Abstract
Background
The phenylpropanoid metabolites are an extremely diverse group of natural products biosynthesized by plants, fungi, and bacteria. Although these compounds are widely used in human health care and nutrition services, their availability is limited by regional variations, and isolation of single compounds from plants is often difficult. Recent advances in synthetic biology and metabolic engineering have enabled artificial production of plant secondary metabolites in microorganisms.
Results
We develop an Escherichia coli system containing an artificial biosynthetic pathway that yields phenylpropanoic acids, such as 4-coumaric acid, caffeic acid, and ferulic acid, from simple carbon sources. These artificial biosynthetic pathways contained a codon-optimized tal gene that improved the productivity of 4-coumaric acid and ferulic acid, but not caffeic acid in a minimal salt medium. These heterologous pathways extended in E. coli that had biosynthesis machinery overproducing tyrosine. Finally, the titers of 4-coumaric acid, caffeic acid, and ferulic acid reached 974 mg/L, 150 mg/L, and 196 mg/L, respectively, in shake flasks after 36-hour cultivation.
Conclusions
We achieved one gram per liter scale production of 4-coumaric acid. In addition, maximum titers of 150 mg/L of caffeic acid and 196 mg/L of ferulic acid were achieved. Phenylpropanoic acids, such as 4-coumaric acid, caffeic acid, and ferulic acid, have a great potential for pharmaceutical applications and food ingredients. This work forms a basis for further improvement in production and opens the possibility of microbial synthesis of more complex plant secondary metabolites derived from phenylpropanoic acids.
Background
Phenylpropanoic acids, such as 4-coumaric acid, caffeic acid, and ferulic acid, are natural phenolic compounds derived from the phenylpropanoid pathway [1]. Phenylpropanoic acids have attracted increasing attention for their various pharmaceutical properties as well as a valuable monomer for the production of liquid crystal polymers, which can be used for electronic applications [2-4]. Although these compounds are widely used in human health care and industrial material, at present they are mainly obtained by extraction from plants, and extraction yields are low because most of these metabolites accumulate at low levels in plant cells. Furthermore, their isolation from plant material, especially as pure compounds, remains a challenge. Nowadays, large-scale production of plant metabolites or precursors via microbial approaches provides a promising alternative to chemical synthesis and extraction from plant sources [5-7].
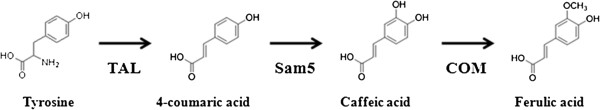
4-Coumaric acid, caffeic acid and ferulic acid are aromatic compounds containing a phenyl ring with a C3 side chain. These phenylpropanoic acids are the pivotal intermediates of the plant phenylpropanoid pathway starting from the deamination of tyrosine. Tyrosine ammonia-lyase (TAL) catalyzes nonoxidative deamination of the primary amino acid tyrosine to 4-coumaric acid (Figure 1). 4-Coumaric acid is converted into caffeic acid by a hydroxylation step at the 3-position of the benzyl ring. The 4-coumarate 3-hydroxylase (C3H) in plants is composed mainly of cytochrome P450 dependent monooxygenases. The purification and characterization of this enzyme is quite challenging because of its instability and membrane-bound property [8]. Recently, a microbial C3H (Sam5) from the actinomycete Saccharothrix espanaensis was reported and its function in Escherichia coli was characterized [9]. 4-Coumaric acid is easily converted into caffeic acid by E. coli containing the Sam5 expression vector, which was also demonstrated in our previous report [10]. Ferulic acid is biosynthesized from caffeic acid by the enzyme caffeate O-methyltransferase [10,11].
Figure 1.
Proposed ferulic acid biosynthetic pathway starting with L-tyrosine. TAL, tyrosine ammonia lyase; Sam5, 4-coumarate hydroxylase; COM, caffeic acid methyltransferase.
Like many other plant secondary metabolites, the production yields of phenylpropanoic acids, notably caffeic acid and ferulic acid, are still low because of the complex and strict regulation of their biosynthesis in plant cells [12,13]. Chemical and enzymatic methods have also been used to produce those phenylpropanoic acid derivatives. Recently, there has been increasing interest in microbial production of phenylpropanoic acid derivatives by reconstructing their biosynthetic pathways in microorganisms. Sachan et al. reported the co-production of caffeic acid and p-hydroxybenzoic acid in Streptomyces caeruleus by feeding 4-coumaric acid [14]. The conversion of tyrosine to caffeic acid (50.2 mg/L) in E. coli was achieved by the co-expression of the enzymes encoded by the TAL from Rhodobacter and E. coli native 4-hydroxyphenylacetate 3-hydroxylase (4HPA3H) [15]. We also reported the production of ferulic acid in E. coli by the sequential co-expression of the enzymes encoded by the sam5 and tal genes from S. espanaensis and an O-methyltransferase (COM) gene from Arabidopsis thaliana[10].
Although previous studies have already made significant gains in demonstrating the feasibility of microbial production in E. coli, the biosynthetic efficiency is greatly limited by the heterologous gene expression and/or host cellular primary precursor productivities. Because tyrosine serves as the main precursor for phenylpropanoids, strains exhibiting an enhanced capacity for its synthesis provide a natural platform for exploring the potential of microbial phenylpropanoid production from glucose [16,17]. In this study, we made an E. coli strain capable of high-level tyrosine production containing feedback-inhibition aroG and tyrA genes by modifying a previously reported expression system [18,19]. The serial artificial biosynthetic gene expression sets encoding TAL, TAL & Sam5, and TAL & Sam5 & COM, graft into the E. coli that is overproducing tyrosine (Figure 2). This method led to the efficient production of serial phenylpropanoic acids, 4-coumaric acid, caffeic acid and ferulic acid. The other three artificial expression sets containing the codon-optimized tal gene tested the improvement gene expression effect with respect to phenylpropanoic acid production. In these systems, engineered E. coli cultured in a growth medium, even through a simple fermentation in minimal media without precursor supplementation, produced the plant-specific 4-coumaric acid, caffeic acid and ferulic acid to yield 974 mg/L, 150 mg/L, and 196 mg/L, respectively. This report describes a convenient and efficient system for production of 4-coumaric acid, caffeic acid and ferulic acid by microorganisms. There is an obvious economic incentive to develop strains capable of converting cheaper feedstocks to high value compounds.
