Abstract
A substantial part of the Earths' soil organic carbon (SOC) is stored in Arctic permafrost peatlands, which represent large potential sources for increased emissions of the greenhouse gases CH4 and CO2 in a warming climate. The microbial communities and their genetic repertoire involved in the breakdown and mineralisation of SOC in these soils are, however, poorly understood. In this study, we applied a combined metagenomic and metatranscriptomic approach on two Arctic peat soils to investigate the identity and the gene pool of the microbiota driving the SOC degradation in the seasonally thawed active layers. A large and diverse set of genes encoding plant polymer-degrading enzymes was found, comparable to microbiotas from temperate and subtropical soils. This indicates that the metabolic potential for SOC degradation in Arctic peat is not different from that of other climatic zones. The majority of these genes were assigned to three bacterial phyla, Actinobacteria, Verrucomicrobia and Bacteroidetes. Anaerobic metabolic pathways and the fraction of methanogenic archaea increased with peat depth, evident for a gradual transition from aerobic to anaerobic lifestyles. A population of CH4-oxidising bacteria closely related to Methylobacter tundripaludum was the dominating active group of methanotrophs. Based on the in-depth characterisation of the microbes and their genes, we conclude that these Arctic peat soils will turn into CO2 sources owing to increased active layer depth and prolonged growing season. However, the extent of future CH4 emissions will critically depend on the response of the methanotrophic bacteria.
Keywords: microbial communities, metagenomics, metatranscriptomics, Arctic peat soils, soil organic carbon, methane
Introduction
Recent estimates point out that 277 Pg of soil organic carbon (SOC) are stored in Arctic peatlands (Tarnocai et al., 2009), which corresponds to 1/3 of the CO2 in the atmosphere. These peat soils have acted as carbon sinks since the Holocene (Post et al., 1982; Harden et al., 1992). In contrast, they are substantial sources of methane (CH4), releasing ∼35 Tg per year, ∼6% of the global CH4 emissions (Cao et al., 1996). CH4 is a more potent greenhouse gas than carbon dioxide (CO2), having 25 times the global warming potential of CO2 on a 100-year timescale (Forster et al., 2007). Arctic and especially high-Arctic regions are already exposed to, and predicted to experience, a strong temperature increase until the end of the century (4–8 °C higher annual surface air temperatures) (IPCC, 2007), which is expected to lead to expanded frost-free vegetation periods and increased active layer depths in permafrost soils (Tarnocai et al., 2009).
The major factors in SOC degradation in peatlands, like in mineral soils, are microorganisms of the bacterial, archaeal and eukaryotic (fungi) domains of life, participating in a cascade of aerobic and anaerobic degradation steps, eventually resulting in the emission of CH4 and CO2. Major parts of the peat carbon are plant polymers such as cellulose and hemicellulose. The degradation of these to oligomeric and monomeric sugars is considered one of the most important steps in anaerobic degradation of SOC (Kotsyurbenko, 2005) and is catalysed by a diverse set of hydrolytic extracellular enzymes produced by microorganisms. Further steps in the anaerobic degradation are carried out through anaerobic respiration (like denitrification), fermentative and methanogenic pathways, while microbial methane oxidation constitutes the biological filter for methane emissions from peat. Phenolic substances accumulate to high concentrations in the anaerobic, water-logged layers of peat soils, partly owing to low activity of phenol oxidases, which require oxygen for function. The inhibiting effect of phenolic substances has been suggested as a major factor for the low SOC degradation rates in peat soils (Fenner and Freeman, 2011).
Several studies have targeted the microorganisms involved in CH4 formation (for example, Ganzert et al., 2007; Kotsyurbenko et al., 2007; Metje and Frenzel, 2007; Høj et al., 2008) and oxidation (for example, Wartiainen et al., 2003; Liebner et al., 2009; Martineau et al., 2010; Graef et al., 2011) in Arctic soils. Broader studies have also been conducted, for example, on the biogeography of soil bacterial communities in different climatic zones (Chu et al., 2010), the dynamics of Arctic soil microbial communities in relation to the composition of plant communities (Zak and Kling, 2006) and the availability of SOC (Waldrop et al., 2010 and Coolen et al., 2011). In recent years, metagenomics and metatranscriptomics have developed into powerful tools in microbial ecology that enable non-targeted studies of the genetic potential, gene expression and the composition of soil microbial communities (for example, Tringe et al., 2005 and Urich et al., 2008). Until now only two metagenomic studies on Arctic soils have been conducted, comparing the community in the permafrost with its overlaying active layer (Yergeau et al., 2010) and investigating the response of a permafrost microbial community to thaw (Mackelprang et al., 2011).
It is in the active layers, where the SOC degradation in Arctic peat soils occurs, that more knowledge about the processes and organisms involved is required to understand these and to predict the magnitude of CH4 and CO2 emissions from Arctic peat in a warmer climate. We have in this study applied, for the first time, a combined metagenomic and metatranscriptomic approach on high-Arctic peat soils to obtain on-site information about the genomic potential for SOC transformation and to identify the active microorganisms driving these processes. We compared the Arctic peat metagenomes with metagenomes from temperate soils and proposed a model of SOC degradation in Arctic peat, including anaerobic respiratory and fermentative pathways, and methanogenic and methanotrophic microorganisms. The data allow some predictions about future CH4 and CO2 emissions from Arctic peat soils.
Materials and methods
Study sites and sampling
We have investigated the active layers of two high-Arctic fens on Svalbard (Norway). Solvatn (N78°55.550 E11°56.611) and Knudsenheia (N78°56.544 E11°49.055) were sampled in August 2009 at the peak of the growing season (Supplementary Figure S1 and S2). Both sites are located in the vicinity of the research settlement Ny-Ålesund. Solvatn is situated on a marine terrace right next to the settlement, while Knudsenheia is located ∼5 km northwest of Ny-Ålesund. For both sites, Knudsenheia and Solvatn, two biological replicates were prepared. Three peat blocks (20 × 30 × 20 cm) were cut from each of the replicate sites, S1 and S2 (Solvatn) and K1 and K2 (Knudsenheia; see Supplementary Figures S1–S3), and transported to the laboratory within 2 h after sampling. The active layer depths was ∼40 cm at both sites. The peat blocks were subsequently stored for 2–3 h at 4 °C until further processing. The three blocks were separated into upper (more oxic) and lower (more anoxic) layers, and the corresponding layers, within each replicate site, were pooled to account for heterogeneity of each replicate site (Supplementary Figure S3). The layer separation was based on visual distinction of the layers based on its colour and structure. The lower layers were processed under nitrogen atmosphere to avoid extensive oxygen contamination. After pooling, samples were immediately shock frozen in liquid nitrogen. The whole procedure took ∼5–10 min. Soil pH was measured using the KCl extraction method on fresh peat samples. Water content was estimated by drying peat over night at 120 °C and reweighing the samples. To estimate organic matter contents, the dried samples were combusted at 450 °C overnight. The samples were reweighed, and the ash weight was calculated and added to the weight of organic matter. Concentrations of volatile fatty acids and ethanol were determined by high pressure liquid chromatography analysis of pore water (Metje and Frenzel, 2005) extracted from soil samples before freezing. Results were integrated using peak simple version 393, and 1 mℳ standards.
Nucleic acids extraction, reverse transcription and 454 sequencing
From each layer of the biological duplicates (Supplementary Figure S3), six or more parallel extractions of nucleic acids were performed using a modified version of the Griffith's protocol (Urich et al., 2008). In samples for metatranscriptomic analysis, DNA was digested using the RQ1 DNase treatment (Promega, Madison, WI, USA), followed by RNA purification using the MEGAclear kit (Ambion, Austin, TX, USA). Samples were subjected to reverse transcription using the Superscript II double-strand complementary DNA synthesis kit (Invitrogen, Carlsbad, CA, USA), following the manufacturer's protocol, with the exception that both first- and second-strand synthesis was carried out for 4 h. RNA template addition was in the range of 500–1000 ng. Concentrations of double-stranded complementary DNA were estimated by SYBR Green I (Invitrogen) assay (Leininger et al., 2006). Nucleic acids for metagenomic analysis were subjected to RNase A treatment for 20 min at 37 °C, followed by phenol:chloroform:isoamylalcohol extraction and chloroform:isoamylalcohol extraction. DNA concentrations were estimated using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Madison, WI, USA). Roche 454 GS FLX Titanium sequencing (454 Life Sciences, Branford, CT, USA) was carried out at the CEES at the University of Oslo.
