIn: Briggs WR, Spudich JL, (eds).Handbook of Photosensory Receptors WILEY-VCH Verlag GmbH & Co: Weinheim; 1–24. [Google Scholar]
- Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW. Three Prochlorococcus cyanophage genomes: Signature features and ecological interpretations. PLoS Biol. 2005;3:e144. doi: 10.1371/journal.pbio.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan MB, Lindell D, Lee JA, Thompson LR, Bielawski JP, Chisholm SW. Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. 2006;4:e234. doi: 10.1371/journal.pbio.0040234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wraight CA, Clayton RK. The absolute quantum efficiency of bacteriochlorophyll photooxidation in reaction centers. Biochim Biophys Acta. 1974;333:246. doi: 10.1016/0005-2728(74)90009-7. [DOI] [PubMed] [Google Scholar]
- Zeidner G, Bielawski JP, Shmoish M, Scanlan DJ, Sabehi G, Béjà O. Potential photosynthesis gene recombination between Prochlorococcus & Synechococcus via viral intermediates. Environ Microbiol. 2005;7:1505–1513. doi: 10.1111/j.1462-2920.2005.00833.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.