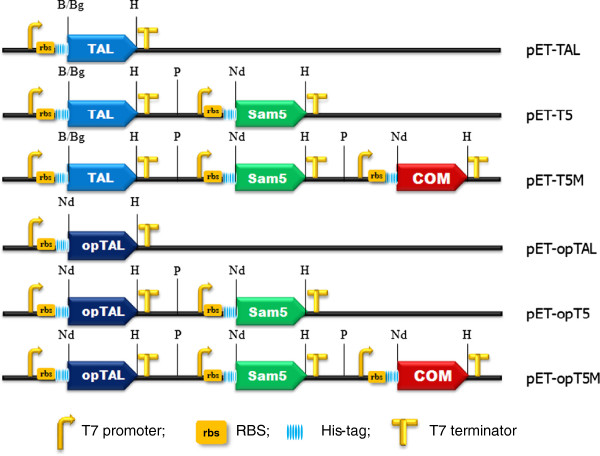
Figure 2.
Organization of the artificial gene clusters used for production of each phenylpropanoic acid in E. coli. Three heterologous enzymes must be expressed in E. coli to mediate the synthesis of ferulic acid from tyrosine. All constructs contained the T7 promoter and the RBS in front of each gene and the T7 terminator located in the rear of each gene. opTAL is a codon-optimized TAL enzyme. B, BamHI; Bg, BglII; H, HindIII; P, PstI; Nd, NdeI.
Results and discussion
Comparison of in vivo activities of TAL and codon-optimized TAL (opTAL)
TALs identified from various sources can catalyze the direct formation of 4-coumaric acid from tyrosine. We have already succeeded in synthesizing 4-coumaric acid in E. coli from a simple medium without the addition of tyrosine using TAL from S. espanaensis[10]. The production of ferulic acid in E. coli was also achieved by the co-expression of the enzymes encoded by the artificial biosynthetic genes, tal, sam5, and com. However, the production yield of ferulic acid using our previous strategy was less than 10 mg/L [10]. Normally, actinomycete genes are often difficult to express in E. coli. They might contain codons and alterations of mRNA structural elements that are rarely used in E. coli[20,21]. In practice with the pET system and other high-level E. coli expression systems, the presence of a small number of rare codons often does not severely depress target protein synthesis. To address this possibility, we tried to make the ‘opTAL’ protein using the codon-optimized program for overcoming the codon bias of the actinomycete gene for enhanced protein expression in E. coli.
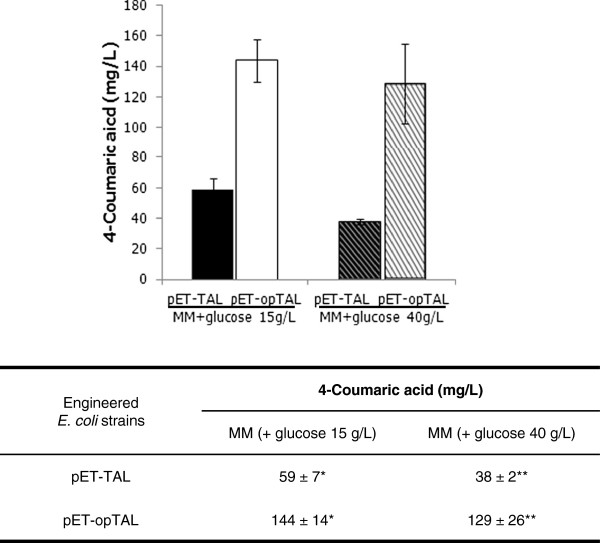
The genes encoding the two TALs were cloned and expressed in E. coli using the plasmids pET-TAL and pET-opTAL (Figure 2). The transformant cells were found in a large amount of TAL in both the insoluble and soluble fractions, when cultured at 37°C (Additional file 1: Figure S1). The transformant cells were cultured at 26°C to avoid the formation of inclusion bodies from the proteins. Even by culturing at 26°C, a considerable amount of the 56 kDa TAL was still recovered in the insoluble fraction (data not shown). To evaluate the performance of the two TALs in E. coli, we assessed 4-coumaric acid production by culturing the recombinant strains in M9 minimal media (with 15 g/L or 40 g/L glucose) supplemented with 1 mM IPTG. Analysis of the product after 36 hours showed that the production of the 4-coumaric acid reached up to 144 ± 14 mg/L from the pET-opTAL strain in a 15 g/L glucose medium, which was 244% higher than the production from the parental strain, pET-TAL (59 ± 7 mg/L) (Figure 3). The codon-optimized TAL showed a marked effect in the native tyrosine pathway of E. coli. On the other hand, 4-coumaric acid synthesis in a 40 g/L glucose medium was 38 ± 2 mg/L (pET-TAL) and 129 ± 26 mg/L (pET-opTAL). However, there was not a statistical significantly difference between the titers for the 15 g/L and 40 g/L glucose samples in media (α = 0.05) (Additional file 1: Table S1). Therefore, the following experiments were performed with an M9 minimal medium with 15 g/L glucose.
Figure 3.