Bioinformatic analysis
Metatranscriptomic and metagenomic sequences were first filtered using LUCY (Chou and Holmes, 2001), removing short (<150 bp) and low-quality sequences (>0.2% error probability). Metatranscriptomic sequences stemming from ribosomal RNA (rRNA) and putative messenger RNAs (mRNAs) were separated by comparing all sequences against a combined database of small and large subunits of rRNA (SSU and LSU rRNA) using BLASTN (Altschul et al., 1997) and MEGAN (Huson et al., 2007; Urich et al., 2008). Sequences with a bit score <50 were assigned as putative mRNA tags. Metagenomic and putative mRNA tags in the metatranscriptomes were subjected to the 454 Replicate filter (Gomez-Alvarez et al., 2009) for the removal of artificially replicated sequences. The filter was applied to remove exact duplicate sequences from the metatranscriptomes (settings: sequence identity cutoff: 1.0, length difference requirement: 1.0 and number of beginning base pairs to check: 3). For the metagenomes, the default settings were applied.
SSU ribo-tags were taxonomically assigned by MEGAN analysis of a BLASTN file against a SSU rRNA reference database (parameters: min. bit score 150, min. support 1, top percent 10; 50 best blast hits) (Urich et al., 2008; Lanzen et al., 2011). SSU ribo-tags assigned to the order Methylococcales were assembled into ribo-contigs using CAP3 (Huang and Madan, 1999), using two subsequent rounds of assembly with (1) a minimum overlap of 150 bp with a minimum similarity threshold of 99% and mismatch and gap scores of −130 and 150, and (2) minimum overlap of 150 bp and minimum 97% similarity threshold, respectively. Contigs were uploaded in ARB (http://www.arb-home.de/), and contigs longer than 1300 bp together with selected reference sequences were used to generate a maximum likelihood tree with the RAxML (Stamatakis et al., 2005) algorithm implemented in ARB using default settings.
The metagenome sequences and the mRNA fraction of the metatranscriptome were functionally annotated for initial screening using the metagenomics (MG)-RAST server with maximum e-value for a significant match set to 1e−4 (Meyer et al., 2008). The sequences were also taxonomically binned by MEGAN analysis (parameters: min. bit score 50, min. support 1, top percent 2; 50 best blast hits) of BLASTX files against the RefSeq protein database (e-value <1e−4). Screening for specific functional genes and transcripts were carried out using custom reference databases generated from the UniProtKB/Swiss-Prot or UniProtKB/TrEMBL databases. BLASTX searches were carried out with an e-value threshold of 1e−10. All sequences below threshold were selected as query sequences for a BLASTX search with the same parameters against the RefSeq database. Only those that were assigned the same function in the second round were considered positives.
In the metagenomes, the genes encoding carbohydrate-active enzymes were annotated. The metagenomic reads were translated into all six frames, each frame into separate ORFs (open reading frames), avoiding any ‘*' characters marking stop codons in a resulting ORF. All ORFs equal to 40 amino acids or larger were screened for assignable conserved protein domains. Glycoside hydrolases (GH), proteins with a carbohydrate-binding motif, carbohydrate esterases, polysaccharide lyases and other carbohydrate-active enzymes were inspected by reference HMMs (Hidden Markov Models) using HMMER tools (http://hmmer.janelia.org/) with the PFAM database HMMs (PFAM release 25, http://pfam.janelia.org). All database hits with e-values below a threshold of 10–4 were counted. For the GH44 family, no PFAM HMM is available. Therefore, representative sequences were selected from the CAZy website (http://www.cazy.org), the sequence regions corresponding to the family were determined and used to create a HMM for the GH44 family. The screening was performed on the HPC computer STALLO at the University of Tromsø (http://docs.notur.no/uit). A selection of sequences was also used as query sequences in BLASTP searches against the RefSeq database of the NCBI. Within the chosen threshold of the HMMER, all identified carbohydrate-active enzymes gave either a corresponding hit or no hit in the RefSeq database. For the taxonomic assignment of gene and transcript sequences, the corresponding BLAST outputs were uploaded in MEGAN (parameters: min. bit score 35, min. support 1, top percent 10; 50 best blast hits).
Statistical analysis
Significant differences between the frequencies of conserved protein domains in ORFs of different soils were evaluated statistically by using the R package (R_Development_Core_Team, 2009) using the χ2 contingency table test. The contingency table holds the frequency counts of hits and non-hits for a certain PFAM domain category of two different soils. The total frequency count is given by all hits found for any domain in the PFAM database. In cases where the frequencies are too low to meet the rules of the test, the probabilities of observing a sample statistic as high as the test statistic were calculated by Monte Carlo simulations with 100 000 replicates.
Data deposition
The sequence data generated in this study was deposited in the Sequence Read Archive of NCBI and are accessible through accession number SRP014474.
Results and discussion
The active layers of the two permafrost fens at Solvatn and Knudsenheia were slightly acidic (pH5–6), had a water content of ∼70–90%, an organic matter content of 40–90% (Supplementary Table S1) and were characterised by a moss cover dominated by Calliergon richardsonii (Solheim et al., 1996; Wartiainen et al., 2003; Høj et al., 2005). The mosses were interspersed by grasses (Dupontia pelligera), which were heavily suppressed by grazing Barnacle geese. High concentrations of acetate, lactate and ethanol indicated anaerobic metabolisms through fermentative and acetogenic pathways, even in the top layers (Supplementary Table S1). Remarkably high yields of nucleic acids (DNA and RNA) per gram dry peat were obtained from all samples and layers (Supplementary Table S2), which suggests a high microbial abundance and activity at the peak of the growing season. The yields, especially of RNA, decreased with depth, indicative of decreasing microbial activity. Replicated metatranscriptome data sets were obtained from all upper layers (Supplementary Figure S3) whereas the reverse transcription was severely inhibited in the lower layers, presumably owing to phenolic substances (Supplementary Table S2). Metagenomic libraries were obtained from all layers of one duplicate from Solvatn and Knudsenheia (K1 and S2, Supplementary Figure S3). 454 Titanium pyrosequencing of the metatranscriptomes and metagenomes resulted in 120 000–190 000 sequences per library, with one exception (S2b complementary DNA, Supplementary Table S3). The number of rRNA sequences in these libraries ranged from 100 000 to 150 000 while the number of putative mRNA sequences ranged from 7000 to 8500.