Comparison of TAL activity. Concentrations of 4-coumaric acid in strains with pET-TAL or pET-opTAL cultures in an M9 medium containing glucose 15 g/L (Black bar and white bar) or 40 g/L (bo1d diagonal bar and thin diagonal bar). Each cultivation was done at least in triplicate, and the standard deviations are shown. Error bars indicate standard errors of the means. The codon-optimized TAL activity and glucose effect were compared by single-factor ANOVA (P < 0.05) using Microsoft Excel. Different stars codes (*, **) indicate significant differences (*, P = 9.44E-5; **, P = 1.62E-4).
Effects of codon-optimized TAL on caffeic acid and ferulic acid production
The functionality of this codon-optimized TAL (opTAL) with Sam5 and Sam5 plus COM within E. coli was evaluated by monitoring the accumulation of caffeic acid and ferulic acid in the culture supernatant. We have already succeeded in synthesizing ferulic acid using the artificial biosynthetic pathway containing the parental TAL [10]. Thus, to obtain caffeic acid from a simple medium without the addition of tyrosine, we made an additional expression vector, pET-T5. In addition, we constructed the same sequential expression vectors, pET-opT5 and -opT5M, containing the codon-optimized tal gene. The sequential expression vectors were also equipped with their own T7 promoter, ribosome binding sites (RBS) and terminator to facilitate strong and inducible expression within the E. coli, as in the parental vectors (Figure 2).
To assess the caffeic acid biosynthesis, recombinant E. coli strains with pET-T5 or pET-opT5 were cultured in M9 minimal media supplemented with 15g/L glucose as the sole carbon source (Table 1). Upon IPTG induction, the caffeic acid secreted in the culture media was analyzed using high-performance liquid chromatography (HPLC) after 36 hours. In this case, the synthesis of caffeic acid in E. coli cells with pET-T5 reached 42 ± 19 mg/L and with pET-opT5 reached 14 ± 2 mg/L. Contrary to our expectations, the caffeic acid synthesis in E. coli cells with pET-opT5 did not improve. Rather, the titer showed a pattern of decrease, even if the difference was not significant (P = 0.066). The low caffeic acid productivity in comparison with the previous 4-coumaric acid was perhaps due to poor expression of Sam5 proteins coupled with high expression of codon-optimized TAL protein. To address this possibility, the engineered strains were tested for protein expression via SDS–PAGE analysis (Additional file 1: Figure S2). However, a considerable amount of the 59 kDa Sam5 protein was detected as having opTAL protein (56 kDa), and the expression level of the parental TAL protein (different His-tagged sequences; 57.4 kDa) showed a tendency to overlap with the Sam5 protein itself (Additional file 1: Figure S2). Thus, the low production of caffeic acids in the opTAL-containing pathway (pET-opT5) was not a result merely of poor protein expression, but may instead be related to inherent deficiencies in enzyme activity. We suspected that Sam5 activity may present a major bottleneck in these strains and culture conditions.
Table 1.
Production of caffeic acid and ferulic acid by E. coli strains with pET-T5 and pET-opT5 or pET-T5M and pET-opT5M
| Engineered E. coli strains | 4-Coumaric acid (mg/L) | Caffeic acid (mg/L) | Ferulic acid (mg/L) |
|---|---|---|---|
| pET-T5 |
ND |
42 ± 19a |
- |
| pET-opT5 |
26 ± 8 |
14 ± 2a |
- |
| pET-T5M |
ND |
ND |
28 ± 10b |
| pET-opT5M | ND | ND | 73 ± 15b |
The engineered strains are cultured in an M9 medium containing glucose 15 g/L. Each batch cultivation was done at least in triplicate, and the standard deviations are shown. Different letter codes (a and b) indicate significant differences (a, P = 0.0664; b, P = 0.00226). The code ‘ND’ indicated that something was not detected in our HPLC analysis.
On the other hand, ferulic acid synthesis in E. coli cells harboring pET-T5M and pET-opT5M was 28 ± 10 mg/L and 73 ± 15 mg/L, respectively, which was a 260% improvement in the codon-optimized TAL construct. An unexpected improvement of the ferulic acid titer compared with the caffeic acid titer in each codon-optimized TAL construct was observed (14 mg/L up to 73 mg/L). The reason for this titer improvement is still unknown, but it is possible that the metabolic flow to ferulic acid may be alleviating any restraint during accumulation of caffeic acid in the cell.
Construction of tyrosine overproducer and improvement of 4-coumaric acid production in the tyrosine overproducing strain
In order to investigate the effect of intracellular tyrosine concentration on phenylpropanoid biosynthesis, the production levels of 4-coumaric acid, caffeic acid and ferulic acid from the tyrosine overproducing strain were evaluated. The biosynthesis of aromatic amino acids like tyrosine was very tightly regulated by the concentration of the final products. We first generated an E. coli strain that disrupted the tyrR gene, the product of which represses the expression genes involved in aromatic amino-acid biosynthesis [22]. In addition, the feedback-inhibition-resistant (fbr) 3-deoxy-d-arabino-heptulosonate-7-phosphate (DAHP) synthase (AroGfbr) and chorismate mutase/prephenate dehydrogenase (TyrAfbr) were overexpressed as previously reported by Lütke-Eversloh and Stephanopoulos [18]. Thus, the tyrosine over-producing E. coli strain contains an in-frame deletion in tyrR and a plasmid (pAD-AG) expressing the aroGfbr and tyrAfbr. The engineered E. coli cells produced tyrosine in the M9 modified medium at a yield of ~400 mg/L, when glucose was used as the carbon source (data not shown). By contrast, the E. coli wild-type cells produced low quantities of tyrosine in culture, which was not detectable in our experimental conditions. The tyrosine production yield was comparable to the E. coli T1 strain (346 mg/L) as previously reported by Lütke-Eversloh and Stephanopoulos [18]. The E. coli T1 strain was made by introducing the constitutive promoter PLtet−O1 for the expression of the aroGfbr and tyrAfbr genes in the low-copy-number (~5 copies/cell) plasmid pCL1920, but our two fbr genes were cloned on the inducible T7 promoter of the medium-copy-number (20–30 copies/cell) plasmid pACYCDuet-1.
Under the same experimental conditions as before, strains overproducing tyrosine acquired a substantial capacity for 4-coumaric acid, caffeic acid and ferulic acid synthesis (Figure 4 and Table 2). As seen in Figure 4A and Table 2, tyrosine-overproducing E. coli (pAD-AG/ΔtyrR) with pET-TAL and pET-opTAL produced more than 974 ± 30 mg/L and 805 ± 41 mg/L of 4-coumaric acid, respectively. The production increased by 1650% and 559% over the parental strains, respectively. The similar production between native and codon-optimized TALs meant that the protein expression level was not a critical factor for the 4-coumaric acid production affecting the excessive tyrosine pool in our culture conditions.
Figure 4.
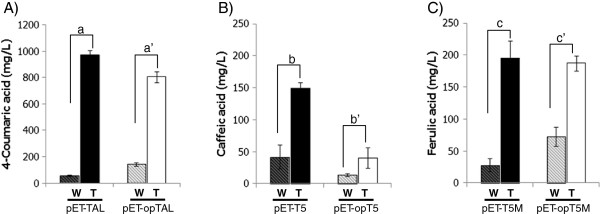
Comparison of 4-coumaric acid (A), caffeic acid (B) and ferulic acid (C) between wild-type and engineered tyrosine overproducing E. coli strains. (A) Production of 4-coumaric acid with pET-TAL and pET-opTAL; (B) Production of caffeic acid with pET-T5 and pET-opT5; (C) Production of ferulic acid with pET-T5M and pET-opT5M. W, wild-type E. coli; T, tyrosine overproducing E. coli. Error bars indicate standard errors of the means. Different letters codes (a to c’) indicate significant differences (a, P = 2.06E-7; a’, P = 2.71E-12; b, P = 8.48E-4; b’, P = 4.3E-4; c, P = 1.67E-6; c’, P = 3.09E-7).
Table 2.
Production of caffeic acid and ferulic acid by engineered tyrosine overproducing E. coli strains
| Engineered tyrosine overproducing E. coli (pAD-AG/ΔtyrR) | 4-Coumaric acid (mg/L) | Caffeic acid (mg/L) | Ferulic acid (mg/L) |
|---|---|---|---|
| pET-TAL |
974 ± 30a |
- |
- |
| pET-opTAL |
805 ± 41a |
- |
- |
| pET-T5 |
307 ± 8 |
150 ± 8b |
- |
| pET-opT5 |
511 ± 69 |
40 ± 16b |
- |
| pET-T5M |
113 ± 12 |
22 ± 3 |
196 ± 26c |
| pET-opT5M | 150 ± 17 | 46 ± 7 | 187 ± 12c |
Each batch cultivation was done at least in triplicate, and the standard deviations are shown. Different letter codes (a to c) indicate significant differences (a, P = 0.0216; b, P = 7.17E-07; c, P = 0.5176).
Caffeic acid and ferulic acid production in the tyrosine overproducing strain
The titers of caffeic acid did not significantly improve (150 ± 8 mg/L and 40 ± 16 mg/L, respectively) in comparison to the 4-coumaric acid titers, despite 357% and 285% improvement over the parental strains (Table 2). As previously mentioned, the same artificial pathways (pET-T5 and pET-opT5) produced 42 mg/L and 14 mg/L of caffeic acid in the native tyrosine pathway of E. coli (Table 1). The titer of caffeic acid (150 ± 8 mg/L) with TAL (pET-T5) was higher than the titer (50 mg/L) of the pathway using 4-hydroxyphenylacetate 3-hydroxylase and TAL as recently reported by Lin et al. [15]. However, the caffeic acid titers with codon-optimized TAL and Sam5 co-expression reached only 4.9% of that of the 4-coumaric acid seen with the codon-optimized TAL expression alone (40 mg/L vs. 805 mg/L). In addition, the tyrosine overproducers containing pET-T5 and -opT5 also accumulated 307 mg/L and 511 mg/L of 4-coumaric acid, respectively (Table 2), and this result meant that the TAL enzymes operated normally, but that Sam5 did not convert 4-coumaric acid to caffeic acid well. Although caffeic acid concentrations were observed to be a somewhat low titer (40 ± 16 mg/L), the presence of any significant 4-coumaric acid accumulation (511 ± 69 mg/L) suggested that this discrepancy may be related to enhanced protein synthesis from the codon-optimized tal gene rather than 4-coumaric acid production and consumption.
On the other hand, ferulic acid synthesis levels in tyrosine-overproducing E. coli cells with pET-T5M and pET-opT5M were 196 ± 26 mg/L and 187 ± 12 mg/L, respectively. At the same time, the accumulation of 4-coumaric acid and caffeic acid also was identified at an expected amount (113 and 22 mg/L and 150 and 46 mg/L, respectively). As was the case for the previous result in wild-type E. coli, the titers of ferulic acid also show an increase over caffeic acid in the tyrosine-overproducing strain.
Conclusions
We successfully established artificial biosynthetic pathways for precursor phenolic acids of phenylpropanoid and constructed E. coli strains for the de novo production of 4-coumaric, caffeic, and ferulic acids. The TALs from S. espanaensis and a codon-optimized synthetic gene expressed the native tyrosine pathway of E. coli and produced 59 ± 7 mg/L and 144 ± 14 mg/L of 4-coumaric acid from glucose sources (Figure 3). Further improvement of 4-coumaric acid production was accomplished by alleviating feedback inhibition and deregulating aromatic amino acids flux into tyrosine biosynthesis. Finally, the titers of 4-coumaric acid reached 974 ± 30 mg/L and 805 ± 41 mg/L in shake flasks after 36-hour cultivation (Table 2). We confirmed that codon optimization of the tal gene had a good effect on de novo production of 4-coumaric acid in a native tyrosine pathway, but the effect was offset by a sufficient tyrosine supply. More notably, this augmented precursor pool had a direct impact on 4-coumaric acid production, which increased 6.7-fold to yield a final titer of 974 ± 30 mg/L.