Community structure of active microorganisms
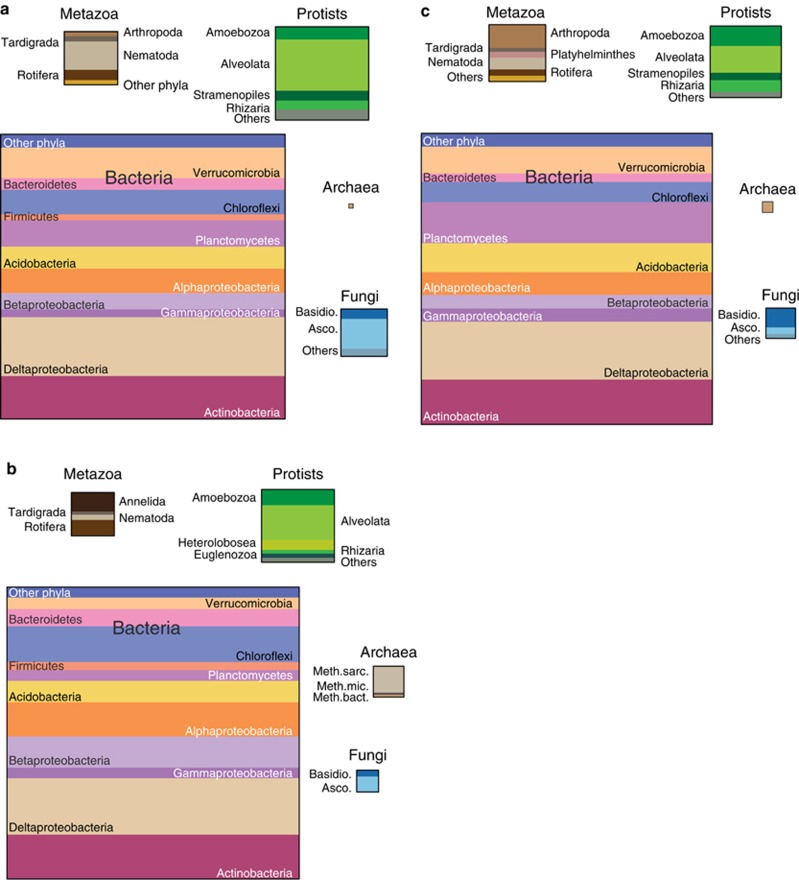
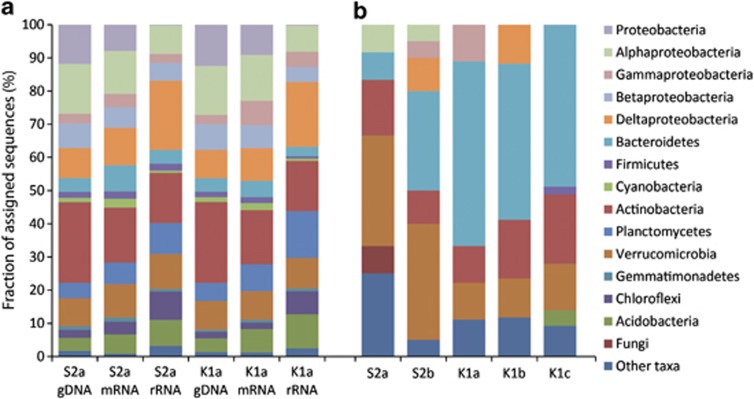
The community composition of bacteria, archaea and eukaryotes, as determined from the small subunit rRNA fragments (SSU ribo-tags) of the metatranscriptomes (up to 74 000 ribo-tags), revealed that the biological replicates of both sites had a similar composition of taxa at domain, phylum and class levels of resolution (Supplementary Figure S4). Ribo-tags are considered a measure of living biomass, as they stem from ribosomes, most likely indicative of active organisms (Urich et al., 2008; Urich and Schleper, 2011). Bacteria comprised most ribo-tags; ∼70%–80% in all upper layers and ∼84% in the lower, anoxic layer of Solvatn, S2b (Figure 1). Eukaryotic ribo-tags comprised ∼15%–30%, whereas the fraction of Archaea was surprisingly small in all the upper layers (∼0.01%–0.13%), but higher in S2b (∼1%). The majority of archaeal sequences stemmed from methanogenic archaea. Similar low relative abundances of Archaea have been reported from Alaskan permafrost soils (for example, Waldrop et al., 2010; Wilhelm et al., 2011). The bacterial community composition in both peat soils was very similar, with most ribo-tags stemming from eight phyla, most notably the Proteobacteria (∼37%–45% of bacterial ribo-tags), Actinobacteria (∼15%), Planctomycetes (∼9%–14%), Verrucomicrobia (∼9%–11%), Acidobacteria (∼8%–10%) and the Chloroflexi (∼7%–9%). Within the Proteobacteria, Deltaproteobacteria (mainly Myxococcales) were the most abundant class (∼19%–21% of bacterial ribo-tags). Alphaproteobacteria made up ∼8%, Betaproteobacteria comprised ∼5% while Gammaproteobacteria represented on average 3.2% of the total community. The major eukaryotic taxon was the Protist kingdom Alveolata, putative grazers of bacteria, which represented ∼17%–20% of eukaryotic ribo-tags in both layers of Solvatn and 6%–8% in the top layer of Knudsenheia (Figure 1). Other abundant Protists were the Amoebozoa, Rhizaria (both predators of bacteria) and Stramenopiles. Interestingly, the Protists constituted the major eukaryotic group, exceeding even the fraction of fungal (∼2%–8%) and metazoan (∼11%–13%) ribo-tags (Figure 1). This is in contrast to temperate and boreal mineral soils, where the fungi and metazoa comprise the majority of either eukaryotic ribo-tags (Urich et al., 2008) or biomass (Schaefer, 1990; Berg and Bengtsson, 2007) and the Protists are much less abundant. Our data indicate that the bacterial energy channel might be more important than the fungal energy channel in these peat soils. Metazoan ribo-tags originated from typical micro- and meso-faunal groups of higher trophic levels, including Nematoda, Tardigrada, Rotifera and Arthropoda. Ascomycota, and to a minor extent Basidiomycota, dominated the small fungal community. At Solvatn, a change in abundance with depth was observed for several taxa (Figure 1). While Alphaproteobacteria, Betaproteobacteria and Chloroflexi were present in larger fractions in the lower layer, Planctomycetes and Verrucomicrobia decreased with depth. Most notably, however, was the increase in the fraction of Archaea and the decrease of Protists and fungi with depth. The ribo-tags originated mostly from actively transcribing organisms, indicated by a taxonomic binning of the mRNA fraction, which showed similar community profiles for the bacterial taxa (Figure 2a).
Figure 1.
Three-domain community profile of the microbiota in Svalbard peatlands. The figures are based on the ribo-tags fraction of the metatranscriptome. The size of the boxes is proportional to the fraction of ribo-tags of the respective taxa. (a) Top layer of Solvatn peat generated from two biological replicate data sets (S1a and S2a). (b) Lower layer of Solvatn generated from one data set (S2b). (c) Top layer of Knudsenheia peat generated from two biological replicate data sets (K1a and K2a). Asco, Ascomycota; Basidio, Basidiomycota; Meth.bac., Methanobacteriales; Meth.mic., Methanomicrobiales; Meth.sarc., Methanosarcinales.
Figure 2.
Taxonomic assignment of metatranscriptomic and metagenomic sequences. The community structure is displayed at phylum level resolution with Proteobacteria split into classes. Sequences assigned to Proteobacteria refer to sequences that could not be assigned to class level resolution. S2 and K1 indicates where the samples were collected, in Solvatn and Knudsenheia respectively, while a, b and c indicates the depth of the sample from top and down (Supplementary Figure S3). (a) Taxonomic assignment of ribo-tags, mRNA and metagenomic DNA (gDNA) to the domain bacteria. (b) Taxonomic assignment of the metagenomic sequences encoding polysaccharide-degrading enzymes (cellulases, endohemicellulases and debranching enzymes). All sequences assigned to these categories were pooled together and taxonomically binned using MEGAN (see Materials and methods for details).
There are few studies addressing Arctic peat soils to compare our metatranscriptomic data with. 16S rRNA gene clone library, and metagenomic studies of permafrost mineral soils have shown that the Actinobacteria, Proteobacteria and Acidobacteria dominate these environments (Hansen et al., 2007; Liebner et al., 2008; Steven et al., 2008; Chu et al., 2010; Yergeau et al., 2010; Wilhelm et al., 2011), while clone library, T-RFLP and FISH studies of a sub-Arctic acidic sphagnum peat report a dominance of Alphaproteobacteria, Betaproteobacteria, Acidobacteria, Verrucomicrobia and Planctomycetes (Dedysh et al., 2006; Pankratov et al., 2011). A recent study of permafrost peat showed that the active layer communities were dominated by Actinobacteria, Proteobacteria and Chloroflexi, while the permafrost microbiota also contained large populations of Bacteroidetes and Firmicutes (Mackelprang et al., 2011). Interestingly, taxa such as the Verrucomicrobia and Planctomycetes, which were among the most abundant taxa in the Svalbard peat soils as well as the acidic Sphagnum peat, were not reported as major taxa in the studies on mineral soils.
Genomic potential for degradation of plant polymers
Plant polymer and phenolic compound degradation are key processes of SOC decomposition in peat soils. We analysed the genomic potential for these processes, by screening the metagenomes for genes encoding conserved protein family (PFAM) domains of relevant enzymes, that catalyse the hydrolysis of abundant plant polymers such as cellulose and hemicelluloses, and the degradation of phenolic compounds (for example, lignin) (Table 1). Despite the rather uniform plant cover, dominated by the moss C. richardsonii, we identified a large variety of CAZy (carbohydrate-active enzyme) families, including 76 GH families, 36 carbohydrate-binding module, 3 phenol oxidase, 3 polysaccharide lyase and 5 carbohydrate esterase families (Supplementary Table S4). GH5 were the most abundant among the families of cellulases. Genes encoding endohemicellulases (especially GH families 10 and 26 that target xylan), debranching enzymes and phenol oxidases involved in the degradation of phenolic compounds were also abundant (Table 1).