An artificial pathway leading from tyrosine to caffeic acid was then constructed and introduced into two kinds of E. coli. The caffeic acid levels increased to 150 mg/L and 40 mg/L in the tyrosine overproducer from just 42 mg/l and 14 mg/L in the native strain, respectively. The ferulic acid levels increased to 187 mg/L from 40 mg/L of caffeic acid in the tyrosine overproducer and to 73 mg/L from just 14 mg/L in the native strain.
As seen in Table 1, expressing all three genes that involved the codon-optimized tal gene led to a 2.6-fold increase in ferulic acid production comprising with native TAL expression in the parental strain (28 mg/L up to 73 mg/L). Similarly, the titer of 4-coumaric acid also increased 2.4-fold (59 mg/L up to 144 mg/L) in the same experimental conditions (Figure 3). However, in the tyrosine overproducing strain, the 4-coumaric acid (805 mg/L vs. 974 mg/L) and ferulic acid (187 mg/L vs. 196 mg/L) titers were similar to native and codon-optimized constructs. Therefore, we suggest that at least in our system the titer of an artificial biosynthetic pathway is more dependent on a higher metabolite (precursors and intermediates) concentration and/or flux than on each enzyme’s excess expression rate.
Finally, we achieved almost one gram per liter scale production of 4-coumaric acid. Maximum titers of 150 mg/L of caffeic acid and 196 mg/L of ferulic acid were achieved. Phenylpropanoic acids, such as 4-coumaric acid, caffeic acid, and ferulic acid, have great potential for pharmaceutical applications and food ingredients.
Methods
Bacterial strains, plasmids, and chemicals
E. coli DH5α and E. coli C41 (DE3) [23] were used for general DNA manipulation and expression of biosynthetic genes, respectively. A pCR®-TOPO vector (Invitrogen, Carlsbad, CA) was used for polymerase chain reaction (PCR) cloning. pET-28a(+) was purchased from Novagen (San Diego, CA). L-Tyrosine, 4-coumaric acid, caffeic acid, ferulic acid, and IPTG were purchased from Sigma-Aldrich (St. Louis, MO). Restriction enzymes (NEB), ExTaq (Takara Biochemicals Inc.), pfu (Solgent, Korea), an AccuPower® Ligation kit (Bioneer, Korea), and a Quick & Easy E. coli gene deletion kit (Gene Bridges, German) were used according to the instructions provided by the manufacturers.
Codon optimization and synthesis of TAL, AroGfbr, and TyrAfbr
Codon optimization and synthesis of tal from S. espanaensis was performed with the GeneGPS™ program (DNA2.0). From here forward, synthetic genes/proteins are denoted by superscript “op” DNA sequences. Both AroG and TyrA feedback-inhibition resistance (fbr) derivatives were used to overcome the end-product inhibition of the respective enzymes. The AroGfbr enzyme had an Asp-146-Asn substitution [24], and the TyrAfbr enzyme comprised a Met-53-Ile in the chorismate mutase domain and Ala-354-Val substitution in the prephenate dehydrogenase domain [19]. Codon optimization and synthesis of the aroGfbr and tyrAfbr genes were also performed by DNA2.0.
Heterologous pathway construction and assembly
A list of plasmids and strains used in this study can be found in Table 3. The plasmids were assembled by a serial stepwise cloning process as previously reported [10]. Briefly, the four genes (tal, optal, sam5, and com) were independently cloned into pET-28a(+). To construct an expression vector containing the three genes that were each under the control of an independent T7 promoter, we amplified the 1.76-kb DNA fragment containing the TAL coding region using pET-TAL as a template with primer TAL-N and CPac (the sequence is located downstream of the T7 terminator region of the pET vector and contains the designed PacI site; Additional file 1: Table S2). Using the pET-Sam5 as a template, the 2.54-kb DNA fragment containing the sam5 coding region was PCR-amplified with primer NPac (the sequence was located upstream of the T7 promoter region of the pET vector and contained the designed PacI site; Additional file 1: Table S2) and CPac. The amplified fragments were digested with each restriction enzyme and cloned between PacI- and HindIII-digested pET-28a(+) by a three-fragment ligation, resulting in pET-T5. To construct a pET-T5M vector, the 2.54-kb DNA fragment containing the sam5 coding region was PCR-amplified with primer NPac (the sequence was located upstream of the T7 promoter region of the pET vector and contained the designed PacI site; Additional file 1: Table S2) and CPac. In addition, the 1.88-kb DNA fragment containing the COM coding region was PCR-amplified using pET-COM as a template with primer NPac and COM-R. The amplified fragments were digested with each of the restriction enzymes and cloned between BamHI- and HindIII-digested pET-28a(+) by a four-fragment ligation, resulting in pET-T5M. The pET-opTAL, pET-opT5, and pET-opT5M vectors were constructed the same way as the previous vectors, pET-TAL, pET-T5, and pET-T5M, respectively. Gene sequences and orientations were verified by sequencing after each round of cloning.
Table 3.
Plasmids and strains used in this study
| Plasmid or strain | Relevant Characteristics | Source |
|---|---|---|
| Plasmids | ||
| pET-28a(+) |
f1 ori, T7 promoter, KanR |
Novagen |
| pET-22b(+) |
f1 ori, T7 promoter AmpR |
Novagen |
| pET-TAL |
pET-28a(+) carrying TAL from Saccharothrix espanaensis |
Choi.et al.
[10] |
| pET-opTAL |
pET-28a(+) carrying codon-optimized S. espanaensis TAL |
This study |
| pET-Sam5 |
pET-28a(+) carrying Sam5 from S. espanaensis |
Choi.et al.
[10] |
| pET-COM |
pET-28a(+) carrying COM from Arabidopsis thaliana |
Choi.et al.
[10] |
| pET-T5 |
pET-28a(+) carrying S. espanaensis TAL and Sam5 |
This study |
| pET-opT5 |
pET-28a(+) carrying codon-optimized TAL and S. espanaensis Sam 5 |
This study |
| pET-T5M |
pET-28a(+) carrying S. espanaensis TAL, S. espanaensis Sam5, and A. thaliana COM |
Choi.et al.
[10] |
| pET-opT5M |
pET-28a(+) carrying codon-optimized TAL, S. espanaensis Sam5, and A. thaliana COM |
This study |
| pACYCDuet-1 |
p15A ori, double T7 promoter, CmR |
Novagen |
| pET28-tyrA* |
pET-28a(+) carrying codon-optimized tyrAfbr |
This study |
| pET22-aroG* |
pET-22b(+) carrying codon-optimized aroGfbr |
This study |
| pAD-AG |
pACYDuet-1 carrying codon-optimized tyrAfbr and aroGfbr |
This study |
| Strains | ||
|
E. coli DH5a |
cloning host |
Invitrogen |
|
E. coli C41(DE3) |
derivative strain of E. coli BL21(DE3) |
Miroux B & Walker JE
[23] |
|
ΔtyrR |
tryR gene in-frame deletion mutant of E. coli C41(DE3) |
This study |
| pAD-AG/ΔtyrR | E. coli C41(DE3) ΔtryR :: pAD-AG; tyrosine overproducing strain | This study |
In-frame deletion of the tyrR gene
The genetic design of the L-tyrosine overproducing strain was followed as previously described by Lütke-Eversloh and Stephanopoulos [18]. The in-frame deletion of tyrR, a repressor gene of aromatic amino acid biosynthesis, was constructed by RED/ET recombination with a Quick & Easy E. coli Gene deletion kit (Gene Bridges) using the manufacturer’s protocols. The PCR recombination product was generated using the FRT-PGK-gb2-neo-FRT fragment as a template for the FRT-flanked kanamycin cassette and the following primers: tyr-Rf (5′-GTCATATCAT CATATTAAT TGTTCTTTTT TCAGGTGAAG GTTCCCATGC GTAATTAACC CTCACTAAAG GGCG-3′) and tyr-Rr (5′-ATCAGGCATA TTCGCGCTTA CTCTTCGTTC TTCTTCTGAC TCAGACCATA TAATACGACT CACTATAGGG CTC-3′). The kanamycin selection marker was removed from the chromosome by transforming the cells with an FLP recombinase expression plasmid, 706-FLP (Gene Bridges). The clones growing on the kanamycin plate still contained the selection marker cassette, while all other clones containing in-frame deletion lost the selection marker. The in-frame deletion mutant (ΔtyrR) was verified through PCR using the following primers: tyr-v1 (5′- AACCT CGCCT CGGGG ATTTC −3′) and tyr-v2 (5′- AGCGC GTGCC GTTGT GGTTA −3′). The PCR product was sequenced and verified.
Construction of L-tyrosine overproducing strain
The L-tyrosine over-producing E. coli strain was achieved by overexpression of aroGfbr and tyrAfbr genes in the in-frame deletion ΔtyrR mutant. To elevate the expression levels of the aroGfbr and tyrAfbr genes, the RBS and the T7 promoter were positioned in front of both genes. A synthetic aroGfbr DNA fragment (from DNA2.0) was digested with restriction enzymes NdeI- and HindIII, and the appropriate fragments were ligated into pET-22b(+) to form pET22-aroG* (Table 3). The pET28-tyrA*, which contained a codon-optimized 1.1-kb tyrAfbr coding region, was digested with restriction enzymes and cloned between NcoI- and HindIII restriction enzyme sites of MCS1 of pACYCDuet-1, resulting in pAD-tyrA*. Then, the 1.1-kb DNA fragment containing the aroGfbr coding region of the pET22-aroG* was digested with restriction enzymes NdeI and XhoI, including the HindIII site of the stop codon, and cloned between the NdeI- and XhoI restriction enzyme sites of MCS2 of pAD-tyrA*, resulting in pAD-AG (Table 3). The L-tyrosine over-producing E. coli strain (pAD-AG/ΔtyrR) was achieved by a ΔtyrR mutant carrying pAD-AG.
Culture conditions for production
Recombinant E. coli C41 (DE3) strains harboring plasmids were precultured overnight at 37°C in a Luria–Bertani (LB) medium containing 50 μg/mL kanamycin. The overnight culture was inoculated (1.5%) into a fresh LB medium supplemented with the same concentration of kanamycin. The culture was grown at 37°C to an optical density of 600 nm (OD600) of 0.6. IPTG was added to the final concentration of 1 mM, and the culture was incubated for 5 hours. The cells were harvested by centrifugation, suspended, and incubated at 26°C for 36 hours in a modified M9 medium (M9 medium supplemented with 15 g/L glucose, 25 g/L CaCO3, appropriate antibiotics (50 μg/mL kanamycin for maintenance of pET-derived plasmids and 34 μg/mL chloramphenicol for pACYC-derived plasmid (pAD-AG)), and 1 mM IPTG) [10,25]. The samples were collected after 36 hours and analyzed by HPLC.