Table 1. PFAMs that target plant polymers, identified in metagenomes from high-Arctic peatlands of Svalbard.
| S2a | S2b | K1a | K1b | K1c | |
|---|---|---|---|---|---|
| Cellulases | |||||
| GH5 | 4 (2.3) | 4 (1.8) | 4 (2.1) | 8 (3.9) | 18 (5.8) |
| GH6 | 2 (1.2) | 0 (0) | 1 (0.5) | 1 (0.5) | 2 (0.7) |
| GH7 | 0 (0) | 1 (0.5) | 0 (0) | 1 (0.5) | 0 (0) |
| GH9 | 1 (0.5) | 2 (0.9) | 4 (2.1) | 1 (0.5) | 6 (2.0) |
| GH44 | 2 (1.2) | 2 (0.9) | 3 (1.6) | 2 (1.0) | 0 (0) |
| GH45 | 1 (0.6) | 0 (0) | 0 (0) | 0 (0) | 1 (0.3) |
| GH48 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| Sum | 10 (5.9) | 9 (4.1) | 12 (6.3) | 13 (6.3) | 27 (8.8) |
| Debranching enzymes | |||||
| GH51 | 5 (2.9) | 8 (3.6) | 3 (1.6) | 5 (2.4) | 13 (4.2) |
| GH54 | 1 (0.6) | 3 (1.4) | 0 (0) | 0 (0) | 0 (0) |
| GH62 | 0 (0) | 0 (0) | 1 (0.5) | 0 (0) | 0 (0) |
| GH67 | 2 (1.2) | 1 (0.5) | 0 (0) | 0 (0) | 2 (0.7) |
| GH78 | 1 (0.6) | 2 (0.9) | 2 (1.1) | 10 (4.9) | 15 (4.9) |
| Sum | 9 (5.3) | 14 (6.4) | 6 (3.1) | 15 (7.3) | 30 (9.7) |
| Endohemicellulases | |||||
| GH8 | 0 (0) | 0 (0.0) | 1 (0.5) | 0 (0) | 2 (0.7) |
| GH10 | 2 (1.2) | 6 (2.7) | 1 (0.5) | 5 (2.4) | 7 (2.3) |
| GH11 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0) | 1 (0.3) |
| GH12 | 0 (0.0) | 1 (0.5) | 0 (0.0) | 0 (0) | 0 (0) |
| GH26 | 4 (2.3) | 4 (1.8) | 0 (0) | 1 (0.5) | 12 (3.9) |
| GH28 | 0 (0.0) | 1 (0.5) | 4 (2.1) | 2 (1.0) | 8 (2.6) |
| GH53 | 0 (0.0) | 3 (1.4) | 3 (1.6) | 2 (1.0) | 4 (1.3) |
| Sum | 6 (3.5) | 15 (6.8) | 9 (4.7) | 10 (4.9) | 34 (11.0) |
| Oligosaccharide-degrading enzymes | |||||
| GH1 | 9 (5.3) | 10 (4.6) | 6 (3.1) | 14 (6.8) | 20 (6.5) |
| GH2 | 5 (2.9) | 0 (0.0) | 4 (2.1) | 4 (1.9) | 15 (4.9) |
| GH3 | 5 (2.9) | 9 (4.1) | 6 (3.1) | 9 (4.4) | 18 (5.8) |
| GH29 | 5 (2.9) | 4 (1.8) | 4 (2.1) | 11 (5.3) | 13 (4.2) |
| GH35 | 2 (1.2) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 3 (1.0) |
| GH38 | 0 (0.0) | 0 (0.0) | 1 (0.5) | 3 (1.5) | 4 (1.3) |
| GH39 | 2 (1.2) | 5 (2.3) | 1 (0.5) | 2 (1.0) | 7 (2.3) |
| GH42 | 3 (1.8) | 2 (0.9) | 2 (1.1) | 6 (2.9) | 11 (3.6) |
| GH43 | 8 (4.7) | 7 (3.2) | 5 (2.6) | 8 (3.9) | 10 (3.2) |
| GH52 | 0 (0.0) | 0 (0.0) | 0 (0.0) | 2 (1.0) | 0 (0.0) |
| Sum | 39 (23) | 37 (17) | 29 (15) | 59 (29) | 101 (33) |
| Phenolic compound-degrading enzymes | |||||
| Laccase | 1 (0.6) | 8 (3.6) | 7 (3.7) | 11 (5.3) | 12 (3.9) |
| Dioxygenase | 2 (1.2) | 8 (3.6) | 3 (1.6) | 2 (1.0) | 5 (1.6) |
| Peroxidase | 4 (2.3) | 5 (2.3) | 3 (1.6) | 8 (3.9) | 1 (0.3) |
| Sum | 7 (4.1) | 21 (9.6) | 13 (6.8) | 21 (10.2) | 18 (5.8) |
| Tot. ass. seq. | 17 067 | 21 980 | 19 104 | 20 581 | 30 834 |
The metagenomic profile is divided into five categories according to the functional role of the protein families in plant polymer degradation, a format previously applied in Allgaier et al. (2010) and Pope et al. (2010). Numbers reflect the number of significant hits (see Materials and methods for details). Shown in brackets are the numbers in 0/000 of the total number of sequences assigned to a PFAM. Tot. ass. seq., the number of sequences that were assigned to a PFAM (See Materials and methods for details). See Supplementary Table S4 for PFAM accession numbers and description of protein families.
We wanted to know if the microbiotas of our moss-dominated, carbon-accumulating high-Arctic peat soils contain a different genetic potential for the degradation of plant polymers and recalcitrant phenolic compounds than the microbiota of soils from other climate zones, having a vascular plant cover. Therefore, we compared our metagenomic data sets with metagenomes from temperate grasslands and farmlands, subtropic rain forest soils and Arctic permafrost mineral soils (see Supplementary Table S5 for details about the soil data sets), focussing on the PFAMs relevant for plant polymers and phenolic compound degradation. The functional potential was similar in all soils, and genes for the same PFAMs dominated the metagenomes (Supplementary Table S6). A χ2 test did not detect statistically significant differences between the metagenomes from arctic peat and the other metagenomes (Supplementary Table S7), regardless of the variations in geography or plant coverage.
The chemical composition of moss cell walls is generally considered to be simpler than the one from vacular plants; mosses have a different and less complex hemicellulose composition, and do not contain lignin, but a simpler and less abundant phenolic compound called lignan (Pena et al., 2008; Sarkar et al., 2009; Popper et al., 2011), with the exception of Sphagnum species containing an abundant lignin-like polymer (Ligrone et al., 2008). It was therefore surprising that the microbiotas harboured such a similar genomic potential. This result might to some extent be explained by methodological constraints. Fon instance, the PFAMs for phenolic compound degradation do not distinguish between lignin and lignan as substrate. Also, the enzymes themselves might have a broad substrate specificity. Finally, the presence of many similar types of polymers in both mosses and vascular plants should be reflected in an overall similar genetic repertoire for degradation in the respective soil microbiotas. It might well be that the actual composition of plant polymers is only reflected in the gene expression patterns of the microbiota and not in its genetic potential. However, the rather small number of mRNAs in the metatranscriptome prevented us from studying this.
Another major constraint for SOC degradation, besides the short growing season, is the accumulation of phenolic compounds in the water-logged, predominantly anaerobic peat soils, which inhibits enzymatic activity (Fenner and Freeman, 2011). Expression of phenol oxidases was detected in the top layer of Solvatn. However, the genetic potential for phenol oxidases was observed in all peat layers, indicating that a degradation of these inhibitory compounds can occur if the peat soils get oxygenated, possibly through more pronounced drought-rewetting cycles at higher temperatures, like it has recently been shown for temperate peatlands (Fenner and Freeman, 2011).