Detection and quantification of the products
For the quantification of L-tyrosine, 4-coumaric acid, caffeic acid, and ferulic acid, 1 mL of cell-free culture supernatants were filtered through 0.2 μm Cellulose membrane syringe filters (Sartorius) and used for HPLC analysis with a Dionex Separations module connected with a Photodiode Array detector (Dionex) set. The compounds were separated by elution with an acetonitrile-water gradient (water containing 0.05% trifluoroacetic acid (TFA)). The L-tyrosine was separated on a YMC C18 column (150 × 4.6 mm, 4 μm). The following gradient was used at a flow rate of 1 mL/min: 5% to 80% acetonitrile for 25 min, 80% to 100% acetonitrile for 3 min, 100% acetonitrile for 3 min, 100% to 5% acetonitrile for 3 min, and 5% acetonitrile for an additional 3 min. The 4-coumaric acid was separated on a YMC C18 column. The following gradient was used at a flow rate of 1 mL/min: 20% to 60% acetonitrile for 25 min, 60% to 100% acetonitrile for 1 min, 100% acetonitrile for 5 min, 100% to 20% acetonitrile for 2 min, and 20% acetonitrile for an additional 5 min. The caffeic acid was separated on a YMC C18 column. The following gradient was used at a flow rate of 1 mL/min: 15% acetonitrile for 5 min, 15% to 60% acetonitrile for 15 min, 60% to 100% acetonitrile for 5 min, 100% acetonitrile for 5 min, 100% to 15% acetonitrile for 2 min, and 15% acetonitrile for an additional 5 min. The ferulic acid was separated on a SunFire™ C18 column (250 × 4.6 mm, 5 μm; Waters, USA). The following gradient was used at a flow rate of 1 mL/min: 20% to 25% acetonitrile for 15 min, 25% to 100% acetonitrile for 5 min, 100% acetonitrile for 5 min, 100% to 20% acetonitrile for 5 min, and 20% acetonitrile for an additional 5 min. Quantification for the four above-mentioned compounds was based on the peak areas of absorbance at 250 nm (L-tyrosine), 300 nm (4-coumaric acid) and 320 nm (caffeic acid and ferulic acid). The concentrations were determined through the use of the corresponding chemical standards (Sigma).
The data shown in this study were generated from triplicate independent experiments. The titers of each production were compared by single-factor ANOVA (P < 0.05) using the single-factor ANOVA tool. The Tukey honestly significant difference (HSD) test was used to determine the significance of differences between group means. The data were analyzed using Microsoft Office Excel 2007 and IBM SPSS Statistics.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SK and OC performed the experiments and wrote the manuscript. JL co-performed the experiments on the metabolite analysis. BH and TU contributed general advice, particularly on the metabolite analysis and resource support. YH designed all the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Figure S1. Comparison of the TAL protein expression of pET-opTAL and pET-TAL in E. coli. Lane 1: E. coli transformation containing pET-opTAL expression vector lysate after IPTG induction; Lane 2: lysate before induction; Lane 3: E. coli transformation containing pET-TAL expression vector lysate after IPTG induction; Lane 4: lysate before induction. Figure S2. SDS-PAGE analysis of co-expression of TAL and Sam5 enzymes in E. coli. Lane 1: E. coli transformation containing pET-opT5 expression vector lysate before IPTG induction; Lane 2: lysate after induction; Lane 3: E. coli transformation containing pET-T5 expression vector lysate before IPTG induction; Lane 4: lysate after induction. Sam5 protein is 59 kDa, TAL protein from pET-opT5 expression vector is 56 kDa, TAL protein from pET-T5 expression vector is 57.4 kDa. The difference in the TAL proteins comes from the length of the His-tagged peptide sequences. Table S1. The results of the Tukey test for the data from Figure 3. Table S2. Oligo nucleotide primers used in this study.
Contributor Information
Sun-Young Kang, Email: kangsun111@kribb.re.kr.
Oksik Choi, Email: dhrtlrl7@kribb.re.kr.
Jae Kyung Lee, Email: scarscar@nate.com.
Bang Yeon Hwang, Email: byhwang@chungbuk.ac.kr.
Tai-Boong Uhm, Email: tbuhm@chonbuk.ac.kr.
Young-Soo Hong, Email: hongsoo@kribb.re.kr.
Acknowledgements
This work was supported in part by Basic Science Research program (2012–0001421) and Global Frontier Project (ISBC #2011-0031947) funded by the NRF and by the Next-Generation BioGreen 21 Program (SSAC, PJ007999) funded by the RDA, Republic of Korea.
References
- Winkel-Shirley B. Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold E. The Science of Flavonoids. New York: Springer; 2007. [Google Scholar]
- Qi WW, Sariaslani FS, Tang X-S. Methods for the production of tyrosine, cinnamic acid and para-hydroxycinnamic acid. World Patent WO 02/090523; 2002. [Google Scholar]
- Kaneko T, Thi TH, Shi DJ, Akashi M. Environmentally degradable, high-performance thermoplastics from phenolic phytomonomers. Nat Mater. 2006;5:966–970. doi: 10.1038/nmat1778. [DOI] [PubMed] [Google Scholar]
- Fowler ZL, Koffas MA. Biosynthesis and biotechnological production of flavanones: current state and perspectives. Appl Microbiol Biotechnol. 2009;83:799–808. doi: 10.1007/s00253-009-2039-z. [DOI] [PubMed] [Google Scholar]
- Leonard E, Yan Y, Fowler ZL, Li Z, Lim CG, Lim KH, Koffas MA. Strain improvement of recombinant Escherichia coli for efficient production of plant flavonoids. Mol Pharm. 2008;5:257–265. doi: 10.1021/mp7001472. [DOI] [PubMed] [Google Scholar]
- Horinouchi S. Combinatorial biosynthesis of plant medicinal polyketides by microorganisms. Curr Opin Chem Biol. 2009;13:197–204. doi: 10.1016/j.cbpa.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Kim YH, Kwon T, Yang HJ, Kim W, Youn H, Lee JY, Youn B. Gene engineering, purification, crystallization and preliminary X-ray diffraction of cytochrome P450 p-coumarate-3-hydroxylase (C3H), the Arabidopsis membrane protein. Protein Expr Purif. 2011;79:149–155. doi: 10.1016/j.pep.2011.04.013. [DOI] [PubMed] [Google Scholar]
- Berner M, Krug D, Bihlmaier C, Vente A, Muller R, Bechthold A. Genes and enzymes involved in caffeic acid biosynthesis in the actinomycete Saccharothrix espanaensis. J Bacteriol. 2006;188:2666–2673. doi: 10.1128/JB.188.7.2666-2673.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi O, Wu CZ, Kang SY, Ahn JS, Uhm TB, Hong YS. Biosynthesis of plant-specific phenylpropanoids by construction of an artificial biosynthetic pathway in Escherichia coli. J Ind Microbiol Biotechnol. 2011;38:1657–1665. doi: 10.1007/s10295-011-0954-3. [DOI] [PubMed] [Google Scholar]
- Do CT, Pollet B, Thevenin J, Sibout R, Denoue D, Barriere Y, Lapierre C, Jouanin L. Both caffeoyl Coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta. 2007;226:1117–1129. doi: 10.1007/s00425-007-0558-3. [DOI] [PubMed] [Google Scholar]
- Paek KY, Murthy HN, Hahn EJ. Establishment of adventitious root cultures of Echinacea purpurea for the production of caffeic acid derivatives. Methods Mol Biol. 2009;547:3–16. doi: 10.1007/978-1-60327-287-2_1. [DOI] [PubMed] [Google Scholar]
- Faulds CB, Williamson G. A major bioactive component of plant cell walls, ferulic acid, influences feruloyl esterase production in Aspergillus niger. Biochem Soc Trans. 1996;24:386S. doi: 10.1042/bst024386s. [DOI] [PubMed] [Google Scholar]
- Sachan A, Ghosh S, Sen SK, Mitra A. Co-production of caffeic acid and p-hydroxybenzoic acid from p-coumaric acid by Streptomyces caeruleus MTCC 6638. Appl Microbiol Biotechnol. 2006;71:720–727. doi: 10.1007/s00253-005-0197-1. [DOI] [PubMed] [Google Scholar]
- Lin Y, Yan Y. Biosynthesis of caffeic acid in Escherichia coli using its endogenous hydroxylase complex. Microb Cell Fact. 2012;11:42. doi: 10.1186/1475-2859-11-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos CN, Koffas M, Stephanopoulos G. Optimization of a heterologous pathway for the production of flavonoids from glucose. Metab Eng. 2011;13:392–400. doi: 10.1016/j.ymben.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Katsuyama Y, Miyahisa I, Funa N, Horinouchi S. One-pot synthesis of genistein from tyrosine by coincubation of genetically engineered Escherichia coli and Saccharomyces cerevisiae cells. Appl Microbiol Biotechnol. 2007;73:1143–1149. doi: 10.1007/s00253-006-0568-2. [DOI] [PubMed] [Google Scholar]
- Lutke-Eversloh T, Stephanopoulos G. L-tyrosine production by deregulated strains of Escherichia coli. Appl Microbiol Biotechnol. 2007;75:103–110. doi: 10.1007/s00253-006-0792-9. [DOI] [PubMed] [Google Scholar]
- Lutke-Eversloh T, Stephanopoulos G. Feedback inhibition of chorismate mutase/prephenate dehydrogenase (TyrA) of Escherichia coli: generation and characterization of tyrosine-insensitive mutants. Appl Environ Microbiol. 2005;71:7224–7228. doi: 10.1128/AEM.71.11.7224-7228.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane JF. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr Opin Biotechnol. 1995;6:494–500. doi: 10.1016/0958-1669(95)80082-4. [DOI] [PubMed] [Google Scholar]
- Kim S, Lee SB. Rare codon clusters at 5'-end influence heterologous expression of archaeal gene in Escherichia coli. Protein Expr Purif. 2006;50:49–57. doi: 10.1016/j.pep.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Pittard J, Camakaris H, Yang J. The TyrR regulon. Mol Microbiol. 2005;55:16–26. doi: 10.1111/j.1365-2958.2004.04385.x. [DOI] [PubMed] [Google Scholar]
- Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Tsujimoto K, Kurahashi O. Mutational analysis of the feedback sites of phenylalanine-sensitive 3-deoxy-D-arabino-heptulosonate-7-phosphate synthase of Escherichia coli. Appl Environ Microbiol. 1997;63:761–762. doi: 10.1128/aem.63.2.761-762.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyahisa I, Kaneko M, Funa N, Kawasaki H, Kojima H, Ohnishi Y, Horinouchi S. Efficient production of (2S)-flavanones by Escherichia coli containing an artificial biosynthetic gene cluster. Appl Microbiol Biotechnol. 2005;68:498–504. doi: 10.1007/s00253-005-1916-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Comparison of the TAL protein expression of pET-opTAL and pET-TAL in E. coli. Lane 1: E. coli transformation containing pET-opTAL expression vector lysate after IPTG induction; Lane 2: lysate before induction; Lane 3: E. coli transformation containing pET-TAL expression vector lysate after IPTG induction; Lane 4: lysate before induction. Figure S2. SDS-PAGE analysis of co-expression of TAL and Sam5 enzymes in E. coli. Lane 1: E. coli transformation containing pET-opT5 expression vector lysate before IPTG induction; Lane 2: lysate after induction; Lane 3: E. coli transformation containing pET-T5 expression vector lysate before IPTG induction; Lane 4: lysate after induction. Sam5 protein is 59 kDa, TAL protein from pET-opT5 expression vector is 56 kDa, TAL protein from pET-T5 expression vector is 57.4 kDa. The difference in the TAL proteins comes from the length of the His-tagged peptide sequences. Table S1. The results of the Tukey test for the data from Figure 3. Table S2. Oligo nucleotide primers used in this study.