We aimed to identify the microorganisms possesing the genes encoding these hydrolytic enzymes by using taxonomic binning with the MEGAN software (See Materials and methods for details). However, this type of taxonomic binning is problematic, owing to the unequal representation of reference genomes for many taxa in the public databases (Urich et al., 2008) and horizontal gene transfer. We therefore compared the community profiles from SSU ribo-tags with the taxonomically assigned metagenomic and metatranscriptomic sequences from the peat top layers using MEGAN (Figure 2a). These profiles were similar for many taxa, suggesting that genes and transcripts can be used as taxonomic markers with some level of confidence. The genes assigned to the three major categories of polysaccharide degradation (cellulases, endohemicellulases and debranching enzymes; see Table 1) were taxonomically annotated. The bacterial phyla Bacteroidetes, Actinobacteria and Verrucomicrobia possessed the majority (>70%) of these genes (Figure 2b). The profiles of the two peat soils differed, with the Knudsenheia metagenomes having a higher fraction of genes from Bacteroidetes and Actinobacteria, while Solvatn had a higher fraction of genes assigned to Verrucomicrobia. Surprisingly, few genes were taxonomically assigned to fungi (Figure 2b). This was in accordance with the low abundance of fungal, and especially basidiomycotal ribo-tags (Figure 1). The latter have been reported as particularly potent decomposers of SOC (Waldrop et al., 2010). Low fungal abundances in permafrost soils and the influence of plant community composition on fungal population size have been shown before (Zak and Kling, 2006; Yergeau et al., 2010; Waldrop et al., 2010).
Anaerobic respiration and fermentation
We investigated the genomic potential for and the expression of key enzymes in anaerobic respiration and fermentation, processes which drive the anaerobic decomposition of SOC. Especially the genes encoding dissimilatory nitrate reductases were abundant (Table 2). This suggests that denitrification is important, which is supported by the high nitrate concentrations detected (70 and 84 μg per gram soil in Solvatn and Knudsenheia, respectively; Alves, 2011). An important role of denitrification has also been suggested by recent metagenomic studies of permafrost affected soils (Yergeau et al., 2010; Mackelprang et al., 2011). Despite the high abundance of denitrification genes, only nirS (nitrite reductase) and norB (nitric oxide reductase) transcripts were identified (Table 2); however, the small size of the mRNA pools in the metatranscriptomic data sets prevented further statements. Also sulphate reduction might have a role, as genes of the key enzyme DSR were found and their abundance increased with soil depth in Knudsenheia (Table 2). The majority of the identified anaerobic respiration genes were assigned to taxa within Actinobacteria and Proteobacteria (Supplementary Figure S5), although the genes were broadly distributed among several different taxa, as previously described for denitrifyers (Philippot and Hallin, 2005) and sulphate reducers (Wagner et al., 2005). Genes and transcripts of several fermentative pathways were detected (Table 2), explaining the high concentration of fermentation products in the peat soils (Supplementary Table S1). Likewise, genes encoding the terminal hydrogenase of H2-evolving fermentations (hydA) were found. Their abundance increased with depth, as did the abundance of formyltetrahydrofolate synthetase genes (fhs), which encodes the key enzyme of the Acetyl-CoA pathway of homoacetogenesis. The majority of hydA and fhs genes were assigned to Firmicutes and Actinobacteria, suggesting an important role of taxa within these phyla in the anaerobic degradation of SOC in the Svalbard peat (Supplementary Figure S6). Taken together, these findings indicate a gradual transition from aerobic to anaerobic degradation of SOC with depth.
Table 2. Metagenomic and metatranscriptomic reads assigned to enzyme classes that catalyse key steps in anaerobic respiration and fermentation, and fermentative pathways.
| Pathway/function | E.C. | Gene | Metagenomes | Metatranscriptomes | |||||
|---|---|---|---|---|---|---|---|---|---|
| |
|
|
S2a |
S2b |
K1a |
K1b |
K1c |
S1a+S2a |
K1a+K2a |
| Nitrate reduction | 1.9.6.1 | napA | 1 | 6 | 7 | 4 | 1 | 0 | 0 |
| Nitrate reduction | 1.7.99.4 | narG | 10 | 6 | 14 | 19 | 28 | 0 | 0 |
| Nitrite reduction | 1.7.2.1 | nirK | 1 | 1 | 2 | 3 | 3 | 0 | 0 |
| Nitrite reduction | 1.7.2.1 | nirS | 0 | 1 | 0 | 2 | 1 | 0 | 2 |
| Nitric oxide reduction | 1.7.99.7 | norB | 1 | 4 | 3 | 2 | 4 | 0 | 1 |
| Nitrous oxide reduction | 1.7.99.6 | nosZ | 3 | 3 | 1 | 2 | 1 | 0 | 0 |
| Sulphate reduction | 1.8.99.3 | dsrA | 0 | 1 | 1 | 4 | 11 | 0 | 0 |
| Fermentation: [FeFe] hydrogenase | 1.12.7.2 | hydA | 2 | 9 | 0 | 7 | 8 | 0 | 0 |
| Homoacetogenesis | 6.3.4.3 | fthfs | 6 | 12 | 7 | 10 | 23 | 0 | 0 |
| Methanogenesis | 2.8.4.1 | mcrA | 0 | 0 | 1 | 0 | 2 | 0 | 0 |
| Acetoin, butanediol metabolism | 55 | 92 | 60 | 85 | 133 | 8 | 8 | ||
| Acetone, butanol, ethanol synthesis | 243 | 298 | 319 | 280 | 346 | 17 | 17 | ||
| Acetyl-CoA fermentation to butyrate | 310 | 362 | 406 | 329 | 407 | 16 | 22 | ||
| Butanol biosynthesis | 235 | 293 | 315 | 263 | 308 | 9 | 19 | ||
| Fermentations: lactate | 52 | 76 | 56 | 74 | 104 | 2 | 3 | ||
| Fermentations: mixed acid | 107 | 157 | 121 | 131 | 190 | 5 | 8 | ||
| Tot. ass. sequences | 69 176 | 63 527 | 85 664 | 54 889 | 72 166 | 6315 | 6205 | ||
First column shows function or main pathway, second column shows the enzyme class entry of the ExPASY bioinformatics resource, while third column shows the gene abbreviation. Merged biological replicates for the two sites Solvatn (S1a+S2a) and Knudsenheia (K1a+K2a) are listed under metatranscriptomes. Total ass. sequences, the number of sequences assigned a function by MEGAN analysis. Sequences assigned to pathways were annotated using (MG)-RAST (see Materials and methods for details).
Methane cycling
The fraction of methanogenic archaea increased with depth (Figure 1 and Supplementary Figure S6), reflecting the presumably more anoxic conditions and higher concentrations of fermentation intermediates (Supplementary Table S1). Three orders of methanogenic archaea were found, Methanobacteriales, Methanomicrobiales and Methanosarcinales, of which the first two are hydrogenotrophic, producing CH4 from H2 and CO2, whereas the Methanosarcinales are metabolically more versatile, carrying out hydrogenotrophic, acetoclastic and methylotrophic methanogenesis. These findings are similar to previous studies of methanogens from Solvatn (Høj et al., 2005; Høj et al., 2006). In Solvatn, the increased abundance of acetotrophic methanogens with depth correlated with a decrease in the concentration of acetate, indicating that the acetotrophic Methanosarcinales acted as the sink for acetate. In general, the high concentrations of fermentation products like ethanol, especially in the lower layers of Knudsenheia, indicated a low efficiency in the terminal fermentation steps, possibly owing to low activity of, or a lack of, established interactions between fermentative syntrophic bacteria and their methanogenic counterparts.
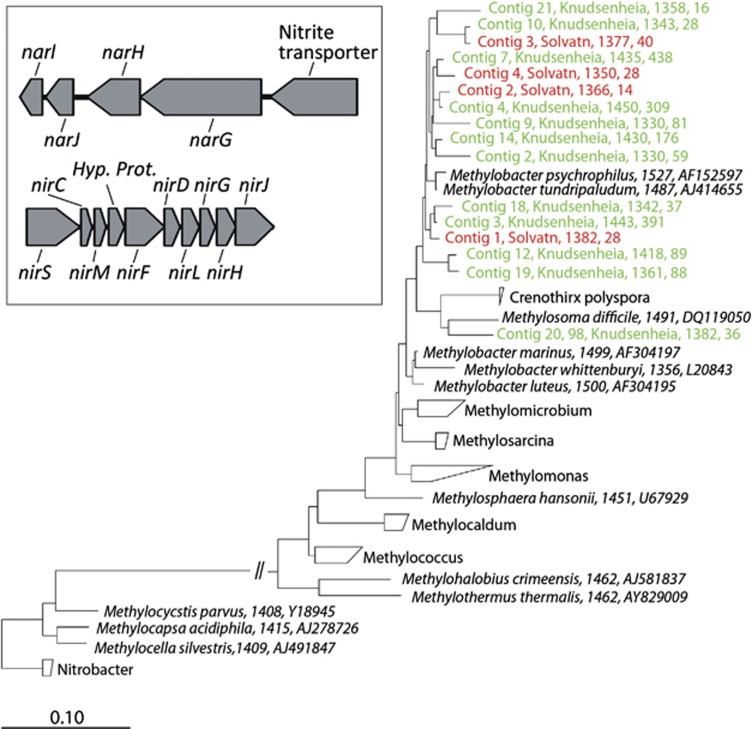
Type I methanotrophs of the order Methylococcales were abundant in both peat soils (0.1%–2.4% of ribo-tags). We did not detect active type II methanotrophs, anaerobic methanotrophic archaea (ANME), the recently described anaerobic methanotroph Candidatus Methylomirabilis oxyfera of the NC10 candidate phylum (Ettwig et al., 2010) or the Verrucomicrobium Methylacidiphilium (Islam et al., 2008). We assembled long, sometimes full-length, SSU rRNAs from the ribo-tags of the type I methanotrophs to determine the phylogenetic position (See Materials and methods (Urich et al., 2008; Radax et al., 2012)). The vast majority of the assembled ribo-contigs was closely related to M. tundripaludum (Figure 3), an arctic methanotroph originally isolated from Solvatn (Wartiainen et al., 2006). We could taxonomically assign 104 mRNA tags to genes encoded in the genome of M. tundripaludum (Svenning et al., 2011). Although this is a small number, it allowed a superficial view into the gene expression and metabolism of M. tundripaludum in situ. Remarkably, as much as 33% of the transcripts encoded subunits of the key enzyme particulate methane monooxygenase (Table 3). This highlights the importance of particulate methane monooxygenase for M. tundripaludum cells, as this enzyme catalyses the initial step of both the carbon and energy metabolism through the oxidation of methane to methanol. Transcripts of the methanol dehydrogenase that catalyses the second step of complete oxidation of methane to CO2 and the ribulose monophosphate pathway for carbon assimilation, as well as transcripts encoding enzymes of the aerobic respiratory chain, were also identified (Table 3). Recently, M. tundripaludum was shown to be the dominating methanotroph in a SIP-RNA experiment with Solvatn peat (Graef et al., 2011). It was also shown to be the dominant methanotroph in two soils from the Siberian and the Canadian Arctic (Liebner et al., 2009; Martineau et al., 2010). Although these are few studies, they point to the importance of methanotrophs closely related to M. tundripaludum having a key role in the biological CH4 filter in Arctic soils. The seemingly ecological advantage of M. tundripaludum in these often water-logged Arctic soils might be owing to its N2-fixation capability (Wartiainen et al., 2006; Svenning et al., 2011), as well as to the presence of nitrate reductase (nar) and nitrite reductase (nir) operons in its genome (Figure 3) (Svenning et al., 2011). It is tempting to speculate that M. tundripaludum could employ the NAR and NIR enzymes in denitrifying anaerobic methane oxidation, utilising a pathway similar to that of Candidatus M. oxyfera (Ettwig et al., 2010). Alternatively, it could have the ability to grow anaerobically on simple organic compounds using denitrification. Experiments are underway to test these hypotheses.
Figure 3.
M. tundripaludum in Svalbard peat soils. The phylogenetic tree shows assembled SSU rRNA contigs of type I methanotrophs. Most of the nearly full-length ribo-contigs (14 out of 16) are >97% identical to M. tundripaludum. Ribo-contig description includes the following: contig id, site (Knudsenheia-green, Solvatn-red), contig length and the number of single ribo-tags that went into the assembly. The reference sequence description (black) includes sequence length and accession number. The length of the bar indicates 0.10 changes per nucleotide. The tree was constructed using the ARB software (See Materials and methods for details). The insert shows the nitrate reductase (open reading frames (ORFs) 3936–3940) and nitrite reductase operons (ORFs 3936–3940) identified in the genome of M. tundripaludum.
Table 3. mRNAs assigned to M. tundripaludum in the Knudsenheia metatranscriptome.
| Annotation | AC | Function | No. of reads |
|---|---|---|---|
| pmoC | ZP_07656564.1 | Methane oxidation | 8 |
| pmoA | ZP_07656563.1 | Methane oxidation | 7 |
| pmoB | ZP_07656562.1 | Methane oxidation | 6 |
| CsbD family protein | ZP_07656622.1 | Stress response | 2 |
| MgtE intracellular region | ZP_07654467.1 | Magnesium transport | 2 |
| Transport-associated protein | ZP_07656600.1 | Transport | 2 |
| Methanol dehydrogenase | ZP_07652313.1 | Methanol oxidation | 1 |
| 6-Phospho 3-hex. | ZP_07653633.1 | RuMP pathway step 2 | 1 |
| Succinate dehydrogenase | ZP_07652392.1 | Electron transport Complex II | 1 |
| NADH dehydrogenase | ZP_07653777.1 | Electron transport Complex I | 1 |
| NADH:ubiquinone oxidoreductase | ZP_07655325.1 | Electron transport Complex I | 1 |
| Elec. trans. prot. SenC | ZP_07652162.1 | Biogenesis of respiratory systems | 1 |
| Nitrogenase iron protein | ZP_07654124.1 | Nitrogen fixation | 1 |
Abbreviations: elec. trans. prot. SenC, electron transport protein SenC; mRNA, messenger RNA; RuMP, ribulose monophosphate.
Functionally annotated transcripts related to energy, carbon and nitrogen metabolism are shown. mRNA sequences were taxonomically assigned to M. tundripaludum by MEGAN (see Materials and methods for details).
Conclusion
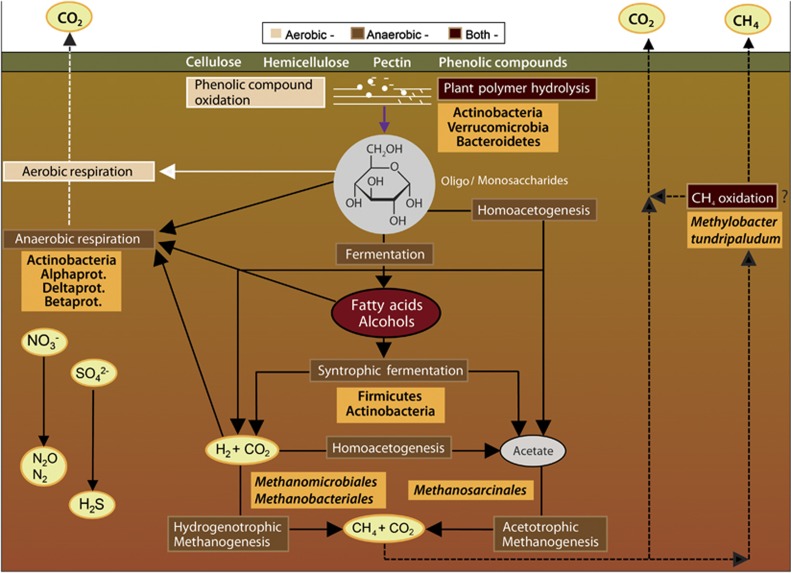
We have described the metabolic potential and activity of microorganisms involved in SOC degradation in high-Arctic peat soils (Figure 4). This first combined metagenomic and metatranscriptomic study on any soil (to our knowledge), revealed taxa that are important to specific processes such as hydrolysis of plant polymers (for example, Verrucomicrobia and Bacteroidetes), fermentations (for example, Firmicutes), methanogenesis and methanotrophy. Actinobacteria seemed to be particularly important, having a metabolic potential for carrying out several of the key steps in SOC degradation (Figure 4). Based on their relative abundance, fungi appeared to not have an important role. However, as warming will lead to an extended growing season, this might result in a change of the plant covers from mosses to vascular plants and in more oxygenated active layers, where fungi might become more important for SOC decomposition. Dryer and more oxygenated active layers will then likely lead to increased peat decomposition, owing to lowered concentrations of phenolic substances, caused by increased activity of aerobic microorganisms that synthesise phenol oxidases (Fenner and Freeman, 2011). These combined effects might increase the rate of SOC degradation above that of carbon sequestration, and turn these peat soils into CO2 sources. Increased active layer depth and higher soil temperatures will likely result in bigger habitat size for anaerobic bacteria and methanogenic archaea, in the cases where the peat soils remain water-saturated. Here, CH4 production likely will increase, although this might well be balanced by methanotrophic bacteria, of which M. tundripaludum seems to be particularly important. However, to which extent such key populations can adapt to the changing conditions remains a crucial question.
Figure 4.
Schematic overview of the main degradation pathways of plant polymers in the high-Arctic peatlands of Svalbard. The pathways are divided into three categories; aerobic (beige), anaerobic (light brown) and processes occurring under both conditions (dark brown). Key microbial taxa for the different degradation steps are presented (orange boxes). The figure is adapted from Figure 1 in (Conrad, 1999).
Acknowledgments
We thank Peter Frenzel, Vigdis Torsvik and Ricardo Alves for important contributions during the fieldwork in Ny-Ålesund, Svalbard. We thank Christoph Bayer for bioinformatic support and Susanne Liebner for help with sequence analysis and phylogeny. We thank Ali Hahn for high pressure liquid chromatography analyses. Christa Schleper is thanked for valuable discusssions and comments. We thank Frøydis Strand for assistance with figure design and preparation. Ave Tooming-Klunderud is thanked for 454 pyro-sequencing. The sequencing service was provided by the Norwegian High-Throughput Sequencing Centre, a national technology platform supported by the ‘Functional Genomics' and ‘Infrastructure' programmes of the Research Council of Norway and the Southeastern Regional Health Authorities (http://www.sequencing.uio.no). Our research in Arctic microbial ecology is currently funded through The Research Council of Norway Grant 191696/V49.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allgaier M, Reddy A, Park JI, Ivanova N, D'Haeseleer P, Lowry S, et al. Targeted discovery of glycoside hydrolases from a switchgrass-adapted compost community. PLoS One. 2010;5:9. doi: 10.1371/journal.pone.0008812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves RJE.2011Ammonia-oxidizing archaea from High Arctic soilsMaster thesis, University of Lisbon: Lisbon, Portugal [Google Scholar]
- Berg MP, Bengtsson J. Temporal and spatial variability in soil food web structure. Oikos. 2007;116:1789–1804. [Google Scholar]
- Cao M, Marshall S, Gregson K. Global carbon exchange and methane emissions from natural wetlands: application of a process-based model. J Geophys Res. 1996;101:399–414. [Google Scholar]
- Chou HH, Holmes MH. DNA sequence quality trimming and vector removal. Bioinformatics. 2001;17:1093–1104. doi: 10.1093/bioinformatics/17.12.1093. [DOI] [PubMed] [Google Scholar]
- Chu HY, Fierer N, Lauber CL, Caporaso JG, Knight R, Grogan P. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ Microbiol. 2010;12:2998–3006. doi: 10.1111/j.1462-2920.2010.02277.x. [DOI] [PubMed] [Google Scholar]
- Conrad R. Contribution of hydrogen to methane production and control of hydrogen concentrations in methanogenic soils and sediments. FEMS Microbiol Ecol. 1999;28:193–202. [Google Scholar]
- Coolen MJL, van de Giessen J, Zhu EY, Wuchter C. Bioavailability of soil organic matter and microbial community dynamics upon permafrost thaw. Environ Microbiol. 2011;13:2299–2314. doi: 10.1111/j.1462-2920.2011.02489.x. [DOI] [PubMed] [Google Scholar]
- Dedysh SN, Pankratov TA, Belova SE, Kulichevskaya IS, Liesack W. Phylogenetic analysis and in situ identification of bacteria community composition in an acidic Sphagnum peat bog. Appl Environ Microbiol. 2006;72:2110–2117. doi: 10.1128/AEM.72.3.2110-2117.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwig KF, Butler MK, Le Paslier D, Pelletier E, Mangenot S, Kuypers MMM, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- Fenner N, Freeman C. Drought-induced carbon loss in peatlands. Nat Geosci. 2011;4:895–900. [Google Scholar]
- Forster P, Ramaswamy V, Artaxo P, Berntsen T, Betts R, Fahey DW, et al. 2007Changes in atmospheric constituents and in radiative forcingIn: Solomon S, Qin D, Manning M, Chen Z, Marquis M, Averyt KB, Tignor M, Miller HL (eds). Climate Change 2007: The Physical Science Basis. Contribution of Working Group I to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change Cambridge University Press: UK [Google Scholar]
- Ganzert L, Jurgens G, Munster U, Wagner D. Methanogenic communities in permafrost-affected soils of the Laptev Sea coast, Siberian Arctic, characterized by 16S rRNA gene fingerprints. FEMS Microbiol Ecol. 2007;59:476–488. doi: 10.1111/j.1574-6941.2006.00205.x. [DOI] [PubMed] [Google Scholar]
- Gomez-Alvarez V, Teal TK, Schmidt TM. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 2009;3:1314–1317. doi: 10.1038/ismej.2009.72. [DOI] [PubMed] [Google Scholar]
- Graef C, Hestnes AG, Svenning MM, Frenzel P. The active methanotrophic community in a wetland from the high Arctic. Environ Microbiol Rep. 2011;3:466–472. doi: 10.1111/j.1758-2229.2010.00237.x. [DOI] [PubMed] [Google Scholar]
- Hansen AA, Herbert RA, Mikkelsen K, Jensen LL, Kristoffersen T, Tiedje JM, et al. Viability, diversity and composition of the bacterial community in a high Arctic permafrost soil from Spitsbergen, Northern Norway. Environ Microbiol. 2007;9:2870–2884. doi: 10.1111/j.1462-2920.2007.01403.x. [DOI] [PubMed] [Google Scholar]
- Harden JW, Mark RK, Sundquist ET, Stallard RF. Dynamics of soil carbon during deglaciation of the laurentide ice sheet. Science. 1992;258:1921–1924. doi: 10.1126/science.258.5090.1921. [DOI] [PubMed] [Google Scholar]
- Huang XQ, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Høj L, Olsen RA, Torsvik VL. Archaeal communities in high Arctic wetlands at Spitsbergen, Norway (78 degrees N) as characterized by 16S rRNA gene fingerprinting. FEMS Microbiol Ecol. 2005;53:89–101. doi: 10.1016/j.femsec.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Høj L, Olsen RA, Torsvik VL. Effects of temperature on the diversity and community structure of known methanogenic groups and other archaea in high Arctic peat. ISME J. 2008;2:37–48. doi: 10.1038/ismej.2007.84. [DOI] [PubMed] [Google Scholar]
- Høj L, Rusten M, Haugen LE, Olsen RA, Torsvik VL. Effects of water regime on archaeal community composition in Arctic soils. Environ Microbiol. 2006;8:984–996. doi: 10.1111/j.1462-2920.2006.00982.x. [DOI] [PubMed] [Google Scholar]
- IPCC 2007. Climate Change 2007—The Physical Science Basis Contribution of Working Group I to the Fourth Assessment Report of the IPCC (ISBN 978 0521 88009-1 Hardback; 978 0521 70596-7 Paperback).
- Islam T, Jensen S, Reigstad LJ, Larsen Ø, Birkeland N-K. Methane oxidation at 55 °C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc Natl Acad Sci USA. 2008;105:300–304. doi: 10.1073/pnas.0704162105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsyurbenko OR. Trophic interactions in the methanogenic microbial community of low-temperature terrestrial ecosystems. FEMS Microbiol Ecol. 2005;53:3–13. doi: 10.1016/j.femsec.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Kotsyurbenko OR, Friedrich MW, Simankova MV, Nozhevnikova AN, Golyshin PN, Timmis KN, et al. Shift from acetoclastic to H-2-dependent methanogenes is in a West Siberian peat bog at low pH values and isolation of an acidophilic Methanobactetium strain. Appl Environ Microbiol. 2007;73:2344–2348. doi: 10.1128/AEM.02413-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzen A, Jorgensen SL, Bengtsson MM, Jonassen I, Ovreas L, Urich T. Exploring the composition and diversity of microbial communities at the Jan Mayen hydrothermal vent field using RNA and DNA. FEMS Microbiol Ecol. 2011;77:577–589. doi: 10.1111/j.1574-6941.2011.01138.x. [DOI] [PubMed] [Google Scholar]
- Leininger S, Urich T, Schloter M, Schwark L, Qi J, Nicol GW, et al. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature. 2006;442:806–809. doi: 10.1038/nature04983. [DOI] [PubMed] [Google Scholar]
- Liebner S, Harder J, Wagner D. Bacterial diversity and community structure in polygonal tundra soils from Samoylov Island, Lena Delta, Siberia. Int Microbiol. 2008;11:195–202. [PubMed] [Google Scholar]
- Liebner S, Rublack K, Stuehrmann T, Wagner D. Diversity of aerobic methanotrophic bacteria in a permafrost active layer soil of the lena delta, Siberia. Microb Ecol. 2009;57:25–35. doi: 10.1007/s00248-008-9411-x. [DOI] [PubMed] [Google Scholar]
- Ligrone R, Carafa A, Duckett JG, Renzaglia KS, Ruel K. Immunocytochemical detection of lignin-related epitopes in cell walls in bryophytes and the charalean alga Nitella. Plant Syst Evol. 2008;270:257–272. [Google Scholar]
- Mackelprang R, Waldrop MP, DeAngelis KM, David MM, Chavarria KL, Blazewicz SJ, et al. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature. 2011;480:368–U120. doi: 10.1038/nature10576. [DOI] [PubMed] [Google Scholar]
- Martineau C, Whyte LG, Greer CW. Stable isotope probing analysis of the diversity and activity of methanotrophic bacteria in soils from the canadian high Arctic. Appl Environ Microbiol. 2010;76:5773–5784. doi: 10.1128/AEM.03094-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metje M, Frenzel P. Effect of temperature on anaerobic ethanol oxidation and methanogenesis in acidic peat from a northern wetland. Appl Environ Microbiol. 2005;71:8191–8200. doi: 10.1128/AEM.71.12.8191-8200.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metje M, Frenzel P. Methanogenesis and methanogenic pathways in a peat from subarctic permafrost. Environ Microbiol. 2007;9:954–964. doi: 10.1111/j.1462-2920.2006.01217.x. [DOI] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, et al. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:8. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratov TA, Ivanova AO, Dedysh SN, Liesack W. Bacterial populations and environmental factors controlling cellulose degradation in an acidic Sphagnum peat. Environ Microbiol. 2011;13:1800–1814. doi: 10.1111/j.1462-2920.2011.02491.x. [DOI] [PubMed] [Google Scholar]
- Pena MJ, Darvill AG, Eberhard S, York WS, O'Neill MA. Moss and liverwort xyloglucans contain galacturonic acid and are structurally distinct from the xyloglucans synthesized by hornworts and vascular plants. Glycobiology. 2008;18:891–904. doi: 10.1093/glycob/cwn078. [DOI] [PubMed] [Google Scholar]
- Philippot L, Hallin S. Finding the missing link between diversity and activity using denitrifying bacteria as a model functional community. Curr Opin Microbiol. 2005;8:234–239. doi: 10.1016/j.mib.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Pope PB, Denman SE, Jones M, Tringe SG, Barry K, Malfatti SA, et al. Adaptation to herbivory by the Tammar wallaby includes bacterial and glycoside hydrolase profiles different from other herbivores. Proc Natl Acad Sci USA. 2010;107:14793–14798. doi: 10.1073/pnas.1005297107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper ZA, Michel G, Herve C, Domozych DS, Willats WGT, Tuohy MG, et al. 2011Evolution and diversity of plant cell walls: from algae to flowering plantsIn: Merchant SS, Briggs WR, Ort D (eds).Annual Review of Plant Biology Vol 62Annual Reviews: Palo Alto; 567–588. [DOI] [PubMed] [Google Scholar]
- Post WM, Emanuel WR, Zinke PJ, Stangenberger AG. Soil carbon pools and world life zones. Nature. 1982;298:156–159. [Google Scholar]
- R_Development_Core_Team . R: A language and environment for statistical computing. Austria. R Foundation for Statistical Computing; 2009. Vienna. [Google Scholar]
- Radax R, Rattei T, Lanzen A, Bayer C, Rapp HT, Urich T, et al. Metatranscriptomics of the marine sponge Geodia barretti: tackling phylogeny and function of its microbial community. Environ Microbiol. 2012;14:1308–1324. doi: 10.1111/j.1462-2920.2012.02714.x. [DOI] [PubMed] [Google Scholar]
- Sarkar P, Bosneaga E, Auer M. Plant cell walls throughout evolution: towards a molecular understanding of their design principles. J Exp Bot. 2009;60:3615–3635. doi: 10.1093/jxb/erp245. [DOI] [PubMed] [Google Scholar]
- Schaefer M. The soil fauna of a beech forest on limestone: trophic structure and energy budget. Oecologia. 1990;82:128–136. doi: 10.1007/BF00318544. [DOI] [PubMed] [Google Scholar]
- Solheim B, Endal A, Vigstad H. Nitrogen fixation in Arctic vegetation and soils from Svalbard, Norway. Polar Biol. 1996;16:35–40. [Google Scholar]
- Stamatakis A, Ludwig T, Meier H. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics. 2005;21:456–463. doi: 10.1093/bioinformatics/bti191. [DOI] [PubMed] [Google Scholar]
- Steven B, Pollard WH, Greer CW, Whyte LG. Microbial diversity and activity through a permafrost/ground ice core profile from the Canadian high Arctic. Environ Microbiol. 2008;10:3388–3403. doi: 10.1111/j.1462-2920.2008.01746.x. [DOI] [PubMed] [Google Scholar]
- Svenning MM, Hestnes AG, Wartiainen I, Stein LY, Klotz MG, Kalyuzhnaya MG, et al. Genome Sequence of the Arctic methanotroph Methylobacter tundripaludum SV96. J Bacteriol. 2011;193:6418–6419. doi: 10.1128/JB.05380-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnocai C, Canadell JG, Schuur EAG, Kuhry P, Mazhitova G, Zimov S.2009Soil organic carbon pools in the northern circumpolar permafrost region Global Biogeochem Cycles 23GB2023, 11PP. [Google Scholar]
- Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- Urich T, Lanzen A, Qi J, Huson DH, Schleper C, Schuster SC. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One. 2008;3:13. doi: 10.1371/journal.pone.0002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urich T, Schleper C.2011The ‘double RNA' approach to simultaneously assess the structure and function of environmental microbial communities by meta-transcriptomicsIn: de Bruijn FJ (ed).Handbook of Molecular Microbial Ecology Wiley-Blackwell: Hoboken, NJ, USA [Google Scholar]
- Wagner M, Loy A, Klein M, Lee N, Ramsing NB, Stahl DA, et al. Functional marker genes for identification of sulfate-reducing prokaryotes. Methods Enzymol. 2005;397:469–489. doi: 10.1016/S0076-6879(05)97029-8. [DOI] [PubMed] [Google Scholar]
- Waldrop MP, Wickland KP, White R, Berhe AA, Harden JW, Romanovsky VE. Molecular investigations into a globally important carbon pool: permafrost-protected carbon in Alaskan soils. Glob Change Biol. 2010;16:2543–2554. [Google Scholar]
- Wartiainen I, Hestnes AG, McDonald IR, Svenning MM. Methylobacter tundripaludum sp nov., a methane-oxidizing bacterium from Arctic wetland soil on the Svalbard islands, Norway (78 degrees N) Int J Syst Evol Microbiol. 2006;56:109–113. doi: 10.1099/ijs.0.63728-0. [DOI] [PubMed] [Google Scholar]
- Wartiainen I, Hestnes AG, Svenning MM. Methanotrophic diversity in high arctic wetlands on the islands of svalbard (Norway)—denaturing gradient gel electrophoresis analysis of soil DNA and enrichment cultures. Can J Microbiol. 2003;49:602–612. doi: 10.1139/w03-080. [DOI] [PubMed] [Google Scholar]
- Wilhelm RC, Niederberger TD, Greer C, Whyte LG. Microbial diversity of active layer and permafrost in an acidic wetland from the Canadian High Arctic. Can J Microbiol. 2011;57:303–315. doi: 10.1139/w11-004. [DOI] [PubMed] [Google Scholar]
- Yergeau E, Hogues H, Whyte LG, Greer CW. The functional potential of high Arctic permafrost revealed by metagenomic sequencing, qPCR and microarray analyses. ISME J. 2010;4:1206–1214. doi: 10.1038/ismej.2010.41. [DOI] [PubMed] [Google Scholar]
- Zak DR, Kling GW. Microbial community composition and function across an arctic tundra landscape. Ecology. 2006;87:1659–1670. doi: 10.1890/0012-9658(2006)87[1659:mccafa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.